Abstract
Aedes aegypti Act4 is a paralog of the Drosophila melanogaster indirect flight muscle actin gene Act88F. Act88F has been shown to be haploinsufficient for flight in both males and females (amorphic mutants are dominant). Whereas Act88F is expressed in indirect flight muscles of both males and females, expression of Act4 is substantially female-specific. We therefore used CRISPR/Cas9 and homology directed repair to examine the phenotype of Act4 mutants in two Culicine mosquitoes, Aedes aegypti and Culex quinquefasciatus. A screen for dominant female-flightless mutants in Cx. quinquefasciatus identified one such mutant associated with a six base pair deletion in the CxAct4 coding region. A similar screen in Ae. aegypti identified no dominant mutants. Disruption of the AeAct4 gene by homology-dependent insertion of a fluorescent protein marker cassette gave a recessive female-flightless phenotype in Ae. aegypti. Reproducing the six-base deletion from Cx. quinquefasciatus in Ae. aegypti using oligo-directed mutagenesis generated dominant female-flightless mutants and identified additional dominant female-flightless mutants with other in-frame insertions or deletions. Our data indicate that loss of function mutations in the AeAct4 gene are recessive but that short in-frame deletions produce dominant-negative versions of the AeAct4 protein that interfere with flight muscle function. This makes Act4 an interesting candidate for genetic control methods, particularly population-suppression gene drives targeting female viability/fertility.
Author summary
The yellow fever mosquito and the Southern house mosquito are important vectors of infectious diseases. Given their widespread presence across tropical and subtropical regions of the world and the increased risk of spread due to global warming there is a growing need for population control. Gene drives aim to spread a genetic element within target genes required for mosquito reproduction to disrupt their function and crash a population. Female-specific genes provide interesting candidates for population control since female mosquitoes determine the reproductive capacity of a population as well as being the actual vectors of disease. Here we describe a study on Actin-4 loss in both Aedes aegypti and Culex quinquefasciatus, where we observe female-specific disruption of flight ability and propose it as a candidate for genetic methods of population suppression.
Introduction
Insect-borne diseases have a huge impact on human health worldwide, in particular mosquito-borne diseases which result in over 1 million deaths every year. Around 17% of the world’s infectious disease disability adjusted life year (DALY) burden is estimated to be due to insect-borne diseases, of which 90% is attributed to mosquito-transmitted diseases [1].
With the advent of promising gene drive strategies, such as homing endonuclease based or CRISPR-Cas9 daisy chain drives [2–4], there is a growing need to study potential target genes in key target mosquito species such as the yellow fever mosquito, Aedes aegypti (L.), and the Southern house mosquito, Culex quinquefasciatus (Say). Ae. aegypti is an anthropophilic mosquito present in Central and South America, Africa, South East Asia, and Oceania [5]. It is the principal vector of human dengue which has an estimated incidence of 390 million infections per year worldwide [6]. It is also an important vector of other arbovirus diseases such as yellow fever [7], Chikungunya [8] and Zika [9]. Cx. quinquefasciatus is a widespread mosquito found in tropical and subtropical regions of the world and is a vector of both human and wildlife pathogens [10]. This species is the main vector of the avian malaria parasite Plasmodium relictum [11,12], as well as the human parasitic worm (Wuchereria bancrofti) [13,14] and is also able to transmit arboviruses such as West Nile virus [15,16].
A leading design of gene drive for population suppression has a ‘homing’ drive located in and disrupting a target gene resulting in a corresponding increase between frequency of both the gene drive and the loss-of-function alleles [2,3,17]. If the target gene is required for viability or fertility, this will lead to population suppression. Reduced ability of the gene drive to spread can be minimized by employing a recessive mutant phenotype, e.g. only females, and if the target gene is required only in somatic cells while homing is restricted to germline cells. Other designs require dominant lethal phenotypes, again preferably female-specific [18]. Few genes affecting female viability/fertility are known in mosquitoes, though analysis of ovarian expression identified some candidates in Anopheles gambiae [19]. We investigated a different functional process, flight, as a potential source of target genes. Ae. aegypti mate in flight, and the female wingbeat frequency is a primary mating recognition cue, therefore flightless females are functionally sterile [20,21] as well as being unable to disperse from their larval pools, evade predators or blood feed, let alone a second time ensuring lack of transmission.
Mechanical power during insect flight is provided by the action of indirect flight muscles (IFMs) in the adult thorax [22]. Actin filaments in these fibrillar muscles are formed by polymerization of a particular actin isoform; Actin-3 in Drosophila melanogaster [23] encoded by Act88F [24]. Actin-4, present in both Ae. aegypti and Cx. quinquefasciatus, is a paralog of Act88F but with actin-coding transcripts expressed in IFMs of females only [25,26]. The promoter of AeAct4 has been previously exploited to develop a RIDL strain displaying a repressible female flightless phenotype [27]. The same strategy has been shown to work in Aedes albopictus, and also that Act4 promoters from either species are functional in the other species [25].
Classical mutagenesis studies indicated that Act88F is haploinsufficient for flight in both male and female D. melanogaster adults [23,28]. Haploinsufficient genes are rare in nature; apart from Act88F, the few examples known in the model organism D. melanogaster are primarily ribosomal protein genes [29]. Both haplosufficient and haploinsufficient genes for female viability or fertility are potentially useful for genetic control. Haplosufficient genes would make useful target genes in a daisy chain drive configuration whilst haploinsufficient genes could be part of an underdominance based drive [4,30]. We therefore investigated the phenotype of mutations in AeAct4 (AAEL001951) in Ae. aegypti and CxAct4 (CPIJ012572) in Cx. quinquefasciatus.
Methods
Mosquito rearing
The Ae. aegypti Liverpool (LVP) strain originated from West Africa and has been maintained at the Liverpool School of Tropical Medicine since 1936 [31]. The Cx. quinquefasciatus Pel strain was originally established from larvae collected in Peliyagoda, Sri Lanka in 1984 [32]. An exu-Cas9 Ae. aegypti line [33] was used in a set of microinjections due to its Cas9 germline activity. The exu-Cas9 line was highly enriched through the removal of any marker negatives for at least 3 generations.
Larvae were reared on dry fish food (TetraMin, Tetra GmbH, Germany) and adults were provided 10% sucrose ad libitum. Insectary rearing conditions were maintained at 27°C (± 2°C), 70% (± 10%) relative humidity and 12:12 light/dark cycle. For egg collection, adult females were fed on defibrinated horse blood (TCS Biosciences Ltd, Buckingham, UK) at 37°C using a Hemotek membrane feeding system (Hemotek Ltd, Blackburn, UK).
Microinjection
Microinjection into Ae. aegypti and Cx. quinquefasciatus pre-blastoderm embryos was carried out following published protocols [34,35]. Clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 homology directed repair (HDR) injections were carried out as described [36], except injected G0 males were crossed instead of discarded. Cas9 microinjections used recombinant Cas9 protein, which was obtained commercially (PNA Bio, Newbury Park, CA, US). Injection mixes consisted of 40 ng/μl of each synthetic single guide RNA (sgRNA), 400 ng/μl of Cas9 protein, 700 ng/μl of dsDNA HDR donor plasmid, and injection buffer [37]. Male G0 adults were individually crossed to 5 females in 12 ounce clear plastic deli pots (Ambican, London, UK) and then pooled in groups of 20 G0 males per cage for blood-feeding and egg collection. Female G0 adults were crossed to males in a 1:1 ratio in pools of 100 G0 females per cage for blood-feeding and egg collection. 15x15x15cm Bugdorm cages (NHBS Ltd, Devon, UK) were used for pooling individuals after being crossed. G1 larvae were screened for mCherry fluorescence using a Leica MZ10 microscope (Leica Biosystems, Wetzlar, Germany). Transgenic lines were established from single G1 positives. The exu-Cas9 line was used for one set of microinjections in Ae. aegypti and did not require the described co-injection of recombinant Cas9 protein. Considering that exu-Cas9 adults were not all homozygotes, eggs collected for microinjection were obtained from individual females identified as expressing the fluorescent marker.
sgRNA design and plasmid construction
The Ae. aegypti Actin-4 gene sequence, AAEL001951, has been previously identified [26] and CPIJ012572 is annotated as a one-to-one ortholog in Vectorbase [38].
CRISPR/Cas9 sgRNAs were designed using CHOP-CHOP online prediction tool [39] to target approximately the first 200 bp of the coding region to maximize the likelihood of generating null alleles. Candidate sgRNAs (S1 Table) were assessed for in vitro efficacy through cutting of an Act4 template by Cas9 protein (S2 Table). LA1429, the single stranded oligo DNA nucleotides (ssODN) donor for HDR, was designed following standard protocols for HDR in Ae. aegypti [36].
AGG1070, the AeAct4 HDR 3xP3-mCherry dsDNA donor, was built through three-way Golden Gate cloning [40] of the 3xP3-mCherry-SV40 marker cassette (amplified from an existing plasmid, AGG1069) and the two AeAct4 homology arms (2Kb each) that were amplified from LVP genomic DNA. Primer sequences are provided in S4 Table. Plasmid sequences are provided in S5 Table.
Analysis of CRISPR/Cas9 induced mutations
Genomic DNA was extracted from individual mosquitoes using the NucleoSpin Tissue kit (Macherey-Nagel, Düren, Germany). The putative mutated region was PCR amplified to detect CRISPR/Cas9 mediated indels or HDR integrations using primers shown in S3 Table. Selected amplicons were Sanger sequenced (Eurofins, Ebersberg, Germany) and the chromatograms were analyzed to determine the nature of the mutations.
Flightless phenotype analysis
G1 adults were assessed for flight ability in 15x15x15cm Bugdorm cages (NHBS Ltd, Devon, UK). A sucrose source (10% sucrose-soaked cotton wool) was placed at the bottom of cages to be accessible without requiring flight ability. Ability to fly was assayed by flight, or lack thereof, upon tapping the cage as previously described in mosquitoes [25,27]. Initial selection of flightless individuals was not stringent, bad fliers and individuals with a range of wing or other defects were isolated. These were considered ‘flightless’ in the final count if they were fully flightless for ~7 days. Nonetheless, all individuals from the initial selection were also analyzed molecularly.
Transmission electron microscopy
AeAct4hdr1 females (heterozygous) were crossed to AeAct4hdr1 males in order to obtain double knock-out and thus non-flying females. Legs, wings, heads and abdomens were removed from 5–7 day-post-emergence adult females and individual thoraces were prepared for TEM. Thoraces were placed in 2% glutaraldehyde in phosphate buffer for 3 hours, after which time the fixative was replaced with 1% aqueous osmium tetroxide. After 90 minutes samples were dehydrated in a series of ethanol dilutions as follows; 70% ethanol for 45 minutes, 90% ethanol for 30 minutes, 3 x 100% ethanol for 15 minutes each. After a 15 minute wash in propylene oxide, samples were placed in a 1:1 mix of propylene oxide and freshly made Agar 100 epoxy resin for 60 minutes before a 2 hour infiltration period in pure epoxy resin. Thoraces were mounted in flat embedding molds before being placed in the polymerization oven at 60°C for 16–24 hours. Every step before polymerization was carried out at room temperature on a rotator to agitate the samples in the solutions. Glutaraldehyde, osmium tetroxide, propylene oxide and Agar 100 epoxy resin were all sourced from Agar Scientific Ltd, UK. Thick sections (0.5μm) were cut using a Leica UC7 ultramicrotome, stained with aqueous 1% toluidine blue and viewed in a transmission light microscope (Leica Microsystems Ltd.) to identify areas of indirect flight muscle. Thin sections (70 nm) of the correct areas were cut, collected on 200 mesh thin bar copper grids and stained with uranyl acetate and lead citrate using a Leica EM Stain before being imaged in a FEI Tecnai12 TEM with a Tietz F214 CCD camera or Thermo Fisher Talos L120C TEM with Ceta camera.
Statistical analysis
The Chi-squared test was conducted using Microsoft Excel to determine the statistical significance between observed and expected progeny distributions.
Results
Generation of flightless phenotype in Cx. quinquefasciatus
Recent advances in the use of CRISPR/Cas9 mutagenesis in Ae. aegypti have facilitated genetic analysis of individual genes of interest in non-model organisms including mosquitoes [33,36]. In vivo mutagenesis of Act4 in Cx. quinquefasciatus (CxAct4) was carried out by co-injecting 915 G0 eggs with sgRNAs 1–4 and Cas9 protein (Table 1). G0 survivors were outcrossed individually to wild-type (Pel) adults to be able to observe potential mutations in a heterozygous genetic background. All G0 adults were able to fly normally, showing no evident phenotype despite their presumed mosaic nature. Despite the low G0 survival rate (0.7%) and the few G1 adults obtained (8 males and 8 females, from a single egg raft from one G0 founder) compared to published rates [34,41,42], 3 flightless females were identified. Sanger sequencing of CxAct4 in flightless G1 females indicated these flightless females were heterozygous for a 6bp deletion (TGCCTA) of nucleotides 156 through 161 from the start codon, with the A of the ATG being 1 (156_161delTGCCTA). Interestingly, analysis of CxAct4 in 5 flying males revealed the same in-frame deletion in 1 individual. The identified mutation affects three different codons, resulting in the deletion of two amino acids (A53_Y54del), as well as use of an alternative codon (GAC instead of GAT) for amino acid D52. This mutation appears to be dominant for lack of flight in Cx. quinquefasciatus females.
Table 1. CRISPR Cas9/sgRNA Act4 mutagenesis in Cx. quinquefasciatus and Ae. aegypti.
| Mosquito species (strain) | Injected components | Injected G0 eggs | Adult G0 survivors (%) | G0 mosaic females (%) | G0 founders (%) | G1 flightless female adults/total female G1 (%) | G1 sequence confirmed Act4 mutant flightless females (%) | Mutation Identified |
|---|---|---|---|---|---|---|---|---|
| Cx. quinquefasciatus (Pel) | CxAct4 sgRNAs (1–4), Cas9 protein | 915 | 6 (0.7) | 0 (0) | 1 (17) | 3/8 (38) | 3/3 (100) | 156_161delTGCCTA (A53_Y54del) |
| Ae. aegypti (Liverpool) | AeAct4 sgRNAs (1, 2), Cas9 protein | 1802 | 67 (4) | 0 (0) | 0 (0) | 1/3713 (0.03) | 0/1 (0) | - |
| Ae. aegypti (Liverpool) | AeAct4 sgRNAs (3, 4), Cas9 protein | 736 | 68 (9) | 0 (0) | 0 (0) | 1/3422 (0.03) | 0/1 (0) | - |
Act4 CRISPR/Cas9 mutagenesis in 2 mosquito species. Potential mutants were screened using a flight assay. sgRNA concentration was 40 ng/μl and Cas9 protein 300 ng/μl.
An equivalent Act4 in vivo mutagenesis attempt was carried out in Ae. aegypti by co-injecting embryos with sgRNAs and Cas9 protein. All injection survivor G0 adults were able to fly normally, showing no evidence of a mosaic effect. One flightless G1 female was isolated from each set of injections (Table 1). However, following analysis of Sanger sequencing of the AeAct4 locus no mutation was identified.
AeAct4 gene disruption leads to recessive female-specific loss of flight ability
Disruption of Act4 in Ae. aegypti was achieved via insertion of a marker cassette through homology directed repair (HDR) (Fig 1A). The HDR donor construct (AGG1070) consisted of a 3xP3-mCherry-SV40 marker cassette flanked by 2kb homology arms corresponding to the immediate upstream and downstream regions of the outermost AeAct4 sgRNA cut sites [43]. AeAct4 sgRNAs targeted the first 220 nucleotides of the coding region to increase likelihood of generating non-functional protein. 600 G0 eggs were co-injected with a donor plasmid (AGG1070), sgRNAs (1–4), and Cas9 protein (Table 2). All G0 adults were able to fly normally, showing no evidence of a mosaic effect. G1 larvae were screened, for presence of the fluorescent marker. Both marker positive and negative female G1 adults were analyzed for their flight ability.
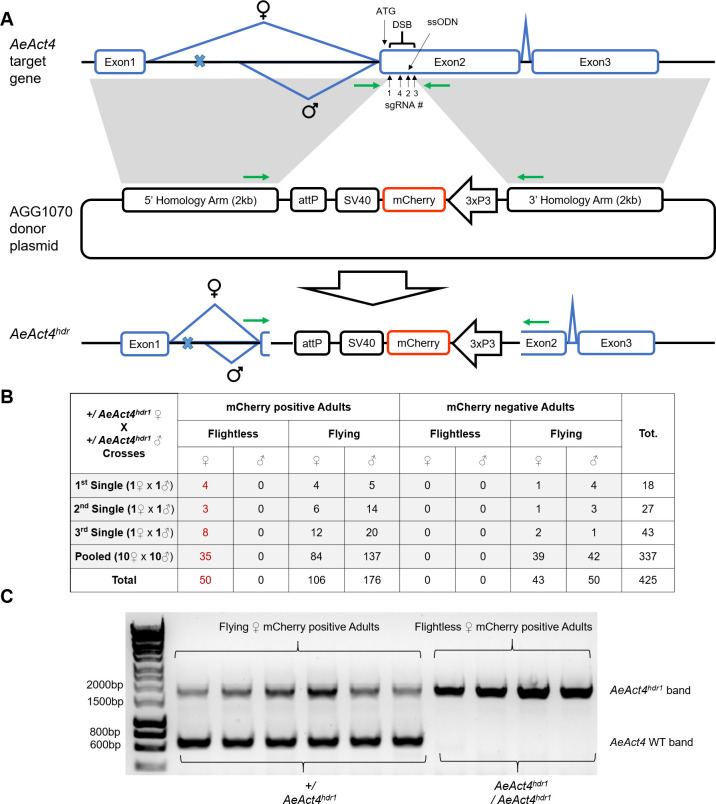
Fig 1. AeAct4 gene disruption by homology-directed repair results in female-specific recessive lack of flight ability.
(A) Injected Cas9 protein and sgRNAs targeting AeAct4 induce a double stranded break (DSB) at the target site. A donor plasmid (AGG1070) having 2kb homology arms corresponding to the immediate upstream and downstream regions of the outermost cut sites was injected as a template for HDR. A 3xP3-mCherry-SV40 cassette serves as a marker for integration. Male-specific alternative splicing incorporates a number of early start and stop codons (marked by blue x) which ablate AeAct4 translation. Green arrows represent primer pair used for amplicons shown in (c). (B) 50 flightless females were identified out of a total of 156 marker-positive female progeny (32%). (C) Molecular confirmation of 6 flying and 4 flightless mCherry positive female adults, individually analyzed by PCR using the primers indicated in (A). AeAct4hdr1 and WT amplicons were generated. Based on this PCR assay, of the marker-positive females analyzed, all fliers were heterozygous for AeAct4hdr1 and all flightless females were homozygous.
Table 2. HDR mediated CRISPR Cas9/sgRNA AeAct4 mutagenesis in Ae. aegypti.
| Knock-out only (no HDR marker) | HDR mediated knock-in | |||||||
|---|---|---|---|---|---|---|---|---|
| Injected components | Injected G0 eggs | Adult G0 survivors (%) | G0 mosaic females (%) | G0 founder pools | flightless G1 females /total G1 females (%) | G1 AeAct4 mutant flightless females (%) | G0 founder pools | G1 marker positive adults/total G1 (%) |
| AGG1070 AeAct4 donor plasmid, Cas9 protein, AeAct4 sgRNAs (1–4) |
600 | 97 (16.2) | 0 (0) | 5 | 8/3557 (0.2) | 2/8 (25) | 1 | 14/7198 (0.2) |
AeAct4 inactivation by CRISPR/Cas9 mediated knock-in of a marker cassette. An initial fluorescence marker screen identified knock-ins from WT or knock-outs. sgRNA concentration was 40 ng/μl, Cas9 protein 400 ng/μl and donor plasmid 700 ng/μl.
14 G1 larvae were identified by expression of the mCherry marker. The resulting transgenic line was named AeAct4hdr1. All the AeAct4hdr1 marker positive G1 adults were able to fly normally. Transgene heterozygotes were interbred and their progeny analyzed for flight ability (Fig 1B). All male progeny were able to fly normally, however a total of 50 out of 156 marker-positive females were identified as flightless. This is not significantly different from the 1:2:1 ratio expected in females (WT:AeAct4hdr1 heterozygotes:AeAct4hdr1 homozygotes) from the hypothesis that all genotypes are viable but homozygous females cannot fly, i.e. that AeAct4 disruption induces a recessive female flightless phenotype (X2 = 1.34, d.f. = 2, p = 0.51). The genetic integration into AeAct4 gene was confirmed by sequence analysis of PCR products generated from AeAct4hdr1 individuals: 4 flying and 6 flightless female G2 adults (Fig 1C). PCR across the insertion gave two amplicons (WT and AeAct4hdr1 allele) in all flying G2 individuals (heterozygotes) whilst only the AeAct4hdr1 amplicon appears in flightless G2s (homozygotes). The phenotype of this insertion, and probably the null phenotype of AeAct4, is therefore recessive female-specific loss of flight ability.
Additionally, from the marker negative G1 adults, 8 flightless females were identified, of which 2 carried mutations (34_40delGACAACG and 207_209delATA) in the AeAct4 locus. The two confirmed mutations in non-marker flightless females appear to be generated by sgRNA 1 and sgRNA 3 respectively. The 6 flightless females which do not show mutations in AeAct4 could correspond to the rate of flightless adults observed in WT samples. The other 2 flightless females carried different mutations which may or may not be responsible for the flightless phenotype. The 207_209delATA mutant could be a dominant negative version since it is in frame whilst 34_40delGACAACG is an out of frame 7bp deletion and is unlikely to result in a dominant negative.
The 6bp in-frame deletion generated in Cx. quinquefasciatus is dominant negative for flight in Ae. aegypti females
Since the single CxAct4 mutant was dominant, yet amorphic mutants of AeAct4 are recessive, we speculated that the CxAct4 mutant might represent a dominant negative mutant. To investigate this, we conducted HDR mutagenesis of AeAct4, aiming to replicate the CxAct4 deletion (156_161delTGCCTA) in Ae. aegypti. A 120bp single stranded oligonucleotide donor (ssODN) was designed (LA1422) to contain 60bp upstream and downstream of 156_161delTGCCTA but not the deleted sequence itself. 580 Ae. aegypti G0 eggs from an exu-Cas9 line [33] were co-injected with AeAct4 sgRNA2 and ssODN (Table 3). Of 187 adult G0 survivors (32.2% survival rate), 85 were females. After analyzing their flight ability, 5 of these G0 females (5.9%) were not able to fly suggesting at least a mosaic effect. The other G0 adults were backcrossed to wild-type (LVP) mosquitoes. G1 individuals were analyzed for flight ability and 27 flightless G1 females were identified, from 7 of the 9 G0 pools. The mutation rate observed when exu-Cas9 line was used for injections is notably higher than the rate observed when Cas9 protein was used (Table 2 knockout only G1 flightless 0.2%, and Table 3 G1 flightless females 1.4%), although the donor template for HDR was single stranded in this case. A sample of these G1 individuals (n = 17) were analyzed through Sanger sequencing. 14 of the 17 flightless females showed mutations in the AeAct4 locus; the other 3 flightless females and all of the 17 flying females appeared to be wild-type in the sequenced region. Out of these 14 mutants, 6 individuals from 2 different pools had the expected in-frame 6bp deletion (156_161delTGCCTA). Each of the other 8 mutant G1 flightless females had other in-frame mutations at the sgRNA2 target site (Table 4).
Table 3. 156_161delTGCCTA ssODN HDR CRISPR/Cas9/sgRNA AeAct4 mutagenesis in Ae. aegypti.
| 17 flightless and 17 flying female G1s were processed for AeAct4 sanger sequencing | ||||||||
|---|---|---|---|---|---|---|---|---|
| Injected components | No. Injected | G0 survivors (%) | G0 mosaic females (%) | G1 flightless female adults/total female G1s (%) | G0 founders (%) | 156_161delTGCCTA AeAct4 mutant flightless females/total G1 females (%) | AeAct4 mutant flightless females/total flightless female G1s sequenced (%) | AeAct4 mutant flying females/total flying female sequenced (%) |
| LA1422 ssODN AeAct4 donor oligo, AeAct4 sgRNA 2 | 580 | 187 (32.2) | 5 (5.9) | 27/1978 (1.4) | ≥2 (1) | 6/1978 (0.3) | 14/17 (82) | 0/17 (0) |
The 156_161delTGCCTA 6bp deletion found in Cx. quinquefasciatus was replicated in Ae. aegypti via ssODN HDR CRISPR/Cas9 mutagenesis of an exu-Cas9 line [33]. Potential flightless G0 mosaics and G1 mutants. 17 flightless and 17 flying G1s were selected for Sanger sequencing of AeAct4. All flying G1s had wild type sequence. 14 out of the 17 selected flightless G1s had in-frame indels, 6 of which showed the intended 156_161delTGCCTA deletion. These 6 individuals originated from 2 different G0 pools. G0s were crossed in pools; hence, the minimum value for founders. To assess the rate of targeted mutation, rather than all mutation, G0s were only considered founders if they resulted in G1s with successful HDR events reproducing the donor template 6bp deletion; founders of other mutagenesis events are not included. sgRNA concentration was 40 ng/μl and donor oligo 125 ng/μl.
Table 4. Potential antimorphic mutations from ssODN HDR CRISPR Cas9/sgRNA AeAct4 mutagenesis in Ae. aegypti.
| AeAct4 Mutation | Amino acids changed | Description | In frame? | G1 flightless females |
|---|---|---|---|---|
| 122_157delACCAGGGTGTGATGGTCGGTATGGGTCAAAAAGATG | H41_A53delinsP (delHQGVMVGMGQKDA/insP) | 36bp deletion | Yes | 2A |
| 127_162delGGTGTGATGGTCGGTATGGGTCAAAAAGATGCCTAC | G43_Y54del (delGVMVGMGQKDAY) | 36bp deletion | Yes | 2B |
| 159_164delCTACGT | Y54_V55del | 6bp deletion | Yes | 4A, 4B, 4C |
| 156_161delTGCCTAinsCGGCAC | A53_Y54delinsGT | 6bp substitution | Yes | 7A |
| 159_161delCTA | Y54del | 3bp deletion | Yes | 8A |
| 160_170delTACGTCGGTGAinsGACAATTT | Y54_D57delinsDNF (delYVGD/insDNF) | 11bp substituted with 8bp | Yes | 8D |
Additional potentially dominant negative mutations from ssODN mutagenesis are shown. 8 flightless females had mutations other than the intended 156_161delTGCCTA deletion. G1 flightless females are numbered by the G0 pool with a different letter for each flightless individual from the same G0 pool.
Loss of Act4 in homozygous null Ae. aegypti females leads to disrupted muscle fibers in female indirect flight muscles
Indirect flight muscles are arranged perpendicularly within the thorax. These muscle fibers are composed of groups of largely parallel myofibrils interspersed with numerous mitochondria. Indirect flight muscles of dissected female adults (AeAct4hdr1 homozygotes, n = 3 and AeAct4hdr1 heterozygotes, n = 1) as well as LVP adults (n = 2) were imaged using transmission electron microscopy (TEM, Fig 2).
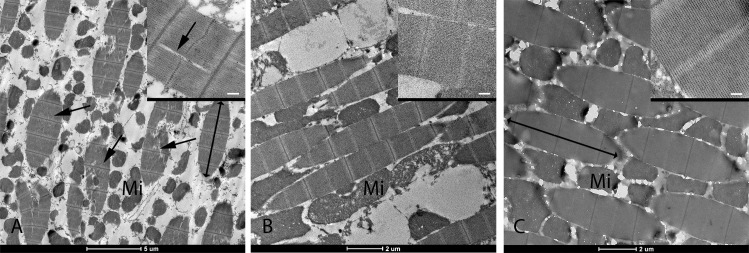
Fig 2. Transmission electron microscope images of the indirect flight muscles of Ae. aegypti female adults arranged within the thorax.
Filaments in the myofibrils appear to be disrupted in AeAct4hdr1 homozygotes (A, arrows and inset) but not in AeAct4hdr1 heterozygotes (B, inset) or WT (C, inset). Individual myofibrils are indicated with double-headed arrows, and mitochondria (Mi) in the muscle fiber cells are clearly visible. Insert scale bars = 200 nm.
The myofibrils in homozygotes (Fig 2A) do not display the normal regular pattern of tightly bundled actin and myosin filaments running parallel and close to each other. Overall, the filaments show a more disrupted and irregular arrangement, when compared to those of the heterozygotes (Fig 2B) or WT (Fig 2C).
Discussion
We have shown that disruption of the Act4 gene by knock-in of a large insertion leads to a recessive, female-specific, loss-of-flight phenotype, which we propose is the amorphic phenotype of this gene. This notably differs from the phenotype of the Drosophila orthologue Act88F by being recessive and female specific. Numerous dominant flightless mutants of Act88F have been described [24,28,44]. These dominant mutations were then classified on the basis of phenotype in the presence of two wild type alleles of Act88F as hypomorphic (including amorphic) or antimorphic. Hypomorphic alleles—flight ability restored in the presence of two wild type alleles in addition to the mutant allele—include premature stop codons at Trp80 and Gln122 [24,45], whereas more C-terminal truncations and many missense mutations were still flightless in the presence of two or more wild type alleles and therefore classified as antimorphic. Act88F and AeAct4 proteins are very similar; both 376 amino acids in length, only 17 amino acids differ from each other, they share 95% sequence similarity. On this basis we expected hypomorphic mutations in the region of AeAct4 targeted to confer a dominant flightless phenotype, as for Act88F but female-specific since the expression of actin-coding AeAct4 transcripts is restricted to females [26,27]. However, HDR knock-in of a fluorescent marker into the 5’ end of AeAct4 coding region gave a recessive female-specific flightless phenotype instead. Consistent with this, CRISPR/Cas9 mutagenesis of AeAct4 did not generate significant numbers of flightless G0 or G1 flightless individuals. This also suggests that a degree of mosaicism for AeAct4 does not prevent flight, either because wild type muscle cells can compensate or because the syncytial nature of muscle cells means that enough Act4 can be produced even if some nuclei are unable to do so.
Phenotypic analysis via dissection and examination of muscle fibers in AeAct4hdr1 heterozygote females showed no obvious visible defects, as seen in the TEM, which correlates with the observed flying phenotype. However, AeAct4hdr1 homozygote flightless females showed defects in myofibril structure in IFMs. The regular wild-type pattern of parallel filaments was disrupted, however the overall muscle structure of IFMs was maintained in homozygotes. Similar analysis of Act88F mutants showed that, perhaps surprisingly, neither Act88F nor the IFM-specific myosin heavy chain isoform are required for the development of muscle fibers [46].
Though the knock-out (amorph or strong hypomorph) phenotype is recessive, we additionally generated a series of dominant mutants. We observed a dominant female-specific flightless phenotype in Cx. quinquefasciatus carrying a CRISPR/Cas9-generated six-base pair deletion in Act4. We further showed that the same deletion gave a similar phenotype when replicated in Aedes aegypti by CRISPR/Cas9 oligo-directed mutagenesis. Although this does not prove that CxAct4 is also haplosufficient, that hypothesis seems plausible. We additionally recovered a series of other dominant mutants, all being in-frame indels with net deletions of 3-36bp in the targeted region which are likely antimorphic. The fact that all of these mutations are in frame could be due to the fact that out of frame mutations are less likely to encode full length Act4-like proteins and hence be hypomorphic or amorphic meaning they would not have been detected in the mutant screen as performed since mutants would have been heterozygous.
A key motivation for analyzing the Act4 mutant phenotype was to assess its relevance in genetic control. Loss of flight ability means that females cannot fly away from their larval habitats to seek a blood meal or to avoid predators. Aedes aegypti males—which do not bite or transmit disease—use female wingbeat frequency as a primary mate-recognition signal, therefore flightless females are also functionally sterile [20]. Flight is only relevant to adults, so the phenotype acts after any density-dependence effects acting on immature stages. For these reasons, dominant female-specific loss of flight has been proposed as a desirable engineered trait for genetic control [25,27]. Germline restriction of gene drive components is an important goal as unwanted somatic expression could lead to undesirable effects in fertility or viability which could prevent efficient gene drive. Therefore recessive female-specific lethal or sterile phenotypes are potentially useful for population suppression drives [2], especially where the gene function is not required in the germline or neighboring tissues. Further studies are required to assess any qualitative difference in flight ability in heterozygous females (which can fly by our assay) as well as the mating competitiveness of AeAct4 mutant males to evaluate the potential of AeAct4 in genetic control of mosquito populations as well as potential fertility effects in gene drive carriers.
Thus far, much attention has focused on genes involved in female fertility [19,47] but flight should provide another independent pathway to target, with no possibility of cross-resistance. Act4 may also have desirable features in respect of cut-site resistance, a significant problem for CRISPR/Cas9-based homing drives [48,49]. A gene drive targeting a female-fertility gene (AGAP007280) in An. gambiae initially spread in experimental populations but then rapidly dropped out of the population [19,50]. Sequence analysis showed that this was due to accumulation of short in-frame deletions at the cut-site, presumably generated by end-joining repair of the CRISPR/Cas9-induced double-strand break. These deletions appeared to be functional, i.e. fertile, while preventing further cutting by CRISPR/Cas9 and so were strongly selected. In contrast, we found short indels in the targeted region of Act4 to be dominant-negative. The targeting of alleles where mutations are likely to be deleterious has been suggested as a strategy to reduce resistance allele formation in homing drives [51], hence, Act4 would be an interesting target in this sense.
The success of future genetic control strategies for the control of insect species is largely dependent on our growing understanding of their biology. In particular, these strategies will benefit from identifying those genes which are essential for reproduction or survival as the aim is to suppress, or if possible locally eradicate, vector populations. Since flight is a key part of mosquito reproduction and survival it is worth pursuing a better understanding of the specific genes and proteins involved.
Supporting information
Target sequences for the sgRNAs used in this study. Predicted cut sites are between the nucleotide numbers shown, counting the A of the ATG start codon as nucleotide 1.
(DOCX)
In vitro cleavage by sgRNAs was carried out at 37°C for indicated periods. Cutting efficiency of each sgRNA within each species is ranked and shown in brackets.
(DOCX)
Mutation assay primers to detect CRISPR/Cas9 mediated indels or HDR integrations in Act4.
(DOCX)
*point where the 6 bases for the 156_161delTGCCTA deletion have been removed. The ssODN oligo consists of 60 nucleotides upstream and downstream of the deletion.
(DOCX)
Plasmid sequences corresponding to the 3xP3-mCherry-SV40 marker cassette used and the final AeAct4 HDR 3xP3-mCherry nDNA donor.
(DOCX)
Acknowledgments
We thank Rebekah Ireland and other members of the Arthropod Genetics Group for technical assistance. We also thank Dr Jaroslaw Krzywinski for providing us with the Ae. aegypti ‘Liverpool’ strain that originated from Liverpool School of Tropical Medicine and Hygiene. The Cx. quinquefasciatus ‘Pel’ strain was a kind gift from Professor Steven Sinkins.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
DN was funded by an MRC Case studentship (https://mrc.ukri.org/skillscareers/studentships/how-we-fund-studentships/industrial-case-studentships/). IF was supported by an NERC iCASE studentship (https://nerc.ukri.org/funding/available/postgrad/focused/industrial-case/). MAEA and LA were supported through an award from DARPA’s Safe Genes program to MIT N66001-17-2-4054 (https://www.darpa.mil/program/safe-genes) and SB by Wellcome Trust Collaborative Award 200171/Z/15/Z (https://wellcome.ac.uk/) to LA. OSA and ML were supported in part by funding from the Defense Advanced Research Project Agency (DARPA) Safe Genes Program Grant HR0011-17-2- 0047 (https://www.darpa.mil/program/safe-genes). The views, opinions and/or findings expressed are those of the author and should not be interpreted as representing the official views or policies of the U.S. Government. This project was supported through strategic funding from the UK Biotechnology and Biological Sciences Research Council (BBSRC) to The Pirbright Institute BBS/E/I/00007033, BBS/E/I/00007038 and BBS/E/I/00007039 (https://bbsrc.ukri.org/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.McGraw EA, O'Neill SL. Beyond insecticides: new thinking on an ancient problem. Nat Rev Microbiol. 2013;11(3):181–93. Epub 2013/02/16. 10.1038/nrmicro2968 . [DOI] [PubMed] [Google Scholar]
- 2.Burt A. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc Biol Sci. 2003;270(1518):921–8. Epub 2003/06/14. 10.1098/rspb.2002.2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esvelt KM, Smidler AL, Catteruccia F, Church GM. Concerning RNA-guided gene drives for the alteration of wild populations. Elife. 2014;3 Epub 2014/07/19. 10.7554/eLife.03401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noble C, Min J, Olejarz J, Buchthal J, Chavez A, Smidler AL, et al. Daisy-chain gene drives for the alteration of local populations. Proc Natl Acad Sci U S A. 2019;116(17):8275–82. Epub 2019/04/04. 10.1073/pnas.1716358116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kraemer MU, Sinka ME, Duda KA, Mylne A, Shearer FM, Brady OJ, et al. The global compendium of Aedes aegypti and Ae. albopictus occurrence. Sci Data. 2015;2:150035 Epub 2015/07/16. 10.1038/sdata.2015.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, et al. The global distribution and burden of dengue. Nature. 2013;496(7446):504–7. Epub 2013/04/09. 10.1038/nature12060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monath TP, Vasconcelos PF. Yellow fever. J Clin Virol. 2015;64:160–73. Epub 2014/12/03. 10.1016/j.jcv.2014.08.030 . [DOI] [PubMed] [Google Scholar]
- 8.Pialoux G, Gauzere BA, Jaureguiberry S, Strobel M. Chikungunya, an epidemic arbovirosis. Lancet Infect Dis. 2007;7(5):319–27. Epub 2007/04/24. 10.1016/S1473-3099(07)70107-X . [DOI] [PubMed] [Google Scholar]
- 9.Song BH, Yun SI, Woolley M, Lee YM. Zika virus: History, epidemiology, transmission, and clinical presentation. J Neuroimmunol. 2017;308:50–64. Epub 2017/03/14. 10.1016/j.jneuroim.2017.03.001 . [DOI] [PubMed] [Google Scholar]
- 10.Farajollahi A, Fonseca DM, Kramer LD, Marm Kilpatrick A. "Bird biting" mosquitoes and human disease: a review of the role of Culex pipiens complex mosquitoes in epidemiology. Infect Genet Evol. 2011;11(7):1577–85. Epub 2011/08/31. 10.1016/j.meegid.2011.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LaPointe DA, Goff ML, Atkinson CT. Comparative susceptibility of introduced forest-dwelling mosquitoes in Hawai'i to avian malaria, Plasmodium relictum. J Parasitol. 2005;91(4):843–9. 10.1645/GE-3431.1 [DOI] [PubMed] [Google Scholar]
- 12.van Riper C III. The Epizootiology and Ecological Significance of Malaria in Hawaiian Land Birds. Ecol Monogr. 1986;v. 56(no. 4):pp. 327–44-1986 v.56 no.4. 10.2307/1942550 .5183298 [DOI] [Google Scholar]
- 13.Bockarie MJ, Pedersen EM, White GB, Michael E. Role of vector control in the global program to eliminate lymphatic filariasis. Annu Rev Entomol. 2009;54:469–87. Epub 2008/09/19. 10.1146/annurev.ento.54.110807.090626 . [DOI] [PubMed] [Google Scholar]
- 14.Davis NC. An Investigation of Possible Vectors of Wuchereria bancrofti (Cobbold) in Bahia, Brazil. The Journal of Parasitology. 1935;21(1):21–6. 10.2307/3271791 [DOI] [Google Scholar]
- 15.Richards SL, Anderson SL, Lord CC. Vector competence of Culex pipiens quinquefasciatus (Diptera: Culicidae) for West Nile virus isolates from Florida. Trop Med Int Health. 2014;19(5):610–7. Epub 2014/06/06. 10.1111/tmi.12277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sardelis MR, Turell MJ, Dohm DJ, O'Guinn ML. Vector competence of selected North American Culex and Coquillettidia mosquitoes for West Nile virus. Emerg Infect Dis. 2001;7(6):1018–22. Epub 2001/12/19. 10.3201/eid0706.010617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M, Yang T, Kandul NP, Bui M, Gamez S, Raban R, et al. Development of a confinable gene drive system in the human disease vector Aedes aegypti. Elife. 2020;9 Epub 2020/01/22. 10.7554/eLife.51701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas DD, Donnelly CA, Wood RJ, Alphey LS. Insect population control using a dominant, repressible, lethal genetic system. Science. 2000;287(5462):2474–6. Epub 2000/03/31. 10.1126/science.287.5462.2474 . [DOI] [PubMed] [Google Scholar]
- 19.Hammond A, Galizi R, Kyrou K, Simoni A, Siniscalchi C, Katsanos D, et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat Biotechnol. 2016;34(1):78–83. Epub 2015/12/08. 10.1038/nbt.3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wheeler RE. A Simple Apparatus for Forced Copulation of Mosquitoes. Mosquito News. 1962;22:402–3. [Google Scholar]
- 21.Cator LJ, Arthur BJ, Harrington LC, Hoy RR. Harmonic convergence in the love songs of the dengue vector mosquito. Science. 2009;323(5917):1077–9. Epub 2009/01/10. 10.1126/science.1166541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawrence PA. Cell lineage of the thoracic muscles of Drosophila. Cell. 1982;29(2):493–503. Epub 1982/06/01. 10.1016/0092-8674(82)90166-0 . [DOI] [PubMed] [Google Scholar]
- 23.Hiromi Y, Hotta Y. Actin gene mutations in Drosophila; heat shock activation in the indirect flight muscles. EMBO J. 1985;4(7):1681–7. Epub 1985/07/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okamoto H, Hiromi Y, Ishikawa E, Yamada T, Isoda K, Maekawa H, et al. Molecular characterization of mutant actin genes which induce heat-shock proteins in Drosophila flight muscles. EMBO J. 1986;5(3):589–96. Epub 1986/03/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Labbe GM, Scaife S, Morgan SA, Curtis ZH, Alphey L. Female-specific flightless (fsRIDL) phenotype for control of Aedes albopictus. PLoS Negl Trop Dis. 2012;6(7):e1724 Epub 2012/07/18. 10.1371/journal.pntd.0001724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munoz D, Jimenez A, Marinotti O, James AA. The AeAct-4 gene is expressed in the developing flight muscles of female Aedes aegypti. Insect Mol Biol. 2004;13(5):563–8. Epub 2004/09/18. 10.1111/j.0962-1075.2004.00519.x . [DOI] [PubMed] [Google Scholar]
- 27.Fu G, Lees RS, Nimmo D, Aw D, Jin L, Gray P, et al. Female-specific flightless phenotype for mosquito control. Proc Natl Acad Sci U S A. 2010;107(10):4550–4. Epub 2010/02/24. 10.1073/pnas.1000251107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mogami K, Hotta Y. Isolation of Drosophila flightless mutants which affect myofibrillar proteins of indirect flight muscle. Mol Gen Genet. 1981;183(3):409–17. Epub 1981/01/01. 10.1007/BF00268758 . [DOI] [PubMed] [Google Scholar]
- 29.Marygold SJ, Roote J, Reuter G, Lambertsson A, Ashburner M, Millburn GH, et al. The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 2007;8(10):R216 Epub 2007/10/12. 10.1186/gb-2007-8-10-r216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reeves RG, Bryk J, Altrock PM, Denton JA, Reed FA. First steps towards underdominant genetic transformation of insect populations. PLoS One. 2014;9(5):e97557 Epub 2014/05/23. 10.1371/journal.pone.0097557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macdonald WW. The Selection of a Strain of aëdes Aegypti Susceptible to Infection with Semi-Periodic Brugia Malayi. Annals of Tropical Medicine & Parasitology. 1962;56(3):368–72. 10.1080/00034983.1962.11686134 [DOI] [PubMed] [Google Scholar]
- 32.Karunaratne SH, Hemingway J, Jayawardena KG, Dassanayaka V, Vaughan A. Kinetic and molecular differences in the amplified and non-amplified esterases from insecticide-resistant and susceptible Culex quinquefasciatus mosquitoes. J Biol Chem. 1995;270(52):31124–8. Epub 1995/12/29. 10.1074/jbc.270.52.31124 . [DOI] [PubMed] [Google Scholar]
- 33.Li M, Bui M, Yang T, Bowman CS, White BJ, Akbari OS. Germline Cas9 expression yields highly efficient genome engineering in a major worldwide disease vector, Aedes aegypti. Proc Natl Acad Sci U S A. 2017;114(49):E10540–E9. Epub 2017/11/16. 10.1073/pnas.1711538114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allen ML, O'Brochta DA, Atkinson PW, Levesque CS. Stable, germ-line transformation of Culex quinquefasciatus (Diptera: Culicidae). J Med Entomol. 2001;38(5):701–10. Epub 2001/10/03. 10.1603/0022-2585-38.5.701 . [DOI] [PubMed] [Google Scholar]
- 35.Lobo NF, Clayton JR, Fraser MJ, Kafatos FC, Collins FH. High efficiency germ-line transformation of mosquitoes. Nature Protocols. 2006;1(3):1312–7. 10.1038/nprot.2006.221 [DOI] [PubMed] [Google Scholar]
- 36.Kistler KE, Vosshall LB, Matthews BJ. Genome engineering with CRISPR-Cas9 in the mosquito Aedes aegypti. Cell Rep. 2015;11(1):51–60. Epub 2015/03/31. 10.1016/j.celrep.2015.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coates CJ, Jasinskiene N, Miyashiro L, James AA. Mariner transposition and transformation of the yellow fever mosquito, Aedes aegypti. Proc Natl Acad Sci U S A. 1998;95(7):3748–51. Epub 1998/05/09. 10.1073/pnas.95.7.3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giraldo-Calderon GI, Emrich SJ, MacCallum RM, Maslen G, Dialynas E, Topalis P, et al. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 2015;43(Database issue):D707–13. Epub 2014/12/17. 10.1093/nar/gku1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Labun K, Montague TG, Gagnon JA, Thyme SB, Valen E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016;44(W1):W272–6. Epub 2016/05/18. 10.1093/nar/gkw398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Engler C, Kandzia R, Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 2008;3(11):e3647 Epub 2008/11/06. 10.1371/journal.pone.0003647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anderson ME, Mavica J, Shackleford L, Flis I, Fochler S, Basu S, et al. CRISPR/Cas9 gene editing in the West Nile Virus vector, Culex quinquefasciatus Say. PLoS One. 2019;14(11):e0224857 Epub 2019/11/13. 10.1371/journal.pone.0224857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li M, Li T, Liu N, Raban RR, Wang X, Akbari OS. Methods for the generation of heritable germline mutations in the disease vector Culex quinquefasciatus using clustered regularly interspaced short palindrome repeats-associated protein 9. Insect Mol Biol. 2020;29(2):214–20. Epub 2019/11/07. 10.1111/imb.12626 . [DOI] [PubMed] [Google Scholar]
- 43.Berghammer AJ, Klingler M, Wimmer EA. A universal marker for transgenic insects. Nature. 1999;402(6760):370–1. Epub 1999/12/10. 10.1038/46463 . [DOI] [PubMed] [Google Scholar]
- 44.Drummond DR, Hennessey ES, Sparrow JC. Characterisation of missense mutations in the Act88F gene of Drosophila melanogaster. Mol Gen Genet. 1991;226(1–2):70–80. Epub 1991/04/01. 10.1007/BF00273589 . [DOI] [PubMed] [Google Scholar]
- 45.An HS, Mogami K. Isolation of 88F actin mutants of Drosophila melanogaster and possible alterations in the mutant actin structures. J Mol Biol. 1996;260(4):492–505. Epub 1996/07/26. 10.1006/jmbi.1996.0417 . [DOI] [PubMed] [Google Scholar]
- 46.Beall CJ, Sepanski MA, Fyrberg EA. Genetic dissection of Drosophila myofibril formation: effects of actin and myosin heavy chain null alleles. Genes Dev. 1989;3(2):131–40. Epub 1989/02/01. 10.1101/gad.3.2.131 . [DOI] [PubMed] [Google Scholar]
- 47.Kyrou K, Hammond AM, Galizi R, Kranjc N, Burt A, Beaghton AK, et al. A CRISPR-Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat Biotechnol. 2018;36(11):1062–6. Epub 2018/09/25. 10.1038/nbt.4245 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.KaramiNejadRanjbar M, Eckermann KN, Ahmed HMM, Sanchez CH, Dippel S, Marshall JM, et al. Consequences of resistance evolution in a Cas9-based sex conversion-suppression gene drive for insect pest management. Proc Natl Acad Sci U S A. 2018;115(24):6189–94. Epub 2018/05/31. 10.1073/pnas.1713825115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marshall JM, Buchman A, Sanchez CH, Akbari OS. Overcoming evolved resistance to population-suppressing homing-based gene drives. Sci Rep. 2017;7(1):3776 Epub 2017/06/21. 10.1038/s41598-017-02744-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hammond AM, Kyrou K, Bruttini M, North A, Galizi R, Karlsson X, et al. The creation and selection of mutations resistant to a gene drive over multiple generations in the malaria mosquito. PLoS Genet. 2017;13(10):e1007039 Epub 2017/10/05. 10.1371/journal.pgen.1007039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Champer J, Liu J, Oh SY, Reeves R, Luthra A, Oakes N, et al. Reducing resistance allele formation in CRISPR gene drive. Proc Natl Acad Sci U S A. 2018;115(21):5522–7. Epub 2018/05/08. 10.1073/pnas.1720354115 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Target sequences for the sgRNAs used in this study. Predicted cut sites are between the nucleotide numbers shown, counting the A of the ATG start codon as nucleotide 1.
(DOCX)
In vitro cleavage by sgRNAs was carried out at 37°C for indicated periods. Cutting efficiency of each sgRNA within each species is ranked and shown in brackets.
(DOCX)
Mutation assay primers to detect CRISPR/Cas9 mediated indels or HDR integrations in Act4.
(DOCX)
*point where the 6 bases for the 156_161delTGCCTA deletion have been removed. The ssODN oligo consists of 60 nucleotides upstream and downstream of the deletion.
(DOCX)
Plasmid sequences corresponding to the 3xP3-mCherry-SV40 marker cassette used and the final AeAct4 HDR 3xP3-mCherry nDNA donor.
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.