ABSTRACT
Circular RNAs (circRNAs) are a class of widely expressed noncoding RNA with significant regulatory potential discovered in recent years. The purpose of this study was to investigate the effects of hsa_circ_0001785 on the proliferation, migration and invasion of breast cancer (BC) cells in vivo and in vitro and the potential underlying molecular mechanism. In the present study, the expressions of hsa_circ_0001785 in five BC cells (T47D, MCF-7, MDA-MB-453, MDA-MB-231 and BT-549) and one normal breast cell (MCF-10A) were the first to examined by qRT-PCR. Then, we studied the biological function of hsa_circ_0001785 in BC by in vivo and in vitro experiments. CCK-8, clone formation, wound-healing and Transwell assays were performed to analyze the cellular proliferation, migration and invasion in vitro. The subcutaneous tumor model of nude mice was used for in vivo experiment. In addition, we determined that hsa_circ_0001785 acted as competing endogenous RNAs (ceRNAs) in BC by RNA immunoprecipitation (RIP) and dual-luciferase reporter assays. Results showed that the expressions of hsa_circ_0001785 were decreased in BC cells. Hsa_circ_0001785 overexpression inhibited the proliferation, migration, invasion of BC cells and tumor growth in nude mice. RIP and dual-luciferase reporter assay demonstrated that hsa_circ_0001785 could regulate the SOCS3 by sponging miR-942. In general, circular RNA hsa_circ_0001785 inhibits the proliferation, migration and invasion of BC cells by modulating the miR-942/SOCS3 signaling axis.
KEYWORDS: Circular RNAs, hsa_circ_0001785, competing endogenous RNAs, breast cancer (BC), proliferation
Introduction
Breast cancer (BC) is the most common cancer among women in the world, and it is also the second most common cancer in the world [1,2]. Approximately 1.67 million women are diagnosed with BC each year, accounting for about 25% of all cancer patients, of which about 500,000 deaths, which is the leading cause of death in women worldwide [3,4]. Moreover, with the increase in global life expectancy, the incidence and mortality of BC have been increasing [5]. Although the improvement of early diagnosis and late treatment strategy, the fatality rate caused by BC has decreased significantly. However, multiple drug resistance and recurrence are still the main obstacles in the treatment of BC [6]. And because the pathogenesis of BC is complex and, its molecular characteristics and clinical manifestations are diverse, the prognosis of patients is widely inconsistent, which can not achieve the desired results. Therefore, it is of great significance to study the molecular mechanism of BC and find early diagnostic markers and novel treatment targets for more effective treatment of BC.
Increasing evidence shows that circular RNAs (circRNAs) participate in the regulation of gene expression in a variety of ways, and thus indirectly affect the occurrence and development of various cancers, including colorectal cancer, lung cancer, cervical cancer, liver cancer, ovarian cancer and pancreatic cancer. It may become a new biomarker and therapeutic target for cancer treatment [7,8]. CircRNAs are a new type of non-coding RNA that is highly expressed in eukaryotic cells. Additionally, it is characterized by a covalently closed circular structure without 3 ‘and 5ʹ ends, which is formed by reverse cleavage of precursor messenger RNA (Pre-mRNA) and not easy to be degraded by exonuclease [9,10]. Studies have shown that circRNAs play an important role in the progression of BC, which have the potential to become a tumor marker for screening BC because of its high conservatism and tissue specificity [11]. CircRNAs mainly sponge miRNA through acting as a ceRNA to reduce the inhibition of miRNA on its target genes and then exert biological function. For instance, Wang et al. reported that circMYO9B was highly expressed in BC cells, and circMYO9B inhibited cell proliferation, migration and invasion by regulating the miR-4316/FOXP4 axis in BC [12]. In addition, hsa_circ_0001982 was upregulated in BC tissues and cells, and hsa_circ_0001982 suppressed the progression of BC by sponging miR-143 to regulate the expression of PDL1 [13]. A recent study illustrated that some circRNAs were aberrantly expressed in peripheral blood of human BC patients compared with that in healthy volunteers. Of notice, hsa_circ_0001785 was significantly decreased in peripheral blood of BC patients, and the expression level of hsa_circ_0001785 was positively correlated with histological tumor grade, Tumor-Node-Metastasis (TNM) stage and lymph node metastasis [14]. These findings indicate that circ_0001785 has better diagnostic value and higher diagnostic accuracy than other circRNAs. However, the specific role and potential mechanism of circ_0001785 in BC have not been studied.
In the present study, we systematically studied the role and regulatory mechanism of hsa_circ_0001785 in BC by in vivo and in vitro experiments. The experimental results suggest that the hsa_circ_0001785 was expressed at low levels in BC cells. Overexpression of hsa_circ_0001785 significantly inhibited proliferation, migration and invasion of BC cell. In addition, hsa_circ_0001785 reduced the progression of BC by sponging miR-942 to upregulate the expression of SOCS3. We believe that circ_0001785 may be a new serum marker for early screening and prognosis of BC.
Materials and methods
Cell culture
In this study, five human BC cells (T47D, MCF-7, MDA-MB-453, MDA-MB-231 and BT-549) and one normal human breast epithelial cell (MCF-10A) were purchased from the American Type Culture Collection (ATCC; Rockville, MDUSA). The four BC cells (T47D, MCF-7, MDA-MB-453, MDA-MB-231) were routinely cultured in Dulbecco’s modified Eagle’s medium (DMEM; Hyclone, Logan, UT, USA) containing 10% fetal bovine serum (FBS; Hyclone) in a humidified atmosphere of 95% O2 and 5% CO2 at 37°C. BT-549 cells were cultured in RPMI-1640 (Invitrogen) medium with 10% FBS. All other cell culture conditions remain the same. MCF-10A cells were cultured in Dulbecco modified Eagle medium-F-12 medium (DMEM/F12; Life Technologies) containing 10% FBS, 10 μg/ml insulin and 20 ng/ml EGF at 37°C in a 5% CO2 and 95% relative humidity environment. All cells were subcultured when they were filled with 80% in the culture bottles. The cells in the logarithmic phase were taken for follow-up experiment.
Cell transfection
MiR-942 mimic (miR-942 mimic), miR-942 inhibitor (miR-942 inhibitor) and negative control (miR-NC) were obtained from Guangzhou RiboBio Co., Ltd. Hsa_circ_0001785 overexpression vector (pcDNA-circ_0001785) and the empty control vector (pcDNA-NC) were constructed by Shanghai GenePharma Co., Ltd. These plasmids were transfected into cells with Lipofectamine 2000 reagent (Invitrogen) following the instructions of manufacturers. The transfection efficiency was detected after 48 h.
qRT-PCR analysis
Cells of each group were harvested after 48 h of transfection, and the total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, USA). According to the specific operation method, cDNA was synthesized using the TaqManTM MicroRNA reverse transcription kit (Life Technologies). The CFX96™Real-Time system (Bio-Rad) was used to amplify the PCR products and detected the expression of the target gene. The primer sequences were as follows: circ_0001785, forward 5ʹ-AAGAACATGGGTCTGGGAAA-3ʹ and reverse 5ʹ-CCGAGGTCTTTCATTCTTGC-3ʹ; GAPDH, forward 5ʹ-AGAAGGCTGGGGCTCATTTG-3ʹ and reverse 5ʹ-AGGGGCCATCCACAGTCTTC-3ʹ; U6, forward 5ʹ-CTCGCTTCGGCAGCACA-3ʹ and reverse 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ. This experiment was repeated 3 times, and GAPDH or U6 was used as the internal reference. The relative expression of the target gene was calculated by 2−ΔΔCt (Livak & Schmittgen, 2001) method.
CCK-8 assay
Cells in the logarithmic growth phase were inoculated into 96-well plates at a density of 5 × 103 cells/well. Each group was set to 3 repeated holes. After 24, 48 and 72 h of culture, 10 μl CCK-8 solution was added to each well. After 2 h of routine culture, the absorbance at 450 nm wavelength was detected using a microplate reader.
Colony formation assay
The treated cells were inoculated into a 6-well plate at a density of 1500 cells/well and cultured in a CO2 incubator for 10 d. After incubation, the culture medium was removed and washed twice with PBS, cells were fixed with 5% paraformaldehyde for 20 min and stained with 5% crystal violet. The incubation was continued at room temperature for 20 min. The colony formation was observed and quantified by an inverted microscope after drying at room temperature.
Cell migration analysis
The scratch wound-healing assay was used to detect cell migration. Cells were digested with 0.25% trypsin and the cell concentration was adjusted to 1 × 105 cells/ml. The cells were seed into 6-well plates and incubated in a 5% CO2 incubator at 37°C. When the cells had grown to 90% confluence, 10 μl sterilized micropipette tip was used to make a scratch wound on the 6-well plate. The cells were washed with PBS for 3 times. After 24 h of culture, the migration cells were observed under an inverted microscope.
Cell invasion analysis
Transwell assay was conducted to detect the cell invasion. Transwell chambers were precoated with Matrigel matrix (Corning) for invasion assay. Cells (5x104) were inoculated on the upper chamber containing a serum-free medium. Meanwhile, 600 μl of DMEM medium containing 10% FBS was added to the lower chamber. The cells were routinely cultured and three repeated holes were set up for each group. After 24 h of culture, the liquid in the chamber was discarded and the cells in the upper chamber were removed with cotton swabs. After staining with 0.1% crystal violet for 15 min, cells were observed and counted under a microscope.
Mouse xenograft assays
Female nude mice (BALB/c, SPF grade, weighing 16–20 g and 4-week old) were purchased from the Beijing Vital River Laboratory Animal Technologies Co. Ltd (Beijing, China). For details regarding this method please refer to the relevant literature [15]. In short, we amplified the sequence of circ_0001785 and cloned it into the overexpression vector pLCDH-ciR (circ_0001785), while, the pLCDH-ciR empty vector (Mock) was used as a control. The retroviral transfer plasmid pLCDH-ciR-puro was purchased from Geneseed Biotech (Guangzhou, China). After transfection, the transfection efficiency was detected by qRT-PCR. One μg/ml puromycin (Sigma-Aldrich, St. Louis, MO, USA) was used to screen the stable cell lines for 2 weeks. In addition, lentiviruses overexpressing miR-942 (miR-942) and corresponding negative control (NC) were obtained from Shanghai GenePharma Co., Ltd. Afterward, 1 × 106 cells were inoculated subcutaneously into fossa axillaris of these nude mice. The growth of the tumor and the body weight of nude mice were monitored every day. After 14 d, the tumor was dissected and weighted. All the animal experiments followed the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health (NIH). This study was approved by the Ethics Committee of the Second Affiliated Hospital of Shantou University Medical College.
RNA immunoprecipitation (RIP) assay
To determine the relationship between hsa_circ_0001785 and miR-942. We cotransfected the cells with MS2bs-circ_0001785 WT, MS2bs-circ_0001785 MT or control MS2bs-Rluc and miR-942. 48 h after transfection, RNA immunoprecipitation (RIP) experiments were performed using the Megna RIP RNA-binding Protein Immunoprecipitation Kit (Millipore). Next, in order to determine the interaction among circ_0001785, miR-942 and SOCS3. We lysed the cells with the lysate containing protease inhibitors (Life Technologies) and RNase inhibitors (Life Technologies). Then, cell lysate was incubated with anti-Argonaute2 (anti-Ago2) or anti-IgG (negative control) overnight at 4°C, followed by the addition of Protein A magnetic beads to get the immunoprecipitation complex. The expressions of circ_0001785, miR-942 and SOCS3 were detected by qRT-PCR.
Dual-luciferase reporter assay
Wild type (WT) and Mutant (MT) plasmids containing circ_0001785 and SOCS3 3ʹUTR fragments of miR-942 binding sequence were amplified from human genomic DNA by qRT-PCR. Then, the plasmids mentioned above were cotransfected with miR-942 mimic or mimic negative control (miR-NC) into cells by Lipofectamine 2000. After 48 h of transfection, the cells were collected, and the fluorescence value of the cells in each group was analyzed using the Luciferase Assay Kit (Promega, USA) according to the manufacturer’s protocol.
Western blot analysis
Cells of each group were harvested and the total protein was extracted with RIPA lysate. The protein concentration was measured using BCA Protein Assay Kit (Beyotime). The proteins were separated by SDS-PAGE and electrotransferred to PVDF membrane (Millipore, France). After blocking with 5% skimmed milk powder at room temperature for 2 h, the membranes were incubated with primary antibodies against KI67 (cat. no. ab15580; 1:50 dilution; Abcam), PCNA (cat. no. ab92552; 1:1000 dilution; Abcam), MMP-7 (cat. no. ab207299; 1:1000 dilution; Abcam), MMP-9 (cat. no. ab76003; 1:1000 dilution; Abcam), SOCS3 (cat. no. ab16030; 1:1000 dilution; Abcam) and GAPDH (cat. no. ab181602; 1:10,000 dilution; Abcam) overnight at 4°C. Then, the membranes washed with TBST for 3 times and then incubated with HRP-conjugated secondary antibody (1:5000; Santa Cruz Biotechnology, Inc.) at 37°C for 1 h. Immune bands were detected by enhanced chemiluminescence (ECL). Quantitative analysis of protein bands was analyzed by Image-J software. GAPDH was used as the internal control.
Statistical analysis
The experimental data were analyzed by SPSS22.0 software (SPSS, Chicago, USA), and measurement data were expressed by the mean ± standard deviation. Student t-test was used for comparison between groups and One-Way ANOVA followed by Tukey’s post hoc test was used for multigroup comparison. Values of p < 0.05 were considered as statistically significant.
Results
Circ_0001785 is lowly expressed and circ_0001785 overexpression inhibits the breast cancer cell proliferation
We first detected the expression levels of circ_0001785 in five BC cells (T47D, MCF-7, MDA-MB-453, MDA-MB-231 and BT-549) by qRT-PCR. As shown in Figure 1(a), the expression level of circ_0001785 in BC cells was significantly downregulated compared with normal cell (MCF-10A). Among all the BC cells, the expression levels of circ_0001785 in T47D and MDA-MB-231 cells were the lowest. Therefore, T47D and MDA-MB-231 cells were selected for the following experiments. Due to circ_0001785 was downregulated, we successfully constructed the overexpression plasmid of circ_0001785 (pcDNA-circ_0001785) and confirmed the overexpression efficiency by qRT-PCR (Figure 1(b)). Subsequently, we determined the effect of circ_0001785 on the viability of T47D and MDA-MB-231 cells by CCK-8 and clone formation assays. The results of CCK-8 assay revealed that circ_0001785 overexpression significantly inhibited the viability of T47D and MDA-MB-231 cells compared with that in the control group (Figure 1(c)). The results of colony formation assay indicated that circ_0001785 overexpression significantly decreased the number of colonies formed by T47D and MDA-MB-231 cells (Figure 1(d,e)). Overall, circ_0001785 overexpression significantly inhibited the proliferation of T47D and MDA-MB-231 cells.
Figure 1.
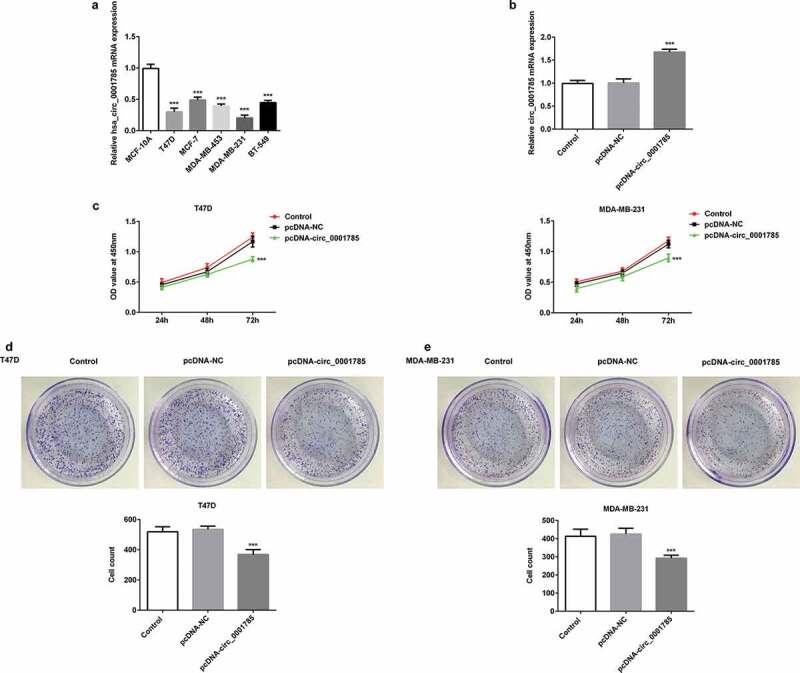
Circ_0001785 is lowly expressed and circ_0001785 overexpression inhibits the breast cancer cell proliferation. (a) Circ_0001785 expression in various breast cancer cell lines. ***p < 0.001 vs. MCF-10A. (b) The transfection efficiency of circ_0001785 overexpressed plasmid was measured by qRT-PCR. ***p < 0.001 vs. pcDNA-NC. (c) The effect of circ_0001785 overexpression on T47D and MDA-MB-231 cells proliferation was measured by CCK-8 assay at 24, 48 and 72 h. ***p < 0.001 vs. pcDNA-NC. (d-e) Colony formation assay was performed to determine the effect of circ_0001785 overexpression on the proliferation abilities of T47D and MDA-MB-231 cells. ***p < 0.001 vs. pcDNA-NC
Circ_0001785 overexpression inhibits the breast cancer cell migration and invasion
Invasiveness is one of the most obvious features of BC cells. We detected the migration and invasion of BC cells by scratch wound-healing and Transwell assays. The migration and invasion assays indicated that circ_0001785 overexpression inhibited wound healing and reduced T47D cell invasion in vitro (Figure 2(a,b)). Compared with the control group, circ_0001785 overexpression significantly reduced the migration and invasion rates of T47D cells (Figure 2(c)). Similar results were noted in MDA-MB-231 cells following the same experimental conditions, circ_0001785 overexpression significantly suppressed the migration and invasion of MDA-MB-231 cells (Figure 2(d-f)). In addition, we measured the protein expressions of invasion-associated proteins (MMP-7/-9). As shown in Figure 2(g,h), circ_0001785 overexpression significantly decreased the expressions of MMP-7 and MMP-9 in T47D and MDA-MB-231 cells. These results indicated that circ_0001785 overexpression reduced the migration and invasion of T47D and MDA-MB-231 cells.
Figure 2.
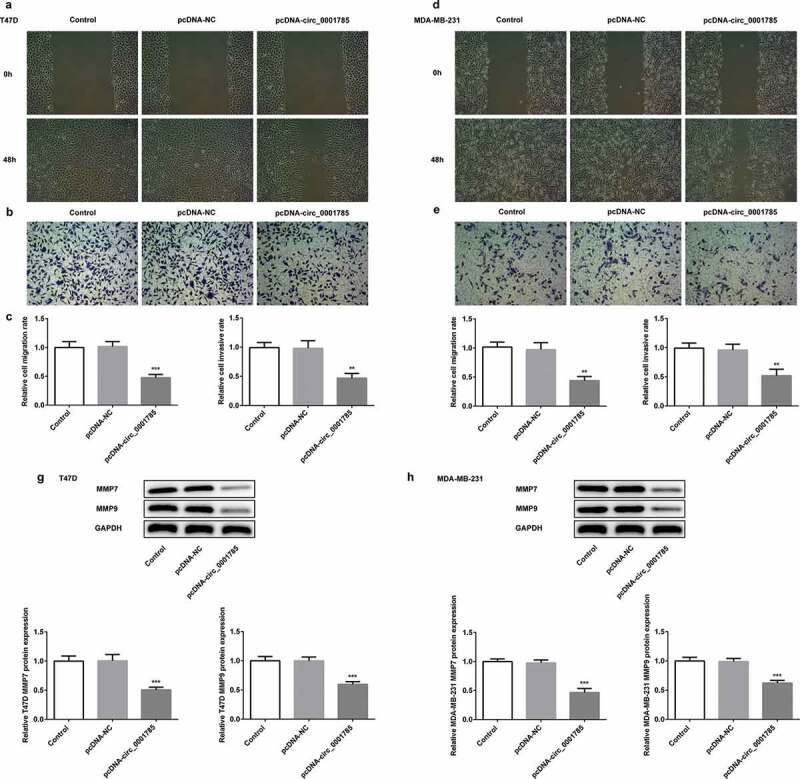
Circ_0001785 overexpression inhibits the breast cancer cell migration and invasion. (a-b) The representative pictures of T47D cell migration and invasion. (c) Effect of circ_0001785 overexpression on cell migration and invasion in T47D cells. (d-e) The representative pictures of MDA-MB-231 cell migration and invasion assays. (f) Effect of circ_0001785 overexpression on cell migration and invasion in MDA-MB-231 cells. (g-h) Effect of circ_0001785 overexpression on MMP7 and MMP9 expressions in T47D and MDA-MB-231 cells. GAPDH was used as a loading control. **p < 0.01 and ***p < 0.001 vs. pcDNA-NC
Circ_0001785 inhibits breast cancer progression in vivo
To further study the effect of circ_0001785 in vivo, we carried out mouse xenograft experiments. As shown in Figure 3(a), there were no significant changes in the body weight between Mock group and circ_0001785 group in nude mice inoculated with T47D cells, which indicated that external factors and in vivo transfection reagents had little effect on the body weight of nude mice. Moreover, circ_0001785 overexpression significantly inhibited the tumor formation ability of mice. Next, we dissected tumor tissues from tumor-bearing mice 14 d after the injection of T47D cells and found that the xenograft tumor in the circ_0001785 group was significantly smaller compared with the Mock group (Figure 3(b)). Similarly, the nude mice xenograft experimental results of MDA-MB-231 cells were basically consistent with that of T47D cells (Figure 3(c,d)). Subsequently, we examined the related protein expressions of proliferation and invasion in tumor tissues by western blot analysis. As shown in Figure 3(e,f), circ_0001785 overexpression significantly decreased the expressions of KI67, PCNA, MMP-7 and MMP-9 in T47D cells. Similar results were noted in MDA-MB-231 cells following the same experimental conditions (Figure 3(g,h)). These findings suggested that circ_0001785 overexpression decreased the protein levels of KI67, PCNA, MMP-7 and MMP-9 and suppressed the tumor growth in BC.
Figure 3.
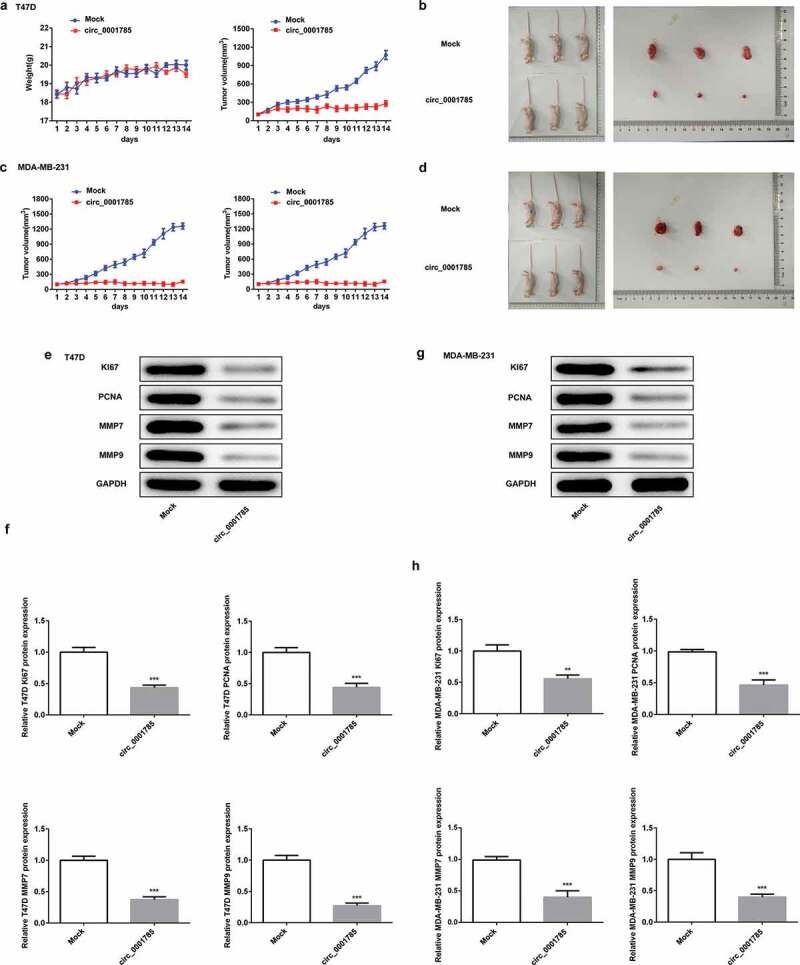
Circ_0001785 inhibits breast cancer progression in vivo. (a) The circ_0001785 overexpression and control T47D cells were inoculated in nude mice, and mouse weight and tumor volume were monitored every day. (b) Tumor mass of T47D tumor xenograft-induced nude mice after 14 d of treatment. (c) The circ_0001785 overexpression and control MDA-MB-231 cells were inoculated in nude mice, and mouse weight and tumor volume were monitored every day. (d) Tumor mass of MDA-MB-231 tumor xenograft-induced nude mice after 14 d of treatment. (e-f) Expressions of KI67, PCNA, MMP7 and MMP9 in tumor tissues inoculated with T47D cells were detected by western blot analysis. GAPDH was used as the loading control. (g-h) Expressions of KI67, PCNA, MMP7 and MMP9 in tumor tissues inoculated with MDA-MB-231 cells were detected by western blot analysis. GAPDH was used as the loading control. **p < 0.01 and ***p < 0.001 vs. Mock
Circ_0001785 serves as a ceRNA via sponging miR-942 in breast cancer
We then determined whether circ_0001785 exert the function of ceRNA in BC. We first detected the subcellular localization of circ_0001785 in BC cells. As shown in Figure 4(a), circ_0001785 was predominantly localized in the cytoplasm. Through the analysis of the CIRCinteractome database, score criteria (reference context+scorepercentile >90), the top five miRNAs binding to circ_0001785 were miR-1200, miR-1251, miR-605, miR-610 and miR-942 (Figure 4(b)). Then, the dual-luciferase reporter assay verified that these five miRNAs significantly decreased the luciferase activity of the reporter vector containing the wild-type 3’-UTR of circ_0001785, but had only a minimal effect on the luciferase activity of the mutant circ_0001785 luciferase reporter vector. Among them, miR-942 reduced the luciferase activity to the maximum extent (Figure 4(c)). This result suggested that circ_0001785 could stably combine with miR-942. Then, we detected the expressions of these five miRNAs in BC cells and found that the expression of miR-942 exhibited the highest expression (Figure 4(d)). Therefore, we chose miR-942 as the target of circ_0001785 for further research. We next examined the expression relationship between circ_0001785 and miR-942. Circ_0001785 overexpression significantly reduced the expression of miR-942 in BC cells (Figure 4(e)). Similarly, we examined the effect of circ_0001785 on the expression of miR-942 in vivo. The results of tumor tissue analysis in nude mice indicated that circ_0001785 overexpression could also decrease the expression of miR-942 (Figure 4(f)). Finally, to further verify the direct combination of circ_0001785 and miR-942, we used RIP analysis based on MS2bs. Results showed that miR-942 was mainly enriched in MS2bs-circ_0001785 WT group, indicating that circ_0001785 specifically bound to miR-942 (Figure 4(g)). Taken together, circ_0001785 served as a ceRNA via sponging miR-942 in BC and could negatively modulate the expression of miR-942.
Figure 4.
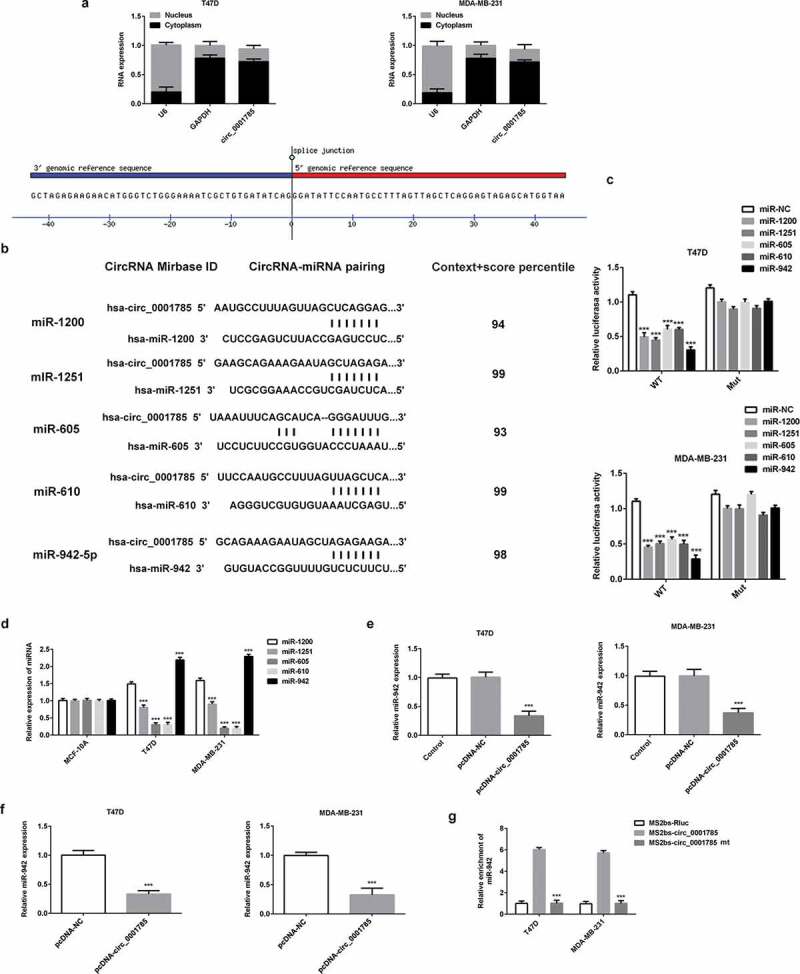
Circ_0001785 serves as a ceRNA via sponging miR-942 in breast cancer. (a) Expressions of circ_0001785, U6 and GAPDH were detected. (b) Circ_0001785 target genes predicted using CircInteractome and five miRNAs (miR-1200, miR-1251, miR-605, miR-610, miR-942) were screened. (c) Dual-luciferase reporter gene assay in T47D and MDA-MB-231 cells cotransfected with wild-type/mutant-type circ_0001785 and five predicted miRNAs mimics/negative control. ***p < 0.001 vs. miR-NC. (d) Expressions of five miRNAs in T47D and MDA-MB-231 cells were examined by western blot analysis. ***p < 0.001 vs. miR-1200. (e) Effect of Circ_0001785 overexpression on the expression of miR-942 in T47D and MDA-MB-231 cells transfected with empty vector or circ_0001785 overexpression plasmid by qRT-PCR. ***p < 0.001 vs. pcDNA-NC. (f) MiR-942 expression levels in tumor tissues transfected with empty vector or circ_0001785 overexpression plasmid by qRT-PCR. ***p < 0.001 vs. pcDNA-NC. (g) RIP assay was performed to confirm the direct binding between circ_0001785 and miR-942. ***p < 0.001 vs. MS2bs-circ_0001785
Circ_0001785 regulated the breast cancer cell proliferation by targeting miR-942
To further investigate the effect of miR-942 on circ_0001785 in BC cells, we successfully constructed miR-942 mimic and miR-942 inhibitor, and confirmed the transfection effects by qRT-PCR (Figure 5(a)). Then, we detected the biological function of miR-942 in BC cells by CCK-8 and clone formation assays. CCK-8 assay showed that miR-942 overexpression reduced the inhibitory effect of circ_0001785 overexpression on cell viability compared with that in the control group, whereas these changes could be reversed by the downregulation of miR-942 (Figure 5(b)). Meanwhile, the colony formation assays showed that miR-942 overexpression significantly increased the number of cell colony formation, compared with that in the pcDNA-circ_0001785 + miR-NC group. However, the downregulation of miR-942 showed the opposite trend (Figure 5(c,d)). Taken together, circ_0001785 regulated the BC cell proliferation by targeting miR-942.
Figure 5.
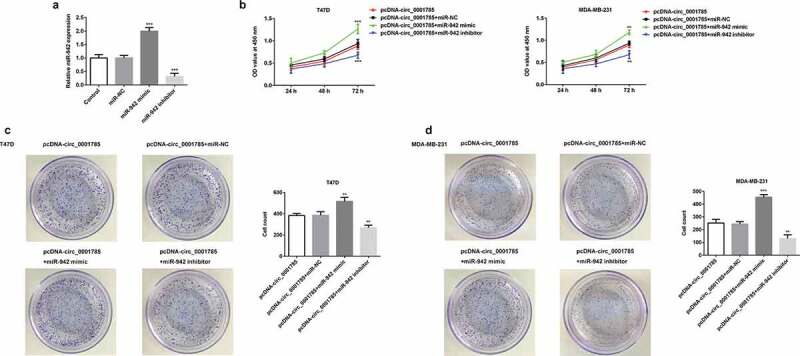
Circ_0001785 regulated the breast cancer cell proliferation by targeting miR-942. (a) The transfection efficiencies of miR-942 overexpression and knockdown were measured by qRT-PCR. ***p < 0.001 vs. miR-NC. (b) The effect of miR-942 overexpression and knockdown on T47D and MDA-MB-231 cells proliferation were measured by CCK-8 assay at 24, 48 and 72 h. **p < 0.01 and ***p < 0.001 vs. pcDNA-circ_0001785+ miR-NC. (c-d) Colony formation assay was performed to determine the effect of miR-942 overexpression and knockdown on the proliferation abilities of T47D and MDA-MB-231 cells. **p < 0.01 and ***p < 0.001 vs. pcDNA-circ_0001785+ miR-NC
Circ_0001785 regulated the breast cancer cell migration and invasion by targeting miR-942
Next, we examined the regulatory relationship between circ_0001785 and miR-942 through migration and invasion assays. Compared with the pcDNA-circ_0001785 + miR-NC group, miR-942 overexpression significantly promoted cell migration and invasion in T47D and MDA-MB-231. These alterations were reversed by the downregulation of miR-942 (Figure 6(a-f)). The protein expressions of MMP-7/-9 were increased in the pcDNA-circ_0001785 + miR-942 mimic compared with the pcDNA-circ_0001785 + miR-NC group, whereas decreased in pcDNA-cicr_0001785+ miR-942 inhibitor group (Figure 6(g,h)). These results indicated that circ_0001785 regulated the BC cell migration and invasion by targeting miR-942.
Figure 6.
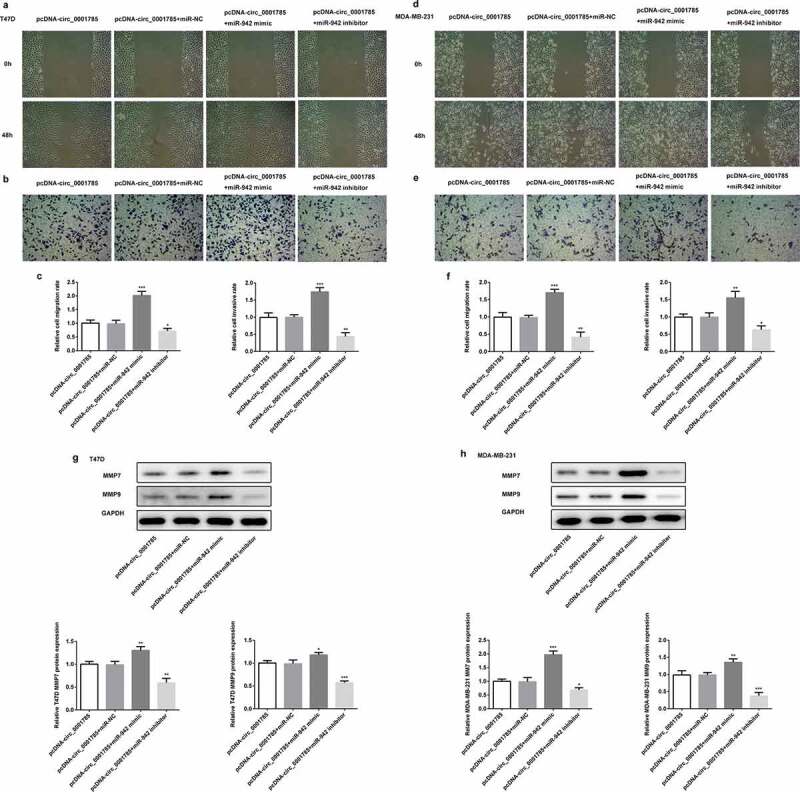
Circ_0001785 regulated the breast cancer cell migration and invasion by targeting miR-942. (a-b) The representative pictures of T47D cell migration and invasion. (c) Effect of miR-942 overexpression and knockdown on cell migration and invasion in T47D cells. (d-e) The representative pictures of MDA-MB-231 cell migration and invasion assays. (f) Effect of miR-942 overexpression and knockdown on cell migration and invasion in MDA-MB-231 cells. (g-h) Effect of miR-942 overexpression and knockdown on MMP7 and MMP9 expressions in T47D and MDA-MB-231 cells. GAPDH was used as a loading control. *p < 0.05, **p < 0.01 and ***p < 0.001 vs. pcDNA-circ_0001785+ miR-NC
Circ_0001785 inhibits breast cancer progression in vivo by targeting miR-942
Then, we studied the effect of circ_0001785 on miR-942 in vivo by the mouse xenograft experiments. The tumor volume was increased in the circ_0001785 + miR-942 group compared with the circ_0001785 + NC group. In addition, the xenograft tumor in the circ_0001785 + miR-942 group was significantly enlarged compared with the circ_0001785 group + NC group (Figure 7(a-d)). Subsequently, miR-942 overexpression significantly increased the expressions of KI67, PCNA, MMP-7 and MMP-9 in T47D and MDA-MB-231 cells (Figure 7(e,h)). These results suggested that circ_0001785 inhibits BC progression in vivo by targeting miR-942.
Figure 7.
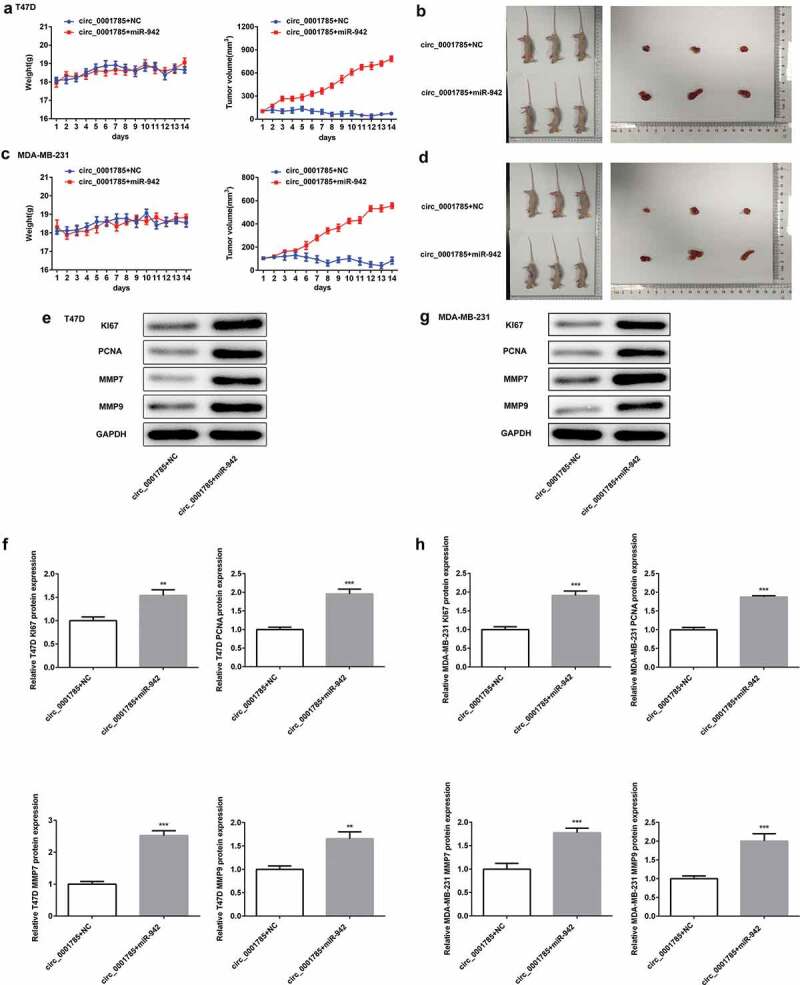
Circ_0001785 inhibits breast cancer progression in vivo by targeting miR-942. (a) The miR-942 mimic and control T47D cells were inoculated in nude mice, and mouse weight and tumor volume were monitored every day. (b) Tumor mass of T47D tumor xenograft-induced nude mice after 14 days of treatment. (c) The miR-942 mimic and control MDA-MB-231 cells were inoculated in nude mice, and mouse weight and tumor volume were monitored every day. (d) Tumor mass of MDA-MB-231 tumor xenograft-induced nude mice after 14 d of treatment. (e-f) Expressions of KI67, PCNA, MMP7 and MMP9 in tumor tissues inoculated with T47D cells were detected by western blot analysis. GAPDH was used as the loading control. (g-h) Expressions of KI67, PCNA, MMP7 and MMP9 in tumor tissues inoculated with MDA-MB-231 cells were detected by western blot analysis. GAPDH was used as the loading control. **p < 0.01 and ***p < 0.001 vs. circ_0001785+ NC
SOCS3 is a target gene of miR-942
So far little is known about the target genes of miR-942. Thus, we predicted the target genes of miR-942-5p by Targetscan software and identified SOCS3 was the potential target gene (Figure 8(a)). Previous studies have shown that high levels of SOCS3 were associated with good prognosis of BC patients [16], and miR-942-5p was predicted to be targeted with SOCS3. Therefore, we intended to further study the relationship between miR-942 and SOCS3 in BC. We first measured the expression level of SOCS3 by qRT-PCR and western blot analysis in BC cells. Compared with normal human breast epithelial cells (MCF-10A), the expression levels of SOCS3 in T47D and MDA-MB-231 cells were significantly decreased (Figure 8(b,c)). And the dual-luciferase reporter assay demonstrated that the luciferase activity in the cells cotransfected with miR-942 mimic and wild-type SOCS3 was significantly decreased, while that in the cells transfected with mutant SOCS3 was almost unchanged (Figure 8(d)). The above results showed that miR-942 could directly target SOCS3 in BC cells.
Figure 8.
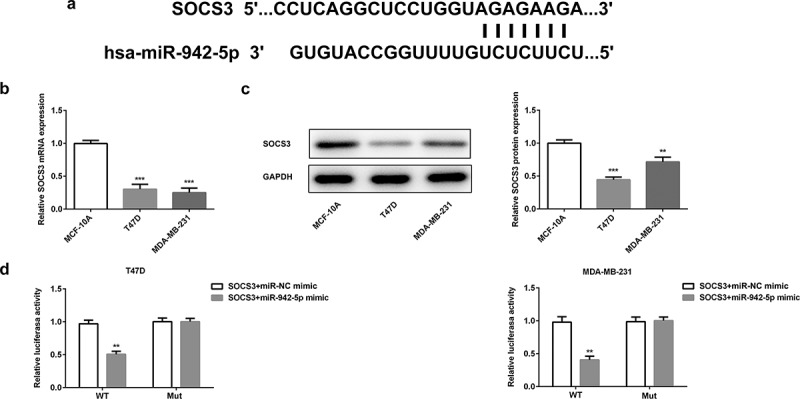
SOCS3 is a target gene of miR-942. (a) TargetScan predicted that SOCS3 was a target gene of miR-942. (b) The expressions of SOCS3 in T47D and MDA-MB-231 cells were detected by qRT-PCR. ***p < 0.001 vs. MCF-10A. (c) The expression of SOCS3 in T47D and MDA-MB-231 cells were detected by western blot analysis. **p < 0.01 and ***p < 0.001 vs. MCF-10A. (d) The relationship between miR-942-5p and SOCS3 was determined by a dual-luciferase reporter assay. **p < 0.01 vs. SOCS3+ miR-NC mimic
Circ_0001785 regulates SOCS3 expression via inhibiting miR-942
Finally, we determined whether circ_0001785 could regulate the expression of SOCS3 by sponging miR-942 in BC. The effect of miR-942-5p and circ_0001785 on the expression of SOCS3 was detected by qRT-PCR and western blot analysis. As shown in Figure 9(a,b), miR-942 overexpression significantly downregulated the expression of SOCS3 both at mRNA level and protein level. This result was completely opposite to the effect of circ_0001785 overexpression on SOCS3. Circ_0001785 overexpression significantly promoted the expression of SOCS3 in BC cells. These findings indicated that circ_0001785 positively regulated the expression of SOCS3 while miR-942 negatively regulated SOCS3. We next performed Ago2 RIP experiments to explore the ceRNA function of circ_0001785. Results demonstrated that circ_0001785, miR-942 and SOCS3 were all enriched to Ago2 in T47D and MDA-MB-231 cells (Figure 9(c)). Finally, we reconfirmed the effect of circ_0001785 and miR-942 on the expression of SOCS3 by cotransfection of pcDNA-circ_0001785 and miR-942 mimic. In T47D and MDA-MB-231 cells, miR-942 overexpression significantly reduced the promoting effect of circ_0001785 overexpression on SOCS3 expression (Figure 9(d)). In mice tumor tissue, miR-942 overexpression also suppressed the promoting effect of circ_0001785 overexpression on SOCS3 expression (Figure 9(e)). All these results indicated that circ_0001785 could regulate SOCS3 expression via sponging miR-942 in BC.
Figure 9.
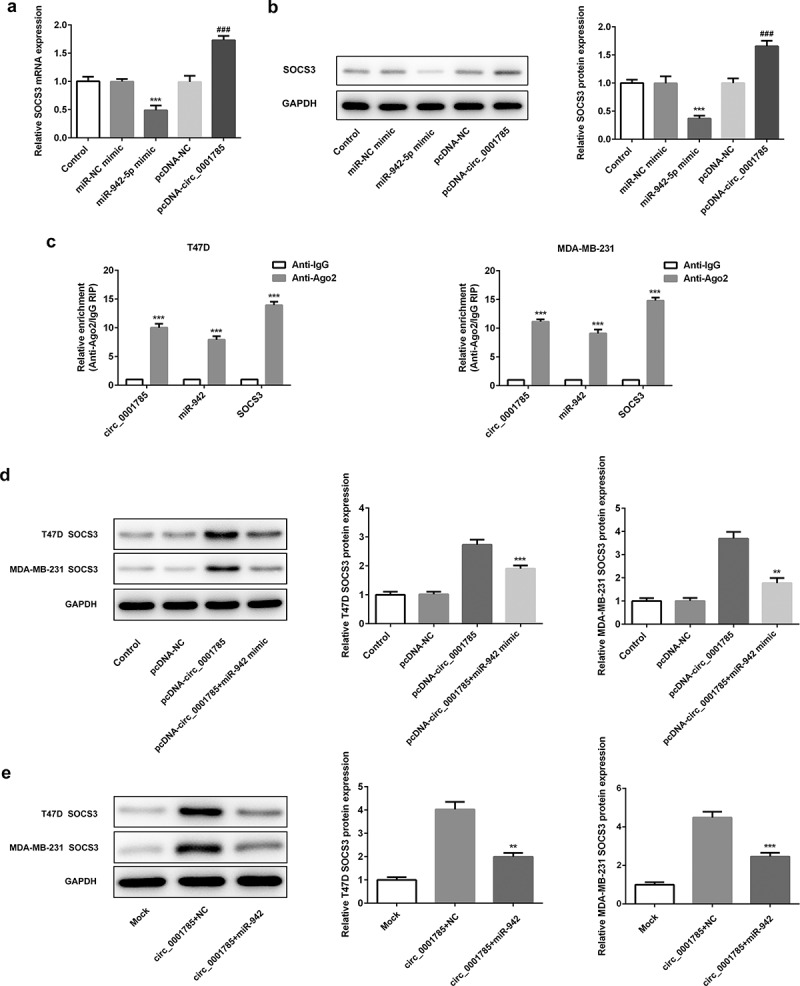
Circ_0001785 regulates SOCS3 expression via inhibiting miR-942. (a) The expressions of SOCS3 in breast cancer cells transfected with miR-942 overexpression plasmid and circ_0001785 overexpression vector were detected by qRT-PCR. ***p < 0.001 vs. miR-NC mimic; ###p < 0.001 vs. pcDNA-NC. (b) The expressions of SOCS3 in breast cancer cells transfected with miR-942 overexpression plasmid and circ_0001785 overexpression vector were detected by western blot analysis. ***p < 0.001 vs. miR-NC mimic; ###p < 0.001 vs. pcDNA-NC. (c) RIP assay revealed the enrichment of circ_0001785, miR-942 and SOCS3 on Ago2. ***p < 0.001 vs. Anti-IgG. (d) Expressions of SOCS3 in T47D and MDA-MB-231 cells were detected by western blot analysis. **p < 0.01 and ***p < 0.001 vs. pcDNA-circ_0001785. (e) Expressions of SOCS3 in tumor tissue from mice were detected by western blot analysis. **p < 0.01 and ***p < 0.001 vs. circ_0001785+ NC
Discussion
CircRNAs are a kind of noncoding RNAs with covalently closed-loop structures, which is highly conserved and widely exists in a variety of eukaryotic organisms. In recent years, the role of circRNAs in the research of cancer treatment has become a hot topic [8]. Yin et al. found that abnormal expressions of circRNAs in the peripheral blood of BC, in which the expression level of hsa_circ_0001785 in plasma of BC patients was lowest, and the patients with high expression of hsa_circ_0001785 had a better prognosis. Meanwhile, the levels of hsa_circ_0001785 in plasma samples were closely related to histological grade, TNM stage and tumor distant metastases [14]. These findings indicated that hsa_circ_0001785 has high diagnostic value and accuracy. Therefore, we chose hsa_circ_0001785 as subjects in the present study. Here, we found that the expression of circ_0001785 was downregulated in BC cells, and circ_0001785 overexpression significantly suppressed cell proliferation, migration and invasion in BC. We believed that circ_0001785 may play an important role in BC.
MicroRNAs (miRNAs) are a class of small noncoding RNAs that are 19–25 nucleotides in length [17]. They could combine with the target mRNA to regulate the translation or degradation of mRNA and then regulate gene expression at the post-transcriptional level, which is involved in the process of cell growth, development, differentiation and metabolism, and is of great significance to the occurrence and development of tumors [18]. Numerous studies showed that circRNAs could act as ceRNAs through competitive binding miRNA by microRNA response element (MRE) reducing the inhibition of miRNA on its corresponding target genes [19,20]. In BC, circRNA_000911 inhibited the progression of BC by sponging miR-499a to regulate Notch1 [21]; cicrcGFRA1 promoted the proliferation of triple-negative BC cells by acting as a sponge for miR-34a [22]; circ_0006528 affected the drug resistance of BC cells by targeting miR-7-5p/Raf1 axis [23]. In the present study, we used bioinformatics software to predict and screen that miR-942 was the target gene of circ_0001785. As a known cancer-promoting factor, miR-942 could promote the occurrence and development of a wide variety of cancers, including liver cancer [24], colorectal cancer [25] and ovarian cancer [26]. To our knowledge, miR-942 regulated the progression of BC by targeting FOXA2 [27]. Moreover, circEPSTI1 promoted the proliferation and invasion of BC cells by sponging miR-942. However, the interaction between circRNAs and miR-942 in BC has not been studied [26]. In the current study, we verified that circ_0001785 could directly target miR-942 through dual-luciferase reporter and RIP assays. Additionally, miR-942 was highly expressed in BC cells and tumor tissues from mice. Circ_0001785 negatively regulated the expression of miR-942. In further biological function experiments, circ_0001785 could regulate cell proliferation, migration and invasion by targeting miR-942 in BC.
Then, we further studied the downstream target for SOCS3. We determined that SOCS3 was the downstream target of miR-942 through TargetScan (http://www.targetscan.org/vert_71). Related studies have shown that high levels of SOCS3 were correlated with early tumor stage and good clinical prognosis of BC [16]. In addition, JZ et al. demonstrated that circTADA2As suppressed the cell proliferation, invasion and migration of BC through sponging miR-203a-3p to regulate SOCS3 [28]. These findings show that SOCS3 has a broad prospect for research in BC. In our experiment, SOCS3 was downregulated in BC cells and tumor tissues from mice. Circ_0001785 overexpression increased the expression of SOCS3 while miR-942 overexpression could reverse it. Taken together, circ_0001785 could modulate the progression of BC by sponging miR-942 and regulating the expression of SOCS3.
Conclusions
We demonstrated that circ_0001785 has low expression in BC. Circ_0001785 overexpression significantly inhibited cell proliferation, migration/invasion and the tumor growth. In addition, circ_0001785 could suppress the progression of BC by decoying miR-942 to upregulate SOCS3. We believed that circ_0001785 could be a potential biomarker for early diagnosis and therapeutic target for BC.
Funding Statement
No funding was received.
Disclosure statement
No potential conflict of interest was reported by the authors.
Ethics committee approval and patient consent
This present study was performed based on the principles expressed in the Declaration of Helsinki. All animal experiments were approved by the Ethics Committee of the Second Affiliated Hospital of Shantou University Medical College and conducted in accordance with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals.
References
- [1].Adeloye D, Sowunmi OY, Jacobs W, et al. Estimating the incidence of breast cancer in Africa: a systematic review and meta-analysis. J Glob Health. 2018;8(1):010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].McAnena PF, McGuire A, Ramli A, et al. Breast cancer subtype discordance: impact on post-recurrence survival and potential treatment options. BMC Cancer. 2018;18(1):203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Harbeck N, Penault-Llorca F, Cortes J, et al. Breast cancer. Nat Rev. 2019;5(1):66. [DOI] [PubMed] [Google Scholar]
- [4].Mitra S, Dash R.. Natural products for the management and prevention of breast cancer. Evidence-Based Complementary Altern Med. 2018;2018:8324696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Power EJ, Chin ML, Haq MM.. Breast cancer incidence and risk reduction in the hispanic population. Cureus. 2018;10(2):e2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hu W, Tan C, He Y, et al. Functional miRNAs in breast cancer drug resistance. Onco Targets Ther. 2018;11:1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lai Z, Yang Y, Yan Y, et al. Analysis of co-expression networks for circular RNAs and mRNAs reveals that circular RNAs hsa_circ_0047905, hsa_circ_0138960 and has-circRNA7690-15 are candidate oncogenes in gastric cancer. Cell Cycle. 2017;16(23):2301–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Jiang L, Sun D, Hou J, et al. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1–7 [DOI] [PubMed] [Google Scholar]
- [9].Guo JU, Agarwal V, Guo H, et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15(7):409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna. 2013;19(2):141–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lü L, Sun J, Shi P, et al. Identification of circular RNAs as a promising new class of diagnostic biomarkers for human breast cancer. Oncotarget. 2017;8(27):44096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wang N, Gu Y, Li L, et al. Circular RNA circMYO9B facilitates breast cancer cell proliferation and invasiveness via upregulating FOXP4 expression by sponging miR-4316. Arch Biochem Biophys. 2018;653:63–70. [DOI] [PubMed] [Google Scholar]
- [13].Huang X, Xie X, Wang H, et al. PDL1 And LDHA act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res. 2017;36(1):1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yin WB, Yan MG, Fang X, et al. Circulating circular RNA hsa_circ_0001785 acts as a diagnostic biomarker for breast cancer detection. Clin Chim Acta. 2018;487:363–368. [DOI] [PubMed] [Google Scholar]
- [15].Yu J, Yang M, Zhou B, et al. CircRNA-104718 acts as competing endogenous RNA and promotes hepatocellular carcinoma progression through microRNA-218-5p/TXNDC5 signaling pathway. Clin Sci. 2019;133(13):1487–1503 [DOI] [PubMed] [Google Scholar]
- [16].Sasi W, Jiang WG, Sharma A, et al. Higher expression levels of SOCS 1, 3, 4, 7 are associated with earlier tumour stage and better clinical outcome in human breast cancer. BMC Cancer. 2010;10(1):178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Pichler M, Calin GA. MicroRNAs in cancer: from developmental genes in worms to their clinical application in patients. Br J Cancer. 2015;113(4):569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bartel DP. Metazoan microRNAs. Cell. 2018;173(1):20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wang X, Fang L. Advances in circular RNAs and their roles in breast Cancer. J Exp Clin Cancer Res. 2018;37(1):206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang H, Xiao YI, Wu LI, et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-000911/miR-449a pathway in breast carcinogenesis. Int J Oncol. 2018;52(3):743–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].He R, Liu P, Xie X, et al. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res. 2017;36(1):145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gao D, Zhang X, Liu B, et al. Screening circular RNA related to chemotherapeutic resistance in breast cancer. Epigenomics. 2017;9(9):1175–1188 [DOI] [PubMed] [Google Scholar]
- [24].Hua Z, Ma K, Liu S, et al. LncRNA ZEB1-AS1 facilitates ox-LDL-induced damage of HCtAEC cells and the oxidative stress and inflammatory events of THP-1 cells via miR-942/HMGB1 signaling. Life Sci. 2020;247:117334. [DOI] [PubMed] [Google Scholar]
- [25].Shan Z, An N, Qin J, et al. Long non-coding RNA Linc00675 suppresses cell proliferation and metastasis in colorectal cancer via acting on miR-942 and Wnt/β-catenin signaling. Biomed Pharmacother. 2018;101:769–776. [DOI] [PubMed] [Google Scholar]
- [26].Xie J, Wang S, Li G, et al. circEPSTI1 regulates ovarian cancer progression via decoying miR-942. J Cell Mol Med. 2019;23(5):3597–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhang J, Zhang Z, Sun J, et al. MiR-942 regulates the function of breast cancer cell by targeting FOXA2. Biosci Rep. 2019;39(11):BSR20192298. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [28].Xu JZ, Shao CC, Wang XJ, et al. circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis. 2019;10(3):1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]


