ABSTRACT
Stem cell and tissue engineering-based therapies for acute liver failure (ALF) have been limited by the lack of an optimal cell source. We aimed to determine the suitability of human parthenogenetic embryonic stem cells (hPESCs) for the development of strategies to treat ALF. We studied the ability of human parthenogenetic embryonic stem cells (hPESCs) with high whole-genome SNP homozygosity, which were obtained by natural activation during in vitro fertilization (IVF), to differentiate into functional hepatocyte-like cells in vitro by monolayer plane orientation. hPESCs were induced on a single-layer flat plate for 21 d in complete medium with the inducers activin A, FGF-4, BMP-2, HGF, OSM, DEX, and B27. Polygonal cell morphology and binuclear cells were observed after 21 d of induction by using an inverted microscope. RT-qPCR results showed that the levels of hepatocyte-specific genes such as AFP, ALB, HNF4a, CYP3A4, SLCO1B3, and ABCC2 significantly increased after induction. Immunocytochemical assay showed CK18 and Hepa expression in the induced cells. Indocyanine green (ICG) staining showed that the cells had the ability to absorb and metabolize dyes. Detection of marker proteins and urea in cell culture supernatants showed that the cells obtained after 21 d of induction had synthetic and secretory functions. The typical ultrastructure of liver cells was observed using TEM after 21 d of induction. The results indicate that naturally activated hPESCs can be induced to differentiate into hepatocellular cells by monolayer planar induction.
KEYWORDS: Human parthenogenetic embryonic stem cells, homozygosity, hepatic differentiation, acute liver failure
Introduction
Acute liver failure (ALF) is a serious threat to human health and is associated with high mortality. Currently, the main clinical strategies to treat ALF are pharmaceutical treatment, artificial liver support, and liver transplantation, which remains the most effective way to treat ALF. However, several challenges are associated with transplantation, including organ source, safety, and cost of treatment. Embryonic stem cells (ESCs), which have unlimited renewal and development capacities, are one of the main sources of artificial liver cells and could be used in novel treatments for ALF.1–2 However, the expression of genes from both parents in ESs results in heterozygous human leucocyte antigen (HLA) sites, which could lead to immune rejection, thereby limiting the clinical application of ES-derived cell/tissue engineering.
Human parthenogenetic embryonic stem cell (hPESC) lines are embryonic stem cell lines derived from a single gamete, which only expresses maternal imprinted genes. The use of hPESCs eliminates the ethical complications associated with the use of human embryonic stem cells (hESCs). Further, hPESCs are capable of infinite self-renewal and totipotency and are derived from haploids activated by parthenogenesis and homozygous alleles, which could prevent immune rejection.3 Parthenogenetic activation could be natural or artificial. Whole-genome SNP distribution indicates that hPESCs obtained by natural activation in IVF are mostly homozygous, and better than hPESCs obtained from artificial parthenogenetic activation.
To our knowledge, there have been no studies on the directional differentiation of naturally activated hPESCs into hepatocytes. The aim of this study was to determine whether directional differentiation of hPESCs could yield hepatocytes that satisfy the requirements of clinical cell transplantation.
Materials and methods
Cell culture
hPESCs (cell line chHES-32) were obtained from the National Engineering Research Center of Human Stem Cells (Changsha, China). The passage number of the hPESCs used in this study ranged from 35 to 38. Human foreskin fibroblasts (hFFs) were obtained by primary isolation and culture, and hPESCs were successfully cultured in vitro on these hFFs, which served as the feeder layer,4 in hPESC medium (DMEM/F12 medium supplemented with 15% knockout serum replacement, 1% Glutamax, 1% nonessential amino acids, 0.1 mM β-mercaptoethanol [all from Invitrogen/Gibco], and 4 ng/ml bFGF [R&D System, MN, USA]), under a standard gas atmosphere of humidified air with 5% CO2. Karyotype analysis indicated that they had a normal karyotype.
HepG2 cells were cultured in DMEM complete medium (Gibco) containing 10% fetal bovine serum (Gibco) under the same conditions as the hPESCs.
Hepatic differentiation
hPESCs were divided into group A (experimental group) and group B (control group). Hepatic differentiation was induced in group A as follows: hPESCs were cultured in DMEM/F12 medium supplemented with 100 ng/ml Activin A (R&D System, MN, USA) for 2 d. On the following 4 d, FGF-4 (20 ng/ml; R&D System, MN, USA) and BMP-2 (10 ng/ml; R&D System, MN, USA) were added to this medium. For the next 6 d, the differentiated cells were cultured in medium containing HGF (R&D System, MN, USA) at a final concentration of 20 ng/ml. The differentiated cells were matured in medium containing OSM (20 ng/ml; Sigma) and DEX (100 nM; Sigma) for 5 d, and further differentiated and maintained in medium supplemented with B27 (2%; GIBCO, NY, USA), OSM (10 ng/ml), and Dex (100 nM) on the last 4 d. In group B, DMEM/F12 complete medium without any inducing factors was used.
Real-time RT-PCR
HepG2 cells and cell samples from groups A and B were collected 0, 2, 9, 16, and 21 d after induction of differentiation. Total RNA was isolated from cells by using TRIzol Reagent (Invitrogen) and reverse-transcribed using the RevertAid First Strand cDNA Synthesis Kit (Thermo) according to the manufacturers’ protocols.
Real-time RT-PCR analysis was performed on an ABI Prism 7500 Sequence Detection System using the SYBR® Premix Ex Taq™ II (Takara, Japan). Primer information is shown in Table 1. The relative expression was calculated using the 2−ΔΔCT method.
Table 1.
List of primers used for RT-qPCR
| Gene name | Primer Sequence(5ʹ-3ʹ) |
|---|---|
| β-actin | F- TGGCACCCAGCACAATGAA |
| R- CTAAGTCATAGTCCGCCTAGAAGCA | |
| SOX17 | F- CTGCAGGCCAGAAGCAGTGTTA |
| R- CCCAAACTGTTCAAGTGGCAGA | |
| GSC | F-ACCTCCGCGAGGAGAAAGTG |
| R- GACGACGACGTCTTGTTCCA | |
| AFP | F- TGCAGCCAAAGTGAAGAGGGAAGA |
| R-CATAGCGAGCAGCCCAAAGAAGAA | |
| ALB | F-GAGACCAGAGGTTGATGTGATG |
| R-AGTTCCGGGGCATAAAAGTAAG | |
| HNF4α | F-TGCGACTCTCCAAAACCCTC |
| R-ATTGCCCATCGTCAACACCT | |
| CYP3A4 | F-GTGTGGGGCTTTTATGATGGTC |
| R-AAGGCCTCCGGTTTGTGAAG | |
| CYP3A7 | F-GCATGAGGTTTGCTCTCGTG |
| R-CAGGCTCCACTTACGGTCTC | |
| SLCO1B3 | F- TGGAAGGGTCTACTTGGGCT |
| R-TTCATTGTCCGATGCCTTGGT | |
| SLC10A1 | F-GCCATGAAGGGGGACATGAA |
| R-ATCATGCCAAGGGCACAGAA | |
| ABCC2 | F-CCTTGGGCTACCTATGGCTC |
| R-TCTGTGAGTACAAGGGCCAG | |
| ABCB4 | F-TCTTTCAAGCAGTAGCCACG |
| R-GCCGAGAGTATCTTTGCCCA |
Immunocytochemical (ICC) assay
After discarding the cell culture medium, cells were washed once with PBS, fixed with ice-cold acetone, permeabilized with 1% Triton X-100 and washed with PBS, blocked with peroxidase-blocking reagent and washed with PBS, incubated at room temperature with CK18 and Hepa primary antibody working solution for 55 minutes, incubated with secondary antibody for 25 minutes, washed, DAB stained, and photographed after hematoxylin re-staining. The expression of CK18 and Hepa in human fetal liver tissue was used as a positive control.
ICG uptake assay
Cells were incubated with 1 mg/mL ICG at 37°C for 60 minutes, and the color of the cells was observed under the microscope. The cells were washed with PBS, placed in complete culture medium for 60 minutes, and photographed to determine the ability to absorb and metabolize dyes.
Analysis of cell culture supernatant
After 21 d of induction, cell culture supernatants from each group were sent to the Laboratory of the First People’s Hospital of Yunnan Province for detection of AFP content by chemiluminescence, ALB content by the bromocresol green method, and urea content by the enzymatic method.
Periodic acid-schiff (PAS) staining for glycogen
The PAS staining system was purchased from Solarbio (Beijing, China). HepG2 and cell samples from groups A and B on the 21st day of induction were collected. Culture dishes containing cells were fixed in 4% paraformaldehyde, and the assay was performed according to the manufacturer’s instructions.
TEM detection
After low-speed centrifugation, the induced cells were collected. After glutaraldehyde immobilization, the induced cell samples were sent to the Electron Microscope Room of the Scientific Research Laboratory Center of Kunming Medical University, to be examined by transmission electron microscopy (TEM) to detect typical structural characteristics of hepatocytes. Human normal liver tissue was used as a positive control.
Statistical analysis
Quantitative data were obtained in triplicate from at least three independent donors and presented as means ± standard error and inferential statistics (p-values). Statistical significance was evaluated using paired two-tailed Student’s t-tests. P < .05 was considered statistically significant.
Results
Effect of induced differentiation on cell morphology
The hPESCs (chHES-32) used in this study are derived from naturally activated parthenogenetic blastocysts, and their pluripotency has been reported previously.4,5 After 40 passages, the hPESCs maintained a normal diploid karyotype of 46, XX (Figure 1a). Under an inverted microscope, the undifferentiated hPESCs appeared closely arranged and grew in a nested mass (Figure 1b). After 4 d of differentiation, the cells became loose (Figure 1c). On the seventh day of differentiation, the original small, compact, round cell morphology changed significantly, and the cells began to acquire a polygonal shape (Figure 1d–e). From the 11th day after differentiation, the cell volume increased further, and most of the cells showed the epithelial morphology of hepatocytes, with the occasional appearance of fibroblastic morphology (Figure 1f–g). By the 21st day of differentiation, the cells became uniform and were arranged in an epithelioid manner, with a few binucleate cells (Figure 1h).
Figure 1.
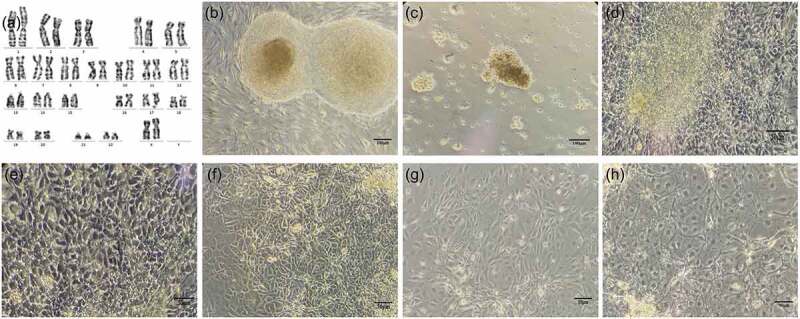
Morphological changes in hPESCs during induced differentiation. (a) After 40 passages, hPESCs maintained a normal diploid karyotype of 46, XX. (b) Undifferentiated hPESCs showed dense nest-like growth and clear contour. (c) On the fourth day of induction, the cells were scattered in small clumps at the bottom of the dish. (d–h) cell morphology on the 7th, 9th, 11th, 14th, and 21st day of induction, respectively. Cell morphology gradually changed to polygonal and epithelioid. At the end of differentiation, binucleate cells were occasionally observed
Effect of induced differentiation on gene expression
In order to confirm the stable expression of hepatocyte genes, we used qRT-PCR to analyze the expression of the endoderm genes SOX17 and Gsc,6,7 and hepatocyte genes AFP, ALB, HNF4A, cytochrome P450 3A4 (CYP3A4), cytochrome P450 3A7 (CYP3A7), SLCO1B3, SLC10A1, ABCC2, and ABCB4 (Figure 2). Because these genes are minimally expressed in hPESCs, undifferentiated hPESCs were selected as a reference. The experimental group was designated as group A, and relative gene expression in this group on d 0, 2, 9, 16, and 21 of induction were recorded as A0, A2, A9, A16, and A21, respectively. The control group was designated as group B, and relative gene expression level in this group on d 0, 2, 9, 16, and 21 was recorded as B0, B2, B9, B16, and B21, respectively. Gene expression levels in HepG2 cells were used as positive controls; although HepG2, a cancer cell line, is not an ideal representative of liver cells, it is widely used as a cellular liver model. SOX17 (Figure 2a) and GSC (Figure 2b) expression was negligible in the induced cells until d 2, increased sharply on d 2, then decreased and remained stable. This suggests that endoderm gene expression is the highest on the second day of induction, which is conducive to the next step of differentiation to hepatocytes. In the control group, there was almost no SOX17 and GSC expression until d 2, and the expression increased significantly on the second day and remained stable until the end of the experiment. This showed that the bFGF and hPESC culture system on a feeder layer could effectively inhibit the spontaneous differentiation of hPESCs, which is consistent with previous reports.
Figure 2.
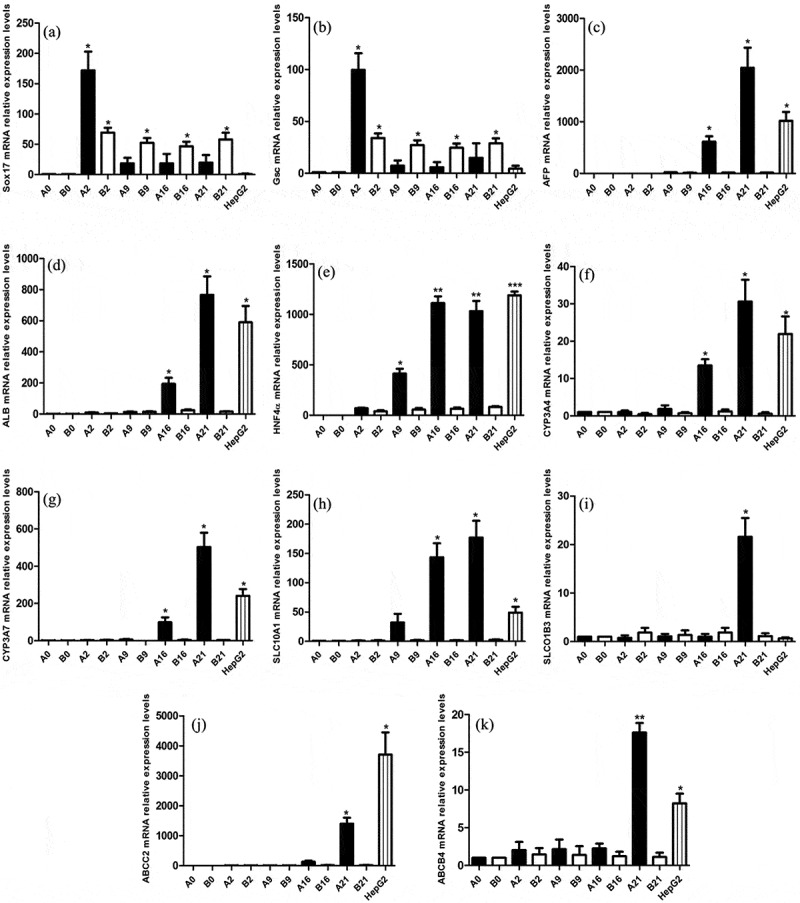
Changes in endoderm and hepatocyte marker expression during induced differentiation. qRT-PCR analysis of the endodermic marker genes Sox17 and Gsc, and the hepatocellular signature genes AFP, ALB, HNF4a, CYP3A4, CYP3A7, SLC10A1, SLCO1B3, ABCC2, and ABCB4. β-Actin was used as an internal control. A0, A2, A9, A16, and A21 indicate samples from the experimental group at d 0, 2, 9, 16 and 21 of differentiation respectively. Samples of the control group at the corresponding time points are indicated with the letter “B”. HepG2 was the positive control. Values are expressed as means ± S.E.M. of three independent experiments. *p < .05, **p < .001 compared to control
In the experimental group, the expression of HNF4α (Figure 2e), a common hepatic lineage marker gene, increased significantly from the ninth day, and subsequently remained high. The primary hepatocyte marker AFP (Figure 2c), mature hepatocyte marker ALB (Figure 2d), metabolic enzyme genes CYP3A4 (Figure 2f), and CYP3A7 (Figure 2g), and liver cells ingesting transportation-related protein SLC10A1 (Figure 2h) showed almost no expression before d 16. Their expression levels in the experimental group increased after induction for 16 d and reached the highest level on d 21. Slco1b3 (Figure 2i), ABCC2 (Figure 2j), and ABCB4 (Figure 2k), which are important transporter genes in hepatocytes, were found to be significantly expressed at the last stage of induction.
Expression of hepatocyte marker proteins in induced cells
We detected CK18 and HEPA expression by using immunocytochemistry (Figure 3), with fetal liver as a positive control (Figure 3c–d). Positive staining (yellow or brownish yellow) for CK18 (Figure 3a) and HEPA (Figure 3b) was observed in the cytoplasm of the experimental group. No positive staining (Figure 3e–f) was observed in the control group.
Figure 3.
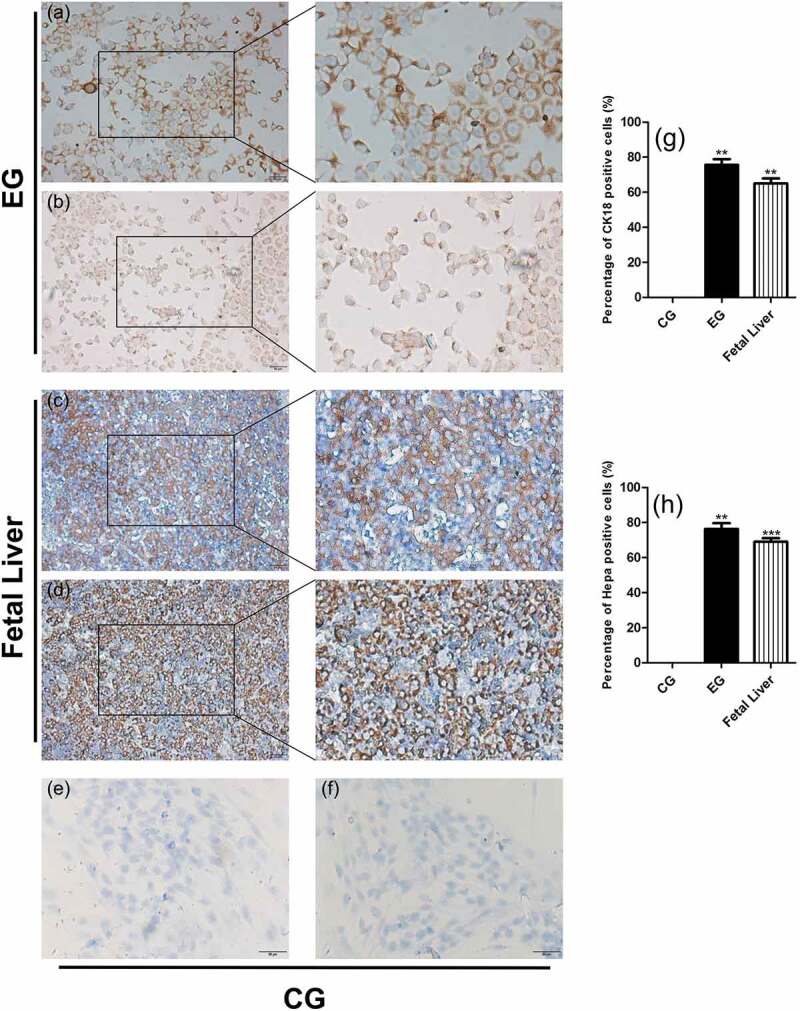
Expression of hepatocyte marker proteins CK18 and Hepa in induced cells on d 21. CG represents the control group, EG represents the experimental group, and fetal liver tissue was used as the positive control. (a) CK18 staining in the experimental group. (b) Hepa staining in the experimental group. (c) CK18 staining in fetal liver. (d) Hepa staining in fetal liver. (e) CK18 staining in the control group. (f) Hepa staining in the control group. (g) Proportion of CK18-positive cells. (h) Proportion of Hepa-positive cells. Values were obtained from three independent experiments, and the two-tailed test was used. *p < .05, **p < .001 compared to control. Bars: 50 μm
Cell function after induction of differentiation
To detect hepatocyte function in cells undergoing 21 d of directional induction and differentiation, we studied the absorption and excretion of ICG, glycogen synthesis, and AFP, ALB, and urea content in the cell supernatants. ICG uptake and metabolism are usually used as an index of liver-cell-specific function.8 We observed ICG-positive cells (in which the cytoplasm turned green) after ICG dye was added to the culture medium, indicating that the cells had the ability to absorb ICG dye. After the cells were washed and placed in normal medium for 1 hour, the original color of the cytoplasm was recovered (Figure 4a–b). Thus, the cells had the ability to discharge the ICG dye. Normal hepatocytes can synthesize and accumulate glycogen, and cellular glycogen appears red after PAS staining. After 21 d of induction, a large number of hepatocyte-like cells with red cytoplasm were observed after PAS staining (Figure 6a–b). This suggests that the induced cells could perform glycogen synthesis and storage.
Figure 4.
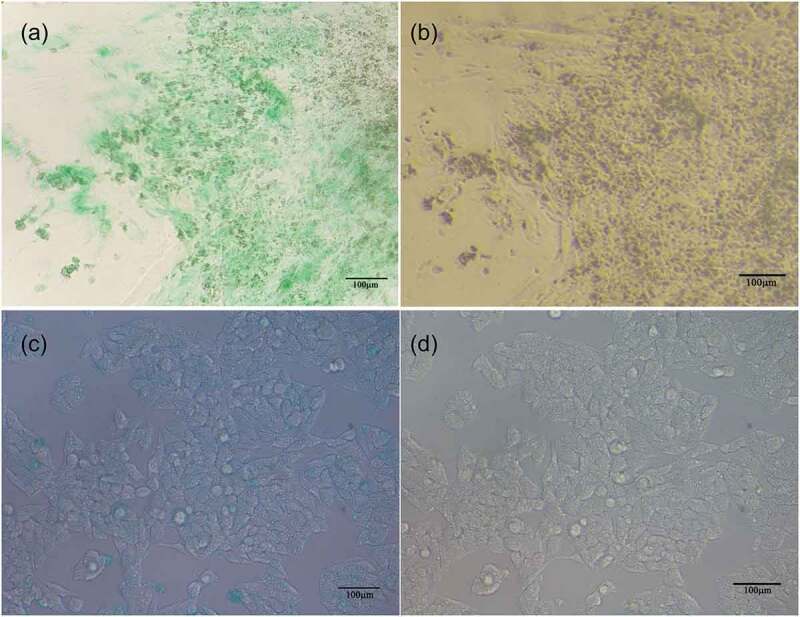
Absorption and excretion of ICG dye in induced cells on d 21. Hepatoid cells obtained by induced differentiation were able to (a) absorb and (b) expel ICG. (c) and (d) represent the absorption and discharge of ICG, respectively
Figure 6.
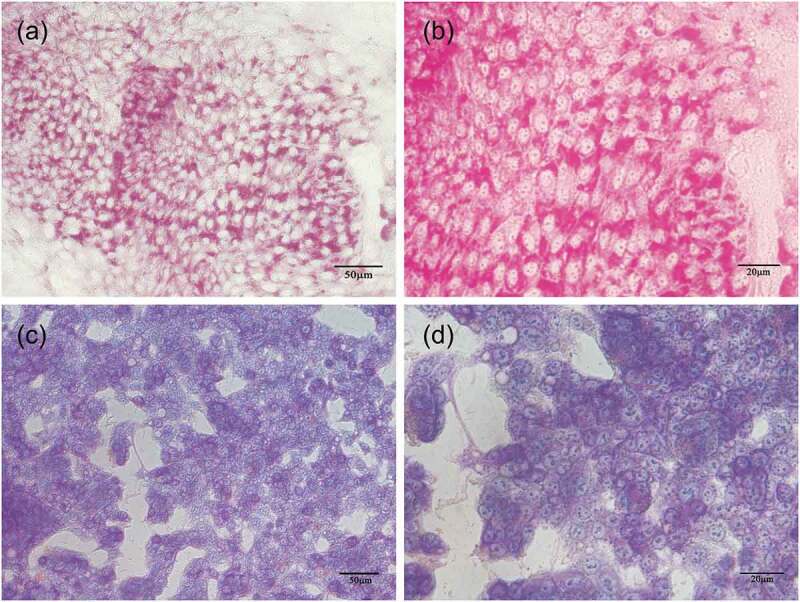
Glycogen synthesis ability in hPESCs after 21 d of induced differentiation. (a) PAS staining of hepatoid cells after induced differentiation. (b) enlarged image of (a). (c) PAS staining of HepG2. (d) enlarged image of (c)
Normal hepatocytes have the ability to synthesize and secrete ALB and urea. We found that the Alb (Figure 5b) and urea (Figure 5c) content was significantly higher in the culture supernatant of induced hepatocytes than in the control group and in the supernatant of HepG2 cells, which may be because HepG2 originated from cancer cell lines and the genome instability leads to the decline of liver cell function. However, the AFP content in the experimental group was also much higher than that in the control group (Figure 5a), indicating that the hepatocyte-like cells obtained resemble fetal hepatocytes.
Figure 5.
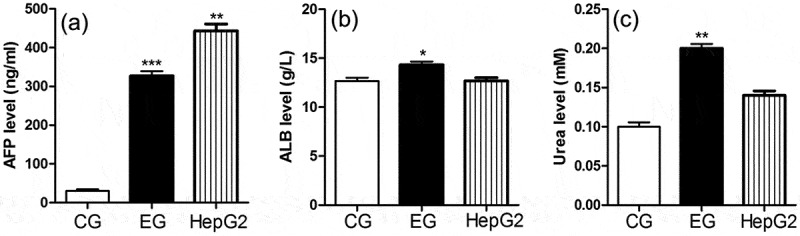
AFP, ALB, and urea secretion in induced cells on d 21. HepG2 was the positive control. (a–c) show AFP, ALB, and urea content, respectively in the culture supernatant. Results were obtained from three independent experiments. CG: control group, EG: experimental group. *p < .05, **p < .001 compared to control
Ultrastructure of cells after induction
To determine whether the ultrastructure of the induced cells was consistent with that of normal hepatocytes (large number of mitochondria and endoplasmic reticula) we carried out transmission electron microscopy with normal liver tissue as a positive control (Figure 7b). TEM showed a large number of mitochondria, endoplasmic reticula, and lysosomal organelles (Figure 7a) in the induced cells, which corresponds with the typical ultrastructure of hepatocytes. These structural features were not observed in the cells of the control group (Figure 7c). These results suggest that the ultrastructure of the induced cells is similar to that of normal hepatocytes.
Figure 7.
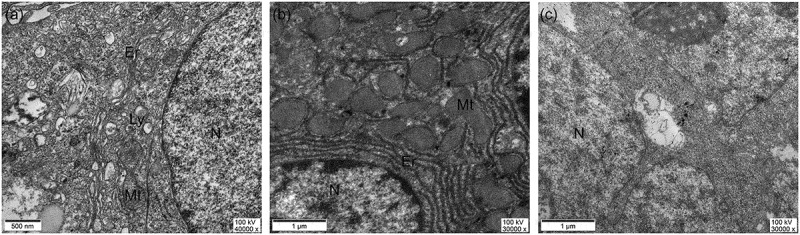
Ultrastructure of cells obtained 21 d after induction of differentiation。N: Nucleus, Mt: Mitochondrion, Ly: Lysosome, Er: Rough endoplasmic reticulum (a) Cell samples obtained after 21 d of induction, (b) adult liver tissue sections, (c) cell samples from the control group
Discussion
ESCs can be induced to differentiate into target cells by artificial directional induction9,10 via two main routes: via embryoid bodies (EB) or without EB.11,12 The EB pathway is a classical method of ES directional differentiation and involves the following basic steps: amplifying ES, removing LIF, forming EB by suspension culture, and inducing their differentiation into target cells. The purpose of the EB pathway is to simulate embryonic development in vivo and provide a microenvironment for ES differentiation. However, the quality of EB obtained is closely related to inoculation density and inoculation size, and directly affects the success of subsequent directional differentiation. The three-dimensional structure of EB accommodates a large number of cells at different differentiation stages. However, it also increases the probability of cell contact inhibition, which prevents optimal absorption of nutrients from the culture medium, leading to aging and death. Moreover, this method is cumbersome and mostly inefficient. Directional differentiation of hESCs can also be achieved without the EB pathway.13 The study of directed ESC differentiation is still in the early stages. Directed differentiation is typically achieved by gradually adding specific cytokines required for the process of embryonic development. For example, to differentiate ESCs into liver cells in vitro, we first add FGF, then HGF, for directed differentiation and development of embryonic liver.
As described by DUAN,14 we used direct induction without the EB pathway, and the combined action of seven inducing factors such as Activin A on hPESCs. We obtained functional liver-like cells that resembled normal hepatocytes in gene and protein expression, cell morphology, and function.
To date, stem cells such as ESCs, MSCs, and iPSCs have been used for directional differentiation into hepatocytes. However, the application of ESCs is subject to ethical restrictions and is associated with immune rejection.15 MSCs have limited sources, and are difficult to obtain. These problems can be avoided with the use of iPSCs,16 which are obtained from the reprogramming of differentiated somatic cells. However, the low reprogramming efficiency of iPSCs has always been a challenge in basic research and clinical application. In addition, the safety of induced cells for clinical use is unclear. Retroviral vectors were initially used for induction.17,18 However, viral vectors and transgenes are permanently integrated into the host genome and could affect cell function and differentiation,17 induce tumor formation, and affect the clinical application of induced cells.19 In addition, iPSCs are potentially immunogenic, animal experiments with induced cells from homologous donors showed immune responses in hosts.20 Gene instability is another challenge associated with iPSCs; chromosome abnormalities and small mutations have been observed in stem cells.21–27 Most of these chromosomal changes can be detected in adult cells prior to reprogramming and are independent of the reprogramming process.28,29 High-resolution analysis of single nucleotide polymorphisms (SNPs) showed that iPSCs have more copy number variations than adult cells from their source do.21 More accurate analysis showed that iPSCs require epigenetic and gene modifications. In this study, we attempted to obtain functional hepatocytes by using hPESCs as the initial differentiation material.
HPESCs are cell lines obtained by activation of oocytes in the second meiotic phase (M II) without sperm fertilization.30 The ethical issues associated with ESC research do not apply to these cells because they are derived from parthenogenetic embryos without developmental potential.31,32 Further, hPESCs are derived from a single gamete, and its MHC allele is homozygote. Theoretically, this would minimize immune rejection during cell transplantation. The genetic characteristics of the cell line are stable,31–37 thereby allowing long-term proliferation and maintenance of undifferentiated characteristics. Compared with MHC-heterozygous stem cell lines, hPESCs can also be used to treat unrelated individuals. The availability of more MHC-homozygous cell lines in embryonic stem cell banks could be of value for clinical and research applications. In summary, hPESCs could be a good source of target cells and merit extensive study.
In this study, seven inducible factors were sequentially applied to hPESCs. After 21 d of induction and differentiation, the cell morphology changed from a compact nest-like small circle to a loosely arranged polygon with occasional double or multiple nuclei. At the mRNA level, hepatocyte lineage genes such as AFP, ALB, and HNF4a, cytochrome P450 genes such as CYP3A4 and CYP3A7, and the transporter genes SLC10A1, SLCO1B3, ABCC2, and ABCB4 were highly expressed. CK18 and Hepa proteins were detected, and AFP, ALB, and urea were synthesized and secreted. The results of ICG uptake assay also indicate that the cells obtained from this study had the ability of metabolic clearance of dyes. ICG is an organic anion. Its uptake in the liver is mainly completed by organ anion transporting polypeptide 1B3 (OATP1B3/SLCO1B3) and Na+-taurocholate co-transporting polypeptide (NTCP/SLC10A1) in hepatocytes. Its excretion is mainly carried out by the multidrug resistance-associated protein 2 (MRP2/ABCC2) carrier system expressed on the bile capillaries.38,39 However, the process of polar transport across the hepatocyte membrane is very complex. The exact process of ICG uptake and secretion in hepatocytes is not completely clear,40 but it can be clearly confirmed that the function of hepatocyte transport exists.41 Glycogen synthesis and storage were also observed in the differentiated cells. TEM showed plenty of mitochondria, endoplasmic reticula, lysosomes, and other organelles in the cells, which is consistent with the ultrastructure of normal hepatocytes. These results were consistent with those reported worldwide.13,42–44
There are obvious differences in SNP distribution in the whole genome of hPESCs obtained by different parthenogenetic activation methods. Most of the sites of hPESCs from natural activation sources are homozygous, compared to those in cells obtained by chemical activation or electrical activation in the pursuit of low rejection rate of engineering cells. This study is the first reported attempt to use an embryonic cell line obtained by natural parthenogenetic activation in IVF to induce differentiation of functional hepatocytes. Using the single-layer plane induction method, the equipment requirements are low and the process is simple, which is highly advantageous. In this study, we developed a novel strategy to obtain functional hepatocytes, which could address the lack of sources for clinical hepatocyte transplantation.
However, there were several drawbacks associated with the hepatocytes obtained by this induction method: 1) After induction, the hepatocyte-like cells obtained by direct induction had the characteristics of hepatocytes, but did not proliferate and gradually aged and died, which limits the possibility of clinical application. 2) The level of AFP secreted by the cells that we eventually obtained was much higher than that of normal hepatocytes, and the difference between ALB secretion in the experimental and negative control groups was not as high as expected. These results suggest that the cells obtained by this method may not be mature enough and remain at the level of early hepatocytes. 3) The differentiated cells expressed both AFP and ALB markers, suggesting cell heterogeneity. 4) It is also imperative to find a low-cost, efficient, and simple method to quickly obtain target cells for subsequent experiments. The solution to these problems still needs the unremitting efforts of researchers.
Acknowledgments
This work was supported by Grants from the Applied Basic Research Project of Yunnan Province (2013FZ179, 2014FB040), National Natural Science Foundation of China (81660302), and Youth Foundation of the second hospital of Tianjin Medical University (Grant No. 2019ydey02). We sincerely thank the Institute of Basic Medical Sciences of the First People’s Hospital of Yunnan Province, the Electron Microscope Room of Kunming Medical University, and the National Engineering Research Center of Human Stem Cells for their and support in this research.
Funding Statement
This study was supported by the Youth Foundation of the second hospital of Tianjin Medical University (Grant No. 2019ydey02), National Natural Science Foundation of China (81660302), and Applied Basic Research Project of Yunnan Province(2013FZ179, 2014FB040).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Russo FP, Parola M.. Stem cells in liver failure. Best Pract Res Clin Gastroenterol. 2012;26(1):35–45. doi: 10.1016/j.bpg.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Cho CH, Parashurama N, Park EY, Suganuma K, Nahmias Y, Park J, Tilles AW, Berthiaume F, Yarmush ML. Homogeneous differentiation of hepatocyte-like cells from embryonic stem cells: applications for the treatment of liver failure. Faseb J. 2008;22(3):898–909. doi: 10.1096/fj.06-7764com. [DOI] [PubMed] [Google Scholar]
- 3.Isaev D, Garitaonandia I, Abramihina TV, Zogovic-Kapsalis T, West RA, Semechkin AY, Müller AM, Semechkin RA. In vitro differentiation of human parthenogenetic stem cells into neural lineages. Regen Med. 2012;7(1):37–45. doi: 10.2217/rme.11.110. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z, Li W, Chen T, Yang J, Wen Z, Yan X, Shen T, Liang R. Activin A can induce definitive endoderm differentiation from human parthenogenetic embryonic stem cells. Biotechnol Lett. 2015;37(8):1711–17. doi: 10.1007/s10529-015-1829-x. [DOI] [PubMed] [Google Scholar]
- 5.Lin G, Ouyang Q, Zhou X, Gu Y, Yuan D, Li W, Liu G, Liu T, Lu G. A highly homozygous and parthenogenetic human embryonic stem cell line derived from a one-pronuclear oocyte following in vitro fertilization procedure. Cell Res. 2007;17(12):999–1007. doi: 10.1038/cr.2007.97. [DOI] [PubMed] [Google Scholar]
- 6.Xu X, Browning VL, Odorico JS. Activin, BMP and FGF pathways cooperate to promote endoderm and pancreatic lineage cell differentiation from human embryonic stem cells. Mech Dev. 2011;128(7):412–27. doi: 10.1016/j.mod.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santamaria P, Rodriguezpiza I, Clementecasares X, Yamanouchi J, Muleroperez L, Aasen T, Raya A, Izpisua Belmonte JC. Turning human epidermis into pancreatic endoderm. Rev Diabet Stud. 2010;7(2):158–67. doi: 10.1900/RDS.2010.7.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamada MD, Yoshikawa M, Kanda S, Kato Y, Nakajima Y, Ishizaka S, Tsunoda Y. In vitro differentiation of embryonic stem cells into hepatocyte‐like cells identified by cellular uptake of indocyanine green. Stem Cells. 2002;20(2):146–54. doi: 10.1634/stemcells.20-2-146. [DOI] [PubMed] [Google Scholar]
- 9.Diekmann U, Naujok O, Blasczyk R, Müller T. Embryonic stem cells of the non‐human primate Callithrix jacchus can be differentiated into definitive endoderm by Activin‐A but not IDE‐1/2. J Tissue Eng Regen Med. 2015;9(4):473–79. doi: 10.1002/term.1709. [DOI] [PubMed] [Google Scholar]
- 10.Toivonen S, Lundin K, Balboa D, Ustinov J, Tamminen K, Palgi J, Trokovic R, Tuuri T, Otonkoski T. Activin A and Wnt-dependent specification of human definitive endoderm cells. Exp Cell Res. 2013;319(17):2535–44. doi: 10.1016/j.yexcr.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Rambhatla L, Chiu C, Kundu P, Peng Y, Carpenter MK. Generation of hepatocyte-like cells from human embryonic stem cells. Cell Transplant. 2003;12(1):1–11. doi: 10.3727/000000003783985179. [DOI] [PubMed] [Google Scholar]
- 12.Turovets N, Damour KA, Agapov V, Turovets I, Kochetkova O, Janus J, Semechkin A, Moorman MA, Agapova L. Human parthenogenetic stem cells produce enriched populations of definitive endoderm cells after trichostatin A pretreatment. Differentiation. 2011;81(5):292–98. doi: 10.1016/j.diff.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Cai J, Zhao Y, Liu Y, Ye F, Song Z, Qin H, Meng S, Chen Y, Zhou R, Song X, et al. Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology. 2007;45(5):1229–39. doi: 10.1002/hep.21582. [DOI] [PubMed] [Google Scholar]
- 14.Duan Y, Ma X, Zou W, Wang C, Bahbahan IS, Ahuja TP, Tolstikov V, Zern MA. Differentiation and characterization of metabolically functioning hepatocytes from human embryonic stem cells. Stem Cells. 2010;28(4):674–86. doi: 10.1002/stem.315. [DOI] [PubMed] [Google Scholar]
- 15.Daley GQ, Richter LA, Auerbach JM, Benvenisty N, Charo RA, Chen G, Deng HK, Goldstein LS, Hudson KL, Hyun I, et al. Ethics. The ISSCR guidelines for human embryonic stem cell research. Science. 2007;315(5812):603–04. doi: 10.1126/science.1139337. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 17.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 19.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448(7151):313–17. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 20.Zhao T, Zhang ZN, Rong Z, Xu Y. Immunogenicity of induced pluripotent stem cells. Nature. 2011;474(7350):212–15. doi: 10.1038/nature10135. [DOI] [PubMed] [Google Scholar]
- 21.Laurent LC, Ulitsky I, Slavin I, Tran H, Schork A, Morey R, Lynch C, Harness JV, Lee S, Barrero MJ, et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8(1):106–18. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayshar Y, Ben-David U, Lavon N, Biancotti JC, Yakir B, Clark AT, Plath K, Lowry WE, Benvenisty N. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cel. 2010;7(4):521–31. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 23.Sareen D, Mcmillan E, Ebert AD, Shelley BC, Johnson JA, Meisner LF, Svendsen CN. Chromosome 7 and 19 trisomy in cultured human neural progenitor cells. PLoS One. 2009;4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gore A, Li Z, Fung HL, Young JE, Agarwal S, Antosiewicz-Bourget J, Canto I, Giorgetti A, Israel MA, Kiskinis E, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471(7336):63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hussein SM, Batada NN, Vuoristo S, Ching RW, Autio R, Närvä E, Ng S, Sourour M, Hämäläinen R, Olsson C, et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471(7336):58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- 26.Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O’Malley R, Castanon R, Klugman S, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471(7336):68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pasi CE, Dereli-Öz A, Negrini S, Friedli M, Fragola G, Lombardo A, Van Houwe G, Naldini L, Casola S, Testa G, et al. Genomic instability in induced stem cells. Cell Death Differ. 2011;18(5):745–53. doi: 10.1038/cdd.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng L, Hansen NF, Zhao L, Du Y, Zou C, Donovan FX, Chou BK, Zhou G, Li S, Dowey SN, et al. Low incidence of DNA sequence variation in human induced pluripotent stem cells generated by nonintegrating plasmid expression. Cell Stem Cell. 2012;10(3):337–44. doi: 10.1016/j.stem.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Young MA, Larson DE, Sun CW, George DR, Ding L, Miller CA, Lin L, Pawlik KM, Chen K, Fan X, et al. Background mutations in parental cells account for most of the genetic heterogeneity of induced pluripotent stem cells. Cell Stem Cell. 2012;10(5):570–82. doi: 10.1016/j.stem.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yangqi O, Ge L, Xiaoying Z, Guang-xiu LU. Comparison of the differentiation capability between human parthenogenetic embryonic stem cells and normal embryonic stem cells. Acta Anatomica Sinica. 2010;41:785–89. [Google Scholar]
- 31.Lampton PW, Crooker RJ, Newmark JA, Warner CM. Expression of major histocompatibility complex class I proteins and their antigen processing chaperones in mouse embryonic stem cells from fertilized and parthenogenetic embryos. Tissue Antigens. 2008;72(5):448–57. doi: 10.1111/j.1399-0039.2008.01132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sritanaudomchai H, Ma H, Clepper L, Gokhale S, Bogan R, Hennebold J, Wolf D, Mitalipov S. Discovery of a novel imprinted gene by transcriptional analysis of parthenogenetic embryonic stem cells. Hum Reprod. 2010;25(8):1927–41. doi: 10.1093/humrep/deq144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu Z, Zhu W, Yu Y, Jin D, Guan Y, Yao R, Zhang YA, Zhang Y, Zhou Q. Derivation and long-term culture of human parthenogenetic embryonic stem cells using human foreskin feeders. J Assist Reprod Genet. 2010;27(6):285–91. doi: 10.1007/s10815-010-9408-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turovets N, Semechkin A, Kuzmichev LN, Janus J, Agapova L, Revazova E. Derivation of human parthenogenetic stem cell lines. Methods Mol Biol. 2011;767:37–54. [DOI] [PubMed] [Google Scholar]
- 35.Kim K, Ng K, Rugg-Gunn PJ, Shieh JH, Kirak O, Jaenisch R, Wakayama T, Moore MA, Pedersen RA, Daley GQ. Recombination signatures distinguish embryonic stem cells derived by parthenogenesis and somatic cell nuclear transfer. Cell Stem Cell. 2007;1(3):346–52. doi: 10.1016/j.stem.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 36.Revazova ES, Kuzmichev LN, Turovets NA, Kochetkova OD, Kindarova LB, Kuzmichev LN, Janus JD, Pryzhkova MV. Patient-specific stem cell lines derived from human parthenogenetic blastocysts. Cloning Stem Cells. 2007;9(3):432–49. doi: 10.1089/clo.2007.0033. [DOI] [PubMed] [Google Scholar]
- 37.Hao J, Zhu W, Sheng C, Yu Y, Zhou Q. Human parthenogenetic embryonic stem cells: one potential resource for cell therapy. Sci China Life Sci. 2009;52:599–602. [DOI] [PubMed] [Google Scholar]
- 38.Huang L, Vore M. Multidrug resistance P-Glycoprotein 2 is essential for the biliary excretion of indocyanine green. Drug Metab Dispos. 2001;29:634–37. [PubMed] [Google Scholar]
- 39.de Graaf W, Hausler S, Heger M, van Ginhoven TM, van Cappellen G, Bennink RJ, Kullak-Ublick GA, Hesselmann R, van Gulik TM, Stieger B. Transporters involved in the hepatic uptake of (99m)Tc-mebrofenin and indocyanine green. J Hepatol. 2011;54(4):738–45. doi: 10.1016/j.jhep.2010.07.047. [DOI] [PubMed] [Google Scholar]
- 40.Cusin F, Azevedo LF, Bonnaventure P, Desmeules J, Daali Y, Pastor CM. Hepatocyte concentrations of indocyanine green reflect transfer rates across membrane transporters. Basic Clin Pharmacol Toxicol. 2017;120:171–78. [DOI] [PubMed] [Google Scholar]
- 41.Torok G, Erdei Z, Lilienberg J, Apáti Á1, Homolya L. The importance of transporters and cell polarization for the evaluation of human stem cell-derived hepatic cells. PLoS One. 2020;15(1). doi: 10.1371/journal.pone.0227751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song Z, Cai J, Liu Y, Zhao D, Yong J, Duo S, Song X, Guo Y, Zhao Y, Qin H, et al. Efficient generation of hepatocyte-like cells from human induced pluripotent stem cells. Cell Res. 2009;19(11):1233–42. doi: 10.1038/cr.2009.107. [DOI] [PubMed] [Google Scholar]
- 43.Park Y, Chen Y, Ordovas L, Verfaillie CM. Hepatic differentiation of human embryonic stem cells on microcarriers. J Biotechnol. 2014;39–48. doi: 10.1016/j.jbiotec.2014.01.025. [DOI] [PubMed] [Google Scholar]
- 44.Mun SJ, Ryu JS, Lee MO, Son YS, Oh SJ, Cho HS, Son MY, Kim DS, Kim SJ, Yoo HJ, et al. Generation of expandable human pluripotent stem cell-derived hepatocyte-like liver organoids. J Hepatol. 2019;71(5):970–85. doi: 10.1016/j.jhep.2019.06.030. [DOI] [PubMed] [Google Scholar]


