SUMMARY
Soluble envelope (Env) trimers, stabilized in a prefusion-closed conformation, can elicit neutralizing responses against HIV-1 strains closely related to the immunizing trimer. However, to date such stabilization has succeeded with only a limited number of HIV-1 strains. To address this issue, here we develop ADROITrimer, an automated procedure involving structure-based stabilization and consensus repair, and generate “RnS-DS-SOSIP”-stabilized Envs from 180 diverse Env sequences. The vast majority of these RnS-DS-SOSIP Envs fold into prefusion-closed conformations as judged by antigenic analysis and size exclusion chromatography. Additionally, representative strains from clades AE, B, and C are stabilized in prefusion-closed conformations as shown by negative-stain electron microscopy, and the crystal structure of a clade A strain MI369.A5 Env trimer provides 3.5 Å resolution detail into stabilization and repair mutations. The automated procedure reported herein that yields well-behaved, soluble, prefusion-closed Env trimers from a majority of HIV-1 strains could have substantial impact on the development of an HIV-1 vaccine.
Graphical Abstract
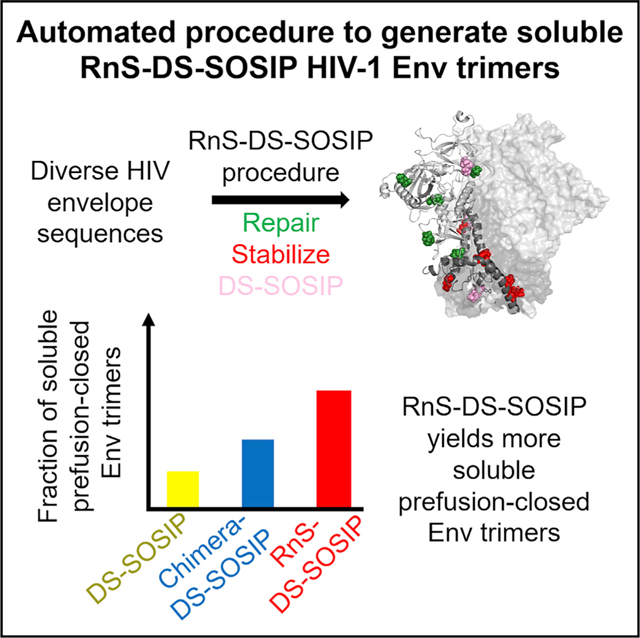
In Brief
A general method to stabilize HIV-1 envelope (Env) trimers in its prefusion-closed conformation may have substantial vaccine utility. Rawi et al. report an automated pipeline based on DS-SOSIP and repair-and-stabilization (RnS) strategies, which successfully stabilizes more than 70% of a diverse cross-clade panel of 180 strains in the prefusion-closed conformation.
INTRODUCTION
The mature HIV-1 envelope (Env) spike is a metastable trimer of gp120 and gp41 heterodimeric subunits, which are derived from a gp160 precursor (reviewed in Harrison, 2015; Wyatt and Sodroski, 1998). Trimer instability and Env sequence variability have been substantial barriers to the development of an HIV vaccine that aims to induce broadly neutralizing antibodies (reviewed in de Taeye et al., 2016; Pancera et al., 2017; Ward and Wilson, 2017), and stabilization of Env into a near native or prefusion-closed state has been deemed highly important to induce such antibodies. Moreover, low expression levels can decrease prospects for successful protein subunit vaccine development or the application of vector-based approaches. Therefore, stable and well-behaved Env trimers that fold efficiently and that arrest conformational changes in a prefusion-closed state are desired.
Several stabilization strategies have been described that yield prefusion-closed HIV Env trimers, including SOSIP mutations (Binley et al., 2000; Sanders and Moore, 2017; Sanders et al., 2002), which comprise an engineered disulfide (SOS) between gp120 and gp41 (Binley et al., 2000) and an Ile-to-Pro substitution (IP) in a critical region of gp41 (Sanders et al., 2002). When incorporated into BG505, a clade A transmitted/founder virus transmitted from a mother to an infant six weeks after birth (Wu et al., 2006) that exhibited broad antigenicity and neutralization sensitivity (Hoffenberg et al., 2013), the BG505 SOSIP.664 Env trimer yields soluble, fully cleaved Env trimers in a prefusion-closed conformation (Ringe et al., 2013; Sanders et al., 2013). Although screening of additional strains could identify others of appropriate antigenicity (de Taeye et al., 2015), the BG505 strain appeared especially suited to presentation of near-native trimers, and SOSIP.664 stabilized Env trimers have been widely used by the field to elicit neutralizing titers against sequence-matched neutralization-resistant HIV isolates in standard vaccine-test species (Cheng et al., 2015; Chuang et al., 2017; de Taeye et al., 2015; Feng et al., 2016; Klasse et al., 2016; Martinez-Murillo et al., 2017; Pauthner et al., 2017; Saunders et al., 2017).
Despite promising results with SOSIP Env variants, the fraction of prefusion-closed trimer is less than optimal (Julien et al., 2015), and the resultant trimers open in the presence of CD4. Structural analysis of the BG505 SOSIP.664 trimer (Julien et al., 2013; Lyumkis et al., 2013; Pancera et al., 2014), however, has provided atomic-level coordinates as templates for structure-based stabilization (Chuang et al., 2017; de Taeye et al., 2015; Guenaga et al., 2017; Kulp et al., 2017; Kwon et al., 2015; Steichen et al., 2016; Sullivan et al., 2017; Torrents de la Peña et al., 2017) and chimeric strategies (He et al., 2018; Joyce et al., 2017). Furthermore, the addition of a disulfide between residues 201 and 433 (DS-SOSIP) has been found to prevent CD4-induced conformational changes (Kwon et al., 2015), and more recently, a general paradigm to improve the quality and yield of prefusion-closed Env trimers has been proposed using a “repair-and-stabilization” (RnS) approach (Rutten et al., 2018). In addition to identification of new stabilization mutations, Rutten et al. (2018) proposed the use of the repair strategy to substitute rare amino acid mutations of Env with consensus amino acid types, to make the Env more “transmitted/founder”-like, which have better folding characteristics.
Here we combined DS-SOSIP with RnS in an automated fashion to create a pipeline that starts with an Env sequence and results in an RnS-DS-SOSIP-stabilized sequence. We used both antigenic analysis and size exclusion chromatography of expressed Env trimers to determine whether they were in prefusion-closed conformations and further expressed and characterized a subset of the Envs by negative-stain electron microscopy (EM), as well as a clade A strain by X-ray crystallography. Overall, we observed the vast majority of the tested Env sequences to form well-behaved soluble prefusion-closed trimers.
RESULTS
Automated Pipeline for Designing Repaired and Stabilized DS-SOSIP
We developed an automated computational pipeline, automated design of prefusion-closed HIV trimers (ADROITrimer), which designs prefusion-closed HIV-1 Env trimers of any given HIV-1 strain (Figure 1; STAR Methods). The first step involved aligning wild-type Env sequences with the corresponding clade reference alignment to identify positions of rare mutations (defined by sequence variants that occurred in <2% of HIV sequences). In the second step, these rare mutations were replaced by consensus amino acids for those positions in the clade-specific reference alignment or group M reference alignment. The average number of repair mutations introduced for each sequence was 9.7 (~1.5% of total residues) (Figure S1A). On average, clades D/CD sequences had the most repair mutations, while clades A/AC/ACD/AD had the fewest repair mutations (Figure S1B). In the third step, seven residues were substituted with amino acid variants—535N, 556P, 588E, 589V, 651F, 655I, and 658V (HXB2 numbering)—which we found to increase trimeric yields for diverse Envs (e.g., for strains Du422, ZM233M, and ZM246F) (Rutten et al., 2018). In the final step, DS-SOSIP mutations (DS: 201C, 433C; SOSIP: 501C, 605C, and 559P) (Kwon et al., 2015) along with cleavage-enhancing six Arg (replacing residues 508–511) and truncation of the protein after residue 664 (Sanders et al., 2013) were incorporated to yield RnS-DS-SOSIP-stabilized Env sequences. ADROITrimer requires only two inputs: the wild-type Env sequence in FASTA format and its clade classification, and outputs the RnS-DS-SOSIP repaired and stabilized sequence.
Figure 1. ADROIT Automatically Designs RnS-DS-SOSIP-Stabilized Constructs from HIV-1 Env Input Sequences.
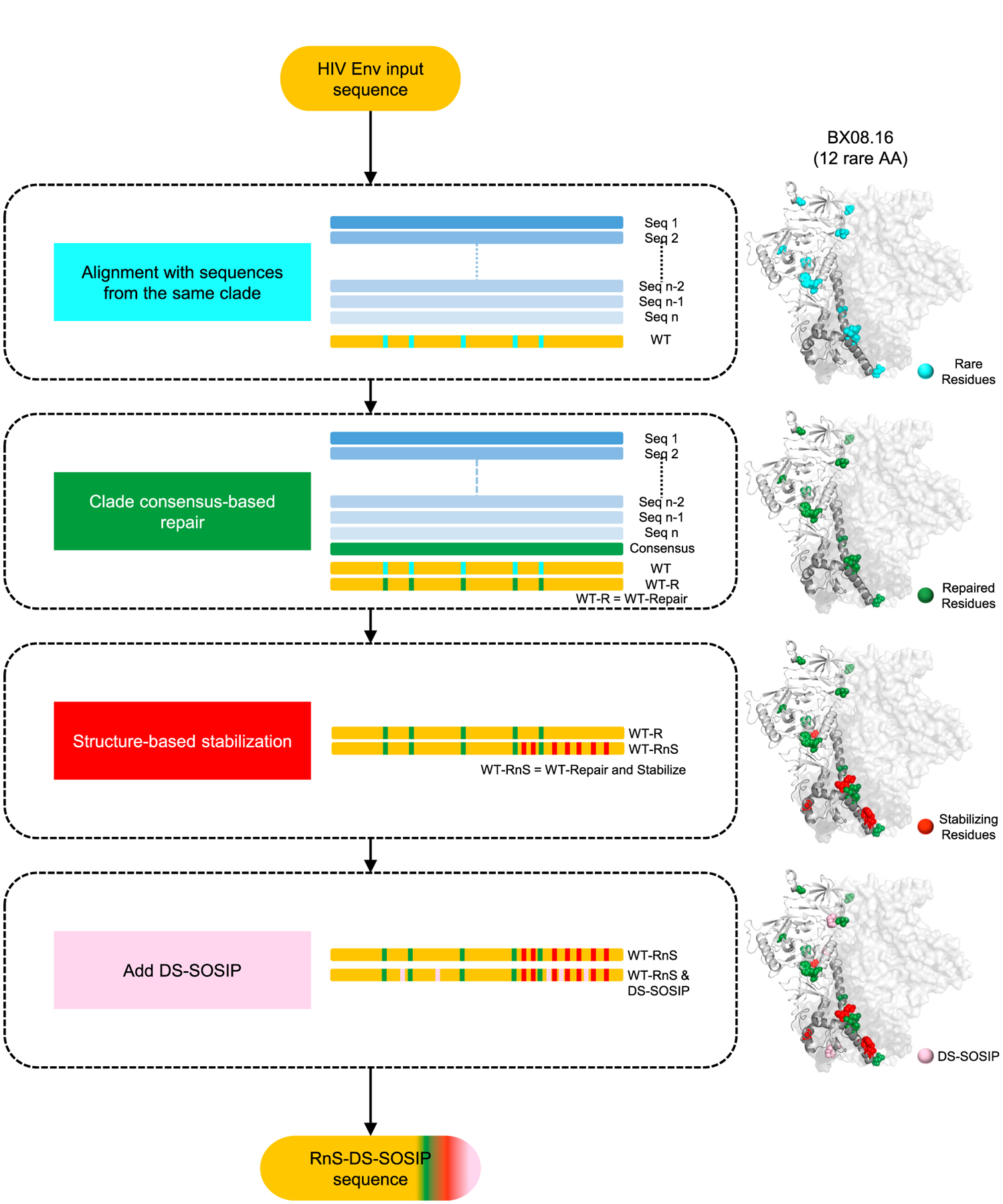
Flowchart depicting the multi-step approach to repair and stabilize HIV-1 Env sequences to form prefusion-closed trimers. Types of mutations introduced are highlighted on a structural model of the BX08.16 Env. The structure-based stabilization mutants include the following mutations: 535N, 556P, 588E, 589V, 651F, 655I, and 658V (HXB2 numbering). ADROITrimer is available at https://github.com/RedaRawi/ADROITrimer.
Antibodies Specific for the Prefusion-Closed Conformations of Env Trimer Recognize 134 of the 180 RnS-DS-SOSIP Env Trimers
To test ADROITrimer, we created 180 RnS-DS-SOSIP Env trimers (Data S1), on the basis of the published panel of diverse Env sequences that we previously used to assess DS-SOSIP (Kwon et al., 2015) and BG505-chimeric (Joyce et al., 2017) approaches to stabilize soluble Env trimer. 293T cells were transfected with plasmids encoding the ADROITrimer-defined Env sequences, and the supernatants were assessed for the presence of Env trimer as well as reactivity to a panel of broadly neutralizing antibodies by using ELISA (see STAR Methods). Two of these antibodies, PGT145 (Walker et al., 2011) and CAP256-VRC26.25 (the latter named for donor-lineage.clone and herein referred to as VRC26.25) (Doria-Rose et al., 2015), specifically bind Env in prefusion-closed conformations. Notably, 134 of the RnS-DS-SOSIP HIV-1 Env constructs showed ELISA responses that were at least 75% of the response relative to the BG505 DS-SOSIP control (Figure 2A; Data S2) for either PGT145 or VRC26.25 binding. As PGT145 and VRC26.25 neutralize only 144 of the 180 strains, the success rate for RnS-DS-SOSIP stabilization on strains neutralization sensitive to either of these two antibodies was 119 of 144 strains (82.6%) (Table S1A). Interestingly, 15 strains that were neutralization resistant to both PGT145 and VRC26.25 satisfied this antigenic criterion, indicating these strains were stabilized in the prefusion-closed conformation with RnS-DS-SOSIP. (Table S1A). For the 15 strains in question, critical binding residues for antibodies PGT145 and CAP256-VRC26.25 were introduced by the repair process for 9 of the strains (glycan 160 for strains 0815.V3.C3, 3637.V5.C3, BR07.DG, and QH0692.42; 166R for strains 3817.v2.c59, CNE30, DU172.17, ZM135.10a; 169K for strain NKU3006.ec1), and with 4 of the strains (6471.V1.C16, MN.3, NKU3006.ec1, and BR07.DG), the ELISA signal was less than that of BG505 DS-SOSIP for either antibody, although it did satisfy the antigenic criteria. To assess phylogenetic clustering of the successful prefusion-closed-stabilized strains, we analyzed the phylogenetic distribution of the 180 tested strains, and observed that the prefusion-closed-stabilized strains were distributed over all clades (Figure 2B), with clade-specific success rates ranging from 53% (clade AG) to 90% (recombinant clade C/BC) (Figure S2A). Notably, the average sequence identity between a non-prefusion-closed construct and the “closest” prefusion-closed construct was 0.87, similar to the average pairwise sequence similarity within a clade (Figures S2C and S2D).
Figure 2. Antigenic Screening and Analytical SEC of RnS-DS-SOSIP Constructs.
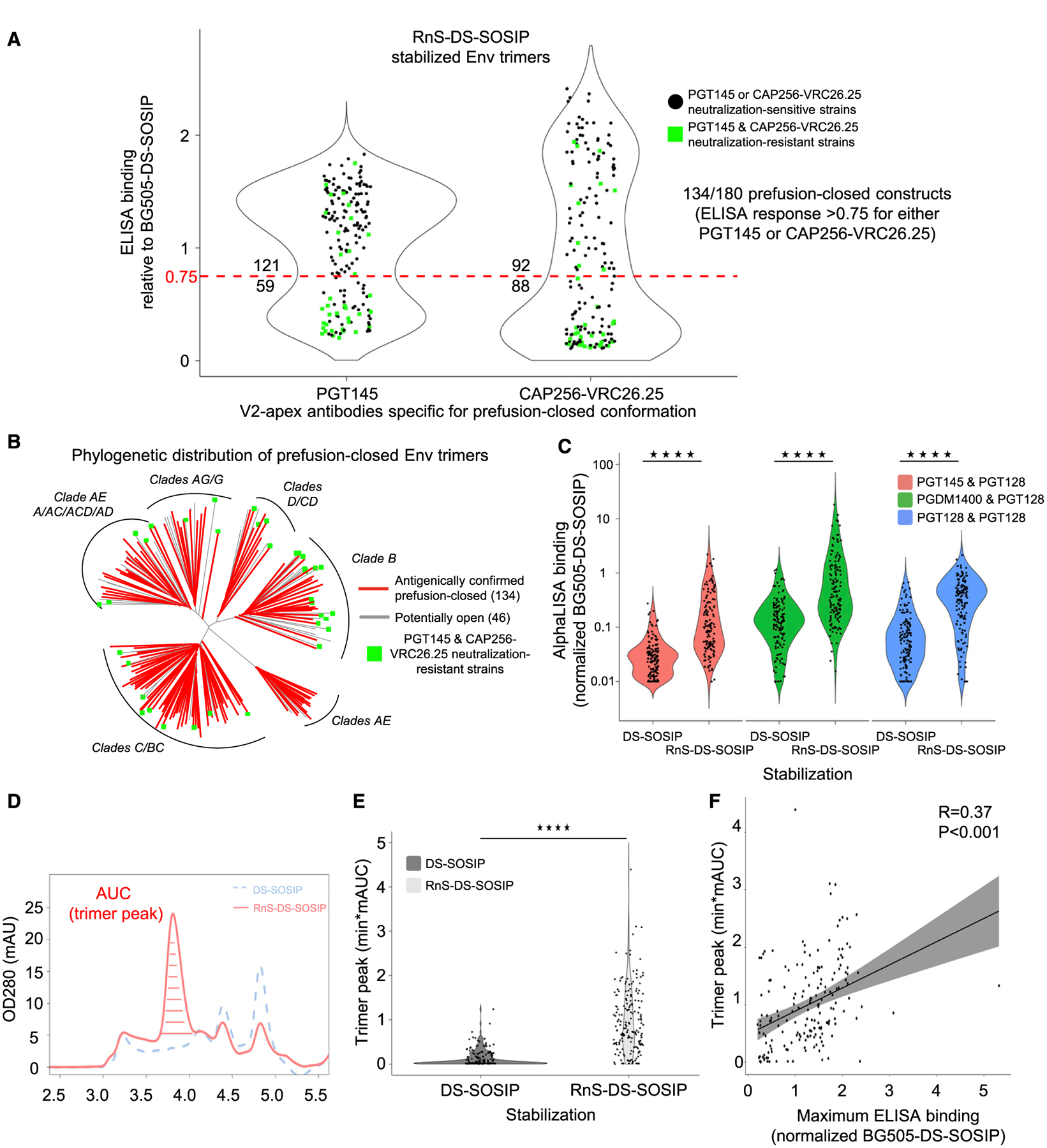
(A) Binding response toward PGT145 and CAP256-VRC26.25, relative to BG505 DS-SOSIP, for RnS-DS-SOSIP of 180 Env strains, including 36 PGT145 and CAP256-VRC26.25 neutralization-resistant strains. Dotted red line indicates 75% binding response relative to BG505 DS-SOSIP. The values displayed above and below the dotted red line indicate the number of Env molecules with binding response greater or lower than 75% of that for BG505 DS-SOSIP. One hundred thirty-four of 180 RnS-DS-SOSIP satisfied the antigenic criteria for prefusion-closed Env. Values are average of duplicate measurements.
(B) Phylogenetic tree of 180 HIV-1 strains, with those that can be stabilized in prefusion-closed conformation with RnS-DS-SOSIP highlighted in red. PGT145 and CAP256-VRC26.25 neutralization-resistant strains are denoted with green squares.
(C) Violin plots highlighting significantly increased AlphaLISA binding responses to 177 RnS-DS-SOSIP versus DS-SOSIP-only stabilized constructs with pre-fusion-closed detecting antibody combinations. Values shown are average of two biological repeats. Statistical significance was assessed using the two-tailed Mann-Whitney test (****p < 0.0001).
(D) Representative size exclusion chromatograms of cell culture supernatant for DS-SOSIP and RnS-DS-SOSIP-stabilized Env construct of clade C HIV-1 strain 25711–2.4. Peak 1 contains aggregates, peak 2 contains trimers, peak 3 contains gp140 monomers, peak 4 contains gp120 monomers, and peak 5 contains gp41 monomers.
(E) Violin plot depicting significantly higher trimer peak AUC values from analytic SEC for RnS-DS-SOSIP (light gray) compared with DS-SOSIP (dark gray) stabilized constructs. A total of 179 HIV-1 strains were assessed. Statistical significance was assessed using the Mann-Whitney test (****p < 0.0001).
(F) Correlation plot between maximum ELISA binding (either PGT145 or CAP256-VRC26.25) and trimer peak AUC from analytic AUC depicting significant Pearson correlation (R = 0.37, p < 0.001) (n = 179).
See also Figures S2 and S3, Table S1A, and Data S1 and S2.
We also used AlphaLISA to assess the recognition by broadly neutralizing antibodies PGT145, PGDM1400 (Sok et al., 2014), and PGT128 (Walker et al., 2011) for 177 of 180 strains, when expressed with DS-SOSIP-only stabilization or the RnS-DS-SOSIP stabilization. Overall, a significantly higher level of recognition by the RnS-DS-SOSIP stabilization was observed (Figure 2C), with the binding ratio increasing 4- to 7-fold for RnS-DS-SOSIP versus DS-SOSIP-only stabilized constructs, with the higher ratios seen with antibodies PGT145 and PGDM1400, which preferentially recognize prefusion-closed forms of the HIV Env (Figure S3).
Analytical SEC Confirmed RnS-DS-SOSIP-Stabilized Constructs to Yield Significantly Higher Amounts of Trimer
As many strains in our 180-strain panel, especially from clade B (Bricault et al., 2019), were not neutralized by antibodies PGT145 and VRC26.25, which we used as the criterion to determine prefusion-closed antigenicity (Figure 2B), this might be underestimating the improvement indicated by 96-well ELISA and AlphaLISA experiments. To evaluate the improvement in folding of soluble trimers, as judged by size exclusion chromatographic behavior, we used high-throughput ultra-high performance liquid chromatography (UHPLC)-based analytical size exclusion chromatography to compare the trimer content of the transiently transfected DS-SOSIP-only and RnS-DS-SOSIP-stabilized Env trimers (Figure 2D). With this chromatography, aggregates, trimers, gp140 monomers, gp120 monomers, and gp41 monomers are well resolved.
For each pair of Env trimers, the trimer area under the curve (AUC; mAU · min) was measured for the elution peaks, and the amount of RnS-DS-SOSIP-stabilized Env trimer was found to be significantly higher than with DS-SOSIP-only stabilization (Figure 2E). In 166 of the 179 Env trimers (~93%), the RnS mutations led to an either small or large increase in the AUC (not shown). For the RnS-DS-SOSIP trimers, 143 of 179 tested strains had AUCs that were higher than half of the AUC value for BG505 DS-SOSIP, suggesting that ~80% of the RnS-DS-SOSIP Envs can be regarded as well-behaving trimers with reasonable yields (Table S1B). Clade-based analysis of the trimer fraction furthermore showed the improvement in trimer yield to extend to all tested clades (Figure S2B). We also observed a positive correlation between the AUC metric and the antigenicity score (R = 0.37, p < 0.0001) (Figure 2F). Overall, analytical SEC indicated increased yields of well-behaved Env trimer from stabilization by RnS-DS-SOSIP versus by DS-SOSIP-only.
Antigenic Analysis and Negative-Stain EM Confirmed Prefusion-Closed Conformation for Select RnS-DS-SOSIP-Stabilized Constructs
To further characterize the repaired and stabilized Env trimers, RnS-DS-SOSIP variants for three strains (clade AE C1080.c3, clade B REJO.67, and clade C 6838.v1) were selected for larger scale expression and purification (Figure 3A). The RnS-DS-SOSIP variants of these strains demonstrated prefusion-closed antigenicity in the 96-well ELISA screening, while their DS-SOSIP versions did not have prefusion-closed antigenicity (Joyce et al., 2017). We used a modified procedure involving affinity chromatography with the broadly neutralizing CD4-binding site antibody, 3BNC117, which had an HRV3C-protease cleavage site inserted into its hinge region. The 3BNC117(HRV3C site-inserted)-bound trimers were captured on Protein A resin, and the Fab-bound trimers were liberated by HRV3C (PreScission Protease) cleavage.
Figure 3. Antigenic Analysis and Negative-Stain EM Confirmed Prefusion-Closed Conformation for Selected RnS-DS-SOSIP-Stabilized Constructs.
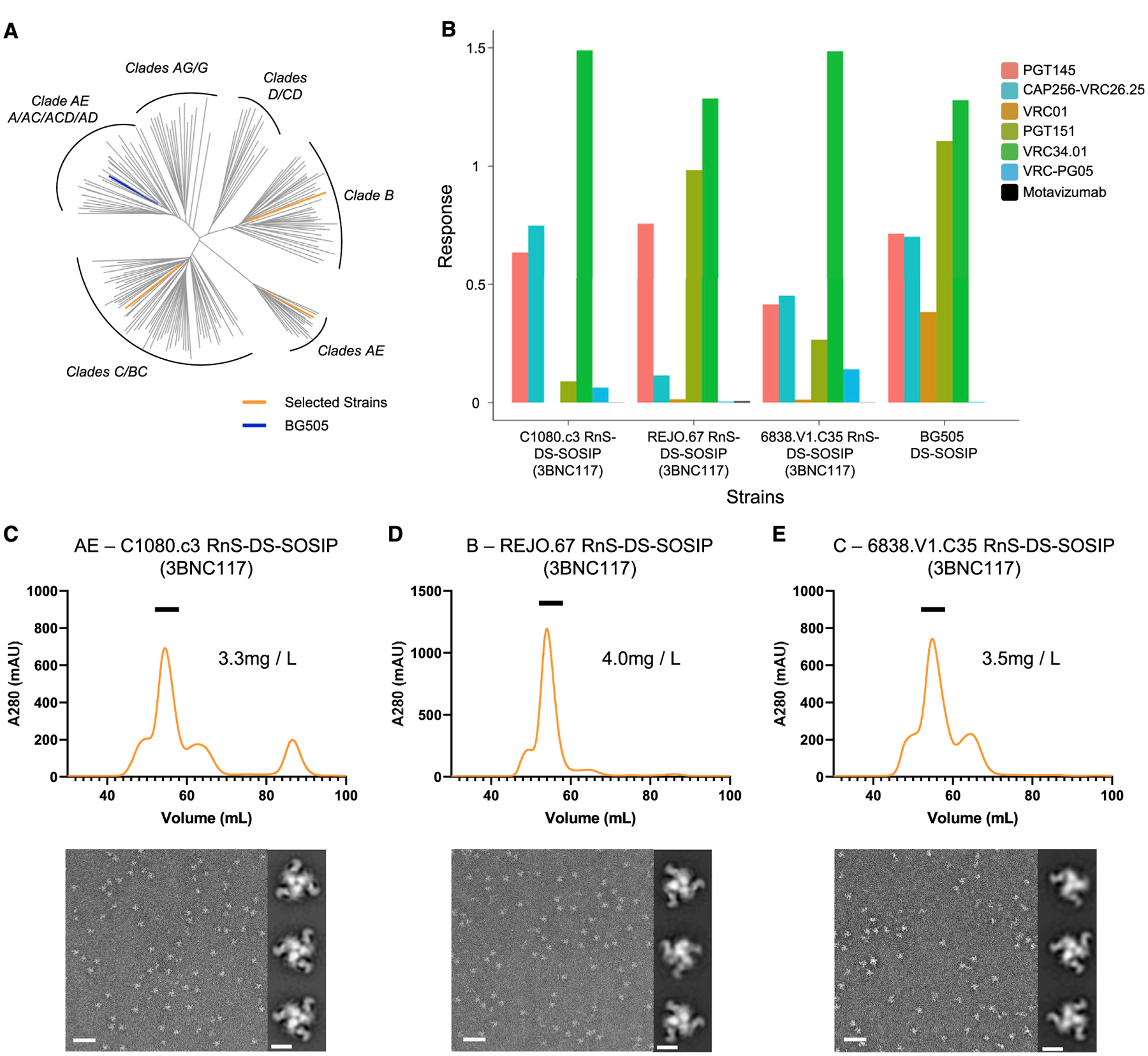
(A) HIV-1 Env constructs of three selected RnS-DS-SOSIP constructs highlighted on a phylogenetic tree of the 180 HIV-1 strains.
(B) All three RnS-DS-SOSIP-stabilized constructs yielded binding to a panel of diverse broadly neutralizing HIV-1 antibodies, including prefusion-closed conformation specific antibodies PGT145 or CAP256-VRC26.25, as measured using Octet.
(C) SEC curve and negative-stain EM results for clade AE C1080.c3 RnS-DS-SOSIP-stabilized construct.
(D) SEC curve and negative-stain EM results for clade B REJO.67 RnS-DS-SOSIP-stabilized construct.
(E) SEC curve and negative-stain EM results for clade C 6838.V1.C35 RnS-DS-SOSIP-stabilized construct. Scale bars: 50 nm (representative EM micrographs) and 10 nm (two-dimensional [2D] class average images).
The antibody binding characteristics of the Fab-bound liberated trimers were characterized using bio-layer interferometry. The Fab-bound liberated trimers were able to bind apex antibodies PGT145 and VRC26.25, indicating a substantial degree of prefusion-closed trimers (Figure 3B). Additionally, the gp41-gp120 interface appears to be properly formed, as indicated by the binding of VRC34.01 (Kong et al., 2016b). Of note, the VRC01 antibody did not bind, consistent with the CD4-binding site being blocked by Fab 3BNC117. The Fab-bound trimer was clearly the most substantial peak by SEC analysis (Figures 3C–3E, upper panels), and negative-stain EM images were consistent with Fab-bound prefusion-closed trimers, with additional density appearing as a pinwheel around the trimer (Figures 3C–3E, lower panels).
To compare the yield and antigenicity of these three strains between RnS-DS-SOSIP and DS-SOSIP contexts, we purified the DS-SOSIP and RnS-DS-SOSIP variants of these three Envs with lectin-based chromatography, followed by gel filtration. Overall, the yield for RnS-DS-SOSIP was higher than that for DS-SOSIP for all three strains, ranging from 1.5-fold to 7-fold (Figure S4A). In addition, RnS-DS-SOSIP showed substantial increases in PGT145 and VRC26.25 binding for the C1080.c3 and REJO.67 strains compared with the DS-SOSIP counterparts (Figure S4B).
Properties and Crystal Structure of MI369.A5 RnS-DS-SOSIP
To provide a structural explanation of the improvement bestowed by the RnS-DS-SOSIP procedure, we determined the crystal structure of MI369.A5 RnS-DS-SOSIP at 3.5 Å resolution, using lattice-forming antibodies as crystallization aids (Lai et al., 2019) (Figure 4). MI369.A5 is a clade A strain, and MI369.A5 RnS-DS-SOSIP showed superior antigenicity based on ELISA screening, while MI369.A5 DS-SOSIP did not exhibit a prefusion-closed antigenicity. Overall, the crystal structure had well-defined electron density that covered most of the Env trimer, including most of the mutations introduced by RnS.
Figure 4. Crystal Structure of MI369 RnS-DS-SOSIP.
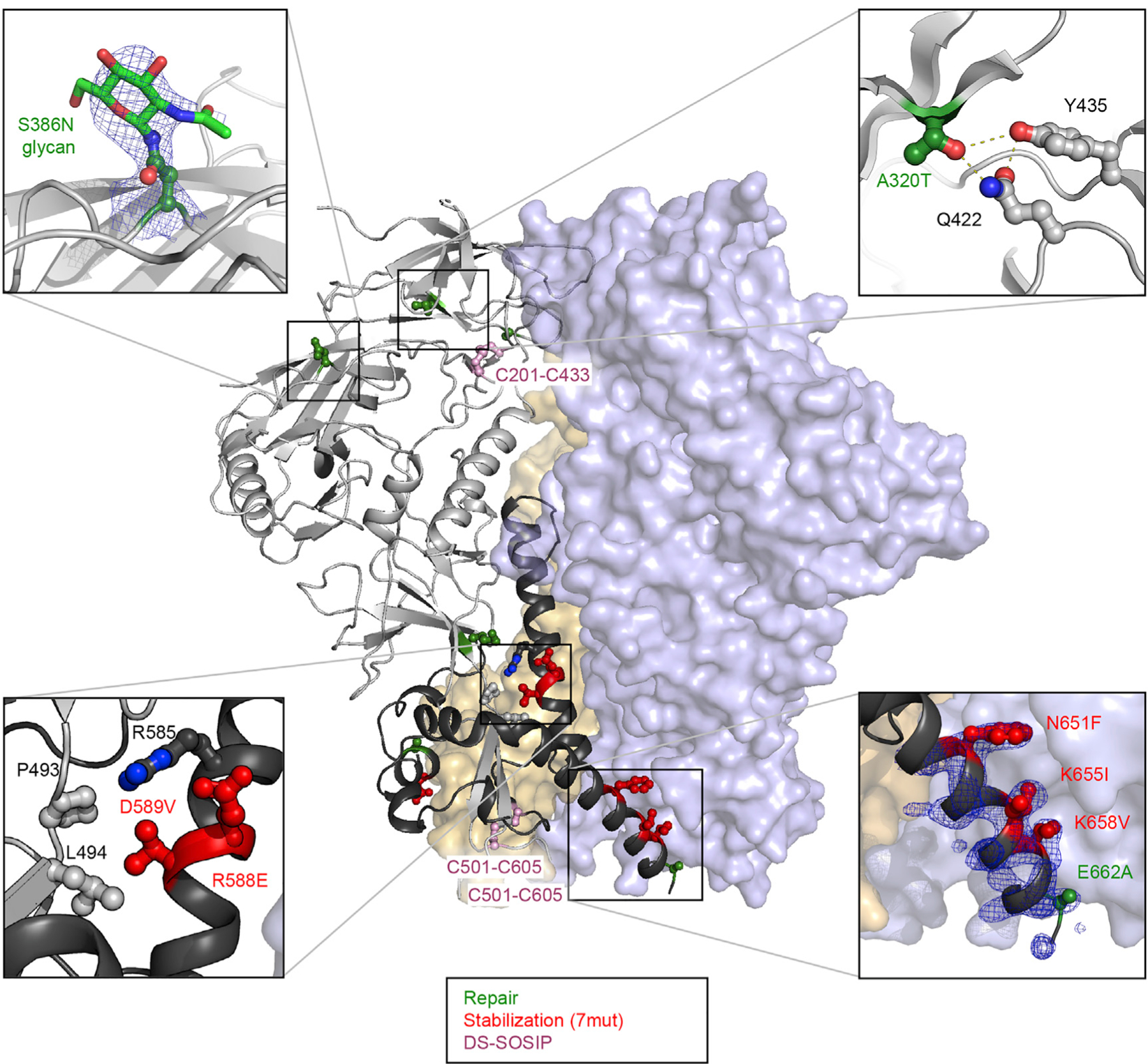
One protomer is shown as ribbon, and the other protomers are shown in surface presentation. The repair, stabilization, and DS-SOSIP mutations are highlighted with balls and sticks and are colored in green, red, and pink, respectively. Panels at the four corners highlight the mutations from repair and stabilization strategies. Blue density in bottom right enlargement is a 2fo-fc map contoured at 1.5σ.
Clear electron density was observed for the side chains of N651F, K655I, K658V, and E662A (Figure 4, lower right panel). These mutations introduced a hydrophobic interface between gp41 of two adjacent protomers and might stabilize the base of the Env trimer. This observation is in line with a previously determined crystal structure of a consensus clade C Env (Rutten et al., 2018). Additionally, the introduction of N535 created a hydrogen bond with the carbonyl of G531, stabilizing the secondary structure of this short helix at the base of the trimer. RnS mutations also appeared to function by stabilizing the gp120-gp41 interface, with E588 hydrogen bonding with R585 and E492 and with V589 interacting hydrophobically with L494 and P493 (Figure 4, lower left). Electron density was also consistent with the introduction of a site for N-linked glycosylation at the base of V3 by the RnS mutation S386N (Figure 4, upper left). The crystal structure also provided the structural basis for understanding the unique stabilizing effects by the repair mutations from ADROITrimer for the MI369.A5 Env trimer. For example, A320T introduced a hydrogen-bonding network with Y435 and Q422 (Figure 4, upper right panel). The crystal structure thus revealed atomic-level structural features of Env trimer stabilization as well as for strain-specific features, specific to the clade A MI369.A5 Env.
RnS-DS-SOSIP Stabilized More Strains in Prefusion-Closed Conformation Than DS-SOSIP and Chimera-DS-SOSIP
In light of the high sequence diversity of Env, each of the RnS-DS-SOSIP stabilization mutations likely contacts different neighboring residues within the 180-strain panel of diverse Envs we assessed for stabilization. On the basis of the PGT145/VRC26.25 antigenic screening criteria used in this study, the RnS-DS-SOSIP approach yielded the best overall performance in stabilizing HIV-1 strains in prefusion-closed conformations compared with DS-SOSIP (Kwon et al., 2015) and Chimera-DS-SOSIP (Joyce et al., 2017).
Among the 179 strains that have been assessed using all three approaches (DS-SOSIP, Chimera-DS-SOSIP, and RnS-DS-SOSIP), we observed 41 DS-SOSIP-, 76 Chimera-DS-SOSIP-, and 133 RnS-DS-SOSIP-stabilized strains to have appropriate PGT145/VRC26.25 prefusion-closed antigenicity, with 34 strains being stabilized using all three approaches (Figures 5A and B). Notable, 61 diverse strains were stabilized only using RnS-DS-SOSIP, and only 8 diverse strains were stabilized by either DS-SOSIP or Chimera-DS-SOSIP but not by RnS-DS-SOSIP (Figure 5B).
Figure 5. Antigenic Screening Yields 133 RnS-DS-SOSIP Prefusion-Closed Env Constructs Compared with 41 and 76 for DS-SOSIP and Chimera-DS-SOSIP.
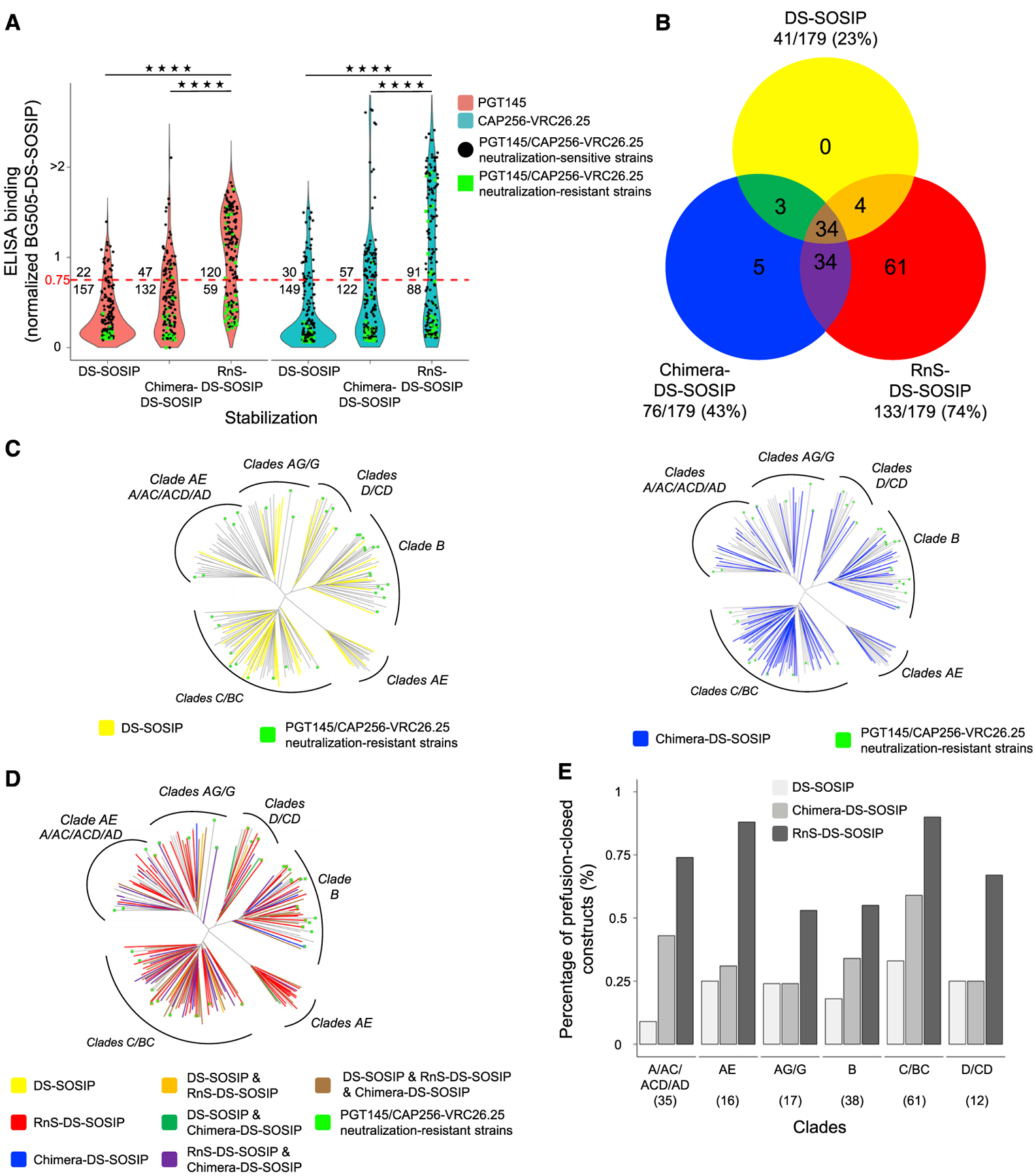
(A) Binding response toward PGT145 and CAP256-VRC26.25, relative to BG505 DS-SOSIP, for DS-SOSIP, Chimera-DS-SOSIP, and RnS-DS-SOSIP of 179 strains, including 36 PGT145 and CAP256-VRC26.25 neutralization-resistant strains. Dotted red line indicates 75% binding response relative to BG505 DS-SOSIP. The values displayed at the top of the graph indicate the number of Env molecules with binding response at least 75% that of BG505 DS-SOSIP Env. Statistical significance was assessed using the two-tailed Mann-Whitney test (****p < 0.0001).
(B) Venn diagram depicting the amount of strains stabilized in prefusion-closed form by either DS-SOSIP, Chimera-DS-SOSIP, or RnS-DS-SOSIP.
(C) Phylogenetic trees of Envs from 179 strains, with those that can be stabilized by DS-SOSIP (left) and Chimera-DS-SOSIP (right) highlighted.
(D) Phylogenetic tree of Envs from 179 strains, with those that can be stabilized by DS-SOSIP, Chimera-DS-SOSIP, and/or RnS-DS-SOSIP highlighted on the basis of the coloring scheme in (B).
(E) Bar plot highlighting the percentage of strains that can be stabilized in prefusion-closed conformation by DS-SOSIP, Chimera-DS-SOSIP, or RnS-DS-SOSIP. Number of strains per clade is depicted in parentheses.
We also analyzed the success of stabilization phylogenetically and by clade (Figures 2B and 5C–5E). Although successful RnS-DS-SOSIP stabilization was generally widespread, it reached only 55% for the clade B strains with the criteria used in this study (even though in 33 of the 38 clade B Env trimers [~87%], the RnS mutations led to an either small or large increase in the AUC [not shown]), for which a higher proportion of the sequences were resistant to both PGT145 and VRC25.26, and among those strains that could be neutralized by PGT145 or VRC25.26, a significant proportion of strains could not be stabilized into prefusion-closed conformation with appropriate antigenicity (Figure 5D). In general, however, the RnS-DS-SOSIP stabilization approach was superior independent of the clade classification, yielding 3-fold and 2-fold improvements over DS-SOSIP and Chimera-DS-SOSIP, respectively (Figure 5E).
DISCUSSION
HIV-1 Env is highly variable in sequence and conformation, two characteristics that impede the development of an efficacious vaccine that seeks to induce broadly neutralizing antibodies against HIV. The extraordinary glycosylation of Env also impedes vaccine development, as human antibodies preferentially recognize immunogenic non-glycosylated Env regions, such as those exposed on open trimers; even minor imperfections in Env folding may hamper the induction of the desired glycan shield-penetrating antibodies. For the development of an effective HIV vaccine that elicits broadly neutralizing antibodies, it may thus be very helpful to be able to produce diverse prefusion-closed trimers with high-quality folding and stability.
Substantial progress has been made in recent years to stabilize the conformation of Env on the basis of disfavoring open or postfusion conformations by introducing disulfide bridges, optimizing packing, or using stabilizing substitutions known from other Envs (Binley et al., 2000; Chuang et al., 2017; de Taeye et al., 2015; Dey et al., 2009; Garces et al., 2015; Guenaga et al., 2015, 2017; Kesavardhana and Varadarajan, 2014; Kong et al., 2016a; Kulp et al., 2017; Kwon et al., 2015; Sanders et al., 2002; Steichen et al., 2016; Sullivan et al., 2017; Torrents de la Peña et al., 2017). Very few of these studies, however, have tested these stabilization mutations on a large variety of HIV-1 strains. In this study, we combined DS-SOSIP (Kwon et al., 2015) and RnS (Rutten et al., 2018) approaches, developed an automated procedure (ADROITrimer) by which the Env sequence from any HIV-1 strain can be converted into a RnS-DS-SOSIP-stabilized Env trimer, and confirmed with antigenic assays and SEC that RnS-DS-SOSIP can stabilize Envs in the prefusion-closed conformation for the vast majority of the 180 tested HIV-1 strains.
We used 535N, 556P, 558E, 589V, 651F, 655I, and 658V for stabilization (Figure 1) which previously were shown to stabilize a number of Envs, such as Du422, ZM233M, and ZM246F (Rutten et al., 2018). To determine if the RnS-DS-SOSIP-stabilized trimers were in the desired prefusion-closed conformation, we used recognition by antibodies PGT145/VRC26.25, which recognize the V2 apex and are considered specific for the prefusion-closed conformation of the Env trimer (Figure 2). This likely underestimated the percentage of Env trimers that could fold into closed conformations, as PGT145 and VRC26.25 neutralized only 144 of the 180 strains in our test panel, and we observed high trimer content in ~80% of the Env strains on the basis of SEC (Figure 3). An alternative antigenic criterion would be low recognition by V3 antibodies specific for the open conformation (e.g., V3 antibodies 447–42D, 3074, and 2557) (Gorny et al., 2004; Jiang et al., 2010; Killikelly et al., 2013), along with good recognition by broadly neutralizing antibodies such as PGT128, PGT151, and VRC01.
We noticed that there were a number of strains resistant to PGT145 and VRC26.25 showing binding toward PGT145 or VRC26.25 when stabilized with RnS-DS-SOSIP. One of the reasons was that for a number of strains for which glycans that are critical to binding of V1V2-tageting antibodies (glycans 156 and 160) were missing, the repair process added these N-linked glycosylation sequons (glycan 156 is missing in 6 of the 180 strains, all 6 strains—0260.v5.c36, SS1196.01, CAP210.E8, ZM109.4, ZM197.7, ZM233.6—glycan 156 was repaired back; glycan 160 is missing in 16 of the 180 strains, 8 of these strains—0815.V3.C3, TH976.17, 89.6.DG, BR07.DG, CNE14, QH0692.42, 3637.V5.C3, 3337.V2.C6—glycan 160 was repaired back). Also, we note that phylogenetic and clade analysis of stabilization (Figure 5) indicated room for further improvement, as a number of strains, especially in clade B, were neutralized by PGT145 or VRC26.25, but the RnS-DS-SOSIP failed to yield pre-fusion-closed Env trimer under the criteria set in this paper. Possibly RnS-DS-SOSIP could be combined with additional stabilization to increase further the percentage of Env strains stabilized in prefusion-closed state. Additional improvements that could be explored relate to purification, as a number of procedures use affinity chromatography along with an unfolding/refolding step to remove the elute trimer (Dey et al., 2018; Sanders et al., 2013). We used lectin followed by SEC (Figure 3), and for the strains we selected for large-scale production (Figure 3), none of the Envs could be purified by 2G12/3 M MgCl2 elution, likely because of an inability to refold after elution. However, they could be purified as Fab 3BNC117-bound prefusion-closed trimers through introduction of an HRV3c-cleavage site (McLellan et al., 2011, 2013). This purification method would be useful in cases in which the remaining Fab would not interfere with downstream experiments, such as cases in which a Env:Fab complex is desired.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Gwo-Yu Chuang (gwo-yu.chuang@nih.gov).
Materials Availability
Plasmids generated in this study will be made available on request but we require a completed Materials Transfer Agreement. Plasmids for C1080.c3 RnS-DS-SOSIP, REJO.67 RnS-DS-SOSIP, 6838.V1.C35 RnS-DS-SOSIP, and MI369 RnS-DS-SOSIP are being deposited to Addgene.
Data and Code Availability
ADROITrimer is available for download at https://github.com/RedaRawi/ADROITrimer. The accession number for the coordinates of MI369.A5 RnS-DS-SOSIP in complex with a variant of antibody 3H+109L (Garces et al., 2015) and a variant of antibody 35O22 (Pancera et al., 2014) reported in this paper is PDB: 6WIX.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Lines
Freestyle 293 cells were purchased from GIBCO (cat#R79007). Expi293F were purchased from GIBCO (cat#A14527). 293T cells were purchased from ATCC (cat#CRL-3216). The above cell lines were used directly from the commercial sources and cultured according to manufacturer suggestions.
METHOD DETAILS
ADROITrimer program
ADROITrimer requires two inputs from the user to generate RnS-DS-SOSIP stabilized constructs, in particular, (i) the target HIV-1 env sequence, and (ii) the clade assignment of the target sequence. The HIV-1 filtered web alignment of 2016 obtained from the Los Alamos National Laboratory’s HIV database (https://www.hiv.lanl.gov/) was used as reference alignment. In an initial step, ADROITrimer counts how many sequences within the reference alignment have the same clade assignment as the target sequence. If greater than or equal to 50 sequences have the target sequence clade assignment, then the clade specific alignment is used. If less than 50 sequences match the target clade, then the full reference alignment is used. Second, the target sequence was aligned to the new clade reference alignment using MUSCLE (Edgar, 2004). Subsequently, a target sequence residue was designated as rare, if either it’s prevalence is below 2% within the clade reference alignment, or if it’s prevalence is between 2% and 7.5% and the residue was buried or partly buried (i.e., buried surface area was less than 30% of the highest surface area, estimated by the HIV-1 Env BG505 structure, PDB: 5FYL; buried surface area was calculated using NACCESS (http://wolf.bms.umist.ac.uk/naccess/)). In the repair step the rare residues are replaced with the consensus residues, if their minimum sequence identity is at least 60%. In the stabilization step, seven residues are mutated – 535N, 556P, 558E, 589V, 651F, 655I, and 658V (HXB2 numbering) – which were previously reported to increase trimeric yield (Rutten et al., 2018). In the final step, DS-SOSIP mutations (DS: 201C, 433C; SOSIP: 501C, 605C, and 559P) (Kwon et al., 2015; Sanders et al., 2013) are added. Finally, the cleavage site (residues 508–511) are replaced with six Arg, residues 31–35 are replaced with those from BG505 strain (“AENL”), and residues after residue 664 (Sanders et al., 2013) are truncated.
HIV Env construct expression vector
The designed RnS-DS-SOSIP constructs, with a D7324 epitope-tag sequence added at the C terminus, were cloned into the mammalian expression vector VRC8400 (Catanzaro et al., 2007).
96-well expression and antigenic analysis
24 hours prior to DNA-transient transfection, 100 μL per well of log-phase growing HEK293T cells were seeded into a 96-well micro-plate at a density of 2.5×105 cells/ml in optimized expression medium (RealFect-Medium, ABI Scientific, VA), and incubated at 37°C, 5% CO2. Prior to transfection, 40 μL per well of spent medium was removed. For transient transfection, DNA-TrueFect-Max complex was prepared by mixing 0.25 μg plasmid DNA in 10 μL per well of Opti-MEM medium (Invitrogen, CA) with 0.75 μL of TrueFect-Max transfection reagent (United BioSystems, VA) in 10 μL of Opti-MEM medium, and incubating for 15 min, and then mixed with growing cells in the 96-well plate and incubated at 37°C, 5% CO2. In day one post transfection, 30 μL per well of enriched expression medium (CelBooster Cell Growth Enhancer Medium for Adherent cell, ABI Scientific, VA) was fed. After four days post transfection, the antigenic analysis of immunogens was characterized by 96-well-formatted ELISA. Briefly, D7324 antibody-coated 96-well ELISA plates were prepared by incubating 100 μL per well of 2 μg/ml of sheep anti-HIV-1 gp120 antibody (D7324) (Aalto Bio Reagents, Ireland, catalog # D7324) in phosphate buffered saline (PBS) in 96-Well Flat-Bottom Immuno Plate (Nunc, Thermo, IL) overnight at 4°C, followed by the removal of the coating solution and incubation of 200 μL per well of CelBooster Cell Growth Enhancer Medium (ABI Scientific, VA) for 2 hours at room temperature (RT). After washing with PBS + 0.05% Tween 20, 30 μL per well of the expressed supernatant mixed with 70 μL per well of PBS was incubated for two hours at room temperature (RT). After washing, 100 μL per well of 10 μg/ml primary antibody in 50% CelBooster Cell Growth Enhancer Medium and PBS with 0.02% tween 20 was incubated for 1 hour at RT. After washing, 100 μL per well of Horseradish peroxidase (HRP)-conjugated goat anti-human IgG antibody (Jackson ImmunoResearch Laboratories Inc., PA), diluted at 1:10,000 in CelBooster Cell Growth Enhancer Medium with 0.02% tween 20, was incubated for 30 min at RT. After washing, the reaction signal was developed using 100 μL of BioFX-TMB (SurModics, MN) at RT for 10 min, and then stopped with 100 μL of 0.5 N H2SO4. The readout was measured at a wavelength of 450 nm, and OD450 values were normalized and analyzed. All samples were performed in duplicate. Following antibodies were assessed: CAP256-VRC26.25 (Doria-Rose et al., 2015), PGT145 (Walker et al., 2011), PGT151 (Falkowska et al., 2014), VRC01 (Zhou et al., 2010), VRC34.01 (Kong et al., 2016b), 3BNC117 (Scheid et al., 2011), N6 (Huang et al., 2016), F105 (Chen et al., 2009), 17b (Kwong et al., 1998), 17b + CD4, and 447–52D (Killikelly et al., 2013).
Analytical SEC
An ultra high-performance liquid chromatography system (Vanquish, Thermo Scientific) was used for performing the analytical SEC experiment using Expi293F cell culture supernatants. Centrifuged crude cell supernatant supernatants were applied to a Unix-C SEC 300 4.6×150 mm column, with the corresponding guard column (Sepax) equilibrated in running buffer (150 mM sodium phosphate, 50 mM NaCl, pH 7.0) at 0.3 mL/min. The signal of supernatants of non-transfected cells was subtracted from the signal of supernatants of Env transfected cells.
AlphaLISA®
AlphaLISA® (Perkin-Elmer) is a bead-based proximity assay in which singlet oxygen molecules, generated by high energy irradiation of Donor beads, transfers to Acceptor beads, which are within a distance of approximately 200 nm. It is a sensitive high throughput screening assay that does not require washing steps. A cascading series of chemical reactions results in a chemiluminescent signal (Eglen et al., 2008). The HIV constructs were expressed in Expi293F cells, which were cultured for 3 days in 96 well plates (200 μl/well). Crude supernatants were diluted 120 times in AlphaLISA® buffer (PBS + 0.05% Tween-20 + 0.5 mg/mL BSA). Subsequently 10 μl of these dilutions were transferred to a half-area 96-well plate and mixed with 40μl acceptor beads, mixed with donor beads and mAb. The beads were mixed well before use. After 2 hours of incubation at RT, non-shaking, the signal was measured with Neo (BioTek) The donor beads were conjugated to Streptavidin (Cat#: 6760002B, Perkin Elmer), which could bind to the mAb, modified with Biotin using Sortase A. The acceptor beads were conjugated to anto-V5 mAb (Cat#: AL129R, Perkin Elmer), which could bind to the mAb, modified with a V5 peptide using Sortase A. The acceptor beads were conjugated to PGT128 and PGT145, PGDM1400 (Sok et al., 2014) and PGT128 (Walker et al., 2011) were conjugated to donor beads. The average signal of mock transfections (no Env) was subtracted from the AlphaLISA counts measured for the different Env proteins. As a reference the BG505_SOSIP Env plasmid was used. For normalization, the data was divided by the BG505_SOSIP signal.
Expression and purification of select HIV Env constructs by 3BNC117-affinity and HRV3c-column cleavage
HIV Env trimers were expressed as previously described (Pancera et al., 2014; Rutten et al., 2018; Sanders et al., 2013). Briefly, 1L of Freestyle 293 cells (GIBCO) were transiently co-transfected with HIV Env and furin DNA. The cells were allowed to grow for 6 days, after which they were spun down and the media collected. The 3BNC117 - HRV 3C modified antibody was expressed by transient transfection of Expi293F cells (GIBCO) with the heavy and light chains DNAs. The cells were boosted 24 hours after transfection with 80ml per liter of AbBooster Enhancer (ABI Scientific) and the cells were grown at 32°C for 5 additional days. Following clarification, the two protein containing medias were mixed at a ratio of 1:8 of 3BNC117 to HIV Env. The crude proteins were allowed to incubate for 1 hour at 4°C after which Protein A resin (GE Healthcare) was added. After incubation the resin was washed with PBS, and HRV 3C was added. The protease was reacted overnight at 4°C, and the following day the cleaved antibody:Env complex was collected.
Protein complexes were further purified by size-exclusion chromatography using a Superdex-200 column in PBS. Following SEC, proteins were concentrated to 1mg/mL, with an addition of 10% glycerol, and flash frozen in liquid nitrogen. Antigenic characterization against CAP256-VRC26.25 (Doria-Rose et al., 2015), PGT145 (Walker et al., 2011), VRC01 (Zhou et al., 2010), PGT151 (Falkowska et al., 2014), VRC34.01 (Kong et al., 2016b), and VRC-PG05 (Zhou et al., 2018) was performed on an Octet HTX (ForteBio) using Anti-Human IgG Fc Capture (AHC) biosensors, and all samples were used at 50μg/mL. Motavizumab (Wu et al., 2007), an RSV antibody, was used as a negative control.
Purification of select HIV Env constructs by lectin affinity
For purification of unliganded HIV Env, lectin based purification was used as has been previously described (Verkerke et al., 2016). Briefly, the supernatants containing either DS-SOSIP or DS-SOSIP-RnS Env’s were incubated for 2 hours with Galanthus nivalis lectin agarose (Sigma). The resin was washed with PBS, and protein eluted with 300mM alpha methyl-mannopyrannoside in PBS. The HIV Env was applied to a Superdex S-200 gel filtration column in PBS, after which it was concentrated to 1mg/ml for assay and storage.
Negative stain EM
Proteins were diluted to a concentration of about 0.02 mg/ml with buffer containing 10 mM HEPES, pH 7.0, and 150 mM NaCl, adsorbed to a glow-discharged carbon-coated copper grid, washed with the same buffer, and stained with 0.75% uranyl formate. Images were recorded at a nominal magnification of 100,000 (pixel size: 0.22 nm) using SerialEM (Mastronarde, 2005) on an FEI Tecnai T20 electron microscope equipped with a 2k × 2k Eagle CCD camera and operated at 200 kV. Particles were selected from the micrographs automatically using in-house written software (Y.T., unpublished data) and extracted into 128×128-pixel boxes. Reference-free 2D classification was performed using Relion Scheres (Scheres, 2012).
HIV-1 Env Trimer-Fab Complex Preparation and Crystallization
Crystallization and structure determination of MI369 was aided by the use of designed antibody scaffold which have been previously shown to improve crystal growth and diffraction resolution (Lai et al., 2019). Purified MI369 HIV-1 Env was mixed in a 1:1.2:1.2 molar with a variant of 3H+109L (Garces et al., 2015) Fab and a variant of 35O22 (Pancera et al., 2014) scFv, and incubated overnight at room temperature. The antibody:Env complex was purified by size exclusion chromatography on a Superdex S-200 column. Crystal drops were setup by mixing 1μL protein at 10–15mg/ml with 1μL 180mM LiSO4, 90mM Imidazole pH 6.5, 4.5% MPD, 7.4% PEG 3,350. After crystals were grown, they were cryo-protected with mother liquor supplemented with 15% (2R,3R)-(−)-2,3-Butanediol and flash frozen in liquid nitrogen.
X-ray data collection, structure solution, model building, and refinement
Diffraction data was collected at the 22ID beamline of the Advanced Photo Source at the Argonne national lab. The data was indexed and scaled using HKL2000, and then submitted to the Diffraction Anisotropy Server (Strong et al., 2006). This was done to compensate for the high anisotropy of the collected data, with the highest resolution diffraction appearing at 2.67Å, but only being reasonably complete to 3.5Å. Molecular replacement was performed in Phaser, and iterative model building and structure refinement were performed in Coot and Phenix Refine (Liu et al., 2019) respectively. Statistics in Table S2 are given according to a high-resolution shell of 3.5Å, as this is the better descriptor of the quality of the maps and structure.
QUANTIFICATION AND STATISTICAL ANALYSIS
Two tailed Mann-Whitney test was used to determine statistical significance in Figures 2C, 2E, and 5A. Pearson correlation was used to determine the correlation in Figure 2F. R was used for statistical analysis. Additional details can be found in respective figure legends.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| 17b | (Kwong et al., 1998) | N/A |
| 35O22 variant | (Lai et al., 2019) | N/A |
| 3BNC117 | (Scheid et al., 2011) | RRID:AB_2491033 |
| 3H+109L variant | (Lai et al., 2019) | N/A |
| 447–52D | (Killikelly et al., 2013) | RRID:AB_2491016 |
| CAP256-VRC26.25 | (Doria-Rose et al., 2015) | N/A |
| F105 | (Chen et al., 2009) | N/A |
| Motavizumab | (Wu et al., 2007) | N/A |
| N6 | (Huang et al., 2016) | N/A |
| PGDM1400 | (Sok et al., 2014) | N/A |
| PGT128 | (Walker et al., 2011) | RRID:AB_2491047 |
| PGT145 | (Walker et al., 2011) | RRID:AB_2491054 |
| PGT151 | (Falkowska et al., 2014) | N/A |
| VRC-PG05 | (Zhou et al., 2018) | N/A |
| VRC01 | (Zhou et al., 2010) | RRID:AB_2491019 |
| VRC34.01 | (Kong et al., 2016b) | RRID:AB_2819225 |
| Bacterial and Virus Strains | ||
| VRC 180 HIV-1 virus panel | (Joyce et al., 2017) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Unix-C SEC 300 4.6×150 mm | Sepax | Cat#231300–4615 |
| Unix-C SEC 300 4.6×50 mm (guard) | Sepax | Cat#231300–4602 |
| Streptavidin donor beads | Perkin-Elmer | Cat#6760002B |
| Anti-V5 acceptor beads | Perkin-Elmer | Cat#AL129R |
| ExpiFectamine 293 Transfection Kit | ThermoFischer | Cat#A14525 |
| Expi293 Expression Medium | ThermoFischer | Cat#A1435101 |
| Opti-MEM I Reduced Serum Medium | ThermoFischer | Cat#31985062 |
| Turbo293 Transfection Reagent | SPEED Biosystems | PXX1002 |
| GIBCO FreeStyle 293 Expression Medium | GIBCO | 12338018 |
| GIBCO Expi293 Expression Medium | GIBCO | A1435102 |
| TrueFect-Max Transfection Reagent | United BioSystems | TM5501 |
| Expression Medium, CelBooster | ABI Scientific | TM9000, PB2462 |
| Lectin agarose (Galanthus nivalis) | Sigma | L8775 |
| Methyl α-D-mannopyranoside | Sigma | M6882 |
| Deposited Data | ||
| MI369 crystal structure | RCSB Protein Data Bank | PDB: 6WIX |
| Experimental Models: Cell Lines | ||
| Freestyle 293 cells | GIBCO | R79007 |
| Expi293F | GIBCO | A14527 |
| 293T cell line | ATCC | CRL-3216 |
| Recombinant DNA | ||
| RnS-DS-SOSIP of 180 HIV-1 strain | This paper (Data S1) | N/A |
| Software and Algorithms | ||
| R | The R Project for Statistical Computing | https://www.r-project.org/ |
| Muscle | (Edgar, 2004) | https://www.drive5.com/muscle/ |
| Astra 7.3 software package | Wyatt Technology | https://www.wyatt.com/products/software/astra.html |
| Chromeleon 7.2.8.0 software package | Thermo Fisher Scientific | https://www.thermofisher.com/order/catalog/product/CHROMELEON7 |
| SerialEM | (Mastronarde, 2005) | https://bio3d.colorado.edu/SerialEM/ |
| Relion | (Scheres 2012) | https://www3.mrc-lmb.cam.ac.uk/relion/index.php?title=Main_Page |
| HLK2000 | HKL Research, Inc. | https://www.hkl-xray.com/ |
| Phenix | Liebschner et al., 2019 | https://www.phenix-online.org/ |
| ADROITrimer | This paper | https://github.com/RedaRawi/ADROITrimer |
Highlights.
Automated ADROITrimer pipeline enables prefusion-closed stabilization of HIV Env
134 of 180 RnS-DS-SOSIP Env variants are recognized by PGT145 or CAP256-VRC26.25
RnS-DS-SOSIP yields soluble Env trimers for 143 of 179 strains as confirmed by SEC
3.5 Å resolution structure of a clade A RnS-DS-SOSIP reveals stabilization details
ACKNOWLEDGMENTS
We thank J. Stuckey for assistance with figures and the Structural Biology Section and Structural Bioinformatics Core at the NIH Vaccine Research Center for helpful discussions and comments on the manuscript. We thank J. Baalwa, D. Ellenberger, F. Gao, B. Hahn, K. Hong, J. Kim, F. McCutchan, D. Montefiori, L. Morris, E. Sanders-Buell, G. Shaw, R. Swanstrom, M. Thomson, S. Tovanabutra, C. Williamson, and L. Zhang for contributing the HIV-1 Env plasmids used in our 180-strain panel. Support for this work was provided by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), United States; by the Office of AIDS Research, NIH, United States; and by the International AIDS Vaccine Initiative (IAVI) Neutralizing Antibody Consortium, United States. This project has been funded in part with federal funds from Frederick National Laboratory for Cancer Research, NIH, United States, under contract HHSN261200800001E (Y.T.). Use of sector 22 (Southeast Region Collaborative Access team) at the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract W-31-109-Eng-38.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108432.
DECLARATION OF INTERESTS
These studies were funded in part by Janssen Vaccines and Prevention. L.R., S.B., J.J., and J.P.M.L. are employees of Janssen. L.R. and J.P.M.L. are inventors on an international patent application describing trimer stabilizing HIV envelope protein mutations.
REFERENCES
- Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, and Moore JP (2000). A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol 74, 627–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricault CA, Yusim K, Seaman MS, Yoon H, Theiler J, Giorgi EE, Wagh K, Theiler M, Hraber P, Macke JP, et al. (2019). HIV-1 neutralizing antibody signatures and application to epitope-targeted vaccine design. Cell Host Microbe 25, 59–72.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzaro AT, Roederer M, Koup RA, Bailer RT, Enama ME, Nason MC, Martin JE, Rucker S, Andrews CA, Gomez PL, et al. ; VRC 007 Study Team (2007). Phase I clinical evaluation of a six-plasmid multiclade HIV-1 DNA candidate vaccine. Vaccine 25, 4085–4092. [DOI] [PubMed] [Google Scholar]
- Chen L, Kwon YD, Zhou T, Wu X, O’Dell S, Cavacini L, Hessell AJ, Pancera M, Tang M, Xu L, et al. (2009). Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science 326, 1123–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Pancera M, Bossert A, Schmidt SD, Chen RE, Chen X, Druz A, Narpala S, Doria-Rose NA, McDermott AB, et al. (2015). Immunogenicity of a Prefusion HIV-1 Envelope Trimer in Complex with a Quaternary-Structure-Specific Antibody. J. Virol 90, 2740–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang GY, Geng H, Pancera M, Xu K, Cheng C, Acharya P, Chambers M, Druz A, Tsybovsky Y, Wanninger TG, et al. (2017). Structure-based design of a soluble prefusion-closed HIV-1 Env trimer with reduced CD4 affinity and improved immunogenicity. J. Virol 91, e02268–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Taeye SW, Ozorowski G, Torrents de la Peña A, Guttman M, Julien JP, van den Kerkhof TL, Burger JA, Pritchard LK, Pugach P, Yasmeen A, et al. (2015). Immunogenicity of stabilized HIV-1 envelope trimers with reduced exposure of non-neutralizing epitopes. Cell 163, 1702–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Taeye SW, Moore JP, and Sanders RW (2016). HIV-1 envelope trimer design and immunization strategies to induce broadly neutralizing antibodies. Trends Immunol. 37, 221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey B, Svehla K, Xu L, Wycuff D, Zhou T, Voss G, Phogat A, Chakrabarti BK, Li Y, Shaw G, et al. (2009). Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced co-receptor binding site. PLoS Pathog. 5, e1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey AK, Cupo A, Ozorowski G, Sharma VK, Behrens AJ, Go EP, Ketas TJ, Yasmeen A, Klasse PJ, Sayeed E, et al. (2018). cGMP production and analysis of BG505 SOSIP.664, an extensively glycosylated, trimeric HIV-1 envelope glycoprotein vaccine candidate. Biotechnol. Bioeng 115, 885–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose NA, Bhiman JN, Roark RS, Schramm CA, Gorman J, Chuang GY, Pancera M, Cale EM, Ernandes MJ, Louder MK, et al. (2015). New member of the V1V2-directed CAP256-VRC26 lineage that shows increased breadth and exceptional potency. J. Virol 90, 76–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eglen RM, Reisine T, Roby P, Rouleau N, Illy C, Bossé R, and Bielefeld M (2008). The use of AlphaScreen technology in HTS: current status. Curr. Chem. Genomics 1, 2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkowska E, Le KM, Ramos A, Doores KJ, Lee JH, Blattner C, Ramirez A, Derking R, van Gils MJ, Liang CH, et al. (2014). Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity 40, 657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Tran K, Bale S, Kumar S, Guenaga J, Wilson R, de Val N, Arendt H, DeStefano J, Ward AB, and Wyatt RT (2016). Thermostability of well-ordered HIV spikes correlates with the elicitation of autologous tier 2 neutralizing antibodies. PLoS Pathog. 12, e1005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garces F, Lee JH, de Val N, de la Pena AT, Kong L, Puchades C, Hua Y, Stanfield RL, Burton DR, Moore JP, et al. (2015). Affinity maturation of a potent family of HIV antibodies is primarily focused on accommodating or avoiding glycans. Immunity 43, 1053–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Revesz K, Williams C, Volsky B, Louder MK, Anyangwe CA, Krachmarov C, Kayman SC, Pinter A, Nadas A, et al. (2004). The v3 loop is accessible on the surface of most human immunodeficiency virus type 1 primary isolates and serves as a neutralization epitope. J. Virol 78, 2394–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenaga J, Dubrovskaya V, de Val N, Sharma SK, Carrette B, Ward AB, and Wyatt RT (2015). Structure-guided redesign increases the propensity of HIV Env to generate highly stable soluble trimers. J. Virol 90, 2806–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenaga J, Garces F, de Val N, Stanfield RL, Dubrovskaya V, Higgins B, Carrette B, Ward AB, Wilson IA, and Wyatt RT (2017). Glycine substitution at helix-to-coil transitions facilitates the structural determination of a stabilized subtype C HIV envelope glycoprotein. Immunity 46, 792–803.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SC (2015). Viral membrane fusion. Virology 479–480, 498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Kumar S, Allen JD, Huang D, Lin X, Mann CJ, Saye-Francisco KL, Copps J, Sarkar A, Blizard GS, et al. (2018). HIV-1 vaccine design through minimizing envelope metastability. Sci. Adv 4, eaau6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffenberg S, Powell R, Carpov A, Wagner D, Wilson A, Kosakovsky Pond S, Lindsay R, Arendt H, Destefano J, Phogat S, et al. (2013). Identification of an HIV-1 clade A envelope that exhibits broad antigenicity and neutralization sensitivity and elicits antibodies targeting three distinct epitopes. J. Virol 87, 5372–5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Kang BH, Ishida E, Zhou T, Griesman T, Sheng Z, Wu F, Doria-Rose NA, Zhang B, McKee K, et al. (2016). Identification of a CD4-binding-site antibody to HIV that evolved near-pan neutralization breadth. Immunity 45, 1108–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Burke V, Totrov M, Williams C, Cardozo T, Gorny MK, Zolla-Pazner S, and Kong XP (2010). Conserved structural elements in the V3 crown of HIV-1 gp120. Nat. Struct. Mol. Biol 17, 955–961. [DOI] [PubMed] [Google Scholar]
- Joyce MG, Georgiev IS, Yang Y, Druz A, Geng H, Chuang GY, Kwon YD, Pancera M, Rawi R, Sastry M, et al. (2017). Soluble prefusion closed DS-SOSIP.664-Env trimers of diverse HIV-1 strains. Cell Rep. 21, 2992–3002. [DOI] [PubMed] [Google Scholar]
- Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, et al. (2013). Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342, 1477–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien JP, Lee JH, Ozorowski G, Hua Y, Torrents de la Peña A, de Taeye SW, Nieusma T, Cupo A, Yasmeen A, Golabek M, et al. (2015). Design and structure of two HIV-1 clade C SOSIP.664 trimers that increase the arsenal of native-like Env immunogens. Proc. Natl. Acad. Sci. U S A 112, 11947–11952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesavardhana S, and Varadarajan R (2014). Stabilizing the native trimer of HIV-1 Env by destabilizing the heterodimeric interface of the gp41 postfusion six-helix bundle. J. Virol 88, 9590–9604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killikelly A, Zhang HT, Spurrier B, Williams C, Gorny MK, Zolla-Pazner S, and Kong XP (2013). Thermodynamic signatures of the antigen binding site of mAb 447–52D targeting the third variable region of HIV-1 gp120. Biochemistry 52, 6249–6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasse PJ, LaBranche CC, Ketas TJ, Ozorowski G, Cupo A, Pugach P, Ringe RP, Golabek M, van Gils MJ, Guttman M, et al. (2016). Sequential and simultaneous immunization of rabbits with HIV-1 envelope glycoprotein SOSIP.664 trimers from clades A, B and C. PLoS Pathog. 12, e1005864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, He L, de Val N, Vora N, Morris CD, Azadnia P, Sok D, Zhou B, Burton DR, Ward AB, et al. (2016a). Uncleaved prefusion-optimized gp140 trimers derived from analysis of HIV-1 envelope metastability. Nat. Commun 7, 12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong R, Xu K, Zhou T, Acharya P, Lemmin T, Liu K, Ozorowski G, Soto C, Taft JD, Bailer RT, et al. (2016b). Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science 352, 828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp DW, Steichen JM, Pauthner M, Hu X, Schiffner T, Liguori A, Cottrell CA, Havenar-Daughton C, Ozorowski G, Georgeson E, et al. (2017). Structure-based design of native-like HIV-1 envelope trimers to silence non-neutralizing epitopes and eliminate CD4 binding. Nat. Commun 8, 1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YD, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, Joyce MG, Guttman M, Ma X, Narpala S, et al. (2015). Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat. Struct. Mol. Biol 22, 522–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, and Hendrickson WA (1998). Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai YT, Wang T, O’Dell S, Louder MK, Schön A, Cheung CSF, Chuang GY, Druz A, Lin B, McKee K, et al. (2019). Lattice engineering enables definition of molecular features allowing for potent small-molecule inhibition of HIV-1 entry. Nat. Commun 10, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D, Afonine PV, Baker ML, Bunkoczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, et al. (2019). Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol 75, 861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Lai YT, Zhang P, Louder MK, Pegu A, Rawi R, Asokan M, Chen X, Shen CH, Chuang GY, et al. (2019). Improvement of antibody functionality by structure-guided paratope engraftment. Nat. Commun 10, 721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, et al. (2013). Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342, 1484–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Murillo P, Tran K, Guenaga J, Lindgren G, Adori M, Feng Y, Phad GE, Vazquez Bernat N, Bale S, Ingale J, et al. (2017). Particulate array of well-ordered HIV clade C Env trimers elicits neutralizing antibodies that display a unique V2 cap approach. Immunity 46, 804–817.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol 152, 36–51. [DOI] [PubMed] [Google Scholar]
- McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, et al. (2011). Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480, 336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JS, Chen M, Joyce MG, Sastry M, Stewart-Jones GB, Yang Y, Zhang B, Chen L, Srivatsan S, Zheng A, et al. (2013). Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science 342, 592–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, et al. (2014). Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Changela A, and Kwong PD (2017). How HIV-1 entry mechanism and broadly neutralizing antibodies guide structure-based vaccine design. Curr. Opin. HIV AIDS 12, 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauthner M, Havenar-Daughton C, Sok D, Nkolola JP, Bastidas R, Boopathy AV, Carnathan DG, Chandrashekar A, Cirelli KM, Cottrell CA, et al. (2017). Elicitation of robust tier 2 neutralizing antibody responses in nonhuman primates by HIV envelope trimer immunization using optimized approaches. Immunity 46, 1073–1088.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringe RP, Sanders RW, Yasmeen A, Kim HJ, Lee JH, Cupo A, Korzun J, Derking R, van Montfort T, Julien JP, et al. (2013). Cleavage strongly influences whether soluble HIV-1 envelope glycoprotein trimers adopt a native-like conformation. Proc. Natl. Acad. Sci. U S A 110, 18256–18261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutten L, Lai YT, Blokland S, Truan D, Bisschop IJM, Strokappe NM, Koornneef A, van Manen D, Chuang GY, Farney SK, et al. (2018). A universal approach to optimize the folding and stability of prefusion-closed HIV-1 envelope trimers. Cell Rep. 23, 584–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, and Moore JP (2017). Native-like Env trimers as a platform for HIV-1 vaccine design. Immunol. Rev 275, 161–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, Paluch M, Berkhout B, Maddon PJ, Olson WC, et al. (2002). Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol 76, 8875–8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Peña AT, Korzun J, et al. (2013). A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 9, e1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders KO, Nicely NI, Wiehe K, Bonsignori M, Meyerhoff RR, Parks R, Walkowicz WE, Aussedat B, Wu NR, Cai F, et al. (2017). Vaccine elicitation of high mannose-dependent neutralizing antibodies against the V3-glycan broadly neutralizing epitope in nonhuman primates. Cell Rep. 18, 2175–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, et al. (2011). Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333, 1633–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D, van Gils MJ, Pauthner M, Julien JP, Saye-Francisco KL, Hsueh J, Briney B, Lee JH, Le KM, Lee PS, et al. (2014). Recombinant HIV envelope trimer selects for quaternary-dependent antibodies targeting the trimer apex. Proc. Natl. Acad. Sci. U S A 111, 17624–17629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steichen JM, Kulp DW, Tokatlian T, Escolano A, Dosenovic P, Stanfield RL, McCoy LE, Ozorowski G, Hu X, Kalyuzhniy O, et al. (2016). HIV vaccine design to target germline precursors of glycan-dependent broadly neutralizing antibodies. Immunity 45, 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, and Eisenberg D (2006). Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U S A 103, 8060–8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JT, Sulli C, Nilo A, Yasmeen A, Ozorowski G, Sanders RW, Ward AB, Klasse PJ, Moore JP, and Doranz BJ (2017). High-throughput protein engineering improves the antigenicity and stability of soluble HIV-1 envelope glycoprotein SOSIP trimers. J. Virol 91, e00862–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrents de la Peña A, Julien JP, de Taeye SW, Garces F, Guttman M, Ozorowski G, Pritchard LK, Behrens AJ, Go EP, Burger JA, et al. (2017). Improving the immunogenicity of native-like HIV-1 envelope trimers by hyperstabilization. Cell Rep. 20, 1805–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerke HP, Williams JA, Guttman M, Simonich CA, Liang Y, Filipavicius M, Hu SL, Overbaugh J, and Lee KK (2016). Epitope-independent purification of native-like envelope trimers from diverse HIV-1 isolates. J. Virol 90, 9471–9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, et al. ; Protocol G Principal Investigators (2011). Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477, 466–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward AB, and Wilson IA (2017). The HIV-1 envelope glycoprotein structure: nailing down a moving target. Immunol. Rev 275, 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Parast AB, Richardson BA, Nduati R, John-Stewart G, Mbori-Ngacha D, Rainwater SM, and Overbaugh J (2006). Neutralization escape variants of human immunodeficiency virus type 1 are transmitted from mother to infant. J. Virol 80, 835–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Pfarr DS, Johnson S, Brewah YA, Woods RM, Patel NK, White WI, Young JF, and Kiener PA (2007). Development of motavizumab, an ultra-potent antibody for the prevention of respiratory syncytial virus infection in the upper and lower respiratory tract. J. Mol. Biol 368, 652–665. [DOI] [PubMed] [Google Scholar]
- Wyatt R, and Sodroski J (1998). The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280, 1884–1888. [DOI] [PubMed] [Google Scholar]
- Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, et al. (2010). Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329, 811–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Zheng A, Baxa U, Chuang GY, Georgiev IS, Kong R, O’Dell S, Shahzad-Ul-Hussan S, Shen CH, Tsybovsky Y, et al. (2018). A neutralizing antibody recognizing primarily N-linked glycan targets the silent face of the HIV envelope. Immunity 48, 500–513.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ADROITrimer is available for download at https://github.com/RedaRawi/ADROITrimer. The accession number for the coordinates of MI369.A5 RnS-DS-SOSIP in complex with a variant of antibody 3H+109L (Garces et al., 2015) and a variant of antibody 35O22 (Pancera et al., 2014) reported in this paper is PDB: 6WIX.


