Graphical abstract
Abbreviations: BLOSUM, BLOcks SUbstitution Matrix; bZIP, basic leucine-zipper; CBC, CCAAT-binding core complex; Cg, Candida glabrata; CRD, Cysteine-rich domain; CS, Consistency score; Eg, Eucalyptus grandis; Fe, Iron; H, Helix; Hap, Heme activator protein; ISC, Iron-sulfur luster; MAFFT, Multiple Alignment using Fast Fourier Transform; MBD, Metal-binding domain; ML, Maximum-likelihood; NRAMP, Natural Resistance-Associated Macrophage Protein; ROS, Reactive oxygen species; TMD, Transmembrane domain; VIT, Vacuolar iron transporter; VTL, Vacuolar iron transporter-like; YRE, Yap response elements
Keywords: Iron detoxification, Iron regulation, Iron transport, Yeast, Fungi, Plants, Vacuole, Ccc1, VIT1
Abstract
Iron is an essential micronutrient for most living beings since it participates as a redox active cofactor in many biological processes including cellular respiration, lipid biosynthesis, DNA replication and repair, and ribosome biogenesis and recycling. However, when present in excess, iron can participate in Fenton reactions and generate reactive oxygen species that damage cells at the level of proteins, lipids and nucleic acids. Organisms have developed different molecular strategies to protect themselves against the harmful effects of high concentrations of iron. In the case of fungi and plants, detoxification mainly occurs by importing cytosolic iron into the vacuole through the Ccc1/VIT1 iron transporter. New sequenced genomes and bioinformatic tools are facilitating the functional characterization, evolution and ecological relevance of metabolic pathways and homeostatic networks across the Tree of Life. Sequence analysis shows that Ccc1/VIT1 homologs are widely distributed among organisms with the exception of animals. The recent elucidation of the crystal structure of a Ccc1/VIT1 plant ortholog has enabled the identification of both conserved and species-specific motifs required for its metal transport mechanism. Moreover, recent studies in the yeast Saccharomyces cerevisiae have also revealed that multiple transcription factors including Yap5 and Msn2/Msn4 contribute to the expression of CCC1 in high-iron conditions. Interestingly, Malaysian S. cerevisiae strains express a partially functional Ccc1 protein that renders them sensitive to iron. Different regulatory mechanisms have been described for non-Saccharomycetaceae Ccc1 homologs. The characterization of Ccc1/VIT1 proteins is of high interest in the development of biofortified crops and the protection against microbial-derived diseases.
1. Introduction
Iron is an indispensable micronutrient for almost all living organisms since it participates as a cofactor in a wide range of biochemical processes, such as oxygen transport, electron transfer in the respiratory and photosynthetic chains, lipid biosynthesis, DNA replication and repair, as well as ribosome biogenesis and recycling. Although iron is the fourth most abundant element in the Earth’s crust, the predominant form of iron is ferric iron (Fe3+), which solubility is very low at physiological pH. Thus, organisms have developed or acquired from other organisms sophisticated mechanisms to overcome poor iron bioavailability (reviewed in [1], [2], [3], [4], [5]). The same redox properties that make iron an essential element render it potentially toxic. When in excess, iron can participate in Fenton reactions, leading to reactive oxygen species (ROS) generation that can damage proteins, lipids and nucleic acids in the cell (reviewed in [6]). Since there is no regulated mechanism for iron excretion, organisms protect themselves from iron toxicity by regulating its acquisition and distribution, and by storing its excess in the less harmful form Fe3+, that can be mobilized when needed.
The most ancient molecule involved in iron detoxification, protection and storage is hypothesized to be ferritin (reviewed in [7]). Ferritin is present in mammals, plants, bacteria and archaea, but it is absent in fungi, where iron is stored in the vacuole [8]. The tertiary structure of this molecule is highly conserved and consists of a multisubunit hollow sphere that can shelter up to 4000 iron atoms in a compact configuration. Ferritin has ferroxidase activity that allows iron to be stored as ferric iron. In animals, iron release from ferritin is mainly promoted in the lysosomes. Then, iron is transported to the cytosol for reutilization, probably via the divalent metal-ion transporter DMT1, and it is exported from the cell through ferroportin. Thus, these vacuole-equivalent organelles act as recycling compartments rather than as a storage site (reviewed in [9]). Plant ferritin, phytoferritin, is very similar in sequence and structure to mammalian ferritin, but it is not localized in the cytoplasm, being instead distributed into organelles like plastids and mitochondria. It is thought that plant ferritin provides iron to metabolic processes in these organelles as well as protection from oxidative stress (reviewed in [10]). Nonetheless, the most important iron storage site in plants, and similarly in fungi, is the vacuole and tonoplast (reviewed in [11]). In the budding yeast Saccharomyces cerevisiae, extracellular iron can enter vacuoles via steady state endocytosis [12]. In fact, yeast mutants unable to endocytose display growth defects in iron-deficient conditions, an observation that highlights the physiological relevance of endocytosed iron [12]. However, the major mechanism for iron detoxification in yeast cells depends on Ccc1 protein, which localizes to the vacuolar membrane and transports iron from the cytosol into the vacuole, being responsible for ~ 60% of vacuolar iron accumulation under iron-replete conditions [12], [13]. Spectroscopy studies have shown that, in fermenting yeast cells, vacuolar iron is predominantly found as magnetically mononuclear high-spin Fe3+ species, although Fe3+ nanoparticles are also present [14], [15], [16]. Specifically, vacuolar Fe3+ associates to low-molecular-mass polyphosphate chains consisting on 6 to 20 phosphate units coordinated to 1–3 metal ions [17]. In response to iron depletion, Fre6 metalloreductase reduces vacuolar Fe3+ to Fe2+, and then Fet5-Fth1 high-affinity and Smf3 low-affinity iron transporters export Fe2+ to the cytoplasm [18], [19], [20]. During the shift from fermentation to respiration, yeast cells transiently accumulate iron in mitochondria and vacuoles [21]. Vacuolar pH and the oxidation state determine the form of iron stored in the vacuole [13], [14]. A ccc1Δ mutant is extremely sensitive to high iron concentrations and displays severe oxidative damage [12], [13].
Complementation assays with S. cerevisiae ccc1Δ mutant strain allowed the identification in Arabidopsis thaliana of the first plant Ccc1 ortholog, denoted AtVIT1 (Vacuolar Iron Transporter) [22]. AtVIT1 localizes to the tonoplast membranes of the endodermal cells of Arabidopsis seeds, and its disruption leads to iron distribution impairment and poor seed growth [22]. Plant VIT1 and its homologs have emerged as attractive targets for the development of iron biofortified crops to improve crop productivity [2], [23], [24]. In addition to VIT1 orthologs, plants express the so-called VIT1-like (VTL) genes. In A. thaliana, VTL transporters AtVTL1-5 share high similarity with AtVIT1, and most of them are iron regulated and complement the ccc1Δ yeast mutant [25]. Besides plants and fungi, Ccc1/VIT1 homologs have been found in other organisms, including bacteria and protists [26], [27], [28], [29]. Interestingly, the crystal structure of Eucalyptus grandis VIT1 (EgVIT1) homolog has been recently elucidated, allowing the comparison between Ccc1/VIT1 orthologs particularly on the residues required for metal transport [30]. The aim of this mini-review is to explore and discuss the diversity of the Ccc1/VIT1 family of iron transporters, focusing on fungal Ccc1 function and regulation by iron.
2. Ccc1/VIT1 conservation and distribution across the tree of Life
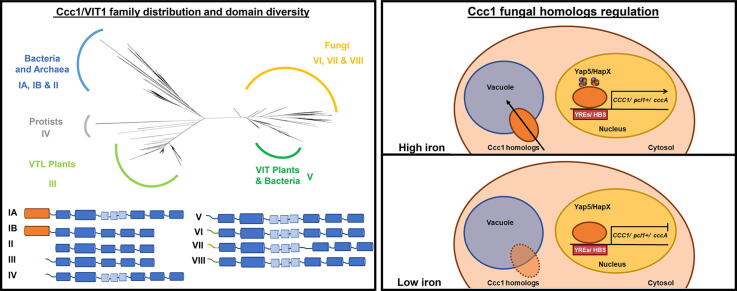
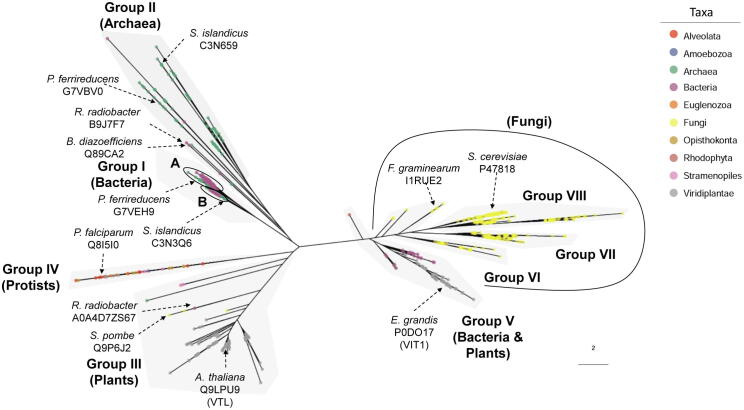
To explore the conservation and diversity of the Ccc1/VIT1 family of proteins, Ccc1/VIT1 homologs were retrieved from the MetaPhORs database [31], [32] using the S. cerevisiae Ccc1 protein as a query (for further details see Supplementary Information). Up to 721 homolog protein sequences were retrieved and 23 public available sequences were manually added (Supplementary Table S1). Ccc1/VIT1 homologs are widely distributed across the Tree of Life with the exception of Metazoa, where ferritin is the main iron storage protein. Ferritin is also the main iron storage site in archaea and bacteria, although these two groups of organisms also contain Ccc1/VIT1 homologs. These Ccc1 protein homologs are functionally related to bacterioferritin, but their main function is the export of iron out of the cell [27], [28], [33]. We classified the Ccc1/VIT1 homologs in eight different groups according to phylogenetic analysis and protein structure (Fig. 1; Supplementary Figure S1). Archaea and bacteria are mostly found in groups I and II. Group III includes VTL protein from angiosperm plants (monocots and dicots), Lycopsida class [25], and a small divergent group composed by the diatom Phaeodactylum tricornutum [29], the algae Chlamydomonas reinhardtii, and Schizosaccharomyces fission yeasts. Group IV is enriched in protist parasites, such as species Plasmodium and Trypanosoma [26], [34]. The topology of the tree indicates that VTL was transferred from an archaean lineage during the origin of the last common eukaryote ancestor [35]. A second major clade includes groups V-VIII, which contains plant VIT1-type [22] and fungi homologs (Fig. 1; Supplemental Figure S1). Group V also contains 21 bacterial sequences and one Rhodophyta algae (Cyanidioschyzon merolae). A plausible scenario is an ancestral horizontal gene transfer from bacteria to the ancestor of eukaryotes, where most fungi (groups VI-VIII) and Viridiplantae retained the gene (Fig. 1, Supplementary Figure S1) [36]. The presence of more than one homolog in several archaea and bacteria species and the high domain diversity in prokaryotes (next section) support a complex evolutionary history for this protein family. In plants, several transpositions and both segmental and tandem duplication events contributed to generate an elevated number of Ccc1/VIT1 proteins with specialized functions (Supplementary Table S1) [36], [37]. In a lesser extent, gene expansions have also occurred in some clades of fungi like the genus Aspergillus and Penicillium, Glomeromycota division, and Mucoromycotina and Agaricomycotina subdivisions.
Fig. 1.
Phylogenetic tree reconstruction of the iron vacuolar transporter Ccc1/VIT1. A maximum likelihood phylogenetic tree was reconstructed using the complete Ccc1/VIT1 protein alignment. The scale bar represents the number of amino acid substitutions. Sequence names of some published proteins are indicated (more details in Supplementary Table S1). Tips are colored according to taxa designation, which taxonomic depth is different for simplicity. The most frequent taxa in each group (i.e. fungi) is displayed. A. thaliana: Arabidopsis thaliana; B. diazoefficiens: Bradyrhizobium diazoefficiens; E. grandis: Eucalyptus grandis; F. graminearum: Fusarium graminearum; P. ferrireducens: Pyrobaculum ferrireducens; P. falciparum: Plasmodium falciparum; R. radiobacter: Rhizobium radiobacter; S. cerevisiae: Saccharomyces cerevisiae; S. islandicus: Sulfolobus islandicus.
3. Ccc1/VIT1 family structure and function
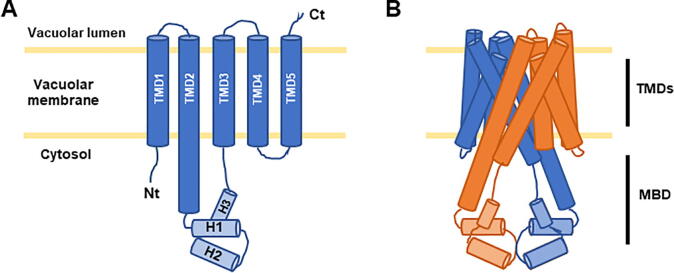
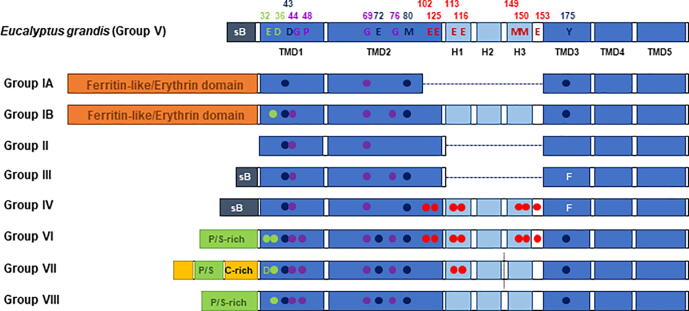
The recently solved crystal structure of the Eucalyptus grandis EgVIT1 protein and its function-structure analysis have allowed the identification of conserved domains and residues within Ccc1/VIT1 proteins that are essential for the mechanism of iron transport [30]. EgVIT1 possesses five transmembrane domains (TMD1-5), with a cytoplasmic loop containing three alpha helices (H1-3) between TMD2 and TMD3. Its amino-terminal region faces the cytosol, whereas its carboxy-terminal extreme localizes towards the vacuolar lumen (Fig. 2). EgVIT1 functions as a dimer and the interface formed by the two protomers constitutes a channel through which the metal ions are translocated. In this cavity, TMD1 and TMD2 provide conserved amino acids with oxygen and sulfur atoms (tyrosine and methionine), as well as carboxylated groups from glutamate and aspartate amino acids, conferring a hydrophilic environment appropriated for metal ion transport (Fig. 3) [30]. Indeed, aspartate 43 and methionine 80 are found in almost all Ccc1/VIT1 sequences (Fig. 3). These two amino acids are present at the entrance of the cavity and are involved in the coordination of the metal ions and in the discrimination between alkaline earth- and transition-metal ions, respectively [30]. Methionine 80 might also participate in preventing proton leakage to the cytosol [30]. The other transmembrane segments are disposed around this cavity. The hydrophilic cavity is opened in the cytosolic face, but it is sealed at the opposite side by hydrophobic residues. To deliver iron to the vacuole, glycine and proline residues generate kink motions that release the seal and allow iron entry into the vacuole [30]. In the same way, glycine residues 44, 69 and 76 of EgVIT1 are present in barely all group sequences (Fig. 3). Studies in Arabidopsis have shown that this highly conserved glycine residue, located within the second TMD, is critical for VIT1 iron transport activity [38].
Fig. 2.
Schematic representation of EgVIT1 membrane topology and dimer structure. A) Transmembrane domains (TMD1-5) are represented by dark blue cylinders. The helices between TMD2 and TMD3 domains, represented by light blue cylinders, conform the MBD. B) An EgVIT1 dimer representation, coloring TMDs and MBD as dark or light colors, respectively. Each protomer is colored in blue or orange. Adapted from [30]. Ct: carboxyl terminus, H: helix, MBD: metal-binding domain, Nt, amino-terminus, TMD: transmembrane domain. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.
Domain organization and amino acid conservation in Ccc1/VIT1 family proteins. Schematic representation of the architecture and key amino acids of the consensus sequence of each group. EgVIT1 group V key amino acids are represented by its IUPAC letter and by the following color code: green for amino acids involved in iron transport from the MBD to the hydrophilic cavity; dark blue, amino acids involved in the transport throughout the cavity; purple, residues implicated in the induction of the seal opening; and red, residues within the MBD. White letters are used when other amino acids are conserved for more than two non-group V protein groups, and circles represent conserved amino acids colored as above for non-group V proteins. Dark and light blue rectangles represent the five TMDs and the three helices, respectively. Orange, grey, green and yellow rectangles symbolize Ferritin-like/erythrin domain, short basic (sB), proline/serine-rich (P/S-rich) and cysteine-rich (C-rich) motives, respectively. Group VII sequences contain a unique short amino acid sequence in the MBD, represented by a dashed line. Numbers indicate the amino acid position in EgVIT1 protein. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Amino acid sequence similarity is low throughout the different species, but Ccc1/VIT1 domains and structure are mostly maintained. Besides such conservation, some domains are highly variable, such as the metal-binding domain (MBD). The MBD is constituted by the carboxy-terminal region of TMD2 and H1 and H3 helices, with both helices forming the cytoplasmic loop between TMD2 and TMD3 (Fig. 2). This domain is rich in glutamate residues and it has been proposed to facilitate iron incorporation from siderophores and other iron-containing molecules [30]. This domain is absent in groups IA, II and III, and seems to be lost independently in these groups. The addition or removal of this region does not affect the topology of the phylogenetic tree of Ccc1/VIT homologs (Supplementary Information). Instead, in some proteobacteria there is an amino-terminal ferritin-like or erythrin domain consisting of a four-α-helix bundle containing the Ex6Y, ExxH, Ex6Y and ExxH motifs and forming a di-iron-binding site. This domain has the ability to oxidize Fe2+ to Fe3+, as a typical ferroxidase catalytic subunit [27], [28], [39]. Curiously, the MDB also protects against H2O2 stress, probably by limiting free iron availability for ROS generation [40]. Mutations in glutamate residues from the erythrin domain impair iron export and the oxidative stress response [28]. Sequences belonging to group IB contain an MBD-like domain consisting of the erythrin domain pattern but lacking well-defined third and fourth helices. Plant VTL proteins lack the MBD and differ in sequence from VIT1 homologs. In A. thaliana, most VTL transporters localize to the tonoplast and participate in iron homeostasis [25]. Interestingly, recent studies have shown that VTL1a from Glycine max, VTL8 from Medicago truncatula, and probably SEN1 from Lotus japonicum localize to the symbiosome membrane and deliver iron from the plant to the bacteroid in order to supply iron-demands derived from the symbiosis interaction [41], [42], [43], [44].
Genetic and biophysical studies have shown that yeast Ccc1 contributes to manganese homeostasis, suggesting that it could facilitate Mn2+ transport from the cytosol to the vacuole [13], [45]. More recently, liposome transport assays with reconstituted EgVIT1 have demonstrated that, in addition to Fe2+, it also transports Co2+ [30]. Iron transport assays with Plasmodium falciparum VIT1 expressed in yeast suggest that Ccc1/VIT1 proteins are selective for Fe2+ over other dimetal ions including Mn2+, Zn2+, Cu2+ and Ca2+ [26]. Liposome transport assays have also shown that EgVIT1 is a H+-coupled antiporter for metal ions, which provides the vacuolar H+-ATPase with an important function in metal storage [30]. Glutamate 72 participates in iron translocation as well as proton movement towards aspartate residue 43 [30]. Nonetheless, aspartate 43 is absent in groups I-IV, suggesting that this is not the principal residue involved in proton-coupled transport, as it happens in other iron transporters such as those from the Natural Resistance-Associated Macrophage Protein (NRAMP) family, where a histidine residue instead of a glutamate plays a key role in proton transport [46]. Tyrosine 175, also involved in iron translocation, is not conserved in groups III and IV, with phenylalanine found instead. In this sense, studies of the crystal structure and protein function of ferroportin have revealed that the benzene ring from phenylalanine also interacts with divalent metals forming a cation-π interaction [47]. Finally, the kink-inducing residue proline 48 is not found in groups I-IV either (Fig. 3). Group III-VIII sequences, including Schizosaccharomyces pombe and P. tricornutum, share high sequence similarity with EgVIT1 and probably a similar mechanism for iron translocation. All these proteins contain a glutamate-rich MBD, but only in certain cases the number of glutamates and their position are strictly conserved (Fig. 3). Methionine residues 149 and 150 are only present in group VI, and group VII possesses a larger MBD (Fig. 3). In the case of P. tricornutum, it has been suggested that VIT1 would act as a Cd2+ transporter since its expression is regulated by this metal [29]. VIT1 proteins of group IV, which includes protist human parasites from the Plasmodium genus, localize mainly in the endoplasmic reticulum [26]. In the case of Plasmodium parasites, VIT1 plays a major role in iron detoxification during infection and is considered as a potential target for anti-malaria drugs [48]. VIT1 protein sequences within group V share high similarity through the plant kingdom and are responsible for iron accumulation around the provasculature in seeds [49]. However, plant VIT1 homologs have experimented functional divergence. For example, in Tulipa gesnariana, iron accumulation into the vacuole by TgVIT1 is responsible for the blue coloration of petals, where it is exclusively expressed [50]. In rice Oryza sativa, OsVIT1 and OsVIT2 homologs are exclusively expressed in flag leaves and are involved in iron distribution between source and sink tissues [23]. Disruption of rice VIT genes decreases flag leaves iron content and increases iron accumulation in seeds, becoming a potential strategy of iron biofortification [23]. Although basic residues and amino acids with carboxylate groups are usually present, the amino-terminal region of Ccc1/VIT1 proteins has diverged and seems unique in each species. The amino-terminal region of fungal Ccc1/VIT1 is rich in serine, proline and cysteine residues (Fig. 3). Interestingly, in S. cerevisiae, genome-wide studies have indicated that some of these serine residues are phosphorylated and together with ubiquitinated lysines could represent potential sites to regulate protein function and stability [51], [52]. Overall, these data strongly suggest that the mechanism of metal transport involving a hydrophilic dimer interface, proton antiporter and kink motions to open the luminal face of the transporter is unique and widely conserved across the Ccc1/VIT1 family, whereas other more variable domains may be involved in fine-tuning its functionality, abundance and subcellular localization.
4. Regulation of yeast CCC1 expression by iron
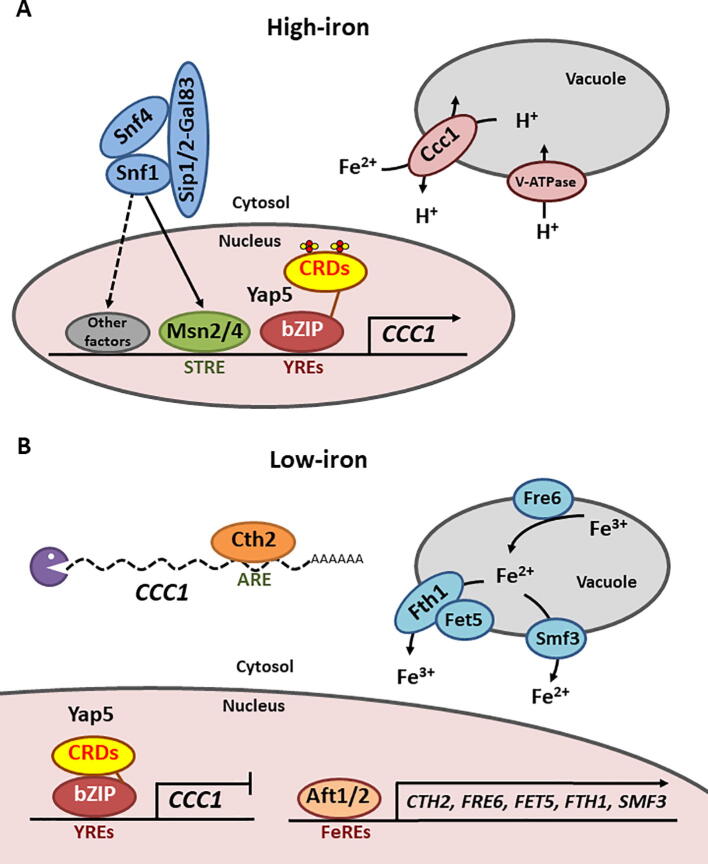
As fungi cannot excrete iron, the vacuolar iron importer Ccc1 represents the most important factor implicated in fungal iron resistance. Consistent with its crucial function in cytosolic iron detoxification, the expression of CCC1 is finely regulated by iron. In response to iron excess, S. cerevisiae Yap5, a member of the yeast activator protein (Yap) subfamily of transcriptional factors (reviewed in [53]), activates the expression of CCC1 ([54]; reviewed in [55]) (Fig. 4). Yap5 contains an amino-terminal basic leucine-zipper (bZIP) DNA-binding domain that associates to two Yap response elements (YREs) within the promoter of CCC1 [54]. Although Yap5 binding to YREs is constitutive and independent of iron levels, the activation of CCC1 transcription only occurs in the presence of excess iron [54]. Yap5 carboxy-terminal region contains an activation domain, which consists of two cysteine-rich domains (CRDs), n-CRD and c-CRD, with four and three conserved cysteine residues, respectively [54]. In vivo and in vitro studies have demonstrated that Yap5 directly binds two [2Fe-2S]-type clusters, which promote a conformational change that could be responsible for transcriptional activation [56] (Fig. 4). The conserved n-CRD domain, with higher affinity for iron than c-CRD, seems to function as a general iron sensor module. Remarkably, Yap5 activation depends specifically on the rate of mitochondrial Fe-S cluster (ISC) biogenesis, but not on cytosolic iron content, since defects in ISC synthesis prevent high-iron CCC1-induced expression [57]. In response to high-iron, Yap5 also activates the expression of TYW1, which encodes for a cytosolic ISC-containing protein probably implicated in iron buffering, and GRX4, which encodes for a monothiol glutaredoxin that limits the transcriptional activity of the low-iron-responsive transcription factors Aft1 and Aft2 [58], [59].
Fig. 4.
Regulation of S. cerevisiae CCC1 in response to iron overload and depletion. A) High-iron conditions. During iron overload, two ISCs associate to the CRDs of DNA-bound Yap5 leading to a conformational change that promotes CCC1 transcription. CCC1 expression is also activated by the Snf1 effectors Msn2/Msn4 and other unknown transcriptional factors. Ccc1 facilitates the import of Fe2+ into the vacuole while exporting H+. Therefore, pH and V-ATPase function are tightly connected to cellular iron homeostasis. B) Low-iron conditions. During iron depletion, Aft1/Aft2 activate the iron regulon, including FRE6, FET5, FTH1 and SMF3 genes, which encode for the vacuolar iron export machinery, and CTH2, which promotes the degradation of CCC1 mRNA. The decrease in ISC synthesis prevents the activation of Yap5, which is constitutively bound to YREs within CCC1 promoter. ARE: AU-rich element; bZIP: basic leucine-zipper domain; CRD: cysteine-rich domain; FeRE: iron regulatory element; STRE: stress response element; V-ATPase: vacuolar ATPase; YRE: Yap5 response element.
Yap5 is not the only transcriptional factor that contributes to CCC1 expression in high-iron conditions, since yap5Δ cells partially activate CCC1 and are less sensitive to iron than ccc1Δ mutants. The overexpression of some members of the Yap family induces the expression of CCC1 in yap5Δ cells [56]. However, the mutagenesis of CCC1 YREs suggests that other cis-regulatory elements and transcription factors beyond the Yap family activate the transcription of CCC1 [58]. A recent report has shown that the low-glucose sensor Snf1 participates in the regulation of CCC1 [60] (Fig. 4). Cells lacking Snf1 kinase activity or other components of the Snf1 complex, such as Snf4, Sip1, Sip2 and Gal83, show defects in CCC1 expression under high-iron conditions that lead to iron toxicity [60]. Previous studies have shown that iron deficiency promotes the phosphorylation of Snf1 [61], but no evidence for changes in Snf1 phosphorylation stage have been reported in excess iron. The Snf1-dependent expression of CCC1 is independent of Yap5 and ISC biogenesis, but requires the general stress transcription factors Msn2 and Msn4, which are targets of Snf1 kinase, and other unidentified regulatory factors [60] (Fig. 4). Similarly to other stresses, Msn4 protein translocates to the nucleus when cells are exposed to excess iron [62]. The relevance of Msn2 and Msn4 in iron homeostasis is corroborated by the sensitivity of msn2Δmsn4Δ double mutants to both iron deficiency and excess [60], [63]. Further studies would be necessary to decipher how iron modulates the activity of the Snf1 kinase complex and its downstream effects on iron homeostasis.
In response to iron deficiency, Aft1 and Aft2 transcriptional factors activate the expression of a group of genes, known as the iron regulon, that participate in iron uptake, iron mobilization from the vacuole and iron metabolism remodeling (reviewed in [64], [65]). The Aft1/Aft2-dependent transcriptional activation of the vacuolar FRE6 metalloreductase and the FET5/FTH1 and SMF3 vacuolar iron transporters facilitates the efflux of iron from the vacuole when iron is scarce [18], [20], [66], [19], [67], whereas another Aft1/Aft2 target, Cth2, limits vacuolar iron storage by promoting the degradation of CCC1 mRNA [54], [68] (Fig. 4). Therefore, both transcriptional and post-transcriptional mechanisms tightly match the expression of CCC1 to cellular iron availability.
Multiple connections link vacuolar and mitochondrial iron homeostasis. For instance, the overexpression of CCC1 suppresses mitochondrial iron overload phenotypes, limits ISC biogenesis and decreases the activity of mitochondrial ISC-dependent enzymes including aconitase [69]. Moreover, the overexpression of the mitochondrial iron importers MRS3 or MRS4 suppresses the sensitivity of ccc1Δ cells to iron, whereas their simultaneous deletion increases vacuolar iron transport and diminishes cytosolic iron [70], [71]. It has been proposed that mrs3Δmrs4Δ mutants generate ROS that increase the activity of the vacuolar Ccc1 transporter [71]. Both ccc1Δ and mrs3Δmrs4Δ mutant cells activate the Aft1/Aft2-dependent low-iron transcriptional response, while cellular iron homeostasis is mostly restored in ccc1Δmrs3Δmrs4Δ mutants [12], [70], [71]. The overexpression of the mitochondrial iron exporters MMT1 and MMT2 leads to phenotypes similar to mrs3Δmrs4Δ mutants, which are also rescued by the deletion of CCC1 [72]. Altogether, these results indicate that subcellular iron pools including mitochondrial, vacuolar and cytosolic iron are highly interdependent. Several studies have linked vacuolar acidification to iron homeostasis. As indicated above, Ccc1/VIT1 seem to function as H+/Fe2+ antiporters whose activity depends on vacuolar pH [13], [30]. Moreover, the loss of vacuolar H+-ATPase activity has been reported to activate the Aft1-dependent low-iron response in iron-repleted cells probably due to oxidative stress, leading to iron homeostasis defects [73], [74]. More recent data have shown that the loss of vacuolar acidification impairs vacuolar cysteine storage, which cause an oxidative damage to mitochondrial ISC biosynthesis components and ISC-containing activities such as aconitase [75], [76]. As a consequence, cells develop an age-related mitochondrial dysfunction characterized by the activation of the iron-deficiency and DNA damage responses, which can be rescued by iron supplementation [75], [76].
5. CCC1 iron regulation in other fungi
Ccc1 function and regulation have been mostly studied in laboratory strains of S. cerevisiae, while little is known about its diversity in other strains and related fungal species. For instance, a genetic analysis with the use of reciprocal hemizygotes has shown that S. cerevisiae CCC1 and YAP5 alleles from a wild Malaysian population contain a partial loss-of-function that renders these yeast strains sensitive to high iron concentrations [77]. Compared to other available S. cerevisiae strain sequences, the Malaysian CCC1 allele displays a single G22R amino acid change within its fungal-specific amino-terminal domain, which role in Ccc1 function has not been deciphered. Moreover, the Malaysian strains also display a gain-of-function allele for the master low-iron regulatory factor AFT1 that improves their adaptation to iron deficiency [77]. These observations suggest that S. cerevisiae Malaysian strains have specialized to low-iron environments and have lost some of their high-iron resistance attributes. The phenotypic analysis of other natural and human-domesticated yeast strains, which genome has been deciphered, will be tremendously useful to understand at the molecular level the evolution and adaptation of S. cerevisiae iron homeostasis network to particular ecological niches.
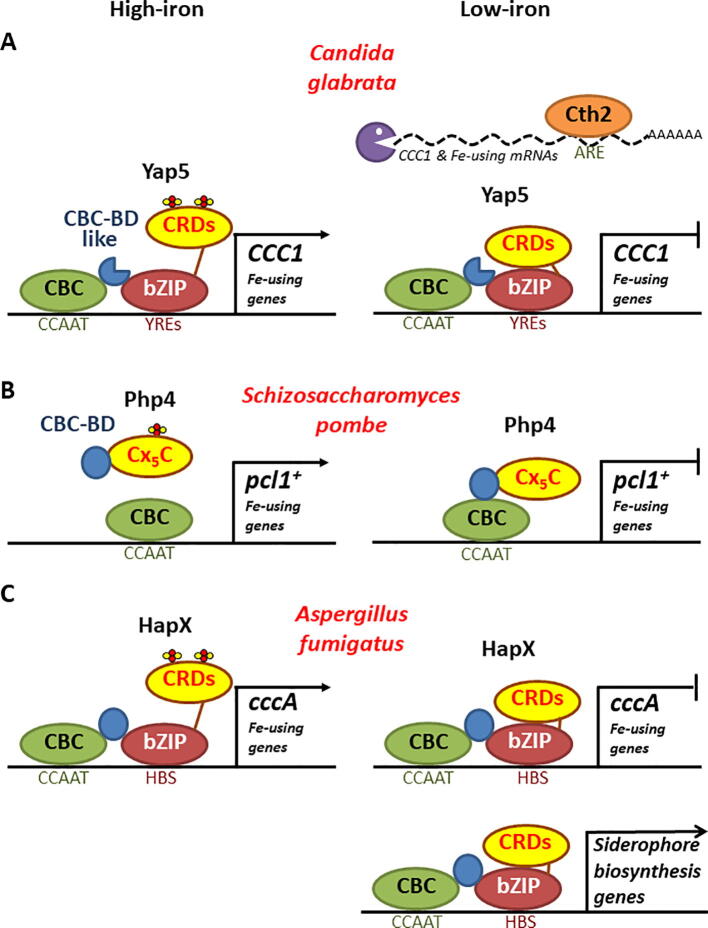
Non-Saccharomycetaceae fungi regulate the expression of CCC1 orthologs through CCAAT-type transcription factors composed of a heterotrimeric CCAAT-binding core complex (CBC), which constitutively associates to CCAAT-consensus promoter sequences, and an iron-sensing Hap-type CBC-regulatory subunit that binds to CBC through a conserved amino-terminal Hap4-like CBC-binding domain (CBC-BD) (reviewed in [1]). S. cerevisiae and its closely related pathogen species Candida glabrata have specialized their CBC-Hap4 regulatory system in the positive control of respiratory genes. However, a recent study suggests that C. glabrata Yap5 also associates to its CBC through a vestigial CBC-BD to activate the expression of CCC1 and iron-dependent processes in response to high-iron conditions [78] (Fig. 5). S. cerevisiae Yap5 also contain a partial CBC-BD, but since a function has not been demonstrated for this region, it has not been represented in Fig. 4. Similar to S. cerevisiae, C. glabrata expresses a Cth2 homolog that promotes CCC1 mRNA down-regulation in response to iron limitation [79] (Fig. 5). The expression of the S. pombe CCC1-homolog gene pcl1+, which deletion confers iron sensitivity, and of genes encoding for non-essential iron-dependent processes is controlled by the Php2/3/5 CBC and its negative regulatory subunit Php4. Php4 lacks a bZIP DNA-binding domain, so during low-iron conditions it associates to CBC through its CBC-BD to repress pcl1+ and iron-consuming genes expression. In response to high-iron conditions, the monothiol glutaredoxin Grx4 delivers a [2Fe-2S] to Php4 that abrogates its interaction with the CBC and allows pcl1+ and iron-using genes transcription to proceed [80] (Fig. 5). Two Php4 cysteine residues (Cx5C) are critical for iron-sensing and pcl1+ regulation (reviewed in [81]). In the pathogen mold Aspergillus fumigatus, there are two homologs of CCC1 (cccA and cccB), but only cccA is responsible for iron storage into the vacuole and contributes to iron resistance [82]. Remarkably, A. fumigatus CBC and its HapX regulatory factor, which contains a CBC-BD, a bZIP domain and four CRDs, function in response to both iron limitation and excess by activating or repressing iron metabolism genes depending on iron bioavailability [83], [84], [85]. When iron is scarce, HapX represses the transcription of genes participating in iron-consuming pathways and activates genes implicated in siderophore biosynthesis. Strikingly in response to high-iron conditions, A. fumigatus CBC and HapX cooperatively associate to the promoter of the vacuolar cccA iron importer and iron-using genes to activate their transcription [83], [84], [85] (Fig. 5). HapX association to both CBC and a variable bipartite HapX DNA-recognition site downstream of the CBC-binding motif are necessary for all described HapX functions [85]. Although A. fumigatus and other fungi synthesize intracellular siderophores that participate in iron distribution and storage, vacuolar CccA still acts as the principal iron detoxification system [82], [86]. The deletion of HapX, which is present in almost all Euascomycetes and Basidiomycetes, attenuates virulence in pathogenic fungi [83], [85]. Candida albicans CBC-Hap43 complex also plays a dual role by activating genes that participate in iron uptake and repressing non-essential iron-consuming processes when iron is scarce [87], [88]. Further studies with C. albicans have reinforced the role of Ccc1 as a connector between vacuolar and mitochondrial iron pools previously observed in S. cerevisiae. Deletion of the unique C. albicans mitochondrial importer, MRS4, and its vacuolar iron exporter SMF3 increase intracellular iron levels, enhance mitochondrial dysfunction, impair filamentous development and adhesion to host epithelial, and consequently reduce virulence [72], [89]. Importantly, the simultaneous deletion of CCC1 rescues all these phenotypes, positioning the Mrs4-Ccc1-Smf3 pathway as an attractive antifungal drug target.
Fig. 5.
Iron regulation of CCC1 and other genes encoding for dispensable iron-dependent processes in C. glabrata, S. pombe and A. fumigatus. A) C. glabrata. Although C. glabrata possesses a transcription iron overload response similar to S. cerevisiae, recent studies have shown that Yap5 associates to CBC through a vestigial CBC-BD to activate the transcription of CCC1 and multiple genes encoding for iron-using proteins in response to high-iron conditions. B) S. pombe. Under iron deficiency, the S. pombe CBC negative regulatory subunit associates to DNA-bound CBC via its CBC-BD and represses the transcription of the S. pombe CCC1 homolog gene, pcl1+, and iron-consuming processes. Under iron-overload conditions, ISC-binding to a Cx5C motif in Php4 abrogates its interaction with the CBC, allowing the expression of pcl1+ and iron-dependent processes. C) A. fumigatus. In A. fumigatus, HapX CBC regulatory subunit exerts both positive and negative effects on CBC activity. During high-iron conditions, ISC-binding to HapX CRDs triggers a conformational change that promotes the expression of CccA and iron-using genes. In response to low-iron conditions, HapX represses the expression of CccA and genes encoding for iron-dependent processes, but promotes iron acquisition by activating the expression of genes implicated in iron siderophore biosynthesis. The association of HapX to both CBC, via its CBC-BD, and two bipartite downstream HapX-binding sites (HBS), via its bZIP domain, are required for all HapX functions. ARE: AU-rich element; bZIP: basic leucine-zipper domain; CBC: CCAAT-binding complex; CBC-BD: CBC-binding domain; CRD: cysteine-rich domain; HBS: bipartite HapX-binding site; YRE: Yap5 response element.
6. Conclusions and remarks
Living organisms have developed sophisticated strategies to deal with iron scarcity at the same time that they protect themselves from high levels of iron. Specifically, prokaryotes and animals synthesize the protein ferritin, which stores iron in a safe and compact but accessible form. Despite ferritin is also present in plants, it plays a secondary role in iron storage. Plants, as well as fungi, mainly detoxify and accumulate excess iron into vacuoles and mobilize it in response to iron limitation. The most studied iron transporters responsible for iron storage into vacuoles are S. cerevisiae Ccc1 and A. thaliana VIT1 [12], [22]. However, homologs to these proteins can be found in all kingdoms with the exception of metazoa. The Ccc1/VIT1 family possesses a complex phylogeny that probably involves several duplications and losses during species evolution. Although amino acid sequence similarity is observed along the protein, with key residues conserved in almost all analyzed organisms, some differences are also consistently found. The most relevant difference is the absence of the MBD in some prokaryotic and plant VTL proteins, and the presence of a ferritin-like domain at the amino-terminus in some bacteria and archaea species (Fig. 3). The conservation of residues involved in the discrimination between metal transition ions and iron transport implies a conserved common mechanism with some particularities in different groups.
The iron-dependent regulation of S. cerevisiae CCC1 mRNA has been widely studied. Both transcriptional (Yap5-dependent) and post-transcriptional (Cth2-dependent) mechanisms modulate CCC1 transcript levels according to cellular iron availability to facilitate vacuolar iron storage in high-iron conditions and vacuolar iron mobilization during iron deficiency. Recent work has discovered that other unexpected (Snf1 and Msn2/Msn4 dependent) and unknown pathways contribute to CCC1 expression and yeast iron resistance [60]. An important aspect of vacuolar iron storage via Ccc1 is its close connection to mitochondrial iron transport with implications not only in iron homeostasis but also in aging [75], [76]. The regulation of Ccc1 homologs in non-Saccharomycetaceae, which includes many pathogenic fungi, seems to differ considerably from S. cerevisiae (reviewed in [1]). In these fungi, CBC DNA-binding complexes associate to Hap-like CBC regulatory subunits to either positively or negatively regulate the transcription of CCC1 homologs. Moreover, little is known about post-translational regulatory mechanisms that influence Ccc1 protein localization, stability or functionality. Further studies are needed to elucidate the intricate regulation of Ccc1 in different fungi and its connections with mitochondrial iron metabolism.
Iron is especially important for microbial virulence. As mentioned above, Plasmodium VIT1 and A. fumigatus HapX have been demonstrated to be required for pathogenicity. Interestingly, the absence of Ccc1/VIT1 transporters in animals might allow the design of specific antifungal or antiparasitic drugs that target Ccc1/VIT1 transporters with lower side effects than other current drugs. Moreover, since Ccc1/VIT1 homologs in bacteria are also involved in H2O2 detoxification, their inhibition could suppose a reinforcement for host defenses [47]. On another hand, plant VIT transporters have an important role in iron distribution in seeds, and their manipulation could represent a promising biofortification tool [2], [23], [24], [90]. New genome sequences are generating datasets that could be used to explore how extended these Ccc1/VIT1 homologs are through the Tree of Life. Together with functional analysis, the pathways that control these proteins might be better understood and new research lines might help to isolate or design organisms able to acquire more iron for nutritional purposes, to unravel new drug targets and to work up actions for the biocontrol of pathogenic fungi involved in the deterioration of food (pre- and post-harvest) or in human-related diseases.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Dr. Lola Peñarrubia for critically reading the manuscript. Work in our laboratory is supported by the Spanish Ministry of Science, Innovation and Universities (MICINN) grant BIO2017-87828-C2-1-P, the Regional Government of Valencia “Generalitat Valenciana” grant PROMETEU/2020/014 and FEDER (Fondo Europeo de Desarrollo Regional) funds to SP, and a predoctoral contract from “Generalitat Valenciana” and FEDER funds to RSD. Computations were performed on Tirant III of the Spanish Supercomputing Network (“Servei d'Informàtica de la Universitat de València”) under the project BCV-2018-2-0002 granted to DP.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.10.044.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Martínez-Pastor M.T., Puig S. Adaptation to iron deficiency in human pathogenic fungi. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2020;1867(10):118797. doi: 10.1016/j.bbamcr.2020.118797. [DOI] [PubMed] [Google Scholar]
- 2.Connorton J.M., Balk J., Rodríguez-Celma J. Iron homeostasis in plants – a brief overview. Metallomics. 2017;9(7):813–823. doi: 10.1039/C7MT00136C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ganz T., Nemeth E. Iron homeostasis in host defence and inflammation. Nat Rev Immunol. 2015;15(8):500–510. doi: 10.1038/nri3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ganz T., Nemeth E. Hepcidin and iron homeostasis. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2012;1823(9):1434–1443. doi: 10.1016/j.bbamcr.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrews N.C., Schmidt P.J. Iron Homeostasis. Annu. Rev. Physiol. 2007;69(1):69–85. doi: 10.1146/annurev.physiol.69.031905.164337. [DOI] [PubMed] [Google Scholar]
- 6.Eid R., Arab N.T.T., Greenwood M.T. Iron mediated toxicity and programmed cell death: A review and a re-examination of existing paradigms. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2017;1864(2):399–430. doi: 10.1016/j.bbamcr.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Arosio P., Ingrassia R., Cavadini P. Ferritins: A family of molecules for iron storage, antioxidation and more. Biochimica et Biophysica Acta (BBA) - General Subjects. 2009;1790(7):589–599. doi: 10.1016/j.bbagen.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Raguzzi F., Lesuisse E., Crichton R.R. Iron storage in Saccharomyces cerevisiae. FEBS Lett. 1988 doi: 10.1016/0014-5793(88)80742-7. [DOI] [PubMed] [Google Scholar]
- 9.Kurz T., Eaton J.W., Brunk U.T. The role of lysosomes in iron metabolism and recycling. The International Journal of Biochemistry & Cell Biology. 2011;43(12):1686–1697. doi: 10.1016/j.biocel.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 10.Briat J.-F., Duc C., Ravet K., Gaymard F. Ferritins and iron storage in plants. Biochimica et Biophysica Acta (BBA) - General Subjects. 2010;1800(8):806–814. doi: 10.1016/j.bbagen.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Sharma S.S., Dietz K.-J., Mimura T. Vacuolar compartmentalization as indispensable component of heavy metal detoxification in plants: Vacuolar functions in HM detoxification. Plant, Cell Environ. 2016;39(5):1112–1126. doi: 10.1111/pce:12706. [DOI] [PubMed] [Google Scholar]
- 12.Li L., Chen O.S., Ward D.M., Kaplan J. CCC1 Is a Transporter That Mediates Vacuolar Iron Storage in Yeast. J. Biol. Chem. 2001;276(31):29515–29519. doi: 10.1074/jbc.M103944200. [DOI] [PubMed] [Google Scholar]
- 13.Cockrell A., McCormick S.P., Moore M.J., Chakrabarti M., Lindahl P.A. Mössbauer, EPR, and Modeling Study of Iron Trafficking and Regulation in Δccc1 and CCC1-up Saccharomyces cerevisiae. Biochemistry. 2014;53(18):2926–2940. doi: 10.1021/bi500002n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cockrell A.L., Holmes-Hampton G.P., McCormick S.P., Chakrabarti M., Lindahl P.A. Mössbauer and EPR Study of Iron in Vacuoles from Fermenting Saccharomyces cerevisiae. Biochemistry. 2011;50(47):10275–10283. doi: 10.1021/bi2014954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes-Hampton G.P., Jhurry N.D., McCormick S.P., Lindahl P.A. Iron Content of Saccharomyces cerevisiae Cells Grown under Iron-Deficient and Iron-Overload Conditions. Biochemistry. 2013;52(1):105–114. doi: 10.1021/bi3015339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishida K., Silver P.A. Induction of biogenic magnetization and redox control by a component of the target of rapamycin complex 1 signaling pathway. PLoS Biol. 2012 doi: 10.1371/journal.pbio.1001269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen T.Q., Dziuba N., Lindahl P.A. Isolated Saccharomyces cerevisiae vacuoles contain low-molecular-mass transition-metal polyphosphate complexes. Metallomics. 2019;11(7):1298–1309. doi: 10.1039/C9MT00104B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Urbanowski J.L., Piper R.C. The Iron Transporter Fth1p Forms a Complex with the Fet5 Iron Oxidase and Resides on the Vacuolar Membrane. J. Biol. Chem. 1999;274(53):38061–38070. doi: 10.1074/jbc.274.53.38061. [DOI] [PubMed] [Google Scholar]
- 19.Portnoy M.E., Liu X.F., Culotta V.C. Saccharomyces cerevisiae Expresses Three Functionally Distinct Homologues of the Nramp Family of Metal Transporters. Mol. Cell. Biol. 2000;20(21):7893–7902. doi: 10.1128/MCB.20.21.7893-7902.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh A., Kaur N., Kosman D.J. The Metalloreductase Fre6p in Fe-Efflux from the Yeast Vacuole. J. Biol. Chem. 2007;282(39):28619–28626. doi: 10.1074/jbc.M703398200. [DOI] [PubMed] [Google Scholar]
- 21.Park J., McCormick S.P., Chakrabarti M., Lindahl P.A. The Lack of Synchronization between Iron Uptake and Cell Growth Leads to Iron Overload in Saccharomyces cerevisiae during Post-exponential Growth Modes. Biochemistry. 2013;52(52):9413–9425. doi: 10.1021/bi4010304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim S.A., Punshon T., Lanzirotti A., Li L., Alonso J.M., Ecker J.R., Kaplan J., Guerinot M.L. Localization of Iron in Arabidopsis Seed Requires the Vacuolar Membrane Transporter VIT1. Science. 2006;314(5803):1295–1298. doi: 10.1126/science:1132563. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y., Xu Y.H., Yi H.Y., Gong J.M. Vacuolar membrane transporters OsVIT1 and OsVIT2 modulate iron translocation between flag leaves and seeds in rice. Plant J. 2012 doi: 10.1111/j.1365-313X.2012.05088.x. [DOI] [PubMed] [Google Scholar]
- 24.Narayanan N., Beyene G., Chauhan R.D., Gaitán-Solis E., Grusak M.A., Taylor N., Anderson P. Overexpression of Arabidopsis VIT1 increases accumulation of iron in cassava roots and stems. Plant Sci. 2015;240:170–181. doi: 10.1016/j.plantsci.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 25.Gollhofer J., Timofeev R., Lan P., Schmidt W., Buckhout T.J. Vacuolar-iron-transporter1-like proteins mediate iron homeostasis in Arabidopsis. PLoS One. 2014 doi: 10.1371/journal.pone.0110468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slavic K., Krishna S., Lahree A., Bouyer G., Hanson K.K., Vera I., Pittman J.K., Staines H.M., Mota M.M. A vacuolar iron-transporter homologue acts as a detoxifier in Plasmodium. Nat Commun. 2016;7(1) doi: 10.1038/ncomms10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhubhanil S., Chamsing J., Sittipo P., Chaoprasid P., Sukchawalit R., Mongkolsuk S. Roles of Agrobacterium tumefaciens membrane-bound ferritin (MbfA) in iron transport and resistance to iron under acidic conditions. Microbiol (United Kingdom) 2014 doi: 10.1099/mic.0.076802-0. [DOI] [PubMed] [Google Scholar]
- 28.Sankari S., O'Brian M.R. A Bacterial Iron Exporter for Maintenance of Iron Homeostasis. J. Biol. Chem. 2014;289(23):16498–16507. doi: 10.1074/jbc.M114.571562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brembu T., Jørstad M., Winge P., Valle K.C., Bones A.M. Genome-Wide Profiling of Responses to Cadmium in the Diatom Phaeodactylum tricornutum. Environ. Sci. Technol. 2011;45(18):7640–7647. doi: 10.1021/es2002259. [DOI] [PubMed] [Google Scholar]
- 30.Kato T., Kumazaki K., Wada M., Taniguchi R., Nakane T., Yamashita K., Hirata K., Ishitani R., Ito K., Nishizawa T., Nureki O. Crystal structure of plant vacuolar iron transporter VIT1. Nat. Plants. 2019;5(3):308–315. doi: 10.1038/s41477-019-0367-2. [DOI] [PubMed] [Google Scholar]
- 31.Chorostecki U., Molina M., Pryszcz L.P., Gabaldón T. MetaPhOrs 2.0: integrative, phylogeny-based inference of orthology and paralogy across the tree of life. Nucleic Acids Res. 2020 doi: 10.1093/nar/gkaa282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pryszcz L.P., Huerta-Cepas J., Gabaldón T. MetaPhOrs: Orthology and paralogy predictions from multiple phylogenetic evidence using a consistency-based confidence score. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkq953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sankari S., O’Brian M.R. Synthetic lethality of the bfr and mbfA genes reveals a functional relationship between iron storage and iron export in managing stress responses in Bradyrhizobium japonicum. PLoS One. 2016 doi: 10.1371/journal.pone.0157250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang G., Ulrich P.N., Storey M., Johnson D., Tischer J., Tovar J.A. Proteomic analysis of the acidocalcisome, an organelle conserved from bacteria to human cells. PLoS Pathog. 2014 doi: 10.1371/journal.ppat.1004555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams T.A., Cox C.J., Foster P.G., Szöllősi G.J., Embley T.M. Phylogenomics provides robust support for a two-domains tree of life. Nat Ecol Evol. 2020;4(1):138–147. doi: 10.1038/s41559-019-1040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao J. Molecular evolution of the Vacuolar Iron Transporter (VIT) Family genes in 14 plant species. Genes (Basel) 2019 doi: 10.3390/genes10020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma S., Kaur G., Kumar A., Meena V., Ram H., Kaur J. Gene expression pattern of vacuolar-iron transporter-like (VTL) genes in hexaploid wheat during metal stress. Plants. 2020 doi: 10.3390/plants9020229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mary V., Schnell Ramos M., Gillet C., Socha A.L., Giraudat J., Agorio A., Merlot S., Clairet C., Kim S.A., Punshon T., Guerinot M.L., Thomine S. Bypassing Iron Storage in Endodermal Vacuoles Rescues the Iron Mobilization Defect in the natural resistance associated-macrophage protein3natural resistance associated-macrophage protein4 Double Mutant. Plant Physiol. 2015;169(1):748–759. doi: 10.1104/pp.15.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andrews S.C. The Ferritin-like superfamily: Evolution of the biological iron storeman from a rubrerythrin-like ancestor. Biochimica et Biophysica Acta (BBA) - General Subjects. 2010;1800(8):691–705. doi: 10.1016/j.bbagen.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 40.Ruangkiattikul N., Bhubhanil S., Chamsing J., Niamyim P., Sukchawalit R., Mongkolsuk S. Agrobacterium tumefaciens membrane-bound ferritin plays a role in protection against hydrogen peroxide toxicity and is negatively regulated by the iron response regulator. FEMS Microbiol Lett. 2012;329(1):87–92. doi: 10.1111/j.1574-6968.2012.02509.x. [DOI] [PubMed] [Google Scholar]
- 41.Brear E.M., Bedon F., Gavrin A., Kryvoruchko I.S., Torres‐Jerez I., Udvardi M.K., Day D.A., Smith P.M.C. GmVTL1a is an iron transporter on the symbiosome membrane of soybean with an important role in nitrogen fixation. New Phytol. 2020;228(2):667–681. doi: 10.1111/nph.16734. [DOI] [PubMed] [Google Scholar]
- 42.Liu S., Liao L.L., Nie M.M., Peng W.T., Zhang M.S., Lei J.N., Zhong Y.J., Liao H., Chen Z.C. A VIT‐like transporter facilitates iron transport into nodule symbiosomes for nitrogen fixation in soybean. New Phytol. 2020;226(5):1413–1428. doi: 10.1111/nph.16506. [DOI] [PubMed] [Google Scholar]
- 43.Walton J.H., Kontra‐Kováts G., Green R.T., Domonkos Á., Horváth B., Brear E.M., Franceschetti M., Kaló P., Balk J. The Medicago truncatula Vacuolar iron Transporter‐Like proteins VTL4 and VTL8 deliver iron to symbiotic bacteria at different stages of the infection process. New Phytol. 2020;228(2):651–666. doi: 10.1111/nph.16735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hakoyama T., Niimi K., Yamamoto T., Isobe S., Sato S., Nakamura Y. The integral membrane protein SEN1 is required for symbiotic nitrogen fixation in Lotus japonicus nodules. Plant Cell Physiol. 2012 doi: 10.1093/pcp/pcr167. [DOI] [PubMed] [Google Scholar]
- 45.Lapinskas P.J., Lin S.-J., Culotta V.C. The role of the Saccharomyces cerevisiae CCC1 gene in the homeostasis of manganese ions. Mol Microbiol. 1996;21(3):519–528. doi: 10.1111/j.1365-2958.1996.tb02561.x. [DOI] [PubMed] [Google Scholar]
- 46.Ehrnstorfer I.A., Manatschal C., Arnold F.M., Laederach J., Dutzler R. Structural and mechanistic basis of proton-coupled metal ion transport in the SLC11/NRAMP family. Nat Commun. 2017;8(1) doi: 10.1038/ncomms14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taniguchi R., Kato H.E., Font J., Deshpande C.N., Wada M., Ito K., Ishitani R., Jormakka M., Nureki O. Outward- and inward-facing structures of a putative bacterial transition-metal transporter with homology to ferroportin. Nat Commun. 2015;6(1) doi: 10.1038/ncomms9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiner J., Kooij T.W.A. Phylogenetic profiles of all membrane transport proteins of the malaria parasite highlight new drug targets. Microb Cell. 2016 doi: 10.15698/mic2016.10.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eroglu S., Karaca N., Vogel-Mikus K., Kavčič A., Filiz E., Tanyolac B. The conservation of VIT1-dependent iron distribution in seeds. Front Plant Sci. 2019 doi: 10.3389/fpls.2019.00907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Momonoi K., Yoshida K., Mano S., Takahashi H., Nakamori C., Shoji K. A vacuolar iron transporter in tulip, TgVit1, is responsible for blue coloration in petal cells through iron accumulation. Plant J. 2009 doi: 10.1111/j.1365-313X.2009.03879.x. [DOI] [PubMed] [Google Scholar]
- 51.Swaney D.L., Beltrao P., Starita L., Guo A., Rush J., Fields S., Krogan N.J., Villén J. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat Methods. 2013;10(7):676–682. doi: 10.1038/nmeth.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Albuquerque C.P., Smolka M.B., Payne S.H., Bafna V., Eng J., Zhou H. A Multidimensional Chromatography Technology for In-depth Phosphoproteome Analysis. Mol Cell Proteomics. 2008;7(7):1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodrigues-Pousada C., Devaux F., Caetano S.M., Pimentel C., da Silva S., Cordeiro A.C. Yeast AP-1 like transcription factors (Yap) and stress response: A current overview. Microb Cell. 2019 doi: 10.15698/mic2019.06.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li L., Bagley D., Ward D.M., Kaplan J. Yap5 Is an Iron-Responsive Transcriptional Activator That Regulates Vacuolar Iron Storage in Yeast. MCB. 2008;28(4):1326–1337. doi: 10.1128/MCB.01219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li L., Ward D.M. Iron toxicity in yeast: transcriptional regulation of the vacuolar iron importer Ccc1. Curr Genet. 2018;64(2):413–416. doi: 10.1007/s00294-017-0767-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rietzschel N., Pierik A.J., Bill E., Lill R., Mühlenhoff U. The Basic Leucine Zipper Stress Response Regulator Yap5 Senses High-Iron Conditions by Coordination of [2Fe-2S] Clusters. Mol. Cell. Biol. 2015;35(2):370–378. doi: 10.1128/MCB.01033-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li L., Miao R., Bertram S., Jia X., Ward D.M., Kaplan J. A Role for Iron-Sulfur Clusters in the Regulation of Transcription Factor Yap5-dependent High Iron Transcriptional Responses in Yeast. J. Biol. Chem. 2012;287(42):35709–35721. doi: 10.1074/jbc.M112.395533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pimentel C., Vicente C., Menezes R.A., Caetano S., Carreto L., Rodrigues-Pousada C. The role of the Yap5 transcription factor in remodeling gene expression in response to Fe bioavailability. PLoS One. 2012 doi: 10.1371/journal.pone.0037434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li L., Jia X., Ward D.M., Kaplan J. Yap5 Protein-regulated Transcription of the TYW1 Gene Protects Yeast from High Iron Toxicity. J. Biol. Chem. 2011;286(44):38488–38497. doi: 10.1074/jbc.M111.286666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li L., Kaplan J., Ward D.M. The glucose sensor Snf1 and the transcription factors Msn2 and Msn4 regulate transcription of the vacuolar iron importer gene CCC1 and iron resistance in yeast. J. Biol. Chem. 2017;292(37):15577–15586. doi: 10.1074/jbc.M117.802504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Puig S., Vergara S.V., Thiele D.J. Cooperation of Two mRNA-Binding Proteins Drives Metabolic Adaptation to Iron Deficiency. Cell Metab. 2008;7(6):555–564. doi: 10.1016/j.cmet.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Du Y., Cheng W., Li W.-F. Expression profiling reveals an unexpected growth-stimulating effect of surplus iron on the yeast Saccharomyces cerevisiae. Mol Cells. 2012;34(2):127–132. doi: 10.1007/s10059-012-2242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Romero A.M., Ramos-Alonso L., Montellá-Manuel S., García-Martínez J., de la Torre-Ruiz M.Á., Pérez-Ortín J.E., Martínez-Pastor M.T., Puig S. A genome-wide transcriptional study reveals that iron deficiency inhibits the yeast TORC1 pathway. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2019;1862(9):194414. doi: 10.1016/j.bbagrm.2019.194414. [DOI] [PubMed] [Google Scholar]
- 64.Ramos-Alonso, Lucía; Romero, Antonia María; Martínez-Pastor, María Teresa; Puig S. Iron Regulatory Mechanisms in Saccharomyces cerevisiae. Front Microbiol 2020;11:582830. https://doi.org/doi: 10.3389/fmicb.2020.582830 [DOI] [PMC free article] [PubMed]
- 65.Philpott C.C., Protchenko O. Response to Iron Deprivation in Saccharomyces cerevisiae. Eukaryot Cell. 2008;7(1):20–27. doi: 10.1128/EC.00354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martins L.J., Jensen L.T., Simon J.R., Keller G.L., Winge D.R. Metalloregulation of FRE1 and FRE2 Homologs in Saccharomyces cerevisiae. J. Biol. Chem. 1998;273(37):23716–23721. doi: 10.1074/jbc.273.37.23716. [DOI] [PubMed] [Google Scholar]
- 67.Rutherford J.C., Jaron S., Ray E., Brown P.O., Winge D.R. A second iron-regulatory system in yeast independent of Aft1p. Proc Natl Acad Sci. 2001;98(25):14322–14327. doi: 10.1073/pnas.261381198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Puig S., Askeland E., Thiele D.J. Coordinated Remodeling of Cellular Metabolism during Iron Deficiency through Targeted mRNA Degradation. Cell. 2005;120(1):99–110. doi: 10.1016/j.cell.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 69.Chen O.S., Kaplan J. CCC1 Suppresses Mitochondrial Damage in the Yeast Model of Friedreich's Ataxia by Limiting Mitochondrial Iron Accumulation. J. Biol. Chem. 2000;275(11):7626–7632. doi: 10.1074/jbc.275.11.7626. [DOI] [PubMed] [Google Scholar]
- 70.Li L., Kaplan J. A Mitochondrial-Vacuolar Signaling Pathway in Yeast That Affects Iron and Copper Metabolism. J. Biol. Chem. 2004;279(32):33653–33661. doi: 10.1074/jbc.M403146200. [DOI] [PubMed] [Google Scholar]
- 71.Li L., Murdock G., Bagley D., Jia X., Ward D.M., Kaplan J. Genetic Dissection of a Mitochondria-Vacuole Signaling Pathway in Yeast Reveals a Link between Chronic Oxidative Stress and Vacuolar Iron Transport. J. Biol. Chem. 2010;285(14):10232–10242. doi: 10.1074/jbc.M109.096859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu N., Dong Y., Cheng X., Yu Q., Qian K., Mao J., Jia C., Ding X., Zhang B., Chen Y., Zhang B., Xing L., Li M. Cellular iron homeostasis mediated by the Mrs4–Ccc1–Smf3 pathway is essential for mitochondrial function, morphogenesis and virulence in Candida albicans. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2014;1843(3):629–639. doi: 10.1016/j.bbamcr.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 73.Milgrom E., Diab H., Middleton F., Kane P.M. Loss of Vacuolar Proton-translocating ATPase Activity in Yeast Results in Chronic Oxidative Stress. J. Biol. Chem. 2007;282(10):7125–7136. doi: 10.1074/jbc.M608293200. [DOI] [PubMed] [Google Scholar]
- 74.Diab H.I., Kane P.M. Loss of Vacuolar H + -ATPase (V-ATPase) Activity in Yeast Generates an Iron Deprivation Signal That Is Moderated by Induction of the Peroxiredoxin TSA2. J. Biol. Chem. 2013;288(16):11366–11377. doi: 10.1074/jbc.M112.419259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen K.L., Ven T.N., Crane M.M., Brunner M.L.C., Pun A.K., Helget K.L., Brower K., Chen D.E., Doan H.a., Dillard-Telm J.D., Huynh E., Feng Y.-C., Yan Z., Golubeva A., Hsu R.A., Knight R., Levin J., Mobasher V., Muir M., Omokehinde V., Screws C., Tunali E., Tran R.K., Valdez L., Yang E., Kennedy S.R., Herr A.J., Kaeberlein M., Wasko B.M. Loss of vacuolar acidity results in iron-sulfur cluster defects and divergent homeostatic responses during aging in Saccharomyces cerevisiae. GeroScience. 2020;42(2):749–764. doi: 10.1007/s11357-020-00159-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hughes C.E., Coody T.K., Jeong M.-Y., Berg J.A., Winge D.R., Hughes A.L. Cysteine Toxicity Drives Age-Related Mitochondrial Decline by Altering Iron Homeostasis. Cell. 2020;180(2):296–310.e18. doi: 10.1016/j.cell.2019.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee H.N., Mostovoy Y., Hsu T.Y., Chang A.H., Brem R.B. Divergence of Iron Metabolism in Wild Malaysian Yeast. G3. 2013;3(12):2187–2194. doi: 10.1534/g3.113.008011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thiébaut A., Delaveau T., Benchouaia M., Boeri J., Garcia M., Lelandais G., Devaux F. The CCAAT-Binding Complex Controls Respiratory Gene Expression and Iron Homeostasis in Candida Glabrata. Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-03750-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gerwien F., Safyan A., Wisgott S., Hille F., Kaemmer P., Linde J., Brunke S., Kasper L., Hube B. A Novel Hybrid Iron Regulation Network Combines Features from Pathogenic and Nonpathogenic Yeasts. mBio. 2016;7(5) doi: 10.1128/mBio.01782-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mercier A., Pelletier B., Labbé S. A Transcription Factor Cascade Involving Fep1 and the CCAAT-Binding Factor Php4 Regulates Gene Expression in Response to Iron Deficiency in the Fission Yeast Schizosaccharomyces pombe. Eukaryot Cell. 2006;5(11):1866–1881. doi: 10.1128/EC.00199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Labbé S., Khan M.G.M., Jacques J.-F. Iron uptake and regulation in Schizosaccharomyces pombe. Curr Opin Microbiol. 2013;16(6):669–676. doi: 10.1016/j.mib.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 82.Gsaller F., Eisendle M., Lechner B.E., Schrettl M., Lindner H., Müller D., Geley S., Haas H. The interplay between vacuolar and siderophore-mediated iron storage in Aspergillus fumigatus. Metallomics. 2012;4(12):1262. doi: 10.1039/c2mt20179h. [DOI] [PubMed] [Google Scholar]
- 83.Schrettl M., Beckmann N., Varga J., Heinekamp T., Jacobsen I.D., Jöchl C. HapX-Mediated adaption to iron starvation is crucial for virulence of Aspergillus fumigatus. PLoS Pathog. 2010 doi: 10.1371/journal.ppat.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gsaller F., Hortschansky P., Beattie S.R., Klammer V., Tuppatsch K., Lechner B.E. The J anus transcription factor H ap X controls fungal adaptation to both iron starvation and iron excess. EMBO J. 2014 doi: 10.15252/embj.201489468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Furukawa T., Scheven M.T., Misslinger M., Zhao C., Hoefgen S., Gsaller F. The fungal CCAAT-binding complex and HapX display highly variable but evolutionary conserved synergetic promoter-specific DNA recognition. Nucleic Acids Res. 2020 doi: 10.1093/nar/gkaa109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schrettl M., Bignell E., Kragl C., Sabiha Y., Loss O., Eisendle M. Distinct roles for intra- and extracellular siderophores during Aspergillus fumigatus infection. PLoS Pathog. 2007 doi: 10.1371/journal.ppat.0030128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen C., Pande K., French S., Tuch B., Noble S. An Iron Homeostasis Regulatory Circuit with Reciprocal Roles in Candida albicans Commensalism and Pathogenesis. Cell Host Microbe. 2011;10(2):118–135. doi: 10.1016/j.chom.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Singh R.P., Prasad H.K., Sinha I., Agarwal N., Natarajan K. Cap2-HAP Complex Is a Critical Transcriptional Regulator That Has Dual but Contrasting Roles in Regulation of Iron Homeostasis in Candida albicans. J. Biol. Chem. 2011;286(28):25154–25170. doi: 10.1074/jbc.M111.233569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xu N., Cheng X., Yu Q., Zhang B., Ding X., Xing L., Li M. Identification and functional characterization of mitochondrial carrier Mrs4 in Candida albicans. FEMS Yeast Res. 2012;12(7):844–858. doi: 10.1111/j.1567-1364.2012.00835.x. [DOI] [PubMed] [Google Scholar]
- 90.Narayanan N., Beyene G., Chauhan R.D., Gaitán-Solís E., Gehan J., Butts P. Biofortification of field-grown cassava by engineering expression of an iron transporter and ferritin. Nat Biotechnol. 2019 doi: 10.1038/s41587-018-0002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.