Abstract
Purpose
Gegen Qinlian decoction (GQD) has been used to treat gastrointestinal diseases, such as diarrhea and ulcerative colitis (UC). A recent study demonstrated that GQD enhanced the effect of PD-1 blockade in colorectal cancer (CRC). This study used network pharmacology analysis to investigate the mechanisms of GQD as a potential therapeutic approach against CRC.
Materials and Methods
Bioactive chemical ingredients (BCIs) of GQD were collected from the Traditional Chinese Medicine Systems Pharmacology (TCMSP) database. CRC-specific genes were obtained using the gene expression profile GSE110224 from the Gene Expression Omnibus (GEO) database. Target genes related to BCIs of GQD were then screened out. The GQD-CRC ingredient-target pharmacology network was constructed and visualized using Cytoscape software. A protein-protein interaction (PPI) network was subsequently constructed and analyzed with BisoGenet and CytoNCA plug-in in Cytoscape. Gene Ontology (GO) functional and the Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway enrichment analysis for target genes were then performed using the R package of clusterProfiler.
Results
One hundred and eighteen BCIs were determined to be effective on CRC, including quercetin, wogonin, and baicalein. Twenty corresponding target genes were screened out including PTGS2, CCNB1, and SPP1. Among these genes, CCNB1 and SPP1 were identified as crucial to the PPI network. A total of 212 GO terms and 6 KEGG pathways were enriched for target genes. Functional analysis indicated that these targets were closely related to pathophysiological processes and pathways such as biosynthetic and metabolic processes of prostaglandins and prostanoids, cytokine and chemokine activities, and the IL-17, TNF, Toll-like receptor, and nuclear factor-kappa B (NF-κB) signaling pathways.
Conclusion
The study elucidated the “multiingredient, multitarget, and multipathway” mechanisms of GQD against CRC from a systemic perspective, indicating GQD to be a candidate therapy for CRC treatment.
1. Introduction
Colorectal cancer (CRC) is a global health burden and is the third most commonly diagnosed malignancy and the second leading cause of cancer deaths worldwide [1]. It constituted approximately 1.8 million new cases and 900,000 deaths annually, according to estimates from the International Agency for Research on Cancer in 2018 [2]. Despite the progress in the treatment of CRC, effects of current therapies including surgery, radiotherapy, chemotherapy, and targeted therapy are still unsatisfactory, especially for patients with metastatic lesions. Therefore, innovative therapeutic agents are needed, which are more effective and less toxic.
Traditional Chinese medicine (TCM) has been widely used in China for millenniums. It has been proved highly effective for a wide range of diseases. During the fight against infectious pneumonia caused by the 2019 novel coronavirus (2019-nCoV), TCM has made vast contributions to the prevention, treatment, and rehabilitation of coronavirus disease 2019 (COVID-19) among Chinese population [3]. This highlights the great value of TCM in the treatment of complicated diseases, especially those with poor response to Western medicine alone.
Gegen Qinlian decoction (GQD) is a well-known TCM formula originally described in the “Treatise on Exogenous Febrile Disease (“Shang Han Lun” in Chinese).” GQD had been used in China for approximately 2,000 years, most commonly for the treatment of gastrointestinal diseases, such as infectious diarrhea. GQD is composed of four herbal components, Radix Puerariae (“Gegen” in Chinese), Scutellariae Radix (“Huangqin” in Chinese), Coptidis Rhizoma (“Huanglian” in Chinese), and licorice (“Gancao” in Chinese). In recent years, studies have shown promising therapeutic effects of GQD in various diseases. A meta-analysis showed that GQD used alone or in combination with Western medicine might have potential benefits in curing ulcerative colitis (UC). UC can lead to the accumulation of high levels of proinflammatory cytokines within the colonic mucosa, resulting in dysplastic lesions and CRC [4, 5]. GQD has been shown to maintain colonic mucosal homeostasis in ulcerative colitis via bidirectionally regulating Notch signaling [6]. GQD also attenuated high-fat diet-induced steatohepatitis via modulation of the gut microbiome and reduced nonalcoholic steatohepatitis-associated liver injuries [7, 8]. In the field of cancer research, GQD has been proved to inhibit the expansion and neoangiogenesis of renal carcinoma by suppressing matrix metalloproteinase-2 [9]. Moreover, GQD was found to enhance the effect of PD-1 blockade in CRC by remodeling the gut microbiota and the tumor microenvironment [10]. These studies suggest potential for the use of GQD in the treatment of CRC. However, more preclinical evidence is needed. It is difficult to illustrate the complex anticancer mechanisms of GQD due to its “multi-”component and “multi-”target characteristics.
Network pharmacology, first proposed by Andrew L Hopkins, integrates a series of disciplines including pharmacology, bioinformatics, chemoinformatics, and systems biology. It offers a new framework for drug design and drug-target relationship prediction and enables unknown mechanisms of drug action to be inferred [11]. The history of “TCM network pharmacology” dates back to 1999, when Li proposed a possible relationship between TCM syndrome and molecular networks [12]. Since then, numerous studies have been conducted to support the concept and practice of TCM network pharmacology [13]. The TCM network pharmacology approach provides a new research paradigm for the discovery of bioactive compounds and elucidation of the mechanisms of herbal formulas [13]. TCM has also been successfully used to identify active compounds and elucidate mechanisms of GQD in the treatment of diseases such as type 2 diabetes and rotavirus enteritis [14, 15]. In the present study, we used a network pharmacology-based approach to investigate the potential mechanisms of how GQD exerts its anticancer effects on CRC. First, CRC-specific genes and bioactive chemical ingredients (BCIs) of GQD were obtained from the Gene Expression Omnibus (GEO) and the Traditional Chinese Medicine Systems Pharmacology Database (TCMSP), respectively. Then, CRC-specific genes related to the BCIs of GQD were screened by chemical-target interaction analysis. A pharmacological network and a protein-protein interaction (PPI) network were subsequently constructed to provide a comprehensive overview of the anti-CRC pharmacological action of GQD. Gene Ontology (GO) functional and Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway enrichment analyses were finally conducted for target genes in the pharmacological network to reveal their functional implications during the anticancer process. The flowchart of the analysis procedures of our study is shown in Figure 1.
Figure 1.

The flowchart of the analysis procedures of the study. Abbreviations: GQD, Gegen Qinlian decoction; BCI, bioactive chemical ingredient; GEO, Gene Expression Omnibus; CRC, colorectal cancer; PPI, protein-protein interaction; TCMSP, Traditional Chinese Medicine Systems Pharmacology.
2. Materials and Methods
2.1. CRC-Specific Genes
The microarray expression profile dataset GSE110224 was downloaded from the National Center for Biotechnology Information (NCBI) GEO (http://www.ncbi.nlm.nih.gov/geo/) database, which is based on the GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array platform. The dataset contains 34 samples, including 17 primary colorectal cancer tissue samples and 17 matched normal tissue samples. The raw data were first preprocessed using the Affy package in R [16]. Then, the data were converted into expression measures, and background correction, quartile data normalization, and probe summarization were performed using the robust multiarray average (RMA) algorithm in R [17]. The paired t-test based on the Linear Models for Microarray data (LIMMA) package in R was used to identify differentially expressed genes (DEGs) between CRC and normal samples [18]. The DEGs with an adjusted P value < 0.05 and a |log2 fold-change (log2 FC)|≥1 were considered significant and defined as CRC-specific genes.
2.2. BCIs of GQD
Chemical compounds were obtained from the TCMSP database (https://tcmspw.com/) [19]. BCIs were screened out according to predicted oral bioavailability (OB) and drug-likeness (DL) values and reserved if OB ≥ 30% and DL ≥ 0.18, which were the suggested drug screening criteria by the TCMSP database.
2.3. CRC-Specific Genes Related to BCIs of GQD
All related targets of BCIs from GQD were obtained using TCMSP. After intersection with CRC-specific genes, only genes that were both GQD-related targets and CRC-specific genes were preserved for further analysis. Only BCIs whose targets were CRC-specific genes were used for pharmacological network construction.
2.4. Construction of the Pharmacological Network
Based on the data generated by previous steps, a pharmacological network was constructed to illustrate the anti-CRC regulation mechanism between BCIs of GQD and their specific targets. The network was visualized using Cytoscape software (https://cytoscape.org/) [20].
2.5. Construction and Analysis of the Protein-Protein Interaction Network
To retrieve all the possible interactions among target genes in the pharmacological network, a PPI network was constructed using the BisoGenet plug-in in Cytoscape [21]. Subsequently, CytoNCA, a Cytoscape plug-in for network centrality analysis, was used to identify crucial genes in the network [22]. Genes with the top 30% highest degree centrality (DC) values were first selected for subnetwork construction using CytoNCA. Then, genes with the top 30% highest betweenness centrality (BC) values in the subnetwork were identified as crucial genes and formed the core network.
2.6. Enrichment Analysis for Target Genes and Target-Pathway Network Construction
GO functional enrichment analysis was performed for target genes in three categories: biological process (BP), cellular component (CC), and molecular function (MF) [23]. Both GO functional and KEGG pathway enrichment analysis for target genes were performed using the R package of clusterProfiler [24,25]. Benjamini–Hochberg correction was performed for multiple testing, and adjusted P value <0.05 was set as the threshold. A target-pathway network was then constructed in Cytoscape to visualize the relationships between target genes and KEGG pathways.
3. Results
3.1. CRC-Specific Genes
Based on the cutoff criteria, a total of 533 DEGs (including 235 upregulated and 298 downregulated genes) were identified between CRC tissues and normal tissues. The top 10 significantly upregulated and downregulated DEGs are listed in Table 1.
Table 1.
The identified top 10 upregulated and downregulated DEGs between CRC tissue samples and normal tissue samples.
| Upregulated DEGs | Downregulated DEGs | ||||
|---|---|---|---|---|---|
| Gene name | Log2FC | Adjusted P value | Gene name | Log2FC | Adjusted P value |
| MMP3 | 4.1101 | 1.41E − 03 | CLCA4 | −4.4741 | 4.45E − 03 |
| REG1A | 3.8436 | 5.89E − 03 | MS4A12 | −4.4636 | 4.03E − 03 |
| FOXQ1 | 3.5684 | 1.79E − 03 | AQP8 | −4.3366 | 1.91E − 03 |
| CXCL11 | 3.4122 | 2.12E − 03 | CA4 | −3.6607 | 2.16E − 03 |
| MMP7 | 3.3618 | 1.56E − 03 | GCG | −3.6245 | 9.42E − 03 |
| REG1B | 3.3276 | 2.67E − 02 | GUCA2A | −3.4325 | 3.83E − 03 |
| MMP1 | 3.2300 | 2.86E −03 | CLDN8 | −3.3370 | 1.14E − 02 |
| TCN1 | 3.2266 | 3.83E − 03 | CHP2 | −3.2031 | 1.58E − 03 |
| SLC6A14 | 2.7141 | 6.57E − 03 | NXPE4 | −3.1622 | 3.12E − 03 |
| CXCL3 | 2.6473 | 5.58E − 03 | MAMDC2 | −3.0890 | 3.21E − 03 |
DEGs, differentially expressed genes; FC, fold change.
3.2. BCIs of GQD
Using the TCMSP database, 489 compounds were retrieved: 18 in Gegen, 143 in Huangqin, 48 in Huanglian, and 280 in Gancao. After filtering with the criteria of OB ≥ 30% and DL ≥ 0.18, 146 bioactive components from GQD were collected. These included 4, 36, 14, and 92 from Gegen, Huangqin, Huanglian, and Gancao, respectively. After excluding duplicates, 140 BCIs were selected for further analysis (as shown in Supplementary Table S1).
3.3. CRC-Specific Genes Related to BCIs of GQD
A total of 240 BCI-related targets were screened out. After intersecting the 240 BCI-related targets with 533 CRC-specific genes, 20 genes were collected as CRC-specific GQD-target genes. After excluding BCIs whose targets were not CRC-specific, 118 effective BCIs were finally used for pharmacological network construction.
3.4. The Pharmacological Network
One hundred and eighteen BCIs of GQD together with 20 CRC-specific GQD-target genes were introduced into Cytoscape to create the pharmacological network (Figure 2). BCIs are displayed as ellipse nodes in the network, and BCIs from different herbs are painted in different colors. BCIs from Gancao, Gegen, Huangqin, and Huanglian are painted in green, brown, purple, and yellow, respectively. Shared BCIs of multiple medicines are painted in red. One hundred and three BCIs with only one target are distributed in two larger circles in the upper half of the figure. Fifteen BCIs with multiple targets are distributed in the smaller circle at the bottom of the figure, such as MOL000098 (quercetin), MOL000173 (wogonin), MOL000354 (isorhamnetin), MOL000392 (formononetin), and MOL000422 (kaempferol). Information from 15 BCIs with multiple targets is listed in Table 2. Twenty CRC-specific GQD-target genes are displayed as V-shaped polygons in blue, including PTGS2, OLR1, NR3C2, HSD3B2, TNFSF15, MMP1, MMP3, MMP9, AKR1C3, CA2, PLAU, IL1B, DUOX2, CCNB1, ABCG2, CXCL11, CXCL10, SPP1, ADH1C, and MAOA. Information from 20 CRC-specific GQD-target genes, including full name, log2FC, adjusted P value, and aliases, is shown in Table 3. The regulation relationships between BCIs and their targets are displayed as lines in figure. As the most important gene, PTGS2 is targeted by 116 BCIs and is emphasized centrally in the upper half of the figure.
Figure 2.
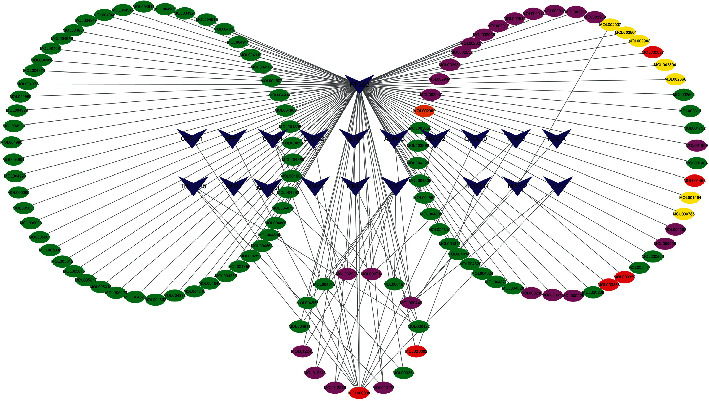
GQD-CRC ingredient-target pharmacology network. Ellipses represent the BCIs of GQD. BCIs of Gancao, Gegen, Huangqin, and Huanglian are painted in green, brown, purple, and yellow, respectively. Shared BCIs of multiple medicines are painted in red. V-shaped polygons in blue represent the target genes of BCIs. The regulation relationships between BCIs and their targets are displayed as lines in figure. Abbreviations: BCI, bioactive chemical ingredient; GQD, Gegen Qinlian decoction; CRC, colorectal cancer.
Table 2.
Information of 15 BCIs with multiple targets in the pharmacological network.
| Molecule ID | Molecule name | Molecule structure | OB (%) | DL | Source |
|---|---|---|---|---|---|
| MOL000098 | Quercetin |
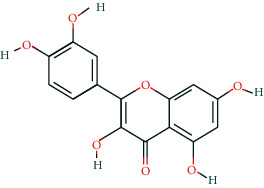
|
46.43 | 0.28 | Huanglian and Gancao |
|
| |||||
| MOL000173 | Wogonin |
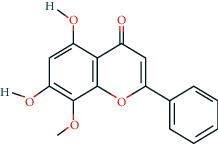
|
30.68 | 0.23 | Huangqin |
|
| |||||
| MOL000354 | Isorhamnetin |
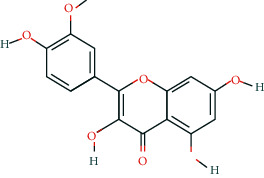
|
49.6 | 0.31 | Gancao |
|
| |||||
| MOL000392 | Formononetin |
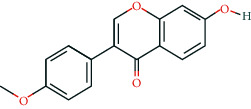
|
69.67 | 0.21 | Gegen and Gancao |
|
| |||||
| MOL000422 | Kaempferol |
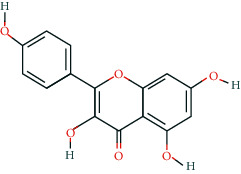
|
41.88 | 0.24 | Gancao |
|
| |||||
| MOL000449 | Stigmasterol |
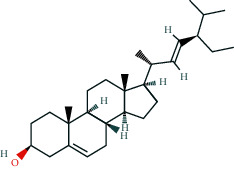
|
43.83 | 0.76 | Huangqin |
|
| |||||
| MOL000497 | Licochalcone a |
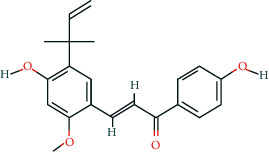
|
40.79 | 0.29 | Gancao |
|
| |||||
| MOL002714 | Baicalein |
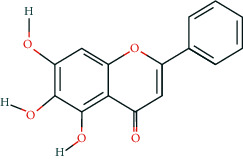
|
33.52 | 0.21 | Huangqin |
|
| |||||
| MOL002928 | Oroxylin a |
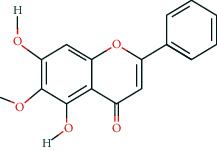
|
41.37 | 0.23 | Huangqin |
|
| |||||
| MOL004815 | (E)-1-(2,4-Dihydroxyphenyl)-3-(2,2-dimethylchromen-6-yl)prop-2-en-1-one |
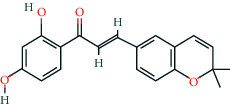
|
39.62 | 0.35 | Gancao |
|
| |||||
| MOL004835 | Glypallichalcone |
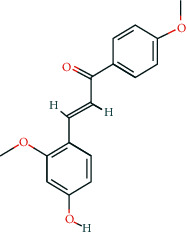
|
61.6 | 0.19 | Gancao |
|
| |||||
| MOL004841 | Licochalcone B |
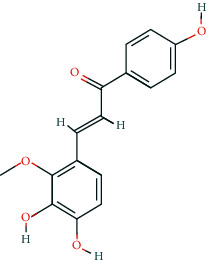
|
76.76 | 0.19 | Gancao |
|
| |||||
| MOL012245 | 5,7,4′-Trihydroxy-6-methoxyflavanone |
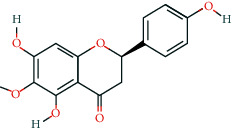
|
36.63 | 0.27 | Huangqin |
|
| |||||
| MOL012246 | 5,7,4′-Trihydroxy-8-methoxyflavanone |
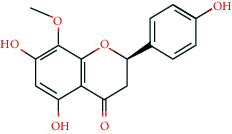
|
74.24 | 0.26 | Huangqin |
|
| |||||
| MOL012266 | Rivularin |
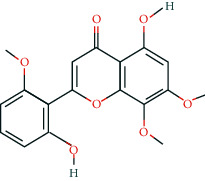
|
37.94 | 0.37 | Huangqin |
BCI, bioactive chemical ingredient; OB, oral bioavailability; DL, drug-likeness.
Table 3.
Information of 20 target genes in the pharmacological network.
| Gene | Full name | Log2FC | Adjusted P value | Aliases |
|---|---|---|---|---|
| PTGS2 | Prostaglandin G/H synthase 2 | 1.7769 | 7.39E − 03 | PGHS-2; COX-2; PHS-2; PGG/HS; hCox-2; GRIPGHS |
| OLR1 | Oxidized low-density lipoprotein receptor 1 | 1.9948 | 4.17E − 03 | LOX1; LOXIN; SLOX1; CLEC8A; SCARE1 |
| NR3C2 | Nuclear receptor subfamily 3 group C member 2 | −1.5129 | 3.92E − 03 | MR; MCR; MLR; NR3C2VIT |
| HSD3B2 | Hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 | −1.1592 | 8.47E − 03 | HSDB; HSD3B; SDR11E2 |
| TNFSF15 | TNF superfamily member 15 | 1.0461 | 1.24E − 02 | TL1; TL1A; VEGI; TNLG1B; VEGI192A |
| MMP1 | Matrix metallopeptidase 1 | 3.2300 | 2.86E − 03 | CLG; CLGN |
| AKR1C3 | Aldo-keto reductase family 1 member C3 | −1.2267 | 6.79E − 03 | DD3; DDX; PGFS; HAKRB; HAKRe; HA1753; HSD17B5; hluPGFS |
| CA2 | Carbonic anhydrase 2 | −1.7184 | 9.85E − 03 | CAC; CAII; Car2; CAII; HEL-76; HEL-S-282 |
| MMP3 | Matrix metallopeptidase 3 | 4.1101 | 1.41E − 03 | SL-1; STMY; STR1; CHDS6; MMP3; STMY1 |
| PLAU | Plasminogen activator, urokinase | 1.2642 | 4.83E − 03 | ATF; QPD; UPA; URK; u-PA; BDPLT5 |
| MMP9 | Matrix metallopeptidase 9 | 1.1245 | 8.08E − 03 | GELB; CLG4B; MMP9; MANDP2 |
| IL1B | Interleukin 1 beta | 2.1239 | 2.27E − 03 | IL-1; IL1F2; IL1beta; IL1-BETA |
| DUOX2 | Dual oxidase 2 | 1.9328 | 2.98E − 02 | TDH6; LNOX2; THOX2; NOXEF2; P138-TOX |
| CCNB1 | Cyclin B1 | 1.0204 | 1.50E − 02 | CCNB |
| ABCG2 | ATP binding cassette subfamily G member 2 | −2.9521 | 2.92E − 03 | MRX; MXR; ABCP; BCRP; BMDP; MXR1; ABC15; BCRP1; CD338; GOUT1; MXR-1; CDw338; UAQTL1; EST157481 |
| CXCL11 | C-X-C motif chemokine ligand 11 | 3.4122 | 2.12E − 03 | IP9; H174; IP-9; b-R1; I-TAC; SCYB11; SCYB9B |
| CXCL10 | C-X-C motif chemokine ligand 10 | 2.1322 | 6.06E − 03 | C7; IFI10; INP10; IP-10; crg-2; mob-1; SCYB10; gIP-10 |
| SPP1 | Secreted phosphoprotein 1 | 1.5076 | 7.22E − 03 | OPN; BNSP; BSPI; ETA-1 |
| ADH1C | Alcohol dehydrogenase 1C (class I), gamma polypeptide | −2.9047 | 7.54E − 03 | ADH3 |
| MAOA | Monoamine oxidase A | −1.1045 | 7.88E − 03 | BRNRS; MAO-a |
FC, fold change.
3.5. Construction and Topological Analysis of the PPI Network
A PPI network comprising 446 nodes and 3,518 edges was generated using BisoGenet (Figure 3(a)). After DC calculation, 130 nodes together with 1,619 edges were selected to form the subnetwork (Figure 3(b)). Ten target genes including CCNB1, SPP1, MMP9, NR3C2, PLAU, MMP3, PTGS2, CA2, MMP1, and IL1B ranked as the top 30% after DC evaluation and were integrated into the subnetwork, with the degree of 140, 90, 38, 32, 26, 26, 25, 19, 18, and 17, respectively. After BC calculation, 41 nodes and 379 edges were further selected for core network construction (Figure 3(c)). Two target genes, CCNB1 and SPP1, gained the top 30% highest BC values and were finally identified as crucial genes.
Figure 3.
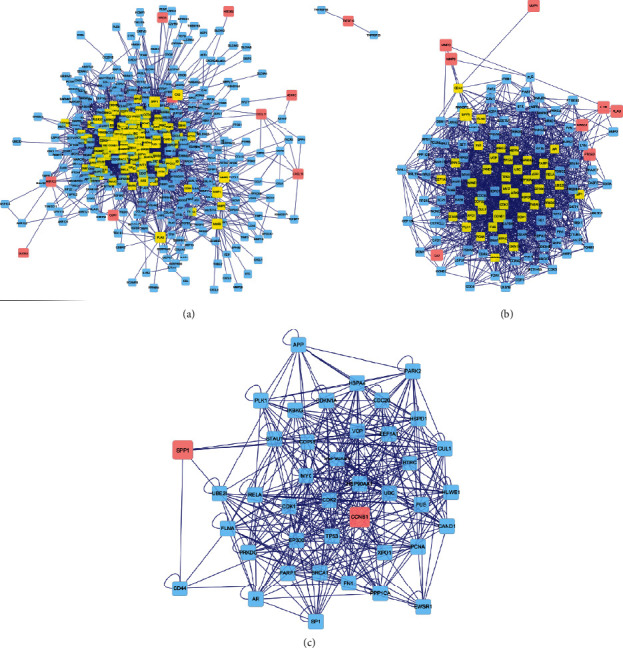
Construction and topological analysis of the PPI network. (a) The PPI network of GQD-CRC target genes generated using BisoGenet, which is comprised of 446 nodes and 3518 edges. Pink nodes represent the CRC-specific GQD-target genes from the pharmacological network. Blue nodes stand for the interacting proteins generated by BisoGenet. The subnetwork with the top 30% highest DC value genes is emphasized in yellow and shown in b. (b) The subnetwork after DC filtration, which is comprised of 130 nodes and 1619 edges. The core network with the top 30% highest BC value genes is emphasized in yellow and shown in c. (c) The core network after BC filtration, which is comprised of 41 nodes and 379 edges. Abbreviations: PPI, protein-protein interaction; GQD, Gegen Qinlian decoction; CRC, colorectal cancer; DC, degree centrality; BC, betweenness centrality.
3.6. Enrichment Analysis for Target Genes and Target-Pathway Network Construction
A total of 171 BP terms and 40 MF terms were enriched for the 20 target genes. The complete BP and MF term lists are shown in Supplementary Tables S2 and S3. Only one CC term (GO: 0070820∼tertiary granule, enriched by OLR1, PLAU, and MMP9 with an adjusted P value of 0.038) was enriched under the threshold. The top 20 BP and MF terms are shown in Figures 4(a) and 4(b). Each bar in Figure 4 represents a GO term, plotted by the number of genes enriched in the term on the horizontal axis. The color of each term represents its adjusted P value. The more red the color of the term, the smaller its adjusted P value.
Figure 4.
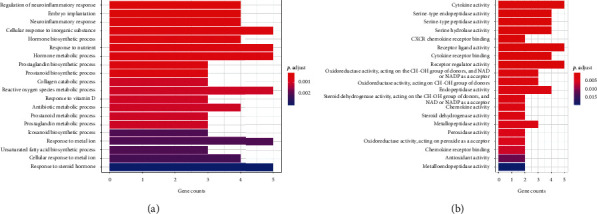
Barplot of GO functional enrichment analysis. (a) The top 20 notable GO-BP terms enriched by target genes in the pharmacological network. (b) The top 20 notable GO-MF terms enriched by target genes in the pharmacological network. Each bar represents a GO term on the vertical axis. The number of genes enriched in each term is recorded on the horizontal axis. Color of each bar represents the adjusted p value of each GO term. More red the color of the term is, smaller its adjusted p value is. Abbreviations: GO, Gene Ontology; BP, biological process; MF, molecular function.
Six KEGG pathways were enriched for target genes, and these are listed in Table 4. The bubble graph of KEGG pathways is shown in Figure 5, with gene ratio on the horizontal axis. The size of each bubble indicates the number of genes enriched in each KEGG pathway. The larger the bubble, the greater the number of genes involved in the pathway. As in the GO barplot, the color of each bubble in Figure 5 represents the adjusted P value of each KEGG pathway. The more red the color of the bubble, the smaller its adjusted P value. The target-pathway network is displayed in Figure 6. Target genes and KEGG pathways are visualized as V-shaped polygons and parallelograms, respectively. The larger the size of the V polygon, the more KEGG pathways the target gene is involved in. Similarly, the larger the size of the parallelogram, the greater the number of target genes the KEGG pathway contains.
Table 4.
KEGG pathways enriched by target genes.
| ID | Description | Adjusted P value |
|---|---|---|
| Hsa04657 | IL-17 signaling pathway | 9.27E − 06 |
| Hsa04668 | TNF signaling pathway | 3.79E − 04 |
| Hsa04620 | Toll-like receptor signaling pathway | 4.21E − 03 |
| Hsa04913 | Ovarian steroidogenesis | 7.08E − 03 |
| Hsa04064 | NF-kappa B signaling pathway | 4.53E − 02 |
| hsa01523 | Antifolate resistance | 4.67E − 02 |
KEGG, Kyoto Encyclopaedia of Genes and Genomes.
Figure 5.
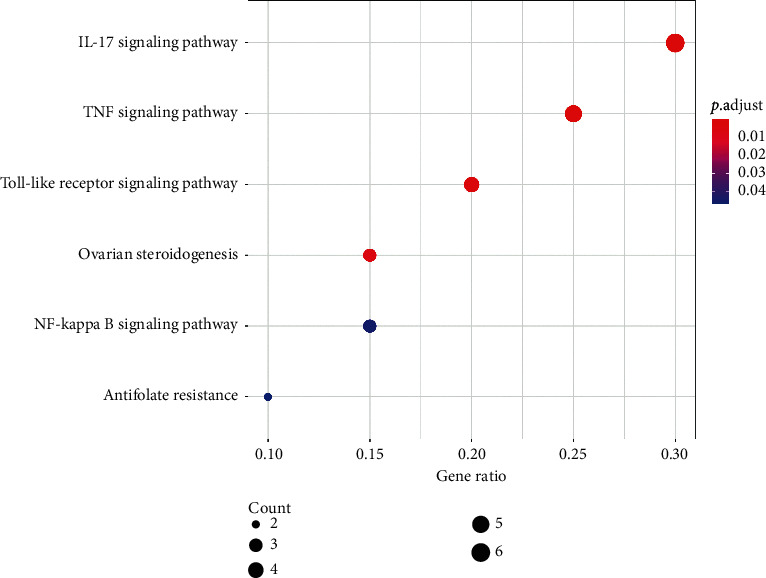
Bubble graph of KEGG pathway enrichment analysis. Each bubble represents a KEGG pathway on the vertical axis. The gene ratio is recorded on the horizontal axis. The size of each bubble indicates the number of genes enriched in each KEGG pathway. Larger the bubble is, more number of genes is involved in the pathway. Color of each bubble represents the adjusted P value of each KEGG pathway. More red the color of the bubble is, smaller its adjusted P value is. Abbreviation: KEGG, Kyoto Encyclopaedia of Genes and Genomes.
Figure 6.
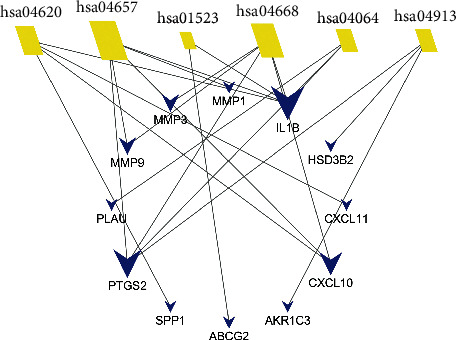
Target-pathway network. Blue V-shaped polygons represent target genes. Yellow parallelograms represent KEGG pathways logoed by its identifier number in KEGG database. Larger the size of the V polygon is, more KEGG pathways the target gene is involved in. Larger the size of the parallelogram is, more number of target genes the KEGG pathway contains. Abbreviation: KEGG, Kyoto Encyclopaedia of Genes and Genomes.
4. Discussion
Colorectal carcinogenesis is a complex and consecutive progression. It involves a multiscale and systemic framework integrating genetic, proteomic, and metabolic networks from responses to DNA damage, gene mutations, population dynamics, inflammation, and metabolism-immune balance [26–28]. The evolution of the genomic landscape through novel sequencing techniques has uncovered major clues into the key mechanisms of CRC. Medicines have been designed to target specific genetic keypoints to block the progression of the disease [29]. However, survival of CRC patients remains unsatisfactory due to the complex crosstalk among these alterations. TCM has unique benefits such as being naturally sourced, multitargeted, and a holistic concept. It has advantages in treating complicated diseases, especially those with a poor response to Western medicine alone. In the present study, we used a network pharmacology-based approach to reveal the pharmacological effects of GQD on CRC, which might provide novel therapeutic strategies for better treatment of CRC.
Network pharmacology analysis showed that 20 target genes were regulated by GQD in CRC, including PTGS2, CCNB1, SPP1, PLAU, MAOA, OLR1, NR3C2, HSD3B2, TNFSF15, AKR1C3, CA2, MMP1, MMP3, MMP9, IL1B, DUOX2, ABCG2, CXCL11, CXCL10, and ADH1C. Most of these have been reportedly associated with CRC.
PTGS2 (prostaglandin G/H synthase 2, also known as PGHS-2; COX-2) is one of the most important genes in the pharmacological network. Many studies have demonstrated that CRC is closely related to PTGS2. Studies have found PTGS2 to be overexpressed in CRC tissues [30], which is consistent with results in the present study (Table 3). The elevation of PTGS2 predicts poor prognosis in colon cancer [31]. PTGS2 produces the inflammatory mediator prostaglandin E2 (PGE2), which is suggested to promote the development and progression of CRC [32–34]. Furthermore, epidemiological evidence indicates that the regular use of aspirin (a PTGS2 inhibitor) reduces the risk of CRC [35]. In the present study, PTGS2 was targeted by as many as 116 BCIs in GQD, indicating it may possess a potentially important PTGS2-related anti-CRC mechanism.
Reduction of NR3C2 (also known as MR) expression was found to be a potential early event involved in CRC progression [36]. Expression of AKR1C3 may be used for the prediction of lymph node metastasis in CRC [37]. High mRNA expression of DUOX2 was significantly associated with better overall survival of CRC patients [38]. ABCG2 was shown to play a potential protective role in CRC by inhibiting the NF-κB signaling pathway to relieve oxidative stress and decrease the inflammatory response [39]. Downregulation of CXCL11 inhibited cell growth and epithelial-mesenchymal transition in CRC [40]. The crucial MMP1, MMP3, and MMP9 genes in the pharmacology network are all matrix metalloproteinase family members. Increasing evidence has demonstrated their oncogenic significance in CRC carcinogenesis [41–43].
After PPI network analysis, CCNB1 and SPP1 were identified as crucial genes of the highest degree. Cyclin B1 (CCNB1) is a well-known gene involved in mitosis. It produces a complex with cyclin-dependent kinase 1 (CDK1), which is necessary for proper control of the G2/M transition phase of the cell cycle [44, 45]. Previous study indicated that CCNB1 is overexpressed in CRC tissues, and inhibition of CCNB1 suppressed the proliferation of CRC cells in vitro and tumorigenicity in vivo [46]. SPP1, also known as OPN, has been shown to regulate multiple functions contributing to CRC progression [47]. Upregulation of SPP1 promoted CRC cell proliferation in vitro and tumor growth in vivo [47]. It also promoted metastasis in CRC by activating the EMT pathway and was associated with poor survival outcomes in CRC [48, 49].
Fifteen BCIs were correlated with multiple target genes in the pharmacological network. Some of these have already been shown to exert anti-CRC properties. Quercetin, a dietary flavonoid, was reported to induce human colon cancer cell apoptosis by inhibiting the NF-κB pathway and inducing apoptosis in KRAS-mutant CRC cells via JNK signaling pathways [50, 51]. Wogonin, a naturally occurring monoflavonoid, induced antiproliferation and G1 arrest via the Wnt/β-catenin signaling pathway in CRC cells [52]. Baicalein was shown to inhibit the proliferation and migration of CRC cells [53–55]. Oroxylin A was reported to suppress the growth and development of CRC via reprogramming of HIF1α-modulated fatty acid metabolism [56]. These findings all suggest the promising potential of GQD in the treatment of CRC.
Based on GO enrichment analysis, BP terms enriched by target genes were mainly concentrated in response to various materials and biochemical processes of different substances. Response to inorganic substance (GO: 0071241), nutrient (GO: 0007584), vitamin D (GO: 0033280), and metal ion (GO: 0010038) are all pivotal in cellular metabolism. Biosynthetic and metabolic processes of prostaglandins and prostanoids (including GO:0006693, GO:0046457, GO:0006692, and GO:0001516) are also important in colorectal tumorigenesis, since prostaglandins and prostanoids have been implicated in various pathological processes in CRC [57–60]. Some enriched MF terms are associated with inflammation, such as GO:0005125: cytokine activity, GO:0005126: cytokine receptor binding, GO:0042379: chemokine receptor binding, GO:0008009: chemokine activity, and GO:0045236: CXCR chemokine receptor binding. Inflammation, a hallmark of CRC, has been discussed in many publications. Cytokines such as interleukin-6 (IL-6), interleukin-7 (IL-7), interleukin-10 (IL-10), and interleukin-17 (IL-17) have been shown to be involved in the development of CRC [61–64]. Chemokines direct leukocyte recruitment and migration under inflammatory conditions [65, 66]. The expression of many chemokines, such as CCL2–4, CXCL1, CXCL5, and CXCL8–10, was reportedly elevated in the CRC microenvironment compared to normal tissues [67, 68].
KEGG enrichment analysis showed that the pharmacological effects of GQD on CRC are closely related to well-known tumor-associated pathways, including the IL-17, tumor necrosis factor (TNF), Toll-like receptor, and NF-κB signaling pathways. Numerous studies have highlighted the important role of the IL-17 signaling pathway in the tumorigenesis, angiogenesis, and metastasis of CRC. Interleukin-17 (IL-17), a proinflammatory cytokine, was significantly upregulated in CRC tissues [69]. IL-17 can promote CRC tumorigenesis by stimulating the production and recruitment of myeloid-derived suppressor cells (MDSCs) [70]. It promotes angiogenesis via stimulating VEGF production in CRC cells [71]. Additionally, IL-17 stimulates the production of PGE2, MMP9, and MMP13, which are involved in the migration of CRC cells [72–74].
TNF-α is one of the most important cell signaling proteins involved in cell growth, differentiation, and apoptosis [75, 76]. It plays a pivotal role in proliferation, angiogenesis, and metastasis in CRC [77]. Serum TNF-α was demonstrated to contribute to CRC susceptibility, and anti-TNF therapy has been considered for CRC treatment [78].
A large body of data demonstrates an important relationship between the Toll-like receptor (TLR) signaling pathway and CRC. The TLR signaling pathway exerts a fundamental role in colorectal epithelium hemostasis and in activating the innate and adaptive immune responses [79]. Activation of the TLR signaling pathway leads to activation of downstream signaling pathways and recruitment of transcription factors such as NF-κB, interferon regulatory factor- (IRF-) 3, AP-1, PI3K/Akt kinases, the mitogen-activated protein kinase (MAPK), and the subsequent generation of cytokines and chemokines [80, 81]. The TLR2 and TLR4 agonists HMGB1 and S100A9 have been proposed as potential biomarkers for CRC [82, 83]. Several TLR-based therapeutic agents have been developed for targeting this pathway and are currently used in clinical trials in patients with CRC [84–86].
The NF-κB signaling pathway is a key regulator of CRC cell proliferation, apoptosis, inflammation, angiogenesis metastasis, and drug resistance [87]. Constitutive NF-κB activation was observed in CRC cell lines and human CRCs [88, 89]. It promotes the proliferation of cancer cells and rescues cancer cells from cell death [90]. Studies suggest that inhibition of the NF-κB pathway can sensitize CRC cells to chemotherapy and radiotherapy, providing more effective strategies for cancer treatment [91, 92].
These findings suggest that the molecular mechanisms of GQD against CRC are closely related to these key target genes, biochemical processes, and important signaling pathways. However, detailed experiments are still needed to confirm these findings.
5. Conclusions
In conclusion, this study is the first to reveal the pharmacological effects of GQD against CRC via network pharmacology analysis. A total of 118 BCIs from GQD were identified, and 20 corresponding genes including PTGS2, NR3C2, CXCL11, CCNB1, and SPP1 were demonstrated to be key targets for GQD in CRC. GO functional and KEGG pathway enrichment analysis indicated that the molecular mechanisms of GQD in CRC were closely related to important biochemical processes and signaling pathways, such as biosynthetic and metabolic processes of prostaglandins and prostanoids, cytokine and chemokine activities, the IL-17 signaling pathway, the TNF signaling pathway, the Toll-like receptor signaling pathway, and the NF-κB signaling pathway. The study provides a research basis for further studies of GQD in the treatment of CRC.
Acknowledgments
The authors greatly appreciate the financial support from Harbin Medical University, which ensured the accomplishment of the work.
Data Availability
The data used to support the findings of this study are included within the article and supplementary information files.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Supplementary Materials
Supplementary Table S1: information for 140 BCIs of GQD. Supplementary Table S2: the significant GO-BP terms enriched by target genes. Supplementary Table S3: the significant GO-MF terms enriched by target genes.
References
- 1.Keum N., Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nature Reviews Gastroenterology & Hepatology. 2019;16(12):713–732. doi: 10.1038/s41575-019-0189-8. [DOI] [PubMed] [Google Scholar]
- 2.International Agency for Research on Cancer. Globocan 2018: Cancer Fact Sheets—Colorectal Cancer. IARC; 2018. https://www.google.com/search?rlz=1C1GCEU_enIN894IN894&q=Lyon&stick=H4sIAAAAAAAAAOPgE-LUz9U3MDNMMjRQ4gAxUwqK0rWMMsqt9JPzc3JSk0sy8_P084vSE_MyqxJBnGKrjNTElMLSxKKS1KJihZz8ZLDwIlYWn8r8vB2sjAAgmgL0VQAAAA&sa=X&ved=2ahUKEwirsoDG1tfsAhU4H7cAHQqDB1cQmxMoATAiegQIHhAD https://gco.iarc.fr/today/data/factsheets/cancers/10_8_9-Colorectum-fact-sheet.pdf. [Google Scholar]
- 3.Zhang D., Zhang B., Lv J.-T., Zhang X.-M., Lin Z.-J. The clinical benefits of Chinese patent medicines against COVID-19 based on current evidence. Pharmacological Research. 2020;157:p. 104882. doi: 10.1016/j.phrs.2020.104882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sa Y., Yi W., Huang H., Mei Z., Feng Z. Efficacy of herbal medicine (gegen qinlian decoction) on ulcerative colitis: a systematic review of randomized controlled trials. Medicine (Baltimore) 2019;98(52) doi: 10.1097/md.0000000000018512.e18512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lopez A., Pouillon L., Beaugerie L., Danese S., Peyrin-Biroulet L. Colorectal cancer prevention in patients with ulcerative colitis. Best Practice & Research Clinical Gastroenterology. 2018;32-33:103–109. doi: 10.1016/j.bpg.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 6.Zhao Y., Luan H., Gao H., Zhang Y., Li R. Gegen qinlian decoction maintains colonic mucosal homeostasis in acute/chronic ulcerative colitis via bidirectionally modulating dysregulated notch signaling. Phytomedicine. 2020;68 doi: 10.1016/j.phymed.2020.153182.153182 [DOI] [PubMed] [Google Scholar]
- 7.Wu Y., Ding P. H., Liu L. J., et al. Gegen qinlian decoction attenuates high-fat diet-induced steatohepatitis in rats via gut microbiota. Evidence Based Complementary Alternative Medicine. 2018;2018:p. 7370891. doi: 10.1155/2018/7370891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang C.-H., Xiao Q., Sheng J.-Q., et al. Gegen qinlian decoction abates nonalcoholic steatohepatitis associated liver injuries via anti-oxidative stress and anti-inflammatory response involved inhibition of toll-like receptor 4 signaling pathways. Biomedicine & Pharmacotherapy. 2020;126 doi: 10.1016/j.biopha.2020.110076.110076 [DOI] [PubMed] [Google Scholar]
- 9.Liu N., Feng Y., Cheung F., Zhang Z., Feng Y. A Chinese medicine formula gegen qinlian decoction suppresses expansion of human renal carcinoma with inhibition of matrix metalloproteinase-2. Integrative Cancer Therapies. 2015;14(1):75–85. doi: 10.1177/1534735414550036. [DOI] [PubMed] [Google Scholar]
- 10.Wang J., Jia Y., Li J., et al. Gegen qinlian decoction enhances the effect of PD-1 blockade in colorectal cancer with microsatellite stability by remodelling the gut microbiota and the tumour microenvironment. Cell Death & Disease. 2019;10(6):p. 415. doi: 10.1038/s41419-019-1638-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hopkins A. L. Network pharmacology. Nature Biotechnology. 2007;25(10):1110–1111. doi: 10.1038/nbt1007-1110. [DOI] [PubMed] [Google Scholar]
- 12.Li S. Possible relationship between traditional Chinese medicine Zheng and molecular networks. Proceedings of the the First Academic Annual Meeting of the China Association for Science and Technology; January 1999; Beijing, China. [Google Scholar]
- 13.Li S., Zhang B. Traditional Chinese medicine network pharmacology: theory, methodology and application. Chinese Journal of Natural Medicines. 2013;11(2):110–120. doi: 10.1016/s1875-5364(13)60037-0. [DOI] [PubMed] [Google Scholar]
- 14.Li H., Zhao L., Zhang B., et al. A network pharmacology approach to determine active compounds and action mechanisms of Ge-Gen-Qin-Lian decoction for treatment of type 2 diabetes. Evidence-Based Complementary and Alternative Medicine. 2014;2014:8. doi: 10.1155/2014/495840.495840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong P., Song L., Gao M., et al. Network pharmacology-based strategy for predicting active ingredients and potential targets of gegen qinlian decoction for rotavirus enteritis. Evidence-Based Complementary and Alternative Medicine. 2020;2020:5. doi: 10.1155/2020/2957567.2957567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gautier L., Cope L., Bolstad B. M., Irizarry R. A. Affy--analysis of affymetrix genechip data at the probe level. Bioinformatics. 2004;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 17.Irizarry R. A., Hobbs B., Collin F., et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 18.Smyth G. K. Limma: linear models for microarray data. In: Gentleman R., Carey V. J., Huber W., Irizarry R. A., Dudoit S., editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Berlin, Germany: Springer; 2005. pp. 397–420. [Google Scholar]
- 19.Ru J., Li P., Wang J., et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. Journal of Cheminformatics. 2014;6:p. 13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin A., Ochagavia M. E., Rabasa L. C., Miranda J., Fernandez-de-Cossio J., Bringas R. BisoGenet: a new tool for gene network building, visualization and analysis. BMC Bioinformatics. 2010;11(1):p. 91. doi: 10.1186/1471-2105-11-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang Y., Li M., Wang J., Pan Y., Wu F.-X. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems. 2015;127:67–72. doi: 10.1016/j.biosystems.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 23.Ashburner M., Ball C. A., Blake J. A., et al. Gene ontology: tool for the unification of biology. Nature Genetics. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu G., Wang L.-G., Han Y., He Q.-Y. ClusterProfiler: an R Package for comparing biological themes among gene clusters. OMICS: A Journal of Integrative Biology. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo Y., Nie Q., MacLean A. L., Li Y., Lei J., Li S. Multiscale modeling of inflammation-induced tumorigenesis reveals competing oncogenic and oncoprotective roles for inflammation. Cancer Research. 2017;77(22):6429–6441. doi: 10.1158/0008-5472.can-17-1662. [DOI] [PubMed] [Google Scholar]
- 27.Tariq K., Ghias K., Ghias K. Colorectal cancer carcinogenesis: a review of mechanisms. Cancer Biology & Medicine. 2016;13(1):120–135. doi: 10.20892/j.issn.2095-3941.2015.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turanli B., Karagoz K., Gulfidan G., Sinha R., Mardinoglu A., Arga K. Y. A network-based cancer drug discovery: from integrated multi-omics approaches to precision medicine. Current Pharmaceutical Design. 2018;24(32):3778–3790. doi: 10.2174/1381612824666181106095959. [DOI] [PubMed] [Google Scholar]
- 29.Florescu-Ţenea R. M., Kamal A. M., Mitruţ P., et al. Colorectal cancer: an update on treatment options and future perspectives. Current Health Sciences Journal. 2019;45(2):134–141. doi: 10.12865/CHSJ.45.02.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roelofs H. M., Te Morsche R. H., van Heumen B. W., Nagengast F. M., Peters W. H. Over-expression of COX-2 mRNA in colorectal cancer. BMC Gastroenterol. 2014;14:p. 1. doi: 10.1186/1471-230x-14-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogino S., Kirkner G. J., Nosho K. N., et al. Cyclooxygenase-2 expression is an independent predictor of poor prognosis in colon cancer. Clinical Cancer Research. 2008;14(24):8221–8227. doi: 10.1158/1078-0432.ccr-08-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Irahara H. The roles of EP4 prostanoid receptors in cancer malignancy signaling. Biological & Pharmaceutical Bulletin. 2016;39(2):149–155. doi: 10.1248/bpb.b15-00840. [DOI] [PubMed] [Google Scholar]
- 33.Fujino H., Seira N., Kurata N., Nakamura H., Regan J. W., Murayama T. Prostaglandin E2-stimulated prostanoid EP4 receptors induce prolonged de novo prostaglandin E2 synthesis through biphasic phosphorylation of extracellular signal-regulated kinases mediated by activation of protein kinase A in HCA-7 human colon cancer cells. European Journal of Pharmacology. 2015;768:149–159. doi: 10.1016/j.ejphar.2015.10.044. [DOI] [PubMed] [Google Scholar]
- 34.Araki S. S., Makar K. W., Li L. Y., et al. Tissue-specific patterns of gene expression in the epithelium and stroma of normal colon in healthy individuals in an aspirin intervention trial. Genomics Data. 2015;6:154–158. doi: 10.1016/j.gdata.2015.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng D., Sha D., Manthravadi S., Sinicrope F. A. Aspirin and colorectal cancer: back to the future. Clinical Cancer Research. 2014;20(5):1087–1094. doi: 10.1158/1078-0432.ccr-13-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Di Fabio F., Alvarado C., Majdan A. A., et al. Underexpression of mineralocorticoid receptor in colorectal carcinomas and association with VEGFR-2 overexpression. Journal of Gastrointestinal Surgery. 2007;11(11):1521–1528. doi: 10.1007/s11605-007-0234-8. [DOI] [PubMed] [Google Scholar]
- 37.Gologan C., Osawa K., Akiyama M. N., et al. Expression of AKR1C3 and CNN3 as markers for detection of lymph node metastases in colorectal cancer. Clinical and Experimental Medicine. 2015;15(3):333–341. doi: 10.1007/s10238-014-0298-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsubara S. Y., Kim J. S., Eun H. S., et al. Expression of NOX family genes and their clinical significance in colorectal cancer. Digestive Diseases and Sciences. 2018;63(9):2332–2340. doi: 10.1007/s10620-018-5121-5. [DOI] [PubMed] [Google Scholar]
- 39.Kang S., Huang Y., Shi M., et al. Protective role of ABCG2 against oxidative stress in colorectal cancer and its potential underlying mechanism. Oncology Reports. 2018;40(4):2137–2146. doi: 10.3892/or.2018.6594. [DOI] [PubMed] [Google Scholar]
- 40.Gao Y. J., Liu D. L., Li S., Li L., Zhu H. Y., Cao G. Y. Down-regulation of CXCL 11 inhibits colorectal cancer cell growth and epithelial-mesenchymal transition. OncoTargets and Therapy. 2018;11:7333–7343. doi: 10.2147/ott.s167872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan K., Zheng J., Yu J., et al. Knockdown of MMP-1 inhibits the progression of colorectal cancer by suppressing the PI3K/Akt/c-myc signaling pathway and EMT. Oncology Reports. 2020;43(4):1103–1112. doi: 10.3892/or.2020.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zucker S., Vacirca J. Role of matrix metalloproteinases (MMPs) in colorectal cancer. Cancer and Metastasis Reviews. 2004;23(1-2):101–117. doi: 10.1023/a:1025867130437. [DOI] [PubMed] [Google Scholar]
- 43.Yang X.-Z., Cui S.-Z., Zeng L.-S., et al. Overexpression of Rab1B and MMP9 predicts poor survival and good response to chemotherapy in patients with colorectal cancer. Aging. 2017;9(3):914–931. doi: 10.18632/aging.101200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng D. O. Principles of CDK regulation. Nature. 1995;374(6518):131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 45.Krek W., Nigg E. A. Differential phosphorylation of vertebrate p34cdc2 kinase at the G1/S and G2/M transitions of the cell cycle: identification of major phosphorylation sites. The EMBO Journal. 1991;10(2):305–316. doi: 10.1002/j.1460-2075.1991.tb07951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fang Y., Yu H., Liang X., Xu J., Cai X. Chk1-induced CCNB1 overexpression promotes cell proliferation and tumor growth in human colorectal cancer. Cancer Biology & Therapy. 2014;15(9):1268–1279. doi: 10.4161/cbt.29691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Irby R. B., McCarthy S. M., Yeatman T. J. Osteopontin regulates multiple functions contributing to human colon cancer development and progression. Clinical & Experimental Metastasis. 2004;21(6):515–523. doi: 10.1007/s10585-004-2873-4. [DOI] [PubMed] [Google Scholar]
- 48.Xu C., Sun L., Jiang C. H., Gu L., Liu Y., Xu Q. SPP1, analyzed by bioinformatics methods, promotes the metastasis in colorectal cancer by activating EMT pathway. Biomedicine & Pharmacotherapy. 2017;91:1167–1177. doi: 10.1016/j.biopha.2017.05.056. [DOI] [PubMed] [Google Scholar]
- 49.Zhou E. K., Yi J. W., Chai Y. J., Park K. J. Upregulation of the adipokine genes ADIPOR1 and SPP1 is related to poor survival outcomes in colorectal cancer. Journal of Surgical Oncology. 2018;117(8):1833–1840. doi: 10.1002/jso.25078. [DOI] [PubMed] [Google Scholar]
- 50.Zhang X.-A., Zhang S., Yin Q., Zhang J. Quercetin induces human colon cancer cells apoptosis by inhibiting the nuclear factor-kappa B Pathway. Pharmacognosy Magazine. 2015;11(42):404–409. doi: 10.4103/0973-1296.153096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Y., Wang T., Chen D. Q., et al. Quercetin preferentially induces apoptosis in KRAS-mutant colorectal cancer cells via JNK signaling pathways. Cell Biology International. 2019;43(2):117–124. doi: 10.1002/cbin.11055. [DOI] [PubMed] [Google Scholar]
- 52.Ma L., Lu N., Dai Q., et al. Wogonin induced G1 cell cycle arrest by regulating Wnt/β-catenin signaling pathway and inactivating CDK8 in human colorectal cancer carcinoma cells. Toxicology. 2013;312:36–47. doi: 10.1016/j.tox.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 53.Zeng Q., Zhang Y., Zhang W., Guo Q. Baicalein suppresses the proliferation and invasiveness of colorectal cancer cells by inhibiting Snail induced epithelial mesenchymal transition. Molecular Medicine Reports. 2020;21(6):2544–2552. doi: 10.3892/mmr.2020.11051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rui X., Yan X., Zhang K. Baicalein inhibits the migration and invasion of colorectal cancer cells via suppression of the AKT signaling pathway. Oncology Letters. 2016;11(1):685–688. doi: 10.3892/ol.2015.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Z., Hou R., Gao S., Song D., Feng Y. Baicalein inhibits proliferation activity of human colorectal cancer cells HCT116 through downregulation of ezrin. Cellular Physiology and Biochemistry. 2018;49(5):2035–2046. doi: 10.1159/000493714. [DOI] [PubMed] [Google Scholar]
- 56.Ni T., He Z., Dai Y., et al. Oroxylin a suppresses the development and growth of colorectal cancer through reprogram of HIF1α-modulated fatty acid metabolism. Cell Death & Disease. 2017;8(6) doi: 10.1038/cddis.2017.261.e2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang D., Fu L., Sun H., Guo L., DuBois R. N. Prostaglandin E2 promotes colorectal cancer stem cell expansion and metastasis in mice. Gastroenterology. 2015;149(7):1884–1895. doi: 10.1053/j.gastro.2015.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H., Liu K., Boardman L. A., et al. Circulating prostaglandin biosynthesis in colorectal cancer and potential clinical significance. EBioMedicine. 2015;2(2):165–171. doi: 10.1016/j.ebiom.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao R., Kawada K., Sakai Y. Prostaglandin E2/EP signaling in the tumor microenvironment of colorectal cancer. International Journal of Molecular Sciences. 2019;20(24):p. 6254. doi: 10.3390/ijms20246254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gustafsson A., Andersson M., Lagerstedt K., Lönnroth C, Nordgren S, Lundholm K. Receptor and enzyme expression for prostanoid metabolism in colorectal cancer related to tumor tissue PGE2. International Journal of Oncology. 2010;36(2):469–478. [PubMed] [Google Scholar]
- 61.Waldner M. J., Foersch S., Neurath M. F. Interleukin-6-a key regulator of colorectal cancer development. International Journal of Biological Sciences. 2012;8(9):1248–1253. doi: 10.7150/ijbs.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krzystek-Korpacka M., Zawadzki M., Neubauer K., et al. Elevated systemic interleukin-7 in patients with colorectal cancer and individuals at high risk of cancer: association with lymph node involvement and tumor location in the right colon. Cancer Immunology, Immunotherapy. 2017;66(2):171–179. doi: 10.1007/s00262-016-1933-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bednarz-Misa R. J., Greenman J., MacDonald A. W, et al. Advanced colorectal cancer is associated with impaired interleukin 12 and enhanced interleukin 10 production. Clinical Cancer Research :An Official Journal of the American Association for Cancer Research. 1998;4(8):1943–1948. [PubMed] [Google Scholar]
- 64.Wu D., Wu P., Huang Q., et al. Interleukin-17: a promoter in colorectal cancer progression. Clinical and Developmental Immunology. 2013;2013 doi: 10.1155/2013/436307.436307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Luster A. D. Chemokines - chemotactic cytokines that mediate inflammation. New England Journal of Medicine. 1998;338(7):436–445. doi: 10.1056/nejm199802123380706. [DOI] [PubMed] [Google Scholar]
- 66.Agace W. W., Roberts A. I., Wu L., Greineder C., Ebert E. C., Parker C. M. Human intestinal lamina propria and intraepithelial lymphocytes express receptors specific for chemokines induced by inflammation. European Journal of Immunology. 2000;30(3):819–826. doi: 10.1002/1521-4141(200003)30:3<819::aid-immu819>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 67.Baier P. K., Eggstein S., Wolff-Vorbeck G., Baumgartner U., Hopt U. T. Chemokines in human colorectal carcinoma. Anticancer Res. 2005;25(5):3581–3584. [PubMed] [Google Scholar]
- 68.Erreni M., Bianchi P., Laghi L. M., et al. Chapter 5 expression of chemokines and chemokine receptors in human colon cancer. Methods in Enzymology. 2009;460:105–121. doi: 10.1016/s0076-6879(09)05205-7. [DOI] [PubMed] [Google Scholar]
- 69.Mirolo G., Yang H., Zhao J., Yuan A., Florholmen J. Elevated proinflammatory cytokine IL-17A in the adjacent tissues along the adenoma-carcinoma sequence. Pathology & Oncology Research. 2015;21(1):139–146. doi: 10.1007/s12253-014-9799-1. [DOI] [PubMed] [Google Scholar]
- 70.Razi S., Baradaran Noveiry B., Keshavarz-Fathi M., Rezaei N. IL-17 and colorectal cancer: from carcinogenesis to treatment. Cytokine. 2019;116:7–12. doi: 10.1016/j.cyto.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 71.Liu J., Duan Y., Cheng X. X., et al. IL-17 is associated with poor prognosis and promotes angiogenesis via stimulating VEGF production of cancer cells in colorectal carcinoma. Biochemical and Biophysical Research Communications. 2011;407(2):348–354. doi: 10.1016/j.bbrc.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 72.Chen L., Wang Y., Jie G., et al. Association between Toll-like receptor 4 and interleukin 17 gene polymorphisms and colorectal cancer susceptibility in Northeast China. Medical Oncology. 2014;31(10):p. 73. doi: 10.1007/s12032-014-0073-x. [DOI] [PubMed] [Google Scholar]
- 73.Chin C.-C., Chen C.-N., Kuo H.-C., et al. Interleukin-17 induces CC chemokine receptor 6 expression and cell migration in colorectal cancer cells. Journal of Cellular Physiology. 2015;230(7):1430–1437. doi: 10.1002/jcp.24796. [DOI] [PubMed] [Google Scholar]
- 74.Shi Q., Feng M., Yu T., Liu X., Zhang P. Intratumoral regulatory T cells are associated with suppression of colorectal carcinoma metastasis after resection through overcoming IL-17 producing T cells. Cellular Immunology. 2014;287(2):100–105. doi: 10.1016/j.cellimm.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 75.Waters J. P., Pober J. S., Bradley J. R. Tumour necrosis factor in infectious disease. The Journal of Pathology. 2013;230(2):132–147. doi: 10.1002/path.4187. [DOI] [PubMed] [Google Scholar]
- 76.Aggarwal B. B., Gupta S. C., Kim J. H. Historical perspectives on tumor necrosis factor and its superfamily: 25 years later, a golden journey. Blood. 2012;119(3):651–665. doi: 10.1182/blood-2011-04-325225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Banday M. Z., Balkhi H. M., Hamid Z., Sameer A. S., Chowdri N. A., Haq E. Tumor necrosis factor-α (TNF-α)-308G/A promoter polymorphism in colorectal cancer in ethnic Kashmiri population - a case control study in a detailed perspective. Meta Gene. 2016;9:128–136. doi: 10.1016/j.mgene.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gutierrez-Hurtado I. A., Puebla-Pérez A. M., Delgado-Saucedo J. I., et al. Association between TNF-α-308G>A and -238G>A gene polymorphisms and TNF-α serum levels in Mexican colorectal cancer patients. Genetics and Molecular Research. 2016;15(2) doi: 10.4238/gmr.15028199. [DOI] [PubMed] [Google Scholar]
- 79.Moradi-Marjaneh R., Hassanian S. M., Fiuji H., Ferns G. A., Avan A., Khazaei M. Toll like receptor signaling pathway as a potential therapeutic target in colorectal cancer. Journal of Cellular Physiology. 2018;233(8):5613–5622. doi: 10.1002/jcp.26273. [DOI] [PubMed] [Google Scholar]
- 80.Soleimanpour L., Botos I., Wang Y., et al. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320(5874):379–381. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kawasaki T., Kawai T. Toll-like receptor signaling pathways. Front Immunol. 2014;5:p. 461. doi: 10.3389/fimmu.2014.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee H., Song M., Shin N., et al. Diagnostic significance of serum HMGB1 in colorectal carcinomas. PLoS One. 2012;7(4) doi: 10.1371/journal.pone.0034318.e34318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee M. J., Lee J. K., Choi J. W., et al. Interleukin-6 induces S100A9 expression in colonic epithelial cells through STAT3 activation in experimental ulcerative colitis. PLoS One. 2012;7(9) doi: 10.1371/journal.pone.0038801.e38801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kaczanowska S., Joseph A. M., Davila E. TLR agonists: our best frenemy in cancer immunotherapy. Journal of Leukocyte Biology. 2013;93(6):847–863. doi: 10.1189/jlb.1012501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gambara G., Cesaris P., Nunzio C., Tubaro A., Filippini A., Riccioli A. Toll-like receptors in prostate infection and cancer between bench and bedside. Journal of Cellular and Molecular Medicine. 2013;17(6):713–722. doi: 10.1111/jcmm.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ziparo E., Eggermont A., Sautès-Fridman C., et al. Trial watch: toll-like receptor agonists for cancer therapy. Oncoimmunology. 2013;2(8) doi: 10.4161/onci.25238.e25238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Soleimani A., Rahmani F., Ferns G. A., Ryzhikov M., Avan A., Hassanian S. M. Role of the NF-κB signaling pathway in the pathogenesis of colorectal cancer. Gene. 2020;726 doi: 10.1016/j.gene.2019.144132.144132 [DOI] [PubMed] [Google Scholar]
- 88.Voboril R., Weberova-Voborilova J. Constitutive NF-kappaB activity in colorectal cancer cells: impact on radiation-induced NF-kappaB activity, radiosensitivity, and apoptosis. Neoplasma. 2006;53(6):518–523. [PubMed] [Google Scholar]
- 89.Sakamoto K., Maeda S., Hikiba Y. H., et al. Constitutive NF- B activation in colorectal carcinoma plays a key role in angiogenesis, promoting tumor growth. Clinical Cancer Research. 2009;15(7):2248–2258. doi: 10.1158/1078-0432.ccr-08-1383. [DOI] [PubMed] [Google Scholar]
- 90.Nakagawa P. Colorectal cancer and NF-κB signaling pathway. Gastroenterology and Hepatology from Bed to Bench. 2011;4(3):127–132. [PMC free article] [PubMed] [Google Scholar]
- 91.Chowdhury R., Gales D., Valenzuela P. S., et al. Bromoethylindole (BEI-9) redirects NF-κB signaling induced by camptothecin and TNFα to promote cell death in colon cancer cells. Apoptosis. 2017;22(12):1553–1563. doi: 10.1007/s10495-017-1427-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Miller Y.-C., Lin W.-C., Chiang Y.-F., et al. Sorafenib sensitizes human colorectal carcinoma to radiation via suppression of NF-κB expression in vitro and in vivo. Biomedicine & Pharmacotherapy. 2012;66(1):12–20. doi: 10.1016/j.biopha.2011.09.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1: information for 140 BCIs of GQD. Supplementary Table S2: the significant GO-BP terms enriched by target genes. Supplementary Table S3: the significant GO-MF terms enriched by target genes.
Data Availability Statement
The data used to support the findings of this study are included within the article and supplementary information files.


