Abstract
In the brain, extracellular matrix (ECM) components form networks that contribute to structural and functional diversity. Maladaptive remodeling of ECM networks has been reported in neurodegenerative and psychiatric disorders, suggesting that the brain microenvironment is a dynamic structure. A lack of quantitative information about ECM distribution in the brain hinders an understanding of region-specific ECM functions and the role of ECM in health and disease. We hypothesized that each ECM protein as well as specific ECM structures, such as perineuronal nets (PNNs) and interstitial matrix, are differentially distributed throughout the brain, contributing to the unique structure and function in the various regions of the brain. To test our hypothesis, we quantitatively analyzed the distribution, colocalization, and protein expression of aggrecan, brevican, and tenascin-R throughout the rat brain utilizing immunohistochemistry and mass spectrometry analysis and assessed the effect of aggrecan, brevican, and/or tenascin-R on neurite outgrowth in vitro. We focused on aggrecan, brevican, and tenascin-R as they are especially expressed in the mature brain, and have established roles in brain development, plasticity, and neurite outgrowth. The results revealed a differentiated distribution of all three proteins throughout the brain and indicated that their presence significantly reduces neurite outgrowth in a 3D in vitro environment. These results underline the importance of a unique and complex ECM distribution for brain physiology and suggest that encoding the distribution of distinct ECM proteins throughout the brain will aid in understanding their function in physiology and in turn assist in identifying their role in disease.
Keywords: aggrecan, brain extracellular matrix, brevican, perineuronal nets, tenascin-R, RRID: AB_90460, RRID: AB_398211, RRID: AB_2207009
The brain extracellular matrix (ECM) provides a microenvironment that is essential for cell survival, plasticity, damage response, and regeneration, and facilitates the organization of distinct brain regions (Dityatev et al., 2010; Kwok et al., 2011). The brain ECM is composed of multiple families of molecules, including hyaluronan (HA), glycosaminoglycans (GAG), proteoglycans (PG), glycoproteins (GP), including tenascin-R (TnR), and chondroitin sulfate proteoglycans (CSPGs), such as aggrecan (Aggr), brevican (Brev), neurocan, versican, and phosphacan (Zimmermann and Dours-Zimmermann, 2008). Several ECM molecules form defined structures, such as perineuronal nets (PNNs), which surround certain neuronal somas and proximal neurites. PNNs are first detected relatively late in development and are involved in synaptic stabilization, neuroprotection, and ionic buffering (Lau et al., 2013). The selective ECM turnover is essential for maintaining brain homeostasis and is regulated by ADAMTs (a disintegrin and metalloproteinase with thrombospondin motifs) and matrix metalloproteases (MMPs) (Zimmermann and Dours-Zimmermann, 2008).
Most brain CSPGs and other ECM proteins such as tenascins have been reported to inhibit neurite outgrowth in vivo and in vitro (Snow et al., 1990; McKeon et al., 1991; Asher et al., 2001; Gilbert et al., 2005; Silver and Silver, 2014). Recently, it has been suggested that CSPGs might exert growth-promoting effects as well, depending on sulfation pattern and neuronal type (Beller and Snow, 2014). In humans ECM changes have been observed in brain disorders, such as Alzheimer’s disease (AD) (Bonneh-Barkay and Wiley, 2009) and schizophrenia (Pantazopoulos et al., 2010), possibly contributing to disease pathology, but cause and consequences have not yet been elucidated. ECM alterations in schizophrenia have been reported in the amygdala, the entorhinal cortex, and specific cortical layers, whereas ECM changes in AD are mainly associated with plaque formation (Bonneh-Barkay and Wiley, 2009; Pantazopoulos et al., 2015). Animal models have proved invaluable to better understand brain diseases (Lipska and Weinberger, 2000; Asher et al., 2001). However, the development and validation of animal models especially for psychiatric disorders remains a challenge. Although there is a great deal of information about the general composition of the brain ECM and the changes associated with brain development (Silver, 1994; Rauch, 2004; Senkov et al., 2014; Silver and Silver, 2014), there is a lack of quantitative studies concerning the distribution of specific ECM proteins throughout the brain, which is critical for understanding the role of the brain ECM in normal and disease states.
We hypothesized that each ECM protein as well as specific ECM structures, such as PNNs and interstitial matrix, are differentially distributed throughout the brain, contributing to region- and cell-specific functions. Characterizing the distribution of ECM proteins throughout the brain will aid in understanding their function in physiology and disease. Moreover, with this information, ECM proteins could be used as a validation marker for rat models of psychiatric disorders. As a first step in characterizing the wide range of ECM molecules in the rat brain, we quantitatively analyzed the distribution of Aggr, Brev, and TnR, due to their established roles in brain development, plasticity, and neurite outgrowth (Bradbury et al., 2002; Lau et al., 2013). They represent members of the lectican family (Aggr and Brev) as well as one GP (TnR), which is, among other purposes, important for the cross-linking of lecticans and bridging of neighboring HA/lectican/link-protein complexes to complete the ECM network (Zimmermann and Dours-Zimmermann, 2008). We expanded our characterization to a wider range of molecules using mass spectrometry (MS). In vitro, we analyzed how Aggr, Brev, and/or TnR affect neurite outgrowth in a 3D environment. Our results show a unique distribution of ECM proteins throughout different brain regions and varying ECM compositions of neighboring PNNs, strongly suggesting that local variation of ECM environments at the cell scale may be a method for locally differentiating the function of neighboring cells. In vitro, Aggr, Brev, and/or TnR reduced neurite outgrowth significantly. Overall, these results indicate that the ECM is a complex, tightly controlled, and uniquely distributed network, suggesting that the cellular microenvironment is an influential component of both normal and disease states.
MATERIALS AND METHODS
Whole brain section harvest, staining, and analysis
Tissue harvest
Adult (4-month-old) female Sprague–Dawley rats (Charles River Laboratories, Boston, MA; RRID:RGD_734476) were anesthetized with 4% isoflurane and oxygen and maintained at this level of anesthesia via nose cone in preparation for thoracotomy. The chest was then opened and the heart rapidly exposed. A needle was inserted into the left ventricle, the right atrium was cut open, and exsanguination was performed with intraventricular infusion of phosphatebuffered saline (PBS) until blood was completely flushed from circulation. This was followed by infusion of approximately 150 ml 4% paraformaldehyde (PFA) in PBS to fix the tissue. The brain was then removed from the skull and incubated overnight at 4°C in 4% PFA. The following day, brains were washed with PBS and incubated in 0.5 M sucrose for 24 hours. The 0.5 M sucrose solution was replaced with 1 M sucrose solution for another 24 hours at 4°C. Brains were then placed in Tissue Freezing Media (OCT Embedding Compound, Electron Microscopy Sciences, Hatfield, PA) and frozen by using liquid nitrogen and isopentane. Frozen samples were stored at −80°C. Brains were cut into 50-μm-thick sections by using a Leica cryostat at −20°C and collected in 0.1 M phosphate buffer (PB) with 0.1% sodium azide. All procedures were approved by the Harvard Animal Care and Use Committee.
Numbers of animals used
For immunohistochemical analysis of this study, we used three adult animals (4 months old, female); for the proteomics analysis (detailed below), we used three adult animals of the same age and gender (4 months old, female) and pooled the respective regions from those animals into one sample (one final sample for each described brain region); for our in vitro experiments (detailed below), we utilized 2-day-old Sprague–Dawley pups (Charles River Laboratories; two for each harvest, two to three separate harvests).
Staining
Sections were rinsed with 0.01 M PBS (0.2 g KCl, 8.7 g NaCl, 200 ml 0.1 M NaPO4, in 1 liter H2O) with 0.2% Triton-X (PBS-TX) and incubated in 80°C citrate buffer, pH 4.5 (0.1 M citric acid anhydrous, 0.2 M Na2HPO4 dibasic), for 20 minutes. Sections were rinsed with PBS-TX and incubated in 0.3% hydrogen peroxide in PBS-TX for 20 minutes followed by a 30-minute incubation in 2% bovine serum albumin (BSA; Jackson ImmunoResearch, West Grove, PA) in PBS-TX. Afterward, the primary antibodies mouse anti-Brevican Clone 2 (Table 1; RRID:AB_398211), goat anti-Tenascin-R (Table 1; RRID:AB_2207009), and rabbit anti-Aggrecan (Table 1; RRID:AB_90460) were applied in PBS-TX with 2% BSA for 2 nights at 4°C followed by 2 hours of incubation with secondary antibody (donkey anti-goat, goat anti-rabbit, goat anti-mouse conjugated to Alexa Fluor 488, 546, or 633; all from Molecular Probes, Grand Island, NY) at room temperature. Due to possible cross-talk, we first incubated the sections with the donkey anti-goat antibody for 2 hours, rinsed them thoroughly to ensure removal of unbound secondary antibody, and then incubated them with the other two secondary antibodies for another 2 hours at RT. In addition to the secondary antibodies, we also incubated the sections with 4′,6-diamidino-2-phenylindole (DAPI; Molecular Probes Life Technologies) as a nuclear counterstain. After the secondary antibody incubation, sections were rinsed in 0.1 M PB and then incubated in copper sulfate solution for 10 minutes to block endogenous lipofucsin autofluorescence (Schnell et al., 1999). Finally, sections were rinsed again with 0.1 M PB and mounted onto glass slides with Prolong Gold Antifade reagent (Molecular Probes Life Technologies). Prepared slides were either imaged immediately or stored at 4°C.
TABLE 1.
Primary Antibodies for Immunofluorescence Labeling
| Target protein | Species | Dilution | Source, cat. no | RRID |
|---|---|---|---|---|
| Brevican | Mouse | 1:100 | B&D Biosciences, #610894, monoclonal | AB_398211 |
| Tenascin-R | Goat | 1:2000 | R&D Systems, #AF3865, polyclonal | AB_2207009 |
| Aggrecan | Rabbit | 1:100 | Millipore, #AB1031, polyclonal | AB_90460 |
| βIII-tubulin | Rabbit | 1:200 | Sigma, T2200, polyclonal | AB_262133 |
Imaging and analysis
Imaging was performed with a VS120 Olympus Slide Scanner and an Olympus confocal microscope (Olympus, Center Valley, PA) with appropriate filter cubes. Signal intensity was analyzed using ImageJ. PNNs were counted manually on 20-μm-thick z-stacks. Colocalization analyses were performed on maximum intensity projection images of 20-μm-thick z-stacks (1-μm steps, 512 × 512 pixels).
Characterization of antibodies
Brevican antibody Clone 2 (monoclonal) was purified from tissue culture supernatant or ascites by affinity chromatography (Table 1). The immunogen is against rat brevican amino acids 232–394. This antibody is routinely tested by western blot analysis by B&D Transduction Labs (San Jose, CA). Other applications were tested at BD Biosciences Pharmingen (San Jose, CA) during antibody development. Usage of this antibody in western blot application recognizes a 140-kDa band in rat cerebrum lysates, which corresponds to the full-length brevican, which can be up to 145 kDa (http://www.bdbiosciences.com/eu/applications/research/cellular-imaging/cellular-imaging-antibodies/purified-mouse-anti-brevican-2brevican/p/610894). The antibody was further tested and referenced in several publications (Yamada et al., 1994; Seidenbecher et al., 1995; Aspberg et al., 1997). In total proteoglycan fractions from bovine brain extracts, this antibody recognizes the 145-kDa full-length protein as well as the 80-kDa core protein in western blot analysis (Yamada et al., 1997).
Tenascin-R is a polyclonal antigen affinity-purified antibody raised in goat (Table 1). The immunogen is against the mouse myeloma cell line NS0-derived recombinant human tenascin-R isoform 1 (Glu34–Phe1358). This antibody has been tested and referenced in several publications (Xiao et al., 1997; Weber et al., 1999; Kecskes et al., 2015). Tenascin-R was detected in immersion-fixed paraffin-embedded sections of human brainstem (https://www.rndsystems.com/products/human-tenascin-r-antibody_af3865). This antibody further recognized Tenascin-R in the retina and optic nerve of adult mice (Xiao et al., 1997). The 160-and 180-kDa isoforms of Tenascin-R were detected in mouse brain protein extracts by western blot analysis, whereas those isoforms were not detected in the Tenascin-R knockout mice (Weber et al., 1999).
Aggrecan is a polyclonal affinity-purified antibody raised in rabbit (Table 1). The immunogen is against GST fusion protein containing amino acids 1177–1326 of mouse aggrecan. This antibody is routinely tested by western blot on mouse brain lysates (http://www.emdmillipore.com/US/en/product/Anti-Aggrecan-Antibody, MM_NF-AB1031) and has been referenced in several publications (Dityatev et al., 2007; Reimers et al., 2007; Giamanco et al., 2010). Bands of 150 kDa and 250 kDa were recognized by the aggrecan antibody in mouse brain lysates (Lendvai et al., 2013).
ßIII-tubulin is a polyclonal affinity-isolated antibody raised in rabbit (Table 1; AB_262133). The immunogen is a synthetic peptide corresponding to amino acid residues 441–450 of human ßIII-tubulin (Ala446 to Ser446 substitution) with N-terminal added cysteine, conjugated to KLH. The sequence is conserved in mammals. The antibody has been tested and referenced in several publications (Bitel et al., 2010; Cai et al., 2010; Gatz et al., 2011).
Collagen-ECM gel preparation, culture, staining, and analysis
Primary neuronal harvest
Cortical neurons were isolated from 2-day-old neonatal Sprague–Dawley rats (Charles River Laboratories). All procedures were approved by the Harvard Animal Care and Use Committee. Animals were sacrificed by decapitation, and cortices were surgically isolated and minced in Hibernate-A solution supplemented with B-27 and Glutamax (HABG) (all Gibco Life Technologies, Grand Island, NY) followed by digestion with papain (Worthington Biochemical Corporation, Lakewood, NJ) for 30 minutes at 37°C. The digested tissue was then transferred to fresh HABG and mechanically disrupted by using silane-treated glass pipettes. The mechanical disruption was done by pipetting the tissue up and down for about 10 times. After the remaining tissue settled down, the supernatant was collected and transferred into a fresh tube. These steps were repeated 2 more times. The collected supernatants were then filtered through a nylon filter of 40-μm pore size (BD Bioscience, San Jose, CA) and centrifuged at 200 g for 3 minutes. The supernatant was removed, and the pellet was suspended in prewarmed Neurobasal A medium supplemented with B-27, Glutamax, and Gentamycin (all Gibco Life Technologies). Cells were counted by using a Moxi Mini Automated Cell Counter (Orflo, Ketchum, ID) and seeded at the desired density. Cultures were incubated under standard conditions at 37°C and 5% CO2. Media was replaced every 72 hours until experiments were executed.
Collagen-ECM gel preparation and culture
Prior to embedding cells in 3D scaffolds, brain ECM proteins (bovine aggregan; Sigma, St. Louis, MO; human brevican and human tenascin-R; R&D Systems, Minneapolis, MN) were activated to bind the collagen scaffold. Solutions of EDC (1-ethyl-3-(3-dimethylaminopropyl)carbodiimide·HCl (Thermo Scientific, San Jose, CA), 0.4 mg/mL sulfo-N-hydroxysulfosuccinimide (NHS; Sigma), and 1.1 mg/mL sodium acetate buffer (0.1 M, pH 5) were prepared. Proteins were activated by adding 5 μl of EDC solution and 5 μl of sulfo-NHS solution to 50 μl of protein solution (1 mg/ml) and incubated for 10 minutes at room temperature. During the incubation time, the collagen solution was prepared. On ice, 800 μl of collagen from bovine skin (6 mg/ml; Sigma) was neutralized with 100 μl of 10× buffer solution (50 mM NaOH, 260 mM NaHCO3, 200 mM HEPES) and 100 μl of 10× DMEM (Sigma). The neutralized collagen solution was added to the activated ECM protein solution and gently mixed. This collagen–ECM solution was then added to 5 × 106 rat cortical neurons (harvested as previously mentioned). Then 40-μl drops of this collagen–ECM neuron suspension were added to each well of 4-well polydimethylsiloxane (PDMS) masks (custom-made) fitting onto 18-mm-diameter coverslips and incubated at 37°C and 5% CO2 for 45 minutes to induce collagen cross-linking. After 45 minutes, medium was added and cells were maintained as mentioned above. For collagen-only scaffolds, the brain ECM protein activation step was omitted.
Staining
Cells in collagen–ECM gels were washed with prewarmed PBS, fixed for 15 minutes with prewarmed 4% PFA, permeabilized for 15 minutes with 0.05% Triton-X-100 in PBS at room temperature, and blocked with 3–5% BSA (Jackson ImmunoResearch) for 30 minutes at room temperature. βIII-tubulin antibody (Table 1) was added and samples were incubated for 1.5 hours at 37°C. A secondary staining solution consisting of goat anti-mouse conjugated to Alexa Fluor 488 (Molecular Probes), and DAPI was added to the cells for 1 hour at 37°C. After another washing step, samples were placed on a glass slides, and ProLong Gold Antifade reagent was added to preserve the samples. Prepared slides were either imaged immediately or stored at 4°C. Imaging was performed with an Olympus confocal microscope with appropriate filter cubes.
Neurite outgrowth analyses
Neurite outgrowth was measured with a custom-made Matlab image analysis code. Confocal stacks (30 slices, 5 μm between consecutive slices) of neurons stained for βIII-tubulin and DAPI were split into two channels. For each channel, several image enhancement operations were performed on each individual image of the stack. First the background was subtracted. To take into account regional variation of background, an opening morphological operation was performed on each image, using a disk of 20-pixel diameter, to create a “background” image that was subtracted from the original image. Then, the brightness and contrast of each individual image of the stack were adjusted. To do this, 0.25% of the pixels at both tails of the intensity histogram were saturated, and the remaining histogram was stretched to fill the whole intensity value (0–65,535, 16-bit) range. The intensity values were then scaled from the range 0–65,535 (16-bit) to the range 0–1. At that point, each image of the stack was thresholded. The threshold value was 0.8 for the DAPI images and varied between 0.5 and 0.75 for the βIII-tubulin images to take into account slight variations in sample staining. A maximum intensity projection was then performed on each channel stack. The resulting DAPI image was segmented, and the number of nuclei were counted; objects out of the range 5–100 μm2 (3–256 pixels) were discarded. The resulting βIII-tubulin image was skeletonized using the morphological operation skeleton embedded in the Matlab image analysis toolbox, and the skeleton zones overlapping the nuclei were subtracted to discard the error due to skeletonization of soma areas. The total number of pixels of the skeletonized image was then counted and converted to micrometers.
Proteomics
Tissue preparation
Tissue samples were harvested from adult Sprague–Dawley rats (Charles River Laboratories). All procedures were approved by the Harvard Animal Care and Use Committee. Adult rats were sacrificed by using CO2 and cervical dislocation. Brains were dissected out of the skull, and the isocortex, the hippocampus, the amygdala, the cerebellum, and the brainstem were further surgically dissected and snap-frozen in liquid nitrogen. Samples were thawed and homogenized by using a hand homogenizer (Cole-Parmer, Vernon Hills, IL) in RIPA buffer (Sigma) and complemented with protease inhibitors (Complete Mini; Roche, Basel, Switzerland) and 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF; Sigma). All steps were conducted on ice. After homogenization, samples were embedded in ice and placed on a rocking platform shaker for 30 minutes. Afterward, samples were centrifuged at 17,000 g for 30 minutes at 4°C. Supernatants were transferred to a fresh Eppendorf tube and centrifuged again at 17,000 g for 30 minutes at 4°C. The resulting supernatants were used for analysis.
TMT labeling and ERLIC off-line fractionation
Samples were measured to 100 μg of total protein in each sample. Samples were digested using a slightly modified filter-aided sample preparation (FASP) protocol as previously published (Wisniewski et al., 2009). Subsequently, all samples were labeled by tandem mass tags (TMT)-10plex reagent (#90061, Thermo Fisher) according to the manufacturer’s protocol. The TMT10plex reagent set enables up to 10 different peptide samples prepared from cells or tissues to be labeled in parallel and then combined for analysis. For each sample, a unique reporter mass in the low-mass region of the highresolution tandem mass spectrometry (MS/MS) spectrum is used to measure relative protein expression levels during peptide fragmentation and MS/MS. The resulting labeled 10 samples were pooled into 1 sample for an off-line fractionation. The high-performance liquid chromatography (HPLC) 1200 Agilent system with fraction collector (Agilent Technologies, Santa Clara, CA) was used for an electrostatic repulsion–hydrophilic interaction chromatography (ERLIC) separation using Poly-WAX LP™ columns (200 × 2.1 mm, 5 μm, 300 Å, PolyLC, Columbia, MD). ERLIC utilizes both hydrophilic interaction and electrostatic forces. By adjusting the pH, salt type, salt concentration, and organic solvent compositions in the mobile phase, an isocratic separation of a mixture of charged analytes (e.g., peptides, amino acids, and nucleotides) can be achieved (Alpert, 2008). Samples were fractionated to a total of 20 fractions on a 70-minute liquid chromatography (LC) gradient. Each fraction was submitted for mass spectrometry analysis.
Mass spectrometry and data analysis
Samples were run on Orbitrap Velos Pro (Thermo Fisher) for all analytical runs. Orbitrap Velos Pro is a hybrid ion trap–Orbitrap mass spectrometer that is ideal for identification of low-level proteins in complex matrices, rapid quantitation of isobarically labeled peptides, and structural elucidation of metabolites. Samples were injected from an autosampler of HPLC (Waters, NanoAquity, Milford, MA) into a trapping column (75 μm column ID, 5 cm packed with C18 media on 5 μm beats on 200 Å poros; Michrom Bioresources, Auburn, CA). After binding in a trapping column, peptides were washed for 15 minutes with buffer A (0.1% formic acid in water) and then eluted to an analytical column (75 μm column ID, 20 cm packed with 3 μm C18 media beats on 100 Å poros) with a gradient from 2 to 32% of buffer B (0.1% formic acid in acetonitrile) over a 90-minute gradient for each fraction. Each sample was run on Orbitrap Velos instrument with a high collisional energy/collisional induced dissociation (HCD/CID) TOP 20 method that isolates and fragments the top ions that came at a particular time of the chromatogram from nanoflow HPLC (HCD fragmentation in Orbitrap was followed by a CID event in the ion trap part of the instrument). HCD/CID is used to sequence the peptides present in the samples. The Orbitrap Velos Pro instrument was set up to run the TOP 20 method for MS/MS in the ion trap with an exclusion function turned on after MS1 scan in Orbitrap with 60 K resolving power at a mass of 400 m/z. For TMT10plex labeling, MS2 data were acquired with 30 K resolving power, and for TMT6plex, MS2 data were collected with 15 K resolving power.
The runs obtained were analyzed by Proteome Discoverer 1.4.1.14 (Thermo Fisher) software with Percolator v2.05 (University of Washington) as a statistical data package. Searches were done against a rat proteome database filtered out of the Uniprot database by species specificity and common contaminants that were added to this database. Searches were done with trypsin enzyme specificity, allowing two missed cleavages. Possible modifications included in the search parameters were as follows: protein N-terminus acetylation, methionine oxidation, and deamidation of asparagine and glutamine amino acids were put as a variable modification as well as for static modifications N-terminus of peptide TMT6plex labeling and lysine side chains with TMT6plex labeling group. The database search criteria were held at a 1% false discovery rate on both protein and peptide levels for all output reported data. All data outputs were filtered to 40% maximum coisolation of MS1, allowing TMT quantitation results to be kept under control from a coisolation effect.
Statistics
Multifactorial one-way analysis of variance (ANOVA) with a Bonferroni post hoc test was used to statistically analyze the results shown in Figures 5–9, 11, 13, and 14. One-way ANOVA tests together with Bonferroni post hoc tests were conducted for statistical comparisons as our data consist of a single dependent variable (e.g., signal intensity or number of PNNs) and multiple independent variables (e.g., multiple brain regions, multiple ECM proteins). The ANOVA test was chosen over the two-tailed Student’s t-test because multiple t-tests cannot be performed on different conditions without inflating type 1 errors. All values depicted in Table 2 are P values resulting from the second step Bonferroni post hoc test.
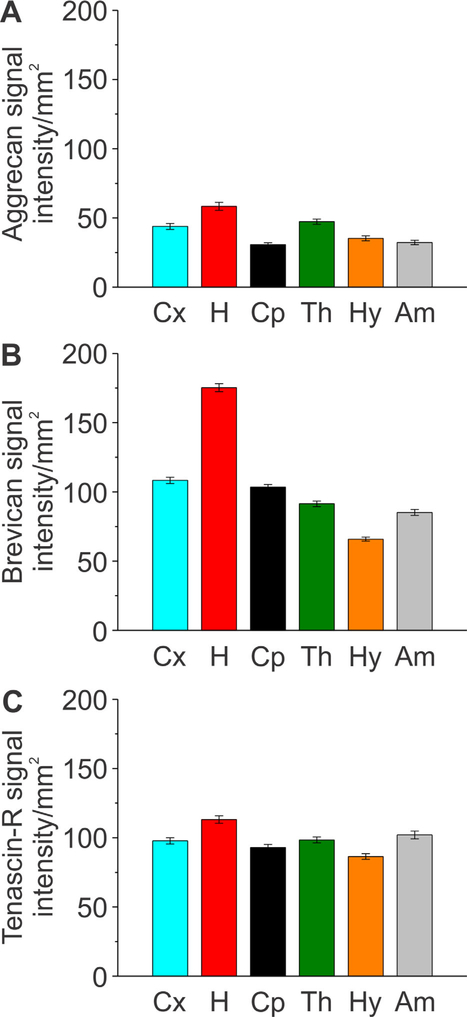
Figure 5.
Quantification of aggrecan, brevican, and tenascin-R distribution throughout distinct regions of the adult rat brain. A–C: Signal intensity of aggrecan, brevican, and tenascin-R was measured for the brain regions outlined in Figure 3. Column color corresponds to the region outline color in Figure 3. Error bars are shown as standard error of the mean (SEM). Significance levels calculated by ANOVA (Bonferroni post hoc test) are shown in Table 2A. Abbreviations: Cx, isocortex; H, hippocampus; Cp, caudate-putamen; Th, thalamus; Hy, hypothalamus; Am, amygdala.
Figure 9.
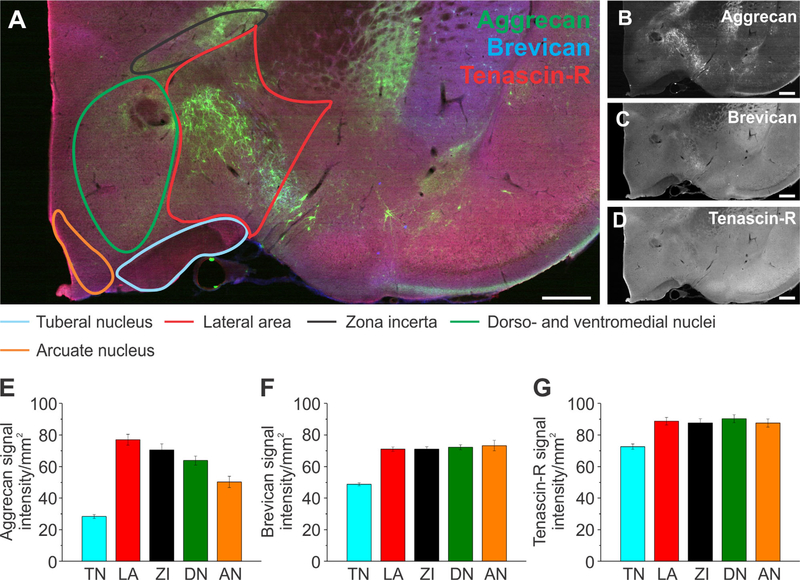
Extracellular matrix distribution in the hypothalamus. A: Adult rat brain slice shows the hypothalamus labeled for aggrecan (green), brevican (blue), and tenascin-R (red) with outlined subregions of the hypothalamus: the tuberal nucleus (blue), lateral area (red), zona incerta (black), dorso- and ventromedial nuclei (green), and arcuate nucleus (orange). B–D: Single channels for each protein are shown in black and white. E–G: Signal intensity measurements for aggrecan (E), brevican (F), and tenascin-R (G) for the subregions outlined in A. Column color corresponds to the region outline color in A. Error bars are shown as standard error of the mean (SEM). Significance levels calculated by ANOVA (Bonferroni post hoc test) are shown in Table 2C. n = 36 slices, 3 brains. Abbreviations: TN, tuberal nucleus; LA, lateral area; ZI, zona incerta; DN, dorso- and ventromedial nuclei; AN, arcuate nucleus. Scale bar = 500 μm in A–D.
Figure 11.
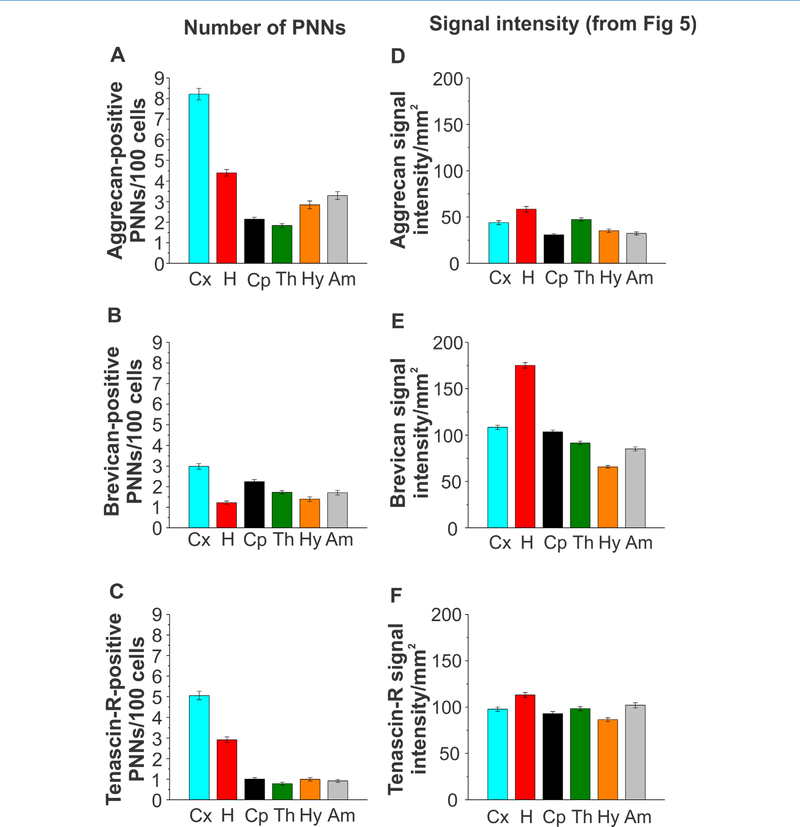
Perineuronal net quantification in distinct brain regions of the adult rat brain. A–C: Numbers of (A) aggrecan-, (B) brevican-, and (C) tenascin-R-positive perineuronal nets in the isocortex, hippocampus, caudate-putamen, thalamus, hypothalamus, and amygdala. D– F: Signal intensity measurements from Figure 5 as a direct comparison with perineuronal net (PNN) numbers is displayed. Error bars are shown as standard error of the mean (SEM). Significance levels calculated by ANOVA (Bonferroni post hoc test) are shown in Table 2D. n = 64 images, 24 slices, 3 brains. Abbreviations: Cx, isocortex; H, hippocampus; Cp, caudate-putamen; Th, thalamus; Hy, hypothalamus; Am, amygdala.
Figure 13.
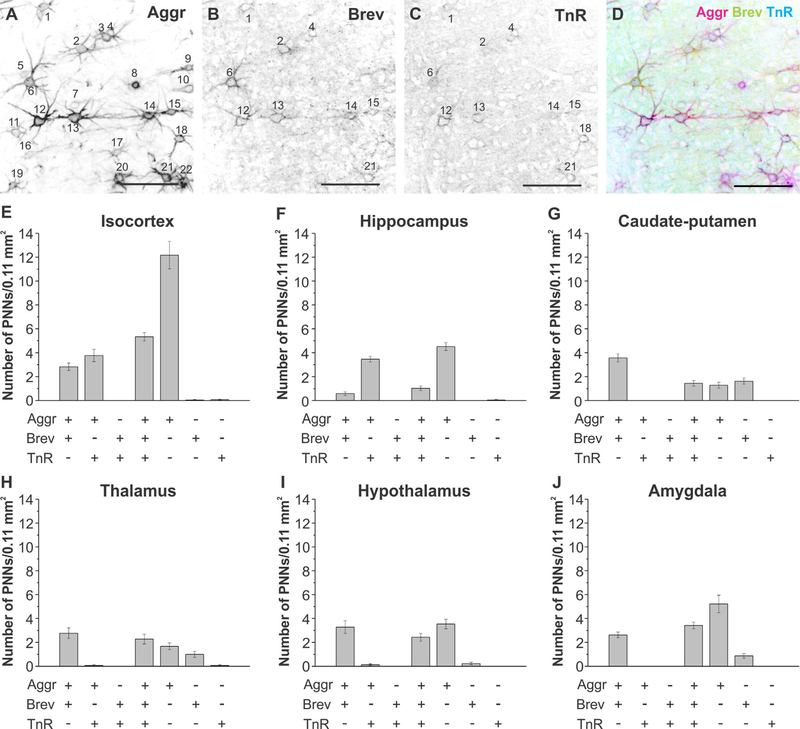
Colocalization analyses of aggrecan, brevican, and tenascin-R in distinct brain regions. A–D: Black and white single channel and merge image of aggrecan-, brevican-, and tenascin-R-positive perineuronal nets. Perineuronal nets were counted in each image, and colocalization was determined. In the depicted images perineuronal nets 1, 2, 4, 6, 12, 13, 14, 15, and 21 are positive for aggrecan, brevican, and tenascin-R, whereas all other nets were positive only for aggrecan. E–J: Colocalization analyses for the (E) isocortex, (F) hippocampus, (G) caudate-putamen, (H) thalamus, (I) hypo-thalamus, and (J) amygdala are depicted. Error bars are shown as standard error of the mean (SEM). Significance levels calculated by ANOVA (Bonferroni post hoc test) are shown in Table 2E. n = 39 images, 18 slices, 3 brains. Abbreviations: Aggr, aggrecan; Brev, brevican; TnR, tenascin-R. Scale bar = 100 μm in E–D.
Figure 14.
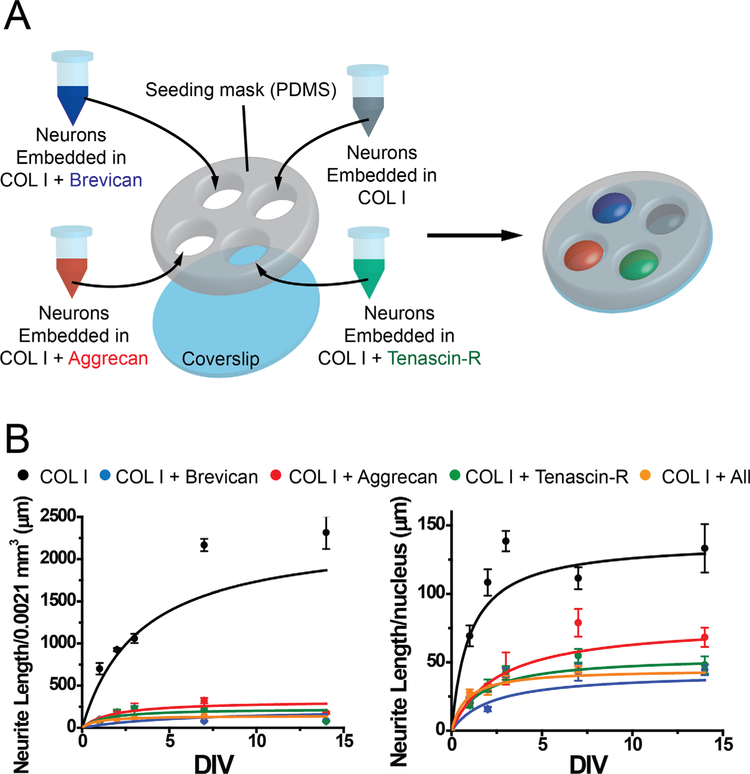
Effect of aggrecan, brevican, and tenascin-R on neurite outgrowth in a 3D environment. A: Schematic overview of sample conditions and preparation of 3D gels in PDMS masks is displayed. Neurons were embedded in collagen I gels either alone (control, gray), or in combination with aggrecan (red), brevican (blue), or tenascin-R (green), or a combination of all three ECM proteins (not shown in the schematic). B: Neurite length analysis for all conditions at day 1, 2, 3, 7 and 14 in vitro are depicted. Z-stacks were taken for each conditions, and neurite length was analyzed in each stack (0.016 mm2, left panel) and normalized to nuclei numbers over all z-stacks analyzed (right panel). n = 10–16 z-stacks, 2–4 samples. Error bars are shown as standard error of the mean (SEM). Significance levels calculated by ANOVA (Bonferroni post hoc test) are shown in Table 2F. Abbreviations: Aggr, aggrecan; Brev, brevican; TnR, yenascin-R; COLI, collagen I; DIV, days in vitro.
TABLE 2.
| A: Figure 5 | ||||||
| Signal intensity | Aggr | Brev | TnR | |||
| Cx vs H | 7.3E-06 | 1.0E-68 | 8.3E-05 | |||
| Cx vs Cp | 6.8E-05 | 1 | 1 | |||
| Cx vs Th | 1 | 1.1E-06 | 1 | |||
| Cx vs Hyp | 0.039 | 3.3E-34 | 0.014 | |||
| Cx vs Amy | 7.5E-04 | 6.2E-12 | 1 | |||
| H vs Cp | 3.3E-19 | 9.7E-76 | 5.6E-08 | |||
| H vs Th | 0.002 | 1.5E-92 | 2.0E-04 | |||
| H vs Hyp | 7.7E-14 | 2.7E-125 | 3.2E-13 | |||
| H vs Amy | 2.4E-17 | 6.0E-101 | 0.014 | |||
| Cp vs Th | 1.5E-07 | 0.002 | 1 | |||
| Cp vs Hyp | 1 | 8.1E-28 | 0.83 | |||
| Cp vs Amy | 1 | 1.2E-07 | 0.11 | |||
| Th vs Hyp | 4.3E-06 | 4.3E-14 | 0.007 | |||
| Th vs Amy | 2.8E-06 | 0.69 | 1 | |||
| Hyp vs Amy | 1 | 1.9E-08 | 8.3E-05 | |||
| B: Figures 6 and 7 | ||||||
| H subdivision | Aggr | Brev | TnR | |||
| SP vs DG | 1.1E-03 | 5.8E-08 | 1 | |||
| SP vs SR | 0.06 | 3.3E-15 | 0.6 | |||
| SP vs SO | 1 | 2.3E-25 | 1 | |||
| DG vs SR | 1.2E-8 | 0.01 | 0.5 | |||
| DG vs SO | 0.01 | 1.3E-10 | 1 | |||
| SR vs SO | 0.006 | 3.1E-04 | 0.09 | |||
| Cx subdivison | Aggr | Brev | TnR | |||
| L1 vs L2/3 | 0.03 | 1 | 1 | |||
| L1 vs L4 | 0.002 | 0.2 | 1 | |||
| L1 vs L5 | 0.002 | 0.3 | 0.002 | |||
| L1 vs L6 | 1 | 0.007 | 0.004 | |||
| L2/3 vs L4 | 1 | 0.02 | 1 | |||
| L2/3 vs L5 | 1 | 1 | 0.04 | |||
| L2/3 vs L6 | 0.04 | 4.7E-04 | 0.08 | |||
| L4 vs L5 | 1 | 9.7E-05 | 0.03 | |||
| L4 vs L6 | 0.004 | 1 | 0.05 | |||
| L5 vs L6 | 0.003 | 6.5E-07 | 1 | |||
| C: Figures 8 and 9 | ||||||
| Thalamus | Aggr | Brev | TnR | |||
| RN vs LDN | 1.6E-12 | 3.8E-11 | 1 | |||
| RN vs CN | 1.1E-38 | 0.001 | 0.05 | |||
| RN vs VN | 0.06 | 1.3E-05 | 0.005 | |||
| LDN vs CN | 2.0E-18 | 1.2E-20 | 1 | |||
| LDN vs VN | 1.3E-06 | 0.07 | 0.6 | |||
| CN vs VN | 2.3E-32 | 4.3E-14 | 0.5 | |||
| Hypothalamus | Aggr | Brev | TnR | |||
| TN vs LHA | 1.5E-20 | 3.3E-13 | 4.8E-05 | |||
| TN vs ZI | 1.9E-16 | 4.0E-13 | 2.0E-04 | |||
| TN vs VMN | 1.9E-12 | 2.1E-14 | 6.2E-06 | |||
| TN vs AN | 1.9E-05 | 2.3E-15 | 2.1E-04 | |||
| LHA vs ZI | 1 | 1 | 1 | |||
| LHA vs VMN | 0.04 | 1 | 1 | |||
| LHA vs AN | 1.1E-07 | 1 | 1 | |||
| ZI vs VMN | 1 | 1 | 1 | |||
| Zi vs AN | 1.0E-04 | 1 | 1 | |||
| VMN vs AN | 0.03 | 1 | 1 | |||
| D: Figure 11 | ||||||
| PNN count | Aggr | Brev | TnR | |||
| Cx vs H | 9.3E-40 | 4.3E-25 | 2.3E-30 | |||
| Cx vs Cp | 3.2E-77 | 3.2E-05 | 9.8E-78 | |||
| Cx vs Th | 8.7E-82 | 8.9E-14 | 9.1E-83 | |||
| Cx vs Hyp | 1.2E-65 | 6.7E-21 | 5.9E-78 | |||
| Cx vs Amy | 3.7E-58 | 3.2E-14 | 6.2E-80 | |||
| H vs Cp | 2.6E-16 | 1.7E-09 | 6.8E-25 | |||
| H vs Th | 3.0E-20 | 0.02 | 9.4E-30 | |||
| H vs Hyp | 2.7E-08 | 1 | 4.3E-25 | |||
| H vs Amy | 2.2E-04 | 0.02 | 6.7E-27 | |||
| Cp vs Th | 1 | 0.01 | 1 | |||
| Cp vs Hyp | 0.09 | 1.2E-06 | 1 | |||
| Cp vs Amy | 1.1E-04 | 0.009 | 1 | |||
| Th vs Hyp | 0.001 | 0.5 | 1 | |||
| Th vs Amy | 2.9E-07 | 1 | 1 | |||
| Hyp Amy | 1 | 0.7 | 1 | |||
| Cx | H | Cp | ||||
| A vs B | 1.7E-40 | 5.7E-40 | 1 | |||
| A vs TnR | 4.9E-20 | 3.4E-13 | 6.8E-15 | |||
| B vs TnR | 2.5E-10 | 2.6E-16 | 5.5E-17 | |||
| Th | Hyp | Amy | ||||
| A vs B | 1 | 8.6E-12 | 4.1E-14 | |||
| A vs TnR | 5.3E-15 | 2.2E-17 | 3.6E-26 | |||
| B vs TnR | 1.6E-12 | 0.1 | 1.3E-04 | |||
| E: Figure 13 | ||||||
| Colocalization | Cx | H | Cp | Th | Hyp | Amy |
| A/B vs A/B/T | 0.01 | 1 | 2.9E-11 | 1 | 0.7 | 1 |
| A/T vs A/B/T | 0.7 | 1.9E-17 | 2.0E-05 | 6.5E-07 | 4.2E-07 | 9.9E-12 |
| A/T vs A/B | 1 | 5.7E-23 | 4.6E-27 | 5.4E-10 | 1.5E-12 | 3.4E-07 |
| B/T vs A/B/T | 4.1E-11 | 0.002 | 2.0E-05 | 2.3E-07 | 7.4E-08 | 9.9E-12 |
| B/T vs A/B | 0.003 | 0.5 | 4.6E-27 | 1.6E-10 | 1.8E-13 | 3.4E-07 |
| B/T vs A/T | 7.6E-06 | 6.8E-31 | 1 | 1 | 1 | 1 |
| A only vs A/B/T | 3.1E-17 | 3.0E-31 | 1 | 1 | 0.1 | 0.002 |
| A only vs A/B | 8.6E-29 | 2.8E-37 | 9.3E-13 | 0.1 | 1 | 4.5E-07 |
| A only vs A/T | 2.7E-24 | 1.1E-03 | 2.3E-04 | 4.8E-04 | 2.2E-15 | 3.2E-24 |
| A only vs B/T | 1.4E-42 | 1.7E-45 | 2.3E-04 | 4.8E-04 | 2.2E-15 | 3.2E-24 |
| B only vs A/B/T | 6.3E-11 | 0.002 | 1 | 0.02 | 1.6E-06 | 8.1E-07 |
| B only vs A/B | 0.003 | 0.5 | 1.3E-09 | 1.5E-04 | 7.6E-12 | 0.003 |
| B only vs A/T | 1.1E-05 | 6.8E-31 | 8.8E-07 | 0.4 | 1 | 1 |
| B only vs B/T | 1 | 1 | 8.8E-07 | 0.2 | 1 | 1 |
| B only vs A only | 2.4E-42 | 1.7E-45 | 1 | 1 | 1.1E-13 | 7.7E-18 |
| T only vs A/B/T | 7.9E-11 | 0.004 | 2.0E-05 | 6.5E-07 | 7.4E-08 | 9.9E-12 |
| T only vs A/B | 0.004 | 0.8 | 4.6E-27 | 5.4E-10 | 1.8E-13 | 3.4E-07 |
| T only vs A/T | 1.3E-05 | 3.4E-30 | 1 | 1 | 1 | 1 |
| T only vs B/T | 1 | 1 | 1 | 1 | 1 | 1 |
| T only vs A only | 3.3E-42 | 8.9E-45 | 2.3E-04 | 0.001 | 2.2E-15 | 3.2E-24 |
| T only vs B only | 1 | 1 | 8.8E-07 | 0.4 | 1 | 1 |
| F: Figure 14 | Neurite length/nuclei | Neurite length/slice | ||||
| ColI vs Aggr | 0.0005 | 0.0002 | ||||
| ColI vs Brev | 3.4E-5 | 0.0001 | ||||
| ColI vs TnR | 9.2E-5 | 0.0002 | ||||
| ColI vs A/B/T | 9.1E-5 | 0.0002 | ||||
| Aggr vs Brev | 1 | 1 | ||||
| Aggr vs TnR | 1 | 1 | ||||
| Aggr vs A/B/T | 1 | 1 | ||||
| Brev vs TnR | 1 | 1 | ||||
| Brev vs A/B/T | 1 | 1 | ||||
| TnR vs A/B/T | 1 | 1 | ||||
P values shown are derived from the second step Bonferroni post hoc test. P values below 0.05 indicate that values are significantly different.
Abbreviations: Cx, isocortex; H, hippocampus; Cp, caudate-putamen; Th, thalamus; Hyp, hypothalamus; Amy, amygdala; Aggr or A, aggrecan; Brev or B, brevican; TnR or T, tenascin-R; SP, stratum pyramidale; DG, dentate gyrus; SR, stratum radiatum; SO, stratum oriens; L, layer; RN, reticular nucleus; LDN, lateral dorsal nucleus; CN, central and medial nuclei; VN, ventral nuclei; TN, tuberal nucleus; LA, lateral area; ZI, zona incerta; DN, dorso- and ventromedial nuclei; AN, arcuate nucleus; PNN, perineuronal net.
RESULTS
Global extracellular matrix distribution throughout distinct brain regions
To determine the distribution of specific ECM proteins throughout the brain, we immunostained adult rodent brain slices for Aggr, Brev, and TnR and quantified the signal intensity of each ECM protein as well as the number of PNNs in the isocortex (Cx), the hippocampus (H), the caudate-putamen (Cp), the thalamus (Th), the hypothalamus (Hy), and the amygdala (Am). Signal intensity levels represent all ECM structures, including PNNs, interstitial matrix, and basement membrane matrix. We cut 50-μm-thick coronal sections (Fig. 1A,B) and for each brain we used consecutive sections (Fig. 1C), which were stained for Aggr (green), Brev (blue), and TnR (red) (Fig. 2). Regions of interest were outlined for each slice as depicted (Fig. 3). Regions of interest were identified according to established cytoarchitectonic criteria (http://atlas.brain-map.org/). Single channel images outline the differential distribution patterns of the various ECM proteins (Fig. 4A–C). Aggr-, Brev-, and TnR-positive PNNs are depicted in Figure 4D–F. Signal intensity analysis of whole brain slices revealed that for all three proteins, the hippocampus showed the highest signal intensity (Fig. 5A–C, n = 70 slices, 3 brains). This result was most apparent for Brev signal intensity (Fig. 5B). This indicates that the hippocampus is densely packed with all three ECM proteins, pointing toward an important role of ECM proteins for memory formation.
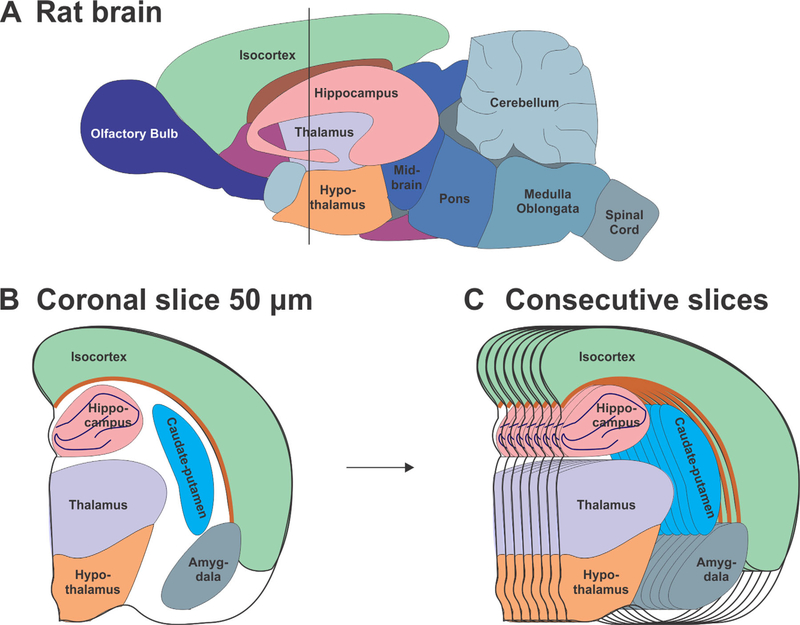
Figure 1.
Rat brain regions used to analyze the distribution of extracellular matrix proteins throughout the rat brain. A: Schematic of the adult rat brain with outlined brain regions. B: 50-μm-thick sections were taken at a depth such that the hippocampus, the thalamus, and the hypothalamus were visible (around Bregma −2.3 mm). C: 22–24 consecutive sections were cut for each brain and used for analysis.
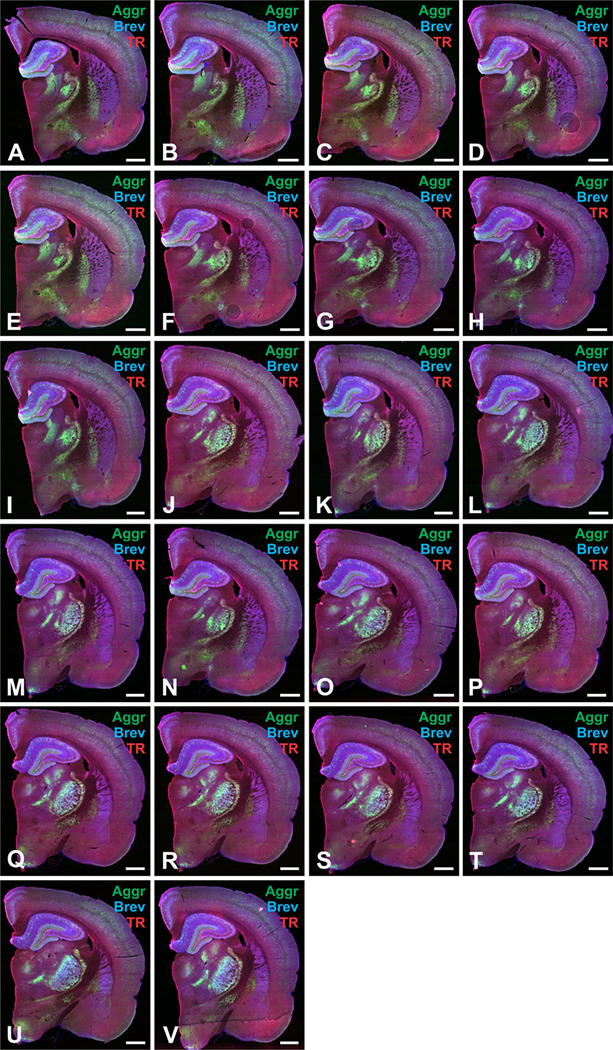
Figure 2.
A–V: Example images of consecutive brain slices stained for extracellular matrix proteins. Consecutive slices used for analysis from one brain were stained for aggrecan (Aggr, green), brevican (Brev, blue), and tenascin-R (TnR, red). Scale bar 5 1 mm in A–V.
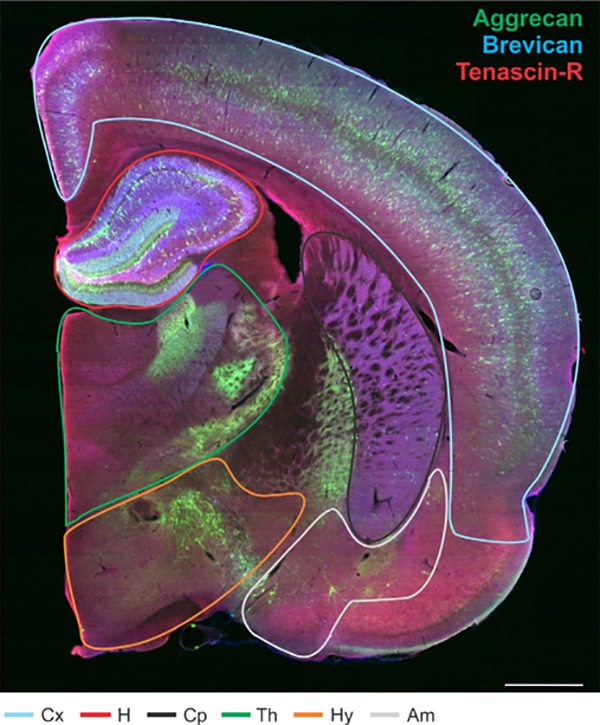
Figure 3.
Extracellular matrix distribution throughout distinct regions of the adult rat brain. Whole adult rat brain section labeled for aggrecan (green), brevican (blue), and tenascin-R (red) with outlined brain regions: isocortex (blue), hippocampus (red), caudate-putamen (black), thalamus (green), hypothalamus (orange), and amygdala (gray). Abbreviations: Cx, isocortex; H, hippocampus; Cp, caudate-putamen; Th, thalamus; Hy, hypothalamus; Am, amygdala. Scale bar = 1 mm.
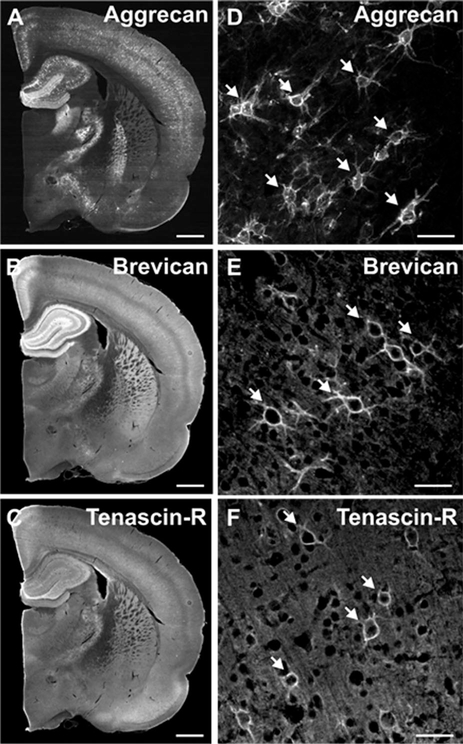
Figure 4.
Distribution of aggrecan, brevican, and tenascin-R throughout distinct regions of the adult rat brain. A–C: Whole adult rat brain sections labeled for aggrecan (A), brevican (B), and tenascin-R (C) are shown in black and white. D–F: Higher magnification images show aggrecan-, brevican-, and tenascin-R positive perineuronal nets. Scale bar = 1 mm (in A–C) and 50 μm (in D–F).
In more detail, Aggr signal intensity was highest in the hippocampus, followed by slightly lower levels in the isocortex and thalamus, with the lowest observed signal intensity in the caudate-putamen, hypothalamus, and amygdala (Fig. 5A). Brev signal intensity was found to be highest in the hippocampus, followed by the isocortex, caudate-putamen, thalamus, amygdala, and hypothalamus (Fig. 5B). The presence of TnR was highest in the hippocampus. The abundance of TnR in the isocortex and amygdala was similar and slightly lower than in the hippocampus. The lowest signal intensities were detected in the caudate-putamen and the hypothalamus (Fig. 5C). A significant difference in signal intensity between several of the analyzed brain regions was observed for all three ECM proteins (Table 2A). Comparison of the distribution patterns for the three analyzed ECM proteins revealed that for all proteins, protein abundance was highest in the hippocampus and lowest in the caudate-putamen and/or hypothalamus. Overall, TnR displayed a more ubiquitous and uniform distribution pattern throughout all brain regions, whereas Aggr and Brev had more pronounced differential distribution patterns. This implies that Aggr and Brev have more specialized functions in distinct brain regions, whereas TnR has similar functions throughout the brain.
Extracellular matrix distribution in substructures of the hippocampus, isocortex, thalamus, and hypothalamus
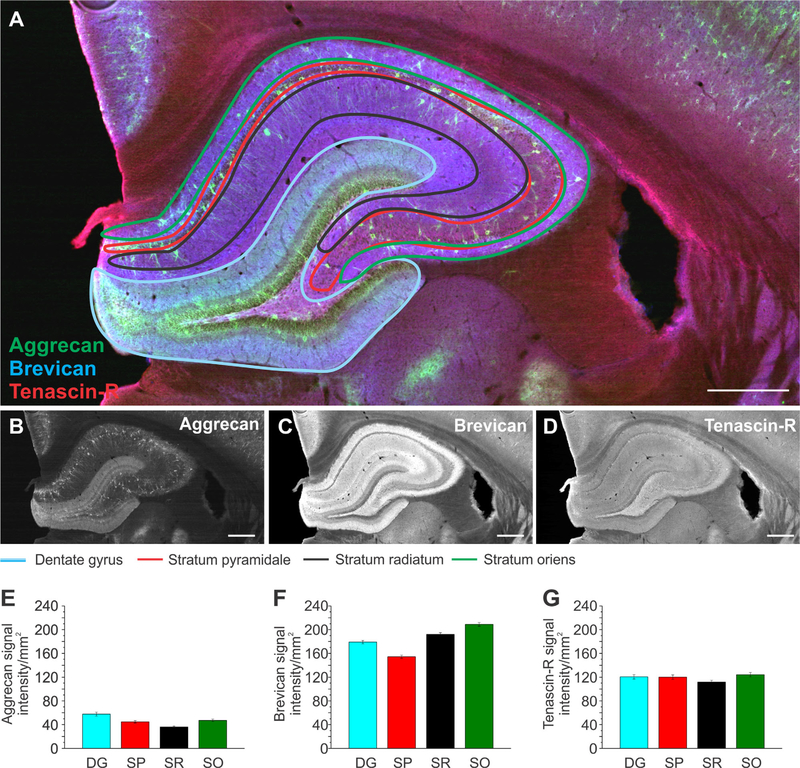
Most of the brain regions analyzed in Figures 3–5 have subregions or distinct layers, which were not taken into account in the first analysis. Different subregions in the brain have specific functions and differ in cell types and projection pathways (Watakabe, 2009; Schultz and Engelhardt, 2014). For example, in humans and rodents the isocortex consists of six layers, each of which has specific connections and projection paths as well as distinct cell populations (Watakabe, 2009). As we hypothesized that the ECM environment plays an important role for brain physiology, the assumption arises that specific ECM proteins may reside in distinct subregions within the brain. Therefore, we divided the hippocampus and the isocortex (Figs. (6 and 7), n = 33 slices, 3 brains) and the thalamus and hypothalamus (Figs. (8 and 9), n = 36 slices, 3 brains) into their respective subregions. The hippocampus was divided into the dentate gyrus (DG), stratum pyramidale (SP), stratum radiatum (SR), and stratum oriens (SO), as depicted in Figure 6A. Signal intensity analyses revealed that Aggr had the highest abundance in the DG and was slightly less abundant in the SP and SO. The lowest presence was observed in the SR (Fig. 6B,E). The SO exhibited high levels of Brev, followed by lower levels in the SR and DG and the lowest levels in the SP (Fig. 6C,F). TnR abundance was similar throughout all subregions of the hippocampus, with a slightly lower presence in the SR compared with the DG, SP, and SO (Fig. 6D,G). A significant difference in signal intensity between several of the analyzed brain regions was observed for Aggr and Brev (Table 2B). Whereas Aggr and Brev differed in their abundance in these subregions, TnR was distributed more equally. Moreover, comparison of the distribution patterns of Aggr and Brev revealed a distinct pattern for each ECM protein. Aggr was highly abundant in the DG and least in the SR, whereas Brev levels were the second highest in the SR and the second lowest in the DG.
Figure 6.
Extracellular matrix distribution in the hippocampus. A: Adult rat brain slice showing the hippocampus labeled for aggrecan (green), brevican (blue), and tenascin-R (red) with its outlined subregions, the dentate gyrus (blue), stratum pyramidale (red), stratum radiatum (black), and stratum oriens (green). B–D: Single channels for each protein are shown in black and white. E–G: Signal intensity measurements for aggrecan (E), brevican (F), and tenascin-R (G) for the subregions outlined in A. Column color corresponds to the region outline color in A. Error bars are shown as standard error of the mean (SEM). Significance levels calculated by ANOVA (Bonferroni post hoc test) are shown in Table 2B. n = 33 slices, 3 brains. Abbreviations: DG, dentate gyrus; SP, stratum pyramidale; SR, stratum radiatum; SO, stratum oriens. Scale bar = 500 μm in A–D.
Figure 7.
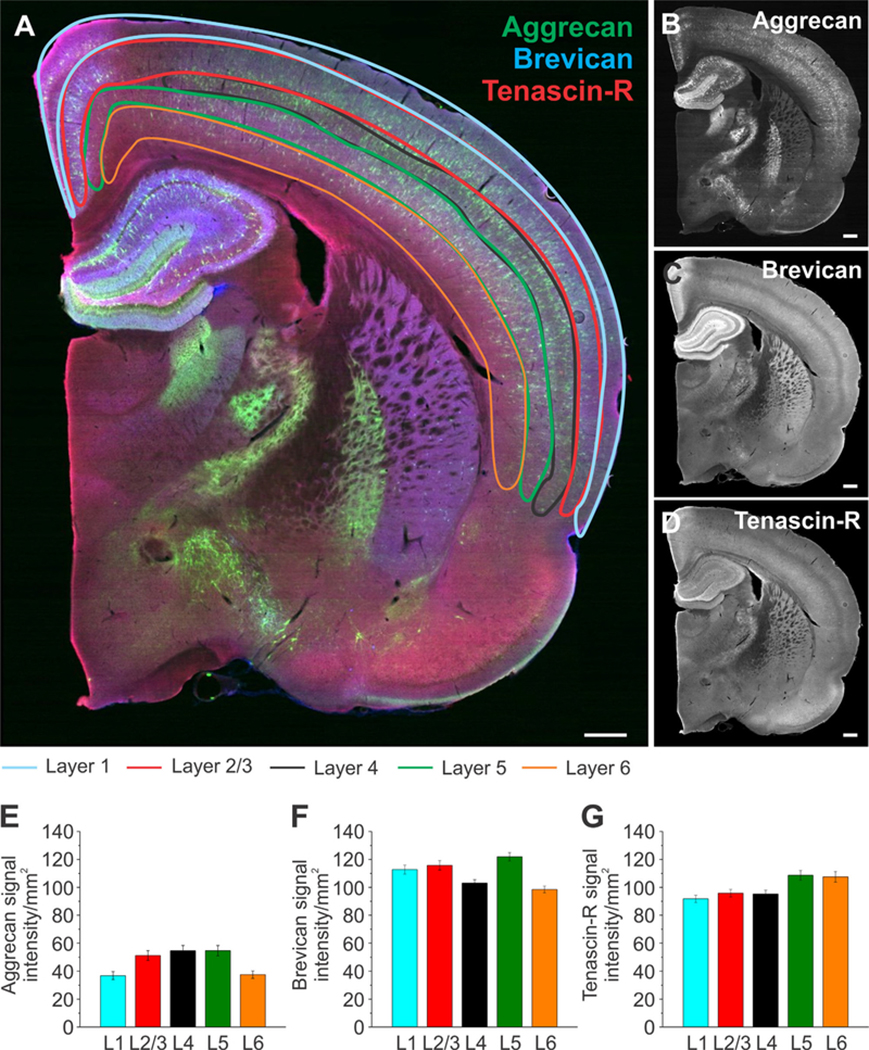
Extracellular matrix distribution in distinct isocortical layers. A: Whole brain slices were labeled for aggrecan (green), brevican (blue), and tenascin-R (red) with outlined subregions of the isocortex: layer 1 (blue), layer 2/3 (red), layer 4 (black), layer 5 (green), and layer 6 (orange). B–D: Single channels for each protein are shown in black and white. E–G: Signal intensity measurements for aggrecan (E), brevican (F), and tenascin-R (G) for the subregions outlined in A. Column color corresponds to the region outline color in A. Error bars are shown as standard error of the mean (SEM). Significance levels calculated by ANOVA (Bonferroni post hoc test) are shown in Table 2B. n = 33 slices, 3 brains. Abbreviation: L, layer. Scale bar = 500 μm in A–D.
Figure 8.
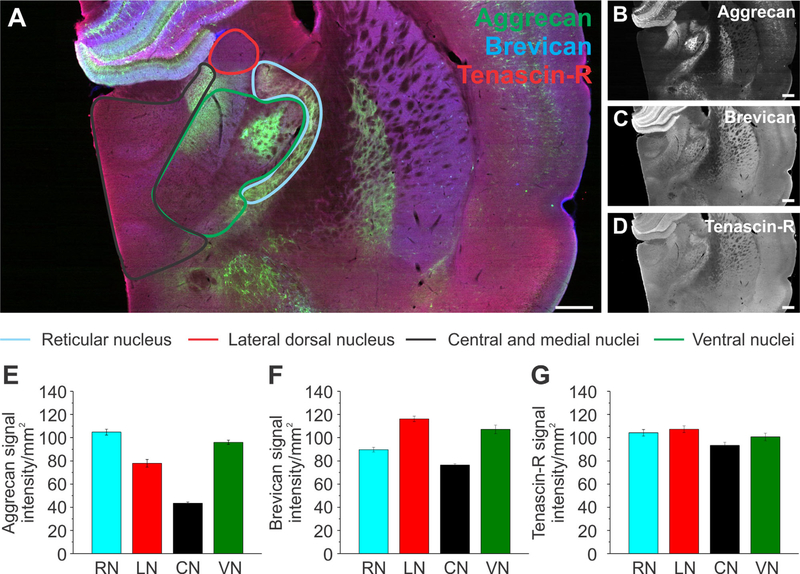
Extracellular matrix distribution in the thalamus. A: Adult rat brain slice depicts the thalamus labeled for aggrecan (green), brevican (blue), and tenascin-R (red) with its outlined subregions, the reticular nucleus (blue), lateral dorsal nucleus (red), central and medial nuclei (including the mediodorsal nuclei, lateral and medial habenula, paraventricular nucleus, intermediodorsal nucleus, central medial nucleus, paracentral nucleus, rhomboid nucleus, submedial nucleus, and the nucleus of reuniens, black), ventral nuclei (including the ventral medial nucleus, ventral anterior lateral complex, ventral posteromedial nucleus, and ventral posterolateral nucleus, green). B–D: Single channels for each protein are shown in black and white. E–G: Signal intensity measurements for aggrecan (E), brevican (F), and tenascin-R (G) for the subregions outlined in A. Column color corresponds to the region outline color in A. Error bars are shown as standard error of the mean (SEM). Significance levels calculated by ANOVA (Bonferroni post hoc test) are shown in Table 2C. n = 36 slices, 3 brains. Abbreviations: RN, reticular nucleus; LA, lateral dorsal nucleus; CN, central and medial nuclei; VN, ventral nuclei. Scale bar = 500 μm in A–D.
The isocortex was divided into layer 1 (L1), layer 2/3 (L2/3), layer 4 (L4), layer 5 (L5), and layer 6 (L6), as depicted in Figure 7A. Aggr signal intensity levels were highest in L5 and 4, followed by L2/3, L1, and L6 (Fig. 7E). Brev signal intensity levels were highest in L5 as well, followed by L2/3, 1, L4, and L6 (Fig. 7F). TnR signal intensity levels were highest in L5 and 6, followed by L2/3, 4, and 1 (Fig. 7G). The distribution pattern was once again different for each ECM protein, except for L5, which had the highest abundance of all of the ECM proteins. Significance values for the observed signal intensity differences between the cortical layers are summarized in Table 2B.
The thalamus was divided into the reticular nucleus (RN), lateral dorsal nucleus (LN), central and medial nuclei (CN) (including the mediodorsal nuclei, lateral and medial habenula, paraventricular nucleus, intermediodorsal nucleus, central medial nucleus, paracentral nucleus, rhomboid nucleus, submedial nucleus, and nucleus of reunions), the ventral nuclei (VN) (including the ventral medial nucleus, ventral anterior lateral complex, and ventral posteromedial nucleus), and the ventral posterolateral nucleus (VN), as depicted in Figure 8A. Aggr was highly abundant in the RN, followed by the VN, LN, and CN (Fig. 8E, n = 36 slices, 3 brains). In contrast, Brev signal intensity was highest in the LA, followed by the VN, RN, and CN (Fig. 8F). TnR displayed a more uniform distribution throughout all subregions, with the exception of a slightly lower abundance in the CN and VN (Fig. 8G). Significance values are summarized in Table 2C. As observed for the hippocampus and the isocortex, the abundance for each ECM protein in the thalamus was subregion specific.
The hypothalamus was divided into the tuberal nucleus (TN), lateral area (LA), zona incerta (ZI), dorsoand ventromedial nuclei (DN), and arcuate nucleus (AN), as depicted in Figure 9A. Aggr signal intensity was highest in the LA, followed by the ZI, DN, and AN, and lowest in the TN (Fig. 9E, n = 36 slices, 3 brains). Significance levels are shown in Table 2C. Brev and TnR signal intensity was very similar in all regions except for the TN, which exhibited the lowest intensity levels (Fig. 9F,G). Overall, our results revealed that ECM distribution was different even within distinct brain subregions, suggesting a more complex region- and subregion-dependent function of the ECM.
Number of perineuronal nets in distinct brain regions
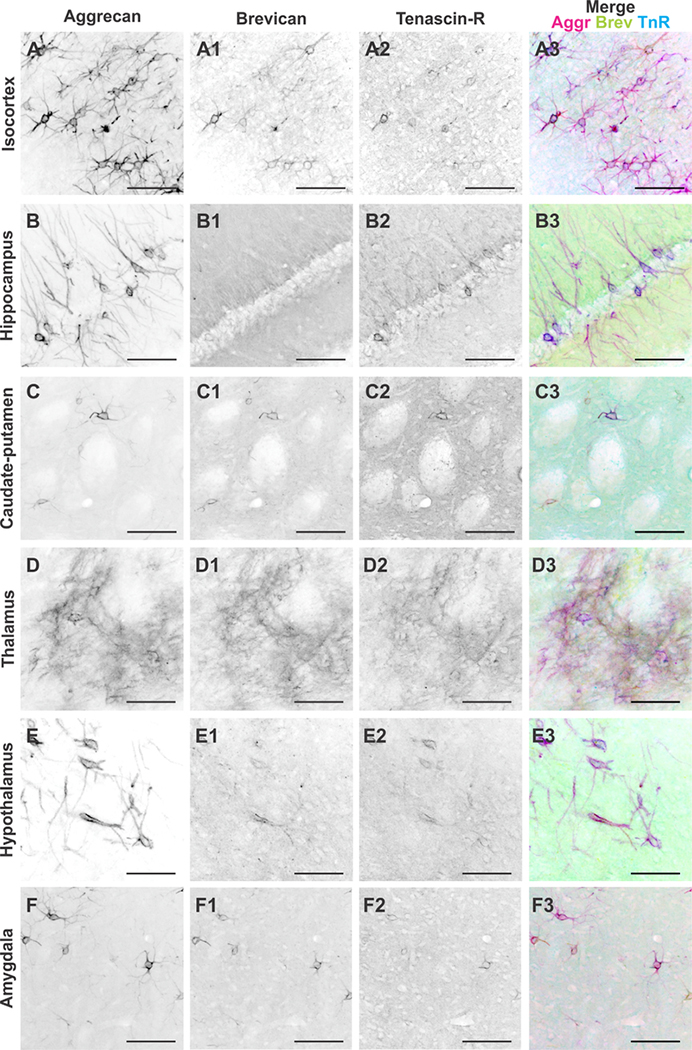
To gain a thorough understanding of ECM components throughout different brain regions, it is necessary to distinguish distinct ECM structures, as they are essential for specific functional roles. One of the most important structural features of the ECM in the brain is the PNN, a specialized structure defined by densely packed ECM proteins surrounding neuronal soma and proximal processes (Lau et al., 2013). We hypothesized that the abundance and composition of PNNs throughout different brain regions would vary, and would be represented by varying numbers of Aggr-, Brev-, and TnR-positive PNNs within each brain region. Therefore, we quantified Aggr-, Brev-, and TnR-positive PNNs in the isocortex, hippocampus, caudate-putamen, thalamus, hypothalamus, and amygdala (Fig. 10, n = 64 images, 24 slices, 3 brains). The depicted immunostained images show a marked difference in PNN abundance, and structural appearance, between brain regions. There is a clear difference in the numbers of Aggr-, Brev-, and TnR-positive PNNs within each specific brain region (Fig. 10A–F). Comparing the abundance of Aggr-positive PNNs across distinct brain regions revealed the highest PNN numbers in the isocortex, followed by the hippocampus, amygdala, hypothalamus, caudate-putamen, and thalamus (Fig. 11A). Brev-positive PNNs were overall less abundant than Aggr-positive PNNs. Most Brev-positive PNNs were found in the isocortex, followed by the caudate-putamen, amygdala, thalamus, hypothalamus, and hippocampus (Fig. 11B). TnR-positive PNNs are also overall less abundant than Aggr-positive PNNs. Most TnR-positive PNNs were found in the isocortex, followed by the hippocampus, caudate-putamen, hypothalamus, amygdala, and thalamus (Fig. 11C). All three ECM proteins showed the highest abundance of PNNs in the isocortex. In contrast, the hippocampus exhibited the second highest abundance of Aggr- and TnR-positive PNNs, but the lowest abundance of Brev-positive PNNs. Most of the observed differences in PNN numbers between regions or between different ECM proteins were statistically significant, as summarized in Table 2D.
Figure 10.
Perineuronal net distribution in distinct brain regions of the adult rat brain. A–F3: Black and white single channel images of aggrecan-, brevican-, and tenascin-R-positive perineuronal nets and merge image of all three channels in the (A–A3) isocortex, (B–B3) hippocampus, (C–C3) caudate-putamen, (D–D3) thalamus, (E–E3) hypothalamus, and (F–F3) amygdala. Abbreviations: Aggr, aggrecan; Brev, brevican; TnR, tenascin-R. Scale bar = 100 μm in A–F3.
Interestingly, the signal intensity distribution of Aggr, Brev, and TnR (Figs. (5 and 11)D–F) was strikingly different in comparison with the numbers of PNNs (Fig. 11D–F). This suggests that specialized ECM structures, such as PNNs, are differently distributed compared with the overall composition of the ECM within the same region. Whereas Aggr PNNs were highly abundant in the isocortex, Aggr signal intensity levels were comparably low in the isocortex (Fig. 11A,D). Similarly, numbers of Brev-positive PNNs were the lowest in the hippocampus, whereas Brev signal intensity levels were highest in the hippocampus (Fig. 11B,E). Numbers of TnR-positive PNNs were quite different among the isocortex, the hippocampus, and the rest of the regions, but TnR signal intensity levels were more equally distributed throughout the different brain regions (Fig. 11C,F). Our results indicate that the distribution of specific ECM structures, PNNs, have a unique distribution pattern compared with the interstitial matrix. This suggests region-dependent distinct functionalities even between specific ECM structures.
ECM protein expression throughout the brain
As a second approach for investigating the distribution of Aggr, Brev, and TnR, as well as a broader range of ECM molecules, we collected tissue samples from the adult rat isocortex, hippocampus, amygdala, cerebellum (Cb), and brainstem (Bs) and analyzed protein levels by mass spectrometry (MS). As depicted in Table 3, we detected various ECM proteins, which are differentially expressed throughout the brain. The highest expression level of Aggr was detected in the Cx, followed by the hippocampus and the amygdala (Table 3), which correlates with the results of the PNN analyses (Fig. 11A). When including the cerebellum and the brainstem were included in our comparison, which were not part of our immunofluorescence studies, these regions showed even higher expression levels of Aggr compared with the isocortex, hippocampus, and amygdala, with highest expression levels in the brainstem (Table 3). Brev expression (only comparing Cx, H, and Am) exhibited the same trend as Aggr, with highest levels in the isocortex, followed by the hippocampus and the amygdala (Table 3). The PNN results for Brev showed the highest numbers in the isocortex, which correlates with the MS data, but PNN numbers were shown to be higher in the amygdala than the hippocampus, which is opposite to the MS data (Fig. 11B, Table 3). In contrast, Brev signal intensity for the hippocampus was higher than the signal detected in the amygdala, which correlates with the MS data (Fig. 7B,E, Table 3). Including the cerebellum and brainstem in our comparison, we observed the highest expression levels of Brev in the brainstem, followed by the cerebellum (Table 3). Protein expression for TnR (only looking at Cx, H, and Am) was highest in the isocortex, followed by the hippocampus and the amygdala (Table 3), which correlates well with the numbers of PNNs that were determined for TnR (Fig. 11C). When the cerebellum and the brainstem were included, we observed the highest TnR expression levels in the brainstem, followed by the cerebellum (Table 3).
TABLE 3.
Extracellular Matrix Protein Expression in Different Regions of the Adult Rat Brain1
| Fold change (normalized to isocortex neonate) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Access. No. | Description | Cx | V(%) | H | V(%) | Am | V(%) | Cb | V(%) | Bs | V(%) |
| B2LYI9 | Tenascin-C | 3.99 | 17 | 4.36 | 24 | 1.92 | 22.8 | 6.15 | 22.5 | 1.09 | 60.4 |
| F1LQ63 | Tenascin-R | 2.91 | 34.6 | 1.84 | 39.9 | 1.52 | 34.3 | 4.56 | 34.4 | 9.56 | 59 |
| F1LM29 | Reelin | 2.61 | 19.9 | 3.56 | 5.3 | 2.05 | 14.7 | 3.98 | 19.1 | 1.75 | 2.6 |
| P55067 | Neurocan core protein | 2.64 | 17.4 | 2.58 | 17.2 | 1.46 | 22 | 3.87 | 18.3 | 2.61 | 20.6 |
| G3V8G4 | Brevican, isoform CRA_a | 3.2 | 30.7 | 2.409 | 16.2 | 1.56 | 37.7 | 3.78 | 23.5 | 5.43 | 33.5 |
| P35053 | Glypican-1 | 3.84 | 7.8 | 2.53 | 20.9 | 2.5 | 7.9 | 6.12 | 25.4 | 2.46 | 6.6 |
| A1A5N6 | Hyaluronan and proteoglycan link protein 1 | 2.47 | 30 | 2.1 | 50.1 | 1.6 | 55.1 | 4.3 | 43.2 | 5.5 | 68.7 |
| G3V8G6 | Hyaluronan and proteoglycan link protein 2 | 4.05 | 8.9 | 2.89 | 2.9 | 1.77 | 0.8 | 4.79 | 37.2 | 11.23 | 56.7 |
| A1L1K8 | Hyaluronan binding protein 4 | 4.3 | 23.1 | 2.1 | 3.8 | 1.84 | 1.7 | 4.5 | 43.4 | 6.0 | 94.8 |
| D3Z9N6 | Versican core protein | 2.56 | 39.2 | 2.03 | 30.7 | 1.89 | 47.6 | 3.46 | 14.5 | 7.52 | 53.8 |
| Q9ERB4-3 | Isoform Vint of versican core protein | 2.51 | 36.3 | 2.02 | 36.7 | 1.74 | 33.6 | 3.44 | 25.7 | 5.98 | 54.8 |
| F1LQI4 | Aggrecan core protein | 3.3 | 6.1 | 2.12 | 1.3 | 1.54 | 9.2 | 4.85 | 7.7 | 22.2 | 45.4 |
| F1M8W5 | Disintegrin and metalloproteinanse domain containing protein 11 | 3.12 | 5.9 | 2.39 | 14.4 | 1.96 | 17.1 | 4.76 | 20.6 | 6.48 | 18.9 |
| D4A6U1 | A disintegrin and metallopeptidase domain 11 | 4.43 | 79.0 | 2.47 | 42.2 | 2.1 | 96.9 | 5.247 | 67.1 | 7.75 | 132.3 |
| B2RYP8 | A disintegrin and metalloprotease domain 8 | 4.33 | 21.9 | 3.02 | 6.0 | 2.29 | 16.4 | 6.49 | 17.4 | 4.62 | 19.0 |
| Q5RKL5 | Metalloreductase STEAP3 | 3.58 | 1.2 | 2.78 | 41.1 | 2.05 | 19.5 | 5.93 | 12.5 | 5.37 | 21.1 |
Tissue from adult rat brain was dissected, and protein expression was analyzed using mass spectrometry. The neonate isocortex was used as a control, and values are shown as fold change normalized to the control. Uniprot accession numbers are shown, as well as the description of the protein, the fold change for each brain region, and the variability.
Abbreviations: Cx, cortex; V, variability; H, hippocampus; Am, amygdala; Cb, cerebellum; Bs, brainstem.
Additional ECM proteins detected included tenascin-C, reelin, neurocan, glypican-1, HA link and binding proteins, and versican. Tenascin-C had the highest expression levels in the cerebellum, followed by the hippocampus, isocortex, amygdala, and brainstem. This distribution was very different from that of TnR (Table 3). Reelin expression was highest in the cerebellum, followed by the hippocampus, isocortex, amygdala, and brainstem. Neurocan expression was highest in the cerebellum, followed by the isocortex, hippocampus, brainstem, and amygdala. Glypican-1 expression was highest in the cerebellum, followed by the isocortex, hippocampus, amygdala, and brainstem. HA link protein 1 and 2 as well as HA binding protein 4 expression was highest in the brainstem, followed by the cerebellum, isocortex, hippocampus, and amygdala. Versican showed the highest expression in the brainstem, followed by the cerebellum, isocortex, hippocampus, and amygdala (Table 3). Generally, the highest expression levels for most of the ECM proteins were observed in the cerebellum, brainstem, and isocortex. Overall, these results suggest a different distribution of each of the ECM proteins throughout distinct brain regions and therefore a unique and brain region-dependent ECM fingerprint. In addition to ECM proteins, we also detected ECM-modifying proteins, such as a disintegrin and metallopeptidase domain 8 and 11 and metalloreductase STEAP3, which all showed the highest expression levels in the brainstem and cerebellum, followed by the isocortex, hippocampus, and amygdala (Table 3), correlating with the general trend observed for ECM proteins. Overall, these results suggest that the regions at the exterior of the brain (brainstem, cerebellum, and isocortex) express the highest amounts of ECM proteins and ECM-related proteins, implying a potential protective role of the ECM in the most vulnerable and exposed brain regions. This distribution may also represent more restricted plasticity in these regions in comparison with the amygdala, thalamus, and hippocampus.
Local distribution of Aggr, Brev, and TnR
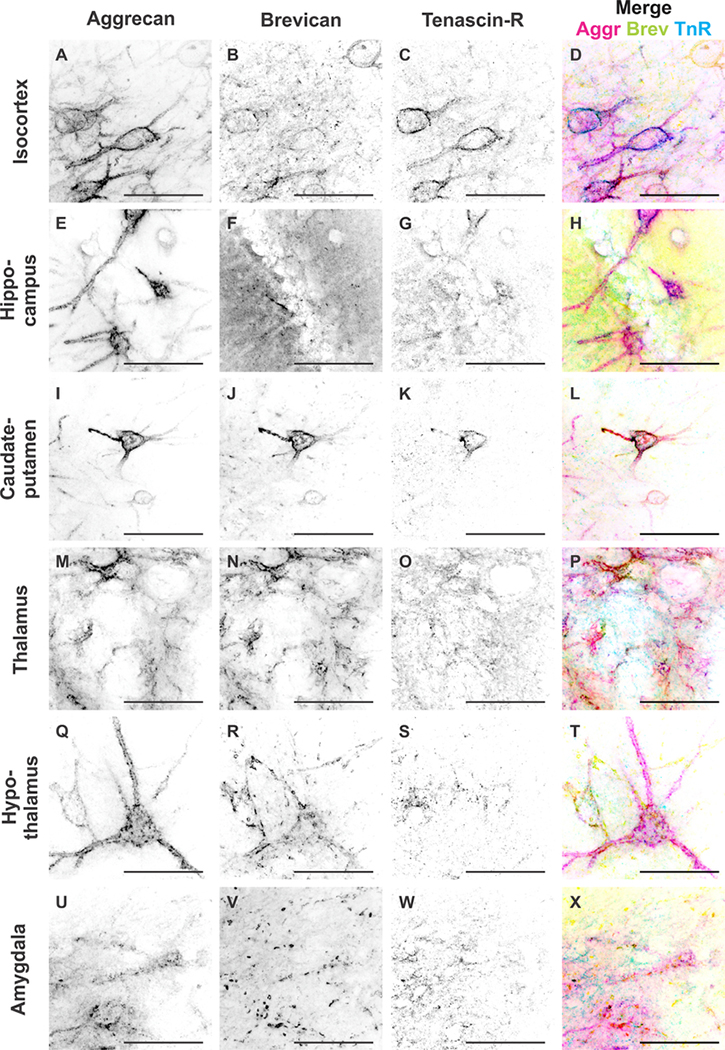
We have shown so far that the distribution of ECM proteins appeared to be distinct depending on the brain region investigated. The question arises as to how distribution of the same ECM proteins might differ locally between neighboring cells, i.e., whether neighboring PNNs have varying ECM compositions. This would suggest that the role a specific ECM protein may play depends on the location as well as on the parallel expression of other ECM proteins. Therefore, we determined the colocalization of those ECM proteins in each brain region (Figs. (12 and 13)). Figure 12 depicts high-magnification images of neighboring PNNs in each of the investigated brain regions. The images accurately depict differences in Aggr, Brev, and TnR composition between neighboring PNNs (Fig. 12). To quantify colocalization patterns, we used 20-μm z-stacks and counted Aggr-, Brev-, and TnR-positive PNNs and colocalization of the various combinations (Fig. 13A–D, n = 39 images, 18 slices, 3 brains). In the isocortex, we observed that the highest numbers of PNNs were positive only for Aggr, but not for Brev or TnR (Fig. 13E). The second highest number of PNNs showed a colocalization of all three ECM proteins, followed by PNNs that were positive for Aggr and TnR but not for Brev, and lastly PNNs that expressed Aggr and Brev but not TnR. Almost no PNNs were found to express only Brev or TnR. In the hippocampus, most PNNs were positive for Aggr only (Fig. 13F), followed by PNNs positive for Aggr and TnR but not for Brev. We observed only a small population of PNNs in the hippocampus that expressed all three ECM proteins, Aggr and Brev, but not TnR, or PNNs positive for only Brev or TnR. The caudate-putamen exhibited the highest numbers of PNNs positive for Aggr and Brev, but not TnR, followed by PNNs positive for Brev only, all three ECM proteins, and lastly Aggr only (Fig. 13G). In the thalamus we determined the highest numbers of PNNs that were positive for Aggr and Brev, but not TnR, followed by PNNs that were positive for all three ECM proteins, Aggr only, and lastly Brev only (Fig. 13H). The hypothalamus showed the highest numbers of PNNs that were positive for Aggr only, followed by Aggr- and Brev-positive PNNs, and lastly PNNs that were positive for all three ECM proteins (Fig. 13I). In the amygdala we observed highest numbers of PNNs that were positive for Aggr only, followed by PNNs positive for all three ECM proteins, Aggr- and Brev-positive PNNs, and lastly Brev only PNNs (Fig. 13J). Differences between PNN compositions were analyzed by using one-way ANOVA, and significance levels are depicted in Table 2E. Overall, the results indicated that, locally, single, neighboring PNNs express different ECM proteins, with most PNNs expressing Aggr, either alone or in combination with both or either of the other two ECM proteins. Further analyses revealed that colocalization patterns vary in distinct brain regions. These results suggest that even at the local and single cell level the ECM is tightly regulated and involved in specific roles.
Figure 12.
Colocalization patterns in neighboring neurons in different brain regions. A–X: High-magnification black and white single channel images of aggrecan-, brevican-, and tenascin-R-positive perineuronal nets and merge image of all three channels for the isocortex (A–D), hippocampus (E–H), caudate-putamen (I–L), thalamus (M–P), hypothalamus (Q–T), and amygdala (U–X). Scale bar = 50 μm in A–X.
Neurite outgrowth of 3D neuronal cultures seeded in different ECM environments
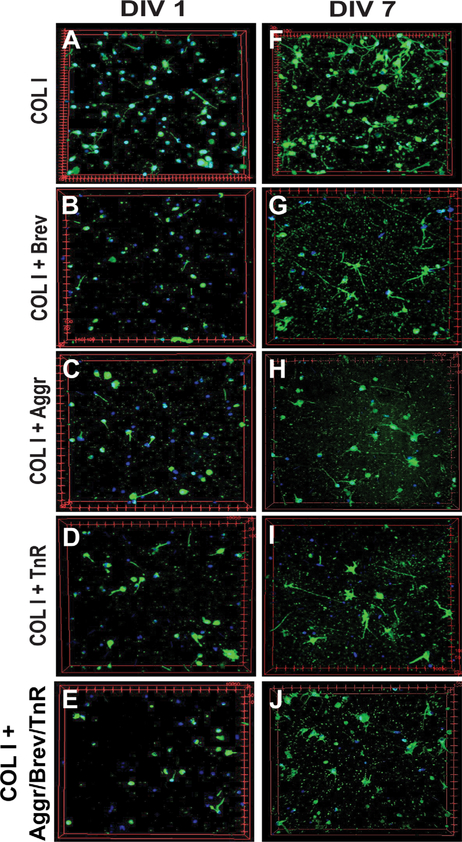
The specific regionalization of ECM proteins at the scale of brain regions, brain subregions, and cells suggests that the composition of the ECM is important for neuronal structure and function. As shown in this study, the ECM environment of two neighboring cells can be completely different, with distinct compositions of PNNs. This strongly suggests that local variation of the ECM environment at the cell scale could be a way to differentiate the function of two neighboring cells. To test whether different ECM environments have an influence on neuronal behavior in vitro, we embedded neonatal cortical neurons in a 3D matrix composed of collagen I (COLI) and brain ECM proteins (Aggr, Brev, and TnR; Fig. 14A). We used a 3D environment for our in vitro experiments, because it was previously shown that a 3D environment is important to mimic proper brain physiology (East et al., 2009; Zhang et al., 2014), and might contribute to cell behavior not observable in a 2D environment. We measured neurite outgrowth in five different environments: 1) COLI, 2) COLI + Aggr, 3) COLI 1 Brev, 4) COLI + TnR, and 5) COLI + a mix of Aggr/Brev/TnR. Example images show visually the differences in neurite outgrowth between the different conditions at day 1 and day 7 in vitro (Fig. 15). Neurite outgrowth experiments revealed that 3D environments containing brain ECM exhibited a significant lower neurite growth than the control (COLI) (Fig. 14B, n = 10–16 z-stacks, 2–4 samples). We first assessed the total neurite length per unit volume (Fig. 14B, left panel). To facilitate the visualization of trends in neurite outgrowth for the different ECM compositions, each dataset was fitted with a Hill function. Hill function has been used to describe population growth for which growth rate dampens along time, similarly to our experiments. The fit took into account the 14 days of growth for COLI only. For COLI + ECM proteins we observed a consistent reduction between day 7 and 14, which is not taken into account by the Hill equation. Therefore, we fitted these samples until day 7 and extrapolated the fit to day 14. The total neurite length per unit volume was significantly higher for the COLI-only environment than for COLI + ECM proteins at each time point. As the total neurite length could be due to both neurite growth and cell proliferation, we normalized the total neurite length by the total number of nuclei (Fig. 14B, right panel). Over 14 days in vitro, neurons in COLI gels showed a neurite outgrowth length of 130 μm per nuclei, whereas COLI gels that were complemented with either Aggr (~70 μm), Brev (~35 μm), or TnR (~50 μm) or a combination of all three proteins (45 μm) showed a significant decrease in neurite length per nuclei, with Brev showing the highest inhibition on neurite outgrowth (Fig. 14B). Significance levels are shown in Table 2F. This confirms previous 2D in vitro as well as in vivo studies that reported an overall inhibiting behavior of brain ECM proteins on neurite outgrowth (Noble et al., 1984; Emerling and Lander, 1996; Asher et al., 2001). Additionally, nuclei numbers for COLI samples increased from 10 nuclei per slice on day 1 to 20 on day 7 and 14, whereas the addition of either ECM protein decreased the number of nuclei overall and over time (data not shown). Our results revealed that the addition of Aggr, Brev, and/or TnR has a significant negative effect on neurite outgrowth and possibly cell attachment, suggesting a potential role of ECM proteins in inhibiting axonal regeneration after injury.
Figure 15.
Effect of aggrecan, brevican, and tenascin-R on neurons in a 3D environment. A–J: Neurons were cultured for 1–14 days in vitro; example images are shown for day 1 and day 7 for all conditions. Neurons were fixed and stained for ßIII-tubulin (green) and DAPI (nucleus, blue). Each image represents a volume of 645 μm × 645 μm × 75 μm. Abbreviations: Aggr, aggrecan; Brev, brevican; TnR, tenascin-R; COLI, collagen I; DIV, days in vitro.
DISCUSSION
To date there have only been studies showing the presence of specific ECM proteins in distinct brain regions (Zimmermann and Dours-Zimmermann, 2008; Morawski et al., 2012); quantitative analyses of CSPGs in one distinct brain region (Pantazopoulos et al., 2008); one study that offered a semiquantitative analyses of the distribution of Aggr and HA throughout the mouse brain (Costa et al., 2007); and one study that investigated PNNs containing chondroitin unsulfated proteoglycans throughout the rodent brain (Bertolotto et al., 1996). Furthermore, some studies analyzed the distributison of PNNs in the rat brain using well-established protocols, such as colloidal iron hydroxide staining for the detection of polyanionic components and the plant lectins Vicia villosa agglutinin and Wisteria floribunda agglutinin, which both show affinity for N-acetylgalactosamine and thereby stain CSPGs in general (Seeger et al., 1994). Although these staining methods are widely used to detect PNNs, PNNs are known as heterogeneous structures that vary in composition between cells and brain regions, as recently demonstrated (Berretta et al., 2015). Therefore, the present study focused on the distribution of specific ECM proteins as well as on the quantification of those specific proteins in distinct brain regions. We believe that there is a lack of quantitative studies concerning the distribution of ECM proteins throughout the brain, making it difficult to interpret the roles of these molecules in various brain disorders, and to accurately model their function and composition in vitro.
It should be noted that there are several established techniques to detect extracellular matrix proteins and specifically PNNs. Early on and presently, PNNs were visualized by using colloidal iron hydroxide staining (as mentioned above), thereby detecting their polyanionic components (Bruckner et al., 1993). Further visualization methods include the use of antibodies directed to glycoproteins (e.g., tenascins), proteoglycans, markers for HA, or lectins, which recognize N-acetylgalactosamine (Celio and Blumcke, 1994). Commonly used lectins include Vicia villosa and Wisteria floribunda, which bind to N-acetylgalactosamine residues of CSPG–glycosaminoglycan chains and glycoproteins, and therefore are regarded as markers of PNNs (Hartig et al., 1994; Koppe et al., 1997; Celio et al., 1998).
In this study we quantitatively analyzed the distribution, protein expression levels, and colocalization patterns of Aggr, Brev, and TnR in the adult rat brain. Overall, our results show that in the adult rat brain the distribution of distinct ECM proteins varies depending on 1) the brain region, 2) the ECM protein, and 3) the specific ECM structure investigated (PNNs or interstitial matrix). More specifically, Aggr exhibited high numbers of PNNs in the isocortex, but comparably low signal intensity levels, suggesting that Aggr in the isocortex is mainly part of PNNs but less abundant in the interstitial matrix. Brev showed high signal intensity levels in the hippocampus, but almost no PNNs, suggesting that Brev is mainly part of the interstitial matrix in that brain region. Generally, Aggr appeared to be mainly part of PNNs and to a much lesser extent of the surrounding interstitial matrix, whereas the opposite was observed for Brev and TnR. Subregions of distinct brain regions showed a highly differential abundance of Aggr, Brev, and TnR. We observed extremely high intensity levels of Brev in the SR and SO of the hippocampus. This unique distribution implies a specific function in this particular brain subregion. Interestingly, Brev-deficient mice exhibit deficits in the maintenance of hippocampal longterm potentiation (Brakebusch et al., 2002). TnR knockout mice show a similar phenotype with a normal viability but aberrant and structurally altered PNNs, reduced conduction velocity, and some behavioral abnormalities (Bruckner et al., 2000; Zimmermann and Dours-Zimmermann, 2008). Aggr-deficient mice die at birth. The comparably mild phenotype of some of the ECM knockout mice might be explained by possible compensation by related proteins (Zimmermann and Dours-Zimmermann, 2008), but has not yet been verified.
Several of the investigated ECM proteins were expressed at highest levels in the brainstem, the cerebellum, and the isocortex, possibly because these regions are located at the exterior of the brain and are thus the most “exposed” parts of the brain. Therefore they may be the most susceptible to various kinds of mechanical injury and require the most structural support from ECM molecules. Hypothetically, that means that the more ECM is present the more protected are the cells surrounded by it. Interestingly, it has been shown that Aggr-based ECM in the isocortex is largely complementary to the patterns of tau pathology (Morawski et al., 2010). Specific types of interneurons and subclasses of pyramidal cells associated with Aggr-containing PNNs are virtually spared from the formation of neurofibrillary tangles even in severely affected cortical areas (Bruckner et al., 1999; Morawski et al., 2010), suggesting that specialized ECM structures may contribute to the selective resistance of certain neuronal systems against degeneration. Similarly, it has been shown that PNNs protect neurons against iron-induced neurodegeneration (Suttkus et al., 2014). This study further demonstrated that Aggr, link protein, and TnR are essential for the neuroprotective properties of PNNs, whereas the contribution of brevican was negligible. These results indicate that the protection of PNN-ensheathed neurons is mediated by the net structure as well as by the specific combination of net components (Suttkus et al., 2014). Moreover, it has been shown that net-associated neurons survived in the vicinity of damaged pyramidal cells after trimethyltin (TMT) intoxication, and that PNNs were not removed by activated microglia (Schuppel et al., 2002), suggesting that the ECM of PNNs resists destruction after TMT treatment in inflamed neural tissue. This indicates that a permanent reconstitution of matrix components may be one of the factors supporting the viability of distinct neuronal types during neurodegenerative diseases (Schuppel et al., 2002). PNNs have also been reported to protect neurons against oxidative stress (Cabungcal et al., 2013). Interestingly, neurons from different brain regions appear to differ in their susceptibility to injury following brain trauma (Lok et al., 2014). In different types of experimental trauma models, the CA3 region and the DG of the hippocampus as well as Purkinje cells of the cerebellum seem especially more susceptible to delayed neuronal damage (Lok et al., 2014), the reason for which has not yet been elucidated, but it might be intriguing to find out if the ECM distribution plays a role here. In conclusion, PNNs appear to have neuroprotective effects, and differential distribution of PNNs throughout different brain regions may reflect the cells’ susceptibility to more or less severe environments, such as a higher susceptibility to external or internal injury or damage, and possibly differential regulation of synaptic plasticity in different local environments.
There are several reports suggesting that ECM proteins can inhibit axonal outgrowth after injury and thereby prevent recovery (Silver, 1994; Asher et al., 2001; Zimmermann and Dours-Zimmermann, 2008). Alterations of several ECM components have been reported in several brain pathologies and diseases, including brain injury (Asher et al., 2001), AD (Bonneh-Barkay and Wiley, 2009), schizophrenia (Berretta, 2012), autism (Mercier et al., 2012), multiple sclerosis (Bonneh-Barkay and Wiley, 2009), epilepsy (Dityatev, 2010; Pitkanen et al., 2014), and gliomas (Hu et al., 2008), supporting the notion that the cellular microenvironment is an influential component of both healthy and disease states. In subjects with schizophrenia, reductions in PNNs and CSPG- and reelin-enriched ECM aggregates, as well as increases in glial cells expressing CSPGs, were found in the amygdala and the entorhinal cortex (Pantazopoulos et al., 2010; Berretta, 2012), in comparison with widespread decreases in Aggrssand CS-6–labeled PNNs and glial cells (Pantazopoulos et al., 2015). Moreover, another study found a decreased density of PNNs in layers 3 and 5 of the prefrontal cortex in subjects with schizophrenia (Mauney et al., 2013). Together, these studies suggest that ECM abnormalities may contribute to several aspects of the pathophysiology of schizophrenia. Decreases in PNNs in disease states may represent failure of neurons to fully mature and form these structures that restrict plasticity, or increased degradation of these structures by ADAMTs and MMPs, through processes that are normally involved in PNN/ECM remodeling in response to learning, brain injury, or abnormal electrical activity. Along these lines, the results of the current study could also be used to validate current rat schizophrenia models (or other rat psychiatric disorder models), verifying whether similar ECM alterations in those models occur compared with the alterations observed in schizophrenia patients, thereby using the ECM as a disease validation marker.
After brain injury, CSPGs and other ECM proteins such as tenascins are upregulated and define barriers for neurons and axonal regrowth (McKeon et al., 1991; Asher et al., 2000, 2002; Gilbert et al., 2005; Silver and Silver, 2014), but the mechanism behind this has yet to be elucidated (Selles-Navarro et al., 2001; Johnson et al., 2002; Gilbert et al., 2005). Digestion of the chondroitin sulfate–glycosaminoglycans (CS-GAGs) significantly improves axonal regeneration after spinal cord injury (Bradbury et al., 2002), suggesting that targeting ECM proteins might be promising for future regenerative therapies. However, the suppression of a single ECM-associated brain inhibitor typically leads to less than 10% robust regrowth of cut axons (Bradbury and McMahon, 2006; Zheng et al., 2006; Gonzenbach and Schwab, 2008). Therefore, multimodal strategies might be required to achieve an improved regenerative response (Zimmermann and Dours-Zimmermann, 2008). In vitro the same inhibitory behavior of CSPGs has been observed. This functional property appears mostly core protein dependent in versican, neurocan, and phosphocan, whereas no inhibition of Brev and Aggr has been detected after chondroitinase digestion, suggesting that CS chains rather than core proteins are responsible for this function in Brev and Aggr (Snow et al., 1990; Maeda and Noda, 1996; Margolis et al., 1996; Yamada et al., 1997; Schmalfeldt et al., 2000). This inhibitory behavior in vitro had so far only been observed and tested in 2D culture systems (Asher et al., 2001; Cabungcal et al., 2013; Lok et al., 2014). We show here that the presence of Aggr, Brev, and/or TnR significantly reduced neurite outgrowth in a 3D environment over time. It has to be noted that this inhibitory effect might be concentration dependent, which was not been tested in this study. Furthermore, it is noteworthy that the functional aspect of PNNs probably depends on their network-like interactions, which might not be reproduced perfectly in an in vitro environment. Nevertheless, we observed that neurite length per nuclei was reduced by around 2–4-fold when Aggr, Brev, and/or TnR was present (Fig. 14), which reflects the neurite outgrowth-reducing effects of CSPG molecules reported by several studies using various conditions and CS concentrations (Snow et al., 1990; Schmalfeldt et al., 2000; Asher et al., 2001; Cabungcal et al., 2013; Lok et al., 2014). Furthermore, rather than being concentration dependent, this effect may be sulfation dependent, as some studies have reported neurite-promoting effects of specific disulfated forms of CS (Beller and Snow, 2014).
The results of this study indicate that a complex and tightly controlled ECM network with distinct functionalities exists. Furthermore, this network differs in composition between distinct brain regions. If we can elucidate the more specific functions of distinct ECM proteins within different brain regions and cellular structures, we can leverage this knowledge to more specifically target dysfunctional ECM components in disease conditions and further use this information to validate and improve rat brain disease models. Moreover, this knowledge might help us understand the causes and consequences of unique ECM alterations in disease or after injury and might aid in finding potential drug targets.
ACKNOWLEDGMENTS
We thank the Harvard Medical School Department of Neurobiology and the Neurobiology Imaging Facility for the consultation and instrument availability that supported this work. We thank Bogdan Budnik, Director of Proteomics at the Mass Spectrometry and Proteomics Resource Laboratory at Harvard University, for his valuable help and advice concerning the mass spectrometry experiments.
Grant sponsor: Congressionally Directed Medical Research Program through the United States Army; Grant number: W81XWH-11-2-0057; Grant sponsor: Emerald Foundation (Grant entitled “Molecular Mechanisms of Diffuse Axonal Injury”); Grant sponsor: the Belgian American Educational Foundation Inc. (grant to T.G.); Grant sponsor: the Neurobiology Imaging Facility at Harvard Medical School is supported in part by the Neural Imaging Center as part of a National Institute of Neurological Disorders and Stroke (NINDS) P30 Core Center grant; Grant number: NS072030.
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare they have no competing financial interests.
LITERATURE CITED
- Alpert AJ. 2008. Electrostatic repulsion hydrophilic interaction chromatography for isocratic separation of charged solutes and selective isolation of phosphopeptides. Anal Chem 80:62–76. [DOI] [PubMed] [Google Scholar]
- Asher RA, Morgenstern DA, Fidler PS, Adcock KH, Oohira A, Braistead JE, Levine JM, Margolis RU, Rogers JH, Fawcett JW. 2000. Neurocan is upregulated in injured brain and in cytokine-treated astrocytes. J Neurosci 20:2427–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher RA, Morgenstern DA, Moon LD, Fawcett JW. 2001. Chondroitin sulphate proteoglycans: inhibitory components of the glial scar. Prog Brain Res 132:611–619. [DOI] [PubMed] [Google Scholar]
- Asher RA, Morgenstern DA, Shearer MC, Adcock KH, Pesheva P, Fawcett JW. 2002. Versican is upregulated in CNS injury and is a product of oligodendrocyte lineage cells. J Neurosci 22:2225–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspberg A, Miura R, Bourdoulous S, Shimonaka M, Heinegard D, Schachner M, Ruoslahti E, Yamaguchi Y. 1997. The C-type lectin domains of lecticans, a family of aggregating chondroitin sulfate proteoglycans, bind tenascin-R by protein-protein interactions independent of carbohydrate moiety. Proc Natl Acad Sci U S A 94:10116–10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beller JA, Snow DM. 2014. Proteoglycans: road signs for neurite outgrowth. Neural Regener Res 9:343–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berretta S 2012. Extracellular matrix abnormalities in schizophrenia. Neuropharmacology 62:1584–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berretta S, Pantazopoulos H, Markota M, Brown C, Batzianouli ET. 2015. Losing the sugar coating: potential impact of perineuronal net abnormalities on interneurons in schizophrenia. Schizophr Res 167:18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotto A, Manzardo E, Guglielmone R. 1996. Immunohisto-chemical mapping of perineuronal nets containing chondroitin unsulfated proteoglycan in the rat central nervous system. Cell Tissue Res 283:283–295. [DOI] [PubMed] [Google Scholar]
- Bitel CL, Perrone-Bizzozero NI, Frederikse PH. 2010. HuB/C/D, nPTB, REST4, and miR-124 regulators of neuronal cell identity are also utilized in the lens. Mol Vis 16:2301–2316. [PMC free article] [PubMed] [Google Scholar]
- Bonneh-Barkay D, Wiley CA. 2009. Brain extracellular matrix in neurodegeneration. Brain Pathol 19:573–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury EJ, McMahon SB. 2006. Spinal cord repair strategies: why do they work? Nat Rev Neurosci 7:644–653. [DOI] [PubMed] [Google Scholar]
- Bradbury EJ, Moon LD, Popat RJ, King VR, Bennett GS, Patel PN, Fawcett JW, McMahon SB. 2002. Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature 416:636–640. [DOI] [PubMed] [Google Scholar]
- Brakebusch C, Seidenbecher CI, Asztely F, Rauch U, Matthies H, Meyer H, Krug M, Bockers TM, Zhou X, Kreutz MR, Montag D, Gundelfinger ED, Fassler R. 2002. Brevicandeficient mice display impaired hippocampal CA1 longterm potentiation but show no obvious deficits in learning and memory. Mol Cell Biol 22:7417–7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckner G, Brauer K, Hartig W, Wolff JR, Rickmann MJ, Derouiche A, Delpech B, Girard N, Oertel WH, Reichenbach A. 1993. Perineuronal nets provide a polyanionic, gliaassociated form of microenvironment around certain neurons in many parts of the rat brain. Glia 8:183–200. [DOI] [PubMed] [Google Scholar]
- Bruckner G, Hausen D, Hartig W, Drlicek M, Arendt T, Brauer K. 1999. Cortical areas abundant in extracellular matrix chondroitin sulphate proteoglycans are less affected by cytoskeletal changes in Alzheimer’s disease. Neuroscience 92:791–805. [DOI] [PubMed] [Google Scholar]
- Bruckner G, Grosche J, Schmidt S, Hartig W, Margolis RU, Delpech B, Seidenbecher CI, Czaniera R, Schachner M. 2000. Postnatal development of perineuronal nets in wild-type mice and in a mutant deficient in tenascin-R. J Comp Neurol 428:616–629. [DOI] [PubMed] [Google Scholar]
- Cabungcal JH, Steullet P, Morishita H, Kraftsik R, Cuenod M, Hensch TK, Do KQ. 2013. Perineuronal nets protect fast-spiking interneurons against oxidative stress. Proc Natl Acad Sci U S A 110:9130–9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Xiong K, Zhang XM, Cai H, Luo XG, Feng JC, Clough RW, Struble RG, Patrylo PR, Chu Y, Kordower JH, Yan XX. 2010. beta-Secretase-1 elevation in aged monkey and Alzheimer’s disease human cerebral cortex occurs around the vasculature in partnership with multisystem axon terminal pathogenesis and beta-amyloid accumulation. Eur J Neurosci 32:1223–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celio MR, Blumcke I. 1994. Perineuronal nets—a specialized form of extracellular matrix in the adult nervous system. Brain Res Brain Res Rev 19:128–145. [DOI] [PubMed] [Google Scholar]
- Celio MR, Spreafico R, De Biasi S, Vitellaro-Zuccarello L. 1998. Perineuronal nets: past and present. Trends Neurosci 21:510–515. [DOI] [PubMed] [Google Scholar]
- Costa C, Tortosa R, Domenech A, Vidal E, Pumarola M, Bassols A. 2007. Mapping of aggrecan, hyaluronic acid, heparan sulphate proteoglycans and aquaporin 4 in the central nervous system of the mouse. J Chem Neuroanat 33:111–123. [DOI] [PubMed] [Google Scholar]
- Dityatev A 2010. Remodeling of extracellular matrix and epileptogenesis. Epilepsia 51(suppl 3):61–65. [DOI] [PubMed] [Google Scholar]
- Dityatev A, Bruckner G, Dityateva G, Grosche J, Kleene R, Schachner M. 2007. Activity-dependent formation and functions of chondroitin sulfate-rich extracellular matrix of perineuronal nets. Dev Neurobiol 67:570–588. [DOI] [PubMed] [Google Scholar]
- Dityatev A, Schachner M, Sonderegger P. 2010. The dual role of the extracellular matrix in synaptic plasticity and homeostasis. Nat Rev Neurosci 11:735–746. [DOI] [PubMed] [Google Scholar]
- East E, Golding JP, Phillips JB. 2009. A versatile 3D culture model facilitates monitoring of astrocytes undergoing reactive gliosis. J Tissue Eng Regen Med 3:634–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerling DE, Lander AD. 1996. Inhibitors and promoters of thalamic neuron adhesion and outgrowth in embryonic neocortex: functional association with chondroitin sulfate. Neuron 17:1089–1100. [DOI] [PubMed] [Google Scholar]
- Gatz SA, Ju L, Gruber R, Hoffmann E, Carr AM, Wang ZQ, Liu C, Jeggo PA. 2011. Requirement for DNA ligase IV during embryonic neuronal development. J Neurosci 31:10088–10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giamanco KA, Morawski M, Matthews RT. 2010. Perineuronal net formation and structure in aggrecan knockout mice. Neuroscience 170:1314–1327. [DOI] [PubMed] [Google Scholar]
- Gilbert RJ, McKeon RJ, Darr A, Calabro A, Hascall VC, Bellamkonda RV. 2005. CS-4,6 is differentially upregulated in glial scar and is a potent inhibitor of neurite extension. Mol Cell Neurosci 29:545–558. [DOI] [PubMed] [Google Scholar]
- Gonzenbach RR, Schwab ME. 2008. Disinhibition of neurite growth to repair the injured adult CNS: focusing on Nogo. Cell Mol Life Sci 65:161–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig W, Brauer K, Bigl V, Bruckner G. 1994. Chondroitin sulfate proteoglycan-immunoreactivity of lectin-labeled perineuronal nets around parvalbumin-containing neurons. Brain Res 635:307–311. [DOI] [PubMed] [Google Scholar]
- Hu B, Kong LL, Matthews RT, Viapiano MS. 2008. The proteoglycan brevican binds to fibronectin after proteolytic cleavage and promotes glioma cell motility. J Biol Chem 283:24848–24859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Caterson B, Eisenstein SM, Hynds DL, Snow DM, Roberts S. 2002. Human intervertebral disc aggrecan inhibits nerve growth in vitro. Arthritis Rheum 46:2658–2664. [DOI] [PubMed] [Google Scholar]
- Kecskes S, Gaal B, Racz E, Birinyi A, Hunyadi A, Matesz C. 2015. Extracellular matrix molecules exhibit unique expression pattern in the climbing fiber-generating precerebellar nucleus, the inferior olive. Neuroscience 284: 412–421. [DOI] [PubMed] [Google Scholar]
- Koppe G, Bruckner G, Brauer K, Hartig W, Bigl V. 1997. Developmental patterns of proteoglycan-containing extracellular matrix in perineuronal nets and neuropil of the postnatal rat brain. Cell Tissue Res 288:33–41. [DOI] [PubMed] [Google Scholar]
- Kwok JC, Dick G, Wang D, Fawcett JW. 2011. Extracellular matrix and perineuronal nets in CNS repair. Dev Neurobiol 71:1073–1089. [DOI] [PubMed] [Google Scholar]
- Lau LW, Cua R, Keough MB, Haylock-Jacobs S, Yong VW. 2013. Pathophysiology of the brain extracellular matrix: a new target for remyelination. Nat Rev Neurosci 14: 722–729. [DOI] [PubMed] [Google Scholar]
- Lendvai D, Morawski M, Negyessy L, Gati G, Jager C, Baksa G, Glasz T, Attems J, Tanila H, Arendt T, Harkany T, Alpar A. 2013. Neurochemical mapping of the human hippocampus reveals perisynaptic matrix around functional synapses in Alzheimer’s disease. Acta Neuropathol 125: 215–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipska BK, Weinberger DR. 2000. To model a psychiatric disorder in animals: schizophrenia as a reality test. Neuropsychopharmacology 23:223–239. [DOI] [PubMed] [Google Scholar]
- Lok J, K. A, Guo S, Leung W, Maki T, Navaratna D, von Leyen K, Xing C, Wu L, Noviski N, Lo EH. 2014. Neurovascular responses to traumatic brain injury. Transl Stroke Res 5: 29–45. [Google Scholar]
- Maeda N, Noda M. 1996. 6B4 proteoglycan/phosphacan is a repulsive substratum but promotes morphological differentiation of cortical neurons. Development 122:647–658. [DOI] [PubMed] [Google Scholar]
- Margolis RK, Rauch U, Maurel P, Margolis RU. 1996. Neurocan and phosphacan: two major nervous tissue-specific chondroitin sulfate proteoglycans. Perspect Dev Neurobiol 3:273–290. [PubMed] [Google Scholar]
- Mauney SA, Athanas KM, Pantazopoulos H, Shaskan N, Passeri E, Berretta S, Woo TU. 2013. Developmental pattern of perineuronal nets in the human prefrontal cortex and their deficit in schizophrenia. Biol Psychiatry 74: 427–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon RJ, Schreiber RC, Rudge JS, Silver J. 1991. Reduction of neurite outgrowth in a model of glial scarring following CNS injury is correlated with the expression of inhibitory molecules on reactive astrocytes. J Neurosci 11:3398–3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier F, Kwon YC, Douet V. 2012. Hippocampus/amygdala alterations, loss of heparan sulfates, fractones and ventricle wall reduction in adult BTBR T+ tf/J mice, animal model for autism. Neurosci Lett 506:208–213. [DOI] [PubMed] [Google Scholar]
- Morawski M, Bruckner G, Jager C, Seeger G, Arendt T. 2010. Neurons associated with aggrecan-based perineuronal nets are protected against tau pathology in subcortical regions in Alzheimer’s disease. Neuroscience 169:1347–1363. [DOI] [PubMed] [Google Scholar]
- Morawski M, Bruckner G, Arendt T, Matthews RT. 2012. Aggrecan: beyond cartilage and into the brain. Int J Biochem Cell B 44:690–693. [DOI] [PubMed] [Google Scholar]
- Noble M, Fok-Seang J, Cohen J. 1984. Glia are a unique substrate for the in vitro growth of central nervous system neurons. J Neurosci 4:1892–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantazopoulos H, Murray EA, Berretta S. 2008. Total number, distribution, and phenotype of cells expressing chondroitin sulfate proteoglycans in the normal human amygdala. Brain Res 1207:84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantazopoulos H, Woo TU, Lim MP, Lange N, Berretta S. 2010. Extracellular matrix-glial abnormalities in the amygdala and entorhinal cortex of subjects diagnosed with schizophrenia. Arch Gen Psychiatry 67:155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantazopoulos H, Markota M, Jaquet F, Ghosh D, Wallin A, Santos A, Caterson B, Berretta S. 2015. Aggrecan and chondroitin-6-sulfate abnormalities in schizophrenia and bipolar disorder: a postmortem study on the amygdala. Transl Psychiatry 5:e496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitkanen A, Ndode-Ekane XE, Lukasiuk K, Wilczynski GM, Dityatev A, Walker MC, Chabrol E, Dedeurwaerdere S, Vazquez N, Powell EM. 2014. Neural ECM and epilepsy. Prog Brain Res 214:229–262. [DOI] [PubMed] [Google Scholar]
- Rauch U 2004. Extracellular matrix components associated with remodeling processes in brain. Cell Mol Life Sci 61: 2031–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimers S, Hartlage-Rubsamen M, Bruckner G, Rossner S. 2007. Formation of perineuronal nets in organotypic mouse brain slice cultures is independent of neuronal glutamatergic activity. Eur J Neurosci 25:2640–2648. [DOI] [PubMed] [Google Scholar]
- Schmalfeldt M, Bandtlow CE, Dours-Zimmermann MT, Winterhalter KH, Zimmermann DR. 2000. Brain derived versican V2 is a potent inhibitor of axonal growth. J Cell Sci 113:807–816. [DOI] [PubMed] [Google Scholar]
- Schnell SA, Staines WA, Wessendorf MW. 1999. Reduction of lipofuscin-like autofluorescence in fluorescently labeled tissue. J Histochem Cytochem 47:719–730. [DOI] [PubMed] [Google Scholar]
- Schultz C, Engelhardt M. 2014. Anatomy of the hippocampal formation. Front Neurol Neurosci 34:6–17. [DOI] [PubMed] [Google Scholar]
- Schuppel K, Brauer K, Hartig W, Grosche J, Earley B, Leonard BE, Bruckner G. 2002. Perineuronal nets of extracellular matrix around hippocampal interneurons resist destruction by activated microglia in trimethyltin-treated rats. Brain Res 958:448–453. [DOI] [PubMed] [Google Scholar]
- Seeger G, Brauer K, Hartig W, Bruckner G. 1994. Mapping of perineuronal nets in the rat brain stained by colloidal iron hydroxide histochemistry and lectin cytochemistry. Neuroscience 58:371–388. [DOI] [PubMed] [Google Scholar]
- Seidenbecher CI, Richter K, Rauch U, Fassler R, Garner CC, Gundelfinger ED. 1995. Brevican, a chondroitin sulfate proteoglycan of rat brain, occurs as secreted and cell surface glycosylphosphatidylinositol-anchored isoforms. J Biol Chem 270:27206–27212. [DOI] [PubMed] [Google Scholar]
- Selles-Navarro I, Ellezam B, Fajardo R, Latour M, McKerracher L. 2001. Retinal ganglion cell and nonneuronal cell responses to a microcrush lesion of adult rat optic nerve. Exp Neurol 167:282–289. [DOI] [PubMed] [Google Scholar]
- Senkov O, Andjus P, Radenovic L, Soriano E, Dityatev A. 2014. Neural ECM molecules in synaptic plasticity, learning, and memory. Prog Brain Res 214:53–80. [DOI] [PubMed] [Google Scholar]
- Silver DJ, Silver J. 2014. Contributions of chondroitin sulfate proteoglycans to neurodevelopment, injury, and cancer. Curr Opin Neurobiol 27:171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver J 1994. Inhibitory molecules in development and regeneration. J Neurol 242(1 suppl 1):S22–24. [DOI] [PubMed] [Google Scholar]
- Snow DM, Lemmon V, Carrino DA, Caplan AI, Silver J. 1990. Sulfated proteoglycans in astroglial barriers inhibit neurite outgrowth in vitro. Exp Neurol 109:111–130. [DOI] [PubMed] [Google Scholar]
- Suttkus A, Rohn S, Weigel S, Glockner P, Arendt T, Morawski M. 2014. Aggrecan, link protein and tenascin-R are essential components of the perineuronal net to protect neurons against iron-induced oxidative stress. Cell Death Dis 5:e1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watakabe A 2009. Comparative molecular neuroanatomy of mammalian neocortex: what can gene expression tell us about areas and layers? Dev Growth Differ 51:343–354. [DOI] [PubMed] [Google Scholar]
- Weber P, Bartsch U, Rasband MN, Czaniera R, Lang Y, Bluethmann H, Margolis RU, Levinson SR, Shrager P, Montag D, Schachner M. 1999. Mice deficient for tenascin-R display alterations of the extracellular matrix and decreased axonal conduction velocities in the CNS. J Neurosci 19:4245–4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Nagaraj N, Mann M. 2009. Universal sample preparation method for proteome analysis. Nat Methods 6:359–362. [DOI] [PubMed] [Google Scholar]
- Xiao ZC, Bartsch U, Margolis RK, Rougon G, Montag D, Schachner M. 1997. Isolation of a tenascin-R binding protein from mouse brain membranes. A phosphacanrelated chondroitin sulfate proteoglycan. J Biol Chem 272:32092–32101. [DOI] [PubMed] [Google Scholar]
- Yamada H, Watanabe K, Shimonaka M, Yamaguchi Y. 1994. Molecular cloning of brevican, a novel brain proteoglycan of the aggrecan/versican family. J Biol Chem 269: 10119–10126. [PubMed] [Google Scholar]
- Yamada H, Fredette B, Shitara K, Hagihara K, Miura R, Ranscht B, Stallcup WB, Yamaguchi Y. 1997. The brain chondroitin sulfate proteoglycan brevican associates with astrocytes ensheathing cerebellar glomeruli and inhibits neurite outgrowth from granule neurons. J Neurosci 17: 7784–7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Pekkanen-Mattila M, Shahsavani M, Falk A, Teixeira AI, Herland A. 2014. A 3D Alzheimer’s disease culture model and the induction of P21-activated kinase mediated sensing in iPSC derived neurons. Biomaterials 35: 1420–1428. [DOI] [PubMed] [Google Scholar]
- Zheng B, Lee JK, Xie F. 2006. Genetic mouse models for studying inhibitors of spinal axon regeneration. Trends Neurosci 29:640–646. [DOI] [PubMed] [Google Scholar]
- Zimmermann DR, Dours-Zimmermann MT. 2008. Extracellular matrix of the central nervous system: from neglect to challenge. Histochem Cell Biol 130:635–653. [DOI] [PubMed] [Google Scholar]