ABSTRACT
The findings that an RNA is not necessarily either coding or non-coding, or that a precursor RNA can produce different types of mature RNAs, whether coding or non-coding, long or short, have challenged the dichotomous view of the RNA world almost 15 years ago. Since then, and despite an increasing number of studies, the diversity of information that can be conveyed by RNAs is rarely searched for, and when it is known, it remains largely overlooked in further functional studies. Here, we provide an update with prominent examples of multiple functions that are carried by the same RNA or are produced by the same precursor RNA, to emphasize their biological relevance in most living organisms. An important consequence is that the overall function of their locus of origin results from the balance between various RNA species with distinct functions and fates. The consideration of the molecular basis of this multiplicity of information is obviously crucial for downstream functional studies when the targeted functional molecule is often not the one that is believed.
KEYWORDS: Bifunctional RNA, non-coding RNA, protein-coding, dual function RNA, splicing, circRNA, small ORF
Introduction
Almost three decades ago, unprecedented research efforts to sequence the human genome and identify the genes that it contains led to the striking evidence that the vast majority of the genome does not contain information to make proteins. Even more surprising, most of these so-called non-coding sequences were shown to be competent for transcription and non-coding transcripts to constitute the bulk of the human transcriptome [1,2].
By definition, a non-coding RNA (ncRNA) is an RNA molecule that is not translated into a protein product and is thus distinguished from messenger RNAs (mRNA). Abundant and functionally important classes of ncRNAs include structural RNAs like transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), small regulatory RNAs such as microRNAs (miRNAs), small interfering RNAs (siRNAs), PIWI-interacting RNAs (piRNAs), small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), small cajal body RNAs (scaRNAs) [3]. We should also mention an extensive list of so-called long ncRNAs (lncRNA) whose functions remain largely unclear for most of them [4]. Nonetheless, ncRNAs have been assigned functions in most aspects of cell biology including transcription, chromatin remodelling, splicing, nuclear import and chromosome architecture, and to function as scaffolding, guiding, signalling or decoy for small regulatory RNAs or proteins [3,5]. Not surprisingly, they are often deregulated in human diseases, notably in a range of cancers or inherited disorders [6,7], although they have been more rarely causally linked to the emergence of disease phenotypes.
Since the release of the first draft, the annotation of the human genome has been quite dynamic and new data continues to enrich the transcriptome every day. Even recently, the number of protein-coding genes has been revised downwards [8]. It is important to remember that the transcriptional potential of eukaryotic genomes is very pervasive and widely intertwined [1,2]. For example, independent transcription units can be hosted in introns of larger ones or overlap in the antisense orientation. In addition, the use of alternative promoters or termination sites is well-known to regulate transcriptional output in a tissue- or stage-specific manner through the production of various isoforms. In that respect, the multifunctionality of a given genomic sequence, whereby a single locus can release more than one type of transcripts, i.e. coding, non-coding, long or short, sense or antisense, has been well documented [9,10,11] and is not the topic of this review. Importantly, this diversity is mostly regulated at the level of transcription depending on cellular context or environmental cues. An important consequence is that the transcripts thus produced may not co-exist in the same cellular context.
However, difficulties in genomic annotation are also complicated by mounting evidence that multifunctionality can also apply to transcripts themselves. Indeed, certain precursor RNAs can release more than one class of transcripts, i.e. coding and non-coding or two non-coding RNAs, and more strikingly, that some mature transcripts can perform more than one function [reviewed in [12] and Figure 1]. The first example of an RNA with dual function was probably the Steroid Receptor RNA activator (SRA) [13,14], and the term ‘bifunctional’ was then coined by Marcel Dinger and John Mattick [15]. Ever since, this duality of information conveyed by some of the transcripts has actually been reported in almost all organisms, from lower eukaryotes through plants to mammals [see Table 1 for prominent examples]. Hence, the view that an RNA should be either coding or non-coding is rather binary and the duality in RNA functions is far from being anecdotal. Unfortunately, and as often, this awareness is somewhat mitigated by the multiplication of various denominations such as ‘dual-function RNAs’ [16,17], ‘coding and non-coding RNAs’ [cncRNAs [16,18,19]] or ‘long non-coding chimeric RNA’ [lnccRNA [20]].
Figure 1.
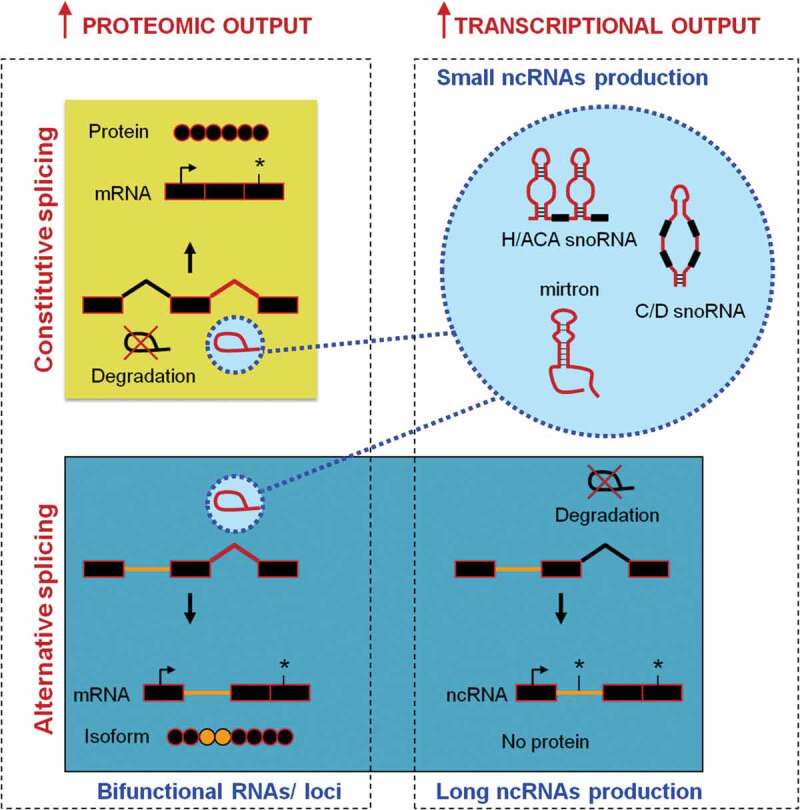
Diversification of proteomic and transcriptional outputs through constitutive or alternative splicing. More than 95% of introns are rapidly degraded after splicing (top left panel), but some can escape degradation and then represent precursors of short ncRNA [148] (top right panel). Retained introns can also favour the formation of protein isoforms (bottom left panel) or, if it disturbs the ORF, it can promote the formation of a long ncRNAs (bottom right panel). Exons and introns are represented by boxes and lines, respectively. mRNA, messenger RNA; ncRNA, non-coding RNA; H/ACA snoRNA, H/ACA box small nucleolar RNA; C/D snoRNA, C/D small nucleolar RNA
Table 1.
Examples of RNAs with dual functions
| Organism | Type | Role | Refs |
|---|---|---|---|
| Bacteria | |||
| RNAIII (S. aureus) | RNA | Modulates the production of toxins and enzymes by base-pairing on mRNA targets | [132] |
| Protein | Controls the switch between early and late expression of several exotoxins | [133] | |
| SgrS (E. coli) | RNA | Regulate sugar stress (prolonged stress) | [134] |
| Protein | Regulate sugar stress (early response) | [134] | |
| SR1 (Bacillus) | RNA | Regulation of arginine catabolism | [135] |
| Protein | SR1P binds and stabilizes the gapA operon mRNAs | [135] | |
| tmRNA (Bacteria) | RNA | Function as a tRNA by adding alanine to a stalled polypeptide chain | [135] |
| Protein | Function as a mRNA by incorporating a degradation tag to the polypeptide chain | [135] | |
| miPEP171b (M. truncatula) | RNA | Downregulates target genes involved in root development | [136] |
| Protein | Enhances the accumulation of its corresponding mature miRNA | [136] | |
| miPEP165a (A. thaliana) | RNA | Downregulates target genes involved in root development | [136] |
| Protein | Enhances the accumulation of its corresponding mature miRNA | [136] | |
| Plants | |||
| Enod40 | RNA | Confers specificity for the recognition of mRNA targets of MtRBP1 | [137] |
| Protein | Affects MtRBP1 localization during nodule development | [137] | |
| Insects | |||
| Oskar (Drosophila) | RNA | Scaffold for the complexes essential for development of the oocyte (early stage) | [138] |
| Protein | Scaffold for the complexes essential for development of the oocyte (late stage) | [138] | |
| Fish | |||
| Sqt (Danio rerio) | RNA | Deliver/sequester maternal factors for dorsal specification | [139] |
| Protein | Deliver/sequester maternal factors for dorsal specification | [139] | |
| Amphibians | |||
| VegT (Xenopus) | RNA | Organization of the cytokeratin cytoskeleton | [140] |
| Protein | Transcription factor of the T-box family | [140] | |
| Mammals (mainly human and mice) | |||
| p53 | RNA | Stimulates p53 translation and prevents p53 protein from Mdm2-induced degradation | [115] |
| Protein | Tumour suppressor | [115] | |
| SRA1 | RNA | Co-activator of transcription factors | [12] |
| Protein | Inhibitor of the co-activation function of SRA RNA | [12] | |
| circ-ZNF609 | RNA | Serves as a sponge for miR-150-5p and controls cell proliferation | [141] |
| Protein | Control of myoblast proliferation | [120] | |
| ASCC3 | RNA | Counteracts the function of the protein-coding isoform | [38] |
| Protein | Negative regulator of the host defence response | [38] | |
| LINC00961/SPAR | RNA | has as an anti-angiogenic role by inhibiting the function of Tβ4 protein | [108] |
| Protein | Involved in muscle regeneration/has a pro-angiogenic role by binding the SYNE1 protein | [107] | |
| HIC | RNA | Activates P-TEFb by displacing 7SK RNA | [142] |
| Protein | Inhibits P-TEFb transcription | [143] | |
| Irs-1 | RNA | Inhibits myoblasts differentiation through complementary sequence to Rb mRNA | [144] |
| Protein | Coordinates skeletal muscle growth and metabolism | [145] | |
| apoE/apoE-I3 | RNA | apoE-I3 controls apoE expression in neurons | [146] |
| Protein | Involved in lipid transport system | [146] | |
| PNUTS | RNA | regulates EMT migration and invasion in vitro through its miR-205 interaction | [39] |
| Protein | functions as a proto-oncogene by sequestering PTEN | [147] | |
We propose here to go over remarkable contexts where any attempt at categorization into coding or non-coding is likely to be reductive. We will go over bifunctional precursor RNAs (pre-RNA) that can release coding and non-coding, or two non-coding, functions depending on post-transcriptional maturation processes. We will also mention single RNA molecules that can operate at least two functions [for reviews see [12,15,21,22]]. This includes messenger RNAs (mRNA) shown to operate as functional RNAs, and certain lncRNAs initially classified as non-coding but shown to release small peptides or re-annotated as coding RNAs. Following the logic, any functional ncRNA that serves as a precursor to a smaller regulatory RNA should be considered as bifunctional (Figure 1). Examples will include certain snoRNAs that function as precursors of miRNAs. Likewise, snoRNAs being exclusively produced from intron splicing, at least in humans, their host precursor is a bifunctional RNA since its splicing releases a mature mRNA and a snoRNA with distinct mechanisms of action and locations.
Understanding the combination of functions and fates that can be carried by a transcript, being it a mature RNA or a pre-RNA, is not only futile nor is it intended to add confusion by naming new categories of RNAs. On a practical note, the annotation of transcripts with coding, non-coding or mixed potential is important for genome or transcriptome manipulations for which the impact may be wider than expected or simply not the desired one. This knowledge is also crucial for genomic studies that aim to understand the determinants of human traits or predisposition to disease.
When a precursor-RNA produces two or more transcripts with distinct fates
We already mentioned cases where the dual coding/non-coding function is carried by the products of a gene and its related pseudogene (reviewed in [12,23]), yet it potentially involves thousands of pairs of gene/pseudogene transcripts that remain to be characterized. In a more remarkable way, this duality of functions can also be released by the same transcription unit, i.e. carried by the same pre-RNA, after steps of post-transcriptional maturation.
Intron retention and the production of lncRNAs
We have already discussed alternative splicing (AS) as a means used by organisms to enhance their proteome, through the production of diverse mRNAs which fate is to be translated into protein variants, but also their transcriptome, by producing both coding and non-coding RNAs in fewer cases [12,24].
The first example was the SRA RNA first identified as an ncRNA with trans-activating functions of the activity of hormone receptor complexes [25]. Further characterization identified dozens of transcripts that classically differ in their initiation and termination sites or by their exon content through exon skipping [13,14,26]. However, the striking finding was that these transcripts also differed in their coding capacity, with the most remarkable disruption of the ORF being through the retention of the first intron. In sum, fully spliced isoforms encode an SRAP protein whereas intron-retaining isoforms form the SRA ncRNA [13,14,27]. Although it is yet unknown how AS of SRA intron 1 is regulated and how the intron-retaining isoforms escape surveillance machineries like the Nonsense-Mediated Decay (NMD), the overall function of the SRA1 locus results from the balance between coding and non-coding isoforms, at least in a muscle context [12,27]. Whereas SRAP was shown to have an antagonistic impact on the function of its cognate SRA RNA, a switch towards non-coding isoforms accompanies muscle differentiation and accelerates reprogramming of non-muscle cells towards the muscle fate [27]. Evidence for the functional importance of this switch is the finding that it does not occur in cells from patients with splicing defects causing the Myotonic Dystrophy type 1 (DM1) [27]. Importantly, because coding and non-coding isoforms co-exist in the cell, addressing their individual functions required to mutate the ORF without affecting the secondary structure of the ncRNA or to destroy the secondary structures without affecting the ORF [27]. Even truer than for classical transcription units, the extensive knowledge of the variety of mature transcripts than can be produced through post-transcriptional maturation of a pre-RNA is key to prevent false assumptions from non-targeted functional approaches.
This was also probably the first example providing evidence that AS of intron is not necessarily a faulty mechanism, although it is less widely used than in plants [28]. Indeed, intron retention (IR) is usually thought to be an error in the splicing mechanism that leads to RNA degradation by triggering the NMD pathway [29]. How some intron-retaining RNAs do escape NMD is currently unclear, but the stable ncRNAs thus produced may operate functions as RNA molecules, or at least have an impact on cellular processes. It was suggested that IR could also impact translation efficiency if the retained intron contains miRNA binding sites [30,31], or if it introduces a uORF (upstream of the ORF) or a rigid structure upstream of the start codon [reviewed in [32]]. Since then, hundreds of mRNA with IR have been bioinformatically predicted [24,33] or identified as stable entities in various cell types [14,34] and reviewed in [12,21,35]. In human cells, IR was proposed to fine-tune gene transcriptomes [36] and, more importantly, to orchestrate the establishment of lineage-affiliated expression programmes that accompany cell differentiation [35,37].
Another example where AS serves to coordinate gene expression programs in response to environmental cues is during the stress response. Upon UV irradiation, the rate of transcriptional elongation is reduced and associated with a shift in expression from long mRNAs to shorter isoforms through the incorporation of an alternative last exon (ALE) [38]. This AS mechanism differs from IR but, as shown for the Activating Signal Cointegrator 1 Complex Subunit 3 (ASCC3), it also promotes an expression switch from a long coding mRNA to a shorter isoform that functions as a nuclear ncRNA [38]. As shown for SRA/SRAP [27], long and short ASCC3 isoforms have opposite effects on transcription recovery after DNA damage. This duality of information carried by short and long ALE isoforms may also apply to other UV-induced ALE transcripts with yet unknown contribution to the DNA damage response [38].
It is also worth mentioning cases where deregulated AS caused by alterations of splicing mechanisms in pathological conditions promotes the formation of an ncRNA instead of a coding mRNA, with dramatic consequences for cellular phenotypes. Alterations of AS are hallmarks of cancer cells and, beyond the consequences for the formation of protein isoforms with gain or loss of function, they can also lead to the release of functional ncRNA. For example, the locus PNUTS/PPP1R10 encodes a protein phosphatase 1 (PP1) regulatory subunit also known as PNUTS (Phosphatase Nuclear Targeting Subunit) involved in cell cycle progression, DNA repair and apoptosis by regulating the activity of the protein phosphatase 1 (PP1). In cancer cells, the unmasking of an alternative splice site located in an exon disrupts the ORF and releases a lncRNA. The lncRNA-PNUTS acts as a sponge for a key determinant of the epithelial phenotype, miR-205, thereby promoting Epithelial-Mesenchymal Transition (EMT) and tumour progression [39].
As a whole, these examples underline the importance of AS as a developmental switch, which allows a single transcription unit to produce multiple transcripts with distinct functions and fates depending on the needs of the cell. Yet, it is important to remind that these transcripts can co-exist in the cell and that the overall function results from the balance between the levels of the different isoforms and the distinct functions they perform. Hence, addressing the function of bifunctional RNAs requires to uncouple the often-antagonistic coding and non-coding functions. Inferring their function(s) without knowing the full range of transcripts produced in a given context and their intertwining is likely to lead to false mechanistic assumptions and release of poor biomarkers of pathophysiological situations.
Splicing of introns and the release of short ncRNAs
In contrast to generating RNAs with new functionality when retained in an mRNA, introns can also function as precursors of smaller ncRNAs when they are excised from the pre-mRNA. This is the case for all known mammalian snoRNAs [40], for non-canonical miRNAs like mirtrons or simtrons [41], or yet unknown categories of intron-derived small RNAs. We collectively named these transcripts SID for Short Intron-Derived ncRNAs [42].
SnoRNAs are abundant, short, nucleoli-residing, small ncRNA, best known for guiding post-transcriptional modifications of other ncRNAs such as rRNAs, tRNAs and snRNAs [40]. They constitute so far the majority of known SIDs. Unlike in yeast and plants where snoRNAs can have their own promoter, all mammalian snoRNAs are embedded within introns, usually on a one per intron basis. They are transcribed from their host genes as portions of the pre-RNA, and the functional snoRNAs are then produced by exonucleolytic trimming after splicing [40,43] (Figure 1). Curiously, snoRNA host transcripts were never referred to as RNAs with dual function, although they clearly are since most host genes generate protein products in addition to their snoRNAs. Maybe is it because snoRNAs were initially identified in introns of ribosomal protein genes or genes encoding translation factors or nucleolar proteins, thus both contributing to the same general biosynthetic process. However, this is no longer a general rule with the identification of more widespread functions in pre-mRNA processing [44] and the identification of non-rRNA targets of snoRNAs [45], stressing the need to distinguish coding from non-coding functions carried by the same snoRNA host transcript in functional studies. More striking is the identification of non-protein-coding hosting transcripts. Indeed, a non-negligible fraction of human snoRNAs [about 22% (Table 2); calculated from UCSC main table ‘Genes and Gene Predictions’ intersected with DASHR collection [46]] lies within small introns of ncRNAs, with the remarkable case of so-called snoRNA host genes (SNHG) that can shelter many snoRNAs in the same transcript [47]. Prominent examples include SNORD115 and SNORD116 clusters of dozens of snoRNAs produced from the same SNHG14 ncRNA. SNHG ncRNAs do not have clear functions (Supplementary Table 1) other than being dedicated to the production of snoRNAs, although their deregulation has been associated with carcinogenesis [47,48]. A prominent example is the growth arrest-specific 5 (GAS5) lncRNA, which carries almost one snoRNA in each of its 11 introns, and functions as a tumour suppressor [49]. Human GAS5 was proposed to also carry snoRNA-independent functions as a ribo-suppressor of glucocorticoid receptors binding to their genomic targets, as a decoy for miRNA or even as a small peptide-encoding ncRNA, although these features lack conservation among species [48,49]. Hence, human GAS5 is typically a lncRNA with multiple functions although several questions remain to be formally addressed like the existence of small peptides from GAS5 translatable small ORFs (sORF) in vivo, and more importantly, the decoupling of the relative contribution of snoRNAs or ribo-mimic functions of GAS5 to understand which elements confer the tumour suppressor function of the GAS5 gene.
Table 2.
Number of small ncRNAs identified in intronic regions of lncRNAs. All numbers were inferred from the DASHR database and were calculated from UCSC main table ‘Genes and Gene Predictions’ intersected with DASHR collection [46]. *, These intronic snoRNA are included in snoRNA host-genes (SNHG); miRNA, microRNA; rRNA, ribosomal RNA; scRNA, small cytoplasmic RNA; snoRNA, small nucleolar RNA; snRNA, small nuclear RNA; tRNA, transfer RNA
| Type of ncRNA | Total number | Within intron of lncRNA |
% |
|---|---|---|---|
| miRNA | 1,582 | 198 | 12.51 |
| rRNA | 1,723 | 204 | 11.84 |
| scRNA | 1,291 | 170 | 13.17 |
| snoRNA | 402 | 88 * | 21.89 |
| snRNA | 4,278 | 533 | 12.46 |
| tRNA | 623 | 52 | 8.35 |
| Mean: | 13.37 ± 4.51 |
The second most represented class of SIDs is the class of non-canonical miRNAs such as simtrons and mirtrons, which are produced independently of the microprocessor DiGeorge syndrome Critical Region 8 (DGCR8) enzyme [41,50,52]. The release of simtrons is concomitant to splicing and involves cleavage by the RNAse III enzyme Drosha recruited through the U1 general splicing factor directly on the pre-mRNA [41,53]. Mirtrons are also processed by Drosha but post-splicing, and thus require debranching of the intron-lariat by Debranching RNA enzyme 1 (DBR1) [50,51]. Hence, simtrons and mirtrons, and a few others described recently [42], are dependent on both the transcription of their host genes and on intron cleavage/splicing. Cases of intron-derived miRNAs have been reported as targeting their own host RNA in regulatory feedback loops [54], suggesting that duos of SIDs/host transcripts are involved in the same biological processes or functions, although it remains poorly documented. In Drosophila, a recent report showed how a mirtron, its host and target transcripts form a complex network of regulatory loops to control synaptic homoeostasis and neural activity [55]. We can speculate that this type of regulation is far from being anecdotal since nearly 500 mirtrons have been identified in humans [56]. Since miRNAs have many potential target transcripts whatever their origin and biogenesis, they are likely to have more pleiotropic functions than their host transcript, stressing that the evaluation of their release in certain normal or pathological cellular contexts must be discriminated from the expression levels of their host transcript.
Another remarkable case of small ncRNAs that are directly produced through splicing of a precursor pre-RNA is the class of circular RNAs, which, by definition, stand out from others by their circular structure. After the splicing reaction, an intron lariat is produced, which normal fate is to be debranched and degraded, although they can be trimmed by an exonuclease to produce an intronic circular RNA (ciRNA) [57]. Although the function of these RNAs is not yet fully understood, 300 ciRNAs have been predicted in humans [58]. In addition, pre-RNAs can also generate covalently closed circular RNAs (circRNAs) through back-splicing, a reverse splicing process where the donor splice site reacts with an upstream acceptor splice site [see definitions in [59,60]]. CircRNAs can be composed of exons (EcircRNAs), introns (IcircRNAs), or both (EIcircRNAs). Recent studies have shown that this type of ncRNAs could act as miRNA sponges or transcriptional regulators [60]. Similar to the above-mentioned SIDs, circRNAs coexist with the mature long RNA, producing a duo of RNA molecules whose cooperation or antagonist functions remain to be addressed. With the advent of high throughput sequencing, about 100,000 circular RNAs have been identified and characterized in humans [61]. According to circBase and a circRNA expression resource of 20 human tissues [61,62], circRNAs are essentially tissue-specific and only a small subset is expressed in a given cell type, generally at high levels and are often more abundant than their linear host RNAs [61,62]. Although the functions of most circRNAs as decoys and whether they are translated or influence the functions of their linear counterparts is still debated, their abundance and restriction to a given cell type is strikingly reminiscent of that of miRNAs [46], pointing to promising roles in gene regulatory networks.
Transfer RNAs are also first transcribed as a precursor (pre-tRNA) and subjected to splicing [63] in all species. We can mention the striking example of the pre-tRNAtrpwhich produces a functional C/D box snoRNA after splicing of its intron (Figure 2) in the archae Haloferax volcanii [64]. Interestingly, the associated small nucleolar ribonucleoprotein complex (snoRNP) serves as a guide for the 2ʹ-O-methylation (2ʹOMe) of its own host transcript, which is a post-transcriptional editing typically involved in the proper functioning of tRNAs [65]. However, the deletion of the intron or the absence of the tRNATrp editing have no impact on the viability of H. volcanii in normal conditions, although it was suggested that it may manifest itself under certain stress or competitive conditions [66].
Figure 2.
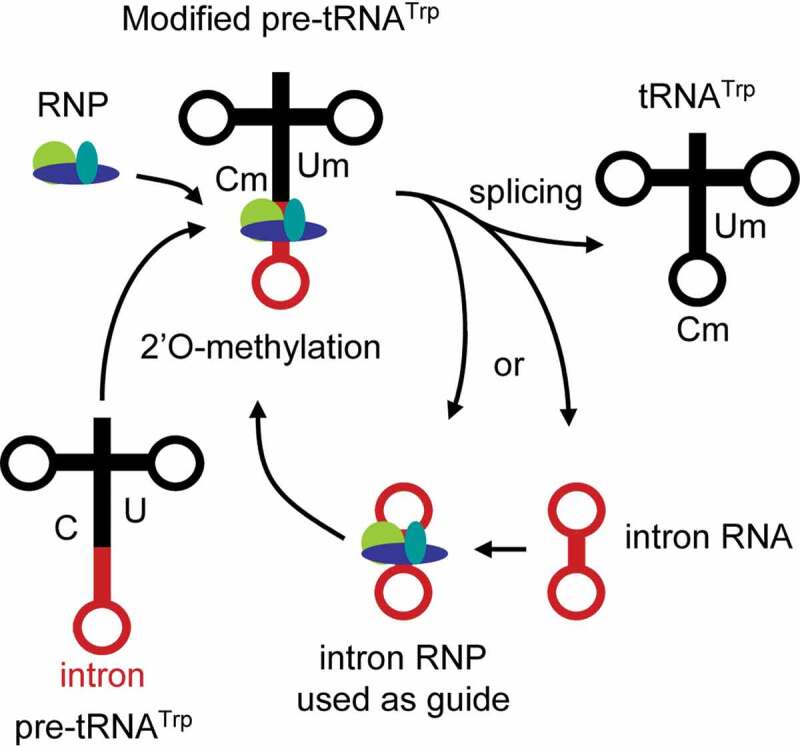
The 2ʹO-methylation of H. volcanii pre-tRNATrp is guided by its own intron. Thick arrows indicate the pre-tRNATrp processing pathway from nucleotide methylation and splicing to the production of a tRNATrp and the excised intron. RNP, ribonucleoprotein complex; C, cytosine; Cm, methylated cytosine; U, uracil; Um, methylated uracil. Adapted from [64]
As a whole, certain pre-RNA can produce two types of transcripts, i.e. coding mRNAs and small regulatory SIDs or circular RNAs. The case of the above-mentioned SRA is even more remarkable as it can release a coding mRNA and a SID, or an mRNA and a long ncRNA that retains the intron containing the SID [42]. Likewise, certain tRNA genes can release a tRNA and a small regulatory RNA. Hence, it appears that precursor RNAs are not always mere transient conveyers of genetic information as they contain multiple pieces of information and can generate a panel of mature RNAs. In that respect, introns represent an obvious contributor to transcriptional diversification as their retention in, or excision from, the pre-RNA will greatly increase the output of a single transcription unit. It is also not surprising that more examples of bifunctional pre-RNA are found in higher eukaryotes, especially mammals, since the number of introns tends to increase with the increased developmental complexity of organisms [67].
Splicing of exons and the release of short ncRNAs
In addition to AS of introns, exon skipping is also well known to create normal or pathological post-transcriptional diversity. The H19 locus is fascinating and probably the first example of an ncRNA hosting a small regulatory RNA in an exon. H19 is transcribed in a 2.3 kb long lncRNA, exclusively from the allele inherited from the mother, which acts as a trans-regulator of an imprinted gene network involved in the control of foetal and early postnatal growth in mice [68]. In fact, transcription within the H19 locus is extremely complex. In addition to the H19 transcript, two transcripts in the antisense orientation have been described, also from the maternal allele: a coding transcript HOTS (H19 opposite tumour suppressor, 6 kb long) [69] and a very long ncRNA named 91 H (120 kb long) [70]. Adding to the complexity, the H19 transcript itself also contains a highly conserved stem-loop structure embedded within its first exon and shown to release hsa-miR-675 [71,72]. H19 is highly expressed in most foetal tissues and in the placenta, but the processing of miR-675 seems to be restricted to the latter where it operates growth suppressing functions [71]. This tight control may be overcome in physiopathological conditions to allow rapid inhibition of cell proliferation [71]. However, whether H19 functionality resides solely in its role as an abundant pri-miRNA, or if H19 operates independent functions as suggested by the disparity between H19 and miR-675 expression and findings of miRNAs sponge activity in muscle cells [73], is still debated. There are probably many other such examples since exonic miRNAs represent 3.9% of total miRNAs, even if it may represent less than a hundred of associated transcripts and hence, candidate bifunctional RNAs [74]. Yet, and in contrast to miRNAs produced through splicing of introns discussed above, it remains to be tested whether and in which contexts exon skipping would promote the release of a miRNA and a functional RNA knowing that i) about half of the exonic miRNA are embedded within exons of spliced lncRNAs whose functions are often elusive and ii) Drosha processing of an exonic miRNA is likely to impair the production of the spliced host mRNA with coding functions [74].
Precursor RNAs producing distinct non-coding functions
It is commonly thought that most lncRNAs are expressed at low levels and are poorly conserved at the sequence level across species. However, certain classes of lncRNAs are clearly as abundant as mRNAs transcribed from housekeeping genes, with a common denominator of escaping degradation and accumulating as surprisingly stable transcripts since they lack terminal structures typical of Pol II transcripts including the polyA tail [75]. The reason is obvious for circular RNAs that have no end or snoRNAs generated through splicing, but this is more remarkable for longer transcripts. Prime examples include MALAT1 (NEAT2) and MEN ε/β (NEAT1) transcribed from adjacent loci. Metastasis Associated Lung Adenocarcinoma Transcript 1 RNA (MALAT1) is a lncRNA originally described as being upregulated in cancer cells [76], in fact among the most abundant long ncRNAs in mouse and human tissues. This rarely spliced transcript is also not poly-adenylated owing to its 3ʹend being cleaved by the endonuclease RNase P at the level of an evolutionary conserved tRNA-like structure, which simultaneously generates the non-polyadenylated long MALAT1 transcript and a tRNA-like small ncRNA [76,77]. The thus-generated 3′ end of MALAT1 folds into a triple helix that prevents exonucleolytic degradation, which is further processed to generate a mature 61-nt transcript known as mascRNA (MALAT1-associated small cytoplasmic RNA). MALAT1 is retained in nuclear speckles where it associates with serine/arginine-rich (SR) proteins and splicing factors [78]. The knock-down of this lncRNA impacts the localization of splicing factors in nuclear speckles, and thus, MALAT1 is thought to play a role in the regulation of splicing [78] although it was proposed to regulate gene expression via multiple mechanism [75]. In clear contrast, mascRNA is exported to the cytoplasm although it is unlikely that it reads the genetic code as its anticodon loop is poorly conserved. A dichotomy of the immunoregulatory functions of MALAT1-mascRNA system has been reported following selective ablation of mascRNA in monocytes [79], but the exact biological function of mascRNA remains undefined. Nevertheless, the primary MALAT1 transcript is the first example of a pre-RNA processed into two mature ncRNAs with distinct fates other than via the intron splicing process [77].
The multiple endocrine neoplasia-β locus (MEN1) is also able to generate lncRNAs (MEN-ε and -β) that are essential organizational components of nuclear paraspeckles [80]. Whereas MEN-ε is subjected to canonical cleavage/polyadenylation, the mature 3′ end of the longer isoform MEN-β is generated via the same mechanism as for MALAT1 together with a tRNA-like RNA (menRNA). Although the latter is structurally unstable in most mouse and human cells, it is stable in other species for unclear reasons and unknown functional consequences, but suggesting that MEN-β could operate dual functions in these species [80,81].
Strikingly, more than 100 loci in vertebrate genomes present a MALAT1 3′-end triple helix structure and its immediate downstream tRNA-like structure [82]. Their functional dichotomy remains to be demonstrated but they deserve further attention when the possible pathogenic relevance of their non-coding transcripts is addressed.
To add to the most recent sources of regulatory small RNAs, the finding that small ncRNAs can themselves be further processed into smaller RNA species is a direct consequence of the rapid progress in RNA sequencing and bioinformatic analysis [83]. More than 25,000 fragments derived from tRNAs (tRFs) [84,85,86] or hundreds from snoRNAs (sdRNAs) [87,90], around 14 to 40nt long depending on species, were indeed found in RNAseq data and first thought to be degradation products. There is now mounting evidence that their biogenesis is conserved and that they are processed to generate stably accumulating fragments in a non-random and regulated manner. These small RNAs fragments share similar features with miRNAs and were actually found to be associated with Argonaute (AGO) proteins suggestive of a role in translational repression, although they may interfere with translation through other mechanisms [91]. The processing of a subset of tRNAs into tRFs was actually thought to provide a rapid response to downregulate RNA translation during the stress response [92]. An interesting finding is that tRFs target endogenous retroviruses and inhibit their retrotransposition through a miRNA-like silencing of transposon reverse transcription as demonstrated in mice [93]. Because all organisms have tRNAs, it is possible that this is in fact a highly conserved mechanism to control the deleterious mobility of transposons in contexts where they escape epigenetic silencing, at early embryonic development stages, for example when the epigenetic landmarks of parental genomes are reset [93]. At these stages, the functions of tRNAs and their derived tRFs are clearly distinct. As for sdRNAs, examples of processed snoRNAs have been described for both H/ACA and C/D boxes snoRNAs [88,94,95]. As reviewed in [89], there is a significant overlap between snoRNA and miRNA processing enzymes, including AGO and DICER, their functional binding partners and even their subcellular localizations, which renders the distinction between their respective activities somewhat tricky. There are a few reports that do provide support for the functions of both snoRNA and miRNA co-existing within the same molecule. Yet, one has to admit that this is essentially based on the use of mimics for the sdRNA-miRNA-like fragment and evidence that the miRNA-like and its parental snoRNA are enriched in distinct subcellular locations, whereas loss-of-function experiments are likely to affect both types of molecules altogether. Nonetheless, target genes for these sdRNAs have been predicted and validated for a few of them, with prominent examples pertaining to the regulation of the p53/Mdm2 (Mouse double minute 2 homolog) feedback loop central to tumour suppression [[96], reviewed in [87]].
In sum, tRNAs and snoRNAs are among the best-known and best-studied small ncRNAs. However, they also hide unexpected functions through their processing into smaller ncRNAs that add to the complexity of regulatory networks of genomes functions. With the resulting processed small RNA fragments operating miRNA functions, an adverse consequence is that the dual function, or more precisely the individual function of the tRNA or snoRNA entities in the processes under consideration, is largely overlooked compared to that of the miRNA-like fragments. Nevertheless, their abnormal levels is an index of a pathological situation, and because small RNAs are easily detectable in body fluids, they are useful biomarkers for human diseases [87,97]. In addition, it could be considered that if their physiological function in most healthy physiological contexts may be marginal, the stress-induced maturation that they have in common [92,98] is actually a mechanism that promotes the production of distinct entities to orchestrate protective cellular functions.
When a single transcript performs multiple functions
Long non-coding RNAs with small ORFs
Remarkably enough, some of the RNAs that are currently considered as ‘true’ ncRNAs may actually have the potential to be translated [99]. In recent years, the last criterion for distinguishing coding from non-coding RNAs has been shaken up with findings that lncRNAs can be associated with polysomes [100]. In fact, large-scale ribosome profiling estimated that nearly 40% of known human lncRNAs are associated with ribosomes in the cytoplasm [101]. However, whether certain lncRNAs are actually translated or this is used as a strategy to regulate their abundance in the cell is still far from being clear. Today, the real challenge is to identify these short peptides translated from lncRNAs in vivo and determine whether they have a function, or an impact, in a biological process. Some of them have been studied in depth and their associated functions in the cell have been described. This is the case for Myoregulin (MLN) and Dwarf open reading frame (DWORF) micropeptides. MLN and DWORF have 46 and 34 amino acids and are encoded by the lncRNAs LINC00948 and LOC100507537, respectively [102,103]. They are both involved in muscle relaxation mediated by the re-import of Ca2+ into the sarcoplasmic reticulum by the sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) pump (Figure 3). MLN acts as a SERCA pump inhibitor and therefore prevents muscle relaxation, whereas DWORF increases SERCA pump activity by displacing SERCA inhibitors and thus facilitates muscle relaxation [102,103]. We can also mention the most recent example of the lncRNA LINC00116 which encodes a highly conserved 56-amino-acid mitochondrial micropeptide, Mitoregulin, which has been implicated in the regulation of respiratory efficiency in cardiac and skeletal muscle cells [104]. Micropeptides have so far evaded annotation efforts because of the technical difficulties to identify micropeptides, but also probably because the expression of at least a subset of these bifunctional micropeptide-encoding lncRNAs is restricted to specific cell types [22,105]. Consequently, their host transcripts, as many others, may have just been misannotated as lncRNAs, awaiting the identification of associated peptides. Many of the corresponding transcripts have now been reclassified as coding transcripts in the latest version of GENCODE (v33) with an NM_ nomenclature. They have not been formally studied in terms of function, localization and downregulation in conditions where their coding portion is deleted, so the dual function of such transcripts is still debated. Of course, this would require being able to uncouple the RNA functionality from its coding potential as it has already been performed in other instances [12,106].
Figure 3.
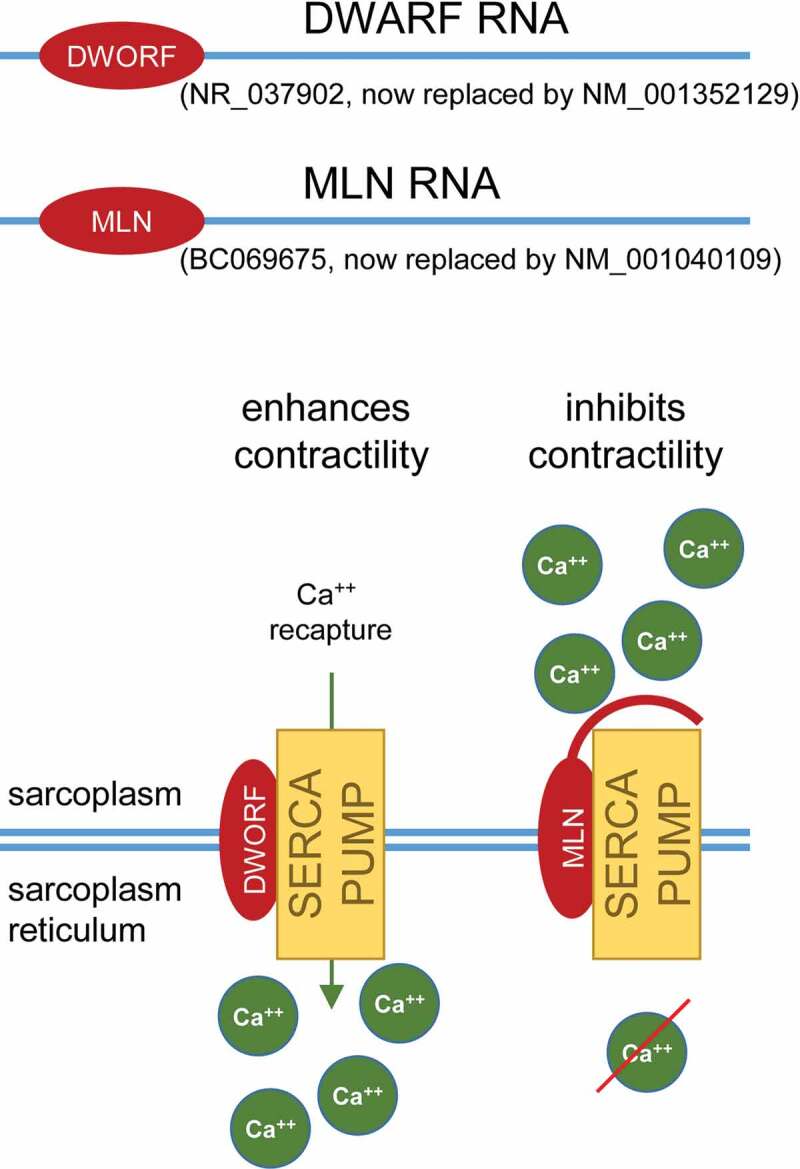
The micropeptides Myoregulin (MLN) and Dwarf open reading frame (DWORF). DWARF RNA and MLN RNA were first identified as long non-coding RNAs (lncRNAs; Refseq numbers NR_037902 and BC069675 respectively). It appears that these two lncRNAs can encode DWORF and MLN micropeptides, respectively. Subsequently, their RefSeq category has been revised: NR_037902 became NM_001352129 and BC069675 were replaced by NM_001040109. DWORF and MLN are both involved in muscle contraction. The first one enables muscle relaxation by activating the SERCA calcium pump and thus the re-import of Ca2+ into the sarcoplasmic reticulum. Conversely, the second one maintains muscle contraction by preventing the re-import of Ca2+ by inhibiting the SERCA pump
Among the examples cited in Table 1, LINC00961 has been reclassified as a protein-coding gene because it was shown to encode a 90-amino-acid micropeptide called Small Regulatory Polypeptide of Amino-Acid Response (SPAAR) involved in muscle regeneration [107]. In the context of angiogenesis, SPAAR and LINC00961 have a pro- and an anti-angiogenic role, respectively. Whereas SPAAR interacts with the pro-angiogenic Spectrin Repeat Containing Nuclear Envelope Protein 1 (SYNE1) protein involved in the connection of organelles to actin filament and endothelial cell migration, LINC00961 binds to and negatively regulates the function of Thymosin beta 4 (Tβ4) protein involved in the rearrangement of actin filament network and the induction of angiogenesis [108]. Thus, LINC00961 can be considered as a bifunctional RNA for which both the lncRNA and its encoded micropeptide contribute to the regulation of angiogenesis. Of note, it would be of interest to know whether a switch of expression of the two entities occurs when angiogenesis is needed and what is the mechanism underlying this regulation. In any case, the example of the pair LINC00961/SPAAR supports the idea that some micropeptide-encoding lncRNAs hide multiple functions that have not yet been uncovered in the appropriate cellular context.
In a more large-scale study, van Heesch et al.have established the translatome of the human heart where they found that 129 lncRNAs over the 783 expressed were also translated into micropeptides [109]. Surprisingly, among these, 27 have assigned non-coding functions like the lncRNAs NEAT1 [110] and JPX (Just Proximal to XIST) [111], and 4 are known SNHG such as the already mentioned GAS5. Moreover, 22 micropeptides encoded from lncRNAs were localized to mitochondria and associate with mitochondrial processes, although further investigation is needed to fully dissect the role of these micropeptides in mitochondrial processes [109]. However, this study stresses the fact that a number of translated lncRNAs are likely to possess both coding and non-coding roles, and that this previously unrecognized biology of lncRNAs is likely to prompt a multitude of follow-up studies.
In sum, the discovery of translatable sORF in so-called lncRNAs also goes against a rigid and dichotomic classification of RNA molecules into strictly coding or non-coding [99,103,112]. Instead, it lends support to a model where translation, just as previously reported for transcription, might also be a rather pervasive mechanism [113].
Messenger RNAs that perform non-coding functions
Just like ncRNAs can produce peptides, some RNAs have been assigned functions distinct from their role as intermediates in protein synthesis, and are also typically bifunctional. This is the case for SRA or p53 (tumour protein 53) transcripts [27,106]. This is not surprising because mRNAs have stable secondary structures just like ncRNA do [12,114], at least predicted with RNAfold (http://unafold.rna.albany.edu/), which allow transcripts to interact with proteins, and in some reported cases, to regulate their function or localization. As an example, SRA mRNA interacts with MyoD, the master transcription factor of myogenic differentiation, leading to MyoD transcriptional activation and activated muscle differentiation/reprogramming [27]. Likewise, p53 mRNA interacts with the E3 ubiquitin-protein ligase mdm2 and prevents the latter from promoting the degradation of the p53 protein. Hence, it is tempting to speculate that other mRNAs could actually operate dual functions in a given cell type or in a particular cellular context as it is the case for p53 mRNA in response to stress [106,115].
It is interesting to note that the ncRNAs and their associated protein(s) are mainly involved in the same biological processes, or even in the same pathways. Indeed, if we focus on the functions attributed to the examples of ncRNA/protein pairs mentioned in Table 1, 14 out of the 18 operate in the same pathways, although it may be in different space-time frames. They can also be involved in the regulation of one another as shown for the duo SRAP/SRA RNA [27]. The molecular basis of this kind of switch of activity is not clear, but it could occur in specific circumstances when the integrity of the genome is threatened and the cell’s defence strategies need to be strengthened or modulated.
A cascade of events:protein-coding genes that also produce ‘ncRNAs’ with a coding capacity
As mentioned above, circRNAs originate from back-splicing of a pre-RNA, and were originally described as ncRNAs with, to date, mainly a miRNA sponge effect [116]. However, hundreds of circRNAs were recently shown to be associated with ribosomes [117], although they lack the 5ʹ cap owing to their splicing origin. Nevertheless, N-6 methyladenosine (m6A) RNA editing or short internal ribosome entry sites-like (IRES-like) elements were suggested to serve as translation start sites [118,119]. As an example, the circRNA originating from the back-splicing of ZNF609 exon 2 has been involved in the proliferation of myoblasts [120]. It also encodes a peptide owing to the presence of an IRES-like sequence [120]. Further research is still needed to determine the possible functions of this circRNA-encoded peptide, and in a way that would discriminate them from the functions of the circRNA. Likewise, the CTNNB1 (β-catenin) gene produces a circRNA called circβ-catenin, which in turn codes for a new β-catenin shorter isoform [121]. This isoform serves as a decoy and competitively prevents GSK3β-mediated degradation of the full-length β-catenin protein, the overall consequence being the activation of the Wnt/β-catenin pathway involved in cancer progression [121]. In fact, the production of smaller protein isoforms through back-splicing and circularization of selected exons from their linear mRNA transcripts may be a more widespread mechanism with the demonstration that hundreds of cirRNAs do harbour a canonical start codon [120]. Whether these shorter isoforms would all compete with the longer isoforms produced from the same locus deserves attention. Despite unknowns regarding the cellular contexts that control the competition between canonical splicing and back-splicing and lead to variable expression patterns of circRNAs and linear RNAs [122], abnormal levels of cirRNAs have been implicated in tumorigenesis, possibly through the production of small protein isoforms [123].
From multitasking to multifunctional RNAs throughout evolution?
One of the most preserved processes during evolution, from lower to higher eukaryotes, i.e. from yeast to human, is probably the splicing of pre-mRNAs, whereby introns are removed from the original transcript leading to the juxtaposition of coding exons. Splicing has also been described in prokaryotes, although this is a rare event, mainly occurring in tRNAs [124]. In mammals, splicing is performed by the core spliceosome complex composed of snRNAs, the main ones being snRNA U1, U2, U4, U5 and U6, and of about 50 proteins, among which the PRP8 protein (pre-mRNA processing factor 8) is the largest and most highly conserved protein of the spliceosome [125] (Figure 4). In bacteria, the whole mechanism is driven by a single molecule, the intron itself, which belongs to the self-catalytic Group II introns [126]. In that case, Group II introns combine the structure and function of snRNAs into six RNA domains (I to VI) with an ORF that encodes the intron-encoded protein (IEP) equivalent to eukaryotic PRP8 [127] (Figure 4). Hence, group II introns from bacteria are inherently all bifunctional RNAs. Yet, one could consider that this is multitasking since the same RNA molecule, i.e. the intron, carries and not releases per se, both RNU RNAs and IEP protein.
Figure 4.
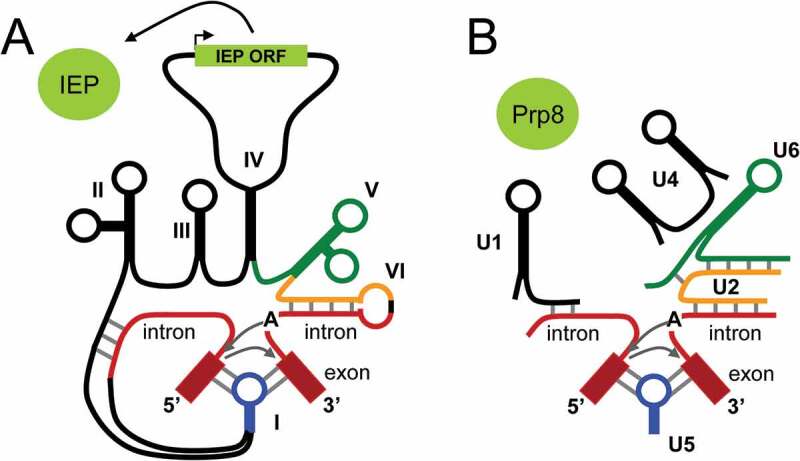
The special case of group II self-catalytic introns. (A) In prokaryotes, group II introns are composed of several domains that confer their self-catalytic activity. Domains I to VI are hypothesized to have evolved into separate activities in eukaryotes, namely snRNAs and the IEP homolog, PRP8 protein. (B)In eukaryotes, splicing of introns requires distinct effector RNAs. Grey arrows represent the 2 steps of the splicing reaction i.e. the nucleophilic attack of the branch point A and of the free 3ʹOH of the exon. 5ʹ and 3ʹ stand for 5ʹ- and 3ʹ-ends of exon; IEP, intron-encoded proteins; ORF, open reading frame. Adapted from Vosseberg [127]
Even in ancient living organisms like viruses, the RNA contains much more than just the genetic information translated into functional proteins since they also carry essential elements for their replication [128]. In addition, their untranslated extremities also seem to have a structure similar to that of an intrinsic tRNA [129].
Prokaryotic genomes are dominated by coding sequences, although dozens of regulatory ncRNAs are present and expressed. Conversely, eukaryotic genomes are mostly non-coding, which has led to the suggestion that the evolution of organisms towards increasing complexity has led to the emergence of numerous ncRNAs with various functions, the expression of which is thought to be restricted to different cell types and contexts [67,130]. The emergence of non-protein regulatory molecules could have then favoured the formation of increasingly complex regulatory networks, allowing for increasing sophistication in the way genomes are regulated in space and time to achieve more complex organismal developmental processes, in particular related to the development of new cognitive functions [67,131].
In fact, all the mentioned observations suggest that the multi-functionality of RNAs could in fact result from a combination of the first two hypotheses with the following scenario: RNAs were essential and omnipresent during the emergence of life, and were gradually supplanted by other more effective and stable macromolecules, including DNA and proteins. Although they remained present, they became available for additional functions as living organisms became more complex and required increasingly sophisticated regulatory networks. If this scenario holds true, some RNAs must have appeared recently during evolution. This is indeed the case in bacteria: most bacterial regulatory RNAs are expressed exclusively in some species and are absent in others, despite their phylogenetic proximity. Thus, RNAs are essential macromolecule in the emergence, evolution and complexification of living organisms, but bifunctional RNAs probably allow high reactivity to changes in the environmental conditions to which host organisms are subjected and for rapid and effective micro-evolution.
Conclusion
It is clear now that the flow of genetic information does not simply go from DNA to proteins through RNA intermediates, or from DNA to a vast repertoire of long and short non-coding RNAs. We went over specific examples where a same genetic locus or pre-RNA can produce coding and non-coding transcript, long or short, and shake the original belief that portions of the genome are either coding or non-coding. Whether these are just discrete cases or the manifestation of a more pervasive phenomenon remains to be properly evaluated. Nonetheless, one has to admit that the information contained in a given portion of the genome can be multiple. As mentioned, all the components produced may co-exist in the cell in a tightly controlled equilibrium and potentially in the same regulatory loops or pathways. There are also cases where a switch of functions operates during development or differentiation for instance. We have also mentioned in many places that cellular stress is a widespread mechanism in most species that can cause a change between distinct RNA entities. Why switching from an mRNA to an ncRNA or a long to small ncRNA may have to do with the need to produce regulatory molecules in specific sub-cellular locations, with increased or decreased stability or number of partners and targets. With splicing being mainly co-transcriptional, it certainly offers a versatile system to rapidly switch gears in a developmental process or in response to genomic insults. Although our vision of the mechanisms and actors behind these changes in the nature of the RNAs produced by a genetic locus is still fragmentary, it is clear that alterations of the equilibrium between these components are altered in pathological situations.
In contrast to the concept of pervasive transcription born over a decade ago, or of pervasive translation proposed fairly recently, the concept of multiple information carried by genes and RNAs only slowly attracts attention. This probably reflects the difficulties in searching for and testing this multiplicity of information when non-targeted functional assays are likely to be misleading. This would be however important in contexts where it appears that both entities from the same transcription unit (mRNA and ncRNA or two ncRNAs) often operate in the same biological processes, or even in the same biological pathways. If the balance between these entities contributes to a global function and its dynamics influences cell fate, then, knowing which molecules are at play would become important for functional experiments. Although it may still seem eccentric to want to know if the function of a genomic locus is through proteins, long or short RNAs, or a dynamic combination of these, it may become less anecdotal in disease situations and in the hunting of diagnostic biomarkers or druggable targets.
The concept of bifunctional RNA is gaining momentum with an increasing number of reported cases. We would like to speculate that studying these ‘chimeras’ between coding and non-coding RNA will help to decipher the evolutionary links between these two groups of molecules.
Funding Statement
B.B. was supported by the Association Française Contre les Myopathies (AFM-Téléthon) under fellowship #21363, C.F. is supported by the Institut National de la Santé et de la Recherche Médicale (INSERM) and F.H. is supported by the Centre National de la Recherche Scientifique (CNRS). This work was supported by AFM (Association Française contre les Myopathies), under Grant #22341.
Author contributions
B.B., C.F. and F.H. wrote the paper.
Disclosure statement
The authors declare no conflict of interest.
References
- [1].Amaral PP, Dinger ME, Mercer TR, et al. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. [DOI] [PubMed] [Google Scholar]
- [2].ENCODE Project Consortium . An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hombach S, Kretz M.. Non-coding RNAs: classification, biology and functioning. Adv Exp Med Biol. 2016;937:3–17. [DOI] [PubMed] [Google Scholar]
- [4].St Laurent G, Wahlestedt C, Kapranov P.. The Landscape of long noncoding RNA classification. Trends Genet. 2015;31:239–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cipolla GA, de Oliveira JC, Salviano-Silva A, et al. Long non-coding RNAs in multifactorial diseases: another layer of complexity. Noncoding RNA. 2018;4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bao Z, Yang Z, Huang Z, et al. LncRNADisease 2.0: an updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2019;47:D1034–D1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Abascal F, Juan D, Jungreis I, et al. Loose ends: almost one in five human genes still have unresolved coding status. Nucleic Acids Res. 2018;46:7070–7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Carninci P, Kasukawa T, Katayama S, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. [DOI] [PubMed] [Google Scholar]
- [10].Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–423. [DOI] [PubMed] [Google Scholar]
- [11].Gerstein MB, Bruce C, Rozowsky JS, et al. What is a gene, post-ENCODE? History and updated definition. Genome Res. 2007;17:669–681. [DOI] [PubMed] [Google Scholar]
- [12].Ulveling D, Francastel C, Hube F. When one is better than two: RNA with dual functions. Biochimie. 2011;93:633–644. [DOI] [PubMed] [Google Scholar]
- [13].Chooniedass-Kothari S, Emberley E, Hamedani MK, et al. The steroid receptor RNA activator is the first functional RNA encoding a protein. FEBS Lett. 2004;566:43–47. [DOI] [PubMed] [Google Scholar]
- [14].Hube F, Guo J, Chooniedass-Kothari S, et al. Alternative splicing of the first intron of the steroid receptor RNA activator (SRA) participates in the generation of coding and noncoding RNA isoforms in breast cancer cell lines. DNA Cell Biol. 2006;25:418–428. [DOI] [PubMed] [Google Scholar]
- [15].Dinger ME, Pang KC, Mercer TR, et al. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput Biol. 2008;4:e1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kumari P, Sampath K. cncRNAs: bi-functional RNAs with protein coding and non-coding functions. Semin Cell Dev Biol. 2015;47-48:40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Raina M, King A, Bianco C, et al. Dual-Function RNAs. Microbiol Spectr. 2018;6. DOI: 10.1128/microbiolspec.ARBA-0023-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dhamija S, Menon MB. Non-coding transcript variants of protein-coding genes - what are they good for? RNA Biol. 2018;1–7. DOI: 10.1080/15476286.2018.1511675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sampath K, Ephrussi A. CncRNAs: RNAs with both coding and non-coding roles in development. Development. 2016;143:1234–1241. [DOI] [PubMed] [Google Scholar]
- [20].Qin F, Zhang Y, Liu J, et al. SLC45A3-ELK4 functions as a long non-coding chimeric RNA. Cancer Lett. 2017;404:53–61. [DOI] [PubMed] [Google Scholar]
- [21].Hube F, Francastel C. Mammalian introns: when the junk generates molecular diversity. Int J Mol Sci. 2015;16:4429–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hube F, Francastel C. Coding and non-coding RNAs, the frontier has never been so blurred. Front Genet. 2018;9:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Muro EM, Mah N, Andrade-Navarro MA. Functional evidence of post-transcriptional regulation by pseudogenes. Biochimie. 2011;93:1916–1921. [DOI] [PubMed] [Google Scholar]
- [24].Ulveling D, Francastel C, Hube F. Identification of potentiallynew bifunctional RNA based on genome-wide data-mining of alternative splicing events. Biochimie. 2011;93:2024–2027. [DOI] [PubMed] [Google Scholar]
- [25].Lanz RB, McKenna NJ, Onate SA, et al. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. [DOI] [PubMed] [Google Scholar]
- [26].Emberley E, Huang GJ, Hamedani MK, et al. Identification of new human coding steroid receptor RNA activator isoforms. Biochem Biophys Res Commun. 2003;301:509–515. [DOI] [PubMed] [Google Scholar]
- [27].Hube F, Velasco G, Rollin J, et al. Steroid receptor RNA activator protein binds to and counteracts SRA RNA-mediated activation of MyoD and muscle differentiation. Nucleic Acids Res. 2011;39:513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chaudhary S, Khokhar W, Jabre I, et al. Alternative splicing and protein diversity: plants versus animals. Front Plant Sci. 2019;10:708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Barash Y, Calarco JA, Gao W, et al. Deciphering the splicing code. Nature. 2010;465:53–59. [DOI] [PubMed] [Google Scholar]
- [30].Schmitz U, Pinello N, Jia F, et al. Intron retention enhances gene regulatory complexity in vertebrates. Genome Biol. 2017;18:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Thiele A, Nagamine Y, Hauschildt S, et al. AU-rich elements and alternative splicing in the beta-catenin 3ʹUTR can influence the human beta-catenin mRNA stability. Exp Cell Res. 2006;312:2367–2378. [DOI] [PubMed] [Google Scholar]
- [32].Jacob AG, Smith CWJ. Intron retention as a component of regulated gene expression programs. Hum Genet. 2017;136:1043–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Middleton R, Gao D, Thomas A, et al. IRFinder: assessing the impact of intron retention on mammalian gene expression. Genome Biol. 2017;18:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Edwards CR, Ritchie W, Wong JJ, et al. A dynamic intron retention program in the mammalian megakaryocyte and erythrocyte lineages. Blood. 2016;127:e24–e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wong JJ, Au AY, Ritchie W, et al. Intron retention in mRNA: no longer nonsense: known and putative roles of intron retention in normal and disease biology. Bioessays. 2016;38:41–49. [DOI] [PubMed] [Google Scholar]
- [36].Braunschweig U, Barbosa-Morais NL, Pan Q, et al. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 2014;24:1774–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wong JJ, Ritchie W, Ebner OA, et al. Orchestrated intron retention regulates normal granulocyte differentiation. Cell. 2013;154:583–595. [DOI] [PubMed] [Google Scholar]
- [38].Williamson L, Saponaro M, Boeing S, et al. UV irradiation induces a non-coding RNA that functionally opposes the protein encoded by the same gene. Cell. 2017;168:843–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Grelet S, Link LA, Howley B, et al. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat Cell Biol. 2017;19:1105–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. [DOI] [PubMed] [Google Scholar]
- [41].Havens MA, Reich AA, Duelli DM, et al. Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic Acids Res. 2012;40:4626–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Hube F, Ulveling D, Sureau A, et al. Short intron-derived ncRNAs. Nucleic Acids Res. 2017;45:4768–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Dieci G, Preti M, Montanini B. Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics. 2009;94:83–88. [DOI] [PubMed] [Google Scholar]
- [44].Falaleeva M, Pages A, Matuszek Z, et al. Dual function of C/D box small nucleolar RNAs in rRNA modification and alternative pre-mRNA splicing. Proc Natl Acad Sci U S A. 2016;113:E1625–E1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Dudnakova T, Dunn-Davies H, Peters R, et al. Mapping targets for small nucleolar RNAs in yeast. Wellcome Open Res. 2018;3:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Leung YY, Kuksa PP, mlie-Wolf A, et al. DASHR: database of small human noncoding RNAs. Nucleic Acids Res. 2016;44:D216–D222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Eddy SR. Noncoding RNA genes. Curr Opin Genet Dev. 1999;9:695–699. [DOI] [PubMed] [Google Scholar]
- [48].Smith CM, Steitz JA. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5ʹ-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol. 1998;18:6897–6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Goustin AS, Thepsuwan P, Kosir MA, et al. The growth-arrest-specific (GAS)-5 long non-coding RNA: A Fascinating lncRNA widely expressed in cancers. Noncoding RNA. 2019;5:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Berezikov E, Chung WJ, Willis J, et al. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Da Fonseca BHR, Domingues DS, Paschoal AR. mirtronDB: a mirtron knowledge base. Bioinformatics. 2019;35:3873–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Janas MM, Khaled M, Schubert S, et al. Feed-forward microprocessing and splicing activities at a microRNA-containing intron. PLoS Genet. 2011;7:e1002330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Gao X, Qiao Y, Han D, et al. Enemy or partner: relationship between intronic microRNAs and their host genes. IUBMB Life. 2012;64:835–840. [DOI] [PubMed] [Google Scholar]
- [55].Amourda C, Saunders TE. The mirtron miR-1010 functions in concert with its host gene SKIP to balance elevation of nAcRbeta2. Sci Rep. 2020;10:1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Svanborg K, Gottlieb C, Bendvold E, et al. Variation in, and inter-relationship between, prostaglandin levels and other semen parameters in normal men. Int J Androl. 1989;12:411–419. [DOI] [PubMed] [Google Scholar]
- [57].Wilusz JE. A 360 degrees view of circular RNAs: from biogenesis to functions. Wiley Interdiscip Rev RNA. 2018;9:e1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Talhouarne GJS, Gall JG. Lariat intronic RNAs in the cytoplasm of vertebrate cells. Proc Natl Acad Sci U S A. 2018;115:E7970–E7977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bogard B, Francastel C, Hube F. A new method for the identification of thousands of circular RNAs. Non-cod RNA Investgat. 2018;2:1–5. [Google Scholar]
- [60].Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Maass PG, Glazar P, Memczak S, et al. A map of human circular RNAs in clinically relevant tissues. J Mol Med (Berl). 2017;95:1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Abelson J, Trotta CR, Li H. tRNA splicing. J Biol Chem. 1998;273:12685–12688. [DOI] [PubMed] [Google Scholar]
- [64].Singh SK, Gurha P, Tran EJ, et al. Sequential 2ʹ-O-methylation of archaeal pre-tRNATrp nucleotides is guided by the intron-encoded but trans-acting box C/D ribonucleoprotein of pre-tRNA. J Biol Chem. 2004;279:47661–47671. [DOI] [PubMed] [Google Scholar]
- [65].Pan T. Modifications and functional genomics of human transfer RNA. Cell Res. 2018;28:395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Joardar A, Gurha P, Skariah G, et al. Box C/D RNA-guided 2ʹ-O methylations and the intron of tRNATrp are not essential for the viability of Haloferax volcanii. J Bacteriol. 2008;190:7308–7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Kapusta A, Feschotte C. Volatile evolution of long noncoding RNA repertoires: mechanisms and biological implications. Trends Genet. 2014;30:439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Gabory A, Jammes H, Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays. 2010;32:473–480. [DOI] [PubMed] [Google Scholar]
- [69].Onyango P, Feinberg AP. A nucleolar protein, H19 opposite tumor suppressor (HOTS), is a tumor growth inhibitor encoded by a human imprinted H19 antisense transcript. Proc Natl Acad Sci U S A. 2011;108:16759–16764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Berteaux N, Aptel N, Cathala G, et al. A novel H19 antisense RNA overexpressed in breast cancer contributes to paternal IGF2 expression. Mol Cell Biol. 2008;28:6731–6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Keniry A, Oxley D, Monnier P, et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14:659–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13:313–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Dykes IM, Emanueli C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genomics Proteomics Bioinformatics. 2017;15:177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Liu B, Shyr Y, Cai J, et al. Interplay between miRNAs and host genes and their role in cancer. Brief Funct Genomics. 2018;18:255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Wilusz JE. Long noncoding RNAs: re-writing dogmas of RNA processing and stability. Biochim Biophys Acta. 2016;1859:128–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Zhang X, Hamblin MH, Yin KJ. The long noncoding RNA Malat1: its physiological and pathophysiological functions. RNA Biol. 2017;14:1705–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Wilusz JE, Freier SM, Spector DL. 3ʹ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Tripathi V, Ellis JD, Shen Z, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Gast M, Schroen B, Voigt A, et al. Long noncoding RNA MALAT1-derived mascRNA is involved in cardiovascular innate immunity. J Mol Cell Biol. 2016;8:178–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Sunwoo H, Dinger ME, Wilusz JE, et al. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Brown JA, Valenstein ML, Yario TA, et al. Formation of triple-helical structures by the 3ʹ-end sequences of MALAT1 and MENbeta noncoding RNAs. Proc Natl Acad Sci U S A. 2012;109:19202–19207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Zhang B, Mao YS, Diermeier SD, et al. Identification and characterization of a class of MALAT1-like genomic loci. Cell Rep. 2017;19:1723–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Pages A, Dotu I, Pallares-Albanell J, et al. The discovery potential of RNA processing profiles. Nucleic Acids Res. 2018;46:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Lee YS, Shibata Y, Malhotra A, et al. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009;23:2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Keam SP, Hutvagner G. tRNA-derived fragments (tRFs): emerging new roles for an ancient RNA in the regulation of gene expression. Life (Basel). 2015;5:1638–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Pliatsika V, Loher P, Magee R, et al. MINTbase v2.0: a comprehensive database for tRNA-derived fragments that includes nuclear and mitochondrial fragments from all the cancer genome atlas projects. Nucleic Acids Res. 2018;46:D152–D159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Abel Y, Rederstorff M. SnoRNAs and the emerging class of sdRNAs: multifaceted players in oncogenesis. Biochimie. 2019;164:17–21. [DOI] [PubMed] [Google Scholar]
- [88].Taft RJ, Glazov EA, Lassmann T, et al. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Scott MS, Ono M. From snoRNA to miRNA: dual function regulatory non-coding RNAs. Biochimie. 2011;93:1987–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Chow RD, Chen S. Sno-derived RNAs are prevalent molecular markers of cancer immunity. Oncogene. 2018;37:6442–6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Kuscu C, Kumar P, Kiran M, et al. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA. 2018;24:1093–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Tao EW, Cheng WY, Li WL, et al. tiRNAs: A novel class of small noncoding RNAs that helps cells respond to stressors and plays roles in cancer progression. J Cell Physiol. 2020;235:683–690. [DOI] [PubMed] [Google Scholar]
- [93].Schorn AJ, Gutbrod MJ, LeBlanc C, et al. LTR-retrotransposon control by tRNA-derived small RNAs. Cell. 2017;170:61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Ender C, Krek A, Friedlander MR, et al. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–528. [DOI] [PubMed] [Google Scholar]
- [95].Brameier M, Herwig A, Reinhardt R, et al. Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res. 2011;39:675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Yu F, Bracken CP, Pillman KA, et al. p53 represses the oncogenic Sno-MiR-28 derived from a SnoRNA. PLoS One. 2015;10:e0129190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Martens-Uzunova ES, Olvedy M, Jenster G. Beyond microRNA–novel RNAs derived from small non-coding RNA and their implication in cancer. Cancer Lett. 2013;340:201–211. [DOI] [PubMed] [Google Scholar]
- [98].Xiao J, Lin H, Luo X, et al. miR-605 joins p53 network to form a p53: miR-605:Mdm2positive feedback loop in response to stress. Embo J. 2011;30:524–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Ruiz-Orera J, Messeguer X, Subirana JA, et al. Long non-coding RNAs as a source of new peptides. Elife. 2014;3:e03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Zeng C, Fukunaga T, Hamada M. Identification and analysis of ribosome-associated lncRNAs using ribosome profiling data. BMC Genomics. 2018;19:414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Anderson DM, Anderson KM, Chang CL, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Nelson BR, Makarewich CA, Anderson DM, et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. 2016;351:271–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Stein CS, Jadiya P, Zhang X, et al. Mitoregulin: A lncRNA-encoded microprotein that supports mitochondrial supercomplexes and respiratory efficiency. Cell Rep. 2018;23:3710–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Choi SW, Kim HW, Nam JW. The small peptide world in long noncoding RNAs. Brief Bioinform. 2018;20:1853–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Candeias MM, Malbert-Colas L, Powell DJ, et al. P53 mRNA controls p53 activity by managing Mdm2 functions. Nat Cell Biol. 2008;10:1098–1105. [DOI] [PubMed] [Google Scholar]
- [107].Matsumoto A, Pasut A, Matsumoto M, et al. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature. 2017;541:228–232. [DOI] [PubMed] [Google Scholar]
- [108].Spencer HL, Sanders R, Boulberdaa M, et al. The LINC00961 transcript and its encoded micropeptide, small regulatory polypeptide of amino acid response, regulate endothelial cell function. Cardiovasc Res. 2020;116:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Heesch S, Witte FSchneider-Lunitz V, et al. The translational landscape of the human heart. Cell. 2019;178:242–260. [DOI] [PubMed] [Google Scholar]
- [110].Clemson CM, Hutchinson JN, Sara SA, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Vallot C, Rougeulle C. Long non-coding RNAs and human X-chromosome regulation: a coat for the active X chromosome. RNA Biol. 2013;10:1262–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Chen J, Brunner AD, Cogan JZ, et al. Pervasive functional translation of noncanonical human open reading frames. Science. 2020;367:1140–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Ingolia NT, Brar GA, Stern-Ginossar N, et al. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014;8:1365–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Ulveling D, Dinger ME, Francastel C, et al. Identification of a dinucleotide signature that discriminates coding from non-coding long RNAs. Front Genet. 2014;5:316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Candeias MM. The can and can’t dos of p53 RNA. Biochimie. 2011;93:1962–1965. [DOI] [PubMed] [Google Scholar]
- [116].Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79. [DOI] [PubMed] [Google Scholar]
- [117].Schneider T, Bindereif A. Circular RNAs: coding or noncoding? Cell Res. 2017;27:724–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27:626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Fan X, Yang Y, Wang Z. Pervasive translation of circular RNAs driven by short IRES-like elements. BioRXiv. 2018;23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Legnini I, Di TG, Rossi F, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Liang WC, Wong CW, Liang PP, et al. Translation of the circular RNA circbeta-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019;20:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Guarnerio J, Bezzi M, Jeong JC, et al. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;165:289–302. [DOI] [PubMed] [Google Scholar]
- [124].Reinhold-Hurek B, Shub DA. Self-splicing introns in tRNA genes of widely divergent bacteria. Nature. 1992;357:173–176. [DOI] [PubMed] [Google Scholar]
- [125].Nguyen TH, Galej WP, Fica SM, et al. CryoEM structures of two spliceosomal complexes: starter and dessert at the spliceosome feast. Curr Opin Struct Biol. 2016;36:48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Novikova O, Belfort M. Mobile group II introns as ancestral eukaryotic elements. Trends Genet. 2017;33:773–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Vosseberg J, Snel B. Domestication of self-splicing introns during eukaryogenesis: the rise of the complex spliceosomal machinery. Biol Direct. 2017;12:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Forterre P. The two ages of the RNA world, and the transition to the DNA world: a story of viruses and cells. Biochimie. 2005;87:793–803. [DOI] [PubMed] [Google Scholar]
- [129].Chujo T, Ishibashi K, Miyashita S, et al. Functions of the 5ʹ- and 3ʹ-untranslated regions of tobamovirus RNA. Virus Res. 2015;206:82–89. [DOI] [PubMed] [Google Scholar]
- [130].Mattick JS. The hidden genetic program of complex organisms. Sci Am. 2004;291:60–67. [DOI] [PubMed] [Google Scholar]
- [131].Mattick JS, Gagen MJ. The evolution of controlled multitasked gene networks: the role of introns and other noncoding RNAs in the development of complex organisms. Mol Biol Evol. 2001;18:1611–1630. [DOI] [PubMed] [Google Scholar]
- [132].Novick RP, Ross HF, Projan SJ, et al. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. Embo J. 1993;12:3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Balaban N, Novick RP. Translation of RNAIII, the Staphylococcus aureus agr regulatory RNA molecule, can be activated by a 3ʹ-end deletion. FEMS Microbiol Lett. 1995;133:155–161. [DOI] [PubMed] [Google Scholar]
- [134].Vanderpool CK, Balasubramanian D, Lloyd CR. Dual-functionRNA regulators in bacteria. Biochimie. 2011;93:1943–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Gimpel M, Preis H, Barth E, et al. SR1–a small RNA with two remarkably conserved functions. Nucleic Acids Res. 2012;40:11659–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Lauressergues D, Couzigou JM, Clemente HS, et al. Primary transcripts of microRNAs encode regulatory peptides. Nature. 2015;520:90–93. [DOI] [PubMed] [Google Scholar]
- [137].Campalans A, Kondorosi A, Crespi M. Enod40, a short open reading frame-containing mRNA, induces cytoplasmic localization of a nuclear RNA binding protein in Medicago truncatula. Plant Cell. 2004;16:1047–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Rongo C, Gavis ER, Lehmann R. Localization of oskar RNA regulates oskar translation and requires Oskar protein. Development. 1995;121:2737–2746. [DOI] [PubMed] [Google Scholar]
- [139].Lim S, Kumari P, Gilligan P, et al. Dorsal activity of maternal squint is mediated by a non-coding function of the RNA. Development. 2012;139:2903–2915. [DOI] [PubMed] [Google Scholar]
- [140].Kloc M, Foreman V, Reddy SA. Binary function of mRNA. Biochimie. 2011;93:1955–1961. [DOI] [PubMed] [Google Scholar]
- [141].Peng L, Chen G, Zhu Z, et al. Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprung’s disease. Oncotarget. 2017;8:808–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Young TM, Tsai M, Tian B, et al. Cellular mRNA activates transcription elongation by displacing 7SK RNA. PLoS One. 2007;2:e1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Young TM, Wang Q, Pe’ery T, et al. The human I-mfa domain-containing protein, HIC, interacts with cyclin T1 and modulates P-TEFb-dependent transcription. Mol Cell Biol. 2003;23:6373–6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Nagano H, Yamagishi N, Tomida C, et al. A novel myogenic function residing in the 5ʹ non-coding region of Insulin receptor substrate-1 (Irs-1) transcript. BMC Cell Biol. 2015;16:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Long YC, Cheng Z, Copps KD, et al. Insulin receptor substrates Irs1 and Irs2 coordinate skeletal muscle growth and metabolism via the Akt and AMPK pathways. Mol Cell Biol. 2011;31:430–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Xu Q, Walker D, Bernardo A, et al. Intron-3 retention/splicing controls neuronal expression of apolipoprotein E in the CNS. J Neurosci. 2008;28:1452–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Kavela S, Shinde SR, Ratheesh R, et al. PNUTS functions as a proto-oncogene by sequestering PTEN. Cancer Res. 2013;73:205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Roberts GC, Smith CW. Alternative splicing: combinatorial output from the genome. Curr Opin Chem Biol. 2002;6:375–383. We apologize to those whose works have not been cited in this article owing to lack of space. [DOI] [PubMed] [Google Scholar]


