Abstract
This study identified Vibrio parahaemolyticus in oyster and seawater samples collected from Delaware Bay from June through October of 2016. Environmental parameters including water temperature, salinity, dissolved oxygen, pH, and chlorophyll a were measured per sampling event. Oysters homogenate and seawater samples were 10-fold serially diluted and directly plated on CHROMagarᵀᴹ Vibrio medium. Presumptive V. parahaemolyticus colonies were counted and at least 20% of these colonies were selected for molecular chracterization. V. parahaemolyticus isolates (n = 165) were screened for the presence of the species-specific thermolabile hemolysin (tlh) gene, the pathogenic thermostable direct hemolysin (tdh)/ thermostable related hemolysin (trh) genes, the regulatory transmembrane DNA-binding gene (toxR), and V. parahaemolyticus metalloprotease (vpm) gene using a conventional PCR. The highest mean levels of the presumptive V. parahaemolyticus were 9.63×103 CFU/g and 1.85×103 CFU/mL in the oyster and seawater samples, respectively, during the month of July. V. parahaemolyticus levels in oyster and seawater samples were significantly positively correlated with water temperature. Of the 165 isolates, 137 (83%), 110 (66.7%), and 108 (65%) were tlh+, vpm+, and toxR+, respectively. Among the V. parahaemolyticus (tlh+) isolates, 7 (5.1%) and 15 (10.9%) were tdh+ and trh+, respectively, and 24 (17.5%), only oyster isolates, were positive for both genes. Potential pathogenic strains that possessed tdh and/or trh were notably higher in oyster (39%) than seawater (15.6%) isolates. The occurrence of total V. parahaemolyticus (tlh+) was not necessarily proportional to the potential pathogenic V. parahaemolyticus. Co-occurrence of the five genetic markers were observed only among oyster isolates. The co-occurrence of the gene markers showed a relatedness potential of tdh occurrence with vpm. We believe exploring the role of V. parahaemolyticus metalloprotease and whether it is involved in the toxic activity of the thermostable direct hemolysin (TDH) protein can be of significance. The outcomes of this study will provide some foundation for future studies regarding pathogenic Vibrio dynamics in relation to environmental quality.
Introduction
Vibrio parahaemolyticus is a gram-negative, halophilic, pathogenic bacterium that negatively impact aquatic ecosystems and human health [1–3]. They are curved rods, motile with a single polar flagellum and belong to the family Vibrionaceae. It is an endemic pathogen in the marine environment that was first identified as a cause of food-borne illness in Japan in 1950 [4, 5]. V. parahaemolyticus is one of the key causes of gastroenteritis leading to diarrhea, headache, vomiting, and abdominal cramps following the consumption of contaminated food or water. In addition, this bacterium can cause septicemia and wound infections [3, 6].
In aquatic ecosystems, organisms like oysters which are filter-feeding mollusks, tend to accumulate different microorganisms from seawater during their filtration [7–9]. Therefore, they are able to accumulate V. parahaemolyticus 100-fold higher than the surrounding water [8, 9]. During the warmer months, V. parahaemolyticus occurrence in oysters can reach 100% [8].
While some V. parahaemolyticus strains are associated with marine animal diseases [10], most strains are investigated as a major concern to human health [11]. V. parahaemolyticus infections are associated with the consumption of seafood, particularly raw or undercooked oysters, and accounted for 59.5% of laboratory-confirmed Vibrionaceae in the United States [11]. In 2006, a total of 177 V. parahaemolyticus infections were reported from New York, Oregon, and Washington states, and the laboratory-confirmed cases were over three-fold higher than the average number in all US states during the same period of 2002–2004 [12]. An outbreak of V. parahaemolyticus involving three people was reported in Maryland, August 2012 [13]. A multistate outbreak of 16 gastrointestinal illnesses linked to oysters were reported in 2019, and four of them were associated with V. parahaemolyticus [14]. The estimated annual mean cost of foodborne illnesses associated with V. parahaemolyticus was over US $40 million [15].
V. parahaemolyticus strains possess tlh species specific gene, which codes for thermolabile hemolysin (TLH) [16, 17]. The virulence of most clinical V. parahaemolyticus isolates are associated with the expression of tdh (thermostable direct hemolysin (TDH)) and/or trh (TDH-related hemolysin-(TRH)) genes [18–20]. Both tdh/trh genes are associated with β hemolysis on Wagatsuma blood agar, which is known as the Kanagawa phenomenon, and both have been used as accepted genetic markers for the detection of pathogenic V. parahaemolyticus in seafood [21–23]. Although, TDH/TRH proteins are the main pathogenic factors in V. parahaemolyticus [24], research also shows that many of the clinical isolates possess neither tdh nor trh genes indicating the potential presence of other virulence-related factors [25, 26].
V. parahaemolyticus harbors a V. parahaemolyticus metalloprotease (vpm) gene that expresses extracellular zinc metalloprotease and shows sufficient proteolytic activity towards type I collagen [27, 28]. V. parahaemolyticus metalloprotease can also degrade host tissue and may promote pathogen invasion [2]. On the other hand, metalloprotease has been investigated and found to be significant as a virulence factor among Vibrio spp. [28]. Metalloprotease was reported to have an important role on the extracellular cleavage and activation process of the V. cholerae enterotoxic hemolysin into mature hemolysin [29–32]. Therefore, exploring the prevalence and co-occurrence of vpm and tdh/trh genes in environmental strains of V. parahaemolyticus can be of significance.
The transmembrane DNA-binding protein, ToxR, is a regulatory protein in V. parahaemolyticus that is encoded by toxR gene. The ToxR protein is strongly associated with the upregulation of the gene encoding the virulence toxin TDH [33]. Genome sequencing of pathogenic V. parahaemolyticus revealed another virulence factor called type III secretion systems (T3SS), T3SS1and T3SS2, by which bacterial proteins (effectors) are injected directly into host cells [34]. An infant rabbit model infected with V. parahaemolyticus revealed that T3SS2 is essential for intestinal colonization [35]. In addition, T3SS2 is also considered as a prime virulence factor of V. parahaemolyticus enterotoxicity [35–37]. It has been reported that ToxR has no role in the production of T3SS2 [38]; however, a later study identified an uncharacterized component of T3SS2 to be critically regulated by ToxR [39]. Furthermore, toxR gene is very important to the bile resistance in the intestine, and the toxR mutant strains have significantly lower minimal bactericidal concentration compared to the wild strains [40, 41]. In addition, toxR gene is required for stress tolerance and colonization of V. parahaemolyticus [40]. On the other hand, similar to the tlh gene, toxR can be a reliable gene for the detection of V. parahaemolyticus, and many studies have used it as V. parahaemolyticus species-specific gene marker [42–44]. Studies also indicate that tlh and toxR genes have a compatible and robust result in terms of reliability and specificity for molecular identification of V. parahaemolyticus [16, 45]. Findings and reports from previous literature highlight the important relationships among the tlh, trh, tdh, vpm and toxR genes in terms of pathogenicity and identification of environmental V. parahaemolyticus associated with human infections. Therefore, our study aimed to screen the above-mentioned genetic markers to further illustrate the prevalence and patterns of these genetic markers in environmental strains of V. parahaemolyticus.
Delaware Bay is the prime oyster ground on the Atlantic coast providing ecological and commercial resources [46]. V. parahaemolyticus outbreaks are one of the leading causes for the closures of commercial shellfish industries on the east coast of the United States [47, 48]. This study was conducted to detect and determine total and potential pathogenic V. parahaemolyticus levels in oyster and seawater samples from Delaware Bay. Direct plating on CHROMagar Vibrio was used since it is a well-established method, allowing V. parahaemolyticus to be simultaneously isolated and differentiated from other Vibrio species, and it has a less inhibitory effect on V. parahaemolyticus growth than TCBS media [49–51]. This study also examined the correlation of V. parahaemolyticus levels in oyster and seawater samples in relation to the physico-chemical parameters. Along with the above-mentioned aims, we reported the co-occurrences of the five genetic markers (tlh, tdh, trh, toxR, and vpm) in the environmental strains of V. parahaemolyticus. To the best of our knowledge there are no published studies on the prevalence and co-occurrence of these genetic markers among V. parahaemolyticus in the Mid-Atlantic region. Furthermore, the regional variation in ecology of V. parahaemolyticus indicates the need of site-specific data, and this study provides a new set of data specific for the Delaware Bay region.
Materials and methods
Study location and sampling
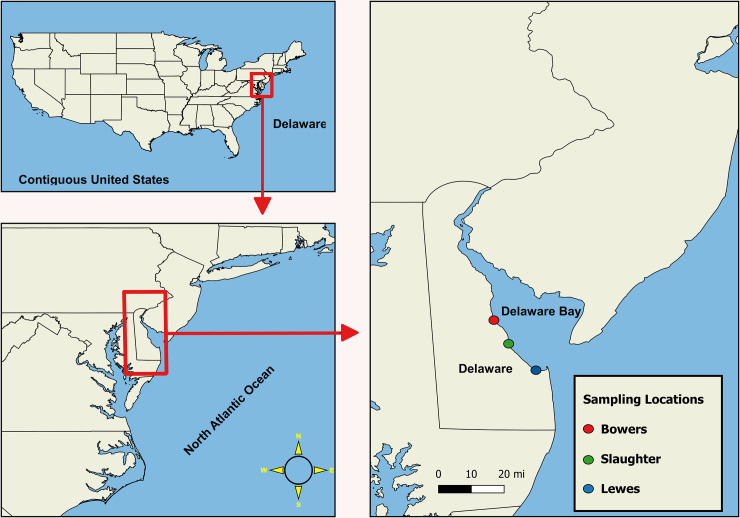
Field sampling collection was granted by the Department of Natural Resources and Environmental Control (DNREC) in 2016 of sampling year. Oysters and seawater samples were collected once a month from June to October 2016 from Bowers Beach (BB) [39°03'25.5"N 75°23'56.8"W] and Lewes, Broadkill (LW) [38°47'26.3"N 75°09'50.2"W] in the Delaware Bay. A third site, Slaughter Beach (SL) [38°56'50.1"N 75°18'52.4"W] was added to this study from August to October 2016 (Fig 1) to assess a wider range of the Delaware Bay area. Ten to twelve oysters from each site were harvested (one site per week) into Ziploc bags (SC Johnson & Sons, Racine, WI, USA), sub-divided into three groups for biological triplicates (A, B, and C), and placed in an insulated cooler with ice packs to maintain the temperature between 2–10°C [52]. One liter of seawater was collected from each site at the same time. Water quality parameters such as water temperature, salinity, turbidity, dissolved oxygen, pH, and chlorophyll a were recorded onsite using YSI 556 Handheld Multiparameter Instrument (YSI Incorporated, Yellow Springs, OH, USA) to assess the relationship between these parameters and the Colony Forming Units (CFUs) of Vibrio parahaemolyticus.
Fig 1. Study locations in Delaware Bay.
Processing of oyster and seawater samples
Ten to twelve oysters were collected from each site and divided into three groups to be analyzed in triplicates. For each replicate 3–4 oysters were cleaned upon arrival at the Aquatic Laboratory using a scrub brush and tap water before they were shucked with sterile knives. Oysters tissues and liquors from each replicate were placed into a sterilized blender jar (Waring Commercial, 7010S) and blended for 90 sec at high speed. Twenty-five grams of the blended tissue was diluted with 225 mL of 0.1% Peptone Water (PW; 1 g of peptone [BD, Bacto™ Peptone, 211677], 10 g of NaCl [Fisher scientific, S271], 1 liter of dH2O, pH 7.4 ± 0.2) and blended again for 60 sec at high speed to prepare the homogenate. This homogenate was labelled as the first (10−1) dilution. The oyster homogenate and seawater samples from each site were aseptically serial diluted in 0.1% PW to a final dilution of (10−6). Following the American Public Health Association Standard [53], one hundred microliters of each dilution [10–1 – 10–6] of both seawater and oyster homogenate samples from each site were aseptically spread plated in duplicate on CHROMagar medium (CHROMagar™ Vibrio, VB912), and incubated for 24 h at 37°C.
Identification and isolation of V. parahaemolyticus
V. parahaemolyticus were identified as mauve colonies on the CHROMagar plates. Each plate with a countable range of 20 to 200 colonies was selected to calculate the number of colony forming units (CFU) of the presumptive V. parahaemolyticus [51]. Using a sterile loop, at least 20% of the mauve colonies from each plate were chosen and inoculated aseptically into a 1.5 mL microcentrifuge tube of Tryptone Soy Broth (TSB; Thermo Fisher Scientific Inc, OXOID, CM0129) supplemented with 1% NaCl, and incubated with shaking (175 rpm) overnight at 37°C (New Brunswick Scientific I 24 Incubator Shaker Series). Microcentrifuge tubes were then centrifuged at 15,000 rpm for 2 min (Eppendorf Centrifuge 5424), and the supernatant was discarded. Equal amounts (600 μL) of Alkaline Peptone Water (APW; 10 g of peptone, 10 g of NaCl, 1 liter of dH2O, pH 8.5 ± 0.2) and TSB + [24% glycerol, BP229, Fisher BioReagents™] were added and the pellet was resuspended and then frozen at -20°C for further molecular analysis. Samples were prepared for PCR by boiling for 10 min, and immediately chilled on ice (2 min) for cell lysis and DNA release.
Molecular analysis (PCR procedures and conditions)
Presumptive V. parahaemolyticus isolates were further typed for the genetic markers tlh, tdh, trh, toxR, and vpm using five sets of primers previously assessed [16, 54]. The PCR reaction mixture (10 μL) consisted of 1 μL of cell lysate as DNA template, 2 μL (1.5 mM MgCl2) of the reaction buffer (5X Green GoTaq® Reaction Buffer; PROMEGA, USA), 0.1 μL (0.5 U) of Taq polymerase (Taq; PROMEGA; USA), 0.4 μL (100 μM) of 2.5 mM deoxynucleotide mix, 0.2 μL (0.2 μM) of each forward and reverse primers (IDT; USA), and 6.1 μL of nuclease free water. The amplification conditions for tlh, tdh, trh, toxR, and vpm genes are shown in (Table 1), and PCR reactions were performed using S1000 thermal cycler (Bio-Rad). One μL of nuclease free water was used for the no template control and 1 μL of V. parahaemolyticus SPRC 10290 cell lysate was used as a positive control [55, 56]. Gel electrophoresis (FB-SB-1316; Electrophoresis System; Fisher Scientific; USA) was used to analyze the PCR amplicons in 1% agarose gels containing 0.5 μg/ml ethidium bromide [Fisher BioReagents]. The gels were overlaid with 1x Tris acetate-EDTA buffer and run at 130 V for 30–45 min. DNA bands were visualized using a gel documentation system (Syngene, G: BOX EF).
Table 1. PCR conditions and primers sequences used in this study.
| Gene | Primer sequences | Cycling conditions |
|---|---|---|
| tlh | F-tlh: ACTCAACACAAGAAGAGATCGACAA | Cycles: 30 |
| R-tlh: GATGAGCGGTTGATGTCCAA | ||
| tdh | F-tdh: TCCCTTTTCCTGCCCCC | Denaturation temp: 95°C/30 sec |
| R-tdh: CGCTGCCATTGTATAGTCTTTATC | Annealing temp: 60°C/45 sec | |
| Extension temp: 68°C/1 min | ||
| trh | F-trh: TTGCTTTCAGTTTGCTATTGGCT | |
| R-trh: TGTTTACCGTCATATAGGCGCTT | ||
| toxR | toxR-4: GTCTTCTGACGCAATCGTTG | Cycles: 35 |
| toxR-7: ATACGAGTGGTTGCTGTCATG | Denaturation temp: 94°C/1 min | |
| vpm | vpm 1: CAGCTACCGAAACAGACGCTA | Annealing temp: 58°C/1 min |
| vpm 2: TCCTATCGAGGACTCTCTCAAC | Extension temp: 72°C/1 min |
Data analysis
For statistical analysis, the CFU values of presumptive V. parahaemolyticus were log10 transformed to normalize the data, and the significance level (P-value) of 0.05 was used. Spearman’s rank correlation analysis was performed to measure the relationship between V. parahaemolyticus levels [log10 CFU/g (or mL)] and the parameters affecting water quality (temperature, salinity, dissolved oxygen (DO), pH, turbidity, and chlorophyll a). Independent samples t-test was used to determine whether V. parahaemolyticus levels [log10 CFU/g (or mL)] among the sample types (oyster and seawater) were significantly different. Statistical analysis was performed using IBM SPSS Statistic software (version 26).
Results and discussion
Physico-chemical water quality parameters
Physico-chemical water quality parameters (Table 2) showed that water temperatures ranged from 14.63°C (LW, October) to 28°C (BB, August). Salinity levels were in the range of 5.37 ppt (LW, October) to 32 ppt (SL, August). The lowest and highest ranges for dissolved oxygen (DO) (3.12 to 8.23 mg/L) were recorded during the months of August and October from BB and LW sites, respectively. The minimum pH value of 6.44 (LW) and maximum of 8.82 (BB) was observed during the month of October. In terms of turbidity and chlorophyll a, the minimum and maximum levels ranged from 19 to 55.35 NTU/FTU and 0.134 to 1.174 μg/L, respectively. Notably, at the LW site and during the month of October, water quality parameters displayed the lowest range of water temperature (14.63°C), minimum level of salinity (5.37 ppt), highest range of dissolved oxygen (8.23 mg/L), and minimum pH value of (6.44). The seasonal variation between temperature and dissolved oxygen previously reported in the Chesapeake Bay shows that the median temperature (°C) is inversely correlated with the dissolved oxygen median (mg/L) [57]. Another study from the same region has also reported the lowest dissolved oxygen level (5.3 mg/L), and the highest temperature (29.4°C) during the month of August [58]. This shows that temperature is inversely correlated with dissolved oxygen concentrations [59].
Table 2. Physico-chemical water quality parameters in relation to study sites and date of collection.
| Site | Date | Temp°C | Salinity ppt | Turbidity NTU/FTU | Dissolved Oxygen mg/L | Chlorophyll a μg/L | pH |
|---|---|---|---|---|---|---|---|
| BB | 06/21/2016 | 24.18 | 20.0 | 29.0 | 6.3 | 1.2 | 8.18 |
| 07/19/2016 | 27.74 | 27.0 | 19.0 | 3.9 | 0.7 | 7.88 | |
| 08/02/2016 | 28.00 | 25.0 | 43.5 | 3.1 | 0.8 | 7.88 | |
| 09/13/2016 | 23.67 | 26.0 | 45.1 | 4.0 | 1.1 | 8.04 | |
| 10/17/2016 | 17.91 | 25.8 | 55.1 | 8.1 | 0.2 | 8.82 | |
| LW | 06/07/2016 | 22.7 | 23.0 | 29.0 | 3.7 | 0.3 | 7.2 |
| 07/06/2016 | 22.98 | 32.0 | 33.0 | 4.3 | 0.4 | 7.84 | |
| 08/08/2016 | 26.43 | 25.0 | 40.8 | 3.4 | 0.2 | 7.55 | |
| 09/06/2016 | 21.32 | 24.0 | 39.0 | 3.4 | 0.1 | 7.75 | |
| 10/10/2016 | 14.63 | 5.40 | 54.8 | 8.2 | 0.5 | 6.44 | |
| SL | 08/30/2016 | 26.74 | 32.0 | 55.4 | 4.0 | 0.8 | 8.06 |
| 09/26/2016 | 20.82 | 26.5 | 55.1 | 4.7 | 0.3 | 7.31 | |
| 10/24/2016 | 14.68 | 16.6 | 20.0 | 7.8 | 0.3 | 7.44 |
BB: Bowers Beach; LW: Lewes, Broadkill; SL: Slaughter Beach.
Concentration of V. parahaemolyticus in oyster and seawater samples
The highest mean levels of presumptive V. parahaemolyticus were 9.63×103 CFU/g in the oyster samples during the month of July from BB site. This was higher than V. parahaemolyticus (CFU) levels (6.0×102 CFU/g) detected by direct plating-colony hybridization procedure in Maryland Chesapeake Bay oysters [58]. According to the United States Food and Drug Administration (FDA) regulations and guidance, V. parahaemolyticus levels (Kanagawa positive or negative) in this study did not exceed the safety limits (≥ 1×104 CFU/g) [60]. Clearly, all presumptive V. parahaemolyticus (CFU) levels, agree well with the strong correlations between water temperature and V. parahaemolyticus densities that are reported in the literature [58, 61–63], indicating that V. parahaemolyticus levels increases with the rise of temperature and vice versa (Table 3). V. parahaemolyticus concentrations from seawater samples were notably lower than oyster samples (Table 3), demonstrating that oysters can concentrate the Vibrio species higher than 10-fold compared to the surrounding water [8, 9]. This results are in agreement with studies conducted on the Pacific, Atlantic and Gulf Coasts of the United States [58, 64, 65]. However, Independent samples t-test indicated that there was no statistically significant difference in mean V. parahaemolyticus log10 CFU/g (or ml) values between sample types (oyster–seawater), t(24) = 1.159, P = 0.258 (P > 0.05). Seawater samples from LW in July, with the highest range of salinity, had low CFU/mL counts compared to BB during the same month indicating that there are parameters other than temperature that may have affected the growth of V. parahaemolyticus [61]. During the month of October, V. parahaemolyticus levels were undetectable (<10 CFU/g (or mL)) in both oyster and seawater samples from LW and SL sites. However, V. parahaemolyticus concentrations in the oyster and seawater samples at site BB were 1.7×10 and 3.3×10 CFU/g or mL, respectively. Although V. parahaemolyticus in oysters and seawater was not detectable in some sampling events, the lowest detectable reading in oysters and seawater in this study was 1.7×10 CFU/g (or mL). Figs 2 and 3 demonstrate the log10 CFU/g (or mL) levels of V. parahaemolyticus in relation to collection time, study site, and water parameters (temperature and salinity) for oyster and seawater samples, respectively.
Table 3. Averages of V. parahaemolyticus CFU/g (or mL) in relation to sample type, study site, and collection time.
| BB-OY | BB-W | LW-OY | LW-W | SL-OY | SL-W | |
|---|---|---|---|---|---|---|
| June | 2017 | 33 | 367 | 83 | -b | - b |
| July | 9633 | 1100 | 1850 | 167 | - b | - b |
| Aug | 980 | 617 | 1133 | 117 | 117 | 20 |
| Sep | 417 | <10a | <10 a | 17 | 17 | 33 |
| Oct | 17 | 33 | <10 a | <10 a | <10 a | <10 a |
OY: Oyster; W: Water; BB: Bowers Beach; LW: Lewes, Broadkill; SL: Slaughter Beach.
a (Not detectable)
b (No sample collection).
Fig 2. Average Vibrio parahaemolyticus levels (log10 CFU/g) in oyster samples in relation to collection time, study sites, and water parameters (temperature and salinity).
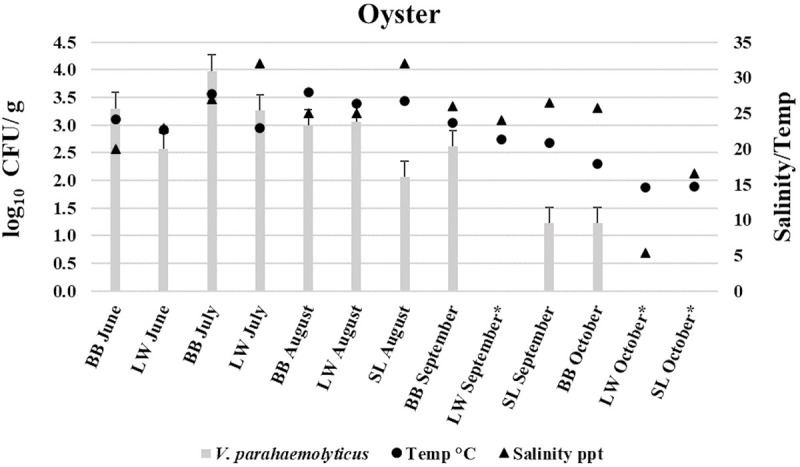
Bowers Beach (BB)—Lewes, Broadkill (LW)—Slaughter Beach (SL). * (Not detectable).
Fig 3. Average Vibrio parahaemolyticus levels (log10 CFU/mL) in seawater samples in relation to collection time, study sites, and water parameters (temperature and salinity).
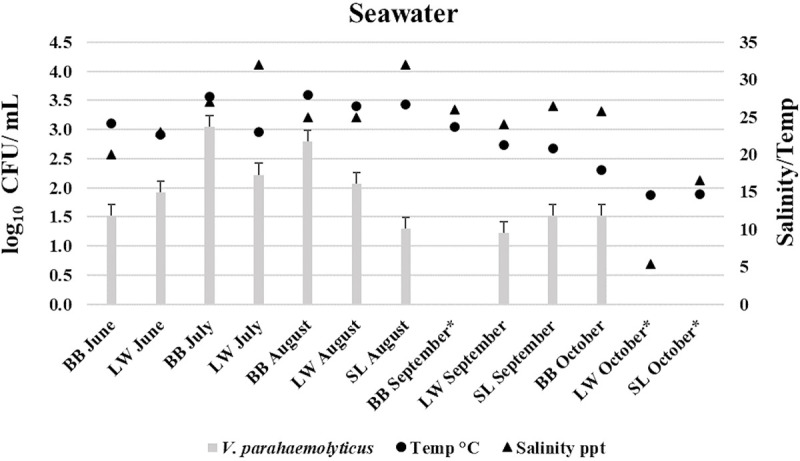
Bowers Beach (BB)—Lewes, Broadkill (LW)—Slaughter Beach (SL). * (Not detectable).
Spearman’s rank correlation analysis showed that water temperature is positively and significantly (P < 0.05) correlated to the total V. parahaemolyticus (log10 CFU/g or mL) levels in oyster and seawater samples (Table 4), which is in agreement with previous studies [58, 61–63]. Salinity had no significant correlation with total V. parahaemolyticus (log10 CFU/g or mL) levels in oyster and seawater samples (Table 4). Several studies have reported conflicting results regarding the correlations between the abundance of V. parahaemolyticus and salinity. Some of these studies have found a correlation between salinity and abundance of V. parahaemolyticus [65–68], while others have not [58, 64, 69–72]. Thus, the insignificance of salinity on the abundance of V. parahaemolyticus identified in this study cannot be generalized. No significant correlation was found between dissolved oxygen and/or turbidity and the abundance of total V. parahaemolyticus (Table 4). This result is in contrast to what has been reported from studies conducted in the Chesapeake Bay [57, 58]. Both pH and chlorophyll a did not significantly correlate with total V. parahaemolyticus (log10 CFU/g or mL) levels (Table 4), and this is consistent with the previous studies from Mid-Atlantic region [57, 58, 64].
Table 4. Correlation between V. parahaemolyticus log10 CFU level and water parameters.
| Water parameters | Spearman’s correlation coefficient, r (OY/W)a | Oyster (Sig.) | Seawater (Sig.) |
|---|---|---|---|
| Temp | (.777/.639)b | (.002)b | (.019)b |
| Salinity | (.416/.423) | (.157) | (.149) |
| Turbidity | (-.393/-.306) | (.184) | (.309) |
| DO mg/l | (-.368/-.528) | (.216) | (.064) |
| pH | (.416/.142) | (.158) | (.644) |
| Chlorophyll a | (.509/.033) | (.076) | (.914) |
aOY: oysters; W: seawater.
bCorrelation is significant at the 0.05 level. Sig. (2-tailed).
Molecular identification and characterization of V. parahaemolyticus
A total of 165 presumptive V. parahaemolyticus isolates (mauve colored on CHROM agar) were further examined for the presence of the species-specific gene (tlh), and 137 (83%) were confirmed to be V. parahaemolyticus (Table 5). Previous investigation revealed that primers targeting the tlh, toxR, and vpm genes were (100%) specific for V. parahaemolyticus strains [16]. The lower occurrence of vpm (66.7%) and toxR (65.5%) genes compared to tlh (83%) gene in this study (Table 6 and Fig 4), suggests that tlh gene may occasionally produce false positive results, as a gene similar to tlh may occur in other Vibrio species specifically V. alginolyticus [7, 73]. Yet, regulatory authorities use tlh gene as a marker to assess the counts of V. parahaemolyticus and reinforce actions to control the outbreaks [74]. This study also showed that (11.7%) of the confirmed V. parahaemolyticus possessed only tlh gene (Fig 5). In contrast, toxR and/or vpm genes were only present in coexistence with tlh, tdh, and/or trh (Fig 5), suggest that toxR and vpm may be more sensitive in detecting V. parahaemolyticus. A high percentage of V. parahaemolyticus (tlh+) were observed among oysters (90.5%) compared to seawater (65.3%) samples (Table 5). At LW and SL sites, V. parahaemolyticus were not-detectable (ND) during the month of October (Table 5). About half of the confirmed V. parahaemolyticus colonies isolated from seawater (43.8%) possessed only the tlh gene indicating the necessity of other gene markers such as toxR and vpm for V. parahaemolyticus strains to survive the internal conditions and colonize oyster gut [2, 40].
Table 5. Occurrence of presumptive and confirmed (tlh+) V. parahaemolyticus isolates on CHROMagar.
| Site | Month | Total Presumptive Vp. (OY, W) | Total Confirmed Vp. tlh+ | Confirmed Vp. Oyster tlh+ | Confirmed Vp. Water tlh+ |
|---|---|---|---|---|---|
| BB | June | 16 (14, 2) | 15 (93.8%) | 13 (92.9%) | 2 (100%) |
| July | 36 (20, 16) | 34 (94.4%) | 20 (100%) | 14 (87.5%) | |
| August | 22 (15, 7) | 18 (81.8%) | 14 (93.3%) | 4 (57.1%) | |
| September | 7 (7, 0) | 7 (100%) | 7 (100%) | 0 | |
| October | 3 (2, 1) | 1 (33.3%) | 1 (50%) | 0 | |
| LW | June | 10 (10, 0) | 8 (80%) | 8 (80%) | 0 |
| July | 40 (33, 7) | 36 (90%) | 31 (93.9%) | 5 (71.4%) | |
| August | 16 (11, 5) | 11 (68.8%) | 7 (63.6%) | 4 (80%) | |
| September | 1 (0, 1) | 0 | 0 | 0 | |
| October | ND | ND | ND | ND | |
| SL | August | 12 (3, 9) | 6 (50%) | 3 (100%) | 3 (33.3%) |
| September | 2 (1, 1) | 1 (50%) | 1 (100%) | 0 | |
| October | ND | ND | ND | ND | |
| Total | 165 (116, 49) | 137 (83%) | 105 (90.5%) | 32 (65.3%) | |
ND: Not detectable; OY: Oyster; W: Water; BB: Bowers Beach; LW: Lewes, Broadkill; SL: Slaughter Beach.
Table 6. Prevalence of V. parahaemolyticus genetic markers.
| Site | Month | tlh OY, W | tdh OY, W | trh OY, W | tdh/trh OY, W | toxR OY, W | vpm OY, W |
|---|---|---|---|---|---|---|---|
| BB | June | 13, 2 | 1, 0 | 5, 1 | 2, 0 | 12, 0 | 11, 0 |
| July | 20, 14 | 2, 2 | 3, 1 | 1, 0 | 14, 4 | 20, 7 | |
| August | 14, 4 | 0, 0 | 1, 0 | 0, 0 | 14, 1 | 12, 0 | |
| September | 7, ND | 0, ND | 0, ND | 0, ND | 6, ND | 6, ND | |
| October | 1, 0 | 0, 0 | 0, 0 | 0, 0 | 1, 0 | 0, 0 | |
| LW | June | 8, 0 | 1, 0 | 0, 0 | 0, 0 | 8, 0 | 7, 0 |
| July | 31, 5 | 1, 0 | 3, 1 | 21, 0 | 29, 3 | 31, 3 | |
| August | 7, 4 | 0, 0 | 0, 0 | 0, 0 | 7, 2 | 7, 2 | |
| September | ND, 0 | ND, 0 | ND, 0 | ND, 0 | ND, 0 | ND, 0 | |
| October | ND, ND | ND, ND | ND, ND | ND, ND | ND, ND | ND, ND | |
| SL | August | 3, 3 | 0, 0 | 0, 0 | 0, 0 | 3, 3 | 2, 2 |
| September | 1, 0 | 0, 0 | 0, 0 | 0, 0 | 1, 0 | 0, 0 | |
| October | ND, ND | ND, ND | ND, ND | ND, ND | ND, ND | ND, ND | |
| Total | 105, 32 | 5, 2 | 12, 3 | 24, 0 | 95, 13 | 96, 14 | |
ND: Not detectable; OY: Oyster; W: Water; BB: Bowers Beach; LW: Lewes, Broadkill; SL: Slaughter Beach.
Fig 4. Percent and number of isolates with detected tdh, trh, toxR, vpm and tlh genes in oysters and seawater.
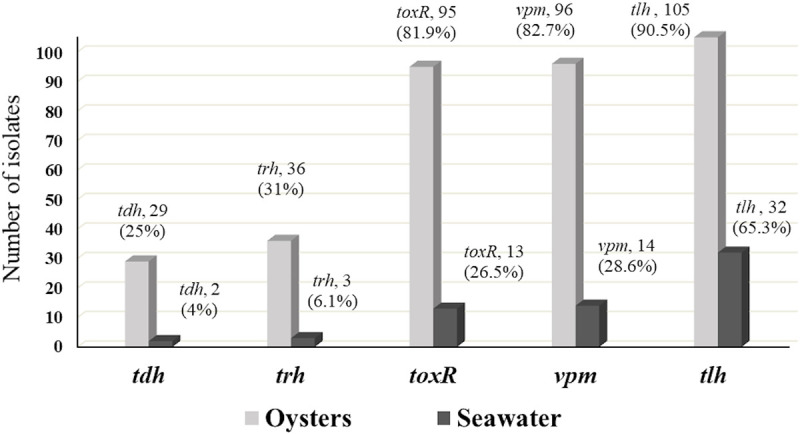
Fig 5. Number of isolates with coexisting genes among total confirmed V. parahaemolyticus isolates.
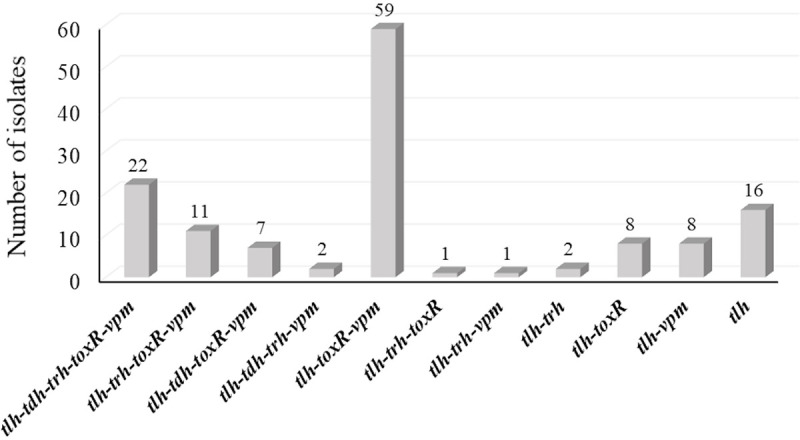
Among V. parahaemolyticus (tlh+), 22.6% and 28.5% were positive for tdh and trh respectively (Table 6).
This relatively low incidence of V. parahaemolyticus (tdh+/trh+) is in agreement with what has been reported in the literature for environmental isolates [58, 75–77]. Isolates that possessed tdh, trh, or both tdh/trh genes account for 33.5% of V. parahaemolyticus (tlh+) and were notably higher in oyster (39%) than seawater (15.6%) (Table 6). More than half (61%) of V. parahaemolyticus (tdh+/trh+) isolated from oysters were detected at LW site in July (Table 6). The occurrence of tdh and/or trh positive V. parahaemolyticus was not observed among the study sites during September and October (Table 6). This observation highlights the importance of understanding the dynamics and seasonal variations of pathogenic V. parahaemolyticus in Delaware Bay. The high frequency of the trh gene (n = 39) compared to the tdh gene (n = 31) agrees well with its occurrence in Gulf Coast and Chesapeake Bay oysters [58], and in South Carolina [51]. The co-occurrence of both tdh/trh genes was observed in 17.5% and 52% of total and potential pathogenic V. parahaemolyticus, respectively, and they were only among oyster isolates (Table 6). This is in contrast with a study in the Mid-Atlantic [78], in which V. parahaemolyticus with both tdh/trh genes were observed more frequently among the water isolates. V. parahaemolyticus (tlh+) as illustrated in Table 6 were not proportional to the potential pathogenic V. parahaemolyticus, and this agrees well with the previous studies [51, 78].
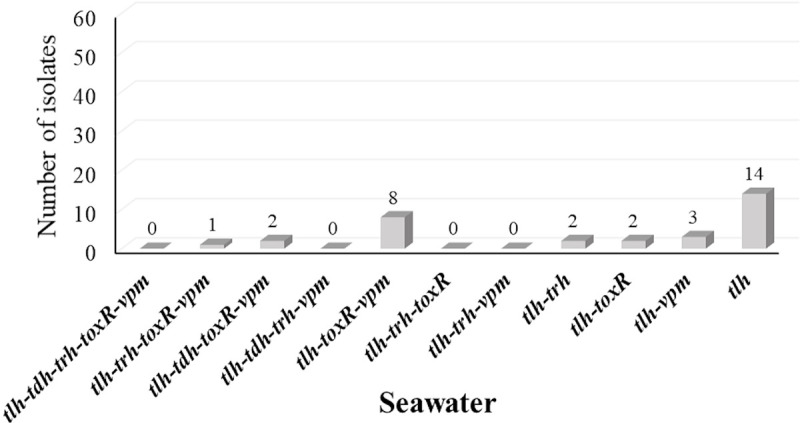
The occurrence of the five genetic markers showed similar patterns among oyster and seawater isolates (Fig 4). The co-occurrence of the five genetic markers among oyster isolates tested were dominated (43.9%) by tlh, toxR and vpm pattern followed by the coexistence of all five genetic markers (18.9%) (Fig 6). On the other hand, co-occurrence of tlh, toxR, and vpm were the second prevalent pattern among seawater isolates (Fig 7). Interestingly and similar to the simultaneous occurrences of tdh/trh, the coexistence of the five genetic markers were observed only among oyster isolates (Figs 6 and 7), and most of them (19/22) were detected in the LW site where the historical average salinity is close to 26 ppt compared to the BB site with a historical average salinity close to 20 ppt [79]. Variation of gene occurrence patterns among the examined isolates suggest the variation of V. parahaemolyticus clones that inhabit Delaware Bay. This study also revealed the relatedness potential of tdh occurrence with vpm, as Figs 5, 6, and 7 demonstrated that whenever tdh was present, vpm was also present but not vice versa. This indicates the importance of understanding the role of V. parahaemolyticus metalloprotease and whether it is involved in the toxic activity of the thermostable direct hemolysin (TDH) protein as is the case with V. cholerae enterotoxic hemolysin [29–32].
Fig 6. Co-existence of genes among confirmed V. parahaemolyticus isolates from oysters.
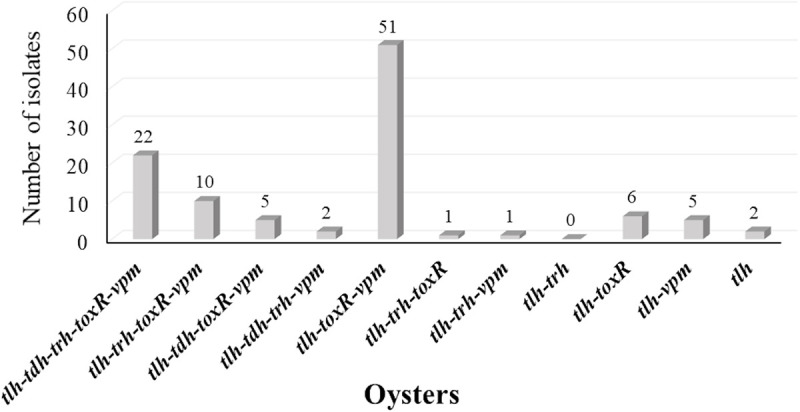
Fig 7. Co-existence of genes among confirmed V. parahaemolyticus isolates from seawater.
Conclusion
This study assessed V. parahaemolyticus levels in oysters and seawater in the Delaware Bay in relation to environmental conditions and the prevalence of key genes. Among the physico-chemical parameters assessed in this study, water temperature was the only factor that significantly positively correlated with total V. parahaemolyticus level in oyster and seawater samples. Occurrence of total V. parahaemolyticus was not necessarily proportional to the occurrence of potentially pathogenic V. parahaemolyticus. The prevalence pattern of the key genes in V. parahaemolyticus isolates from seawater does not reflect the pattern of V. parahaemolyticus isolates from oysters. The low occurrence of V. parahaemolyticus isolates that possessed tdh, trh, toxR, and/or vpm genes in seawater samples compared to oysters confirmed the significance of the bioaccumulation process by oysters as a natural nursery for potential pathogenic V. parahaemolyticus. Although salinity in this study did not significantly correlate with the V. parahaemolyticus level, the historically higher average salinity at Lewes may explain the high frequency of strains from this site that possess all five genes. Utilizing V. parahaemolyticus metalloprotease gene (vpm) as species-specific gene may provide more accurate results when assessing the prevalence and abundance of pathogenic V. parahaemolyticus, and a better understanding of the proportional correlation between the total and potentially pathogenic V. parahaemolyticus. The variation among V. parahaemolyticus isolates that we have reported indicates the difference in growth rates among Delaware Bay oysters.
Future studies may focus on conducting whole genome sequencing for the V. parahaemolyticus isolates to identify the coexistence of the virulence and virulence related genes reported in the literature and illustrate the genetic diversity among V. parahaemolyticus isolates inhabiting Delaware Bay. Future studies may also focus on the role of V. parahaemolyticus metalloprotease on the toxic activity of TDH. This study provided informative data on oyster-Vibrio natural contamination factors that can be applied to the risk management programs. The outcomes of this study provide some foundation for future studies regarding pathogenic Vibrio dynamics in relation to environmental quality.
Supporting information
(XLSX)
Acknowledgments
We would like to thank Dr. Bettina C. Taylor for valuable comments and editing. We also thank Dr. Gary Richards of USDA ARS for his valuable discussion on our findings.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
1- United States Department of Agriculture-National Institute of Food and Agriculture-Capacity Building Grant for Vibrio Project (2014-38821-22430) - funded AA, GO, and SP (https://nifa.usda.gov/). 2- United States Department of Agriculture-Evans-Allen Grant (DELXDSUGO2015 Award) - funded KC (https://nifa.usda.gov/program/agricultural-research-1890-land-grant-institutions). 3- National Science Foundation-Established Program to Stimulate Competitive Research Grant (EPS-1301765), funded GO. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lee CT, Chen IT, Yang YT, Ko TP, Huang YT, Huang JY, et al. The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin. Proc Natl Acad Sci U S A. 2015;112(34):10798–803. 10.1073/pnas.1503129112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luan X, Chen J, Zhang XH, Li Y, Hu G. Expression and characterization of a metalloprotease from a Vibrio parahaemolyticus isolate. Can J Microbiol. 2007;53(10):1168–73. 10.1139/W07-085 [DOI] [PubMed] [Google Scholar]
- 3.Morris JG Jr, Black RE. Cholera and other vibrioses in the United States. N Engl J Med. 1985;312(6):343–50. 10.1056/NEJM198502073120604 [DOI] [PubMed] [Google Scholar]
- 4.Fujino T, Okuno Y, Nakada D, Aoyama A, Fukai K, Mukai T, et al. On the bacteriological examination of shirasu-food poisoning. Medical Journal of Osaka University. 1953;4(2/3):299–304. [Google Scholar]
- 5.Daniels NA, MacKinnon L, Bishop R, Altekruse S, Ray B, Hammond RM, et al. Vibrio parahaemolyticus infections in the United States, 1973–1998. The Journal of infectious diseases. 2000;181(5):1661–6. 10.1086/315459 [DOI] [PubMed] [Google Scholar]
- 6.Iwamoto M, Ayers T, Mahon BE, Swerdlow DL. Epidemiology of seafood-associated infections in the United States. Clin Microbiol Rev. 2010;23(2):399–411. 10.1128/CMR.00059-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klein SL, Gutierrez West CK, Mejia DM, Lovell CR. Genes similar to the Vibrio parahaemolyticus virulence-related genes tdh, tlh, and vscC2 occur in other vibrionaceae species isolated from a pristine estuary. Appl Environ Microbiol. 2014;80(2):595–602. 10.1128/AEM.02895-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morris JG Jr Cholera and other types of vibriosis: a story of human pandemics and oysters on the half shell. Clin Infect Dis. 2003;37(2):272–80. 10.1086/375600 [DOI] [PubMed] [Google Scholar]
- 9.DePaola A, Hopkins LH, Peeler JT, Wentz B, McPhearson RM. Incidence of Vibrio parahaemolyticus in U.S. coastal waters and oysters. Appl Environ Microbiol. 1990;56(8):2299–302. 10.1128/AEM.56.8.2299-2302.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khimmakthong U, Sukkarun P. The spread of Vibrio parahaemolyticus in tissues of the Pacific white shrimp Litopenaeus vannamei analyzed by PCR and histopathology. Microb Pathog. 2017;113:107–12. 10.1016/j.micpath.2017.10.028 [DOI] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention FDASNF. FoodNet 2015 Surveillance Report (Final Data). Atlanta, GA: Centers for Disease Control and Prevention, Foodborne Diseases Active Surveillance Network (FoodNet), Services USDoHaH; 2017. [Google Scholar]
- 12.Centers for Disease Control and Prevention. Vibrio parahaemolyticus infections associated with consumption of raw shellfish—three states, 2006. MMWR Morbidity and mortality weekly report. 2006;55(31):854 [PubMed] [Google Scholar]
- 13.Haendiges J, Rock M, Myers RA, Brown EW, Evans P, Gonzalez-Escalona N. Pandemic Vibrio parahaemolyticus, Maryland, USA, 2012. Emerg Infect Dis. 2014;20(4):718–20. 10.3201/eid2004.130818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Centers for Disease Control and Prevention NCEZID. Multistate Outbreak of Gastrointestinal Illnesses Linked to Oysters Imported from Mexico. Atlanta, GA: Centers for Disease Control and Prevention, National Center for Emerging and Zoonotic Infectious Diseases, Division of Foodborne W, and Environmental Diseases; 2019. 6/21/2019. [Google Scholar]
- 15.U.S. Department of Agriculture ERS. Cost estimates of foodborne illnesses: Cost of foodborne illness estimates for Vibrio parahaemolyticus. Washington, DC2014.
- 16.Luan XY, Chen JX, Zhang XH, Jia JT, Sun FR, Li Y. Comparison of different primers for rapid detection of Vibrio parahaemolyticus using the polymerase chain reaction. Lett Appl Microbiol. 2007;44(3):242–7. 10.1111/j.1472-765X.2006.02074.x [DOI] [PubMed] [Google Scholar]
- 17.Bej AK, Patterson DP, Brasher CW, Vickery MC, Jones DD, Kaysner CA. Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. J Microbiol Methods. 1999;36(3):215–25. 10.1016/s0167-7012(99)00037-8 [DOI] [PubMed] [Google Scholar]
- 18.Okuda J, Ishibashi M, Abbott SL, Janda JM, Nishibuchi M. Analysis of the thermostable direct hemolysin (tdh) gene and the tdh-related hemolysin (trh) genes in urease-positive strains of Vibrio parahaemolyticus isolated on the West Coast of the United States. J Clin Microbiol. 1997;35(8):1965–71. 10.1128/JCM.35.8.1965-1971.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shirai H, Ito H, Hirayama T, Nakamoto Y, Nakabayashi N, Kumagai K, et al. Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect Immun. 1990;58(11):3568–73. 10.1128/IAI.58.11.3568-3573.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taniguchi H, Ohta H, Ogawa M, Mizuguchi Y. Cloning and expression in Escherichia coli of Vibrio parahaemolyticus thermostable direct hemolysin and thermolabile hemolysin genes. J Bacteriol. 1985;162(2):510–5. 10.1128/JB.162.2.510-515.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagley S, Koofhethile K, Rangdale R. Prevalence and potential pathogenicity of Vibrio parahaemolyticus in Chinese mitten crabs (Eriocheir sinensis) harvested from the River Thames estuary, England. J Food Prot. 2009;72(1):60–6. 10.4315/0362-028x-72.1.60 [DOI] [PubMed] [Google Scholar]
- 22.Tada J, Ohashi T, Nishimura N, Shirasaki Y, Ozaki H, Fukushima S, et al. Detection of the thermostable direct hemolysin gene (tdh) and the thermostable direct hemolysin-related hemolysin gene (trh) of Vibrio parahaemolyticus by polymerase chain reaction. Mol Cell Probes. 1992;6(6):477–87. 10.1016/0890-8508(92)90044-x [DOI] [PubMed] [Google Scholar]
- 23.Honda T, Chearskul S, Takeda Y, Miwatani T. Immunological methods for detection of Kanagawa phenomenon of Vibrio parahaemolyticus. J Clin Microbiol. 1980;11(6):600–3. 10.1128/JCM.11.6.600-603.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamazaki W, Kumeda Y, Misawa N, Nakaguchi Y, Nishibuchi M. Development of a loop-mediated isothermal amplification assay for sensitive and rapid detection of the tdh and trh genes of Vibrio parahaemolyticus and related Vibrio species. Appl Environ Microbiol. 2010;76(3):820–8. 10.1128/AEM.02284-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.García K, Torres R, Uribe P, Hernández C, Rioseco ML, Romero J, et al. Dynamics of clinical and environmental Vibrio parahaemolyticus strains during seafood-related summer diarrhea outbreaks in southern Chile. Appl Environ Microbiol. 2009;75(23):7482–7. 10.1128/AEM.01662-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Honda S, Goto I, Minematsu I, Ikeda N, Asano N, Ishibashi M, et al. Vibrio parahaemolyticus infectious disease caused by Kanagawa phenomenon-negative O3: K6 originated from Maldives. Kansenshogaku zasshi The Journal of the Japanese Association for Infectious Diseases. 1987;61(9):1070–8. 10.11150/kansenshogakuzasshi1970.61.1070 [DOI] [PubMed] [Google Scholar]
- 27.Miyoshi S-i, Nitanda Y, Fujii K, Kawahara K, Li T, Maehara Y, et al. Differential gene expression and extracellular secretion of the collagenolytic enzymes by the pathogen Vibrio parahaemolyticus. FEMS Microbiol Lett. 2008;283(2):176–81. 10.1111/j.1574-6968.2008.01159.x [DOI] [PubMed] [Google Scholar]
- 28.Kim S-K, Yang J-Y, Cha J. Cloning and sequence analysis of a novel metalloprotease gene from Vibrio parahaemolyticus 04. Gene. 2002;283(1–2):277–86. 10.1016/s0378-1119(01)00882-4 [DOI] [PubMed] [Google Scholar]
- 29.Benitez JA, Silva AJ. Vibrio cholerae hemagglutinin (HA)/protease: an extracellular metalloprotease with multiple pathogenic activities. Toxicon. 2016;115:55–62. 10.1016/j.toxicon.2016.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Binesse J, Delsert C, Saulnier D, Champomier-Verges M-C, Zagorec M, Munier-Lehmann H, et al. Metalloprotease vsm is the major determinant of toxicity for extracellular products of Vibrio splendidus. Appl Environ Microbiol. 2008;74(23):7108–17. 10.1128/AEM.01261-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagamune K, Yamamoto K, Naka A, Matsuyama J, Miwatani T, Honda T. In vitro proteolytic processing and activation of the recombinant precursor of El Tor cytolysin/hemolysin (pro-HlyA) of Vibrio cholerae by soluble hemagglutinin/protease of V. cholerae, trypsin, and other proteases. Infect Immun. 1996;64(11):4655–8. 10.1128/IAI.64.11.4655-4658.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Booth B, Boesman-Finkelstein M, Finkelstein R. Vibrio cholerae hemagglutinin/protease nicks cholera enterotoxin. Infect Immun. 1984;45(3):558–60. 10.1128/IAI.45.3.558-560.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin Z, Kumagai K, Baba K, Mekalanos J, Nishibuchi M. Vibrio parahaemolyticus has a homolog of the Vibrio cholerae toxRS operon that mediates environmentally induced regulation of the thermostable direct hemolysin gene. J Bacteriol. 1993;175(12):3844–55. 10.1128/jb.175.12.3844-3855.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Makino K, Oshima K, Kurokawa K, Yokoyama K, Uda T, Tagomori K, et al. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. The Lancet. 2003;361(9359):743–9. 10.1016/S0140-6736(03)12659-1 [DOI] [PubMed] [Google Scholar]
- 35.Ritchie JM, Rui H, Zhou X, Iida T, Kodoma T, Ito S, et al. Inflammation and disintegration of intestinal villi in an experimental model for Vibrio parahaemolyticus-induced diarrhea. PLoS Pathog. 2012;8(3):e1002593 10.1371/journal.ppat.1002593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hiyoshi H, Kodama T, Iida T, Honda T. Contribution of Vibrio parahaemolyticus virulence factors to cytotoxicity, enterotoxicity, and lethality in mice. Infect Immun. 2010;78(4):1772–80. 10.1128/IAI.01051-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park K-S, Ono T, Rokuda M, Jang M-H, Okada K, Iida T, et al. Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect Immun. 2004;72(11):6659–65. 10.1128/IAI.72.11.6659-6665.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gotoh K, Kodama T, Hiyoshi H, Izutsu K, Park K-S, Dryselius R, et al. Bile acid-induced virulence gene expression of Vibrio parahaemolyticus reveals a novel therapeutic potential for bile acid sequestrants. PloS one. 2010;5(10). 10.1371/journal.pone.0013365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hubbard TP, Chao MC, Abel S, Blondel CJ, Zur Wiesch PA, Zhou X, et al. Genetic analysis of Vibrio parahaemolyticus intestinal colonization. Proceedings of the National Academy of Sciences. 2016;113(22):6283–8. 10.1073/pnas.1601718113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whitaker WB, Parent MA, Boyd A, Richards GP, Boyd EF. The Vibrio parahaemolyticus ToxRS regulator is required for stress tolerance and colonization in a novel orogastric streptomycin-induced adult murine model. Infect Immun. 2012;80(5):1834–45. 10.1128/IAI.06284-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Provenzano D, Schuhmacher DA, Barker JL, Klose KE. The Virulence Regulatory Protein ToxR Mediates Enhanced Bile Resistance in Vibrio cholerae and Other PathogenicVibrio Species. Infect Immun. 2000;68(3):1491–7. 10.1128/iai.68.3.1491-1497.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lo C, Leung P, Yip S, To T, Ng T, Kam K. Rapid detection of pathogenic Vibrio parahaemolyticus by a sensitive and specific duplex PCR‐hybridization probes assay using LightCycler. J Appl Microbiol. 2008;105(2):575–84. 10.1111/j.1365-2672.2008.03776.x [DOI] [PubMed] [Google Scholar]
- 43.Croci L, Suffredini E, Cozzi L, Paniconi M, Ciccaglioni G, Colombo MM. Evaluation of different polymerase chain reaction methods for the identification of Vibrio parahaemolyticus strains isolated by cultural methods. J AOAC Int. 2007;90(6):1588–97. [PubMed] [Google Scholar]
- 44.Kim YB, Okuda J, Matsumoto C, Takahashi N, Hashimoto S, Nishibuchi M. Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J Clin Microbiol. 1999;37(4):1173–7. 10.1128/JCM.37.4.1173-1177.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taiwo M, Baker-Austin C, Powell A, Hodgson E, Natňs OB, Walker DI. Comparison of toxR and tlh based PCR assays for Vibrio parahaemolyticus. Food Control. 2017;77:116–20. [Google Scholar]
- 46.Canzonier WJ. Delaware Bay Oyster Culture–Past, Present and Potential Future In: Agriculture/Seafood, editor.: N J Aquaculture Association; 2005. [Google Scholar]
- 47.Marquis ND, Record NR, Robledo JAF. Survey for protozoan parasites in Eastern oysters (Crassostrea virginica) from the Gulf of Maine using PCR-based assays. Parasitol Int. 2015;64(5):299–302. 10.1016/j.parint.2015.04.001 [DOI] [PubMed] [Google Scholar]
- 48.DePaola A, Jones JL, Woods J, Burkhardt W, Calci KR, Krantz JA, et al. Bacterial and viral pathogens in live oysters: 2007 United States market survey. Appl Environ Microbiol. 2010;76(9):2754–68. 10.1128/AEM.02590-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Di Pinto A, Terio V, Novello L, Tantillo G. Comparison between thiosulphate-citrate-bile salt sucrose (TCBS) agar and CHROMagar Vibrio for isolating Vibrio parahaemolyticus. Food Control. 2011;22(1):124–7. [Google Scholar]
- 50.FAO/WHO. Risk assessment tools for Vibrio parahaemolyticus and Vibrio vulnificus associated with seafood. 2020;Microbiological Risk Assessment Series No. 20:71p. [Google Scholar]
- 51.Klein SL, Lovell CR. The hot oyster: levels of virulent Vibrio parahaemolyticus strains in individual oysters. FEMS Microbiol Ecol. 2017;93(2). 10.1093/femsec/fiw232 [DOI] [PubMed] [Google Scholar]
- 52.Interstate Shellfish Sanitation Conference, National Shellfish Sanitation Program: Guide for the Control of Molluscan Shellfish: Shellfish Laboratory Evaluation Checklist for PCR Microbiology, (2017).
- 53.American Public Health Association. Recommended procedures for the examination of sea water and shellfish. Recommended procedures for the examination of sea water and shellfish 1970. p. viii, 105–viii,. [Google Scholar]
- 54.Nordstrom JL, Vickery MC, Blackstone GM, Murray SL, DePaola A. Development of a multiplex real-time PCR assay with an internal amplification control for the detection of total and pathogenic Vibrio parahaemolyticus bacteria in oysters. Appl Environ Microbiol. 2007;73(18):5840–7. 10.1128/AEM.00460-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DePaola A, Ulaszek J, Kaysner CA, Tenge BJ, Nordstrom JL, Wells J, et al. Molecular, serological, and virulence characteristics of Vibrio parahaemolyticus isolated from environmental, food, and clinical sources in North America and Asia. Appl Environ Microbiol. 2003;69(7):3999–4005. 10.1128/aem.69.7.3999-4005.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cook DW. Sensitivity of Vibrio species in phosphate-buffered saline and in oysters to high-pressure processing. J Food Prot. 2003;66(12):2276–82. 10.4315/0362-028x-66.12.2276 [DOI] [PubMed] [Google Scholar]
- 57.Davis BJ, Jacobs JM, Davis MF, Schwab KJ, DePaola A, Curriero FC. Environmental determinants of Vibrio parahaemolyticus in the Chesapeake Bay. Appl Environ Microbiol. 2017;83(21):e01147–17. 10.1128/AEM.01147-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parveen S, Hettiarachchi KA, Bowers JC, Jones JL, Tamplin ML, McKay R, et al. Seasonal distribution of total and pathogenic Vibrio parahaemolyticus in Chesapeake Bay oysters and waters. Int J Food Microbiol. 2008;128(2):354–61. 10.1016/j.ijfoodmicro.2008.09.019 [DOI] [PubMed] [Google Scholar]
- 59.National Ocean Service Education. Estuaries: Dissolved Oxygen: National Oceanic and Atmospheric Administration 2020. [Available from: https://oceanservice.noaa.gov/education/kits/estuaries/media/supp_estuar10d_disolvedox.html. [Google Scholar]
- 60.U.S. Food and Drug Administration, Center for Food Safety and Applied Nutrition: Fish and Fishery Products Hazards and Controls Guidance Fourth Edition, Appendix 5: FDA and EPA Safety Levels in Regulations and Guidance, (2020). [Google Scholar]
- 61.Urquhart EA, Jones SH, Jong WY, Schuster BM, Marcinkiewicz AL, Whistler CA, et al. Environmental conditions associated with elevated Vibrio parahaemolyticus concentrations in Great Bay Estuary, New Hampshire. PloS one. 2016;11(5). 10.1371/journal.pone.0155018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takemura AF, Chien DM, Polz MF. Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Frontiers in microbiology. 2014;5:38 10.3389/fmicb.2014.00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ozbay G, Karuna Chintapenta L, Lingham T, Lumor S, Lee J-l, Taylor B, et al. Delaware Inland Bays and Market Oyster (Crassostrea virginica) Quality for Consumption. J Food Qual. 2018;2018. [Google Scholar]
- 64.Johnson CN, Bowers JC, Griffitt KJ, Molina V, Clostio RW, Pei S, et al. Ecology of Vibrio parahaemolyticus and Vibrio vulnificus in the coastal and estuarine waters of Louisiana, Maryland, Mississippi, and Washington (United States). Appl Environ Microbiol. 2012;78(20):7249–57. 10.1128/AEM.01296-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zimmerman A, DePaola A, Bowers J, Krantz J, Nordstrom J, Johnson C, et al. Variability of total and pathogenic Vibrio parahaemolyticus densities in northern Gulf of Mexico water and oysters. Appl Environ Microbiol. 2007;73(23):7589–96. 10.1128/AEM.01700-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tey YH, Jong K-J, Fen S-Y, Wong H-C. Occurrence of Vibrio parahaemolyticus, Vibrio cholerae, and Vibrio vulnificus in the aquacultural environments of Taiwan. J Food Prot. 2015;78(5):969–76. 10.4315/0362-028X.JFP-14-405 [DOI] [PubMed] [Google Scholar]
- 67.Yu W-T, Jong K-J, Lin Y-R, Tsai S-e, Tey YH, Wong H-c. Prevalence of Vibrio parahaemolyticus in oyster and clam culturing environments in Taiwan. Int J Food Microbiol. 2013;160(3):185–92. 10.1016/j.ijfoodmicro.2012.11.002 [DOI] [PubMed] [Google Scholar]
- 68.Martinez-Urtaza J, Lozano-Leon A, Varela-Pet J, Trinanes J, Pazos Y, Garcia-Martin O. Environmental determinants of the occurrence and distribution of Vibrio parahaemolyticus in the rias of Galicia, Spain. Appl Environ Microbiol. 2008;74(1):265–74. 10.1128/AEM.01307-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Froelich B, Ayrapetyan M, Fowler P, Oliver J, Noble R. Development of a matrix tool for the prediction of Vibrio species in oysters harvested from North Carolina. Appl Environ Microbiol. 2015;81(3):1111–9. 10.1128/AEM.03206-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cruz C, Hedderley D, Fletcher G. Long-term study of Vibrio parahaemolyticus prevalence and distribution in New Zealand shellfish. Appl Environ Microbiol. 2015;81(7):2320–7. 10.1128/AEM.04020-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kirs M, Depaola A, Fyfe R, Jones J, Krantz J, Van Laanen A, et al. A survey of oysters (Crassostrea gigas) in New Zealand for Vibrio parahaemolyticus and Vibrio vulnificus. Int J Food Microbiol. 2011;147(2):149–53. 10.1016/j.ijfoodmicro.2011.03.012 [DOI] [PubMed] [Google Scholar]
- 72.Deepanjali A, Kumar HS, Karunasagar I. Seasonal variation in abundance of total and pathogenic Vibrio parahaemolyticus bacteria in oysters along the southwest coast of India. Appl Environ Microbiol. 2005;71(7):3575–80. 10.1128/AEM.71.7.3575-3580.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li R, Chiou J, Chan EW-C, Chen S. A novel PCR-based approach for accurate identification of Vibrio parahaemolyticus. Frontiers in microbiology. 2016;7:44 10.3389/fmicb.2016.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.U.S. Food and Drug Administration, National Shellfish Sanitation Program: Guide for the control of molluscan shellfish (2017). [Google Scholar]
- 75.Yang Y, Xie J, Li H, Tan S, Chen Y, Yu H. Prevalence, antibiotic susceptibility and diversity of Vibrio parahaemolyticus isolates in seafood from South China. Frontiers in Microbiology. 2017;8:2566 10.3389/fmicb.2017.02566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rojas MVR, Matté MH, Dropa M, Silva MLD, Matté GR. Characterization of Vibrio parahaemolyticus isolated from oysters and mussels in São Paulo, Brazil. Revista do Instituto de Medicina Tropical de São Paulo. 2011;53(4):201–5. 10.1590/s0036-46652011000400005 [DOI] [PubMed] [Google Scholar]
- 77.Wagley S, Koofhethile K, Wing JB, Rangdale R. Comparison of V. parahaemolyticus isolated from seafoods and cases of gastrointestinal disease in the UK. Int J Environ Health Res. 2008;18(4):283–93. 10.1080/09603120801911064 [DOI] [PubMed] [Google Scholar]
- 78.Williams TC, Froelich BA, Phippen B, Fowler P, Noble RT, Oliver JD. Different abundance and correlational patterns exist between total and presumed pathogenic Vibrio vulnificus and V. parahaemolyticus in shellfish and waters along the North Carolina coast. FEMS Microbiol Ecol. 2017;93(6):fix071 10.1093/femsec/fix071 [DOI] [PubMed] [Google Scholar]
- 79.Delaware Department of Natural Resources and Environmental Control (DNREC). Surface Water Quality Monitoring Program: Delaware Water Quality Portal; 2020. [Available from: http://demac.udel.edu/waterquality/. [Google Scholar]