ABSTRACT
Blood-tumour barrier (BTB) has been known to significantly attenuate the efficacy of chemotherapy for glioma. In this report, we identified that insulin-like grown factor 2 mRNA-binding protein 2 (IGF2BP2) was over-expressed in glioma microvessel and glioma endothelial cells (GECs). Knockdown of IGF2BP2 decreased the expression of lncRNA FBXL19-AS1 and tight junction-related proteins, thereby promoting BTB permeability. FBXL19-AS1 was over-expressed and more enriched in the cytoplasm of GECs. In addition, FBXL19-AS1 could bind to 3ʹ-UTR of ZNF765 mRNA and down-regulate ZNF765 mRNA expression through STAU1-mediated mRNA decay (SMD). The low expression of ZNF765 was discovered in GECs and verified to increase BTB permeability by inhibiting the promoter activities of tight junction-related proteins. Meanwhile, ZNF765 also inhibited the transcriptional activity of IGF2BP2, thereby forming a feedback loop in regulating the BTB permeability. Single or combined application of silenced IGF2BP2 and FBXL19-AS1 improved the delivery and antitumor efficiency of doxorubicin (DOX). In general, our study revealed the regulation mechanism of IGF2BP2/FBXL19-AS1/ZNF765 axis on BTB permeability, which may provide valuable insight into treatment strategy for glioma.
KEYWORDS: IGF2BP2, FBXL19-AS1, blood-tumour barrier, stau1-mediated decay
Introduction
Glioma is a common primary intracranial tumour that mostly arise from glial cells or precursor cells. 379,848 cases data of primary brain tumours (during 2010–2014) were collected and analysed by CBTRUS (Central Brain Tumour Registry of the United States), and the results indicated that the broad category glioma accounted for approximately 26.5% of primary brain tumours and 80.7% of malignant tumours [1]. The current treatment for glioma is mainly surgery with adjuvant chemoradiotherapy. However, the blood-brain tumour barrier (BTB) greatly limits the efficacy of therapy by impeding drug delivery into the microenvironment of brain tumours [2]. Therefore, selectively opening of BTB and increasing the effectiveness of drug delivery is one of the major therapeutic approaches in the comprehensive treatment of glioma.
Accumulated studies have shown that RNA-binding proteins (RBPs) have an important role in the post-translational regulation of gene expression and the cellular biological behaviours [3]. For instant, human antigen R (HuR) inhibits the expression of RhoB by binding to the 3ʹ-untranslated region (3ʹ-UTR) of rhoB mRNA, thereby promoting the UV-induced apoptosis of human keratinocytes [4]. RBP-IGF2BP2 (insulin-like grown factor 2 mRNA-binding protein 2), 4311 bp in length and located on the human chr3, could promote the development of glioblastoma multiforme (GBM) through activating PI3 K/Akt signalling [5]. In addition, IGF2BP2 could bind to and prevent miR-let-7 from silencing glioblastoma stem cell (GSCs) programmes thereby inhibiting GSCs differentiation [6]. Other researches have also shown that IGF2BP2 is up-regulated in mesenchymal GSCs, which could form complexes with DXH9 in the presence of HIF1A-AS2, further promoting the expression of oncogenes such as HMGA1 [7]. But so far the expression and role of IGF2BP2 in glioma endothelial cells (GECs) have yet to be studied.
Long non-coding RNA (lncRNA) is a subtype of ncRNAs over 200bp in size [8], which has been revealed to be associated with numerous tumours including glioma, ovarian cancer and non-small cell lung cancer [9–11]. FBXL19 (F-box and leucine rich repeat protein 19) is a member of the Skp1-Culi-F-box family and known for its leucine-rich repeats [12]. FBXL19 is involved in the regulation of the pathological process of psoriasis [13]. In mice models of pneumonia, overexpression of FBXL19 inhibits the pro-apoptotic and inflammatory effects of IL-33 and reduces the degree of lung injury [14]. FBXL19-AS1 is a natural non-coding antisense of FBXL19. In colorectal cancer (CRC), the expression of FBXL19-AS1 is up-regulated and positively related with the lymphatic metastasis and TNM stage. Knockdown of FBXL19-AS1 inhibited CRC cell proliferation, migration and invasion, and also facilitated carcinogenesis by regulating miR-203 as a molecular sponge [15]. So far little is known about the function of FBXL19-AS1 in GECs.
Table 1.
Primers used for ChIP experiments
| Target gene | Primers | Product size | |
|---|---|---|---|
| Occludin | Control | Forward: AGTCCTACCCAGCTACCAAGT | 270bp |
| Reverse: TCCTTCTTGGCAACTCACAGT | |||
| ZNF765 binding |
Forward: TGGATGGCAACTAACACCTACA | 211bp | |
| Reverse: ACAGGACACTTTTCTGCTTCCA | |||
| ZO-1 | Control | Forward: GTGCTGGGATTATAAACGTGAGC | 204bp |
| Reverse: TGCCTGTAGTCCCAGCTACT | |||
| ZNF765 binding | Forward: GCACTTCGAAGCCTTAATTGAT | 207bp | |
| Reverse: TGCCAGAAATCCTTAGTAACATCA | |||
| Claudin-5 | Control | Forward: GCAGAAGAGGCCAAGGTAAGA | 201bp |
| Reverse: ACCTAAGACCCTCCATGGCT | |||
| ZNF765 binding | Forward: GCACACTTGCCTTCAGAACC | 205bp | |
| Reverse: CTGGTCTGTGCTCAGTTCCA | |||
| IGF2BP2 | Control | Forward: TCCTAGGCGTCCCGACTC | 239bp |
| Reverse: CAAGGTGGACCGGATGTGAG | |||
| ZNF765 binding | Forward: CGCCCGTACAAACTCTCACA | 105bp | |
| Reverse: CTCTGCCTCCCTCTCTCTCC |
Staufen1-mediated mRNA decay (SMD) is a regulation pathway that meditated by the binding between STAU1 and the STAU1 binding site (SBS) [16]. SMD participates in many physiological processes including wound healing, myogenesis and adipogenesis [17–19]. In the process of SMD, STAU1 recognize and bind to SBS which is formed by imperfect base pairing between an ALU element in the 3ʹ-UTR of target mRNA and another ALU element of a lncRNA [16]. The combination of STAU1 and SBS further recruits UPF1 helicase, reducing the half-life of target mRNA and accelerating its degradation [20].
Zinc-finger proteins (ZNFs) are the most abundant transcription factors in the genome of eukaryotes and could form a stable finger that is stabilized by Zn2+ and takes part in many biological processes, such as gene expression, cell differentiation and tumour development [21]. Ragusa et al. found that the down-regulated expression of ZEB1 (Zinc Finger E-box binding protein 1) could directly promote the inhibition on cell cycle mediated by TP73 and further inducing cell apoptosis [22]. ZNF765 is a member of ZHFs family located on the human 19q13.42. Significant mutation of ZNF765 has been found in chromophobe renal cell carcinoma, suggesting its potential roles in kidney cancer [23].
In this study, we verified the expression of IGF2BP2, FBXL19-AS2 and ZNF765 in GSCs and the potential roles of them in the permeability of BTB. In addition, the interaction and the latent mechanism of IGF2BP2, FBXL19-AS1 and ZNF765 in BTB regulation were illustrated.
Results
IGF2BP2 was up-regulated in GECs, knockdown of IGF2BP2 increased BTB permeability
As shown in Fig. 1A, we obtained vessels from normal brain tissues (NBTs), low-grade glioma tissues (LGGTs, WHO I–II) and high-grade glioma tissues (HGGTs, WHO III–IV) via LCM method. The expression levels of IGF2BP2 were evaluated by qRT-PCR and western blot subsequently. The results showed that the expression level of IGF2BP2 was up-regulated in LGG and HGG groups compared with NBTs group, and higher in HGG group compared with LGG group (Fig. 1B,C). IGF2BP2 was significantly up-regulated in GECs compared with astrocyte-exposed endothelial cells (AECs, Fig. 1D,E). To examine the effect of IGF2BP2 on BTB permeability, we further established GECs with IGF2BP2 stable overexpression and knockdown. The transfection efficiency was shown in Figure S1A. As shown in Fig. 1F,G, there was no significant difference in TEER values and HRP flux between control groups and negative control (NC) group. Overexpression of IGF2BP2 significantly elevated TEER values and decreased HRP flux, while knockdown of IGF2BP2 had opposite effects. The expression levels of junction-associated proteins were elevated with overexpressed IGF2BP2 and decreased with knockdown of IGF2BP2 (Fig. 1H). Consistently, the results of immunofluorescence staining showed that silenced IGF2BP2 damaged the continuity distribution of ZO-1, occludin and claudin-5, whereas overexpression of IGF2BP2 had the contrary effect (Fig. 1I). Similarly, the mRNA and protein expression levels of ZNF765 were decreased in the IGF2BP2 over-expression group, whereas increased in the IGF2BP2 knockdown group (Fig. 1J,K).
Figure 1.
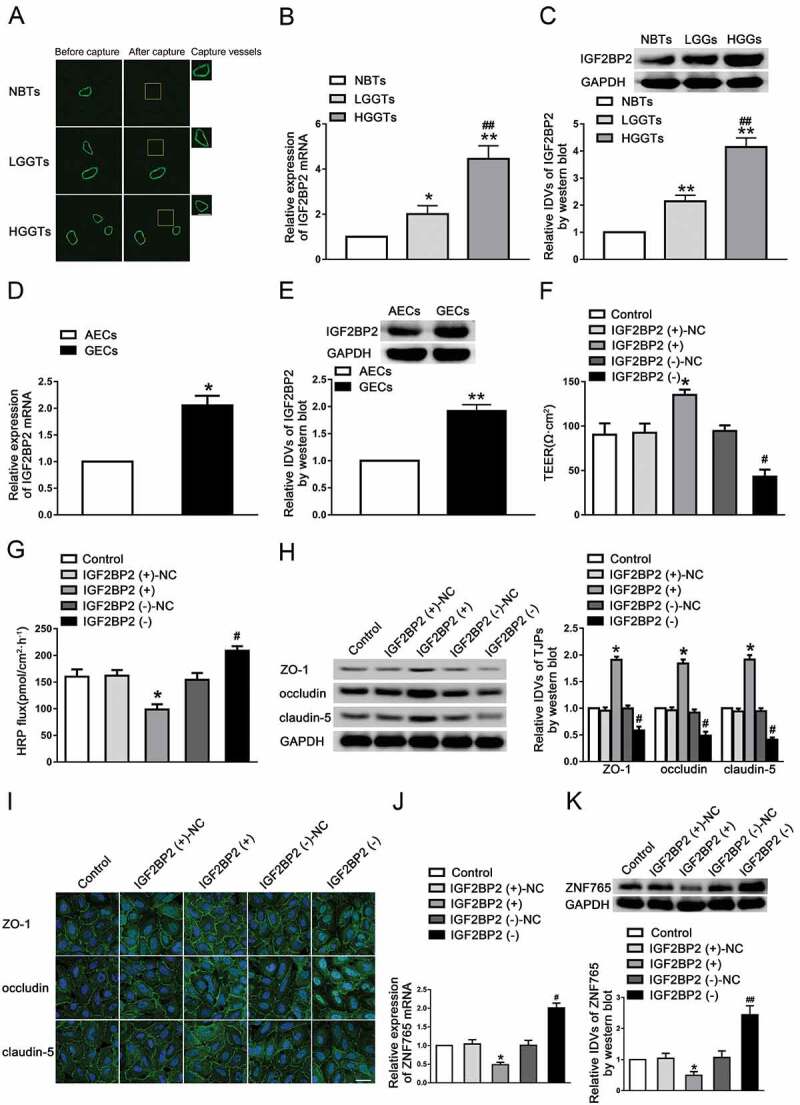
Knockdown of IGF2BP2 increased BTB permeability in vitro
(A) Staining vessel from normal or glioma brain tissues with the UEA-I. LCM capture of UEA-I-stained vessel from human brain sections. Scale bars represent 40 μm. (B,C) The mRNA and protein expression levels of IGF2BP2 were higher in glioma tissues than in normal brain tissues (NBTs), and highest in low-grade glioma tissues (LGGTs). Data represent means ± SD (n = 3, each). *P < 0.05, **P < 0.01 vs. NBTs group, ##P < 0.01 vs. LGGTs group. (D,E) The mRNA and protein expression levels of IGF2BP2 were higher in glioma endothelial cells (GECs) than in astrocyte-exposed endothelial cells (AECs). Data represent mean ± SD (n = 3, each). *P < 0.05, **P < 0.01 vs. AECs group. (F,G) Knockdown of IGF2BP2 decreased TEER values and increased HRP flux. (H) Knockdown of IGF2BP2 decreased the expression levels of ZO-1, occludin and claudin-5. Data represent mean ± SD (n = 3, each). *P < 0.05 vs. IGF2BP2 (+)-NC group, #P < 0.05 vs. IGF2BP2 (-)-NC group. (I) Knockdown of IGF2BP2 damaged the continuity of ZO-1, occludin and claudin-5 distributions. Scale bar represents 30 μm. (J,K) Knockdown of IGF2BP2 decreased the mRNA and protein expression levels of ZNF765. Data represent mean ± SD (n = 3, each). *P < 0.05 vs. IGF2BP2 (+)-NC group, #P < 0.05 vs. IGF2BP2 (-)-NC group. QRT-PCR results were analysed using the relative quantification (2–ΔΔCT) method, and blots were analysed in terms of IDVs. All results were analysed using one-way ANOVA test.
FBXL19-AS1 was highly expressed in GECs, knockdown of FBXL19-AS1 increased BTB permeability
Studies have reported that RBPs can exert biological functions by regulating lncRNAs. In our study, microarray analysis was carried out for searching IGF2BP2-related lncRNAs in GECs. As shown in Fig. 2A, in IGF2BP2 stable knockdown GECs, the expression of lnc-FBXL19-AS1 and lnc-SNHG12 were significantly down-regulated, with FBXL19-AS1 being mostly affected. As shown in Fig. 2B, FBXL19-AS1 was mainly located in the cytoplasm of GECs and highly expressed in GECs compared with AECs (Fig. 2C). Similarly, to further assess the functional roles of FBXL19-AS1 on BTB permeability, GECs with stable overexpression and knockdown of FBXL19-AS1 were also established (Figure S1B). As shown in Fig. 2D,E, the TEER value was markedly enhanced and the HRP flux was significantly decreased in FBXL19-AS1 overexpressed cells compared with NC group respectively, whereas results were reserved in FBXL19-AS1(-) groups. Moreover, the expression levels of ZO-1, occludin and claudin-5 were increased in FBXL19-AS1(+) group while decreased in FBXL19-AS1(-) group (Fig. 2F). The results of immunofluorescence analysis demonstrated in Fig. 2G that the continuous distribution of tight junction-associated proteins was intensified in FBXL19-AS1(+) group but disrupted in FBXL19-AS1(-) group. We then investigated the regulation of FBXL19-AS1 on ZNF765. The results revealed that knockdown of FBXL19-AS1 significantly reduced the mRNA, protein expression levels and the half-life of ZNF765, whereas overexpressed FBXL19-AS1 exerted the itopposite effects (Fig. 2H,J).
Figure 2.
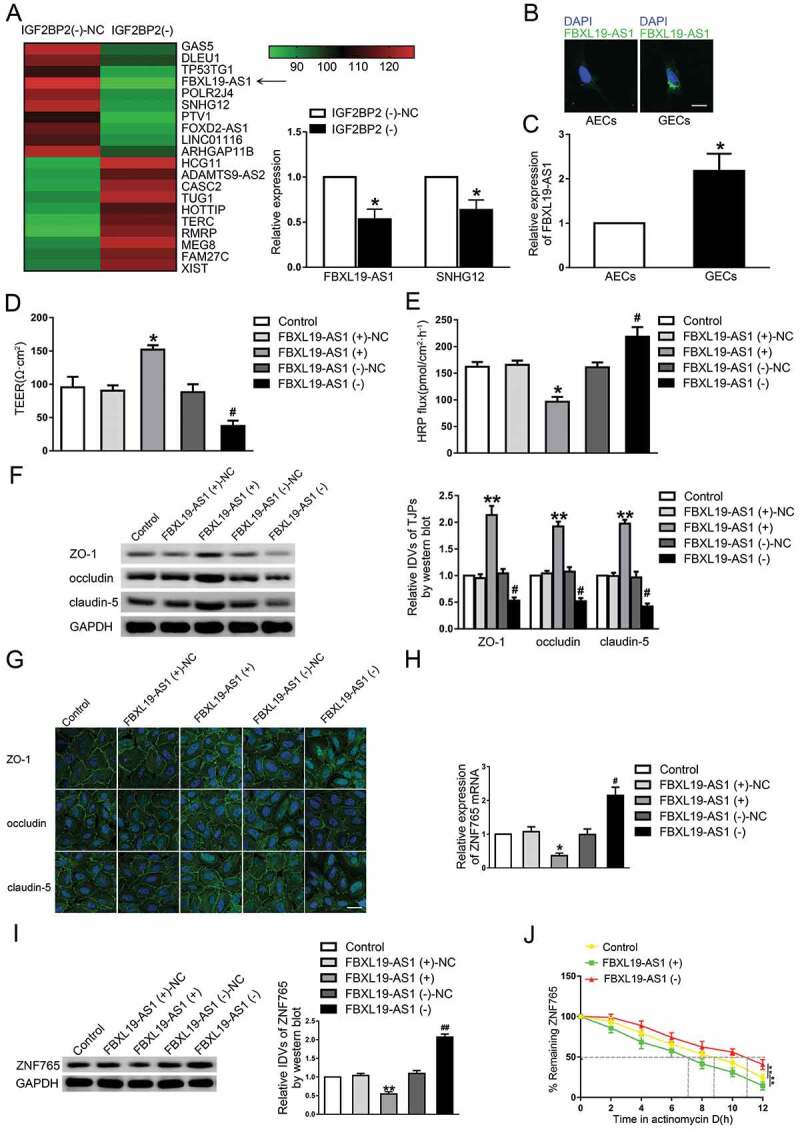
Knockdown of FBXL19-AS1 increased BTB permeability in vitro
(A) LncRNAs microarray analysis of total RNAs from the GECs of silenced IGF2BP2. Red means high expression, green means low expression. QRT-PCR assay was used to evaluated the selected molecules. Data represent mean ± SD (n = 3, each). *P < 0.05 vs. IGF2BP2(-) group. (B) FBXL19-AS1 located in the cytoplasm in AECs and GECs and conspicuously elevated in GECs. Scale bars represent 20 μm. (C) The expression level of FBXL19-AS1 was elevated in GECs that in AECs. Results were analysed using two-tailed paired Student’s t test. Data represent mean ± SD (n = 3, each). *P < 0.05 vs. AECs group. (D,E) Knockdown of FBXL19-AS1 decreased TEER values and increased HRP flux. (F) Knockdown of FBXL19-AS1 decreased the expression levels of ZO-1, occludin and claudin-5. Results were analysed using one-way ANOVA test. Data represent mean ± SD (n = 3, each). *P < 0.05, **P < 0.01 vs. FBXL19-AS1 (+)-NC group, #P < 0.05 vs. FBXL19-AS1 (-)-NC group. (G) Knockdown of FBXL19-AS1 damaged the continuity of ZO-1, occludin and claudin-5 distributions. Scale bar represents 30 μm. (H,I) Knockdown of FBXL19-AS1 decreased the mRNA and protein expression levels of ZNF765. Data represent mean ± SD (n = 3, each). *P < 0.05, **P < 0.01 vs. FBXL19-AS1 (+)-NC group, #P < 0.05, ##P < 0.01 vs. FBXL19-AS1 (-)-NC group. (J) Knockdown of FBXL19-AS1 increased the half-life time of ZNF765. Results were analysed using one-way ANOVA test. Data represent mean ± SD (n = 3, each). **P < 0.01 vs. Control group.
IGF2BP2 bound to and stabilized FBXL19-AS1
Based on the results of microarray analysis, we proved that silenced IGF2BP2 could inhibit the expression of FBXL19-AS1. A potential binding site between IGF2BP2 and FBXL19-AS1 was further discovered using an online bioinformatic software Starbase (Figure S1 C). To further reveal the mechanism of IGF2BP2’s effect on FBXL19-AS1, a series of assays were performed. As shown in Fig. 3A, compared with NC groups respectively, the expression levels of FBXL19-AS1 was significantly higher in IGF2BP2(+) group and lower in IGF2BP2(-) group. RIP assay and RNA pull-down assay were then carried out to validate the binding between IGF2BP2 and FBXL19-AS1. As results presented (Fig. 3B,C), FBXL19-AS1 was markedly enriched in anti-IGF2BP2 group compared with anti-IgG group. The western blot assay was performed using the retrieved proteins following pull-down assays, and the results further verified the association of IGF2BP2 and FBXL19-AS1. At this point we confirmed that IGF2BP2 could bind to FBXL19-AS1 and up-regulate its expression. To better understand the regulation mechanisms, the nascent RNA capture assay and half-life assay were carried out. As shown in Fig. 3D,E, there was no significant change of nascent transcribed FBXL19-AS1 after IGF2BP2 knockdown, but the half-life time of FBXL19-AS1 was significantly reduced. These results indicated that FBXL19-AS1 was regulated by IGF2BP2 on a post-transcriptional level. Then we speculated whether FBXL19-AS1 was involved in the regulatory process of IGF2BP2 on BTB permeability, The cell lines co-transfected with IGF2BP2 and FBXL19-AS1 were conducted and assessed subsequently. As shown in Fig. 3F,G, compared with control groups, there were no significant difference of TEER value and HRP flux was found in IGF2BP2(-)+FBXL19-AS1(+) groups and IGF2BP2(+)+FBXL19-AS1(-) groups. Similarly, the effect of IGF2BP2 overexpression or knockdown on the expression level of tight junction-associated proteins were rescued by FBXL19-AS1 knockdown or overexpression respectively (Fig. 3H). These results validated our hypothesis that FBXL19-AS1 mediated the regulatory process of IGF2BP2 on BTB permeability. Next, the mRNA and protein expression levels of ZNF765 were assessed via qRT-PCR and western blot. The results showed that overexpression or knockdown of FBXL19-AS1 rescued the expression change of ZNF765 caused by IGF2BP2 knockdown of overexpression respectively (Fig. 3I,J). Furthermore, we evaluated the effect of IGF2BP2 and FBXL19-AS1 on ZNF765 distribution. As shown in Figure S2 C, IGF2BP2 and FBXL19-AS1 did not affect the distribution of ZNF765 in GECs.
Figure3.
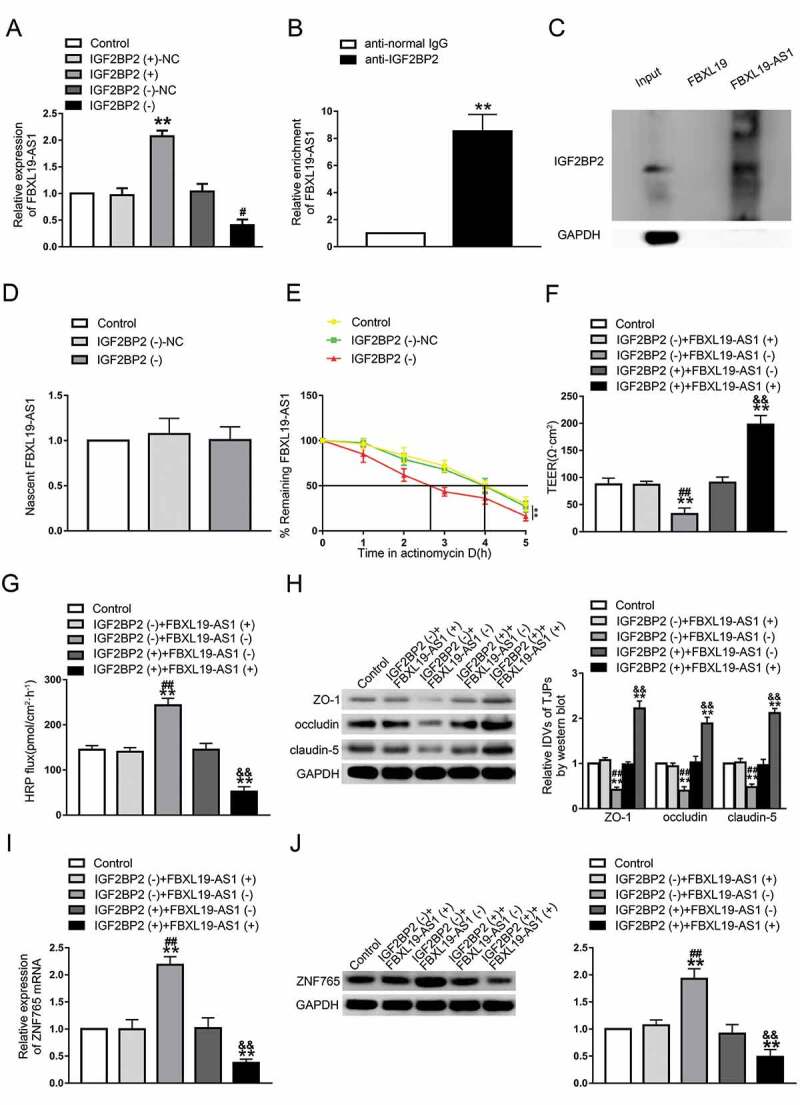
Upregulation of IGF2BP2 increased FBXL19-AS1 stability, FBXL19-AS1 participated in the process of IGF2BP2 regulating BTB permeability
(A). Knockdown of IGF2BP2 decreased the expression level of FBXL19-AS1. Data represent mean ± SD (n = 3, each). **P < 0.01 vs. IGF2BP2 (+)-NC group, #P < 0.05 vs. IGF2BP2 (-)-NC group. (B) RIP assay evaluated the remarkable enrichment of FBXL19-AS1 in anti-IGF2BP2 immunoprecipitates. Results were analysed using two-tailed paired Student’s t test. Data represent mean ± SD (n = 3, each). **P < 0.01 vs. Anti-normal IgG group. (C) IGF2BP2 and GAPDH protein in immunoprecipitation with FBXL19 and FBXL19-AS1 RNA were visualized by western blot. (D) No significant difference of nascent FBXL19-AS1 was found in silenced IGF2BP2 GECs. (E) Knockdown of IGF2BP2 decreased the half-life time of FBXL19-AS1. Results were analysed using two-tailed paired Student’s t test. Data represent mean ± SD (n = 3, each). **P < 0.01 vs. IGF2BP2 (-)-NC group. (F,G) Overexpression of FBXL19-AS1 significantly rescued the inhibitory or increasing effects of IGF2BP2 knockdown on TEER value and HRP exudate respectively. (H) Overexpression of FBXL19-AS1 significantly rescued the inhibitory effect of IGF2BP2 knockdown on the expression levels of ZO-1, occludin and claudin-5. (I,J) Overexpression of FBXL19-AS1 significantly rescued the increasing effect of IGF2BP2 knockdown on the mRNA and protein expression levels of ZNF765. Results were analysed using one-way ANOVA test. Data represent mean ± SD (n = 3, each). **P < 0.01 vs. Control group, ##P < 0.01 vs. IGF2BP2(-)+FBXL19-AS1(+) group, &&P < 0.01 vs. IGF2BP2 (+)+FBXL19-AS1(-) group.
Overexpression of ZNF765 increased BTB permeability by down-regulating the expression levels of ZO-1, occludin and claudin-5
To explore the expression and localization of ZNF765 in GECs, fluorescence in situ hybridization (FISH), qRT-PCR and western blot assays were carried out. As shown in Fig. 4A–C, compared with ZNF765 in GECs, ZNF765 was mainly located in AECs cytoplasm and its expression was higher in AECs. Furthermore, both stable ZNF765 overexpression and silencing GECs were constructed to explore the functions of ZNF765 on BTB permeability (Figure S1D). As Fig. 4D,E presented, the overexpressed ZNF765 decreased TEER values and enhanced the HRP flux, while silenced ZNF765 had the opposite effect. Moreover, the overexpression of ZNF765 also reduced the mRNA and protein expression levels of ZO-1, occludin and claudin-5, which disrupted the continuities of their distributions in GECs to some extent (Fig. 4F–H). The results above suggested that ZNF765 was down-regulated in GECs, and the overexpression of ZNF765 promoted BTB permeability by inhibiting tight junction-associated proteins. Furthermore, the change in IGF2BP2 expression were also observed. As Fig. 4I,J presented, the mRNA and protein expression levels of IGF2BP2 were decreased in ZNF765(+) group and increased after the ZNF765 depletion.
Figure 4.
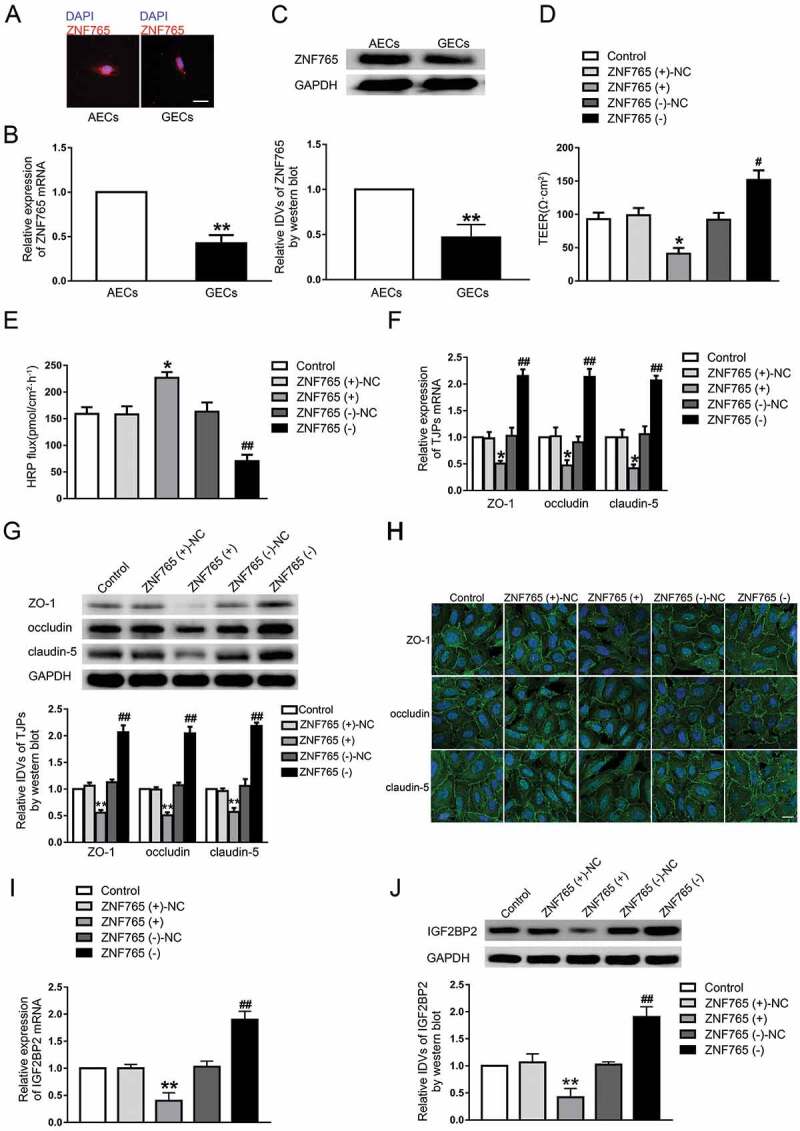
Overexpression of ZNF765 increased BTB permeability in vitro
(A) ZNF765 was mainly located in the cytoplasm in AECs and GECs and conspicuously reduced in GECs. Scale bars represent 20 μm. (B,C) The mRNA and protein expression levels of ZNF765 were decreased in GECs than in AECs. Results were analysed using two-tailed paired Student’s t test. Data represent mean ± SD (n = 3, each). **P < 0.01 vs. AECs group. (D,E) Overexpression of ZNF765 decreased TEER values and increased HRP flux. (F,G) Overexpression of ZNF765 decreased the mRNA and protein expression levels of tight junction-associated proteins (TJPs). (H) Overexpression of ZNF765 damaged the continuity of TJPs distributions. Scale bar represents 30 μm. (I-J) Overexpression of ZNF765 decreased the mRNA and protein expression levels of IGF2BP2. Results were analysed using one-way ANOVA test. Data represent mean ± SD (n = 3, each). *P < 0.05, **P < 0.01 vs. ZNF765 (+)-NC group, #P < 0.05, ##P < 0.01 vs. ZNF765 (-)-NC group.
FBXL19-AS1 down-regulated the expression of ZNF765 through SMD pathway
The microarray analysis of FBXL19-AS1 knockdown in GECs showed that, among these dysregulated transcriptional factors in glioma, ZNF765 and SOX7 expression levels were significantly elevated in FBXL19-AS1 silenced GECs and ZNF765 was mostly affected compared with others (Figure S2A). With the help of bioinformatic tools RepeatMasker and IntaRNA, two Alu elements were found to exist respectively in the 3ʹ-UTR of ZNF765 mRNA and FBXL19-AS1, and were predicted to be pairing with each other imperfectly (Figure S1E). To verify the above predictions, we transfected reporter plasmids of ZNF765 3ʹ-UTR wild type (Wt) and ZNF765 3ʹ-UTR with the mutational predicted binding site (Mut) into 293 T cells, and performed a dual-luciferase reporter assay. As Fig. 5A presented, compared with groups transfected with FBXL19-AS1(+)-NC, co-transfection of FBXL19-AS1(+) and ZNF765 3ʹ-UTR-Wt significantly decreased the activity, whereas co-transfection of FBXL19-AS1(+) and ZNF765 3ʹ-UTR-Mut did not (Fig. 5A). As noted above, this imperfect base pairing may produce a STAU1-binding site (SBS) [17], which made us wonder if FBXL19-AS1 regulated ZNF765 with the synergism of STAU1 and UPF1 via STAU1-mediated decay (SMD). The result of RIP assays showed that the abundant of FBXL19-AS1 and ZNF765 mRNA were enriched in samples immunoprecipitated with STAU1, and reduced by FBXL19-AS1 knockdown (Fig. 5B). After applied with actinomycin D, the half-life of ZNF765 mRNA in GECs was evaluated. Consistently, as shown in Fig. 5C, not only FBXL19-AS1 overexpression, STAU1 knockdown or the SMD factor UPF1 knockdown can also reduce the half-life of ZNF765 mRNA. To further verify our conjecture, the expression levels of ZNF765 were validated in GECs that co-transfected with FBXL19-AS1 overexpression and STAU1 knockdown. The results showed that compared with FBXL19-AS1(+)-NC+STAU1(-)-NC groups, overexpressed FBXL19-AS1 decreased the mRNA and protein expression levels of ZNF765, while silenced STAU1 increased their expression. In addition, compared with NC groups, silenced STAU1 rescued the reducing effect on ZNF765 expression caused by overexpressed FBXL19-AS1(Fig. 5D,E). The results above indicated that FBXL19-AS1 negatively regulated the expression of ZNF765 through SMD pathway.
Figure 5.
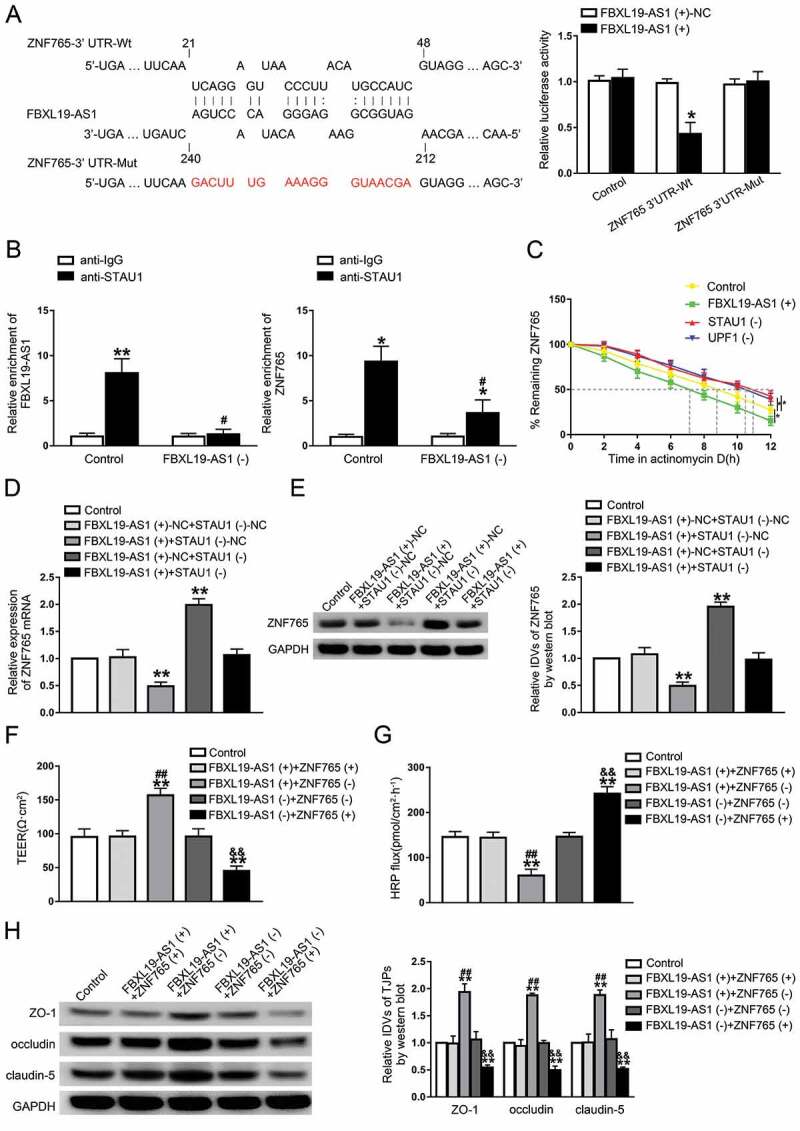
FBXL19-AS1 negatively regulated ZNF765 expression through SMD pathway
(A) Dual-luciferase reporter assays were performed to determine the binding sites of FBXL19-AS1 and ZNF765 3ʹ-UTR. Data represent mean ±SD (n = 3, each). *P < 0.05 vs ZNF765 3ʹ-UTR-Wt + FBXL19-AS1 (-)-NC group. (B) Knockdown of FBXL19-AS1 significantly induced the enrichment of FBXL19-AS1 and ZNF765 mRNA with STAU1 respectively. Results were analysed using two-tailed paired Student’s t test respectively. Data represent mean ± SD (n = 3, each). *P < 0.05, **P < 0.01 vs. anti-IgG group, #P < 0.05 vs. Control+anti-STAU1 group. (C) Overexpression of FBXL19-AS1, knockdown of STAU1 and knockdown of UPF1 reduced the half-life time of ZNF765 mRNA. Results were analysed using one-way ANOVA test. Data represent mean ± SD (n = 3, each). *P < 0.05 vs. Control group. (D,E) Knockdown of STAU1 rescued the inhibitory effect of overexpression FBXL19-AS1 on the mRNA and protein expression levels of ZNF765. Results were analysed using one-way ANOVA test. Data represent mean ± SD (n = 3, each). **P < 0.01 vs. Control group. (F,G) Knockdown of ZNF765 rescued the inhibitory or increasing effect of FBXL19-AS1 knockdown on TEER values or HRP of GECs respectively. (H) Knockdown of ZNF765 rescued the inhibitory effect of FBXL19-AS1 knockdown on the expression of ZO-1, occludin and claudin-5 in GECs. Results were analysed using one-way ANOVA test. Data represent mean ± SD (n = 3, each). **P < 0.01 vs. Control group, ##P < 0.01 vs. FBXL19-AS1 (+)+ZNF765 (+) group, &&P < 0.01 vs. FBXL19-AS1 (-)+ZNF765 (-) group.
FBXL19-AS1 regulated BTB permeability by down-regulating ZNF765
In order to further reveal the function of ZNF765 in FBXL19-AS1-regulated BTB permeability, GECs co-transfected with FBXL19-AS1 and ZNF765 were established. As shown in Fig. 5F,G, compared with control groups, there was no significant difference of TEER values and HRP flux in FBXL19-AS1(+)+ZNF765(+) group or in FBXL19-AS1(-)+ZNF765(-) group, while knockdown or overexpression of ZNF765 enhanced the effect on TEER values and HRP flux caused by FBXL19-AS1 overexpression or knockdown respectively. Consistently, overexpressed or silenced ZNF765 also reversed the impact of FBXL19-AS1 on tight junction-associated proteins expression levels (Fig. 5H). Moreover, the expression level of ZNF765 were significantly decreased in FBXL19-AS1(+)+ZNF765(-) group and elevated in FBXL19-AS1(-)+ZNF765(+) group, while no significant change was observed in FBXL19-AS1(+)+ZNF765(+) and FBXL19-AS1(-)+ZNF765(-) group (Figure S1 F). The above results indicated that overexpression of ZNF765 can reverse the decreasing of BTB permeability caused by overexpressed FBXL19-AS1. FBXL19-AS1 affected BTB permeability by regulating ZNF765.
ZNF765 bound to the promoter regions of ZO-1, occludin, claudin-5, IGF2BP2 and inhibited their activities
As Fig. 4F,G presented, the overexpression of ZNF765 reduced mRNA and protein expression levels of tight junction-associated proteins, leading us to speculate that ZNF765 may regulate them on the transcriptional level. Moreover, overexpressed ZNF765 also contributed to the decrease of IGF2BP2 mRNA and protein expression levels (Fig. 4I,J). With the help of DBTSS HOME bioinformatic software, the promoter region and transcription start site (TSS) sequences of ZO-1, occludin, claudin-5 and IGF2BP2 were acquired. The potential binding sites of ZNF765 were also discovered by using WILMER BIOINFORMATICS database. To confirm our prediction, dual-luciferase reporter assay and chromatin immunoprecipitation (ChIP) assay were carried out. As shown in Fig. 6A, a potential binding site of ZNF765 were found in the promoter region of ZO-1 at 814 bases preceding the Transcription Start Sites (TSS). The luciferase activity in cells that co-transfected with ZNF765 and ZO-1 promoter was significantly reduced compared to cells co-transfected with ZNF765 with a deletion of predicted binding site and ZO-1 promoter. Similarly, as shown in Fig. 6B-D, the deletion of putative binding site of ZNF765 markedly increased the promoter activities of occludin, claudin-5 and IGF2BP2 respectively. Furthermore, ChIP assays were subsequently performed to verify the binding sites. As the results presented (Fig. 6E–G), there was no association of ZNF765 with the negative control region of ZO-1, occludin, claudin-5 and IGF2BP2 that amplified without the predicted binding sites of ZNF765. Whereas the PCR products that amplified with potential binding sites of ZO-1, occludin, claudin-5 and IGF2BP2 by specific primers were observed respectively. These results suggested that ZNF765 could bind to and reduce the promoter activities of ZO-1, occludin and claudin-5. Moreover, combining with the results showed in Fig. 4I,J, we concluded that overexpression of ZNF765 can decreased IGF2BP2 expression through down-regulating its promoter activity in a positive feedback manner.
Figure 6.
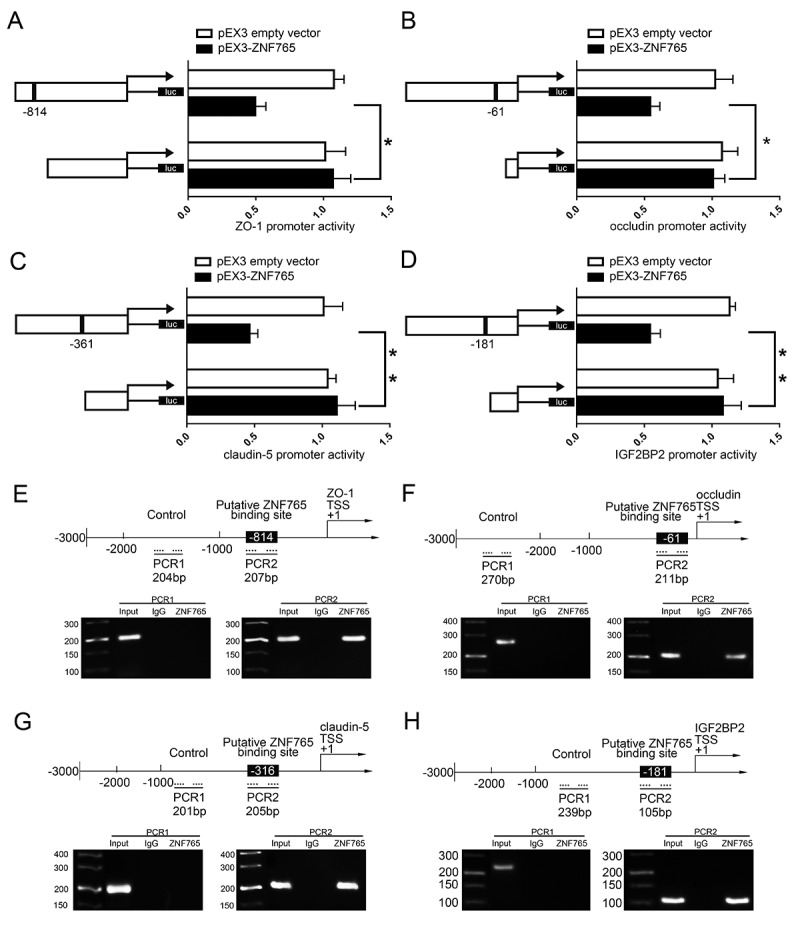
ZNF765 bound to ZO-1, occludin, claudin-5 and IGF2BP2 promoters and inhibited their expressions at the transcriptional level
(A-D) Schematic diagrams of different reporter plasmids and relative dual-luciferase activities of ZO-1, occludin, claudin-5 and IGF2BP2. The left part showed the deletion positions on the promoter fragments of ZO-1, occludin, claudin-5 and IGF2BP2 respectively, and the right part showed that the reporter vector activities of them were increased when depleted the putative binding sites of ZNF765. The results were showed after normalized with the co-transfected reference vector (pRL-TK), and relative to the activity of pEX3 empty vector, which the activity was set to 1. Results were analysed using two-tailed paired Student’s t test. Data represent means ± SD (n = 3, each). *P < 0.05, **P < 0.01 vs. empty group. (E,H) Schematic representation of ZO-1, occludin, claudin-5 and IGF2BP2 promoter region 3,000 bps upstream of the transcription start sites (TSS) which were designated as +1. Chromatin Immunoprecipitation PCR products for putative ZNF765 binding sites and an upstream region not supposed to associate with ZNF765 were depicted with bold lines. Dashed arrows represent the primers employed to each PCR. GECs were utilized to conduct ChIP assays. PCR was conducted with the resulting precipitated DNA.
Combination of IGF2BP2 knockdown and FBXL19-AS1 knockdown increased the effect of DOX in promoting apoptosis of U87 cells
Doxorubicin (DOX) has been widely used as a chemotherapy drug in several diseases including cancer, but barred by BTB and limited in the treatment of brain cancer [24]. To evaluate the potential clinical value of IGF2BP2 knockdown and FBXL19-AS1 knockdown, the apoptosis analysis was performed after establishing the BTB model. As shown in Fig. 7A, compared with the control group, there was no significant difference in IGF2BP2(-) group and FBXL19-AS1(-) group, but treatment with DOX obviously promoted U87 cell apoptosis. In the meantime, compared with DOX group, knockdown of IGF2BP2 or FBXL19-AS1 promoted U87 cells apoptosis caused by DOX, with the combination of simultaneous knockdown of IGF2BP2 and FBXL19-AS1 exhibiting the greatest effect. Furthermore, we used another chemotherapeutic agent carboplatin (CPN) to further demonstrate the effect of IGF2BP2 and FBXL19-AS1 knockdown. As shown in Figure S2D, consistent with the results above, knockdown of IGF2BP2 or FBXL19-AS1 enhanced U87 cells apoptosis caused by CPN, with the combination of simultaneous knockdown of IGF2BP2 and FBXL19-AS1 exhibiting the greatest effect. A flowchart of the regulating process of IGF2BP2/FBXL19-AS1/ZNF765 axis in BTB permeability was also illustrated in Fig. 7B.
Figure 7.
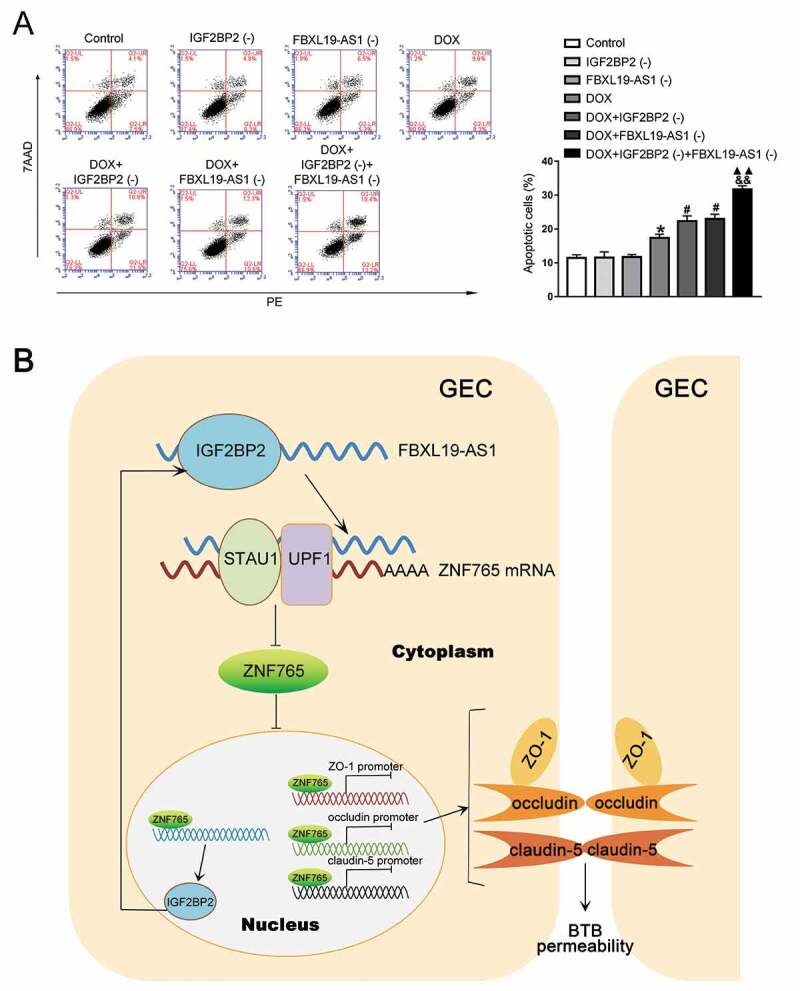
IGF2BP2 knockdown combined with FBXL19-AS1 knockdown increased the effect of DOX in promoting apoptosis of U87 cells
(A) Alone or combination use of IGF2BP2 knockdown and FBXL19-AS1 knockdown increased the promotional effect of DOX on the apoptosis rate of U87 cells. Results were analysed using one-way ANOVA test. Data represent mean ± SD (n = 3, each). *P < 0.05 vs. Control group, #P < 0.05 vs. DOX group, &&P < 0.01 vs. DOX+IGF2BP2 (-) group, ▲▲P < 0.05 vs. DOX+FBXL19-AS1 (-) group. (B) The schematic diagram about the regulation process of IGF2BP2/FBXL19-AS1/ZNF765 axis on BTB permeability.
Discussion
In this study, we observed that the expression levels of RBP-IGFBP2 and lncRNA-FBXL19-AS1 were up-regulated in GECs. Knockdown of IGFBP2 and FBXL19-AS1 significantly down-regulated the expression of tight junction-associated proteins (ZO-1, occludin and claudin-5), and increased BTB permeability via the paracellular pathway. Knockdown of IGFBP2 also increased BTB permeability by destabilizing FBXL19-AS1. Overexpression of FBXL19-AS1 reduced the half-life of transcription factor ZNF765 mRNA via SMD pathway and further accelerated its degradation. ZNF765, lowly expressed in GECs, bound directly to the promoters of ZO-1, occludin and claudin-5 respectively and repressed their transcriptional activities. Overexpression of ZNF765 increased BTB permeability by reducing the expression of tight junction-associated proteins.
In recent years, multiple studies have found that RNA-binding proteins (RBPs) play important roles in the regulation of biological processes on different levels by interacting with RNAs primarily. Dysregulated RBPs affects transcriptome and proteome of cells by disturbing RNA metabolism, and regulated cell growth, proliferation, invasion and apoptosis consequently [25,26]. Previous studies have shown that IGF2BP2 expression is elevated in glioma cells and GSCs [5,6], suggesting that IGF2BP2 may play regulatory roles in glioma microenvironment. Our study found that IGF2BP2 was highly expressed in GECs. Knockdown of IGF2BP2 increased BTB permeability by inhibiting the expression of tight junction-associated proteins ZO-1, occludin and claudin-5. Up-regulating of IGF2BP2 has also been found in different tumours and associated with adverse prognosis in patients, such as pancreatic cancer, acute myelocytic leukaemia and oesophageal adenocarcinoma. In breast cancer, IGF2BP2 promotes the metastatic progression by motivating IGF type 1 receptor and recruiting endothelial cells [27].
As reported in many studies, lncRNAs contributed to the development of inflammatory diseases and cancers, and have been described to play a vital role in regulating blood-brain barrier (BBB) and BTB permeability. For instance, down-regulation of MEG3 in a post cerebral ischaemia-reperfusion injury rat model can decrease BBB permeability [28]. MALAT1 protects BBB integrity against ischaemia via MALAT1/CREB/PGC-1α/PPARγ pathway [29]. HOTAIR, overexpressed in glioma microvascular endothelial cells, participates in the regulation of BTB permeability via HOTAIR/miR-148b-3p/USF1 competing endogenous RNAs (ceRNA) mechanism [30]. Our study for the first time found FBXL19-AS1 expression was up-regulated in GECs. Knockdown of FBXL19-AS1 reduced tight junction-associated proteins expression and TEER value, increasing the HRP flux and BTB permeability. There has been no report about FBXL19-AS1 in other vascular endothelial cells heretofore, while similar studies has been performed on tumours. FBXL19-AS1 expression in non-small cell lung cancer (NSCLC) is up-regulated and can promote the proliferation and metastasis of NSCLC cells through EMT [31]. In addition, FBXL19-AS1 is highly expressed in breast cancer and osteosarcoma, and promotes the proliferation and invasion of tumour cells through ceRNA mechanism by binding miR-718 and miR-346 respectively [32,33].
As post-transcriptional regulators, RBPs are involved in the regulation of RNA targets stability, splicing, nuclear export and translation by binding them [34]. In this study, based on the prediction by STARBASE database, we performed RIP and pull-down assays and verified the potential binding site between IGF2BP2 and FBXL19-AS1. It was further revealed that knockdown of IGF2BP2 in GECs can decrease the half-life and the expression level of FBXL19-AS1. Moreover, knockdown of FBXL19-AS1 reversed the effects of IGF2BP2 on the TEER values, HRP flux and the expression levels of ZO-1, occludin and claudin-5, suggesting that IGF2BP2 exerted effects on BTB permeability by stabilizing FBXL19-AS1. A similar research has found that RBP-LIN28A could stabilize and interact with FBXL19-AS1, and further promote cell migration, invasion and EMT in breast cancer [35]. In liver cancer, IGF2BP1 from IGF2BPs family destabilizes liver-specific lncRNA-HULC at the post-transcriptional level [36].
The mechanism that induced by lncRNAs has been widely reported, with ceRNA mechanism being mostly studied. Recent studies found that the imperfect base paring between an Alu element in a lncRNA and another Alu element in 3ʹ-UTR of a target mRNA can form a STAU1-binding site (SBS) [17]. When the target mRNA translation terminates in the upstream region of SBS, STAU1 that bound to the SBS site recruits UPF1 through protein interactions, thereby inducing mRNA degradation and instability [18,37]. The mechanism described above is SMD, which has recently been revealed in the regulation network centred on lncRNAs. For example, lncRNA-mediated SMD networks presents in ischaemic stroke and is responsible to regulate the pathogenesis of focal cerebral ischaemia[38]. In neural progenitor cells (NPCs), KLF4 interacts with its partner STAU1 and affects the SMD level, thereby regulating target neurogenesis-associated mRNAs in mammalian [16]. The STAU1 that exists in neurons of axons and dendrites can elicit local RNA degradation through SMD pathway and regulate the capacity of protein synthetic in axons and growth cones [39]. In this study, we predicted an Alu element base-pairing between FBXL19-AS1 and ZNF765 mRNA 3ʹ-UTR via bioinformatic software Repeatmasker and IntaRNA. We then verified the binding by dual luciferase assay, and proved the binding site was SBS by RIP experiment. Down-regulated STAU1 or UPF1 increased the expression and half-life of ZNF765 mRNA. Down-regulated STAU1 also rescued the down-regulation of ZNF765 caused by FBXL19-AS1 overexpression. Furthermore, increasing or decreasing the expression of ZNF765 reversed the effect of FBXL19-AS1 overexpression or knockdown on the BTB permeability. These results indicated that FBXL19-AS1 regulated BTB permeability by degrading ZNF765 via SMD pathway. The regulatory effect of SMD in vessels has not been reported.
Our research discovered lower expression of ZNF765 in GECs compared with ECs. Overexpressed ZNF765 decreased ZO-1, occludin and claudin-5 expression and increased BTB permeability. Various biomedical functions of zinc finger family protein have been attracting more and more attention, including maintaining blood-brain barrier and regulating BTB permeability [40,41]. In an ischaemic stroke mice model, absence of a zinc finger protein MCPIP1 increased MMP-9/3 expression and decreased claudin-5 and ZO-1 expression, disrupting the integrity of BBB subsequently [42]. In mice brain endothelial cells under hypoxia condition, the expression of ZEB1 was highly related to ZO-1 mRNA expression [43]. MiR-34 c regulated tight junction-associated proteins expression and BTB permeability by reversely altering its target zinc finger protein MAZ [41]. Similarly, miR-181a modulated BTB permeability by negatively regulating KLF6 [44]. Coincidentally, three potential binding sites of ZNF765 in the promoter region of ZO-1, occludin and claudin-5 were found and verified respectively. Further, the results of luciferase assays showed that ZNF765 could reduce the promoter activities of ZO-1, occludin and claudin-5. These results indicated that ZNF765 increased permeability of BTB by reducing the promoter activity and expression of tight junction-associated proteins. Moreover, we found that ZNF765 could also bind to and inhibit the promoter activity of IGF2BP2, which formed a feedback loop in GECs. In this loop, silenced IGF2BP2 increased ZNF765 expression by decreasing FBXL19-AS1. Up-regulated ZNF765 further inhibited the transcription and expression level of IGF2BP2. Our results may unearth a feedback loop between IGF2BP2 and ZNF765, which may play a key role in the regulating process of BTB permeability.
Because of the existence of BBB, as one of the most widely used anthracyclines antibiotics, doxorubicin (DOX) has been widely applied in the clinical treatment of multiple diseases including glioma, but considerably restricted from entering brain tissues [45]. Finally, through the BTB model in vitro, we found that the combination of knockdown IGF2BP2 and FBXL19-AS1 significantly increased U87 apoptosis rate caused by DOX, compared with the single silencing of IGF2BP2, FBXL19-AS1 or DOX treatment alone. Similarly, the combination of knockdown IGF2BP2 and FBXL19-AS1 also increased U87 apoptosis that caused by carboplatin (CPN), which is widely used as a chemotherapeutic agent that restricted in the process of drug delivery due to BBB [46,47]. The results suggested the combined application of knockdown IGF2BP2, FBXL19-AS1 and overexpression of ZNF765 could increase BTB permeability and promote the inhibitory effect of DOX or CPN on glioma cells subsequently.
In summary, our study verified for the first time that the elevated expression of IGF2BP2 in GECs regulated BTB permeability by binding and stabilizing FBXL19-AS1. The increasing of FBXL19-AS1 elicited the degradation of ZNF765 mRNA via SMD pathway. The down-regulated expression of ZNF765 decreased BTB permeability by promoting expression of tight junction-associated protein ZO-1, occludin and claudin-5, and also repressed transcription of IGF2BP2 thereby forming a positive feedback loop during the regulatory process. Combined treatment of silencing IGF2BP2 and FBXL19-AS1 increased the apoptosis of glioma cells caused by doxorubicin. Our findings reveal the important role of IGF2BP2/FBXL19-AS1/ZNF765 feedback axis in regulating BTB permeability and provides new potential therapeutic strategies for glioma.
Materials and methods
Cell culture
The human brain microvascular endothelial cell line hcMEC/D3 (ECs) was a gift from Dr. Couraud (Institut Cochin, Paris, France) and grown in culture flasks coated with 150 μg/ml Cultrex Rat Collagen I (R&D Systems, Minneapolis, MN, USA). The culture medium included EBM-2 (endothelial basal medium, Lonza, Walkersville, MD, USA) supplemented with 5% FBS (foetal bovine serum) ‘Gold’, 1% penicillin-streptomycin, 1% chemically defined lipid concentrate, 1.4 μmol/l hydrocortisone, 5 μg/ml ascorbic acid, 10 mmol/l HEPES and 1 ng/ml bFGF (basic fibroblast grown factor). Normal human astrocytes, glioblastoma cell lines U87 MG and embryonic kidney 293 T cells were purchased from the Shanghai Institutes for Biological Sciences Cell Resource Center, and grown in high glucose DMEM (Dulbecco’s Modified Eagle Medium, Sigma-Aldrich) with 10% FBS, 100 μg/ml streptomycin and 100 U/ml penicillin. All cells were cultures in a thermostatic incubator at 37°C, 5% CO2 and 100% humidity. Every 48 h, cells were sub-cultured or medium changed.
Laser capture microdissection (LCM)
To be able to acquire the blood vessel sections of normal and glioma tissues accurately, the tissue specimens were obtained and frozen-dissected at the laser beam smaller than 10 μm by using Microtome Cryostat (MICROM International GmbH, Walldorf, Germany). Tissue sections were next performed fluorescent dye-tagged lectin staining using Ulex europaeus lectin 1 (Vector Laboratories, Burlington, Canada) according to the manufacturer’s instructions for visualization. Furthermore, LCM were visualized by using ArcturusXTTM Laser Capture Microdissection Instrument.
Establishment of a BTB model in vitro
The BTB models in vitro were built according to previous study [48]. In short, 48 h after plating U87 MG cells into six-well plates at 2 × 104 per well, ECs were seeded onto the upper of inserts that coated with 150 μg/ml Culrex Rat Collagen I at 2 × 105 per well. Prepared EBM-2 medium was used as the culture for both upper and lower chamber and changed every 48 h, and GECs modelling endothelial cell phenotype of glioma were obtained after 96 h of co-culturing with U87 MG.
Quantitative Real-time PCR assay (qRT-PCR)
To evaluated the expression levels of IGF2BP2, FBXL19-AS1 and ZNF765, we carried out qRT-PCR. Total cellular RNA was extracted using Trizol Reagent (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s protocol, and the Nanodrop Spectrophotometer (Thermo Scientific, Waltham, MA, USA) was used to measure the quality. In addition, One Step PrimeScriptTM Kits (Takara, Japan) were used to detect the expression of IGF2BP2, FBXL19-AS1 and ZNF765 with GAPDH used as the internal control. All results were analysed using the relative quantification (2–ΔΔCT) method. The primer sequences of IGF2BP2, FBXL19-AS2 and ZNF765 were designed as shown in Table 2.
Table 2.
Primers used for qRT-PCR to detect IGF2BP2, FBXL19-AS1 and ZNF765
| Gene Name | Sequence (5ʹ->3ʹ) |
|---|---|
| IGF2BP2 | Forward primer: TGAAGCTGGAAGCGCATATC |
| Reverse primer: CACGAGGCACGATGACTTCT | |
| FBXL19-AS1 | Forward primer: CATGATGCCGGACAGAGTTC |
| Reverse primer: GCAGTGCTCGGTAACAGACG | |
| ZNF765 | Forward primer: TCATTCGCATCTGCCTGAAC |
| Reverse primer: AACCAAGGAAGCATCGTGGA |
Cell transfection
To analyse the regulatory effect of IGF2BP2, FBXL19-AS1 and ZNF765 on BTB permeabilities, full length and shRNA sequence against IGF2BP2, FBXL19-AS1 and ZNF765 were ligated into pIRES2-EGFP vector and pGPU6/GFP/Neo vector respectively (GenePharma, Shanghai, China), and their empty vectors were used as negative control. Endothelial cells were pated on 24-well plates until they were at 70%–80% confluence, and transfected using Lipofectamine LTX Reagent (Life Technologies, Carlsbad, CA, USA) according to manufacturer’s protocol. Stable transfected cell lines were selected by using Geneticin (G418) for 4 weeks, and qRT-PCR was used to evaluate the transfection efficiency.
Half-life measurement
To analyse the stability of ZNF765 and FBXL19-AS1, we performed half-life assays with under treatment of actinomycin D. Actinomycin D is known as a transcriptional inhibitor through directly binding to the transcription initiation complex and blocking the transcription [49]. Cells were treated with 80 μmol/ml Actinomycin D (Sigma-Aldrich, Louis, MO, USA) to halt transcription, harvested and collected at 0, 1, 2, 3, 4 and 5 h after the treatment. Total cellular RNA was isolated and analysed by qRT-PCR with GAPDH mRNA as internal reference.
TEER and HRP flux assays
After successfully constructed the BTB model in vitro, millicell-ERS instrument (Millipore, Billerica, MA, USA) was used to evaluate TEER values. To eliminate the effects of temperature and medium on outcome, TEER values were measured after restoring at room temperature for 30 min and exchanging mediums. Final resistances (Ω·centimeter2) were measured after reducing background electrical resistances using the surface of the transwell insert.
After successfully constructed the BTB model in vitro, HRP flux assays were performed to further measure the permeability of BTB models. 1 ml of culture medium supplemented with 10 μg/ml HRP and 2 ml of culture medium were added to the upper and lower compartments respectively. After incubating at 37°C for 1 h, 5 μl of medium in the lower compartment was collected and analysed by the tetramethylbenzidinee colorimentry with a spectrophotometer at 370 nm. The final amount of HRP permeability was expressed as pmol per cm2/h.
Western blot analysis
The protein expression of IGF2BP2, ZNF765, ZO-1, occludin and claudin-5 were evaluated via western blot assays. Cells were collected and homogenized in RIPA lysis buffer (Beyotime, Jiangsu, China) with PMSF and centrifuged at 17,000 rpm, 4 °C for 10 min to obtain the total proteins. The concentration was quantified by BCA protein kit (Beyotime, Jiangsu, China), separated by electrophoresis on SDS-polyacrylamide gel (SDS-PAGE) and transferred to a nitrocellulose membrane subsequently. After the non-specific binding was being blocked in TTBS with 5% non-fat milk for 2 h, membranes were washed and incubated with specific antibodies overnight at 4 °C. Then, membranes were incubated with goat anti-rabbit or anti-mouse secondary antibodies for 2 h at room temperature. Finally, the blots were visualized by chemiluminescence using the ECL kit (Santa Cruz Biotechnology, Dallas, TX, USA) and digitally scanned via Chemi Imager 5500 V2.03 software. GAPDH band was used as an internal control. The relative integrated density values (IDVs) were analysed using Fluor Chem2.0 software.
Immunofluorescence assays
To confirmed the continuous distribution of tight junction-related proteins, immunofluorescence assays were carried out. Briefly, Cells were cultured and fixed on coverslips coated with 1% gelatin and fixed with 4% paraformaldehyde solution for 30 min at room temperature. Then, cells were permeated with 0.3% Triton X-100 for 10 min if the target protein was ZO-1 or claudin-5, or permeated with methanol for 20 min at −20°C if the target protein was occludin. After washing three times with PBS, cells were blocked in 5% BSA buffer for 2 h at room temperature, and further incubated with antibodies overnight at 4°C. Secondary antibodies were used to incubate cells for 2 h after rewarming to room temperature, and the cells were stained with DAPI for 8 min and washed three times with PBST. The fluorescence was visualized and analysed by Olympus DP71 immunofluorescence microscope.
Dual-luciferase reporter assays
To verify the putative binding between the Alu elements in FBXL19-AS1 and ZNF765, we performed dual-luciferase reporter assay. To construct reporter vector, the ZNF765 3ʹ-UTR sequence was amplified and cloned as wild type (wt) vector, and the ZNF765 3ʹ-UTR with mutated FBXL19-AS1 binding sequence was amplified and cloned as a mutant type (Mut) vector. HEK 293 T cells were seeded in 96-well plates and co-transfected with FBXL19-AS1 overexpression vectors and ZNF765 3ʹ-UTR (Wt or Mut) vectors using Lipofectamine 3000. After 48 h, luciferase activities were evaluated using a dual luciferase reporter system (Promega, Beijing, China). Relative luciferase activity was given as the ratio of firefly/Renilla luciferase activity.
Similarly, the promoter regions of ZO-1, occludin and claudin-5 (−1000 to +100bp from) were amplified and constructed into pGL3 plasmid. Moreover, promoter regions of them with depletion of predicted binding sequences of ZNF765 were amplified and constructed as well. Full-length ZNF765 gene was constructed in pEX3 plasmid. HEK 293 T cells were seeded in 96-well plates and co-transfected with ZNF765 full-length or empty vectors and whole or deleted promoter regions vectors using Lipofectamine 3000, and the relative luciferase activity was evaluated as well.
RNA binding protein immunoprecipitation (RIP) assay
RIP assay was performed using EZ-Magna RIP kit (Millipore, Billerica, MA, USA) according to the manufacturer’s instructions. Endothelial cells were lysed using RNA lysis buffer and incubated with RIP buffer that containing magnetic beads, which conjugated with anti-human IGF2BP2 or anti-human STAU1 antibodies overnight. Anti-human IgG was used as the negative control. The co-precipitated samples were next treated with proteinase K and the immunoprecipitated RNA was isolated subsequently. Finally, the RNA concentration was purified and analysed by qRT-PCR to verify the binding between FBXLAS1 and IGF2BP2 or STAU1.
RNA pull-down assay
RNA pull-down assay was performed to verify the binding between FBXL19-AS1 and IGF2BP2 according to the manufacturer’s instruction of Pierce Magnetic RNA-Protein Pull-Down kit (ThermoFisher, USA). Briefly, the biotin-labelled FBXL19-AS1 or antisense FBXL19 were constructed and incubated with protein extract of GECs and magnetic beads at room temperature. Next, the beads-RNA-protein precipitates were obtained by low speed centrifugalization and purified by washing buffer. Finally, western blot assays were carried out to analyse the protein level after washing by elution buffer and GAPDH was used as the control.
Chromatin immunoprecipitation (ChIP) assay
To verify the predicted binding sites of ZNF765 on the promoter regions of ZO-1, occludin, claudin-5 and IGF2BP2 respectively, simple ChIP Enzymatic Chromatin IP Kit (Cell signalling Technology, Danvers, MA, USA) was used to perform ChIP assay according to the instruction. Briefly, GECs were cross-linked with 1% formaldehyde for 10 min, and added glycine for 5 min at room temperature to terminate the crosslinking. Then the cells were then collected and lysed in lysis buffer that containing 1% PMFS, and incubated with micrococcal nuclease for 20 min at 37°C. Samples were next incubated with ZNF765 antibody or IgG antibody (negative control) overnight at 4°C with gentle shaking. In addition, 5 mol/l NaCl and proteinase K were used for decrosslinking at 65°C for 2 h. In the end, immunoprecipitated DNA was purified and amplified with primers, and then isolated and visualized by 3% agarose gel. The primers that used were showed in Table 2.
Apoptosis analysis
To further evaluate the regulation of knockdown IGF2BP2 and FBXL19-AS1 in vitro BTB models, we performed apoptosis analyses to analyse the apoptosis glioma cell-U87 under different treating conditions. Doxorubicin (DOX) and carboplatin (CPN) are often used as chemotherapy drugs in several diseases including glioma, which can promote the apoptosis of cancer cells, and often limited in delivering across BTB to the brain [24]. In apoptosis analysis, U87 was first seeded into the lower chamber and cultured in DMEM medium for 24 h, and then ECs with knockdown IGF2BP2 and knockdown FBXL19-AS1 alone or combined were seeded into the upper chamber established as BTB models. After 3–4 days, 10 μM DOX or 5 mg/ml CPN (Beyotime Institute of Biotechnology, Jiangsu, China) were added into the upper chamber in study groups respectively, and the apoptosis of U87 were evaluate after 12 h using Annexin V-PE/7AAD kit (Southern Biotech, AL, USA). U87 were collected, washed twice using phosphate-buffered saline, and suspended using Annexin-V binding buffer. Then cells were added with 5 μl VE, 5 μl 7AAD to stain early apoptotic cells and late apoptotic cells respectively, and evaluated by flow cytometry (FACScan, BD Biosciences).
Statistical analysis
Data were derived from at least three independent experiments and presented as mean ± SD. All results were analysed using two-tailed paired Student’s t test and one-way ANOVA test. GraphPad Prism v5.01 software was performed for statistical analysis and P value < 0.05 was defined as statistically significant.
Supplementary Material
Funding Statement
This work was supported by grants from the Natural Science Foundation of China (81872073, 81672511, 81872503), China Postdoctoral Science Foundation (2019M661172), Liaoning Science and Technology Plan Project (2020-BS-097, 2017225020), Project of Key Laboratory of Neuro-oncology in Liaoning Province (112-2400017005) and outstanding scientific fund of Shengjing hospital (No. 201802).
Author contributions
Y.L. and X.L. contributed to the study concept and design, Y.X. designed the experiments. X.L., D.W., C.Y., L.S. and X.R. contributed to the experimental implementation and data analysis. P.W. and R.S. contributed to the manuscript draft. J.Z., Y.Y. and Z.L. contributed to the critical revision of the manuscript for important intellectual content. All authors read and approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Ostrom QT, Gittleman H, Liao P, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2010-2014. Neuro Oncol. 2017;19:v1–v88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Black KL, Ningaraj NS.. Modulation of brain tumor capillaries for enhanced drug delivery selectively to brain tumor. Cancer Control. 2004;11:165–173. [DOI] [PubMed] [Google Scholar]
- [3].Gerstberger S, Hafner M, Tuschl T.. A census of human RNA-binding proteins. Nat Rev Genet. 2014;15:829–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Glorian V, Maillot G, Polès S, et al. HuR-dependent loading of miRNA RISC to the mRNA encoding the Ras-related small GTPase RhoB controls its translation during UV-induced apoptosis. Cell Death Differ. 2011;18(11):1692–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mu Q, Wang L, Yu F, et al. Imp2 regulates GBM progression by activating IGF2/PI3K/Akt pathway. Cancer Biol Ther. 2015;16:623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Degrauwe N, Schlumpf TB, Janiszewska M, et al. The RNA binding protein IMP2 preserves glioblastoma stem cells by preventing let-7 target gene silencing. Cell Rep. 2016;15:1634–1647. [DOI] [PubMed] [Google Scholar]
- [7].Mineo M, Ricklefs F, Rooj AK, et al. The long non-coding RNA HIF1A-AS2 facilitates the maintenance of mesenchymal glioblastoma stem-like cells in hypoxic niches. Cell Rep. 2016;15:2500–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wang L, Yu Z, Sun S, et al. Long non-coding RNAs: potential molecular biomarkers for gliomas diagnosis and prognosis. Rev Neurosci. 2017;28:375–380. [DOI] [PubMed] [Google Scholar]
- [10].Worku T, Bhattarai D, Ayers D, et al. Long non-coding RNAs: the new horizon of gene regulation in ovarian cancer. Cell Physiol Biochem. 2017;44:948–966. [DOI] [PubMed] [Google Scholar]
- [11].Ricciuti B, Mencaroni C, Paglialunga L, et al. Long noncoding RNAs: new insights into non-small cell lung cancer biology, diagnosis and therapy. Med Oncol. 2016;33(2):18. [DOI] [PubMed] [Google Scholar]
- [12].Zhao J, Mialki RK, Wei J, et al. SCF E3 ligase F-box protein complex SCF FBXL19 regulates cell migration by mediating Rac1 ubiquitination and degradation. Faseb J. 2013;27(7):2611–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chandran V. The genetics of psoriasis and psoriatic arthritis. Clin Rev Allergy Immunol. 2013;44:149–156. [DOI] [PubMed] [Google Scholar]
- [14].Zhao J, Wei J, Mialki RK, et al. F-box protein FBXL19-mediated ubiquitination and degradation of the receptor for IL-33 limits pulmonary inflammation. Nat Immunol. 2012;13:651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Shen B, Yuan Y, Zhang Y, et al. Long non-coding RNA FBXL19-AS1 plays oncogenic role in colorectal cancer by sponging miR-203. Biochem Biophys Res Commun. 2017;488(1):67–73. [DOI] [PubMed] [Google Scholar]
- [16].Moon B-S, Bai J, Cai M, et al. Kruppel-like factor 4-dependent Staufen1-mediated mRNA decay regulates cortical neurogenesis. Nat Commun. 2018;9(1):401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gong C, Kim YK, Woeller CF, et al. SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev. 2009;23:54–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cho H, Kim K, Han S, et al. Staufen1-mediated mRNA decay functions in adipogenesis. Mol Cell. 2012;46(4):495–506. [DOI] [PubMed] [Google Scholar]
- [19].Park E, Maquat LE. Staufen-mediated mRNA decay. Wiley Interdiscip Rev RNA. 2013;4:423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Park E, Gleghorn ML, Maquat LE. Staufen2 functions in Staufen1-mediated mRNA decay by binding to itself and its paralog and promoting UPF1 helicase but not ATPase activity. Proc Natl Acad Sci U S A. 2013;110:405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Cassandri M, Smirnov A, Novelli F, et al. Zinc-finger proteins in health and disease. Cell Death Discov. 2017;3(1):17071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ragusa M, Majorana A, Banelli B, et al. MIR152, MIR200B, and MIR338, human positional and functional neuroblastoma candidates, are involved in neuroblast differentiation and apoptosis. J Mol Med (Berl). 2010;88(10):1041–1053. [DOI] [PubMed] [Google Scholar]
- [23].Durinck S, Stawiski EW, Pavía-Jiménez A, et al. Spectrum of diverse genomic alterations define non-clear cell renal carcinoma subtypes. Nat Genet. 2015;47:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Arvanitis CD, Askoxylakis V, Guo Y, et al. Mechanisms of enhanced drug delivery in brain metastases with focused ultrasound-induced blood-tumor barrier disruption. Proc Natl Acad Sci U S A. 2018;115:E8717–E8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Muller-McNicoll M, Rossbach O, Hui J, et al. Auto-regulatory feedback by RNA-binding proteins. J Mol Cell Biol. 2019;11(10):930–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lin C, Miles WO, Beyond CLIP. advances and opportunities to measure RBP-RNA and RNA-RNA interactions. Nucleic Acids Res. 2019;47:5490–5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Png KJ, Halberg N, Yoshida M, et al. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2011;481:190–194. [DOI] [PubMed] [Google Scholar]
- [28].You D, You H. Repression of long non-coding RNA MEG3 restores nerve growth and alleviates neurological impairment after cerebral ischemia-reperfusion injury in a rat model. Biomed Pharmacother. 2019;111:1447–1457. [DOI] [PubMed] [Google Scholar]
- [29].Ruan W, Li J, Xu Y, et al. MALAT1 up-regulator polydatin protects brain microvascular integrity and ameliorates stroke through C/EBPbeta/MALAT1/CREB/PGC-1alpha/PPARgamma pathway. Cell Mol Neurobiol. 2019;39:265–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sa L, Li Y, Zhao L, et al. The Role of HOTAIR/miR-148b-3p/USF1 on regulating the permeability of BTB. Front Mol Neurosci. 2017;10:194. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [31].Yu DJ, Li YH, Zhong M. LncRNA FBXL19-AS1 promotes proliferation and metastasis via regulating epithelial-mesenchymal transition in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2019;23:4800–4806. [DOI] [PubMed] [Google Scholar]
- [32].Ding Z, Ye P, Yang X, et al. LncRNA FBXL19-AS1 promotes breast cancer cells proliferation and invasion via acting as a molecular sponge to miR-718. Biosci Rep. 2019;39. DOI: 10.1042/BSR20182018. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [33].Pan R, He Z, Ruan W, et al. lncRNA FBXL19-AS1 regulates osteosarcoma cell proliferation, migration and invasion by sponging miR-346. Onco Targets Ther. 2018;11:8409–8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Dassi E. Handshakes and fights: the regulatory interplay of RNA-binding proteins. Front Mol Biosci. 2017;4:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhang Y, Xiao X, Zhou W, et al. LIN28A-stabilized FBXL19-AS1 promotes breast cancer migration, invasion and EMT by regulating WDR66. In Vitro Cell Dev Biol Anim. 2019;55:426–435. [DOI] [PubMed] [Google Scholar]
- [36].Hammerle M, Gutschner T, Uckelmann H, et al. Posttranscriptional destabilization of the liver-specific long noncoding RNA HULC by the IGF2 mRNA-binding protein 1 (IGF2BP1). Hepatology. 2013;58:1703–1712. [DOI] [PubMed] [Google Scholar]
- [37].Kim YK, Furic L, Desgroseillers L, et al. Mammalian Staufen1 recruits Upf1 to specific mRNA 3ʹUTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. [DOI] [PubMed] [Google Scholar]
- [38].Liu J, Zhang KS, Hu B, et al. Systematic analysis of RNA regulatory network in rat brain after ischemic stroke. Biomed Res Int. 2018;2018:8354350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kim JY, Deglincerti A, Jaffrey SR. A Staufen1-mediated decay pathway influences the local transcriptome in axons. Translation (Austin). 2017;5:e1414016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Aruga J. Zic Family Proteins in Emerging Biomedical Studies. Adv Exp Med Biol. 2018;1046:233–248. [DOI] [PubMed] [Google Scholar]
- [41].Zhao L, Wang P, Liu Y, et al. miR-34c regulates the permeability of blood-tumor barrier via MAZ-mediated expression changes of ZO-1, occludin, and claudin-5. J Cell Physiol. 2015;230(3):716–731. [DOI] [PubMed] [Google Scholar]
- [42].Jin Z, Liang J, Li J, et al. Absence of MCP-induced protein 1 enhances blood-brain barrier breakdown after experimental stroke in mice. Int J Mol Sci. 2019;20(13):3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Leduc-Galindo D, Qvist P, Tóth AE, et al. The effect of hypoxia on ZEB1 expression in a mimetic system of the blood-brain barrier. Microvasc Res. 2019;122:131–135. [DOI] [PubMed] [Google Scholar]
- [44].Ma J, Yao Y, Wang P, et al. MiR-181a regulates blood-tumor barrier permeability by targeting Kruppel-like factor 6. J Cereb Blood Flow Metab. 2014;34:1826–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lockman PR, Mittapalli RK, Taskar KS, et al. Heterogeneous blood-tumor barrier permeability determines drug efficacy in experimental brain metastases of breast cancer. Clin Cancer Res. 2010;16(23):5664–5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Cote J, Bovenzi V, Savard M, et al. Induction of selective blood-tumor barrier permeability and macromolecular transport by a biostable kinin B1 receptor agonist in a glioma rat model. PLoS One. 2012;7(5):e37485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ningaraj NS, Rao MK, Black KL. Adenosine 5ʹ-triphosphate-sensitive potassium channel-mediated blood-brain tumor barrier permeability increase in a rat brain tumor model. Cancer Res. 2003;63:8899–8911. [PubMed] [Google Scholar]
- [48].Cai H, Liu W, Xue Y, et al. Roundabout 4 regulates blood-tumor barrier permeability through the modulation of ZO-1, occludin, and claudin-5 expression. J Neuropathol Exp Neurol. 2015;74:25–37. [DOI] [PubMed] [Google Scholar]
- [49].Sobell HM. Actinomycin and DNA transcription. Proc Natl Acad Sci U S A. 1985;82:5328–5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


