ABSTRACT
Glutamine: fructose-6-phosphate amidotransferase (GFAT) enzymes catalyse the first committed step of the hexosamine biosynthesis pathway (HBP) using glutamine and fructose-6-phosphate to form glucosamine-6-phosphate (GlcN6P). Numerous species (e.g. mouse, rat, zebrafish, chicken) including humans and Drosophila encode two broadly expressed copies of this enzyme but whether these perform redundant, partially overlapping or distinct functions is not known. To address this question, we produced single gene null mutations in the fly counterparts of gfat1 and gfat2. Deletions for either enzyme were fully lethal and homozygotes lacking either GFAT1 or GFAT2 died at or prior to the first instar larval stage. Therefore, when genetically eliminated, neither isoform was able to compensate for the other. Importantly, dietary supplementation with D-glucosamine-6-phosphate rescued GFAT2 deficiency and restored viability to gfat2−/- mutants. In contrast, glucosamine-6-phosphate did not rescue gfat1−/- animals.
KEYWORDS: Drosophila, GFAT1, GFAT2
Introduction
Glucose-fructose amidotransferase (GFAT) enzymes carry out rate-limiting steps in the Hexosamine Biosynthetic Pathway (HBP). By converting Fruc-6-P and glutamine into GlcN-6-P (glucosamine-6-P) GFAT enzymes define critical biosynthetic activities that produce uridine 5′-diphosphate–N-acetylglucosamine (UDP-GlcNAc), an essential substrate for multiple glycosylation pathways, including O-linked GlcNAcylation and N-linked protein glycosylation (reviewed in [1–5]). These post-translational modifications target a widespread number of proteins and as such, they impact numerous cellular processes, including transcription, translation, metabolism and signal transduction [1,6–8]. Furthermore, perturbations in either O-linked GlcNAcylation and N-linked protein glycosylation pathways have been implicated in metabolic disorders, insulin resistance and certain cancers [6–9].
In Drosophila, HBP enzymes are generally well-conserved and, likewise, O-linked GlcNAc modification is clearly an important regulator of growth, metabolism, and physiology [2]. The fly genome codes for orthologs of glucosamine-phosphate N-acetyltransferase (CG1969), phosphoacetylglucosamine mutase and UDP-N-acetylglucosamine diphosphorylase. The latter two enzymes are clearly essential since mutations in the corresponding genes are lethal (genes are nst and mmy) [10–13]. In flies, the HBP is also critical to form the exoskeleton since it feeds the production of chitin, which is a polymer primarily composed of UDP-GlcNAc [11,12].
The fly genome contains two genes coding for GFAT enzymes, designated gfat1 and gfat2 [2,14,15]. gfat1 mRNA expression were associated with tissues that produce chitin [14] and, in contrast to the single isoform produced from gfat2, the gfat1 locus generates up to nine alternate transcript isoforms (see Flybase.org [15]). In biochemical studies, Drosophila GFAT1 produced enzymatic activity in vitro [14], but the protein coded by gfat2 has not been similarly studied. Here we individually eliminated both genes and determined that each is essential for viability.
Results
As the rate-limiting enzyme of the HBP, glucose-fructose amidotransferase (GFAT) converts Fruc-6-P and glutamine into GlcN-6-P (glucosamine-6-P) [2]. Two GFAT genes encoding two GFAT enzymes are found in the genomes of humans, mouse and Drosophila (see Figure 1). In flies, the predicted GFAT1 and GFAT2 proteins are highly similar to each other (Figure 1). Residues involved in substrate binding [16] are conserved in both Drosophila GFAT1 and GFAT2 (Figure 1) and, furthermore, GFAT1 activity was shown to be regulated by UDP-N-acetylglucosamine and PKA [14]. However, the action of GFAT1 and GFAT2 in development has not been investigated and whether these genes code for distinct or redundant functions was not known. To address this question, we produced single gene null mutations at both of these genes.
Figure 1.
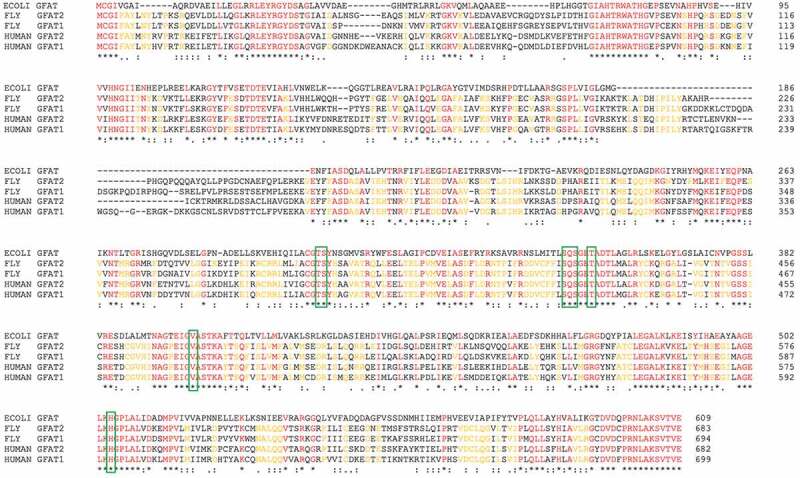
Sequence alignment of GFAT proteins
The amino acid sequences of E. coli, human and fly GFAT1 and GFAT2 are aligned using Clustal Omega. An * (asterisk) indicates positions which have a single, fully conserved residue (red). Amino acids that are conserved among fly and human GFAT 1 and GFAT2 are yellow. A: (colon) indicates conservation between groups of strongly similar properties. A. (period) indicates conservation between groups of weakly similar properties. Residues in green frames are involved in substrate binding [16]
As shown in Figure 2, we generated deletion mutants of both gfat1 and gfat2 using CRISPR technology to replace the corresponding sequences with DsRed. To edit gfat1, guide RNAs located at the beginning of the first intron and in the 3' UTR were chosen in order to delete most of the gene (over 10kb). Similarly, at the gfat2 locus, guide RNAs were chosen to delete the largest exon.
Figure 2.
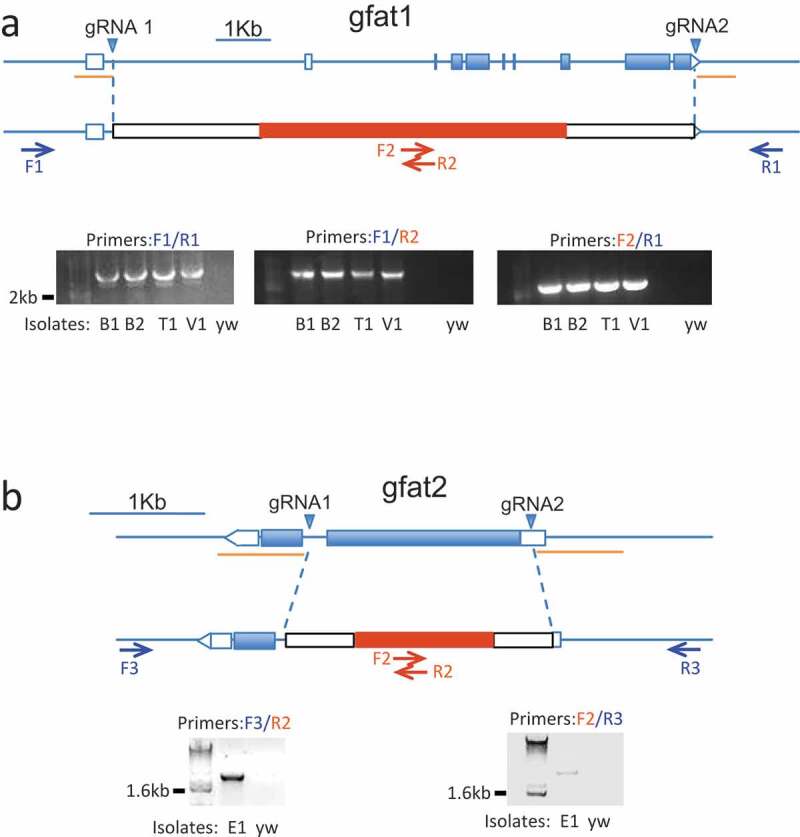
Crispr-mediated elimination of Drosophila gfat genes
In each panel, the wild type genomic structure for gfat1 (A) and gfat2 (B) is indicated above and edits that replace gfat sequences with DsRed and other sequences are below. The positions of guide RNAs (gRNAs, blue triangles) and verifying primers (arrows) for each gene are indicated. The sequences flanking the gRNAs that were cloned into the donor plasmids are labeled as light orange lines. Combinations of primers outside the homology arms (F1/R1 for gfat1 and F3/R3 for gfat2) together with DsRed specific primers (F2/R2) were used to perform PCR analyses as indicated. The diagnostic PCR products of expected sizes are shown in A (3.8kb for F1/R1, 1.9kb for F1/R2 and F2/R1) and in B (2.4kb for both F3/R2 and F2/R3)
For gfat1, five independent isolates were identified and four of these (designated B1, B2, T1, V1) were PCR verified. Similarly, we identified two independent isolates of gfat2 (designated E1 and F1). After genomic sequencing, we found that gfat1B2 and gfat1T1 were identical alleles and, likewise, gfat2E1 and gfat2F1 were found to be identical alleles. Therefore, gfat1B2 and gfat2E1 flies were selected for subsequent studies and, accordingly, were crossed in the yw background for five generations.
The Tb marker can be reliably scored at L2 larval stage or older and, using this marker, we assessed viability. As indicated in Figure 3, only heterozygotes for either gfat1B2 and gfat2E1 survived to the L2 stage or later. Furthermore, as seen in Table 1, gfat2E1 failed to complement Deficiency chromosomes that uncover gfat2. Likewise, gfat1B2 failed to complement a MiMIC insertion at the gfat1 locus. Therefore, we conclude that animals lacking either gfat1 or gfat2 are lethal prior to the L2 stage. To biochemically assess the effect of these mutations, we pooled 24–28 hr. animals (L1 larvae and embryos) produced from heterozygous parents and assayed these lysates for GFAT enzymatic activity. As seen in Figure 3(c), gfat1 and gfat2 samples were each reduced by 28.6% and 30.3% respectively compared to parental lysates, suggesting that both GFAT enzymes mutually contribute to canonical activity (Figure 3(c)).
Figure 3.
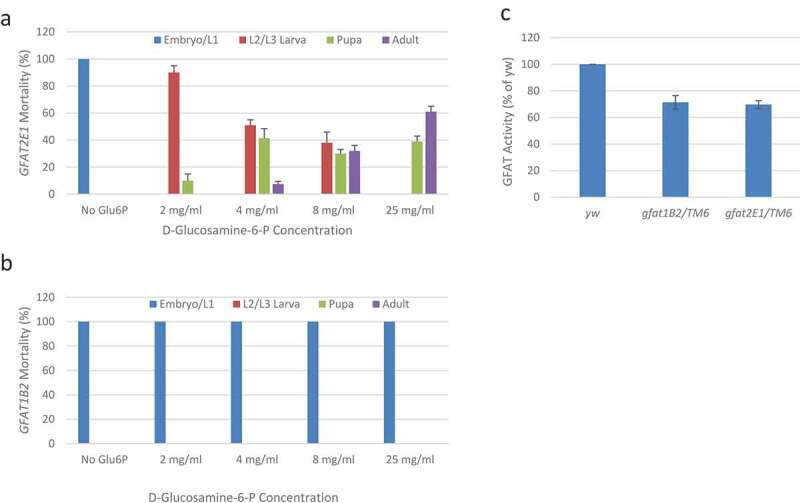
Dietary GlcN-6-P rescues development of animals lacking GFAT2.
In (a), with increasing dietary GlcN-6-P the lethal phase of gfat2E1 homozygotes was extended and, importantly, eclosion was restored. Note, more than 60% of homozygous gfat2E1 animals survive to adulthood when supplemented with 25mg/ml GlcN-6-P. In (b), homozygous gfat1B2 animals were similarly tested and dietary GlcN-6-P had no detectable effects. For both gfat2E1/TM6,Tb and gfat1B2/TM6,Tb, 50% of the embryos become heterozygous adults following the mendelian ratio with or without GlcN-6-P supplement. The number of heterozygotes were used as reference for the calculation of homozygotes survival ratios. In (c) GFAT activity was measured in lysates of pooled 24-28hr progeny generated from heterozygous parents (expected mendelian ratios are 25% homozygous and 50% heterozygous for the indicated genotypes). Specific activities were normalized to the parental yw strain. Note that minor fractions of unhatched embryos were observed in similar numbers of both mutant lines and that TM6 homozygotes should produce wild type levels of enzymatic activity. Error bars represent standard deviations. There were too few replicates to justify further statistical analysis
Table 1.
Complementation analyses for gfat1 and gfat2 crispr deletions.
gfat1B2 and gfat2E1 alleles were tested in trans combinations as indicated. For each cross, three vials were set up, and at least 100 flies were scored. Where crosses are labeled “yes”, the expected mendelian ratio of genotypes was observed. Where crosses are labeled “no“, transheterozygous mutant animals older than the L1 stage were not observed. Note that gfat1B2 did not complement MiMIC gfat1MI11277, and gfat2E1 did not complement the two Deficiencies that uncover gfat2 (indicated). An irrelevant control Deficiency, Df(3R)Exel6209 was complemented by gfat2E1
| Genotype | gfat1B2 | gfat2E1 | gfat2F1 |
|---|---|---|---|
| Mi{MIC}gfat1MI11277 | No | yes | yes |
| gfat1B2 | N/A | yes | yes |
| gfat2E1 | yes | N/A | no |
| gfat2F1 | yes | no | N/A |
| Df(3R)BSC460 (uncovers gfat2) |
ND | no | no |
| Df(3R)BSC881 (uncovers gfat2) |
ND | no | no |
| Df(3R)Exel6209 (control) |
ND | yes | yes |
ND: not determined.
Elimination of GFAT activity, which is pivotal for HBP, should deplete GlcN-6-P in these mutants. Therefore, we tested for dietary rescue using increasing concentrations of GlcN-6-P (see Figure 3 and methods). Without dietary GlcN-6-P, all gfat2E1 homozygotes died as early larvae or unhatched embryos (Figure 3). However, when supplemented at GlcN-6-P concentrations greater than 4 mg/ml, we observed that substantial numbers of adult gfat2E1 homozygotes eclosed. Furthermore, when supplemented with 25 mg/ml GlcN-6-P, over half of these were restored to adult viability. Importantly, as seen in Figure 3, a clear dose response was observed, as increasing concentrations of dietary GlcN-6-P permitted increasing eclosion rates. Adults homozygous for gfat2E1 that were rescued by dietary GlcN-6-P died within 3–4 days, but when crossed to wild type virgin females, we found that rescued gfat2E1 males were clearly fertile. In parallel studies, the same GlcN-6-P supplement was provided to gfat1B2 flies and, at all concentrations, gfat1B2 mutants were unaffected by dietary GlcN-6-P (Figure 3).
Discussion
We applied CRISPR editing to produce single gene deletions of both gfat1 and gfat2 genes in Drosophila and found each is required for viability. In complementation assays we further verified that gfat1 and gfat2 are each homozygous lethal in the 1st instar larval stage (although some fail to hatch and die as late embryos). From these observations we conclude that gfat1 and gfat2 are not functionally redundant. Indeed, it is noteworthy that loss of gfat1 was not compensated by gfat2 and, conversely, loss of gfat2 was not compensated by gfat1. Hence, with regard to developmental functions the action of each must produce unique activities essential for viability. These genetic findings could potentially be explained by non-overlapping expression patterns. For instance, gfat1 and gfat2 could be exclusively expressed in distinct vital tissues and, consistent with this, Dutta et al. showed that GFAT2 is the prevailing isoform in the fly midgut [17]. On the other hand, datasets available from modENCODE indicate that gfat1 and gfat2 share widely expressed patterns with many tissues in common [15]. Notably gfat1 is expressed at levels lower than gfat2 but, excluding the gonads and 3rd instar fat body (where gfat1 RNAs are absent) inspection of these datasets showed that both are present in the 25 anatomical sites reported [15].
As expected we observed reversal of gfat2 lethality using GlcN-6-P and, consistent with this, Mattila et al [18] found that dietary N-acetyl-D-glucosamine (GlcNAc) had a similar impact. An interesting distinction is that Mattila et al [18] delayed the lethal phase (from 1st instar to pupa) while, here, supplementation rescued viable adults that otherwise would have died as L1 larvae. Hence, although both HBP intermediates effectively restored HBP shortages in gfat2 flies, dietary GlcN-6-P could be a more effective supplement compared to GlcNAc [18]. In contrast, 1st instar larvae homozygous for gfat1 were surprisingly unaffected by dietary GlcN-6-P. This result might reflect barriers that prevent dietary GlcN-6-P from accessing tissue(s) where GFAT1 is essential, such as limited dispersion, inadequate absorption, compromised feeding or acute death after hatching. However, as outlined above, public datasets [15] are somewhat at odds with this prediction since they suggest that GFAT1 is often the less abundant counterpart and therefore, if anything, GFAT1 homozygotes should be easier to rescue with dietary GlcN-6-P compared to GFAT2 mutants. An alternative formal possibility is that, in addition to GlcN-6-P [14], GFAT1 might generate a non-canonical product. Though highly speculative, precedents for such promiscuous activity have been documented with other enzymes [19]. In this regard, it is worth noting that the gfat1 locus produces nine alternatively spliced transcripts, in contrast to gfat2, which produces a single RNA. The precise action of gfat1 remains to be determined and, in future studies, tissue specific analyses of intermediate HBP metabolites (as in [20]) may provide useful information.
Material and methods
Generating gfat1B2 and gfat2E1 using Crispr/Cas9
To generate the gfat1B2 and gfat2E1 alleles, guide RNAs were designed using http://tools.flycrispr.molbio.wisc.edu/targetFinder and synthesized as 5ʹ-unphosphorylated oligonucleotides, annealed, phosphorylated and ligated into the BbsI sites of pU6-BbsI-chiRNA plasmid [21]. Homology arms upstream of the 5' gRNA sequences, and downstream of the 3' gRNA sequences (for gfat1, 919bp 5' upstream and 908bp 3' downstream, for gfat2, 923bp 5ʹ upstream and 922bp 3ʹ downstream) were synthesized as gene blocks (IDT) and cloned into pHD-dsRed-attP [22] (Addgene). The sequences of the guide RNAs are:
GFAT1 5ʹgRNA: CAACTTGCGGCCGTATAATAAGG,
GFAT1 3ʹgRNA: ATGTGGAGATTATCATGTAGAGG;
GFAT2 5ʹgRNA: CTGATCTTACACTTCTGAGGCGG;
GFAT2 3ʹgRNA: AAATTCAGCTCATGAAGCGTGGG
Guide RNAs and the donor vector were co-injected into nosP Cas9 attP embryos at the following concentrations: 250 ng/µl pHD-dsRed-attP donor vector and 20 ng/µl of each of the pU6-BbsI-chiRNA plasmids containing the guide RNAs (Rainbow Transgenics Inc.). Mutant flies were identified using dsRed eyes as a marker, and subsequently verified by sequencing.
Lethal phase analyses and dietary supplementation tests
Lethality in earlier stages was imputed from the numbers of heterozygous and homozygous animals scored at later stages. Specifically, embryos laid by gfat1B2/TM6, Tb and gfat2E1/TM6, Tb adults were collected for 4 hours, half of the collection plates were aged at 25°C for 26 hours, and embryos on the other half of the collection plates were transferred to vials at 25°C. The numbers of unhatched embryos and empty egg cases were counted. Larvae were recovered from three separate vials and scored for Tb on each day from day 2 to day 5. After pupation, the numbers of Tb+ and Tb pupae and adults were counted. A minimum of 100 heterozygous adults were scored. For dietary supplementation, gfat1B2/TM6, Tb and gfat2E1/TM6, Tb flies were raised in fly food (Nutri-Fly Instant Formulation, Genesee Scientific, 66–118) containing the indicated concentrations of GlcN-6-P (Sigma-Aldrich, G5509). Homozygous larvae lacking the Tb marker on TM6 were scored. Homozygous pupae were transferred to a fresh vial, and assessed for eclosion rates. Deficiency and MiMIC strains used in the complementation testing were from Bloomington Drosophila Stock Centre.
GFAT activity assay
0–4 hr embryos were aged at 25°C for 24 hours. Hatched and unhatched larva, were homogenized with a pestle and lysed in enzyme assay buffer (20 mM Tris-Cl (pH 7.5), 2.5 mM CaCl2, 50 mM NaCl, 10 mM MgCl2,1 mM dithiothreitol, 10% glycerol and protease inhibitors) with 0.5% Triton X-100. After a 30 min incubation on ice, the lysates were spun at 18,000 RCF for 15 minutes. The resulting supernatants were transferred to a clean tube and used for the activity assay. The GFAT enzyme activity was determined by a spectrophotometric method as described [23], and adapted to a 96-well format. In 100 ul assay volumes, 30ug protein samples were incubated with 10 mM glutamine (Sigma-Aldrich, G7513), 10 mM fructose 6-phosphate (Sigma-Aldrich, F3627), 0.5 mM 3-acetylpyridine adenine dinucleotide (APAD) (Santa Cruz, sc-209519), and 2 units of glutamate dehydrogenase (Sigma-Aldrich, 10197734001) in enzyme assay buffer at 37°C for 1 h. The change in absorbance at 365 nm due to reduction of APAD to APADH was measured in separate duplicate samples over the 1 hr incubation time and standard deviations were calculated. For each lysate, 30ug of heat-inactivated protein (95°C for 10 minutes) was used as a control.
Acknowledgments
We thank Dr. Nicole L. Link and Dr. Oguz Kanca (BCM) for help with GFAT1 mutants, and Dr. Annika Wylie for intellectual contribution.
Funding Statement
This work was supported by grants from the NIH [R01GM072124, R01GM115682, R01CA222579] and the Cancer Prevention Research Institute of Texas [RP170086] to J.M.A.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Yang X, Qian K.. Protein O-GlcNAcylation: emerging mechanisms and functions. Nat Rev Mol Cell Biol. 2017;18(7):452–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Mattila J, Hietakangas V.. Regulation of carbohydrate energy metabolism in drosophila melanogaster. Genetics. 2017;207(4):1231–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ferrer CM, Sodi VL, Reginato MJ. O-GlcNAcylation in cancer biology: linking metabolism and signaling. J Mol Biol. 2016;428(16):3282–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Molinari M. N-glycan structure dictates extension of protein folding or onset of disposal. Nat Chem Biol. 2007;3(6):313–320. [DOI] [PubMed] [Google Scholar]
- [5].Dennis JW, Nabi IR, Demetriou M. Metabolism, cell surface organization, and disease. Cell. 2009;139(7):1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hardiville S, Hart GW. Nutrient regulation of signaling, transcription, and cell physiology by O-GlcNAcylation. Cell Metab. 2014;20(2):208–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446(7139):1017–1022. [DOI] [PubMed] [Google Scholar]
- [8].Hart GW, Slawson C, Ramirez-Correa G, et al. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu Rev Biochem. 2011;80:825–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mereiter S, Balmaña M, Campos D, et al. Glycosylation in the era of cancer-targeted therapy: where are we heading? Cancer Cell. 2019;36(1):6–16. [DOI] [PubMed] [Google Scholar]
- [10].Schimmelpfeng K, Strunk M, Stork T, et al. Mummy encodes an UDP-N-acetylglucosamine-dipohosphorylase and is required during Drosophila dorsal closure and nervous system development. Mech Dev. 2006;123(6):487–499. [DOI] [PubMed] [Google Scholar]
- [11].Tonning A, Helms S, Schwarz H, et al. Hormonal regulation of mummy is needed for apical extracellular matrix formation and epithelial morphogenesis in Drosophila. Development. 2006;133(2):331–341. [DOI] [PubMed] [Google Scholar]
- [12].Araujo SJ, Aslam H, Tear G, et al. mummy/cystic encodes an enzyme required for chitin and glycan synthesis, involved in trachea, embryonic cuticle and CNS development–analysis of its role in Drosophila tracheal morphogenesis. Dev Biol. 2005;288(1):179–193. [DOI] [PubMed] [Google Scholar]
- [13].Mariappa D, Sauert K, Marino K, et al. Protein O-GlcNAcylation is required for fibroblast growth factor signaling in Drosophila. Sci Signal. 2011;4(204):ra89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Graack HR, Cinque U, Kress H. Functional regulation of glutamine:fructose-6-phosphate aminotransferase 1 (GFAT1) of Drosophila melanogaster in a UDP-N-acetylglucosamine and cAMP-dependent manner. Biochem J. 2001;360(Pt 2):401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Thurmond J, Goodman JL, Strelets VB, et al. FlyBase 2.0: the next generation. Nucleic Acids Res. 2019;47(D1):D759–d765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Nakaishi Y, Bando M, Shimizu H, et al. Structural analysis of human glutamine:fructose-6-phosphate amidotransferase, a key regulator in type 2 diabetes. FEBS Lett. 2009;583(1):163–167. [DOI] [PubMed] [Google Scholar]
- [17].Dutta D, Dobson A, Houtz P, et al. Regional cell-specific transcriptome mapping reveals regulatory complexity in the adult drosophila midgut. Cell Rep. 2015;12(2):346–358. [DOI] [PubMed] [Google Scholar]
- [18].Mattila J, Kokki K, Hietakangas V, et al. Stem cell intrinsic hexosamine metabolism regulates intestinal adaptation to nutrient content. Dev Cell. 2018;47(1):112–121.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Intlekofer AM, Wang B, Liu H, et al. L-2-Hydroxyglutarate production arises from noncanonical enzyme function at acidic pH. Nat Chem Biol. 2017;13(5):494–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Turnock DC, Ferguson MA. Sugar nucleotide pools of trypanosoma brucei, trypanosoma cruzi, and leishmania major. Eukaryot Cell. 2007;6(8):1450–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gratz SJ, Cummings AM, Nguyen JN, et al. Genome engineering of drosophila with the CRISPR RNA-Guided Cas9 nuclease. Genetics. 2013;194(4):1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gratz SJ, Ukken FP, Rubinstein CD, et al. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in drosophila. Genetics. 2014;196(4):961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chang Q, Su K, Baker JR, et al. Phosphorylation of human glutamine:fructose-6-phosphate amidotransferase by cAMP-dependent protein kinase at serine 205 blocks the enzyme activity. J Biol Chem. 2000;275(29):21981–21987. [DOI] [PubMed] [Google Scholar]


