Graphical abstract
Keywords: Parkinson’s disease, DNA methylation, Database
Abstract
Parkinson’s disease (PD) is the second most common neurodegenerative disease, of which the histopathological hallmark is the formation of Lewy bodies consisting of α-synuclein as the major component. α-Synuclein can sequester DNA Methyltransferase 1 (DNMT1), the maintenance DNA methylation enzyme, from the nucleus and into the cytoplasm, leading to global DNA hypomethylation in human brain. As DNA methylation is a major epigenetic modification that regulates gene expression and there is no specific database storing PD associated methylation information, PDmethDB (Parkinson’s Disease Methylation Database) aims to curate PD associated methylation information from literature to facilitate the study of the relationship between PD and methylation. Currently, PDmethDB contains 97,077 PD methylation associated entries among 12,308 molecules, 37,944 CpG sites, 31 tissues and 3 species through a review of about 1600 published papers. This includes information concerning the gene/molecule name, CpG site, methylation alteration, expression alteration, tissue, PMID, experimental method, and a brief description about the entry. PDmethDB provides a user-friendly interface to search, browse, download and submit data. PDmethDB supports browsing by molecule, species, tissue, gene region, methylation alteration and experimental methods. PDmethDB also shows the entry gene interaction network including protein–protein interactions and miRNA-targets interactions with a highlight of PD associated genes from DisGeNET database. PDmethDB aims to facilitate the understanding of the relationship between PD and methylation.
Database URL: https://ageing.shinyapps.io/pdmethdb/
1. Introduction
Parkinson’s disease (PD) is the second most common neurodegenerative disease and mainly affects the motor system of patients [1]. Its symptoms include tremor, slowed movement, rigid muscle, and impaired balance [2]. The hallmark of PD is the formation of Lewy bodies with α-synuclein (SNCA) as the major protein component [3]. Although the causes for PD remain unclear, genetic and environmental factors appear to be associated with PD [4]. Meta-analysis of genome-wide association studies (GWAS) have identified 90 genetic variants that account for 16–36% of the heritable risk of PD [5]. The mutations of SNCA (A53T, A30P and E46K) are associated with familial PD [6]. In addition, intron 1 of SNCA is hypomethylated in the substantia nigra of sporadic PD patients [7], [8]. Moreover, SNCA can sequester DNA Methyltransferase 1 (DNMT1), the DNA methylation enzyme, from the nucleus to the cytoplasm, leading to global DNA hypomethylation in human brain [9]. An increasing body of research has aimed to determine the role(s) of methylation alteration in PD pathogenesis [10], [11], [12], [13]. Nonetheless, a gene may have different methylation patterns in different tissues or brain regions. For example, SNCA has two significantly hypermethylated CpG sites in parietal cortex of PD patients than controls, while several significantly hypomethylated CpG sites in occipital cortex and substantia nigra [13]. Moreover, there remains some controversy regarding the relationship between specific methylation alterations and PD. Some researchers observed no significant methylation alteration in the intron 1 region of SNCA in the leukocyte of PD patients by pyrosequencing analysis [14], while others found significant hypomethylation in the same region [15]. All results identified by previous analysis might therefore be helpful to resolve conflicting results, aid in meta-analysis, and facilitate data integration. However, no specific database stores PD associated methylation alteration information to our knowledge.
A current PD database (PDGene) contains only genetic association results [16], [17] or speech recordings [18] from patients. Alternatively, MethHC database contains methylation results but only from cancer [19], while Encode has a wealth of methylation and other epigenetic data, but not from PD patients [20]. Additionally, Gene Expression Omnibus (GEO) [21] and the Michael J. Fox Foundation contain some DNA methylation datasets from PD patients, but require considerable effort to obtain and analyze specific PD associated methylation alterations. Therefore, a user-friendly curated database focused on PD associated methylation alteration is needed.
To facilitate the study of the relationship between PD and methylation alteration, we have developed a manually curated database, Parkinson’s Disease Methylation Database (PDmethDB), to collect and make available PD associated methylation information from related literature. We expect PDmethDB can serve as an important tool and resource for studies in this area.
2. Materials and methods
2.1. Data collection and implementation
In order to collect available PD methylation associated papers, we used keywords, “Parkinson AND methylation”, to search PubMed, Scopus, and Embase databases and got 1019, 884, and 1099 related papers on Sept. 15, 2020 from PubMed, Scopus, and Embase, respectively. Table S1 shows the titles, publication year and PubMed ID (PMID) list of these papers. Then, we manually retrieved PD associated methylation alteration information from these papers. We collected tissue, gene/molecule name, CpG site, gene region (e.g. 5’UTR, TSS1500, TSS200, and Intron), expression alteration (e.g. up-regulation, and down-regulation), methylation alteration (e.g. hypomethylation, and hypermethylation), experimental method (e.g. Illumina Infinium HumanMethylation450 BeadChip, pyrosequencing, and review), species (human, mouse, rat), hyperlink to PubMed records, a brief description about the relationship between the methylation alteration and PD, the title of the paper, the interactors with ‘gene/molecule’ mentioned-above, the alteration of the interactor, hyperlinks to HGNC [22], Ensembl [23], and NCBI [24] database using HGNC approved symbol, Ensembl gene ID, and Entrez gene ID, and detailed information about the gene including Gene_biotype (e.g. protein_coding, lincRNA, and miRNA), chromosome name, band, and strand information. Eventually, we curated a total of 97,077 PD methylation associated entries. The methylation alterations of these entries include ‘Hypermethylation’, ‘Hypomethylation’, ’Methylation alteration’, ‘No significant methylation alteration’, ‘Differential methylation involved in hemispheric asymmetry’ etc. Each entry contains the methylation associated gene or/and CpG site information, totally about 12,308 genes and 37,944 CpG sites. The relationship between PD and methylation for specific entry might exist in specific tissue. Therefore, PDmethDB curated tissue and the relationship information, simultaneously. PDmethDB contains the relationship between PD and methylation from about 32 tissues including ‘Substantia nigra’, ‘Cortex’, ‘Putamen’, ‘Cerebrospinal fluid’, and ‘Leukocyte’ etc. These entries are from 3 species including human, mouse, and rat. Each entry was allocated an accession number (e.g. Entry 1, and Entry 2) according to the publish time of the related paper. Additionally, we collected PD associated genes from DisGeNET v6.0 database [25], protein–protein interactions from HPRD (release 9) database [26], and miRNA-targets interactions from miRTarBase (version 7.0) [27] to construct a gene associated interaction network.
The database was constructed by the “shiny” package [28]. The data tables for searching or browsing results are fulfilled by the “DT” package [29]. The network visualization is fulfilled by the “networkD3” package [30]. The word cloud, pie chart, and bar chart are implemented by the “ECharts2Shiny” package [31]. The submission function is fulfilled by the “shinyjs” package [32].
3. Results
3.1. User interface
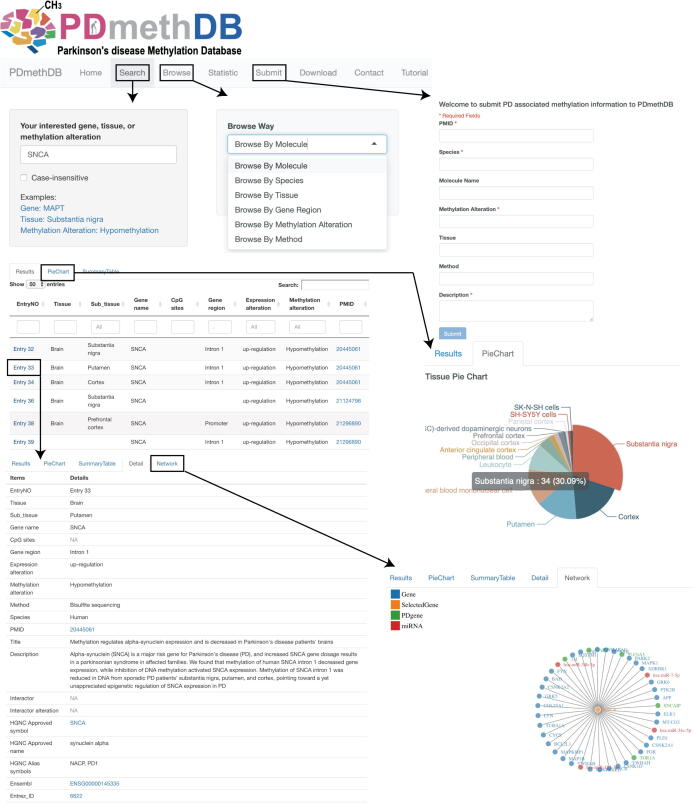
PDmethDB provides interfaces for searching, browsing, submission, downloading data, contact, and network visualization (Fig. 1). In the search page, users can search the PDmethDB using gene symbol (e.g. SNCA, or MAPT), tissue name (e.g. substantia nigra, or cortex), and methylation alteration (e.g. hypomethylation, or hypermethylation). The checkbox named ‘Case-insensitive’ can fulfill case-sensitive and case-insensitive searching. Additionally, PDmethDB provides six ways to browse: Browse by Molecule, Browse by Species, Browse by Tissue, Browse by Gene Region, Browse by Methylation Alteration, and Browse by Method. By clicking the “EntryNO” column of the searching or browsing results, PDmethDB will go to the “Detail” tab. The “Detail” tab shows all information retrieved from the corresponding paper and detailed information about the entry gene (e.g. gene_biotype, chromosome name, and band) and hyperlinks to HGNC, Ensembl, and NCBI database that makes it easier to extend a query regarding a gene. If the gene in the entry has protein–protein interactions in the HPRD database or miRNA-Targets interactions in miRTarBase, PDmethDB will also show the “network” tab which displays the interactions of the entry gene with other molecules (protein or miRNA). Notably, the PD associated genes from DisGeNET that interact with the gene in the entry will be highlighted in the network with the legend “PDgene”. The “PieChart” tab shows four pie charts including Tissue, Methylation Alteration, Gene Region, and Expression Alteration. These pie charts illustrate the proportion of different terms obtained in the search results. Based on the information of the pie charts, users can easily obtain the information about the major tissue used to study the queried gene, the major methylation or expression alteration of the gene, and the major methylation alteration associated gene region of the gene, which might be useful for experimental design. PDmethDB also provides a tab in tabular format named “SummaryTable” to summarize the searching results from the perspective of tissue, methylation alteration, gene region, and expression alteration. In the Statistics page, PDmethDB provides three tabs to show the key features of the data from the perspective of genes, methylation alteration, tissue, and experimental methods. As “Top Genes Word Cloud” shows, the frequently studied methylation altered genes in PD include SNCA, MIR886, MAPT, and CYP2E1. The top differentially methylated signals in PD from the ‘Statistics’ page might facilitate researchers to do literature meta-analysis. PDmethDB also supports a user-friendly submission function and users can submit PD associated methylation alteration information to PDmethDB to make the database even more comprehensive. In the “Download” page, users can download all curated data easily by clicking the download button. We expect the data in this form could be beneficial for investigations initiated by users. In the “Contact” page, PDmethDB provides the email of the database developer and feedback submission box, with the aim to utilize users’ feedbacks or suggestions to make PDmethDB more user-friendly and efficient. PDmethDB provides a “Tutorial” page to guide users on how to use PDmethDB. In summary, PDmethDB aims to be a user-friendly and comprehensive platform to display and provide current information about the relationship between PD and methylation alteration.
Fig. 1.
The interface of PDmethDB. The black rectangle indicates the clickable link button and the arrow leads to the corresponding return page.
3.2. Case study using PDmethDB database
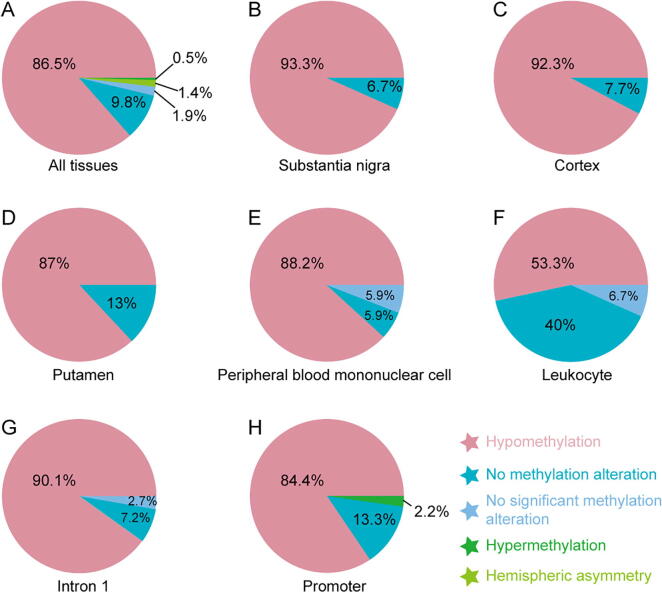
The methylation alteration in PD patients versus controls of SNCA can be observed in different tissues and different gene regions. Based on the information contained in our database, a user can perform analysis flexibly. For example, we performed an analysis of the methylation alteration patterns of SNCA in different tissues and gene regions. Firstly, we searched PDmethDB by “SNCA” and we used the results to reveal the SNCA methylation alteration proportion in PD patients at each tissue and gene region (Fig. 2). Fig. 2A shows the methylation alteration of SNCA in all tissues for which data is available. Percentages refer to returned items in the database for a specific keyword that support the methylation alteration. Of these, 86.5% entries show hypomethylation in PD patients versus controls, 9.8% no methylation alteration, 1.9% methylation alteration but not reaching significance, 1.4% differential methylation involved in hemispheric asymmetry, and 0.5% hypermethylation. In detail, over 87% of entries support hypomethylation of SNCA in substantia nigra, cortex, putamen, and peripheral blood mononuclear cell (Fig. 2B-E). However, only 53.3% entries report hypomethylation of SNCA occurs in leukocyte (Fig. 2F). Fig. 2G, H shows the methylation alteration at different gene regions of SNCA. For intron 1 region of SNCA, 90.1% entries reveal hypomethylation in PD patients versus controls. For the promoter region of SNCA, 84.4% entries mention hypomethylation in PD patients versus controls.
Fig. 2.
Pie charts displaying methylation alteration summaries of SNCA in different tissues or cell types and gene regions. Percentages refer to returned items in the database for a specific keyword that support the methylation alteration. The methylation alteration proportion of SNCA in PD patients versus controls are indicated as all tissues (A), substantia nigra (B), cortex (C), putamen (D), peripheral blood mononuclear cell (E), leukocyte (F), intron 1 (G), and promoter region (H), respectively. The methylation alteration information is defined as PD versus controls: Hypermethylation, No methylation alteration, No significant methylation alteration, and Hypomethylation. Alteration status was curated from the related papers.
3.3. Future extensions
As the development and throughput of sequencing technology improves rapidly, an increasing volume of PD associated methylation information will be generated and PDmethDB will continue to manually curate such information. We will update PDmethDB every six months. In addition, we will incorporate new analysis tools to PDmethDB to extend its utility and content coverage. We will update PDmethDB according to users’ suggestions.
4. Discussion and conclusions
PD is a multifactorial disease that is associated with genetic, environmental, nutritional and ageing risks. Epigenetic changes might occur in response to some of these factors [15], [33]. For some specific individuals, epigenetic drift might be triggered by factors, such as lifestyle and caloric intake. With the existence of an epigenetic clock, some studies focus on the relationship between PD and methylation alteration [34]. Researchers found that the same gene might have different methylation alterations in different tissues or regions. For example, SNCA has two significantly hypermethylated CpG sites in parietal cortex of PD patients than controls, while several significantly hypomethylated CpG sites in occipital cortex and substantia nigra [13]. Therefore, PDmethDB collected the methylation alteration and tissue information, simultaneously. In addition, there are some controversial results about the methylation alteration of a specific gene or region in PD. One study revealed that no significant methylation alteration was found at the intron 1 region of SNCA in post-mortem brain of PD patients compared to controls using the EZ DNA methylation kit [35], while another study showed that hypomethylation occurs in substantia nigra, putamen, and cortex region of sporadic PD patients [8]. PDmethDB collected all these data aiming to provide more comprehensive information about the relationship between the methylation alteration and PD. Additionally, PDmethDB provides the search result pie charts displaying methylation alteration proportion, tissue proportion, experimental method proportion and gene region proportion, which might be helpful to interpret current results and guide design of future experiments. In addition, for researchers who study genes in PD associated biological processes such as inflammation, autophagy, DNA repair, or oxidative stress [34], an inspection on whether a gene linked to these processes contains methylation alteration could be conducted in PDmethDB, and thus provide guidance for future studies.
In summary, PDmethDB provides a platform for storing PD associated methylation alteration information from related papers, with the aim to facilitate the study of the relationship between methylation alteration and PD pathology.
CRediT authorship contribution statement
Changliang Wang: Conceptualization, Methodology, Software, Data curation, Writing - original draft. Liang Chen: Conceptualization, Methodology, Software, Data curation, Writing - original draft. Menglei Zhang: Data curation, Validation, Writing - review & editing. Yang Yang: Data curation, Validation, Writing - review & editing. Garry Wong: Conceptualization, Validation, Writing - review & editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We thank members from Prof. Garry Wong’s lab who provided constructive suggestions and comments. We also thank researchers who shared their results concerning the relationship between PD and methylation and apologize to those whose data was overlooked or omitted.
Funding
This study was supported in part by grants from the Faculty of Health Sciences, University of Macau (MYRG2016-00101-FHS), Li Ka Shing Foundation Cross-Disciplinary Research Grant (2020LKSFG04D, 2020LKSFG07D), the National Natural Science Foundation of China (62002212), and the STU Scientific Research Foundation for Talents (35941918).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.11.015.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Lebouvier T. The second brain and Parkinson’s disease. Eur J Neurosci. 2009;30(5):735–741. doi: 10.1111/j.1460-9568.2009.06873.x. [DOI] [PubMed] [Google Scholar]
- 2.Mazzoni P., Shabbott B., Cortes J.C. Motor control abnormalities in Parkinson's disease. Cold Spring Harb Perspect Med. 2012;2(6) doi: 10.1101/cshperspect.a009282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spillantini M.G. alpha-Synuclein in filamentous inclusions of Lewy bodies from Parkinson's disease and dementia with lewy bodies. Proc Natl Acad Sci USA. 1998;95(11):6469–6473. doi: 10.1073/pnas.95.11.6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein C., Westenberger A. Genetics of Parkinson’s disease. Cold Spring Harb Perspect Med. 2012;2(1) doi: 10.1101/cshperspect.a008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nalls M.A. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–1102. doi: 10.1016/S1474-4422(19)30320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coppede F. Genetics and epigenetics of Parkinson's disease. ScientificWorldJournal. 2012;2012 doi: 10.1100/2012/489830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsumoto L. CpG demethylation enhances alpha-synuclein expression and affects the pathogenesis of Parkinson's disease. PLoS ONE. 2010;5(11) doi: 10.1371/journal.pone.0015522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jowaed A. Methylation regulates alpha-synuclein expression and is decreased in Parkinson's disease patients' brains. J Neurosci. 2010;30(18):6355–6359. doi: 10.1523/JNEUROSCI.6119-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desplats P. Alpha-synuclein sequesters Dnmt1 from the nucleus: a novel mechanism for epigenetic alterations in Lewy body diseases. J Biol Chem. 2011;286(11):9031–9037. doi: 10.1074/jbc.C110.212589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miranda-Morales E. Implications of DNA Methylation in Parkinson’s disease. Front Mol Neurosci. 2017;10:225. doi: 10.3389/fnmol.2017.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang C. Identification of potential blood biomarkers for Parkinson's disease by gene expression and DNA methylation data integration analysis. Clin Epigenetics. 2019;11(1):24. doi: 10.1186/s13148-019-0621-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wullner U. DNA methylation in Parkinson's disease. J Neurochem. 2016;139(Suppl. 1):108–120. doi: 10.1111/jnc.13646. [DOI] [PubMed] [Google Scholar]
- 13.Navarro-Sanchez L. Epigenetic study in Parkinson’s disease: a pilot analysis of DNA methylation in candidate genes in brain. Cells. 2018;7(10):150. doi: 10.3390/cells7100150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song Y. Pyrosequencing analysis of SNCA methylation levels in leukocytes from Parkinson's disease patients. Neurosci Lett. 2014;569:85–88. doi: 10.1016/j.neulet.2014.03.076. [DOI] [PubMed] [Google Scholar]
- 15.Pavlou M.A.S., Outeiro T.F. Epigenetics in Parkinson’s disease. Adv Exp Med Biol. 2017;978:363–390. doi: 10.1007/978-3-319-53889-1_19. [DOI] [PubMed] [Google Scholar]
- 16.Lill C.M. Comprehensive research synopsis and systematic meta-analyses in Parkinson's disease genetics: The PDGene database. PLoS Genet. 2012;8(3) doi: 10.1371/journal.pgen.1002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nalls M.A. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nat Genet. 2014;46(9):989–993. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orozco-Arroyave JR, et al., New Spanish speech corpus database for the analysis of people suffering from Parkinson's disease. Language Resources and Evaluation, 2014: p. 342-347.
- 19.Huang W.Y. MethHC: a database of DNA methylation and gene expression in human cancer. Nucl Acids Res. 2015;43(Database issue):D856–D861. doi: 10.1093/nar/gku1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis C.A. The encyclopedia of DNA elements (ENCODE): data portal update. Nucl Acids Res. 2018;46(D1):D794–D801. doi: 10.1093/nar/gkx1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucl Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruford E.A. The HGNC Database in 2008: a resource for the human genome. Nucl Acids Res. 2008;36(Database issue):D445–D448. doi: 10.1093/nar/gkm881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yates A.D. Ensembl 2020. Nucl Acids Res. 2020;48(D1):D682–D688. doi: 10.1093/nar/gkz966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maglott D. Entrez Gene: gene-centered information at NCBI. Nucl Acids Res. 2007;35(Database issue):D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinero J. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database (Oxford) 2015;2015:p. bav028. doi: 10.1093/database/bav028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keshava Prasad T.S. Human protein reference database–2009 update. Nucl Acids Res. 2009;37(Database issue):D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chou C.H. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucl Acids Res. 2018;46(D1):D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang W. et al., shiny: Web Application Framework for R. 2019.
- 29.Xie Y. et al., DT: A Wrapper of the JavaScript Library 'DataTables'. 2019.
- 30.Allaire JJ, et al., networkD3: D3 JavaScript Network Graphs from R. 2017.
- 31.Deng X, et al., ECharts2Shiny: Embedding Interactive Charts Generated with ECharts Library into Shiny Applications. 2017.
- 32.Attali D, shinyjs: Easily Improve the User Experience of Your Shiny Apps in Seconds. 2018.
- 33.Riess O., Kruger R. Parkinson’s disease–a multifactorial neurodegenerative disorder. J Neural Transm Suppl. 1999;56:113–125. doi: 10.1007/978-3-7091-6360-3_6. [DOI] [PubMed] [Google Scholar]
- 34.van Heesbeen H.J., Smidt M.P. Entanglement of genetics and epigenetics in Parkinson’s disease. Front Neurosci. 2019;13:277. doi: 10.3389/fnins.2019.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guhathakurta S. Hypomethylation of intron1 of alpha-synuclein gene does not correlate with Parkinson’s disease. Mol Brain. 2017;10(1):6. doi: 10.1186/s13041-017-0285-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.