ABSTRACT
Long non-coding RNAs (lncRNAs) have been noted to influence the progression of ossification of posterior longitudinal ligament (OPLL). The work aims to probe the effect of lncRNA SNHG1 on osteogenic differentiation of ligament fibroblastic cells (LFCs). Aberrantly expressed lncRNAs in ossified PLL tissues were screened out by microarray analysis. Gain- and loss-of function experiments of SNHG1 were performed to identify its role in osteogenic differentiation of LFCs. The downstream molecules of SNHG1 were explored. Altered expression of miR-320b was introduced in LFCs as well. The interactions among SNHG1, miR-320b and IFNGR1 were identified. Consequently, SNHG1 was found highly expressed in OPLL patients. Silencing of SNHG1 inhibited BMP-2, RUNX2 and OCN expression and the ALP activity and reduced osteogenic differentiation of LFCs. Importantly, SNHG1 could and upregulate IFNGR1 through serving as a sponge for miR-320b. Over-expression of miR-320b inhibited osteogenic differentiation of LFCs and inactivated the JAK/STAT signaling pathway. Further administration of Fedratinib, a JAK2-specific agonist, increased osteogenic differentiation of LFCs. To conclude, the study suggested that SNHG1 could upregulate IFNGR1 by sequestering miR-320b and activate the JAK/STAT signaling. Silencing of SNHG1 could reduce the osteogenic differentiation and mineralization of LFCs. The study may offer new insights into OPLL treatment.
KEYWORDS: Long non-coding RNA SNHG1, ossification of posterior longitudinal ligament, ceRNA, microrna-320b, IFNGR1, JAK/STAT signaling pathway
Introduction
Ossification of posterior longitudinal ligament (OPLL) is attributed to the degeneration and calcification of tendons and ligaments caused by chronic inflammation [1,2]. Pathologically, OPLL is featured with abnormal ossification in the posterior ligament, and the developing bony spurs can lead to radiculopathy, myelopathy or even quadriplegia through compression or direct injury to the spinal cord or nerve roots [3]. OPLL is more frequently found in the middle-aged and elderly population in Asia [4]. Anterior and posterior approaches are currently two major therapies for OPLL, but the outcomes are still not satisfactory [4]. Fibroblastic cells are the most common cells in connective tissues and are implicated in the progression of exotic ossification in multiple diseases [5]. Preventing osteogenic differentiation of fibroblastic cells might serve as an important and promising therapeutic target for OPLL treatment.
Although emerging studies suggest that the causes of OPLL are largely correlated with aberrant bone and mineral metabolism, during which abnormal gene mutations, hormones, cytokines, and intracellular signal transcription factors are frequently observed [6–8], the underlying mechanism and regulatory networks remain largely unknown. Non-coding RNAs, mainly including long non-coding RNAs (lncRNAs) and microRNAs (miRNAs), are diverse transcriptional outcomes of mammalian genomes that participate in diverse processes coordinating gene expression [9–11]. LncRNAs are longer than 200 nucleotides that play critical roles in gene regulation including mRNA translation in cytoplasm and chromatin modification in nuclei [12]. miRNAs are about 21 nucleotides in length and serve as guide molecules that induce mRNA degradation or block protein translation of the target genes [13]. But there is very limited information concerning the role of lncRNAs and miRNAs in OPLL. Nevertheless, both lncRNA and miRNAs mediate diverse biological conditions including osteogenesis [14–16]. Importantly, the “competitive endogenous RNA (ceRNA)” theory, which was firstly initiated in 2011, proposing that lncRNAs may “talk” with mRNAs through the shared miRNA response elements [17]. The ceRNA network has recently been validated in several human diseases [18,19] including OPLL [7]. In light of these findings, our study was performed to figure out the possible ceRNA network and the molecular mechanism involved in OPLL. Integrated microarray analysis, online predictions, and expression detection between ossified and non-ossified PLLs were performed to identify the possible cytokines. Artificial up- or down-regulation of the target cytokines were introduced to identify their roles in OPLL progression in both cell and animal experiments.
Materials and methods
Ethics statement
The study got the approval of the Clinical Ethical Committee of the First Affiliated Hospital of Zhengzhou University. Each eligible participant signed the informed consent. All experimental procedures were conducted in line with the ethical guidelines for the study of experimental pain in conscious animals.
Clinical tissue sample collection
Normal PLL (from patients with spinal trauma) or OPLL in patients were diagnosed by Computed Tomography (CT) and Preoperative Magnetic Resonance Imaging (MRI). The tissue samples of PLL or OPLL were obtained by surgical resection, and then the ligament fibroblastic cells (LFCs) from patients were collected as previously reported [20]. In brief, the ligament tissues were washed twice in 1% penicillin/streptomycin-supplemented phosphate buffer saline (PBS). Then, the tissues were opened carefully under the microscope (Olympus Optical Co., Ltd, Tokyo., Japan) to avoid any contamination to osteoblasts or other cells. The ligament tissues were chopped and washed by PBS. The tissue plates were sorted on culture dishes (100 mm) and cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS), 1% L-glutamine and 1% penicillin/streptomycin (all purchased from Gibco Company, Grand Island, NY, USA). The tissues were cultured in humid environment at 37°C with 5% CO2. The fibroblast-like cells from tissues were collected using 0.02% EDTA/0.05% trypsin and spread on a T25 culture flask for further analysis. The cell morphology was first observed under a microscope, and the Vimentin expression in cells was determined by immunofluorescence to confirm the extracted cells were LFCs.
A total of 68 pairs of OPLL tissue samples were collected from anterior cervical discectomy and another 18 pairs of PLL tissues were extracted from anterior cervical vertebrectomy. All patients were subjected to anterior cervical decompression and interbody fusion with plate fixation, and the PLL were resected during surgery and collected as tissue samples. The demographic characteristics of patients is listed in Table 1, and the spinal cord signal intensity grading of MRI was performed as a previously study by Yuwaka [21]. Patients were included if: 1) they met the diagnostic criteria for cervical spondylotic myelopathy; 2) the lesioned spinal segments presented good range of motion; 3) they had one or two lesioned segments; 4) they did not have absolute contraindication. Patients were excluded if: 1) they did not show significant instable and developmental spinal stenosis; 2) they were younger than 20 years old or over 80 years old; 3) they had congenital or traumatic acute and severe segmental instability; 4) they had severe osteoporosis, metabolic osteopathy, tumor or infections; 3) they had severe cardiopulmonary diseases that cannot undertake surgery.
Table 1.
Demographic characteristics of the respondents
| Number | Age | Gender | Classification |
|---|---|---|---|
| 1 | 31 | Male | II |
| 2 | 35 | Male | I |
| 3 | 62 | Female | I |
| 4 | 47 | Male | II |
| 5 | 64 | Female | II |
| 6 | 52 | Male | II |
| 7 | 65 | Male | I |
| 8 | 59 | Male | II |
| 9 | 61 | Male | II |
| 10 | 67 | Male | I |
| 11 | 34 | Male | II |
| 12 | 51 | Female | I |
| 13 | 64 | Female | I |
| 14 | 68 | Male | II |
| 15 | 38 | Male | II |
| 16 | 55 | Male | II |
| 17 | 41 | Male | I |
| 18 | 32 | Female | I |
| 19 | 27 | Male | II |
| 20 | 53 | Male | II |
| 21 | 49 | Male | I |
| 22 | 42 | Male | I |
| 23 | 63 | Female | I |
| 24 | 43 | Male | I |
| 25 | 51 | Female | I |
| 26 | 53 | Female | I |
| 27 | 49 | Female | II |
| 28 | 43 | Female | II |
| 29 | 38 | Male | I |
| 30 | 38 | Male | I |
| 31 | 53 | Female | II |
| 32 | 64 | Female | I |
| 33 | 49 | Male | I |
| 34 | 65 | Female | I |
| 35 | 60 | Male | I |
| 36 | 46 | Female | I |
| 37 | 30 | Male | II |
| 38 | 27 | Male | II |
| 39 | 51 | Female | I |
| 40 | 31 | Female | II |
| 41 | 61 | Female | II |
| 42 | 30 | Female | II |
| 43 | 55 | Female | II |
| 44 | 35 | Male | I |
| 45 | 63 | Male | I |
| 46 | 53 | Female | II |
| 47 | 64 | Male | I |
| 48 | 63 | Male | I |
| 49 | 51 | Male | I |
| 50 | 53 | Male | I |
| 51 | 58 | Female | I |
| 52 | 25 | Female | I |
| 53 | 58 | Female | II |
| 54 | 52 | Male | I |
| 55 | 55 | Male | I |
| 56 | 51 | Female | I |
| 57 | 42 | Male | II |
| 58 | 59 | Female | II |
| 59 | 25 | Male | II |
| 60 | 26 | Female | I |
| 61 | 57 | Male | II |
| 62 | 27 | Male | I |
| 63 | 67 | Male | II |
| 64 | 67 | Male | II |
| 65 | 52 | Male | II |
| 66 | 42 | Female | II |
| 67 | 28 | Male | I |
| 68 | 28 | Male | II |
| 69 | 67 | Male | 0 |
| 70 | 62 | Male | 0 |
| 71 | 45 | Male | 0 |
| 72 | 47 | Female | 0 |
| 73 | 36 | Female | 0 |
| 74 | 33 | Female | 0 |
| 75 | 37 | Male | 0 |
| 76 | 41 | Female | 0 |
| 77 | 66 | Male | 0 |
| 78 | 60 | Male | 0 |
| 79 | 47 | Male | 0 |
| 80 | 39 | Male | 0 |
| 81 | 45 | Male | 0 |
| 82 | 49 | Male | 0 |
| 83 | 67 | Male | 0 |
| 84 | 66 | Female | 0 |
| 85 | 28 | Male | 0 |
Cell treatment and grouping
The amplified cDNA of lncRNA small nucleolar RNA host gene 1 (SNHG1) was cloned to the pcDNA3.1-SNHG1 vector (Invitrogen Inc., Carlsbad, CA, USA) using specific primers (Sangon Biotech Co., Ltd., Shanghai, China). The specific small interfering (si) RNA targeting SNHG1 was synthetized by Beijing TianGen Biotech Co., Ltd. (Beijing, China) with the corresponding pcDNA3.1 and scramble siRNA set as negative control (NC). The miR-320b mimic and miR-320b inhibitor was synthesized by GenePharma Co, Ltd. (Shanghai, China). The primers are presented in Table 2.
Table 2.
Primer sequences used for cell transfection
| Primer | Sequence |
|---|---|
| SNHG1 | Sense: 5'-GGGGTACGTTCTCATTTTTTACTGCTCGTG-3’ |
| Anti-sense: 5'-CGGGATCCATGTAATCAATCATTTTATTATTTTCATC-3’ | |
| si-SNHG1 | Sense: 5'-CCAGCAUCUCAUAAUCUAUTT-3’ |
| Anti-sense: 5'-AUAGAUUAUGAGAUGCUGGAA-3’ | |
| miR-320b | Sense: 5'-AAAGCUGGUGAGAGGGAA-3’ |
| Anti-sense: 5'-UUGCUCUCAACCCAGCUUU-3’ | |
| miR-inhibitor | Sense: 5'-UUGCCCUCUCAACCCAGCUUUU-3 |
Note: SNHG1, small nucleolar RNA host gene 1; si-SNHG1, small interfering SNHG1; miR, microRNA.
The LFCs from PLL or OPLL tissues were cultivated in DMEM containing 10% FBS and 1% penicillin/streptomycin. The cells were passaged when the cell confluence reached 80%, and the passage 3 cells were harvested for the subsequent experiments.
Well growing PLL-LFCs were allocated into scramble group (cells were transfected with 100 ng empty vector), SNHG1 group (cells were transfected with 100 ng pcDNA-3.1-SNHG1), SNHG1-mock group (cells were transfected with 100 ng pcDNA-3.1-SNHG1 and 50 ng mock), SNHG1-miR-320b-5p group (cells were transfected with 100 ng pcDNA-3.1-SNHG1 and 50 ng miR-320b-5p mimic) and SNHG1 + Fedratinib group (cells were transfected with 100 ng pcDNA-3.1-SNHG1 and Fedratinib).
Well-growing OPLL-LFCs were allocated into scramble group (cells were transfected with 100 ng empty vector), si-SNHG1 group (cells were transfected with 100 ng pcDNA3.1-siSNHG1), si-SNHG1-mock group (cells were transfected with 100 ng pcDNA3.1-siSNHG1 and 50 ng mock), SNHG1-inhibitor group (cells were transfected with 100 ng pcDNA3.1-siSNHG1 and 50 ng miR-320b inhibitor), DMSO group (cells were treated with DMSO) and Fedratinib group (cells were treated with 20 μM Fedratinib). Well-constructed vectors and the lipofectamine 2000 mixture was transfected into the cells using a lipofectamine 2000 kit (Invitrogen) as per the kit’s protocols. The cells were collected for the subsequent experiments 48 h after transfection. In brief, 50 ng or 100 ng pcDNA vector was loaded into each well and fixed with 250 μL DMEM, and 5 μL lipofectamine 2000 solution was also mixed with 250 μL DMEM. The diluted vectors were mixed with the lipofectamine solution (the total volume in each well was 500 μL). Then, the mixture was added into the cell plates for transfection. After 48 h, the expression of SNHG1 and miR-320b in OPLL-LFCs and PLL-LFCs were determined quantitative reverse transcription polymerase chain reaction (qRT-PCR) to examine the transfection efficiency, and the cells with stable transfection were collected for the subsequent experiments.
qRT-PCR
Total RNA from tissues and cells was collected using the RNAiso Plus (Takara, Otsu, Shiga, Japan) and Trizol LS Reagent (Takara). The qualified RNA for the subsequent experiments was determined by formaldehyde denaturing gel electrophoresis. Next, reverse transcription of RNA was performed using a PrimeScriptTM RT kit (Takara), and then the real-time qPCR was performed to quantify the RNA expression. The primer sequences are shown in Table 3. U6 was set as the internal reference for miRNA while GAPDH for other genes. Relative gene expression was determined using the 2−ΔΔCT method.
Table 3.
Primer sequences in RT-qPCR
| Primer | Sequence |
|---|---|
| SNHG1 | F: 5'-GTGACTTCGCCTGTGATGGA-3’ |
| R: 5'-GGCCTCTATCTGTACCTTTATTC-3’ | |
| BMP-2 | F: 5'-AGCCATTGCACACCTCAC-3’ |
| R: 5'-CGTGGTACCAAGAGGACAGAGT-3’ | |
| OCN | F: 5'-TGAGGACCATCTTCTGCTCA-3’ |
| R: 5'-TGGACATGAAGGCTTTGTCA-3’ | |
| RUNX2 | F: 5'-UUGCCCUCUCAACCCAGCUUUU-3 |
| R: 5'-TGGACATGAAGGCTTTGTCA-3’ | |
| miR-320b | F: 5'-CCTGGGAGTATGTCGATCTATTG-3’ |
| R: 5'-TGGTGTCGTGGAGTCG-3’ | |
| IFNGR1 | F: 5'-AAGTGGCGTGTTCAAGTGTG-3’ |
| R: 5'-GGCTTTGGAGATCTGAGCTG-3’ | |
| U6 | F: 5'-CTCGCTTCGGCAGCACA-3’ |
| R: 5'-AACGCTTCACGAATTTGCGT-3’ | |
| GAPDH | F: 5'-CGGACCAATACGACCAA-3’ |
| R: 5'-AGCCACATCGCTCAGACACC-3’ |
Note: RT-qPCR, reverse transcription quantitative polymerase chain reaction; SNHG1, small nucleolar RNA host gene 1; BMP-2, bone morphogenetic protein-2; OCN, osteocalcin; RUNX2, runt-related transcription factor 2; miR, microRNA; IFNGR1, interferon gamma receptor 1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Sub-cellular localization of SNHG1
Sub-cellular localization of SNHG1 was first predicted on the lncATLAS bioinformation system (http://lncatlas.crg.eu/), which suggested that SNHG1 is mainly localized in cytoplasm. Next, the localization of SNHG1 was validated through fluorescence in situ hybridization (FISH) assay in accordance with the instructions of a RiboTM lncRNA FISH Probe Mix (Green) Kit (Ribo Biotech, Guangzhou, China). Then a nuclear/cytoplastic RNA separation assay was performed using a PARISTM Kit (Life Technologies, Inc., Gaithersburg, MD, USA) to further detect the SNHG1 distribution in cells. All procedures were performed as the kit’s protocol.
Western blot analysis
Western blot analysis was conducted as guided by a previous report [22]. Cells or tissues were resuspended in cold cell lysis buffer containing protease inhibitor (Cell Signaling Technology, Beverly, MA, USA) and then incubated on ice for 15–30 minutes to collect the total protein. Then, the protein was separated on 4%-20% sodium dodecyl sulfate separation gel polyacrylamide gel electrophoresis (Invitrogen) and transferred to polyvinylidene fluoride membranes (Millipore, Corp., Billerica, MA, USA). The membranes were blocked in 5% skimmed milk and 0.05% Tween-20 for 10 min and then incubated with the primary antibodies at 4°C overnight and then with the secondary antibody at 37°C for 2 hours. The antibodies used in the experiments are shown in Table 4.
Table 4.
Antibodies used in western blot analysis and immunohistochemistry
| Antibody | Item. No. | Dilution ratio |
|---|---|---|
| BMP-2 | ab214821 | 1: 2000 |
| RUNX2 | ab192256 | 1: 5000 |
| OCN | ab13420 | 1: 2000 |
| JAK2 | ab108596 | 1: 100 |
| P-JAK2 | ab32101 | 1: 100 |
| STAT-1 | ab92506 | 1: 1000 |
| P-STAT-1 | ab109461 | 1: 5000 |
| STAT-2 | ab231184 | 1: 1000 |
| p-STAT-2 | ab53132 | 1: 5000 |
| β-actin | ab179467 | 1: 5000 |
| Secondary antibody | ab150117 | 1: 5000 |
Note: BMP-2, bone morphogenetic protein-2; RUNX2, runt-related transcription factor 2; OCN, osteocalcin; JAK, janus kinase; STAT, signal transducers and activators of transcription.
Alizarin red staining
The number of calcium nodules differentiated by osteoblasts was determined by alizarin red staining. All experimental procedures were conducted as guided by a previous study [23]. Cells were cultured in 12-well plates and fixed in 4% methanol for 45 min. Next, the samples were washed in distilled water and exposed to alizarin red S (2% aqueous solution, Sigma-Aldrich Chemical Company St Louis, MO, USA) for 5 min, and then washed and observed under the microscope at a 200 × magnification.
Alkaline phosphatase (ALP) staining
The activity of ALP in LFCs was evaluated by ALP staining as previously reported [24]. In brief, cells were washed in 1× PBS and immobilized in 4% paraformaldehyde for 15 min. Next, the cells were washed in deionized water, and then incubated in 166 µmL/4 µmL naphthol/fast blue solution (Sigma) in the dark for 20 min. The stained cells were observed and photographed under a digital camera (DMI 3000)-connected microscope (Leica, Solms, Germany) at a 200 × magnification.
Dual-luciferase reporter gene assay
The binding sequences between miR-320b-5p and SNHG1 or the 3'-untranslated regions (3'-UTR) of interferon-gamma receptor 1 (IFNGR1) were predicted on StarBase (http://starbase.sysu.edu.cn/). Then the SNHG1-wild type (WT) and mutant type (MT) sequences, as well as the IFNGR1-WT and IFNGR1-MT sequences were synthesized (Sangon) and then inserted into the pmiR-REPORTTM luciferase reporter vectors [25]. Well-constructed vectors were co-transfected either with miR-320b-5p mimic or mimic-NC into 293 T cells. Cells were lysed 24 h later, and the relative luciferase activity was assessed using a Dual-Luciferase Reporter Assay System (Promega Corp., WI, USA) [26].
Heterotopic bone formation in vivo
Well growing OPLL-LFCs and PLL-LFCs were made into suspensions and seeded into 7 mm × 5 mm × 2 mm collagen scaffolds (Bio-Oss, Baden-Baden, Germany) as previously reported [27]. Six weeks later, the collagen scaffolds were taken out for immunohistochemistry and hematoxylin and eosin (HE) staining as guided by a previous report [28]. The antibodies used in immunohistochemistry were the same as the ones used in western blot analysis.
Statistical analysis
The SPSS 21.0 (IBM Corp. Armonk, NY, USA) was used for data analysis. Kolmogorov–Smirnov tested the data were in normal distribution. Measurement data were exhibited as mean ± standard deviation (SD) from at least three independent experiments. Differences between every two groups were evaluated using the t test, while differences among multiple groups were compared using one-way or two-way analysis of variance (ANOVA). Tukey’s multiple comparisons test was used for the pairwise comparisons after ANOVA analysis. The p value was obtained from a two-tailed test, and p < 0.05 was considered to show significant difference.
Results
SNHG1 is highly expressed in patients with OPLL
LFCs were extracted from OPLL and PLL tissues, respectively The collected cells were confirmed as LFCs through the microscope observation and Vimentin immunofluorescence (p < 0.05) (Figure 1(a)). Next, the mRNA and protein expression levels of osteogenesis-related factors bone morphogenetic protein-2 (BMP-2), runt-related transcription factor 2 (RUNX2) and osteocalcin (OCN) in OPLL- or PLL-derived LFCs were detected. The results presented that the expression levels of the above factors were notably higher in OPLL-derived LFCs than that in PLL-derived ones (all p < 0.05) (Figure 1(b–c)). Then, the alizarin red staining was performed to identify the number of formed calcium nodules after 12 d of osteo-induction. It was found that the number of mineralized nodules formed by OPLL-derived LFCs was larger than that by PLL-derived ones. The same trends were found by ALP staining, which suggested that the ALP activity was higher in OPLL-derived LFCs than that in by PLL-derived ones (all p < 0.05) (Figure 1(d–e)).
Figure 1.
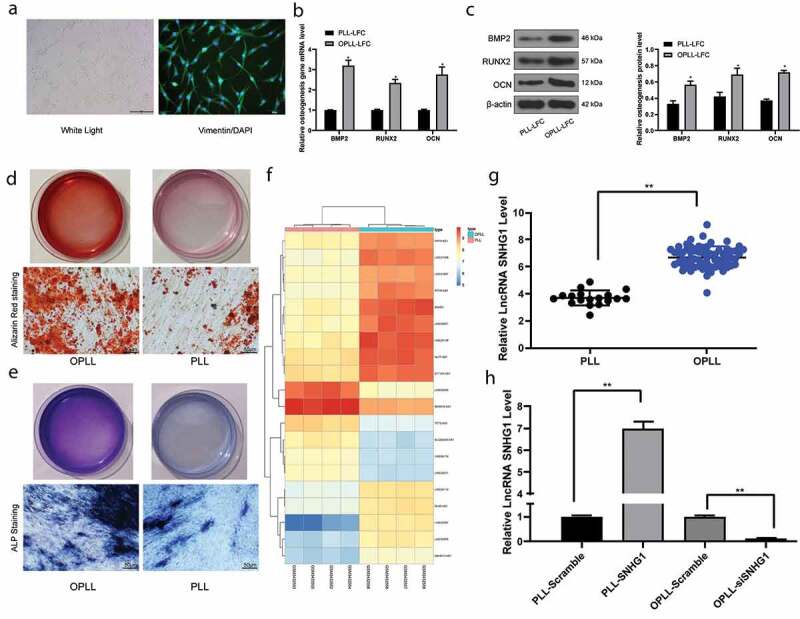
SNHG1 is highly expressed in patients with OPLL. A, PLL- or OPLL-derived LFCs identified by microscope observation and Vimentin immunofluorescence; B-C, mRNA (b) and protein (c) expression of osteogenesis-related factors BMP-2, RUNX2 and OCN determined by QRT-PCR and western blot analysis, respectively; D-E, the osteogenic properties of LFCs determined by Alizarin Red staining (d) and ALP (e) staining; F, the heatmap for differentially expressed lncRNAs between 4 OPLL tissues and 4 healthy PLL tissues determined by Arraystar Human LncRNA microarray V2.0 (Agilent_033010 Probe Name version); G, SNHG1 expression in 68 pairs of OPLL tissues and 18 pairs of PLL tissues identified by QRT-PCR; H, SNHG1 expression in cells after SNHG1 siRNA and SNHG1 overexpressing vector transfection determined by QRT-PCR as well. Data are expressed as the mean ± SD; in panels B and C, data were analyzed using two-way ANOVA, while data in panel H were analyzed using one-way ANOVA, and Tukey’s multiple comparisons test was used for the pairwise comparisons after ANOVA analysis; *, p < 0.05
LncRNAs with differential expression between 4 OPLL tissues and 4 healthy PLL tissues were screened out by a lncRNA microarray with |LogFC| > 1 and p < 0.05 as the criteria. Consequently, a total of 143 differentially expressed lncRNAs were screened out, among which 73 were up-regulated and 69 were down-regulated. A heatmap for top 20 differentially expressed lncRNAs is shown in figure 1(f), which suggests that SNHG1 held the greatest differential expression. Next, SNHG1 expression in 68 OPLL tissues and 18 PLL tissues was identified by qRT-PCR, which suggested that SNHG1 was significantly highly expressed in OPLL patients (p < 0.05) (Figure 1(g)). Then, to further identify the function of SNHG1 on the ossification of LFCs, pcDNA3.1-si-SNHG1 and pcDNA3.1-SNHG1 vectors were constructed and transfected into OPLL-derived or PLL-derived LFCs, respectively. The qRT-PCR results showed that SNHG1 expression in cells was correspondingly upregulated or suppressed, indicating the successful transfection (all p < 0.05) (Figure 1(h)).
Silencing of SNHG1 suppresses osteogenic differentiation ability of LFCs
The mRNA and protein expression of osteogenesis-related factors BMP-2, RUNX2 and OCN in each group of cells was determined after SNHG1 interference. The results of qRT-PCR and western blot analysis suggested that silencing of SNHG1 in OPLL-derived LFCs led to an inhibition in the levels of BMP-2, RUNX2 and OCN. In contrast, over-expression of SNHG1 increased the osteogenic differentiation ability of PLL-derived LFCs. The similar trends were found in alizarin red staining and ALP staining (all p < 0.05) (Figure 2(a–d)).
Figure 2.
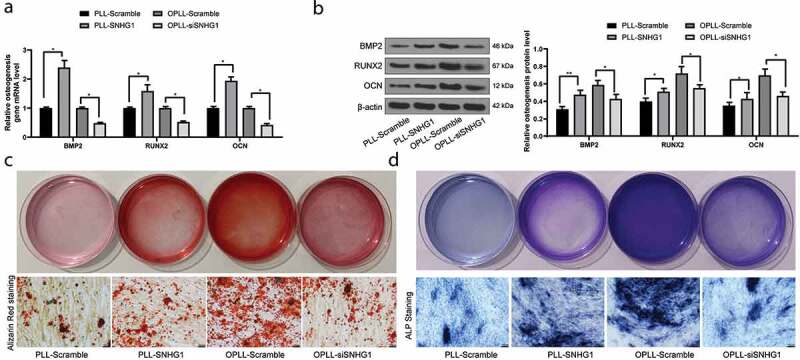
Silencing of SNHG1 inhibits the osteogenic differentiation of LFCs. A-B, mRNA (a) and protein (b) expression of osteogenesis-related factors BMP-2, RUNX2 and OCN determined by QRT-PCR and western blot analysis, respectively; C-D, the osteogenic properties of LFCs determined by alizarin red staining (c) and ALP (d) staining. The data are expressed as the mean ± SD. Two-way ANOVA and Tukey’s multiple comparison test was used to determine statistical significance; *, p < 0.05
SNHG1 competes with IFNGR1 to bind to miR-320b
To further explore the mechanisms involved in the above events, we first analyzed the sub-cellular localization of SNHG1. Data on the lncATLAS bioinformation system (http://lncatlas.crg.eu/) predicted that SNHG1 is mainly localized in cytoplasm (p < 0.05) (Figure 3(a)). Next, the FISH assay and separation of nuclear/cytoplastic RNA assays were performed, and the results suggested that SNHG1 was mainly distributed in the cytoplasm in LFCs (p < 0.05) (Figure 3(b–c)).
Figure 3.
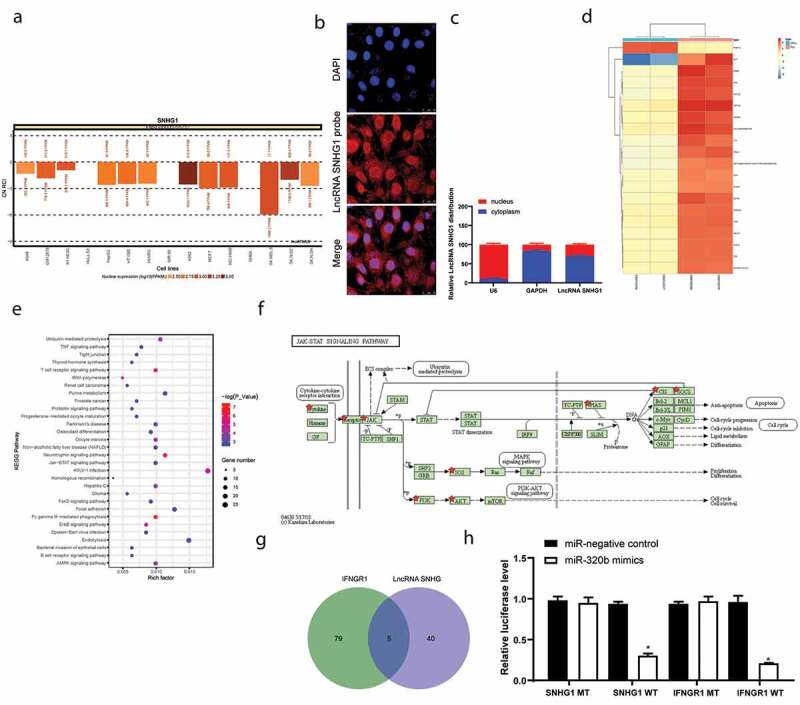
SNHG1 serves as a ceRNA for miR-320b to mediate IFNGR1 expression. A, subcellular localization of SNHG1 predicted on the lncATLAS DataBase (http://lncatlas.crg.eu/); B, FISH experiments with probes targeting SNHG1 were performed to validate the subcellular localization of SNHG1 in LFCs. The cytoplasm was stained with probes targeting SNHG1 (red stain), and the nuclei were stained with DAPI (blue stain); C, nuclear and cytoplasmic expression of SNHG1 in LFCs determined by QRT-PCR; D, heatmap for the top 30 differentially expressed genes between PLL- and OPLL- LFCs analyzed using the Llimma Rstudio based on GSE5464 microarray, with |LogFC ≥ 4.0| and p < 0.05 as the screening criteria; E, KEGG-based clustering analysis was performed on DAVID 6.8 software, and a total of 43 pathways where the differentially genes clustered were figured out; F, genes clustered on the JAK/STAT signal pathway; G, intersected miRNAs that could bind to SNHG1 and IFNGR1 predicted on StarBase; H, binding relationships between SNHG1 and miR-320b and between miR-320b and IFNGR1 validated by dual luciferase reporter gene assay. The data are expressed as the mean ± SD; two-way ANOVA and Tukey’s multiple comparison test was used to determine statistical significance; *, p < 0.05
This finding indicated that SNHG1 might exert functions through ceRNA networks. Thereafter, we explored the differentially expressed genes in patients with OPLL through a GSE5464 chip from the GEO DataBase (https://www.ncbi.nlm.nih.gov/geo/) using a limma Rstudio software. Consequently, a total of 385 differentially expressed genes were screened out with |LogFC| > 4 and p < 0.05 as the screening criteria. Among them, 144 genes were up-regulated while the rest 241 were down-regulated, and a heatmap for the top 30 genes with differential expression is presented in Figure 3(d). Next, according to the Kyoto Encyclopedia of Genes and Genomes (KEGG)-based pathway analysis based on the differentially expressed genes, the JAK-STAT signaling was identified as the pathway where a large number of genes were enriched (Figure 3(e)). This pathway has once been documented to be closely linked to osteogenic differentiation [29]. In addition, a total of 16 genes were enriched on this signaling, among which IFNGR1 was found at the upstream of the pathway (figure 3(f)). Next, to find out the upstream mediator of IFNGR1, we explored the target miRNAs of SNHG1 as well as the miRNAs that mediate IFNGR1 expression on the bioinformatics system StarBase. Consequently, three miRNAs (miR-320b, miR-380 and miR-409) were found to be intersected (Figure 3(g)). Among these three miRNAs, miR-320b has been reported to negatively regulate BMP-2-induced osteogenic differentiation [30]. Moreover, the dual-luciferase reporter gene assay identified that miR-320b could both bind to SNHG1 and INFGR1 (p < 0.05) (Figure 3(h)).
Overexpression of miR-320b inhibits osteogenic differentiation ability of LFCs
To explore the effect of miR-320b on osteogenic differentiation of LFCs, miR-320b inhibitor was administrated in OPLL-LFCs with SNHG1 silencing, while of miR-320b mimic was introduced in SNHG1 overexpressing PLL-LFCs. The results suggested that silencing of miR-320b promoted the osteogenic differentiation of OPLL-LFCs. Correspondingly, reverse trends were found in PLL-LFCs where overexpression of miR-320b inhibited the osteogenic differentiation ability of cells (Figure 4(a–e)).
Figure 4.
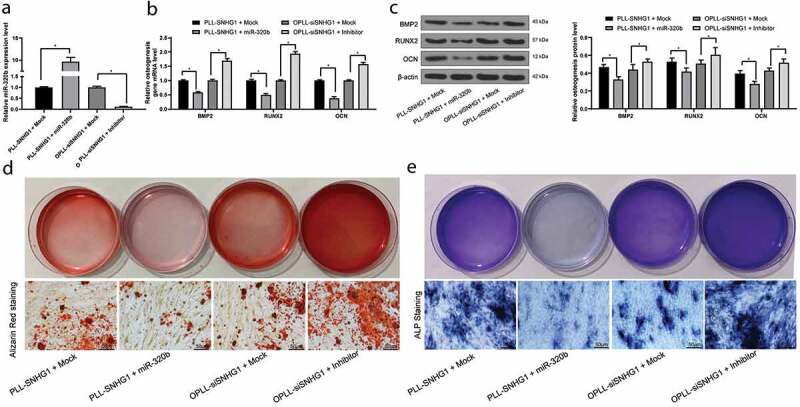
Overexpression of miR-320b inhibits the osteogenic differentiation of LFCs. miR-320b mimic or mock were transfected into PLL-derived LFCs with overexpressed SNHG1, and miR-320b inhibitor or inhibitor control were transfected into OPLL-derived LFCs with silenced SNHG1. A, miR-320b expression in cells after transfection measured by QRT-PCR; B-C, mRNA (b) and protein (c) expression of BMP-2, RUNX2 and OCN in cells determined by QRT-PCR and western blot analysis, respectively; D-E, osteogenic properties of LFCs detected by alizarin red staining (d) and ALP staining (e). The data are expressed as the mean ± SD; in panel A, data were analyzed using one-way ANOVA, while data in panels B and C were analyzed using two-way ANOVA, and Tukey’s multiple comparison test was used to determine statistical significance; *, p < 0.05
JAK-STAT signaling promotes osteogenic differentiation of LFCs
Following the above findings, we next measured the activation status of the JAK-STAT signaling pathway in cells. It was found that overexpression of SNHG1 in PLL-LFCs promoted the phosphorylation of the JAK-STAT pathway-related proteins, while these changes were reversed by the further upregulation of miR-320b. Correspondingly, silencing of SNHG1 in OPLL-LFCs inactivated the JAK-STAT pathway, but the further inhibition of miR-320b partly recovered the phosphorylation of the JAK-STAT pathway-related proteins (Figure 5(a)). Moreover, Fedratinib, a JAK2-specific agonist, was administrated into the OPLL-LFCs with SNHG1 silencing. Then it was found that JAK-STAT activation stimulated osteogenic differentiation of OPLL-LFCs (Figure 5(b–e)).
Figure 5.
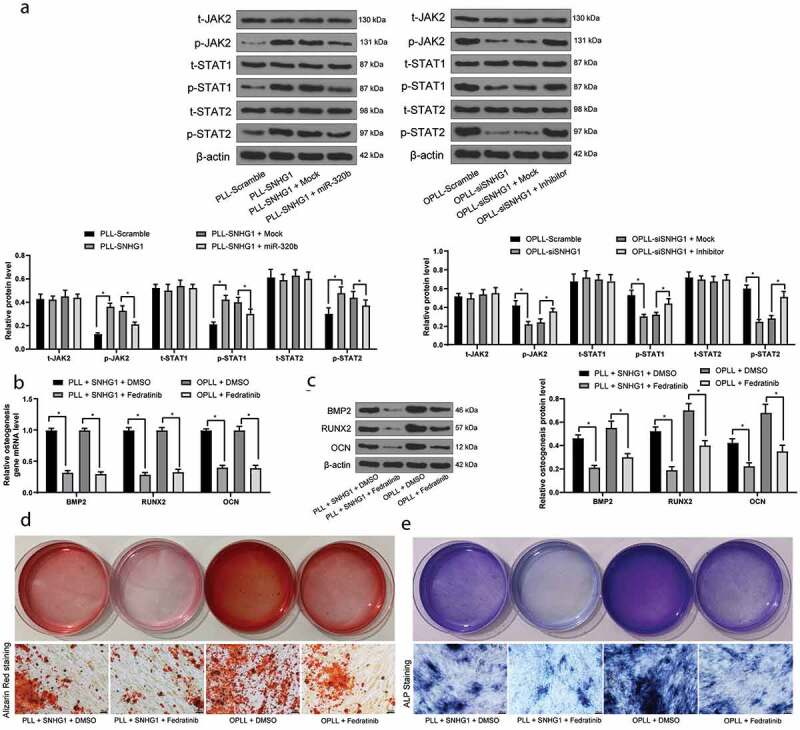
JAK-STAT signaling pathway activation promotes the osteogenic differentiation of LFCs. A, western blot analysis was performed to measure the phosphorylation of JAK2, STAT1 and STAT3 in LFCs; Then, OPLL-derived Fedratinib, a JAK2-specific agonist, was administrated into the LFCs; Next, B-C, the mRNA (b) and protein (c) expression of BMP2, RUNX2 and OCN was determined by QRT-PCR and western blot analysis, respectively; D-E, osteogenic properties of LFCs were detected by alizarin red staining (d) and ALP staining (e). The data are expressed as the mean ± SD; two-way ANOVA and Tukey’s multiple comparison test was used to determine statistical significance; *, p < 0.05
Silencing of SNHG1 reduces osteogenic differentiation of LFCs in vivo
An in vivo heterotopic bone formation assay was also performed to validate the role of SNHG1 in osteogenic differentiation of LFCs. Again, PLL-LFCs were transfected with pcDNA3.1-SNHG1 vector while OPLL-LFCs were transfected with pcDNA3.1-siSNHG1 vectors, and then the cells with stable transfection were loaded onto the Bio-Oss Collagen scaffolds and transplanted into the nude mice. Six weeks later, the scaffolds were taken out, and the immunohistochemical staining results of BMP-2, RUNX2 and OCN suggested that silencing of SNHG1 inhibited the expression of BMP-2, RUNX2 and OCN in mice, while overexpression of SNHG1 reversed the changes. Moreover, the toluidine blue staining found that over-expression of SNHG1 led to an increase in the number of cartilages, while silencing of SNHG1, correspondingly, inhibited the formation of cartilages (Figure 6).
Figure 6.
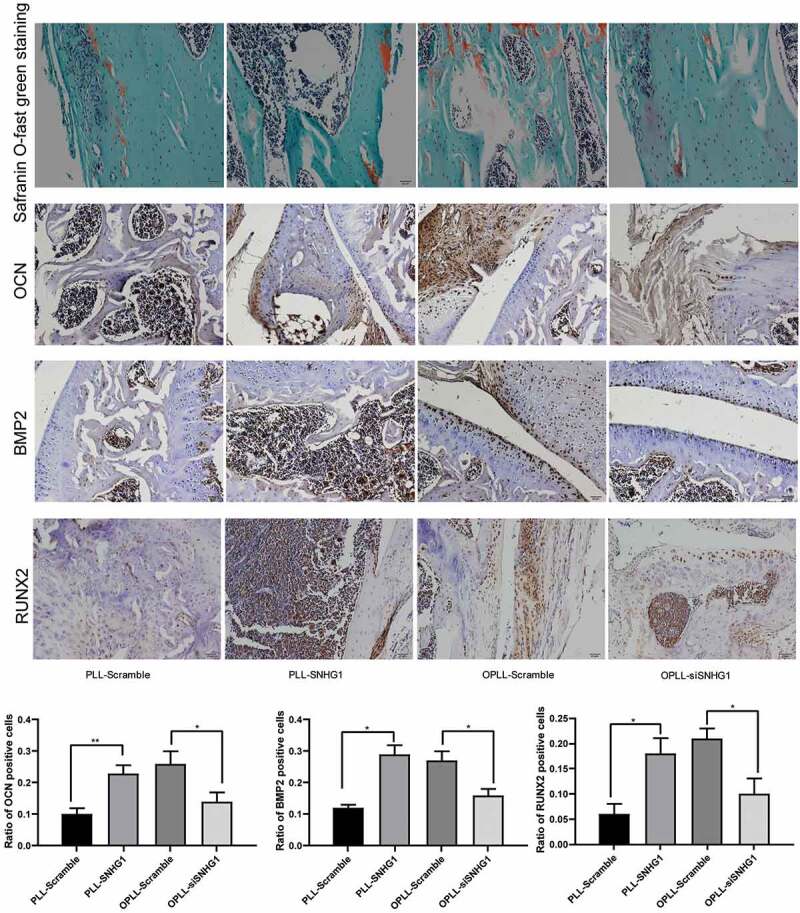
Silencing of SNHG1 inhibited the osteogenic differentiation of LFCs in vivo. HE staining was performed to observe the scaffold morphology and immunohistochemistry staining was performed to identify the expression of OCN, BMP2 and RUNX2 in the scaffolds seeded with different cell types. The data are expressed as the mean ± SD; one-way ANOVA and Tukey’s multiple comparison test was used to determine statistical significance; *, p < 0.05
Discussion
OPLL imposes compression on the spinal cord by both static and dynamic factors and leads to progressive radiculo-myelopathy, leaving a great challenge in the treatment of the disease [31]. LncRNAs exert key functions in gene modulation through the interaction with DNA, RNA or protein, and their regulatory roles in osteogenic differentiation also have been documented [32]. Here, our study identified a novel lncRNA, SNHG1, which could serve as a sponge for miR-320b to increase IFNGR expression, leading to further activation of the JAK/STAT signaling and the osteogenic differentiation of human LFCs.
Firstly, LFCs were collected from both ossified PLL and non-ossified PLLs. The LFCs from ossified PLL tissues presented higher expression of BMP-2, RUNX2 and OCN and ALP activity than that in the non-ossified ones. BMP-2 is widely distributed in mesenchymal stem cells and the ossified matrix as well as chondrocytes close to the cartilaginous areas in the ossified PLL [33]. As a subgroup of the transforming growth factor β family, BMPs are multi-functional growth factors and are well-known for their role in cartilage and bone formation [34]. RUNX2 is a master transcription factor necessary for the process of osteogenesis and can activate the marker genes for osteoblast differentiation [35,36]. As for OCN, it is the most abundant non-collagenous protein in bone matrix and plays crucial role in the process of biomineralization during osteogenic maturation [37]. Moreover, ALP is an osteoblast-specific phenotypic marker [36] whose increase may promote mineralization [33]. These results validated that LFCs from ossified PLL tissues presented stronger osteogenic differentiation tendency than the normal ones.
The studies of lncRNAs in osteogenic differentiation have been concerning their correlations with the marrow mesenchymal stem cells (BMSCs) [32,38]. But the correlation between lncRNAs and osteogenic differentiation of LFCs or about OPLL is yet limited. Importantly, a lncRNA microarray analysis based on the LFCs from different sources suggested that SNHG1 had aberrant high expression in OPLL-derived LFCs. SNHG1 has been largely studied to be implicated in human cancer progression [39] and cell death induced by oxygen-glucose deprivation/reperfusion [40]. Interestingly, the function of SNHG1 on bone formation has been controversial. It was documented to suppress osteogenic differentiation of BMSCs by triggering Nedd4-mediated ubiquitination of the p38 MAPK signaling pathway [41]. On the other hand, downregulation of SNHG1 was found to be implicated in postmenopausal osteoporosis, during which the formation of osteoblast is reduced and the formation of osteoclast is increased [42], indicating SNHG1 is possibly responsible for osteoblast activity. Here, our study evidenced a promoting effect of SNHG1 on osteogenic differentiation of LFCs. Silencing of SNHG1 led to a decline in the levels of BMP-2, RUNX2 and OCN as well as decreased mineralization and ALP activity of LFCs.
The cytoplasm-localization of SNHG1 suggested a potential implication of other RNA transcripts in the above events. Then, a mRNA microarray analysis identified 385 aberrantly expressed miRNAs in patients with OPLL, and many of these mRNAs were enriched on the JAK-STAT signaling pathway according to the KEGG signaling enrichment analysis, and IFNGR1 was the upstream of the pathway. IFNGR1 has rarely been mentioned in osteogenesis, and studies have summarized its important role in host defense against intracellular pathogens and its deficiency is frequently linked to mycobacterial diseases including osteomyelitis [43,44]. In line with our findings, IFNGR1 was suggested to be essential for the downstream signal transduction of JAK2 [45]. Activation of the JAK/STAT pathway was suggested to increase BMP-2 expression and ALP activity, resulting in increased biomineralization and osteoblast differentiation [46]. Similarly, activation of the JAK/STAT/IGF1 axis was found to trigger the osteogenic differentiation of murine multilineage cells [47]. To confirm the interaction between SNHG1 and the JAK/STAT pathway, we then explored the target transcripts of SNHG1 as well as the upstream miRNAs of IFNGR1, and miR-320b was found to be intersected. As aforementioned, lncRNAs may serve as ceRNA for miRNAs to mediate gene expression, and SNHG1 was suggested to mediate differentiation of osteoblasts and osteoclasts through sponging miRNAs [42]. The involvements of miRNAs in osteogenic differentiation potentials of LFCs have been increasingly recognized. For instance, two miRNAs, miR-17-5p and miR-27b-3p, were found to be upregulated by other RNA transcripts in patients with ankylosing spondylitis and promote pathological osteogenic differentiation [48]. As for miR-320b, it has once been documented to be negatively correlated with cartilage repair and bone formation [49]. More relatively, miR-320b was suggested to inhibit the BMP-2-induced osteoblast differentiation and the following downstream master osteogenic transcription factors [30]. Partially in line with these findings, our present study identified that upregulation of miR-320b suppressed the osteogenic differentiation of LFCs triggered by overexpressed SNHG1. Importantly, our study suggested that the phosphorylation of the JAK2/STAT3 signaling pathway was increased in the presence of SNHG1 upregulation but then suppressed by the further miR-320b overexpression. In addition, the osteogenic differentiation of OPLL-LFCs inhibited by si-SNHG1 were recovered following the artificial activation of JAK-STAT. These results collectively evidenced a SNHG1/miR-320b/IFNG1 ceRNA network in the OPLL progression and the implication of the activation of the JAK/STAT signaling pathway.
To sum up, this study suggested that SNHG1 could serve as a sponge for miR-320b to increase IFNGR1 expression and activate the JAK/STAT signaling pathway, resulting in increased mineralization and osteogenic differentiation of LFCs. Silencing of SNHG1 might alleviate the progression of OPLL. In addition, most of the bioinformatic analyses in the current paper were performed using RNA microarrays. We would like to perform an RNA-Seq experiment [50] in future studies to validate the interactions between the transcripts. We hope the findings can offer novel insights into OPLL treatment.
Funding Statement
The authors have no funding to report.
Disclosure statement
All authors declare no conflict of interests in this study.
References
- [1].Cerecedo-Lopez CD, Tafel I, Lak AM, et al. Surgical management of ossification of the posterior longitudinal ligament in the cervical spine. J Clin Neurosci. 2020;72:191. [DOI] [PubMed] [Google Scholar]
- [2].Moon BJ, Choi SK, Shin DA, et al. Prevalence, incidence, comorbidity, and mortality rates of ossification of posterior longitudinal ligament in the cervical spine: a nested case-control cohort study. World Neurosurg. 2018;117:e323–e328. [DOI] [PubMed] [Google Scholar]
- [3].Xu C, Zhang H, Gu W, et al. The microRNA-10a/ID3/RUNX2 axis modulates the development of ossification of posterior longitudinal ligament. Sci Rep. 2018;8(1):9225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Miao J, Sun J, Shi J, et al. A novel anterior revision surgery for the treatment of cervical ossification of posterior longitudinal ligament: case report and review of the literature. World Neurosurg. 2018;113::212–216. [DOI] [PubMed] [Google Scholar]
- [5].Yu F, Cui Y, Zhou X, et al. Osteogenic differentiation of human ligament fibroblasts induced by conditioned medium of osteoclast-like cells. Biosci Trends. 2011;5(2):46–51. [DOI] [PubMed] [Google Scholar]
- [6].Kashii M, Matuso Y, Sugiura T, et al. Circulating sclerostin and dickkopf-1 levels in ossification of the posterior longitudinal ligament of the spine. J Bone Miner Metab. 2016;34(3):315–324. [DOI] [PubMed] [Google Scholar]
- [7].Liao X, Tang D, Yang H, et al. Long non-coding RNA XIST may influence cervical ossification of the posterior longitudinal ligament through regulation of miR-17-5P/AHNAK/BMP2 signaling pathway. Calcif Tissue Int. 2019;105(6):670–680. [DOI] [PubMed] [Google Scholar]
- [8].Sohn S, Chung CK.. Increased bone mineral density and decreased prevalence of osteoporosis in cervical ossification of the posterior longitudinal ligament: a case-control study. Calcif Tissue Int. 2013;92(1):28–34. [DOI] [PubMed] [Google Scholar]
- [9].Donato L, Bramanti P, Scimone C, et al. miRNAexpression profile of retinal pigment epithelial cells under oxidative stress conditions. FEBS Open Bio. 2018;8(2):219–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Koirala D, Lewicka A, Koldobskaya Y, et al. Synthetic antibody binding to a preorganized RNA domain of hepatitis C virus internal ribosome entry site inhibits translation. ACS Chem Biol. 2020;15(1):205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wan ZY, Song F, Sun Z, et al. Aberrantly expressed long noncoding RNAs in human intervertebral disc degeneration: a microarray related study. Arthritis Res Ther. 2014;16(5):465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Liu Y, Li G, Zhang JF.. The role of long non-coding RNA H19 in musculoskeletal system: A new player in an old game. Exp Cell Res. 2017;360(2):61–65. [DOI] [PubMed] [Google Scholar]
- [13].Trionfini P, Benigni A. MicroRNAs as master regulators of glomerular function in health and disease. J Am Soc Nephrol. 2017;28(6):1686–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Huo S, Zhou Y, He X, et al. Insight into the role of long non-coding RNAs during osteogenesis in mesenchymal stem cells. Curr Stem Cell Res Ther. 2018;13(1):52–59. [DOI] [PubMed] [Google Scholar]
- [15].Sun H, Peng G, Ning X, et al. Emerging roles of long noncoding RNA in chondrogenesis, osteogenesis, and osteoarthritis. Am J Transl Res. 2019;11(1):16–30. [PMC free article] [PubMed] [Google Scholar]
- [16].Taipaleenmaki H. Regulation of bone metabolism by microRNAs. Curr Osteoporos Rep. 2018;16(1):1–12. [DOI] [PubMed] [Google Scholar]
- [17].Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Qi X, Zhang DH, Wu N, et al. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710–718. [DOI] [PubMed] [Google Scholar]
- [19].Zhou Z, Zhu Y, Gao G, et al. Long noncoding RNA SNHG16 targets miR-146a-5p/CCL5 to regulate LPS-induced WI-38 cell apoptosis and inflammation in acute pneumonia. Life Sci. 2019;228::189–197. [DOI] [PubMed] [Google Scholar]
- [20].Xu C, Chen Y, Zhang H, et al. Integrated microRNA-mRNA analyses reveal OPLL specific microRNA regulatory network using high-throughput sequencing. Sci Rep. 2016;6:21580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yukawa Y, Kato F, Ito K, et al. Postoperative changes in spinal cord signal intensity in patients with cervical compression myelopathy: comparison between preoperative and postoperative magnetic resonance images. J Neurosurg Spine. 2008;8(6):524–528. [DOI] [PubMed] [Google Scholar]
- [22].Xu C, Zhang Y, Wang Q, et al. Long non-coding RNA GAS5 controls human embryonic stem cell self-renewal by maintaining NODAL signalling. Nat Commun. 2016;7:13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Li N, Cheng W, Huang T, et al. Vascular adventitia calcification and its underlying mechanism. PLoS One. 2015;10(7):e0132506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gao A, Hang R, Huang X, et al. The effects of titania nanotubes with embedded silver oxide nanoparticles on bacteria and osteoblasts. Biomaterials. 2014;35(13):4223–4235. [DOI] [PubMed] [Google Scholar]
- [25].Jiang C, Zhu W, Xu J, et al. MicroRNA-26a negatively regulates toll-like receptor 3 expression of rat macrophages and ameliorates pristane induced arthritis in rats. Arthritis Res Ther. 2014;16(1):R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhang Y, Guo L, Li Y, et al. MicroRNA-494 promotes cancer progression and targets adenomatous polyposis coli in colorectal cancer. Mol Cancer. 2018;17(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Huang Y, Zheng Y, Jia L, et al. Long noncoding RNA H19 promotes osteoblast differentiation via TGF-beta1/Smad3/HDAC signaling pathway by deriving miR-675. Stem Cells. 2015;33(12):3481–3492. [DOI] [PubMed] [Google Scholar]
- [28].Wu Y, Liu H, Shi X, et al. The long non-coding RNA HNF1A-AS1 regulates proliferation and metastasis in lung adenocarcinoma. Oncotarget. 2015;6(11):9160–9172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Luttrell LM, Dar MS, Gesty-Palmer D, et al. Transcriptomic characterization of signaling pathways associated with osteoblastic differentiation of MC-3T3E1 cells. PLoS One. 2019;14(1):e0204197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Laxman N, Mallmin H, Nilsson O, et al. miR-203 and miR-320 regulate bone morphogenetic protein-2-induced osteoblast differentiation by targeting distal-less homeobox 5 (Dlx5). Genes (Basel). 2016;8(1). DOI: 10.3390/genes8010004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ha Y, Moon BJ, You NK, et al. Clinical characteristics and surgical outcome of revision surgery in patients with cervical ossification of the posterior longitudinal ligament. World Neurosurg. 2016;90::164–171. [DOI] [PubMed] [Google Scholar]
- [32].Zhang W, Dong R, Diao S, et al. Differential long noncoding RNA/mRNA expression profiling and functional network analysis during osteogenic differentiation of human bone marrow mesenchymal stem cells. Stem Cell Res Ther. 2017;8(1):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yan L, Gao R, Liu Y, et al. The pathogenesis of ossification of the posterior longitudinal ligament. Aging Dis. 2017;8(5):570–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Haversath M, Catelas I, Li X, et al. PGE(2) and BMP-2 in bone and cartilage metabolism: 2 intertwining pathways. Can J Physiol Pharmacol. 2012;90(11):1434–1445. [DOI] [PubMed] [Google Scholar]
- [35].Bruderer M, Richards RG, Alini M, et al. Role and regulation of RUNX2 in osteogenesis. Eur Cell Mater. 2014;28::269–286. [DOI] [PubMed] [Google Scholar]
- [36].Vimalraj S, Arumugam B, Miranda PJ, et al. Runx2: structure, function, and phosphorylation in osteoblast differentiation. Int J Biol Macromol. 2015;78::202–208. [DOI] [PubMed] [Google Scholar]
- [37].Tsao YT, Huang YJ, Wu HH, et al. Osteocalcin mediates biomineralization during osteogenic maturation in human mesenchymal stromal cells. Int J Mol Sci. 2017;18(1):159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Xie Z, Li J, Wang P, et al. Differential expression profiles of long noncoding RNA and mRNA of osteogenically differentiated mesenchymal stem cells in ankylosing spondylitis. J Rheumatol. 2016;43(8):1523–1531. [DOI] [PubMed] [Google Scholar]
- [39].Thin KZ, Tu JC, Raveendran S. Long non-coding SNHG1 in cancer. Clin Chim Acta. 2019;494::38–47. [DOI] [PubMed] [Google Scholar]
- [40].Hu L, Fang R, Guo M. Knockdown of lncRNA SNHG1 alleviates oxygen-glucose deprivation/reperfusion-induced cell death by serving as a ceRNA for miR-424 in SH-SY5Y cells. Neurol Res. 2020;42(1):47–54. [DOI] [PubMed] [Google Scholar]
- [41].Jiang Y, Wu W, Jiao G, et al. LncRNA SNHG1 modulates p38 MAPK pathway through Nedd4 and thus inhibits osteogenic differentiation of bone marrow mesenchymal stem cells. Life Sci. 2019;228::208–214. [DOI] [PubMed] [Google Scholar]
- [42].Huang S, Zhu X, Xiao D, et al. LncRNA SNHG1 was down-regulated after menopause and participates in postmenopausal osteoporosis. Biosci Rep. 2019;39(11):BSR2019044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Obinata K, Lee T, Niizuma T, et al. Two cases of partial dominant interferon-gamma receptor 1 deficiency that presented with different clinical courses of bacille Calmette-Guerin multiple osteomyelitis. J Infect Chemother. 2013;19(4):757–760. [DOI] [PubMed] [Google Scholar]
- [44].Rottman M, Soudais C, Vogt G, et al. IFN-gamma mediates the rejection of haematopoietic stem cells in IFN-gammaR1-deficient hosts. PLoS Med. 2008;5(1):e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sano S, Wang Y, Yura Y, et al. JAK2 (V617F) -mediated clonal hematopoiesis accelerates pathological remodeling in murine heart failure. JACC Basic Transl Sci. 2019;4(6):684–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Mikami Y, Asano M, Honda MJ, et al. Bone morphogenetic protein 2 and dexamethasone synergistically increase alkaline phosphatase levels through JAK/STAT signaling in C3H10T1/2 cells. J Cell Physiol. 2010;223(1):123–133. [DOI] [PubMed] [Google Scholar]
- [47].Huang E, Zhu G, Jiang W, et al. Growth hormone synergizes with BMP9 in osteogenic differentiation by activating the JAK/STAT/IGF1 pathway in murine multilineage cells. J Bone Miner Res. 2012;27(7):1566–1575. [DOI] [PubMed] [Google Scholar]
- [48].Zhang C, Wang C, Jia Z, et al. Differentially expressed mRNAs, lncRNAs, and miRNAs with associated co-expression and ceRNA networks in ankylosing spondylitis. Oncotarget. 2017;8(69):113543–113557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Shiraishi K, Kamei N, Takeuchi S, et al. Quality evaluation of human bone marrow mesenchymal stem cells for cartilage repair. Stem Cells Int. 2017;2017:8740294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Donato L, Scimone C, Nicocia G, et al. Role of oxidative stress in retinitis pigmentosa: new involved pathways by an RNA-Seq analysis. Cell Cycle. 2019;18(1):84–104. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]


