Graphical abstract
Keywords: Type VI secretion system (T6SS), Adaptor, Toxic effector, Bacterial genome, Contextual gene, Bioinformatics
Highlights
-
•
Each class of adaptors can be used as an effective marker to identify T6SS effectors.
-
•
The PRK06147 homolog may be a novel adaptor.
-
•
1356 putative toxic effectors related to adaptors from 92 families were identified.
Abstract
Toxic effectors secreted by the type VI secretion system (T6SS) facilitate interbacterial warfare, as well as pathogenesis toward humans, animals and plants. However, systematically predicting T6SS effectors remains challenging due to their sequence and functional diversity. In this study, we systematically identified putative T6SS toxic effectors in prokaryotic genomes on the basis of the observation that genes encoding adaptor proteins and genes encoding cognate effector proteins are generally adjacent in the genome. Adaptor proteins are mediators that help to load their cognate effectors onto the T6SS spike complex. The contextual genes of the known adaptor proteins (DUF1795, DUF2169 or DUF4123) all exhibited a high proportion of encoding T6SS spike complex protein (VgrG or PAAR) and effector proteins. On the basis of the genomic context, we found that PRK06147 might be a novel adaptor protein. These four adaptors are widely distributed among the bacterial genomes. From neighbors of 5297 adaptor genes, we identified 1356 putative effector genes from 92 different families, and two-thirds were currently annotated as hypothetical proteins or as having unknown functions. Our results indicate that each class of adaptors can be used as an effective marker to identify T6SS toxic effectors, moreover, this approach can promote the discovery of new effectors.
1. Introduction
The type VI secretion system (T6SS) is a multicomponent nanomachine of bacteria that is analogous to the contractile bacteriophage tail. This machine mediates killing toward neighboring prokaryotic or eukaryotic cells in a contact-dependent manner by delivering toxic effectors [1], [2], [3]. T6SS toxin effectors help bacteria win competitive advantages in intraspecific or interspecific conflicts [4], [5], [6], [7] and are also a critical factor in the pathogenicity of many bacterial pathogens that threaten human health [8], [9]. Many T6SS effectors have been identified, and they exhibit diverse activities on a wide range of targets [10]. The T6SS effectors include nucleases that cleave DNA and RNA [11], [12], [13], [14], [15], [16], [17], pore-forming toxins and phospholipases that target cell membranes [18], [19], glycoside hydrolases and amidases that function on the cell wall [4], [20], [21], [22], [23], [24], NADases that affect the cell energy balance [25], [26], and ADP-ribosyltransferases that target tubulin-like protein to interrupt cell division [27]. However, the known toxin effectors represent only a small fraction of the large number of T6SS effectors in bacteria. Because T6SS effectors exhibit high diversity in sequence and function, identifying unknown T6SS effectors is challenging.
The T6SS machine is a large complex that consists of 13 components. In the T6SS, the VgrG protein (in a trimer form) and the conical PAAR protein form the spike complex, which is located at the top of the T6SS secretion structure and responsible for creating an opening in the target cell envelope [3], [28], [29]. Toxic effectors can be fused to VgrG or PAAR as an extension domain or as an independent protein loaded onto the secretory component through protein–protein interactions [29], [30]. Thus, the VgrG and PAAR proteins, which have an extension domain at the C-terminus, are promising effector candidates. In addition, the N-terminal domain of the multidomain effector proteins may have specific motifs, such as rearrangement hotspots (RHS), YD repeats, MIX motifs and FIX motifs, which can serve as markers for unknown T6SS effectors [31], [32], [33]. However, many T6SS effectors do not contain the abovementioned identification characteristics, especially single-domain proteins. These effectors require additional assistance to load onto the T6SS, and this assistance is provided by proteins known as adaptors or chaperones [34]. The adaptors assist with loading effectors onto the T6SS spike complex, and they are not secreted by the T6SS. In species such as Pseudomonas aeruginosa and Serratia marcescens, the DUF1795 protein is used as an adaptor to bind to the effector protein and it is delivered with the effector to the binding site of VgrG during the assembly process of the T6SS [26], [35], [36]. In Agrobacterium tumefaciens, adaptors characterized by the conserved DUF2169 domain or DUF4123 domain have been identified, and they are necessary for T6SS effector secretion [37]. Moreover, DUF4123 has also been identified as an adaptor in species such as Vibrio cholerae, P. aeruginosa and Aeromonas hydrophila [17], [18], [30], [38], [39].
Adaptor genes are generally adjacent to genes that encode putative T6SS effectors in the genome, which suggests that adaptor genes may have the potential to be employed as markers for identifying and predicting T6SS effectors [34], [39]. In this study, we attempted to identify the homologs of the three known adaptors DUF1795, DUF2169 and DUF4123 on a large scale in all reference or representative prokaryotic genomes, determine the characteristics of their upstream and downstream neighboring genes to identify other unknown kinds of adaptors and comprehensively identify the unknown T6SS effectors related to all adaptor genes in the genome.
2. Materials and methods
2.1. Acquisition of adaptor homologs
Taxonomic information and functional annotation information of the protein domain were acquired from the Conserved Domain Database (CDD) and PFAM database [40], [41]. Three classes of adaptor homologs were defined: DUF1795 (DcrB, cl03356), DUF2169 (cl02212) or DUF4123 (cl16292). All proteins with adaptor domains were identified in the CDD (52,910 position-specific scoring matrices (PSSMs)) by RPS-BLAST, and the retrieval condition was that the E-value did not exceed 0.01. Taxonomic information on the species was obtained from the NCBI taxonomic database.
The NCBI database defines at least one reference or representative genome for each sequenced species, and this forms a collection of 11,495 prokaryotic genomes [42]. These genomes usually have high sequencing quality. The genome sequence information of all reference or representative prokaryotes and the positional information of all coding genes in the genome were obtained from the NCBI reference sequence (RefSeq) database [42]. The identified adaptor proteins were correlated with the genomic protein products via the accession number, and the adaptor-encoding genes in each genome were determined.
2.2. Genomic context analyses of adaptor homologs
Ten upstream genes and ten downstream genes of each adaptor-encoding gene were extracted from all reference or representative prokaryotic genomes. CDD domain annotations of the protein products of all the genes were performed on the 52,910 PSSMs with an E-value threshold of 0.01 [41].
The genes encoding putative spike complex components VgrG and PAAR proteins were identified based on the domain annotation. The identification of toxic effectors was performed based on the 114 families with reported toxic effector functions [31]. Because these families might be multifunctional, we defined that the putative toxic effectors must satisfy the two conditions as followed: (1) the homologs belonged to the 114 families with reported toxic effector functions and (2) the encoding gene was located within the range of 10 genes upstream and 10 genes downstream of the adaptor-encoding gene.
2.3. Recognition of novel adaptors
The 20 gene neighbors of the homologs of the spike complex component (PAAR or VgrG) and the 114 families with reported toxin effector functions were extract from the genomes, and domain annotations were performed on their protein products, which produced two collections. After removing the function-known families, the intersections of the two collections were selected as candidates of new adaptor. The homologs and gene neighbors of the candidates were identified in the reference or representative prokaryotic genomes. The putative novel adaptors met the following two conditions at the same time: (1) at least one of PAAR or VgrG homologs was encoded by adjacent genes located upstream or downstream of the cognate candidate genes and (2) the T6SS toxic effector homologs were encoded by the first two downstream genes.
Like the aforementioned three classes of known adaptors, the homologs of the novel adaptor (PRK06147) were identified in all reference or representative prokaryotic genomes. Ten upstream and ten downstream genes of PRK06147 homologs were extracted from the genome, and domain annotations were performed on their protein products. The genes encoding putative spike complex components and toxic effectors were recognized on the basis of the domain annotation.
2.4. Analysis of adaptor sequence characteristics
All four classes of adaptor homologs were annotated to determine whether they were fused to additional domains. The domains were annotated for all the identified putative toxic effectors to determine whether they were fused to a spike complex component VgrG or PAAR. The domain annotation was performed on the 52,910 PSSMs by RPS-BLAST, and the retrieval condition was set at that the E-value did not exceed 0.01.
2.5. Identification of T6SS and eCIS in genome
TssB (IglA, VipA), TssC (IglB, VipB), TssD (Hcp) and TssM (IcmF) are four conserved components of T6SS [43]. We set that homologs of at least three of these four T6SS components were encoded by the genome as a criterion to determine the existence of T6SS in the genome.
Afp1/5 (Phage_T4_gp19), afp2/3/4 (Phage_sheath_1), afp11 (Baseplate_J) and afp16 (DUF4255) are functionally critical to extracellular contractile injection systems (eCISs) [44]. Similarly, the criterion used for determining the existence of T6SS was applied to determine the existence of eCIS.
We also evaluated the proportion of adaptor homologs locating in the T6SS or eCIS gene cluster in the genome. If at least two of the TssB, TssC, TssD and TssM homologs were encoded by the 10 upstream genes or 10 downstream genes adjacent to the adaptor-encoding gene, the adaptor-encoding gene was considered to be in the T6SS gene cluster. Similarly, if at least two of the Afp1/5, afp2/3/4, afp11 and afp16 homologues were encoded by the adjacent 10 genes upstream or 10 genes downstream of adaptor-encoding gene, the adaptor-encoding gene was considered to be in the eCIS gene cluster.
3. Results
3.1. Adaptors are adjacent to the T6SS spike complex and effector genes in the genome
We searched for all adaptor homologs from prokaryotes containing the DUF1795 (DcrB, cl03356), DUF2169 (cl02212) or DUF4123 (cl16292) domain in the nonredundant NCBI reference sequence (RefSeq) database. Under the condition that the E-value of RPS-BLAST was equal to or less than 0.01, we identified 7475 proteins containing the DUF1795 domain, 4935 proteins containing the DUF2169 domain, and 11,325 proteins containing the DUF4123 domain. The NCBI defines a collection of 11,495 genomes referencing or representing each sequenced prokaryotic species [42]. Our following analyses were performed on these reference or representative prokaryotic genomes.
The adaptor protein mediates the interaction between the T6SS spike complex (PAAR protein or VgrG protein) and the cognate effector protein. Genes encoding the adaptor, the spike complex and effector are predicted to be adjacent in the genome. Therefore, we scanned the adaptor homologs and their gene neighbors (within the range of 10 genes upstream and 10 genes downstream) on a large scale in all the 11,495 reference or representative prokaryotic genomes (Fig. 1). We identified 1826, 1123 and 1941 genes that encoded DUF1795, DUF2169 and DUF4123 domains from these genomes, respectively (Table S1); 423, 966 and 1424 VgrG homologs that were related to DUF1795, DUF2169 and DUF4123 homologs, respectively; and 421, 934 and 735 PAAR homologs that were related to DUF1795, DUF2169 and DUF4123 homologs, respectively. The proportion that the first gene located upstream of the DUF4123 homologs encoded the spike complex was as high as 65.7%, whereas the value of any other position located upstream or downstream (within 10 genes) was not more than 6.5%. Similarly, the proportion that the first gene upstream of the DUF1795 homolog encoded the spike complex was at least 15.3%, while that of the first downstream gene was 19.7%, and any other position was not more than 3.0%. The first upstream gene of the DUF2169 homolog also had the greatest proportion of encoding the spike complex (35.5%), and the values of the three closest genes upstream or downstream all exceeded 10%. Therefore, the closer a gene is to the adaptor homolog, the greater the proportion encoding a T6SS spike complex protein. Interestingly, for the two members of the spike complex, the upstream gene of the adaptor homolog primarily encoded the VgrG protein, while the downstream gene primarily encoded the PAAR protein.
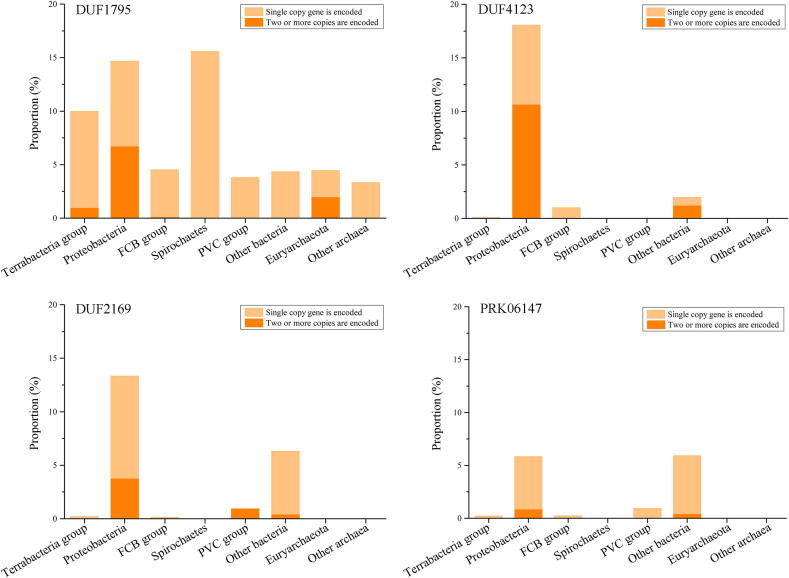
Fig. 1.
Frequency of occurrence of genes encoding spike complex proteins (VgrG or PAAR) or effector proteins located within 10 genes upstream or 10 genes downstream of the adaptor protein genes. The position of the gene encoding the adaptor is taken as 0, the position of the gene upstream of the gene encoding the adaptor is labeled with a negative value, and the position of the gene downstream is labeled with a positive value. VgrG is shown in light green, PAAR is shown in dark green, and the predicted toxin is shown in pink. The analysis was performed on 11,495 reference or representative prokaryotic genomes. These genomes contained 1826, 1123, 1941 and 407 homologs of the DUF1795, DUF2169, DUF4123 and PRK06147, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Furthermore, we also identified homologs of putative T6SS toxic effectors in the gene neighbors of adaptor homologs on the basis of the 114 families with reported toxic effector functions (Fig. 1). These families might be multifunctional, the putative toxic effectors were thus defined to be the proteins of the 114 families encoded by the 10 upstream genes or 10 downstream genes of the adaptor-encoding gene. As a result, we revealed 439, 440 and 494 putative toxic effectors related to the homologs of the DUF1795, DUF2169 and DUF4123, respectively (Table S2). Several typical clusters containing toxins are shown in Fig. 2.
Fig. 2.
Graphical depiction of adaptor‐containing gene clusters drawn to scale in several representative prokaryotic genomes. The bacterial species encoding the genes are indicated on the left. These gene clusters all included genes encoding toxic effectors and immunity proteins.
The proportion of the DUF2169 homologs with a putative toxic effector detected within 10 genes upstream or 10 genes downstream was as high as 32.7%, and the corresponding values for DUF4123 and DUF1795 also reached 21.2% and 18.6%, respectively. The proportion that the first gene located downstream of the DUF1795 homolog encoded a putative toxin was 13.6%, while that of any other position upstream or downstream (within 10 genes) was less than 1.6%. The proportion that the first downstream gene of the DUF2169 homolog encoded a putative toxin was 9.3%, while the corresponding value of the second downstream gene was 12.6%. Similarly, the proportions that the first and second downstream genes of the DUF4123 homolog encoded a putative toxin was 7.6% and 7.8%, respectively. Therefore, compared with those at the other positions, the first two genes located downstream of the adaptor homologs constituted a greater proportion of those encoding toxin effectors.
Remarkably, many putative toxic effectors were fused to the spike complex component PAAR or VgrG (Table S2). The proportion of the DUF1795-associated putative toxic effectors fused to PAAR or VgrG domain was 66.3%, and the corresponding value of the DUF2169 and DUF4123was 55.0% and 14.8%, respectively (Fig. 3A). The toxic effectors encoded by the first two genes downstream of the adaptor homologs were rarely fused to VgrG domain. However, 96.5% of the putative toxic effectors encoded by the first two genes located downstream of the DUF1795 homologs were fused to the PAAR domain (Fig. 3B). The corresponding value of the DUF2169 reached 73.8%, but none of the toxic effectors related to DUF4123 was fused to the PAAR domain. In addition, 73.9% of PAAR associated with DUF1795 were fused to RHS, while for any other adaptor, the proportion did not exceed 5%. RHS proteins are a class of giant proteins representing a major group of secreted polymorphic toxins [45], [46].
Fig. 3.
Proportion of adaptor homologs with putative toxic effectors identified among gene neighbors. (A). Proportions of the DUF1795, DUF2169, DUF4123 and PRK06147 homologs with toxic effectors detected within the 10 upstream or 10 downstream gene neighbors. (B). Proportions of the adaptor homologs with toxic effectors detected in the first two downstream genes. The effectors fused to the VgrG domain, the PAAR domain and not fused to these two domains are shown in light green, dark green and gray colors, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2. The PRK06147 homolog may be a novel adaptor
Based on the high proportion that the gene neighbors located upstream or downstream of the adaptor homologs encode the spike complexes and effector homologs, we scanned the gene neighbors of the spike complexes and effector homologs in all reference or representative prokaryotic genomes to identify novel and unknown adaptors (Fig. 4). We found that in addition to the three known adaptor classes, the closer a gene is to the spike complexes and the effector homologs, the higher the proportion it encodes PRK06147 (cl30694) homologs. The PRK06147 homolog was predicted to be 3-oxoacyl-(acyl carrier protein) synthase. The gene of this family has been reported as being associated with T6SS gene clusters, although the function of this protein in T6SS has not been determined [15], [47], [48].
Fig. 4.
Schematic diagram of the recognition method of a novel adaptor PRK06147. First, we analyzed the gene neighbors of the three classes of known adaptors (DUF1795, DUF2169 and DUF4123). Second, we defined the recognition criterion based on that the closer a gene is to the adaptor homolog, the higher the proportion it encodes a T6SS spike complex protein or putative toxic effector (Fig. 1). Third, we analyzed the gene neighbors of the genes encoding T6SS spike complex components and the known toxic effector homologs, and determined their intersections. Finally, the novel adaptor PRK06147 was recognized. Green represents the spike complex component (PAAR or VgrG), pink represents the toxic effector. A darker color corresponds to a greater proportion of the related gene. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
We identified 407 PRK06147 homologs in the reference or representative prokaryotic genomes (Table S1). The gene neighbors of the PRK06147 homologs were analyzed, which revealed 339 VgrG homologs and 326 PAAR homologs (Fig. 1). The proportion that the first gene located downstream of these PRK06147 homologs encoded a component of the spike complex was as high as 52.3%, and almost all were PAAR. There were 159 putative toxic effectors related to PRK06147 homologs (Table S2). The proportion of PRK06147 homologs with a putative toxic effector detected in the range of 10 upstream genes and 10 downstream genes reached 36.1%, which exceeded that of any of the other three known adaptors (Fig. 3A). Particularly, the proportion that the first gene located downstream of the PRK06147 homologs encoded the T6SS toxic effector homologs was 24.3%. The proportion of putative toxic effectors related to PRK06147 fusing to PAAR or VgrG domain was 54.1%, which was lower than the corresponding values of the DUF1795 and DUF2169. However, 64.5% of the putative effectors encoded by the first two genes downstream of the PRK06147 homologs were fused to PAAR domain, but no putative effectors were fused to VgrG. In conclusion, like the other three classes of known adaptors, PRK06147 homologs were adjacent to the T6SS spike complex and effector genes in the genome. Therefore, we speculated that PRK06147 may be a novel adaptor.
Notably, among the four classes of adaptors, the proportion of any two adaptor homologs being adjacent in the genome (within 10 upstream and 10 downstream genes) was normally less than 5%, except that 74.4% of PRK06147 homologs were adjacent to DUF2169 homologs (Figure S1), of which more than 80% were encoded by the first upstream gene. PRK06147 and DUF2169 were determined to be adjacent in the genome, implying that they were closely related in function. Compared with DUF2169, the PRK06147 homologs and effector homologs were closer in the genome. In addition, many gene clusters encoded only PRK06147 and effector homologs, but did not encode other adaptors (Fig. 2).
3.3. The four classes of adaptors have different sequence characteristics
The four classes of adaptors had no homology in sequence, and were clearly different in protein size (Fig. 5A). The smallest DUF1795 proteins exhibited an average size of only approximately 220 aa, and more than 80% had 100–250 residues. The average size of the DUF4123 proteins was approximately 280 aa, and 90% were in the range of 150–350 aa. The PRK06147 protein was relatively large and exhibited an average size of approximately 380 aa, and 90% were in the range of 300–450 aa. The DUF2169 protein was the largest and exhibited an average size of approximately 545 aa, and large differences between the members were observed, with approximately 60% including 300–450 residues, and approximately 20% including 800–900 residues. We also determined the size of the core region of the domain through RPS-BLAST. The DUF1795 and DUF4123 domains had an average of 132 and 117 residues, respectively, while the DUF2169 and PRK06147 domains had an average of 293 and 296 residues, respectively.
Fig. 5.
Sequence characteristics of adaptors. (A). Protein lengths of the four classes of adaptors. (B). Proportions of the four classes of adaptors containing additional domains.
Most of adaptors were single-domain proteins, and a small portion had additional domains (Fig. 5B). In addition, 33.4% of the DUF2169-containing proteins were multidomain, and almost all the additional domains were located at the C-terminus of the proteins. Meanwhile, 15.2%, 8.3% and 2.7% of the DUF1795-, PRK06147- and DUF4123-containing proteins were multidomain, and the probabilities that their additional domains were located at the N-terminus or C-terminus of the protein were relatively close. There were a wide variety of additional domains detected in the four classes of adaptors, although the additional domains fused with each class of adaptor were specific. The most common additional domains in the DUF2169-containing protein were YjbI (present in 29.3% of the DUF2169 protein) and pentapeptide (present in 11.9%), both of which had pentapeptide repeats with unknown functions (Figure S2). In the DUF1795-, DUF4123- and PRK06147-containing proteins, the most common additional domains were PKc-like (5.8%), FHA (1.1%) and TIGR02270 (3.8%), respectively. These additional domains may be related to the interaction between the toxic effector and the T6SS spike complex mediated by the adaptor, although the specific mechanism has not been determined. In conclusion, the four classes of adaptors had different sequence characteristics, which implies that they might mediate the delivery of toxic effector in different modes.
3.4. Adaptor homologs are widely distributed among bacterial genomes
We analyzed the distributions of the four classes of adaptor-encoding genes in 11,495 reference or representative prokaryotic genomes (Table S3). In bacterial genomes, we identified 11.1%, 5.7%, 7.5% and 2.6% genomes encoding DUF1795, DUF2169, DUF4123 and PRK06147 domains, respectively (Fig. 6). A total of 4.3% of the archaeal genomes encoded the DUF1795 domain, but archaeal genomes encoding the other adaptor domains were not found. Among several major prokaryote groups, more than 10% of the Terrabacteria group, Proteobacteria and Spirochaetes genomes encoded DUF1795, more than 10% of the Proteobacteria genomes encoded DUF2169 and DUF4123, and more than 5% of the Proteobacteria genomes encoded PRK06147. Totally, 32.6% of Proteobacteria encoded at least one adaptor, of which the proportion in Alphaproteobacteria was 13.9%, in Betaproteobacteria was 44.8%, in Gammaproteobacteria was 45.0%, and in delta/epsilon subdivisions was 22.8% (Figure S3). Therefore, adaptor-encoding genes are widely distributed among bacterial genomes, especially Proteobacteria genomes.
Fig. 6.
Distribution and copy number of the four classes of adaptors among prokaryotic phyla. A single copy is represented by yellow, and multiple copies are represented by orange. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Instances of multiple copies of four classes of adaptor-encoding genes in the reference or representative prokaryotic genomes were also counted (Table S4). Up to 57.6% of members in the genomes encoding DUF4123 encoded two or more copies, while for the genomes encoding DUF1795, DUF2169 and PRK06147, the corresponding values reached 28.3%, 27.1% and 13.0%, respectively. The proportions of multiple-copy adaptors encoded among different taxa varied greatly. For example, up to 45.6% of the 658 Proteobacteria genomes, 9.6% of the 481 Terrabacteria groups, and none of the 22 Spirochaetes genomes encoding DUF1795 encoded multiple copies. The maximum copy number of the genes encoding four classes of adaptors in a single genome reached 70 in Chondromyces apiculatus DSM 436, which is a member of the Proteobacteria order Myxococcales. Moreover, all strains with more than 20 copies of the adaptor genes were derived from myxobacteria.
3.5. Most adaptor homologs are related to T6SS
The conserved components of T6SS in the reference or representative prokaryotic genomes were identified. We used at least three of the four conserved component homologs of T6SS (TssB, TssC, TssD and TssM [43]) encoded by the genome as a criterion to determine the existence of T6SS in the genome. As a result, we identified a total of 1729 genomes encoding T6SS, of which 96.8% (1674) were from the Proteobacteria and none were from Terrabacteria. Among the 11,495 reference or representative prokaryotic genomes, 2380 genomes were identified to possess no T6SS, but they encoded PAAR homologs and/or VgrG homologs.
The extracellular contractile injection system (eCIS) central spike resembles T6SS, although it is mechanistically distinct from T6SS and the spike complex is also formed by the PAAR and VgrG protein homologs [49]. We similarly set a criterion that homologs of at least three of the four critical components of eCISs (Afp1/5, afp2/3/4, afp11 and afp16 [44]) were encoded by the genome to determine the existence of eCIS. As a result, a total of 1305 genomes encoding eCIS were identified, of which 617 were from Terrabacteria, 390 were from Proteobacteria, 247 were from the FCB group, and 29 were from Euryarchaeota. Notably, among the 2380 genomes possessing no T6SS but encoding the spike complex, 1036 encoded eCIS. However, 1344 genomes, which were from more than 10 prokaryotic phyla, did not encode the T6SS or the eCIS, but still encoded the spike complex. We speculated that these genomes may encode atypical T6SS or eCIS, or have other unknown secretion mechanisms.
We further analyzed whether the adaptor homologs were related to T6SS or eCIS (Fig. 7A). We identified 5297 adaptor homologs from 2113 reference or representative genomes, of which 1302 genomes encoded T6SS (Table S4). The genomes with T6SS encoded 4336 adaptor homologs, which accounted for 81.9% of the total number of identified adaptor homologs. There were also 487 adaptor homologs from 371 genomes that did not encode T6SS but encoded VgrG or PAAR, and 275 of these genomes were predicted to encode eCIS. In addition, there were 474 adaptor homologs from 440 genomes that had neither T6SS nor spike complexes. Among them, the proportion of the DUF1795 homologs was as high as 94.5% (448), while the DUF2169, DUF4123 and PRK06147 homologs numbered only 8, 10 and 8, respectively. Therefore, we speculated that the DUF1795 homologs might have evolved multiple functions, and might not function as adaptors in many cases. We found that the number of adaptor homologs in the genome was highly positively correlated with the number of VgrG genes (r = 0.73, p < 0.01) and PAAR genes (r = 0.67, p < 0.01), but had a weak correlation with the number of T6SS gene clusters (r = 0.43, p < 0.01) (Fig. 7B and Table S4). In general, the functions of the most adaptors might be related to T6SS, although there might be a small number of adaptors related to the eCIS or other unknown secretion systems that have spike complex components.
Fig. 7.
Four classes of adaptors co-occurrence with T6SS in the reference or representative prokaryotic genomes. (A) Number of four classes of adaptor homologs co-occurred with T6SS, eCIS, or spike complex components in the genome. Dark red: the adaptor homologs are located in the T6SS gene clusters. Light red: the adaptor homologs are not located in the T6SS gene clusters, but the genomes encode the T6SS gene clusters. Dark blue: T6SS is not found in the genomes encoding the adaptor homologs, but the eCIS gene clusters and the spike complex component (VgrG or PAAR) are present. Light blue: T6SS and eCIS are not found in the genome encoding the adaptor homologs, but the spike complex component is present. Gray: T6SS, eCIS and spike complex components are not detected in the genomes encoding the adaptor homolog. (B) Correlation between the total number of four classes of adaptor homologs and number of VgrG (light green), PAAR (dark green) or putative toxic effectors (pink) in the reference or representative prokaryotic genomes. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The proportion of adaptor homologs located in the T6SS or eCIS gene cluster was also evaluated. If at least two of the TssB, TssC, TssD and TssM homologs were encoded by the adjacent 10 genes upstream or 10 genes downstream of an adaptor-encoding gene, the adaptor-encoding gene was considered to be in the T6SS gene cluster. Similarly, if at least two of the Afp1/5, afp2/3/4, afp11 and afp16 homologues were encoded by the adjacent 10 genes upstream or 10 genes downstream of an adaptor-encoding gene, the adaptor-encoding gene was considered to be in the eCIS gene cluster. Our results showed that, among the 5297 identified adaptor homologs, at least 397 adaptor-encoding genes were located in the T6SS gene cluster, and none were located in the eCIS gene cluster. Proportionally, the proportion of the DUF1795 homologs located in the T6SS gene clusters was only 3.9%, while the corresponding values of the DUF2169, DUF4123 and PRK06147 homologs were 16.7%, 5.5% and 8.1%, respectively. Therefore, in most instances, the adaptor-encoding gene was not directly located in the T6SS or eCIS gene cluster.
3.6. Effectors adjacent to the adaptor are diverse
We identified a total of 1356 putative toxin genes in the 20 upstream or downstream genes of the four classes of adaptors (5297 in total) (Table S2). Among these genes, 830 toxins were encoded by the first or second gene downstream of the adaptor gene, accounting for 61.2%. Two-thirds of the protein products of the 1356 genes were currently annotated as hypothetical proteins or proteins of unknown function in public databases. We found that the number of putative toxic effectors in the genome was highly positively correlated with the number of adaptor homologs (r = 0.70, p < 0.01) (Fig. 7B). The putative toxic effector genes were from 653 prokaryotic genomes, of which 621 genomes were thought to also encode the T6SS gene clusters at the same time (Table S4). These 621 genomes included 1319 putative toxic effector genes, accounting for 97.3% of the total. Because the genomes possessed T6SS, we speculated that these genes were putative T6SS effectors. In addition, there were 31 putative toxic effector genes from 28 genomes with no T6SS gene cluster but with spike complex genes.
The identified toxins were from as many as 92 protein families, of which 62 families had identified at least 3 members (Table S2). The majority of these protein toxins were predicted to have enzymatic activity, and most of these acted on nucleic acids. Moreover, 23 families accounted for more than 1% of the putative toxin genes, and 16 were related to nucleic acids (Fig. 8A). The three toxin families with the greatest abundance were Tox-REase-5, AHH and Tox-GHH2, which all had more than 100 homologs and predicted DNase activity. In addition to nucleases, the activities of these toxins also included acting on lipids (such as Abhydrolase), amidases (such as Tae4), and protein-modifying toxins (such as VIP2 and Tox-ART-HYD1).
Fig. 8.
Families of toxic effectors recognized by the adaptor. (A). The families of toxins that accounted for more than 1% of the genetic neighbors of all adaptors. The nucleic acid-related toxins are represented in blue. (B). The effector compositions and relative abundances of the four classes of adaptors. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Interestingly, all four classes of adaptors can be associated with a variety of effectors (Fig. 8B). We identified 260 toxins from 39 families encoded by the first or second downstream gene of the genes encoding DUF1795 homologs; for DUF2169, 247 toxins from 38 families were identified; for DUF4123, 298 toxins from 38 families were identified; and for PRK06147, 210 toxins from 12 families were also identified. Significantly, the four classes of adaptors had different preferences for toxins. The two families with the greatest abundance of the DUF1795-related toxins were AHH (18.1%) and Ntox47 (12.3%), the two most abundant families for DUF2169 were Tox-GHH2 (38.1%) and Ntox15 (10.5%), the two most abundant families for DUF4123 were Tox-REase-5 (37.6%) and DUF2235 (19.1%), and the two most abundant families for PRK06147 were Tox-GHH2 (54.5%) and AHH (23.6%). In short, adaptors can be used as effective markers to identify T6SS toxic effectors in their neighboring genomic sequences.
4. Discussion
The adaptor protein mediates the binding of the effector to the T6SS spike complex, and it is necessary for the delivery of many T6SS toxic effectors [13], [26], [34], [36], [37]. In this study, we comprehensively scanned the adaptor-coding genes on a large scale in all reference or representative prokaryotic genomes and determined that the gene neighbors of the adaptors had a high proportion of putatively encoding T6SS spike complexes and toxic effectors. These adjacent positional relationships in genomes may be related to the interaction among the adaptor, spike complex, and effector [38], [39]. In the mechanism of the DUF4123 adaptor-dependent secretion by the T6SS, the interactions of the effector with PAAR, the adaptor and other chaperone proteins were characterized as a complex, multilayered competitive process, and the operon gene order is important for effector delivery [30]. In all known chaperone-containing T6SS effector modules, chaperone genes were located upstream of effector genes. Only the correctly formed effector-PAAR-chaperone complex can be delivered to the cognate VgrG complex [30]. Different adaptors had disparate sequence characteristics, which implies that they mediated the delivery of toxic effector in various modes.
We examined the gene neighbors of the spike complexes and effector homologs and identified a novel adaptor protein class (PRK06147) based on the feature of the gene order of the adaptor protein and the T6SS effector-related operon. The gene neighbors of PRK06147-encoding genes also had a high proportion of encoding T6SS spike complexes and effector homologs. PRK06147 and DUF2169 were determined to be adjacent in many genomes, implying that they were closely related in function. On various occasions, PRK06147 and DUF2169 might combine to mediate the interaction of the T6SS spike complex with the cognate effector, and compared with DUF2169, PRK06147 homologs and effector homologs were closer in the genome. In addition, many gene clusters encoded only PRK06147 and effector homologs but did not encode other adaptors. Therefore, we speculate that PRK06147 may represent a novel adaptor.
Our analysis showed that the proportion of PRK06147 homologs with a putative toxic effector in the range of proximate 10 genes upstream or 10 genes downstream was 36.1%, and the corresponding values of the DUF2169, DUF4123 and DUF1795 also reached 32.7%, 21.2% and 18.6%, respectively. The identification of toxic effectors was performed based on 114 families with reported toxic effector functions. Obviously, the identified toxic effectors represented only a small number of the toxic effectors encoded by the gene neighbors upstream and downstream of the adaptors, and a large number of toxic effector families have not been reported. We suggest that among the gene neighbors of the adaptors, especially the first two downstream genes, the proportion of the genes encoding toxic effectors after excluding the T6SS component genes and other definite nontoxin genes was actually very high. Notably, the four classes of adaptors had different preferences for toxic effectors. Therefore, the four classes of adaptors can be used as an effective marker to identify toxic effectors.
Based on the characteristics of the adaptor gene neighbors, instead of relying solely on diverse effector sequences, we demonstrate an effective approach of using adaptor-encoding genes as markers to identify the associated downstream effectors. In this way, we identified 1356 putative toxin genes related to adaptors from 92 protein families, and two-thirds of these protein products were still annotated as hypothetical proteins or proteins with unknown functions. These toxic effectors enrich our understanding of the prokaryotic arsenal and help to elucidate how T6SS spike proteins and adaptor proteins mediate effector translocation.
CRediT authorship contribution statement
Ya Liu: Conceptualization, Methodology, Investigation, Visualization, Writing - original draft, Validation. Zheng Zhang: Conceptualization, Methodology, Resources, Investigation, Visualization, Validation, Funding acquisition. Feng Wang: Resources, Validation. Dan-dan Li: Data curation, Validation. Yue-zhong Li: Conceptualization, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (32070030), the Special Investigation on Scientific and Technological Basic Resources (2017FY100300), the National Key Research and Development Program (2018YFA0900400 and 2018YFA0901704), the Key Research and Development Program of Shandong Province (2018GSF121015) (YL), the Natural Science Foundation of Jiangsu Province (BK20190199), the China Postdoctoral Science Foundation (2018M642649 and 2019T120586), the Special Funding for Postdoctoral Innovation Project of Shandong Province (201902014) (ZZ).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.11.003.
Contributor Information
Zheng Zhang, Email: zhangzheng@sdu.edu.cn.
Yue-zhong Li, Email: lilab@sdu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Galán J.E., Waksman G. Protein-injection machines in bacteria. Cell. 2018;172(6):1306–1318. doi: 10.1016/j.cell.2018.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cianfanelli F.R., Monlezun L., Coulthurst S.J. Aim, Load, Fire: The Type VI Secretion System, a Bacterial Nanoweapon. Trends Microbiol. 2016;24(1):51–62. doi: 10.1016/j.tim.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Brackmann M., Nazarov S., Wang J., Basler M. Using Force to Punch Holes: Mechanics of Contractile Nanomachines. Trends Cell Biol. 2017;27(9):623–632. doi: 10.1016/j.tcb.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Russell A.B., Hood R.D., Bui N.K., LeRoux M., Vollmer W., Mougous J.D. Type VI secretion delivers bacteriolytic effectors to target cells. Nature. 2011;475(7356):343–347. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iyer LM, Zhang D, Rogozin IB, Aravind L. Evolution of the deaminase fold and multiple origins of eukaryotic editing and mutagenic nucleic acid deaminases from bacterial toxin systems. Nucleic acids research. 2011;39(22):9473–97. [DOI] [PMC free article] [PubMed]
- 6.Ross B.D., Verster A.J., Radey M.C., Schmidtke D.T., Pope C.E., Hoffman L.R., Hajjar A.M., Peterson S.B., Borenstein E., Mougous J.D. Human gut bacteria contain acquired interbacterial defence systems. Nature. 2019;575(7781):224–228. doi: 10.1038/s41586-019-1708-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vassallo C.N., Wall D. Self-identity barcodes encoded by six expansive polymorphic toxin families discriminate kin in myxobacteria. Proc Natl Acad Sci USA. 2019;116(49):24808–24818. doi: 10.1073/pnas.1912556116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grant S.R., Fisher E.J., Chang J.H., Mole B.M., Dangl J.L. Subterfuge and Manipulation: Type III Effector Proteins of Phytopathogenic Bacteria. Annu Rev Microbiol. 2006;60(1):425–449. doi: 10.1146/annurev.micro.60.080805.142251. [DOI] [PubMed] [Google Scholar]
- 9.Khan M., Subramaniam R., Desveaux D. Of guards, decoys, baits and traps: pathogen perception in plants by type III effector sensors. Curr Opin Microbiol. 2016;29:49–55. doi: 10.1016/j.mib.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Russell A.B., Peterson S.B., Mougous J.D. Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol. 2014;12(2):137–148. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma L.-S., Hachani A., Lin J.-S., Filloux A., Lai E.-M. Agrobacterium tumefaciens Deploys a Superfamily of Type VI Secretion DNase Effectors as Weapons for Interbacterial Competition In Planta. Cell Host Microbe. 2014;16(1):94–104. doi: 10.1016/j.chom.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koskiniemi S., Lamoureux J.G., Nikolakakis K.C., t'Kint de Roodenbeke C., Kaplan M.D., Low D.A., Hayes C.S. Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci. 2013;110(17):7032–7037. doi: 10.1073/pnas.1300627110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alcoforado Diniz J., Coulthurst S.J., Christie P.J. Intraspecies Competition in Serratia marcescens Is Mediated by Type VI-Secreted Rhs Effectors and a Conserved Effector-Associated Accessory Protein. J Bacteriol. 2015;197(14):2350–2360. doi: 10.1128/JB.00199-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitzsimons TC, Lewis JM, Wright A, Kleifeld O, Schittenhelm RB, Powell D, et al. Identification of Novel Acinetobacter baumannii Type VI Secretion System Antibacterial Effector and Immunity Pairs. Infect Immun. 2018;86(8). [DOI] [PMC free article] [PubMed]
- 15.Pissaridou P., Allsopp L.P., Wettstadt S., Howard S.A., Mavridou D.A.I., Filloux A. The Pseudomonas aeruginosa T6SS-VgrG1b spike is topped by a PAAR protein eliciting DNA damage to bacterial competitors. Proc Natl Acad Sci USA. 2018;115(49):12519–12524. doi: 10.1073/pnas.1814181115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gong Y., Zhang Z., Liu Y., Zhou X.-W., Anwar M.N., Li Z.-S., Hu W., Li Y.-Z. A nuclease‐toxin and immunity system for kin discrimination in Myxococcus xanthus. Environ Microbiol. 2018;20(7):2552–2567. doi: 10.1111/1462-2920.14282. [DOI] [PubMed] [Google Scholar]
- 17.Pei T.-T., Li H., Liang X., Wang Z.-H., Liu G., Wu L.-L., Kim H., Xie Z., Yu M., Lin S., Xu P., Dong T.G. Intramolecular chaperone-mediated secretion of an Rhs effector toxin by a type VI secretion system. Nat Commun. 2020;11(1) doi: 10.1038/s41467-020-15774-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyata ST, Unterweger D, Rudko SP, Pukatzki S. Dual expression profile of type VI secretion system immunity genes protects pandemic Vibrio cholerae. PLoS Pathog. 2013;9(12):e1003752. [DOI] [PMC free article] [PubMed]
- 19.LaCourse K.D., Peterson S.B., Kulasekara H.D., Radey M.C., Kim J., Mougous J.D. Conditional toxicity and synergy drive diversity among antibacterial effectors. Nat Microbiol. 2018;3(4):440–446. doi: 10.1038/s41564-018-0113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russell A., Singh P., Brittnacher M., Bui N., Hood R., Carl M., Agnello D., Schwarz S., Goodlett D., Vollmer W., Mougous J. A Widespread Bacterial Type VI Secretion Effector Superfamily Identified Using a Heuristic Approach. Cell Host Microbe. 2012;11(5):538–549. doi: 10.1016/j.chom.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma J., Sun M., Pan Z., Lu C., Yao H. Diverse toxic effectors are harbored by vgrG islands for interbacterial antagonism in type VI secretion system. Biochimica et Biophysica Acta (BBA) - General Subjects. 2018;1862(7):1635–1643. doi: 10.1016/j.bbagen.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 22.English G., Trunk K., Rao V.A., Srikannathasan V., Hunter W.N., Coulthurst S.J. New secreted toxins and immunity proteins encoded within the Type VI secretion system gene cluster of Serratia marcescens. Mol Microbiol. 2012;86(4):921–936. doi: 10.1111/mmi.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srikannathasan V., English G., Bui N.K., Trunk K., O'Rourke P.E., Rao V.A. Structural basis for type VI secreted peptidoglycan DL-endopeptidase function, specificity and neutralization in Serratia marcescens. Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 12):2468–2482. doi: 10.1107/S0907444913022725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hersch S.J., Watanabe N., Stietz M.S., Manera K., Kamal F., Burkinshaw B., Lam L., Pun A., Li M., Savchenko A., Dong T.G. Envelope stress responses defend against type six secretion system attacks independently of immunity proteins. Nat Microbiol. 2020;5(5):706–714. doi: 10.1038/s41564-020-0672-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang J.Y., Bullen N.P., Ahmad S., Whitney J.C. Diverse NADase effector families mediate interbacterial antagonism via the type VI secretion system. J Biol Chem. 2018;293(5):1504–1514. doi: 10.1074/jbc.RA117.000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitney J., Quentin D., Sawai S., LeRoux M., Harding B., Ledvina H., Tran B., Robinson H., Goo Y., Goodlett D., Raunser S., Mougous J. An Interbacterial NAD(P)+ Glycohydrolase Toxin Requires Elongation Factor Tu for Delivery to Target Cells. Cell. 2015;163(3):607–619. doi: 10.1016/j.cell.2015.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ting S.-Y., Bosch D.E., Mangiameli S.M., Radey M.C., Huang S., Park Y.-J., Kelly K.A., Filip S.K., Goo Y.A., Eng J.K., Allaire M., Veesler D., Wiggins P.A., Peterson S.B., Mougous J.D. Bifunctional Immunity Proteins Protect Bacteria against FtsZ-Targeting ADP-Ribosylating Toxins. Cell. 2018;175(5):1380–1392.e14. doi: 10.1016/j.cell.2018.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silverman J.M., Brunet Y.R., Cascales E., Mougous J.D. Structure and Regulation of the Type VI Secretion System. Annu Rev Microbiol. 2012;66(1):453–472. doi: 10.1146/annurev-micro-121809-151619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shneider M.M., Buth S.A., Ho B.T., Basler M., Mekalanos J.J., Leiman P.G. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature. 2013;500(7462):350–353. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burkinshaw B.J., Liang X., Wong M., Le A.N.H., Lam L., Dong T.G. A type VI secretion system effector delivery mechanism dependent on PAAR and a chaperone–co-chaperone complex. Nat Microbiol. 2018;3(5):632–640. doi: 10.1038/s41564-018-0144-4. [DOI] [PubMed] [Google Scholar]
- 31.Zhang D., de Souza R.F., Anantharaman V., Iyer L.M., Aravind L. Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct. 2012;7(1):18. doi: 10.1186/1745-6150-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salomon D., Kinch L.N., Trudgian D.C., Guo X., Klimko J.A., Grishin N.V., Mirzaei H., Orth K. Marker for type VI secretion system effectors. Proc Natl Acad Sci. 2014;111(25):9271–9276. doi: 10.1073/pnas.1406110111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jana B., Fridman C.M., Bosis E., Salomon D. A modular effector with a DNase domain and a marker for T6SS substrates. Nat Commun. 2019;10(1) doi: 10.1038/s41467-019-11546-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Unterweger D., Kostiuk B., Pukatzki S. Adaptor Proteins of Type VI Secretion System Effectors. Trends Microbiol. 2017;25(1):8–10. doi: 10.1016/j.tim.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 35.Cianfanelli FR, Alcoforado Diniz J, Guo M, De Cesare V, Trost M, Coulthurst SJ. VgrG and PAAR Proteins Define Distinct Versions of a Functional Type VI Secretion System. PLoS Pathog. 2016;12(6):e1005735. [DOI] [PMC free article] [PubMed]
- 36.Quentin D., Ahmad S., Shanthamoorthy P., Mougous J.D., Whitney J.C., Raunser S. Mechanism of loading and translocation of type VI secretion system effector Tse6. Nat Microbiol. 2018;3(10):1142–1152. doi: 10.1038/s41564-018-0238-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bondage D.D., Lin J.-S., Ma L.-S., Kuo C.-H., Lai E.-M. VgrG C terminus confers the type VI effector transport specificity and is required for binding with PAAR and adaptor–effector complex. Proc Natl Acad Sci USA. 2016;113(27):E3931–E3940. doi: 10.1073/pnas.1600428113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Unterweger D., Kostiuk B., Otjengerdes R., Wilton A., Diaz-Satizabal L., Pukatzki S. Chimeric adaptor proteins translocate diverse type VI secretion system effectors in Vibrio cholerae. EMBO J. 2015;34(16):2198–2210. doi: 10.15252/embj.201591163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang X., Moore R., Wilton M., Wong M.J.Q., Lam L., Dong T.G. Identification of divergent type VI secretion effectors using a conserved chaperone domain. Proc Natl Acad Sci USA. 2015;112(29):9106–9111. doi: 10.1073/pnas.1505317112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A., Salazar G.A., Tate J., Bateman A. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44(D1):D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marchler-Bauer A., Bo Y., Han L., He J., Lanczycki C.J., Lu S., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R., Gwadz M., Hurwitz D.I., Lu F., Marchler G.H., Song J.S., Thanki N., Wang Z., Yamashita R.A., Zhang D., Zheng C., Geer L.Y., Bryant S.H. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45(D1):D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Durand E., Nguyen V.S., Zoued A., Logger L., Péhau-Arnaudet G., Aschtgen M.-S., Spinelli S., Desmyter A., Bardiaux B., Dujeancourt A., Roussel A., Cambillau C., Cascales E., Fronzes R. Biogenesis and structure of a type VI secretion membrane core complex. Nature. 2015;523(7562):555–560. doi: 10.1038/nature14667. [DOI] [PubMed] [Google Scholar]
- 44.Chen L., Song N., Liu B.o., Zhang N., Alikhan N.-F., Zhou Z., Zhou Y., Zhou S., Zheng D., Chen M., Hapeshi A., Healey J., Waterfield N.R., Yang J., Yang G. Genome-wide Identification and Characterization of a Superfamily of Bacterial Extracellular Contractile Injection Systems. Cell Reports. 2019;29(2):511–521.e2. doi: 10.1016/j.celrep.2019.08.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Busby J.N., Panjikar S., Landsberg M.J., Hurst M.R.H., Lott J.S. The BC component of ABC toxins is an RHS-repeat-containing protein encapsulation device. Nature. 2013;501(7468):547–550. doi: 10.1038/nature12465. [DOI] [PubMed] [Google Scholar]
- 46.Meusch D., Gatsogiannis C., Efremov R.G., Lang A.E., Hofnagel O., Vetter I.R., Aktories K., Raunser S. Mechanism of Tc toxin action revealed in molecular detail. Nature. 2014;508(7494):61–65. doi: 10.1038/nature13015. [DOI] [PubMed] [Google Scholar]
- 47.Alteri CJ, Himpsl SD, Pickens SR, Lindner JR, Zora JS, Miller JE, et al. Multicellular bacteria deploy the type VI secretion system to preemptively strike neighboring cells. PLoS Pathog. 2013;9(9):e1003608. [DOI] [PMC free article] [PubMed]
- 48.Chang Y.W., Rettberg L.A., Ortega D.R., Jensen G.J. In vivo structures of an intact type VI secretion system revealed by electron cryotomography. EMBO Rep. 2017;18(7):1090–1099. doi: 10.15252/embr.201744072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang F., Li N., Wang X., Cheng J., Huang Y., Yang Y., Yang J., Cai B., Wang Y.-P., Jin Q., Gao N. Cryo-EM Structure and Assembly of an Extracellular Contractile Injection System. Cell. 2019;177(2):370–383.e15. doi: 10.1016/j.cell.2019.02.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.