ABSTRACT
Pancreatic cancer (PC) is a leading cause of cancer mortality and is expected to continue increasing incidence. Abnormally expressed microRNAs have been demonstrated tightly correlated with the development and progression of PC. However, the molecular mechanisms remain largely unknown. In this study through combing both the TCGA database and our two verification PC cohorts, we found the consistent reduction of miR-3613-5p in PC tumors, which significantly correlated with reduced cumulative survival rate among PC patients. PC patients with higher lymph node metastasis rate show reduced miR-3613-5p expression. Through further mechanistic investigation, we demonstrate that miR-3613-5p down-regulated CDK6 in repressing the metastasis capacity of PC cells in vitro and in vivo. Elevated CDK6 were also found in PC samples, which also correlate with poor prognosis. Thus, our study found a novel tumor repressor miR-3613-5p in PC progression.
KEYWORDS: Pancreatic cancer, miR-3613-5p, CDK6, metastasis
Introduction
As one of the most common lethal malignancies, pancreatic cancer (PC) represents the fourth highest cause of cancer deaths worldwide, with a 5-year survival rate of only 7% [1,2]. Owing to our current inability to detect the disease in its early stages, most diagnosed patients miss the time window for curative surgery [3]. While PC patients have traditionally faced a dismal prognosis, advances in the diagnosis and treatment have shown a positive impact on the prognosis of this disease over the past several years [4]. The molecular pathogenesis underlying PC development and progression are largely unknown. Deeper understanding of molecular aberrations leading to PC development would, on the one hand, show promise for future developing targeted therapeutic agents, on the other hand, provide an instructive message on clinical PC patients’ classification. Instead of dictated by the routine schedule, high-risk patients may be followed up more frequently after surgery.
MicroRNAs (miRNAs) are approximately 22 nucleotides in length and represent a group of evolutionarily conserved, single-stranded non-coding RNAs [5]. Through binding to the 3′-untranslated regions (3′-UTRs) of target genes, they have been identified played important roles in tumor pathogenesis through targeting mRNAs for cleavage or translational repression [6]. Altered miRNAs have also been identified as an important mechanism leading to the development, progression, and chem-resistance of PC cells [7–9]. Helped by in vivo mouse model analysis, multiple miRNAs with their corresponding targets have confirmed implicate in modulating PC cell biological behavior like proliferation, invasion, and apoptosis [10–13]. For instance, growth-promoting miRNAs like miR-10b enhances epidermal growth factor-dependent invasion and proliferation of PC cells [14], and elevated miR-194 mediates migration, proliferation, and colony formation of PC cells.
In this study, through combing public databases and our PC verification cohorts, we found that miR-3613-5p is reduced in PC tumor samples and also correlated with poor prognosis of PC patients. Further combined with in vitro and in vivo experiments, we found that CDK6 as an important target gene regulated by miR-3613-5p in regulating PC cells migration and invasion. Our study adds an understanding of PC progression, which would provide insight into future PC treatment and prognostic evaluation.
Results
Reduced miR-3613-5p expression in PC correlated with poor tumor prognosis
To identify candidate miRNAs involved in PC pathogenesis, we first analyzed the correlation between miRNA (n = 2558) and tumor prognosis using TCGA database. The result shows that miR-1301-3p and miR-3613-5p are the ranking top two with the highest associations with PC prognosis (Figure 1(a,b)). As with the two miRNAs, miR-3613-5p exhibited a consistent correlation with the cumulative survival rate of PC patients in our two independent verification PC cohorts (Figure 1(c,d)). We also analyzed the expression of miR-3613-5p in both the public database including TCGA and GSE43797 and our verification PC cohort 1, of which the corresponding 80 normal pancreatic tissues were collected. Consistent reduction of miR-3613-5p in PC tumor samples as compared with corresponding normal pancreatic tissues was found (Figure 1(e,g)). Moreover, by analyzing the TCGA database, we found that tumors classified into T3-T4 stage, node positive (N1) and distant metastasis (M1) show reduced miR-3613-5p expression as compared with those in T1-T2 stage, node negative (N0) and no distant metastasis (M0) (Figure 1(h,j)). PC tumors in TNM II–IV stages also revealed lower miR-3613-5p expression as compared with those in TNM I stage (Figure 1(k)). Intriguingly, helped by our two verification cohorts, the correlation between miR-3613-5p and TNM stage and lymph node metastasis in verification PC cohort 1, and lymph node metastasis in verification PC cohort 2 were found (Figure 1(l)). In conclusion, the above results reduced miR-3613-5p expression in PC, which might be a favorable prognostic marker for PC patients.
Figure 1.
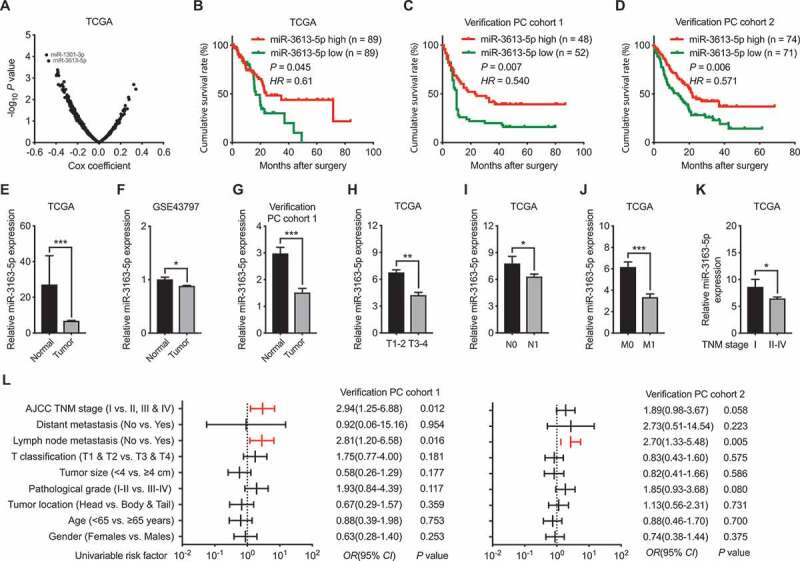
Correlations between miR-3613-5p and the prognosis of PC patients. (a) The correlation between miRNAs (n = 2558) expression with the prognosis of PC patients were analyzed using data from TCGA database. Two miRNAs (miR-1301-3p and miR-3613-5p) with the highest associations were identified. (b) Cumulative survival rate of PC patients from TCGA database analyzed according to the expression level of miR-3613-5p. (c-d) Cumulative survival rates of patients from our PC cohort 1 (c) and cohort 2 (d) with higher or lower miR-3613-5p expression (e-f) Relative miR-3613-5p expression in PC samples as compared with normal controls as analyzed using data from TCGA (e) and GSE43797 (f). (g). Reduced miR-3613-5p expression PC tumors as compared with normal control tissues in our PC cohort1. (h-k) miR-3613-5p expression level in PC samples from TCGA database as classified by T1-T2/T3-T4 (h), N0/N1 (i), M0/M1 (j), and TNM Ⅰ/TNM Ⅱ-Ⅳ (k). (l) Comparison of the TNM stages, lymph node metastasis, T classification, tumor size, tumor differentiation, tumor location, age and gender among the two PC cohorts according to the expression levels of miR-3613-5p. Bar, SEM. * P< 0.05, ** P< 0.01, *** P< 0.001; Student’s t test
miR-3613-5p inhibited PC cell migration and invasion in vitro and in vivo
To explore the role of miR-3613-5p in pancreatic cancer, miR-3613-5p mimic and inhibitor were transfected into the human pancreatic cancer cell lines AsPC-1 and SW1990 cells, respectively. Then, wound-healing assays were performed to determine the impact of miR-3613-5p on cell migration. The results revealed that while AsPC-1 and SW1990 cells with miR-3613-5p mimic transfection manifested a slower recovering rate than that of control cells, suppression of miR-3613-5p level by transfecting with its inhibitor in PC cells lead to faster wound closure rates than control cells (Figure 2(a,b)). Transwell assays showed that elevated miR-3613-5p inhibited PC cell invasion ability and miR-3613-5p reduction otherwise accelerated the invasive capacity of both AsPC-1 and SW1990 cells (Figure 2(c,d)). Finally, PC orthotopic tumor models were used to analyze the development of spontaneous metastasis influenced by miR-3613-5p in vivo. All mice of the control group were accompanied by liver metastasis, it was rarely detected in miR-3613-5p over-expression group (Figure 2(e,h)). Therefore, these findings suggested that downregulated miR-3613-5p in pancreatic cancer cells might help promoting PC cell metastasis.
Figure 2.
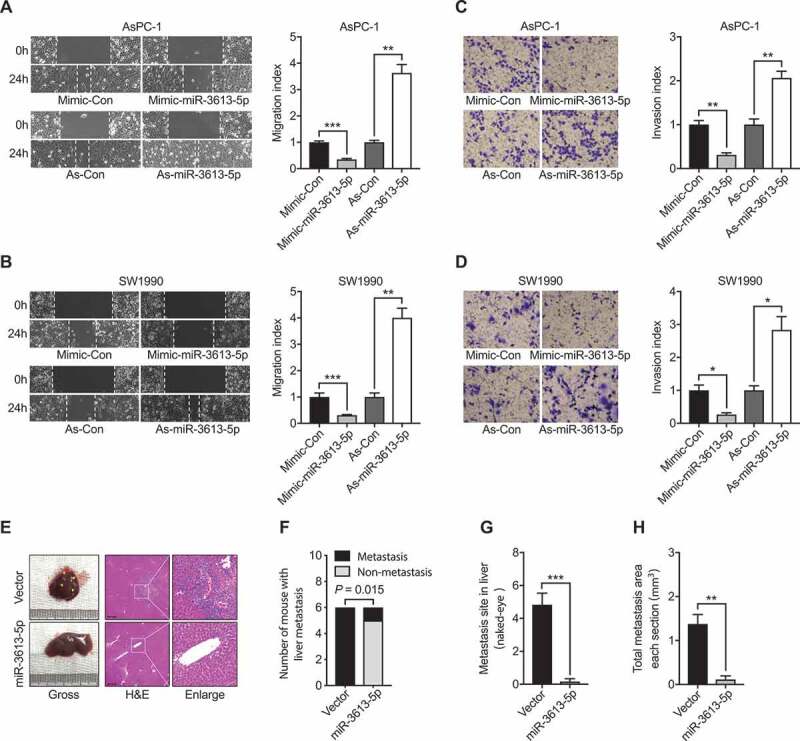
miR-3613-5p modulates the migration and invasion behavior of PC cells. (a-b) Wound-healing assays measured the effect of increasing (mimic-miR-3613-5p) or reducing (As-miR-3613-5p) miR-3613-5p on the migration ability of PC cells including AsPC-1 (a) and SW1990 (b). (c-d) Transwell assays measured the effect of modulating miR-3613-5p on the invasion capacity of PC cells in AsPC-1 (c) and SW1990 (d). (e) Representative images of macroscopic and H&E-stained livers of mice bearing AsPC-1 Control or miR-3613-5p overexpression orthotopic tumors. The yellow triangles indicate liver metastatic foci. Scale bars: 200 μm. (f-h) Quantification of liver metastatic mice (f) and colonies number (g) and area per section (h) originated from AsPC-1 Control or miR-3613-5p overexpression orthotopic tumors as described in E. All in vitro experiments, n = 3; All in vivo experiments, n = 6; bar, SEM; * P< 0.05, ** P< 0.01, *** P< 0.001; Student’s t test
CDK6 acted downstream of miR-3613-5p
To further elucidating the underlying mechanism of miR-3613-5p in suppressing metastasis of human pancreatic cancer cells, four online softwares (starBase, miRmap, TargetScan, and GEPIA) were analyzed to identify the possible target of miR-3613-5p. Five common genes were identified using the four softwares including CDK6, AP3S1, MBNL2, HEARTR5A, and EIF2AK2 (Figure 3(a)). Further verification in AsPC-1 and SW1990 cells we found that CDK6 is the only one gene that showed corresponding changes with varied miR-3613-5p expression. While overexpression miR-3613-5p using mimic-reduced CDK6 expression, antisense-miR-3613-5p on the other hand increased CDK6 level (Figure 3(b,c)). Similar results were obtained in SW1990 cells (Figure 3(d)). Further using western blot assay, we confirmed the regulatory role of miR-3613-5p on CDK6 (Figure 3(e)). Moreover, an inverse relationship between CDK6 and miR-3613-5p expression was also identified in our two PC cohorts by ISH and IHC (Figure 3(f,g)). Dual-luciferase reporter assay was used to confirm whether miR-3613-5p could directly target CDK6. We found that miR-3613-5p faithfully suppressed the luciferase activity in both AsPC-1 and SW1990 cells transfected with a reporter containing wild type 3′-UTR of CDK6 fused downstream of luciferase cDNA. However, this capacity was largely abolished as with cells transfected with a reporter containing mutated 3′-UTR of CDK6 (Figure 3(h,j)). Thus, the above results suggested that miR-3613-5p directly targeted CDK6 by specifically binding on the 3ʹ UTR of CDK6 in PC cells.
Figure 3.
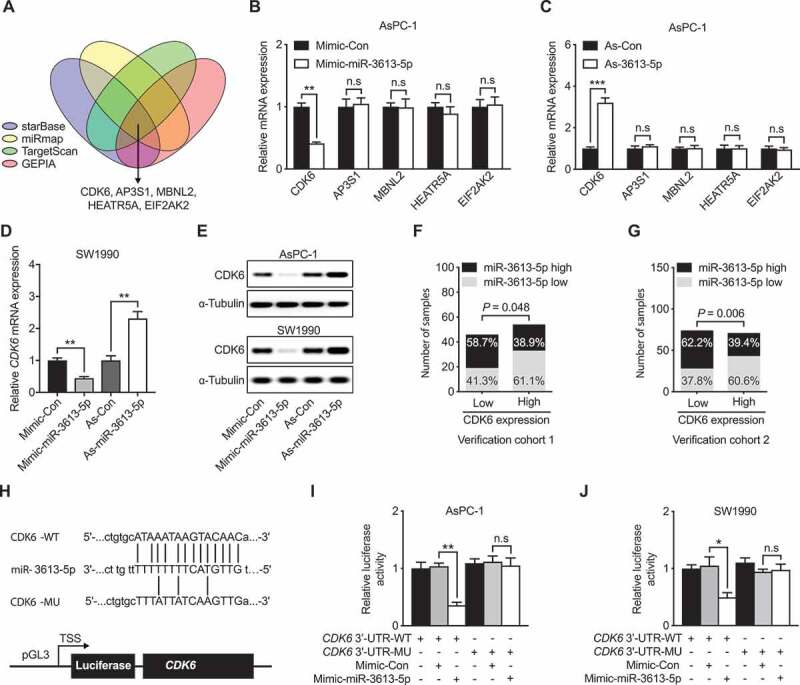
miR-3613-5p directly targets CDK6 and represses its expression. (a) Six common genes were shown could be targeted by miR-3613-5p using four software. (b-c) mRNA expression levels of the six common genes with miR-3656 mimic (b) or As-miR-3613-5p (c) addition in AsPC-1 cells. (d) CDK6 mRNA expression levels in SW1990 cells with miR-3656 mimic or As-miR-3613-5p addition. (e) The protein level of CDK6 in AsPC-1 or SW1990 cells with either miR-3656 mimic or As-miR-3613-5p addition. (f-g) Calculation of the percentages of patients with lower or higher miR-3613-5p expression according to CDK6 expression levels in our PC cohort1 (f) and cohort 2 (g). (h) Schematic diagram showing the predicted binding sequence between CDK6 and miR-3613-5p. The wild-type (WT) and mutated (MU) 3′-UTR of CDK6 were cloned into luciferase reporter constructs. (i-j) The influences of miR-3613-5p mimic addition on the luciferase activity of WT or MU 3′-UTR CDK6 reporter as analyzed in AsPC-1 (i) and SW1990 (j) cells. All n = 3; bar, SEM, n.s, not significant, * P< 0.05, ** P< 0.01, *** P< 0.001; Student’s t-test or χ2 test
miR-3613-5p regulated PC cell migration and invasion through CDK6
Previous study showed that CDK6 is a key factor in regulating cancer cell metastasis and invasion [15,16]. To examine the biological function of CDK6 in PC, the correlation between CDK6 expression and clinic-pathologic characteristics, and prognosis of PC patients were analyzed. Via IHC assay, we found that CDK6 protein was elevated in PC tissues compared to normal pancreatic tissues in our verification PC cohort 1 (Figure 4(a)). Correspondingly, poor prognosis was observed in PC patients with higher CDK6 protein expression (Figure 4(b-d)). Moreover, the correlation between increased CDK6 protein expression and higher lymph node metastasis rate was also identified in both two verification PC cohorts (Figure 4(e)).
Figure 4.
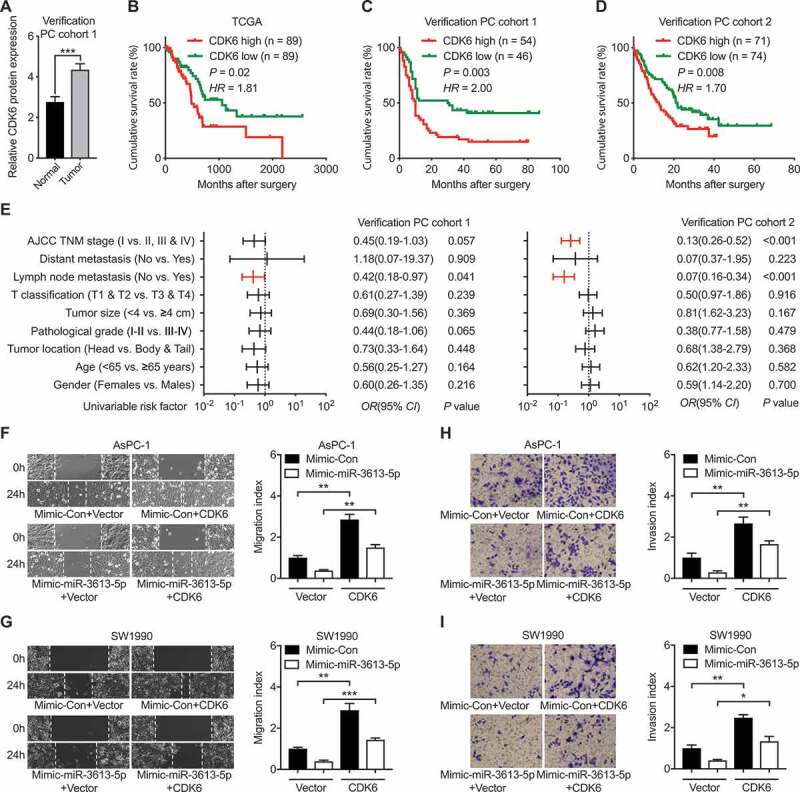
miR-3613-5p promoted PC cells metastasis by manipulating CDK6. (a) Elevated CDK6 expression in tumor samples from our PC cohort 1. (b-d) The correlation between CDK6 expression and the cumulative survival rate of PC patients from the TCGA database (b), PC cohort 1 (c) and PC cohort 2 (d). Data was analyzed by Kaplan-Meier analysis. Statistical significance was determined using the log-rank test. (e) Comparison of the TNM stages, lymph node metastasis, T classification, tumor size, tumor differentiation, tumor location, age and gender among our two PC cohorts according to the expression levels of CDK6. Statistical significance was performed using χ2 test. (f-g) Wound-healing assays measured the effect of CDK6 overexpression on the migration capacity of PC cells with mimic-miR-3613-5p addition. Data are shown for AsPC-1 (f) and SW1990 (g) cells. (h-i) CDK6 overexpression on the invasion capacity of PC cells with mimic-miR-3613-5p addition analyzed by transwell assays in AsPC-1 (h) and SW1990 (i). All n = 3; bar, SEM; * P< 0.05, ** P< 0.01, *** P< 0.001; Student’s t test
To investigate the function of the CDK6, we analyzed the metastatic ability of PC cells after CDK6 overexpression in vitro. Wound healing and transwell assays showed that PC cells with higher CDK6 expression revealed enhanced cell migration and invasion capacity (Figure 4(f-i)). To clarify the reliance of miR-3613-5p on CDK6 in modulating the metastasis behavior of PC cells, we then overexpressed CDK6 in concomitant with miR-3613-5p and then examined the migration and invasion capacity, respectively. Interestingly, CDK6 co-overexpression attenuated the tumor inhibitory role of miR-3613-5p as shown by increased PC cell migration and invasion in both AsPC-1 and SW1990 cells (Figure 4(f-i)). Taken together, these data imply that miR-3613-5p probably suppressed PC cell metastasis through regulating CDK6.
Discussion
In this work, we initially found that miR-3613-5p shows prominent correlations with PC patients' prognosis using data from TCGA database. Combing with our two PC cohorts, we verified the reduction of miR-3613-5p in PC tumors as compared with corresponding normal pancreatic tissues. Moreover, miR-3613-5p also shows a reduction in PC tumors with advanced stages like lymph node positive, suggesting miR-3613-5p probably served in inhibit the acquisition of malignant phenotype in PC cells. Further combing bioinformatics analysis and functional verification, we found CDK6 plays an indispensable role in mediating PC cells migration and invasion under miR-3613-5p. Taken together, our findings indicate that elevated CDK6 caused by lower miR-3613-5p in PC play important role in its malignant transformation.
Differentially expressed miR-3613-5p was initially found in the serum of subjects with endometriosis, chronic inflammatory, and estrogen-dependent disease, as compared with those without, and a combination of miR-3613-5p further improved diagnostic performance of endometriosis [17,18]. Later it was shown that in the plasma exosomes from mesial temporal lobe epilepsy with hippocampal sclerosis, miR-3613-5p possibly affects axon guidance through pathways like actin cytoskeleton, focal adhesion, the calcium-signaling pathway, the MAPK signaling pathway, and the PI3K-Akt signaling pathway [19]. Consistent with our results supporting the anti-tumor effects of miR-3613-5p in PC, a recent study also found that miR-3613-5p, in combination with six other miRNAs including miR-424-5p, miR-139-5p, miR-5586-5p, miR-126-3p, miR-454-3p, and miR-1271-5p in stratifying PC patients with into low- and high-risk groups [20]. Alongside the involvement of miR-3613-5p in PC, it was also shown as a favorable indicator in hepatocellular carcinoma clear cell renal cell carcinoma patients [21,22]. However, recent investigations on the molecular mechanism mediated miR-3613-5p are lacking and mostly based on correlation analysis. In our study, we found a direct regulatory role of miR-3613-5p on CDK6 expression, a star molecular marker in cancer prognosis. Elevated CDK6 were found in PC and also inversely correlated with miR-3613-5p expression. Elevating CDK6 phenocopies the migration and invasion inducing capacity of lowering miR-3613-5p as observed on PC cell lines. Importantly, CDK6 overexpression also efficiently compromised the role of miR-3613-5p overexpression on suppressing PC cells migration and invasion. Dysregulated activation of the CDK6 has been found in many kinds of cancers encompassing breast cancer, non-small cell lung cancer, and other solid tumors [23,24]. The combined usages of CDK4/6 inhibitor palbociclib with aromatase inhibitor letrozole have obtained benefits for breast cancer patients, which also harbor promising opportunities for the treatment of other types of cancers [16,25].
Overall, this study demonstrated that dysregulated miR-3613-5p was related to tumor metastasis of human PC. Downregulation of miR-3613-5p directly targets CDK6, leading to inhibit cell migration and invasion of PC cells. Therefore, miR-3613-5p/CDK6 axis could be identified as a novel therapeutic target future medical combination in PC treatment.
Materials and methods
Tissue samples
Tissue microarray of primary 100 PC and 80 normal pancreatic tissue samples (verification cohort 1) were obtained from Shanghai OutdoBiotech Ltd. Furthermore, a total of 145 formalin-fixed, paraffin-embedded PC tissues were retrieved from PC patients who underwent surgical resection from the department of pathology of Renji hospital. All the clinic-pathological features and follow-up information of the PC patients were collected. This study was approved by the Ethical Committee of Renji hospital, Shanghai Jiao Tong university school of medicine.
Cell culture and transfection
The two PC cell lines (AsPC-1 and SW1990) were purchased from ATCC and were cultured in DMEM medium containing 10% fetal bovine serum (Gibco, USA) and 1% antibiotics mixture of penicillin and streptomycin (Sigma, USA) in a humidified atmosphere of 5% CO2 at 37°C. The miR-3613-5p mimic (5ʹ-UGUUGUACUUUUUUUUUUGUUC-3ʹ), miR-3613-5p inhibitor (antisense, As) (5ʹ-GAACAAAAAAAAAAGUACAACA-3ʹ), mimic control (5ʹ-UUGUACUACACAAAAGUACUG-3ʹ), and inhibitor control (5ʹ-CAGUACUUUUGUGUAGUACAA-3ʹ) were purchased from GenePharma (Shanghai, China). CDK6 overexpression plasmid was obtained from MiaolingBio (Wuhan, China). RNAs and vectors were transfected into PC cells using Lipofectami-ne 2000 (Invitrogen, USA) following the manufacturer’s instruction. After 48 h of transfection, the cells were collected for subsequent experiments.
Cell migration and invasion assays
PC cell migration was assessed by wound-healing assays. Transfected cells grown to 90% in 6-well plates were scratched using a standard 200 μl pipette tip to create the wound. PC cells were washed to remove debris and grown for 24 h to allow wound closure. The wound area was tested and compared to the width at 0 h by using Image J software (NIH, USA). Cell invasion assays were conducted using Matrigel Invasion Chambers following the producer’s protocols (BD Biosciences, USA). Transfected cells were seeded into on the top side of the membrane pre-coated with matrigel (Corning, USA). After 24 h, the invasive PC cells were fixed with 4% paraformaldehyde and stained with 5% crystal violet for 20 min, respectively.
Quantitative PCR (qPCR)
RNA samples were extracted from cells using TRIzol reagent (Invitrogen, USA). After synthesizing cDNAs with miRNA and mRNA cDNA Synthesis Kit (Takara, Japan), PCR amplification was performed with SYBR Premix Ex Taq (Takara) and run with Applied Biosystems ViiA™ 7 Real-Time PCR System (Applied Biosystems, USA). Relative level was calculated by 2−ΔΔCT method [26] and presented relative to the level of GAPDH for CDK6 and rRNA18S for miR-3613-5p. The following primers were used: CDK6, forward: 5ʹ-GGATCTCTGGAGTGTTGGCT-3ʹ, reverse: 5ʹ- TGGTTGGGCAGATTTTGAATGA-3ʹ; GAPDH, forward: 5ʹ-AGCCACATCGCTCAGACAC-3ʹ, reverse: 5ʹ-GCCCAATACGACCAAATCC-3ʹ; miR-3613-5p, forward: 5′-CTTGTTTTTTTTTTCATGTTGT-3′, reverse: 5ʹ-AGTCTCAGGGTCCGAGGTATTC-3ʹ; rRNA18S, forward: 5′-ATTCCGATAACGAACGAGAC-3′, reverse: 5′-TCACAGACCTGTTATTGCTC-3′;
Western blot
Cells were lysed for isolating total protein using RIPA buffer supplemented with a proteinase inhibitor cocktail (Beyotime, China). Protein concentration was quantified by BCA method. Standard Western blot procedure, and the Bio-Rad ChemiDoc MP imaging system (Hercules, USA) were used according to the method described previously [27]. Bands were exposed by electrochemiluminescence (Pierce, USA) and analyzed by Image J Software. The primary antibodies used were as follows: CDK6 (1:500, ab124821, Abcam, UK) and α-Tubulin (1:2000, ab18251, Abcam).
Dual-luciferase reporter gene assay
PC Cells were co-transfected with CDK6-3ʹ-UTR WT/CDK6-3ʹ-UTR MU and miR-3613-5p mimics/control for 24 h. Luciferase activity was finally determined using the dual-luciferase reporter assay system (Promega, USA) according to our procedure described previously [27].
In situ hybridization (ISH) and immunohistochemistry (IHC) assay
ISH was used to detect miR-3613-5p using digoxigenin-labeled antisense probes and IHC was used to detect CDK6 using its primary antibodies in PC tissue microarray. The ISH and IHC staining of PC tissue and its scoring system were performed as described previously [27]. The probes and primary antibodies used were as follows: miR-3613-5p probe sequences, 5ʹ-GAACAAAAAAAAAAGUACAACA-3ʹ; CDK6 antibody (1:300, Cat. #ab124821, Abcam).
Orthotopic tumor model
The human miR-3613-5p construct was created by inserting the sequence of miR-3613-5p into the pCDH-CMV-MCS-EF1-copGFP vector (System Biosciences, USA). AsPC-1 cells stably overexpressing miR-3613-5p were created according to a previously described [27]. Then, 20,000 AsPC-1 cells (transfected with control or miR-3613-5p overexpressing vector) were injected (50 μl, 1:1 PBS:Matrigel) into the tail of the pancreas via a small abdominal incision in the left flank of anesthetized male BALB/c athymic nude mice (4-week-old, each group, n = 6). Tumor growth was weekly monitored by palpation under isoflurane anesthesia in 1 month's time. After 1 month, all mice were sacrificed, and the livers were excised to count the number of metastases, which were also fixed in 10% formalin for further histological quantifications of metastases. All animal studies were conducted strictly in accordance with the guidelines of the Animal Care and Use Committee of Shanghai Jiao Tong University.
Bioinformatics analyses
Prediction of miRNA target genes was performed on starBase [28], miRmap [29], and TargetScan [30]. TCGA data download and prognosis analysis was performed on Broad GDAC Firehose and GEPIA [31].
Statistics analysis
Data were expressed as mean ± standard error of mean (SEM). Group comparisons of were performed with unpaired Student’s t test (two-tailed). Dichotomous variables were compared using χ2 test. Survival probabilities were estimated based on Kaplan-Meier method and analyzed by the log-rank test. P < 0.05 considered the difference was statistically significant. All statistical analysis was used with SPSS17.0 software (IBM, USA).
Funding Statement
This work was supported by the National Natural Science Foundation of China (81803014, 81802424, 81802937, 81600406, and 81602043).
Abbreviations
PC: pancreatic cancer; miRNA: microRNA; 3′-UTR: 3′-untranslated region; qPCR: quantitative PCR, ISH: in situ hybridization; IHC, immunohistochemistry; SEM: standard error of mean.
Data availability statement
Online PC microarray data sets GSE43797, TCGA-PAAD and corresponding clinical data in this study were directly downloaded from GEO database (http://www.ncbi.nlm.nih.gov/geo/) and the open-access tiers of TCGA data portal (https://gdc-portal.nci.nih.gov/).
Declarations
This study was approved by the Ethical Committee of Renji hospital, School of Medicine, Shanghai Jiao Tong University. And all patients involved in this study provided written informed consent.
Disclosure statement
No potential conflicts of interest were disclosed.
References
- [1].Neoptolemos JP, Kleeff J, Michl P, et al. Therapeutic developments in pancreatic cancer: current and future perspectives. Nat Rev Gastroenterol Hepatol. 2018;15:333–348. [DOI] [PubMed] [Google Scholar]
- [2].Siegel RL, Miller KD, Jemal A.. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. [DOI] [PubMed] [Google Scholar]
- [3].Kamisawa T, Wood LD, Itoi T, et al. Pancreatic cancer. Lancet. 2016;388:73–85. [DOI] [PubMed] [Google Scholar]
- [4].Garrido-Laguna I, Hidalgo M.. Pancreatic cancer: from state-of-the-art treatments to promising novel therapies. Nat Rev Clin Oncol. 2015;12:319–334. [DOI] [PubMed] [Google Scholar]
- [5].McGeary SE, Lin KS, Shi CY, et al. The biochemical basis of microRNA targeting efficacy. Science. 2019;366(6472):eaav1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol. 2019;20:5–20. [DOI] [PubMed] [Google Scholar]
- [7].Liu D, Song L, Dai Z, et al. MiR-429 suppresses neurotrophin-3 to alleviate perineural invasion of pancreatic cancer. Biochem Biophys Res Commun. 2018;505:1077–1083. [DOI] [PubMed] [Google Scholar]
- [8].Schultz NA, Andersen KK, Roslind A, et al. Prognostic microRNAs in cancer tissue from patients operated for pancreatic cancer–five microRNAs in a prognostic index. World J Surg. 2012;36:2699–2707. [DOI] [PubMed] [Google Scholar]
- [9].Yang RM, Zhan M, Xu SW, et al. miR-3656 expression enhances the chemosensitivity of pancreatic cancer to gemcitabine through modulation of the RHOF/EMT axis. Cell Death Dis. 2017;8:e3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Qadir MI, Faheem A. miRNA: a diagnostic and therapeutic tool for pancreatic cancer. Crit Rev Eukaryot Gene Expr. 2017;27:197–204. [DOI] [PubMed] [Google Scholar]
- [11].Weidle UH, Birzele F, Nopora A. Pancreatic ductal adenocarcinoma: microRNAs affecting tumor growth and metastasis in preclinical in vivo models. Cancer Genomics Proteomics. 2019;16:451–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhang Y, Tang X, Shi M, et al. MiR-216a decreases MALAT1 expression, induces G2/M arrest and apoptosis in pancreatic cancer cells. Biochem Biophys Res Commun. 2017;483:816–822. [DOI] [PubMed] [Google Scholar]
- [13].Zhu W, Wang Y, Zhang D, et al. MiR-7-5p functions as a tumor suppressor by targeting SOX18 in pancreatic ductal adenocarcinoma. Biochem Biophys Res Commun. 2018;497:963–970. [DOI] [PubMed] [Google Scholar]
- [14].Ouyang H, Gore J, Deitz S, et al. microRNA-10b enhances pancreatic cancer cell invasion by suppressing TIP30 expression and promoting EGF and TGF-beta actions. Oncogene. 2017;36:4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Liu T, Yu J, Deng M, et al. CDK4/6-dependent activation of DUB3 regulates cancer metastasis through SNAIL1. Nat Commun. 2017;8:13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Patnaik A, Rosen LS, Tolaney SM, et al. Efficacy and safety of abemaciclib, an inhibitor of CDK4 and CDK6, for patients with breast cancer, non-small cell lung cancer, and other solid tumors. Cancer Discov. 2016;6:740–753. [DOI] [PubMed] [Google Scholar]
- [17].Cosar E, Mamillapalli R, Ersoy GS, et al. Serum microRNAs as diagnostic markers of endometriosis: a comprehensive array-based analysis. Fertil Steril. 2016;106:402–409. [DOI] [PubMed] [Google Scholar]
- [18].Cosar E, Mamillapalli R, Moridi I, et al. Serum MicroRNA biomarkers regulated by simvastatin in a primate model of endometriosis. Reprod Sci. 2019;26:1343–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yan S, Zhang H, Xie W, et al. Altered microRNA profiles in plasma exosomes from mesial temporal lobe epilepsy with hippocampal sclerosis. Oncotarget. 2017;8:4136–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bai X, Lu D, Lin Y, et al. A seven-miRNA expression-based prognostic signature and its corresponding potential competing endogenous RNA network in early pancreatic cancer. Exp Ther Med. 2019;18:1601–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Qin L, Huang J, Wang G, et al. Integrated analysis of clinical significance and functional involvement of microRNAs in hepatocellular carcinoma. J Cell Physiol. 2019;234:23581–23595. [DOI] [PubMed] [Google Scholar]
- [22].Qin S, Shi X, Wang C, et al. Transcription factor and miRNA interplays can manifest the survival of ccRCC patients. Cancers (Basel). 2019;11(11):1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sherr CJ, Beach D, Shapiro GI. Targeting CDK4 and CDK6: from discovery to therapy. Cancer Discov. 2016;6:353–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yang C, Li Z, Bhatt T, et al. Acquired CDK6 amplification promotes breast cancer resistance to CDK4/6 inhibitors and loss of ER signaling and dependence. Oncogene. 2017;36:2255–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lim JS, Turner NC, Yap TA. CDK4/6 inhibitors: promising opportunities beyond breast cancer. Cancer Discov. 2016;6:697–699. [DOI] [PubMed] [Google Scholar]
- [26].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- [27].Long M, Zhan M, Xu S, et al. miR-92b-3p acts as a tumor suppressor by targeting Gabra3 in pancreatic cancer. Mol Cancer. 2017;16:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hsu SD, Chu CH, Tsou AP, et al. miRNAMap 2.0: genomic maps of microRNAs in metazoan genomes. Nucleic Acids Res. 2008;36:D165–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Online PC microarray data sets GSE43797, TCGA-PAAD and corresponding clinical data in this study were directly downloaded from GEO database (http://www.ncbi.nlm.nih.gov/geo/) and the open-access tiers of TCGA data portal (https://gdc-portal.nci.nih.gov/).


