Abstract
The ACMG/AMP variant classification framework was intended for highly penetrant Mendelian conditions. While it is appreciated that clinically relevant variants exhibit a wide spectrum of penetrance, accurately assessing and expressing the pathogenicity of variants with lower penetrance can be challenging.
The vinculin gene (VCL) illustrates these challenges. Model organism data provides evidence that loss of function of VCL may play a role in cardiomyopathy and aggregate case-control studies suggest low penetrance. VCL loss of function variants, however, are rarely identified in affected probands and therefore there is a paucity of family studies clarifying the clinical significance of individual variants.
This study, which aggregated data from >18,000 individuals who underwent gene panel or exome testing for inherited cardiomyopathies, identified 32 probands with VCL loss-of-function variants and confirmed enrichment in probands with dilated cardiomyopathy (OR= 9.01; CI=4.93–16.45). Our data revealed that the majority of these individuals (89.5%) had pediatric onset of disease. Family studies demonstrated that heterozygous loss of function of VCL alone is insufficient to cause cardiomyopathy but that these variants do contribute to disease risk.
In conclusion, VCL loss-of-function variants should be reported in a diagnostic setting but need to be clearly distinguished as having lower penetrance.
INTRODUCTION
Modern sequencing technology has enabled the inclusion of large numbers of genes in diagnostic testing panels for inherited cardiomyopathy, often incorporating genes which have a role in disease that is not completely understood. The vinculin (VCL) gene is included on many clinically available cardiomyopathy panels (Genetic Testing Registry, https://www.ncbi.nlm.nih.gov/gtr/, accessed 1/14/2020). It encodes an actin-binding cytoplasmic protein that regulates cell-cell and cell-matrix adhesions and is critical for development, tissue remodeling, and cell migration1,2.
The first study linking VCL to a cardiac phenotype in humans was a candidate gene analysis published in 2002, which reported two VCL variants in individuals with dilated cardiomyopathy (DCM)3. Subsequent studies, reporting predominantly missense variants, identified VCL variants in individuals with a range of cardiac phenotypes including hypertrophic cardiomyopathy (HCM), DCM, and sudden unexplained death4–15. Other variant types have been reported in a few individuals and include a stop-loss variant in a proband with DCM, an in-frame deletion in a proband with DCM, a frameshift variant in an individual with HCM, and several deep intronic variants suggested to be modifiers of the HCM phenotype3, 16–18.
In vivo functional data from mouse, zebrafish, and Drosophila studies supported a role of VCL in myocardial function and pointed towards loss of function as a disease mechanism19–26. In mouse models, reduced VCL protein expression lead to a gradient of severity ranging from lethality in germline homozygous knockouts to stress-induced HCM in heterozygotes. Cardiomyocyte-specific knockouts developed DCM/sudden death secondary to HCM 26–28. An aggregate analysis comparing the prevalence of VCL loss-of-function (pLOF) variants in individuals with DCM to a reference population showed moderate effect sizes for this variant class (OR= 21.33; CI=7.23–62.89)27. These data suggest reduced penetrance, but there is insufficient data from family studies to further characterize the clinical impact of individual VCL pLOF variants.
Appropriate clinical characterization of variants of reduced penetrance is challenging. A lack of consensus terminology and interpretation criteria for these variants has led to inconsistent classifications (including “pathogenic”, “risk factor”, “variant of uncertain significance” and sometimes even “benign” if they are common in the general population). Labeling them as “pathogenic” can trigger a perceived high risk for disease, which is of particular concern when testing clinically unaffected individuals who may be unlikely to develop disease and may not require the same screening or management as someone with a highly penetrant variant. Conversely, labeling them as “variants of uncertain significance” may lead to dismissal or may be uninformative in the clinical setting. Only recently a framework for classifying and reporting such variants as “risk alleles” has been proposed28. The guidelines recommend categorizing these variants as established, likely, or uncertain risk alleles according to statistical confidence of the variant’s association to disease in combination with other available evidence. A third option is a hybrid approach combining Mendelian terminology and penetrance language (“pathogenic-reduced penetrance”). The community will need to establish consensus and clear guidance on how to describe these variants.
Clinical genetic laboratories have been interpreting VCL pLOF variants as “variants of uncertain significance” due to a lack of evidence at the individual variant level (13/14 VCL pLOF variants assessed in last 3 years in ClinVar are VUS; https://www.ncbi.nlm.nih.gov/clinvar/; accessed 12/11/19). While burden analysis suggests a role in disease, individual variants have not been sufficiently studied. Family studies are needed to clarify their contribution to disease; however, VCL pLOF variants are extremely rare and gathering sufficient data is difficult for individual laboratories. This study aggregates case level and familial segregation data from several large diagnostic testing and clinical research laboratories to better define the clinical impact of VCL pLOF variants.
METHODS
Clinical Cohort
The study cohort included 18,135 individuals who underwent genetic testing for inherited cardiomyopathies from 2009–2019 via gene panel testing that included VCL. Testing was performed at one of five laboratories: the Laboratory for Molecular Medicine (LMM) (n=2,689), GeneDx (n=6,500), Invitae (n=7,200), the Oxford Molecular Genetics Laboratory (OMGL) (n=895), or Imperial College London/Royal Brompton Hospital (ICL/RBH) (n=851). Specific phenotype information was available for three of the cohorts (LMM, OMGL, and ICL/RBH). Of those cohorts, 3,118 individuals had diagnoses of DCM, 1,372 tested at LMM, 895 tested at OMGL, and 851 tested at ICL/RBH. For the GeneDx and Invitae cohorts, a breakdown of cardiomyopathy phenotypes for all tested individuals was not available. For these cohorts, specific phenotype information was obtained for VCL pLOF heterozygotes included in this study.
Cohort details are provided in Supp. Table S1. Testing methodologies consisted of next generation sequencing, Sanger sequencing, and a chip-based resequencing assay29–31. In addition, two probands (#23–24) underwent exome sequencing as part of the ClinSeq® Study32. These two probands were not included in the statistical analysis. Age at time of testing and sex were the only uniformly available demographic variables. Phenotypic information for individuals with VCL variants was obtained from requisition forms and medical records provided by ordering clinicians. All data was gathered under an IRB–approved protocol (Partners HealthCare) and de-identified.
Identification of Variants
VCL variants impacting either the NM_014000.2 or NM_003373.3 transcript, both of which are expressed in cardiac tissue per the GTEx database (https://www.gtexportal.org/home/), and predicted to result in the introduction of a premature termination codon (nonsense or frameshift variants) or affect a canonical splice site (cds +/−1, 2) were considered to be putative LOF (pLOF) variants. Variants were manually reviewed and those predicted to escape nonsense mediated decay (variants in which the premature termination codon occurred within the last exon or within the last 50 base pairs of the penultimate exon) were excluded. All identified variants were named in accordance with Human Genome Variation Society (HGVS) nomenclature guidelines, and additional variants identified in any of the tested cardiomyopathy genes were classified in accordance with the ACMG/AMP Guidelines33, 34.
Statistical Analysis
The prevalence of pLOF VCL variants in individuals in the reference population was calculated by dividing the sum of VCL variants annotated as pLOF (nonsense, frameshift, and splice donor/acceptor variants) in the Genome Aggregation Database (gnomAD) v2.1 exomes by the mean number of individuals (mean allele number/2). Variants flagged as dubious by gnomAD and variants predicted to escape nonsense mediated decay, or those in which the premature termination codon occurred within the last exon or within the last 50 base pairs of the penultimate exon, were excluded from the analysis. This equated to a prevalence of VCL pLOF variants of 0.00046 (58/125,297) in individuals in gnomAD (Table 1). The frequency of pLOF variants in gnomAD was compared to the frequency of pLOF variants in the whole cohort, 0.0017 (30/18,135), and to the frequency in DCM cases, 0.0042 (13/3,118). Odds ratios were calculated comparing the allele frequency of pLOF variants in gnomAD to the allele frequencies in the overall cardiomyopathy testing cohort and in individuals with DCM. A second analysis was done in the same way, but excluding variants with minor allele frequencies (MAF) greater than 0.00002 (Table 2).
Table 1.
Prevalence of VCL pLOF Variants in Probands vs. gnomAD
| Exomes Samples from the gnomAD Database (v2.1) | Probands Referred for Cardiomyopathy Testing | Probands with Clinical Diagnosis of DCM [LMM + ICL/RBH + OMGL] | |
|---|---|---|---|
| Number of Individuals | 125,297* | 18,135 | 3,118 |
| Number of VCL pLOF Alleles | 58 | 30 | 13 |
| Prevalence of VCL pLOF Variants | 0.00046 | 0.0017 | 0.0042 |
| Odds Ratio Compared to Prevalence in gnomAD (95% CI) | N/A | 3.57 (2.30–5.55) | 9.01 (4.93–16.45) |
| p value | N/A | <0.0001 | <0.0001 |
Variants predicted to escape nonsense mediated decay in gnomAD or the proband cohort and those flagged as dubious in gnomAD v2.1 exomes were excluded from the analysis. DCM: dilated cardiomyopathy; LMM: Laboratory of Molecular Medicine cohort; ICL/RBH: Imperial College London/Royal Brompton Hospital; pLOF; putative loss of function.
Average number of individuals across all pLOF VCL variants.
Table 2.
Prevalence of Rare VCL pLOF Variants (MAF in gnomAD <0.00002*) in Probands vs. gnomAD
| Exomes Samples from the gnomAD Database (v2.1) | Probands Referred for Cardiomyopathy Testing | Probands with Clinical Diagnosis of DCM [LMM + ICL/RBH + OMGL] | |
|---|---|---|---|
| Number of Individuals | 125,297* | 18,135 | 3,118 |
| Number of VCL pLOF Alleles | 39 | 23 | 11 |
| Prevalence of VCL pLOF Variants | 0.00031 | 0.0013 | 0.0035 |
| Odds Ratio Compared to Prevalence in gnomAD (95% CI) | N/A | 4.07 (2.43–6.82) | 11.33 (5.80–22.15) |
| p value | N/A | <0.0001 | <0.0001 |
This analysis is identical to the one presented in Table 3 but excludes two variants that are more common in gnomAD (p.Pro943Argfs*9 and p.Ala573Hisfs*8), as well as the multi exon duplication variant (c.241_622+1dup). Variants predicted to escape nonsense mediated decay in gnomAD or the proband cohort and those flagged as dubious in gnomAD v2.1 exomes were excluded from the analysis. MAF: minor allele frequency; DCM: dilated cardiomyopathy; LMM: Laboratory of Molecular Medicine cohort; ICL/RBH: Imperial College London/Royal Brompton Hospital; pLOF; putative loss of function.
Average number of individuals across all pLOF VCL variants.
RESULTS
In total, 18,135 probands had gene panel testing that included VCL during the study period. Genes included in the panel testing for each proband are provided in Supp. Table S2. Thirty probands were heterozygous for VCL pLOF variants. In addition 2 probands from the ClinSeq cohort who underwent exome sequencing had heterozygous VCL pLOF variants. In total there were 26 unique VCL pLOF variants identified. Variants identified included six affecting canonical splice sites, twelve nonsense variants, seven frameshift variants, and one multi exon duplication. All variants except for the p.Pro943Argfs*9 variant were present in both the NM_014000.2 and NM_003373.3 transcripts.
Twenty-three of the variants were very rare with overall MAF <0.00002 in gnomAD v2.1 exomes and of those, two were seen in more than one proband (p.Arg188*, n=3 and p.Arg570*, n=2). Two additional variants (p.Pro943Argfs*9 and p.Ala573Hisfs*8) were detected in more than one individual in our cohort. These variants had the two highest MAF of all pLOF variants present in gnomAD v2.1 exomes at 0.00004 (9/251,460 chromosomes) for p.Pro943Argfs*9 and 0.00004 (10/251,460 chromosomes) for p.Ala573Hisfs*8. One variant was a multi exon duplication and, therefore, an accurate allele frequency is unavaliable.
Characterization of VCL putative LOF variant heterozygotes
We evaluated the cardiac phenotypes of the 32 probands with pLOF variants in VCL. The most common cardiac phenotype was DCM or left ventricular dilation (n=19, 59.4%, Table 3). The remainder (n=13, 40.6%, Table 4) had other cardiac phenotypes including LVNC (n=2), HCM or LVH (n=3), unspecified cardiomyopathy (n=3), abnormal echo with family history of DCM (n=1), bradycardia and heart block (n=3), and supraventricular tachycardia, tuberous sclerosis, and rhabdomyoma (n=1). Of probands, 59.4% (n=19) were male. In terms of ethnicity, 40.6% (n=13) were Caucasian, 12.5% (n=4) were African American, 9.4% (n=3) were Asian, and 3.1% (n=1) was Hispanic. The remainder (n=10, 31.2%) did not specify ethnicity. Race is not listed in Tables 3 and 4 due to IRB restrictions.
Table 3. Clinical and variant information for VCL pLOF probands and family members with DCM.
All VCL variants are reported in NM_014000.2. Infantile: age at testing ≤ 1 year; Childhood: age at testing >1 year and ≤10 years; Adolescent >10 years and ≤20 years; Adult: age at testing >20 years
| Proband | Family ID. Position in Pedigree | Age at Testing | Sex | Clinical Information | VCL pLOF Variant Identified | Max Allele Frequency in gnomAD (v2.1) | Additional Variant(s) Identified | Classification of Add’l Variant(s) |
|---|---|---|---|---|---|---|---|---|
| 1 | A. II-1 | Infantile | M | DCM and VT | c.1639C>T p.Arg547* | 0.00009 (1/113,732 alleles) in European Population | DSP: c.943C>T p.Arg315Cys | VUS |
| A. II-2 | Infantile | F | Mildly decreased cardiac function | c.1639C>T p.Arg547* | 0.00009 (1/113,732 alleles) in European Population | DSP: c.943C>T p.Arg315Cys – Not Detected | ||
| 2 | B. II-1 | Childhood | F | DCM or myocarditis with adenovirus and RSV infections at time of diagnosis; stable myocardial dysfunction at follow up | c.1713delA p.Ala573Hisfs*8 | 0.0007 (7/10,076 alleles) in Ashkenazi Jewish Population | MYH7: c.2063delT p.Leu688Argfs*38 | LP (recessive) |
| 3 | C. III-1 | Infantile | F | DCM | c.562C>T p.Arg188* | 0.00003 (3/113,666 alleles) in European Population | ||
| 4 | D. II-1 | Infantile | F | DCM | c.313C>T p.Arg105* | 0.00005 (1/18,388 alleles) in East Asian Population | LAMA4: c.2619G>T p.Gln873His | VUS |
| 5 | E. II-1 | Infantile | M | DCM | c.1762C>T p.Gln588* | Absent | ||
| 6 | F. III-1 | Adolescent | M | DCM and VT | c.659dupA p.Asn220Lysfs*21 | 0.00007 (1/15,330 alleles) in European Population | LAMA4: c.4031A>G p.Lys1344Arg | VUS |
| TPM1: c.97G>A p.Glu33Lys | VUS | |||||||
| 7 | G. III-2 | Infantile | M | DCM | c.1544−2A>G | Absent | MYH7: c.2347C>G p.Arg783Gly | LP |
| G. I-2 | Adult | F | DCM | c.1544−2A>G | Absent | MYH7: c.2347C>G p.Arg783Gly | ||
| G. II-2 | Adult | F | DCM | c.1544−2A>G (obligate carrier) | Absent | MYH7: c.2347C>G p.Arg783Gly (obligate carrier) | ||
| 8 | Infantile | M | DCM | c.670dupG p.Glu224Glyfs*17 | Absent | |||
| 9 | Adolescent | M | DCM | c.2828_2829delCT p.Pro943Argfs*9 | 0.0005 (12/24,968 alleles) in African Population | RYR2: c.2786G>A p.Arg929His | VUS | |
| 10 | Infantile | F | DCM | c.3163C>T p.Arg1055* | Absent | |||
| 11 | H. II-1 | Adolescent | F | DCM | c.3115C>T p.Gln1039* | Absent | ||
| H. I-2 | Adult | M | DCM | c.3115C>T p.Gln1039* | Absent | |||
| 12 | Adolescent | M | Moderate LV dilation with normal cardiac function four years later | c.2131+1delG | Absent | |||
| 13 | I. II-1 | Infantile | F | Neonatal DCM that improved to mild LV dilation at follow up | c.2435−1G>A | Absent | SCN5A: c.1844G>A p.Gly615Glu | VUS |
| I. I-2 | Adult | F | Episode of chest pain and normal QT interval | Negative | N/A | SCN5A: c.1844G>A p.Gly615Glu | VUS | |
| I. II-2 | Childhood | M | Asymptomatic but prolonged QT interval | Negative | N/A | SCN5A: c.1844G>A p.Gly615Glu | VUS | |
| I. II-3 | Childhood | F | Asymptomatic but prolonged QT interval | c.2435−1G>A | Absent | SCN5A: c.1844G>A p.Gly615Glu | VUS | |
| I. II-4 | Childhood | M | Asymptomatic but prolonged QT interval | Not Tested | N/A | SCN5A: c.1844G>A p.Gly615Glu | VUS | |
| 14 | Childhood | M | DCM | c.2828_2829delCT p.Pro943Argfs*9 | 0.0005 (12/24,968 alleles) in African Population | Unspecified* | VUS | |
| 15 | Childhood | F | DCM | c.562C>T p.Arg188* | 0.00003 (3/113,666 alleles) in European Population | |||
| 16 | Adult | M | DCM | c.2005C>T p.Arg669* | 0.00005 (1/18,348 alleles) in East Asian Population | Unspecified* | VUS | |
| 17 | Infantile | F | DCM | c.1543+1G>T | Absent | |||
| 18 | Adult | F | DCM | c.241_622+1dup | Absent | ACTC1: c.602C>T p.Ser201Phe | VUS | |
| 19 | Infantile | F | Severe left ventricular dilation with impaired function and acute heart failure | c.2331del p.Lys778Serfs*6 | Absent |
Detailed information for additional variants identified was not available for all probands due to IRB restrictions.
M: male; F: female; pLOF: putative loss of function; DCM: dilated cardiomyopathy; VT: ventricular tachycardia; LV: left ventricular; VUS: variant of uncertain significance; LP: likely pathogenic; RSV: respiratory syncytial virus.
Table 4. Clinical and variant information for VCL pLOF probands with other cardiac phenotypes.
All VCL variants are reported in NM_041000.2. Infantile: age at testing ≤ 1 year; Childhood: age at testing >1 year and ≤10 years; Adolescent >10 years and ≤20 years; Adult: age at testing >20 years;
| Proband | Age at Testing | Sex | Clinical Information | VCL pLOF Variant Identified | Allele Frequency in gnomAD (v2.1) | Additional Variant(s) Identified | Classification of Add’l Variant(s) |
|---|---|---|---|---|---|---|---|
| 20 | Adult | M | Bradycardia and complete AV block with family history of DCM | c.2853delT p.Glu952Serfs*4 | Absent | ||
| 21 | Adult | F | HCM | c.1708C>T p.Arg570* | 0.00009 (1/113,694 alleles) in European Population | ||
| 22 | Adult | F | LVNC | c.562C>T p.Arg188* | 0.00003 (3/113,666 alleles) in European Population | ||
| 23 | Adolescent | M | VT, tuberous sclerosis, and rhabdomyoma | c.2387delC p.Pro796Argfs*3 | Absent | ||
| 24 | Adult | M | HCM and CAD | c.2828_2829delCT p.Pro943Argfs*9 | 0.0005 (12/24968 alleles) in African Population | ||
| 25 | Adult | M | Sinus bradycardia and first degree heart block | c.1225C>T p.Arg409* | 0.00003 (1/34,592 alleles) in Latino Population | ||
| 26 | Adult | M | Sinus bradycardia and first degree heart block | c.1713delA p.Ala573Hisfs*8 | 0.0007 (7/10,076 alleles) in Ashkenazi Jewish Population | ||
| 27 | Infantile | M | LVH | c.2828_2829delCT p.Pro943Argfs*9 | 0.0005 (12/24,968 alleles) in African Population | Unspecified* | VUS |
| 28 | Adult | F | CM (unspecified type) | c.1708C>T p.Arg570* | 0.00009 (1/113,694 alleles) in European Population | Unspecified* | VUS |
| 29 | Infantile | M | Abnormal Echo and family history of DCM | c.3163C>T p.Arg1055* | Absent | Unspecified* | VUS |
| 30 | Adolescent | M | Congenital CM (unspecified type) | c.1744−1G>T | Absent | ||
| 31 | Adult | M | CM (unspecified type) and heart failure | c.3259−1G>T | Absent | Unspecified* | VUS |
| 32 | Adolescent | M | LVNC after pericarditis | c.1225C>T p.Arg409* | 0.00300 (1/34,592 alleles) in Latino Population | DSG2 c.2434G>A p.Gly812Ser | VUS |
| ILK c.680G>A p.Trp227* | VUS |
Detailed information for additional variants identified was not available for all probands due to IRB restrictions.
M: male; F: female; pLOF: putative loss of function; AV: atrioventricular; DCM: dilated cardiomyopathy; HCM: hypertrophic cardiomyopathy; LVNC: left ventricular noncompaction cardiomyopathy; VT: ventricular tachycardia; CAD: coronary artery disease; LVH: left ventricular hypertrophy; CM: cardiomyopathy.
Interestingly, the majority (68.8%; n=22) of probands with VCL pLOF had early onset of disease (age <20 years) with 50% (n=16) being 10 years or younger at the time of testing and 40.6% (n=13) having infantile onset disease (age ≤1 year). This distribution was more pronounced in the DCM cohort, where 89.5% (n=17) had early onset of disease (age <20 years), 57.9% were 10 years or younger (n=11), and 52.6% had infantile onset disease (n=10). While pediatric age at onset is not uncommon for DCM, the age of VCL pLOF variant heterozygotes with DCM in the LMM cohort (mean age: 4.3 ± 6.4 years) is significantly younger compared to DCM individuals who had positive test results associated with pathogenic variants in other DCM genes (mean age: 32.4 ± 22.1 years, n=293, p=0.0001) as well as all individuals with DCM (mean age: 29.3 ± 23.8 years, n=1,372, p=0.0007).
Eight of the probands in the DCM cohort and five probands in the remaining cohort had heterozygous variants of uncertain significance in cardiomyopathy-related genes other than VCL. Two individuals in the DCM cohort also had additional likely pathogenic variants in MYH7. Details of these variants are provided in Supp. Table S3.
Enrichment of VCL LOF variants in individuals with DCM
Walsh et al. reported an enrichment (OR=21.33; CI=7.23–62.89) of VCL pLOF variants in individuals with DCM (n=4) compared to individuals in the ExAC database (http://exac.broadinstitute.org/)27. The four individuals from Walsh et. al. are included in this cohort and this study expands the analysis using the subset of our entire cohort for which phenotype information was available for all tested probands, not just those with VCL variants (LMM, OMGL, and ICL/RBH cohorts). This provided nine additional DCM probands with heterozygous pLOF variants (n=13) out of 3,118 individuals with diagnoses of DCM. When compared to the gnomAD database (58/125,297 individuals with VCL pLOF variants), the data confirm the enrichment, although, with a lower effect size (Table 1: OR=9.01; CI=4.93–16.45). A second analysis was preformed considering only rare variants (MAF <0.0002) in both the gnomAD cohort and the proband cohort. This resulted in the exclusion of the p.Pro943Argfs*9, p.Ala573Hisfs*8, and c.241_622+1dup variants leaving 11/3,118 DCM probands with VCL pLOF variants compared to 39/125,297 individuals in the gnomAD cohort and demonstrated an OR of 11.33 (CI=5.80–22.15) (Table 2).
Family Studies of Probands with VCL Putative LOF variants
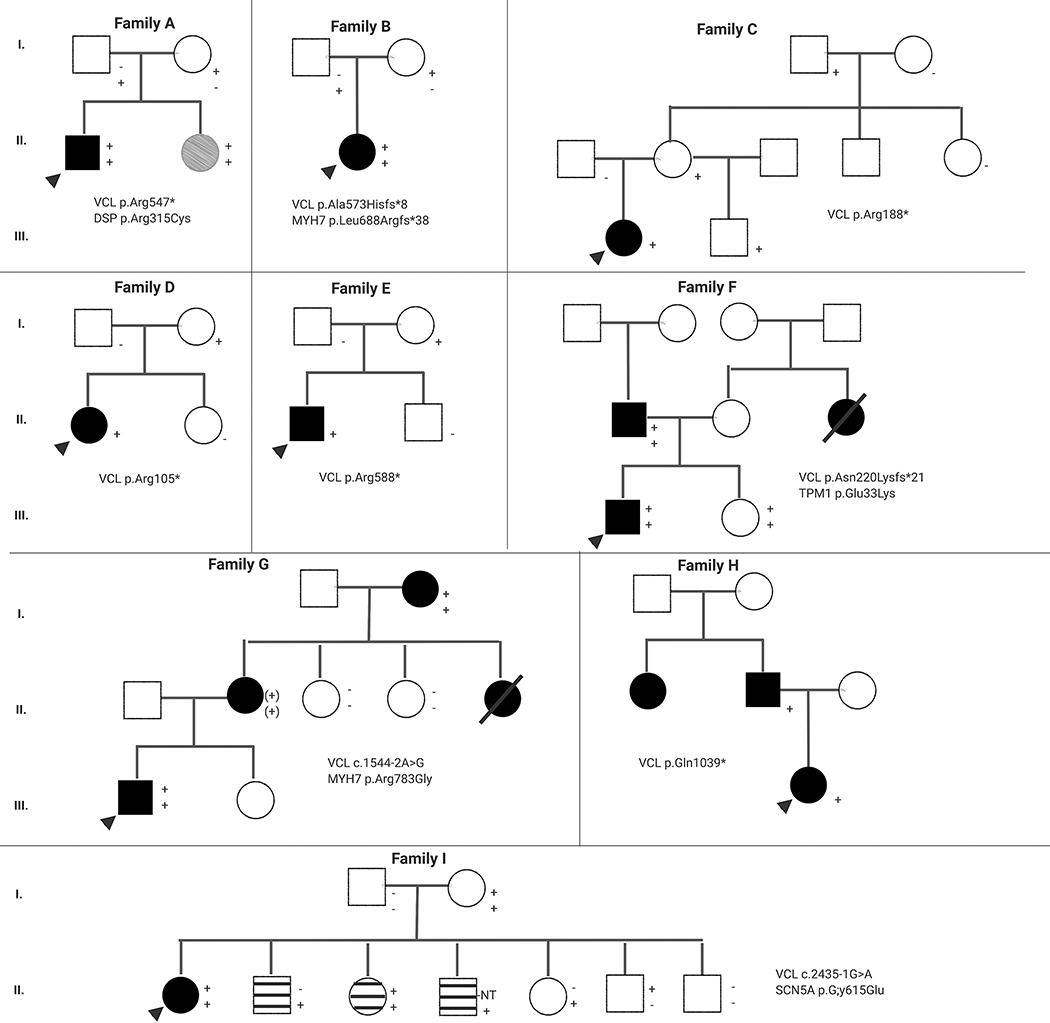
The effect sizes obtained from the association analyses (Walsh et al. and the data presented here) suggested that VCL pLOF variants are not highly penetrant. However, the possibility that they act as modifiers for other clinically significant variants remains. This is relevant in the setting of cardiomyopathy genetic testing where approximately 3–5% of affected probands are found to have more than one rare variant in cardiomyopathy-associated genes and an additive effect of these variants on clinical severity has been observed35–40. To further investigate the role of VCL pLOF variants in inherited cardiomyopathy, available family studies from nine pediatric probands with DCM were analyzed (Figure 1). Five of these probands had additional variants in genes that are either associated with DCM or genes associated with disorders that clinically overlap with DCM.
Figure 1.
Pedigrees of families with dilated cardiomyopathy and VCL putative loss of function variants. Circles: Females; Squares: Males; Black Shape: Affected with DCM; Grey Shape: Mild Cardiac Dysfunction; Striped Shape: Prolonged QT interval; White Shape: Unaffected; + : Tested positive for the variant; (+): Obligate carrier for the variant; − : Tested negative for the variant; NT: Not tested for the variant; Arrow: Indicates the proband
In families A, B, C, D, and E, the probands did not have a family history of disease. In families A and B, the probands had additional variants in MYH7 and DSP, respectively. Both of these variants are unlikely to be disease-causing on their own based on the available evidence. The DSP variant has an allele frequency of 0.0008 (16/19,928 chromosomes) in the East Asian population of gnomAD. The MYH7 variant is a pLOF variant and haploinsufficiency is not a known disease mechanism for autosomal dominant cardiomyopathy. In both families, each parent is heterozygous for one of the two variants and all are reportedly unaffected. As MYH7 and DSP are associated with DCM, it is conceivable that these variants in combination with the VCL pLOF variant resulted in disease, while each variant on its own was insufficient. In families C, D, and E, the VCL pLOF variants were also inherited from reportedly unaffected parents. No additional variants were tested in these families.
Families F, G, and H are consistent with autosomal dominant inheritance of disease with affected individuals in two or more generations. In family F, the VCL pLOF variant and two additional variants of uncertain significance (one in TPM1 and one in LAMA4) were all inherited from the proband’s affected father. A segregation study of family F was recently published, showing that out of 31 family members tested, only those heterozygous for both the VCL pLOF variant and the TPM1 missense variant were affected, although age of onset varied widely (0.5 to 60 years)41. Family G was similar to family F with the VCL pLOF variant and a missense variant in MYH7 identified in the three affected family members who underwent testing. In this family, no individuals were heterozygous for either the VCL or MYH7 variants alone, making it impossible to discern their relative contributions. Of note, the MYH7 variant met ACMG/AMP criteria to be classified as likely pathogenic, but has not been reported in other families. In family H the VCL variant was present in the proband and his affected father and no additional variants were identified.
In family I, the proband had DCM and intermittent prolonged QT interval. He was also heterozygous for a variant of uncertain significance in SCN5A, which segregated with prolonged QT interval in the proband’s siblings. SCN5A is associated with long QT syndrome; however, the variant has conflicting evidence for pathogenicity (Supp. Table S1).
DISCUSSION
The studies presented here add to prior analyses and confirm the enrichment of VCL LOF variants in DCM probands. While the effect size is lower compared to previously published data, this may be explained by the addition of the ICL/RBH cohort to this study, as the average age across these individuals was markedly higher compared to the other cohorts (Supp. Table S1).
This study assembled a series of families to further assess the degree to which VCL pLOF variants contribute to cardiomyopathy. The lack of clear segregation data confirms that VCL pLOF variants do not cause a phenotype with a highly penetrant autosomal dominant pattern of inheritance; although, clinical records including echocardiogram results were not available for all family members, so the possibility of a mild presentation cannot be ruled out in reportedly unaffected individuals. In addition, there was no evidence for autosomal recessive inheritance, as no compound heterozygous or homozygous individuals were identified. The lack of evidence for autosomal recessive inheritance could be due to lethality of biallelic loss of function of VCL, as is observed in mouse models20–22.
Overall, the data are most consistent with VCL pLOF variants acting as genetic modifiers that can lead to more severe disease or the precipitation of disease in the presence of additional risk factors, such as other genetic and/or environmental contributions. An additive effect of more than one variant is a tempting speculation but does not hold true in some of the families as individuals harboring multiple variants appear to present with an wide range of ages at onset (families F and G)41. In addition, individuals with multiple variants did not have younger ages of onset than those with only the VCL LOF variant; although, little can be concluded from this as the majority of the variants are of uncertain clinical significance. It is conceivable that this is the result of a gene-environment interaction, such that disease is more likely to occur in the setting of non-genetic stressors (e.g. viral myocarditis). This view is consistent with data from in vitro and in vivo studies of VCL variants demonstrating that LOF results in some disruption of cardiomyocyte function, but cardiomyopathy only occurs when stress-induced20–22.
We note that the majority of DCM probands with VCL pLOF variants underwent genetic testing at a significantly younger age than the rest of the DCM cohort. Currently, however, there is only conjecture as to how to explain this phenomenon. It is possible that VCL pLOF variants affect cardiac development or pose an increased risk for developing DCM in the presence of stressors specific to the pediatric setting, such as particular types of infections. Having longitudinal data on additional VCL pLOF heterozygotes will be needed to elucidate the role of these variants in the development of cardiomyopathy and shed light on the prognosis of these children.
In addition to DCM, VCL LOF variants were identified in several individuals with other presentations including, LVNC, HCM, and bradycardia with heart block. It is possible that these variants are contributing to these phenotypes; however, due to the limited number of probands it is difficult to draw any definitive conclusions from this data. As VCL LOF variants are also present in the general population, their detection in individuals with other cardiac phenotypes for which broad cardiomyopathy gene panels tend to be ordered could also potentially be coincidental. Additional analyses with larger numbers of probands and informative families are needed to expand upon both the association with DCM and to clarify any potential associations with additional phenotypes. Further studies including in depth analyses of environmental precipitators and additional genetic factors, that may be contributing to disease may help to clarify the outstanding questions from this study, including the enrichment in children. As pediatric cardiomyopathy is less well studied than adult onset disease, exome or genome testing will be the most suitable method for further characterizing VCL pLOF variants as genetic modifiers.
STUDY LIMITATIONS
Limitations include a lack of comprehensive clinical data for probands and family members, including longitudinal health records and ethnicity information. Furthermore, no clinical information was available for the individuals in the gnomAD database and we cannot exclude the possibility that some of these individuals may be affected. In addition, family studies were unavailable in most cases; although, this could be suggestive of an overall lack of family history of disease, as segregation studies are more likely to be carried out when affected family members are available for testing.
CONCLUSIONS
The examination of VCL pLOF variants highlights challenges associated with low penetrance variants in a clinical testing setting. Current practice across clinical laboratories is to classify novel VCL pLOF variants as variants of uncertain significance (13/14 VCL pLOF variants assessed in last three years in ClinVar are VUS; https://www.ncbi.nlm.nih.gov/clinvar/; accessed 12/11/19). The term VUS implies that insufficient or conflicting data exist and that, over time, such variants will evolve to be classified either as benign or pathogenic. Our data, as well as previously published aggregate studies, suggest that VCL pLOF variants act as moderate penetrance risk alleles that, in conjunction with additional genetic and/or environmental factors, predispose to development of DCM, particularly in the pediatric setting. Recently proposed classification terminology for low penetrance variants may be a better fit for VCL pLOF variants18. This framework is currently being refined by the clinical community (Clinical Genome Resource: https://clinicalgenome.org/working-groups/low-penetrance-risk-allele-working-group/). Open questions include what level of penetrance constitutes an adequate threshold for moving from a Mendelian classification terminology to one that describes an existing but lower risk of developing disease. Whichever terminology will be the consensus chosen by the genetic testing community, a clear description of a moderate risk role for VCL pLOF variants as a variant class even in the absence of data on the individual variant level should be included in interpretive evidence summaries on clinical reports.
Supplementary Material
REFERENCES
- 1.Bays JL, DeMali KA. Vinculin in cell-cell and cell-matrix adhesions. Cell Mol Life Sci. 2017;74(16):2999–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Izard T, Brown DT. Mechanisms and Functions of Vinculin Interactions with Phospholipids at Cell Adhesion Sites. J Biol Chem. 2016;291(6):2548–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olson TM, Illenberger S, Kishimoto NY, Huttelmaier S, Keating MT, Jockusch BM. Metavinculin mutations alter actin interaction in dilated cardiomyopathy. Circulation. 2002;105(4):431–437. [DOI] [PubMed] [Google Scholar]
- 4.Lu C, Wu W, Liu F, et al. Molecular analysis of inherited cardiomyopathy using next generation semiconductor sequencing technologies. J Transl Med. 2018;16(1):241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Theis JL, Bos JM, Bartleson VB, et al. Echocardiographic-determined septal morphology in Z-disc hypertrophic cardiomyopathy. Biochem Biophys Res Commun. 2006;351(4):896–902. [DOI] [PubMed] [Google Scholar]
- 6.Vasile VC, Ommen SR, Edwards WD, Ackerman MJ. A missense mutation in a ubiquitously expressed protein, vinculin, confers susceptibility to hypertrophic cardiomyopathy. Biochem Biophys Res Commun. 2006;345(3):998–1003. [DOI] [PubMed] [Google Scholar]
- 7.Mook OR, Haagmans MA, Soucy JF, et al. Targeted sequence capture and GS-FLX Titanium sequencing of 23 hypertrophic and dilated cardiomyopathy genes: implementation into diagnostics. J Med Genet. 2013;50(9):614–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bottillo I, D’Angelantonio D, Caputo V, et al. Molecular analysis of sarcomeric and non-sarcomeric genes in patients with hypertrophic cardiomyopathy. Gene. 2016;577(2):227–235. [DOI] [PubMed] [Google Scholar]
- 9.Zhao Y, Feng Y, Zhang YM, et al. Targeted next-generation sequencing of candidate genes reveals novel mutations in patients with dilated cardiomyopathy. Int J Mol Med. 2015;36(6):1479–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wells QS, Ausborn NL, Funke BH, et al. Familial dilated cardiomyopathy associated with congenital defects in the setting of a novel VCL mutation (Lys815Arg) in conjunction with a known MYPBC3 variant. Cardiogenetics. 2011;1(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng J, Kyle JW, Wiedmeyer B, Lang D, Vaidyanathan R, Makielski JC. Vinculin variant M94I identified in sudden unexplained nocturnal death syndrome decreases cardiac sodium current. Sci Rep. 2017;7:42953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin Y, Williams N, Wang D, et al. Applying High-Resolution Variant Classification to Cardiac Arrhythmogenic Gene Testing in a Demographically Diverse Cohort of Sudden Unexplained Deaths. Circ Cardiovasc Genet. 2017;10(6). [DOI] [PubMed] [Google Scholar]
- 13.Shanks GW, Tester DJ, Nishtala S, Evans JM, Ackerman MJ. Genomic Triangulation and Coverage Analysis in Whole-Exome Sequencing-Based Molecular Autopsies. Circ Cardiovasc Genet 2017;10(5). [DOI] [PubMed] [Google Scholar]
- 14.Narula N, Tester DJ, Paulmichl A, Maleszewski JJ, Ackerman MJ. Post-mortem Whole exome sequencing with gene-specific analysis for autopsy-negative sudden unexplained death in the young: a case series. Pediatr Cardiol 2015;36(4):768–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng J, Kyle JW, Lang D, et al. An East Asian Common Variant Vinculin P.Asp841His Was Associated With Sudden Unexplained Nocturnal Death Syndrome in the Chinese Han Population. J Am Heart Assoc. 2017;6(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haas J, Frese KS, Peil B, et al. Atlas of the clinical genetics of human dilated cardiomyopathy. Eur Heart J. 2015;36(18):1123–1135a. [DOI] [PubMed] [Google Scholar]
- 17.Xu J, Li Z, Ren X, et al. Investigation of Pathogenic Genes in Chinese sporadic Hypertrophic Cardiomyopathy Patients by Whole Exome Sequencing. Sci Rep. 2015;5:16609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mendes de Almeida R, Tavares J, Martins S, et al. Whole gene sequencing identifies deep-intronic variants with potential functional impact in patients with hypertrophic cardiomyopathy. PLoS One. 2017;12(8):e0182946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu W, Baribault H, Adamson ED. Vinculin knockout results in heart and brain defects during embryonic development. Development. 1998;125(2):327–337. [DOI] [PubMed] [Google Scholar]
- 20.Zemljic-Harpf AE, Ponrartana S, Avalos RT, et al. Heterozygous inactivation of the vinculin gene predisposes to stress-induced cardiomyopathy. Am J Pathol. 2004;165(3):1033–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marg S, Winkler U, Sestu M, et al. The vinculin-DeltaIn20/21 mouse: characteristics of a constitutive, actin-binding deficient splice variant of vinculin. PLoS One. 2010;5(7):e11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zemljic-Harpf AE, Miller JC, Henderson SA, et al. Cardiac-myocyte-specific excision of the vinculin gene disrupts cellular junctions, causing sudden death or dilated cardiomyopathy. Mol Cell Biol. 2007;27(21):7522–7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vogel B, Meder B, Just S, et al. In-vivo characterization of human dilated cardiomyopathy genes in zebrafish. Biochem Biophys Res Commun. 2009;390(3):516–522. [DOI] [PubMed] [Google Scholar]
- 24.Han MKL, van der Krogt GNM, de Rooij J. Zygotic vinculin is not essential for embryonic development in zebrafish. PLoS One. 2017;12(8):e0182278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng F, Miao L, Wu Q, Gong X, Xiong J, Zhang J. Vinculin b deficiency causes epicardial hyperplasia and coronary vessel disorganization in zebrafish. Development. 2016;143(19):3522–3531. [DOI] [PubMed] [Google Scholar]
- 26.Maartens AP, Wellmann J, Wictome E, Klapholz B, Green H, Brown NH. Drosophila vinculin is more harmful when hyperactive than absent, and can circumvent integrin to form adhesion complexes. J Cell Sci. 2016;129(23):4354–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walsh R, Thomson KL, Ware JS, et al. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med. 2017;19(2):192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Senol-Cosar O, Schmidt RJ, Qian E, et al. Considerations for clinical curation, classification, and reporting of low-penetrance and low effect size variants associated with disease risk. Genet Med. 2019. [DOI] [PubMed] [Google Scholar]
- 29.Zimmerman RS, Cox S, Lakdawala NK, et al. A novel custom resequencing array for dilated cardiomyopathy. Genet Med. 2010;12(5):268–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alfares AA, Kelly MA, McDermott G, et al. Results of clinical genetic testing of 2,912 probands with hypertrophic cardiomyopathy: expanded panels offer limited additional sensitivity. Genet Med. 2015;17(11):880–888. [DOI] [PubMed] [Google Scholar]
- 31.Pugh TJ, Kelly MA, Gowrisankar S, et al. The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genet Med. 2014;16(8):601–608. [DOI] [PubMed] [Google Scholar]
- 32.Johnston JJ, Lewis KL, Ng D, et al. Individualized iterative phenotyping for genome-wide analysis of loss-of-function mutations. Am J Hum Genet. 2015;96(6):913–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abou Tayoun AN, Pesaran T, DiStefano MT, et al. Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Hum Mutat. 2018;39(11):1517–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burns C, Bagnall RD, Lam L, Semsarian C, Ingles J. Multiple Gene Variants in Hypertrophic Cardiomyopathy in the Era of Next-Generation Sequencing. Circ Cardiovasc Genet. 2017;10(4). [DOI] [PubMed] [Google Scholar]
- 36.Richard P, Charron P, Carrier L, et al. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107(17):2227–2232. [DOI] [PubMed] [Google Scholar]
- 37.Richard P, Isnard R, Carrier L, et al. Double heterozygosity for mutations in the beta-myosin heavy chain and in the cardiac myosin binding protein C genes in a family with hypertrophic cardiomyopathy. J Med Genet. 1999;36(7):542–545. [PMC free article] [PubMed] [Google Scholar]
- 38.Ingles J, Doolan A, Chiu C, Seidman J, Seidman C, Semsarian C. Compound and double mutations in patients with hypertrophic cardiomyopathy: implications for genetic testing and counselling. J Med Genet. 2005;42(10):e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Driest SL, Vasile VC, Ommen SR, et al. Myosin binding protein C mutations and compound heterozygosity in hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44(9):1903–1910. [DOI] [PubMed] [Google Scholar]
- 40.Olivotto I, Girolami F, Ackerman MJ, et al. Myofilament protein gene mutation screening and outcome of patients with hypertrophic cardiomyopathy. Mayo Clin Proc. 2008;83(6):630–638. [DOI] [PubMed] [Google Scholar]
- 41.Deacon DC, Happe CL, Chen C, et al. Combinatorial interactions of genetic variants in human cardiomyopathy. Nat Biomed Eng. 2019;3(2):147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.