Abstract
The dynamic organization of actin cytoskeleton meshworks relies on multiple actin-binding proteins endowed with distinct actin-remodeling activities. Filamin A is a large multi-domain scaffolding protein that cross-links actin filaments with orthogonal orientation in response to various stimuli. As such it plays key roles in the modulation of cell shape, cell motility, and differentiation throughout development and adult life. The essentiality and complexity of Filamin A is highlighted by mutations that lead to a variety of severe human disorders affecting multiple organs. One of the most conserved activity of Filamin A is to bridge the actin cytoskeleton to integrins, thereby maintaining the later in an inactive state. We here review the numerous mechanisms cells have developed to adjust Filamin A content and activity and focus on the function of Filamin A as a gatekeeper to integrin activation and associated adhesion and motility.
Keywords: actin cytoskeleton, filamin, integrin, cell adhesion, cell migration
Introduction
The filamin protein family is represented in nearly all Metazoa. Phylogenetically, the filamin genes diverge from a common single ancestral gene between chordate invertebrate and vertebrate lineages. Filamins comprise a N-terminal actin-binding domain (ABD) composed of two actin-binding calponin homology (CH) domains followed by immunoglobulin like repeats (IgFLN) of high sequence similarity (van der Flier and Sonnenberg, 2001). All C-terminal filamin repeats of filamins characterized so far have the property to homodimerize. The number of filamin repeats differs substantially in invertebrates but is almost constant in vertebrates (Light et al., 2012). The vertebrate genomes contain three filamins, Filamin A, B and C, with an intraspecies sequence identity of over 64% (Kesner et al., 2010). Filamins A and B are ubiquitously expressed, whereas Filamin C is expressed in smooth and striated muscles. Filamins A and B are localized to the cortex and stress fibers, whereas Filamin C is localized to the sarcomeric Z-line complex (van der Ven et al., 2000; Sheen et al., 2002). Disease-associated mutations and knockout mouse models suggest that Filamins A and B are critical for various aspects of skeletal, vasculature, cardio, and cerebral development (Fox et al., 1998; Sheen et al., 2002; Robertson et al., 2003; Krakow et al., 2004; Farrington-Rock et al., 2006, 2008; Feng et al., 2006; Hart et al., 2006; Lu et al., 2007; Zhou et al., 2007; Metais et al., 2018; Yamak et al., 2020), whereas Filamin C is essential for skeletal muscle and heart development (Goetsch et al., 2005; Vorgerd et al., 2005; Dalkilic et al., 2006; Duff et al., 2011; Zhou et al., 2020).
Filamin A, a Hub for Multiple Binding Partners
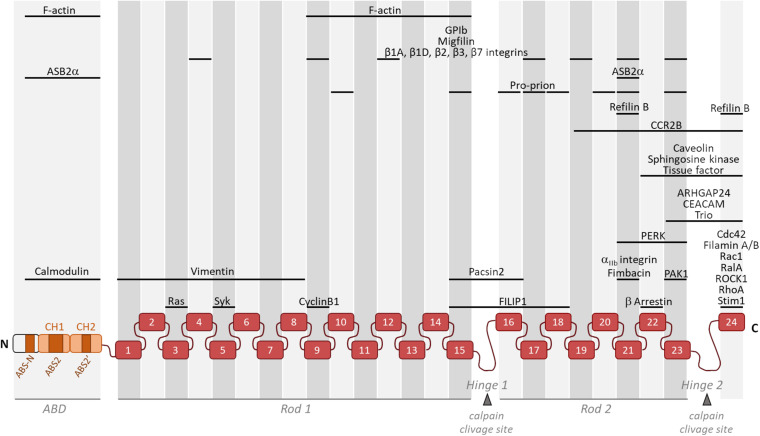
Filamin A interacts with about a hundred binding-partners, many of which being involved in the regulation of signaling pathways converging toward actin cytoskeleton organization (Figure 1). Filamin A has a dual role in controlling the architecture and the mechanics of the actin cytoskeleton. Filamin A is an actin-binding and cross-linking protein whose primary function is to organize the actin cytoskeleton in orthogonal filament arrays (Nakamura et al., 2007). Importantly, the mechanic properties of this filamentous actin (F-actin) network is dependent on Filamin A concentration (Tseng et al., 2004; Gardel et al., 2006; Esue et al., 2009). At high Filamin A concentration, tighter F-actin bundles are observed and the F-actin network undergoes stress stiffening under applied forces (Schmoller et al., 2009). In contrast, at lower relative Filamin A cross-link concentrations, the F-actin cytoskeleton is more dynamic and can soften in response to stress (Tseng et al., 2004). Furthermore, the non-linear elasticity of the actin network is attributed to the flexibility of Filamin A (Kasza et al., 2009; Schmoller et al., 2009). Filamin A also localizes to points of intersection between stress fibers and cortical actin where it plays a role in the isotropic redistribution of applied forces to focal adhesions (Kumar et al., 2019). Three actin-binding sites (ABS) within the Filamin A ABD were recently identified (Figure 1; Iwamoto et al., 2018). The first one, ABS-N, located at the N-ter of the CH domain 1 contributes to F-actin binding while the two others, ABS2 and ABS2′, facilitate binding in the groove between adjacent actin subunits (Iwamoto et al., 2018). While the ABD is necessary and sufficient for F-actin binding (Razinia et al., 2011), a domain within a Filamin A fragment encompassing filamin repeats 9 to 15 is necessary for high avidity F-actin binding (Nakamura et al., 2007).
FIGURE 1.
Schematic representation of monomeric Filamin A illustrating its general structure and the binding location of partners involved in actin cytoskeleton organization and/or cell motility. Filamin A consists of an actin-binding domain (ABD) composed of two actin-binding calponin homology (CH) domains followed by Ig-like repeats (1–24). Two intervening calpain-sensitive hinge domains separate the 24 Ig-like repeats into two rod domains. The three ABS (ABS-N, ABS2, and ABS2′) are indicated. Partners involved in actin cytoskeleton organization and/or cell motility but with unknown Filamin A binding domain(s) (14-3-3, ELP1, FILIP1L, p190RhoGAP, and Lck) are not represented.
In mammals, the ABD of filamins is followed by 24 filamin repeats interrupted by two hinge regions often referred as rod 1 and 2, one between repeats 15 and 16 and another between repeats 23 and 24 (Figure 1). Filamin A domains can be divided into four subgroups (A, B, C, and D) based on amino acid similarities (Ithychanda et al., 2009b). Filamin A repeats of subgroup A (4, 9, 12, 17, 19, 21, and 23) interact with a set of biologically important ligands including platelet receptor glycoprotein Ibα (GPIbα) (Nakamura et al., 2006), migfilin (Lad et al., 2008; Ithychanda et al., 2009a, b), Cystic Fibrosis Transmembrane conductance Receptor (CFTR) (Smith et al., 2010), FilGAP (Nakamura et al., 2009), Pro-prion (Li et al., 2010), Ankyrin repeat containing protein with a SOCS box 2 alpha (ASB2α) (Lamsoul et al., 2011) and β chains of integrins (Kiema et al., 2006; Takala et al., 2008). All subgroup A Filamin A repeats and their binding partners have similar mode of interaction. Indeed, the CD face of Filamin A repeats represents a common interface for Filamin A-ligand interaction (Lad et al., 2007; Heikkinen et al., 2009). Interestingly, Filamin A repeats can also engage in intramolecular contacts (IgFLNa16-17, IgFLNa18-19, and IgFLNa20-21) that may become disrupted by binding of one of the repeats to the integrin β cytoplasmic tail or by mechanical forces (Lad et al., 2007; Heikkinen et al., 2009). Smoothelins A and B, as well as fimbacin can bind to the cryptic CD cleft of Filamin A repeat 21 exposed in mechanically activated Filamin A (Wang and Nakamura, 2019a, b). Filamin A can also interact with small GTPases of the Rho family, Rac, Rho, cdc42, and RalA (Ohta et al., 1999; Bellanger et al., 2000; Vadlamudi et al., 2002; Ueda et al., 2003) and with proteins upstream and downstream of the GTPases (Ohta et al., 2006; Nakamura et al., 2009) known to regulate cytoskeletal dynamics and cell protrusions. In addition, RhoA activity is downregulated through interactions between Filamin A and αIIbβ3 integrins and is critical to proplatelet formation (Donada et al., 2019).
Filamin A is also localized into the nucleus where it plays roles in DNA repair through interaction with BRCA1 and BRCA2 (Yue et al., 2009; Velkova et al., 2010), as well as transcription through interaction with transcription factors such as the androgen receptor (Loy et al., 2003; McGrath et al., 2013; Savoy et al., 2015), Smads (Sasaki et al., 2001) or PEBP2β/CBFβ (Yoshida et al., 2005). Filamin A also associates with the MKL1 transcriptional co-activator, stimulating the activity of the Serum Responsive Factor (SRF) transcription factor and cell migration (Kircher et al., 2015).
How Filamin A integrates the signals triggered by its multiple binding partners and whether such complex molecular interactions might be tuned differentially in different cell types remain key open questions. Nevertheless, the essentiality of Filamin A is highlighted by variants in the gene FLNA that lead to 10 distinct genetic syndromes affecting a wide diversity of organs (Wade et al., 2020). Importantly, pathogenic variants of FLNA could contribute to aberrant cytoskeletal regulation leading either to loss-of-function or gain-of function disorders. Indeed, variants are found in the two CH domains of the ABD, CHD1 and 2, in periventricular nodular heterotopia (PH) and otopalatodigital (OPD) syndromes type 1 and 2, respectively. Variants within CHD1 are likely to disrupt Filamin A interaction with actin (Iwamoto et al., 2018) whereas variants within CHD2 are likely to constitutively expose the CHD1 ABS to ligand (Clark et al., 2009). Interestingly, a mutation in FLNA in a male patient with PH and congenital intestinal pseudoobstruction potentiates αIIbβ3 integrin activation likely through less binding of mutant Filamin A to β3 integrin and facilitated recruitment of Talin by the β3 subunit (Berrou et al., 2017). The relationship between other FLNA pathogenic variants and Filamin A functions is less understood and remains to be investigated.
Filamin A, a Negative Regulator of Integrins
Because the filamin domains involved in binding actin and integrins have the highest content of ancestral residues of any domains (Kesner et al., 2010), integrins are considered among the most important interaction partners of filamins. Integrins are heterodimeric transmembrane receptors formed by α and β subunits. These are single spanning membrane proteins with a large extracellular ectodomain and a short intracellular cytoplasmic tail. Integrins mediate cell to extracellular matrix and cell to cell contacts and integrate external cues to the actin cytoskeleton and signaling pathways (Legate and Fassler, 2009; Humphries et al., 2019; Kechagia et al., 2019). Interactions between integrins and their extracellular ligands are tightly regulated thanks to integrin activators and integrin inhibitors. Importantly, switching integrins between inactive and active conformations is crucial for integrin functions (Bouvard et al., 2013). Integrin activation has largely been documented (Kim et al., 2011; Sun et al., 2019). This process is regulated via either extracellular ligands (outside-in activation) or intracellular binding partners (inside-out activation). Integrin-inactivating proteins such as integrin cytoplasmic domain-associated protein 1 (ICAP1), SHARPIN (SHANK-associated RH domain-interacting protein) and filamins are required for integrin inactivation in different settings (Calderwood et al., 2001; Bouvard et al., 2003; Rantala et al., 2011). The physiological relevance of integrin-inactivating proteins is crucial for integrin function as exemplified by the phenotypes of mice lacking integrin inactivators (Bouvard et al., 2013). Integrin inactivators either stabilize the inactive state of integrins or promote integrin deactivation during cyclical cell-adhesion processes such as migration. Indeed, a substantial proportion of cell surface integrins is in a resting state (Arjonen et al., 2012). Inactive integrins are in a closed conformation, in which the binding of both extracellular ligand and intracellular activators is repressed. In human, there are several integrin β subunits that bind filamins (Kiema et al., 2006; Ithychanda et al., 2009b).
Filamin A is a major gatekeeper to integrin activation. Since the discovery of Filamin A as a binding partner of the β2 integrin subunit 25-years ago (Sharma et al., 1995) and the first evidence that increased Filamin A-β2 integrin interactions restrict cell migration (Calderwood et al., 2001; Bouvard et al., 2003; Rantala et al., 2011), several modes of action of Filamin A as an integrin inactivator have been proposed. They depend on the identity of the integrin α and β chains or could be specific to only a subset of integrin αβ heterodimers. First, binding of Filamin A domains of subgroup A to the C terminus of the integrin β tail (β1, β2, β3, or β7) results in direct competition with talin binding by occupying an overlapping binding site (Kiema et al., 2006; Ithychanda et al., 2009b). Second, Filamin A forms a ternary complex engaging the cytoplasmic tails of both integrin αIIb and β3, thereby stabilizing the inner-membrane clasp and competing with talin recruitment to the β subunit cytoplasmic tail by binding both the C-terminal and membrane-proximal regions of the β3 tail (Liu et al., 2015). These two modes of action of Filamin A restrain the integrin in a resting state. Interestingly, domains within functionally important binding interfaces of both filamin repeats and integrin subunits have diverged in critical residues, indicating that filamin isoforms may bind and regulate integrin αβ heterodimers differentially. Indeed, Kesner et al. (2010) described the substitution in the β strand C of the filamin repeat 21, an ancestral Ser/Thr in Filamins B and C changed to an Ala at residue 2272 in Filamin A during the mammalian period. Furthermore, β1 and β7 integrins have ideally positioned hydrophobic amino acids to bind Filamin tighter than β2 and β3 integrins (Ithychanda et al., 2009b). Furthermore, some of the key residues in the αIIb subunit that are important for interaction with filamin via their side chains, are not conserved in all integrin α subunits, reinforcing the notion that filamins bind integrins differentially.
Because several Filamin A repeats can bind the cytoplasmic tails of β integrins and have the ability to clasp αIIb and β3 cytoplasmic tails, it seems plausible that they can bind simultaneously, and such interactions may promote clustering of inactive integrins (Ithychanda et al., 2009b; Liu et al., 2015). Although the biological significance of these Filamin A clutches remains to be establish, it is tempting to speculate that upon Filamin A removal, pre-clustered integrins would become engaged by multivalent ECM and thereby activated.
Multiple Regulatory Mechanisms Controling Filamin A Activities and Levels
Integrin activation can be achieved through the binding of proteins to Filamin A. Indeed, migfilin can bind Filamin A with a high affinity, uncoupling the Filamin A-integrin link, sequestering Filamin A away from the β integrin cytoplasmic tail and thus counteracting Filamin A-mediated integrin inactivation (Lad et al., 2008; Ithychanda et al., 2009a; Das et al., 2011). This allows the binding of integrin activators, talin and kindlins, to the β integrin cytoplasmic tail, leading to inside-out integrin activation (Tadokoro et al., 2003; Tu et al., 2003; Kiema et al., 2006; Wegener et al., 2007; Moser et al., 2008). Internally generated and externally imposed mechanical forces can also regulate Filamin A interaction with partners by triggering conformational changes that expose otherwise masked partner-binding site, thereby leading to integrin activation (Ehrlicher et al., 2011; Nakamura et al., 2014). Although evidence for the mechanosensing function of Filamin A in Drosophila oogenesis has been provided, its precise role in cell differentiation and morphogenesis in mammals is still lacking (Razinia et al., 2012; Huelsmann et al., 2016). Filamin A is also regulated by phosphorylation. Several kinases such as protein kinase C (Kawamoto and Hidaka, 1984), ribosomal S6 kinase (Ohta and Hartwig, 1996; Woo et al., 2004), p21-activated kinase 1 (PAK1) (Vadlamudi et al., 2002; Hammer et al., 2013), the cyclic adenosine monophosphate (cAMP)-dependent protein kinase A (Jay et al., 2004), Akt (Li et al., 2015), mTOR2 (Chantaravisoot et al., 2015; Sato et al., 2016) and the serine/threonine kinase Ndr2 (Waldt et al., 2018) phosphorylate Filamin A on serine 2152. This phosphorylation event positively regulates cell migration (Woo et al., 2004; Hammer et al., 2013; Li et al., 2015; Sato et al., 2016). Activation of receptor tyrosine kinases was shown to trigger cell rounding and integrin inactivation via increased Filamin A phosphorylation (Vial and McKeown-Longo, 2012; Mai et al., 2014). In addition, G Protein-Coupled Receptors that directly bind Filamin A can also promote Filamin A phosphorylation (Tirupula et al., 2015). Filamin A phosphorylation by the cAMP-dependent protein kinase protects Filamin A against proteolysis by calpains (Chen and Stracher, 1989). Phosphorylation of β2 integrins impairs Filamin A binding, allowing the binding of the 14-3-3 protein to the β2 subunit and adhesion of Jurkat T cells to ICAM-1 (Takala et al., 2008).
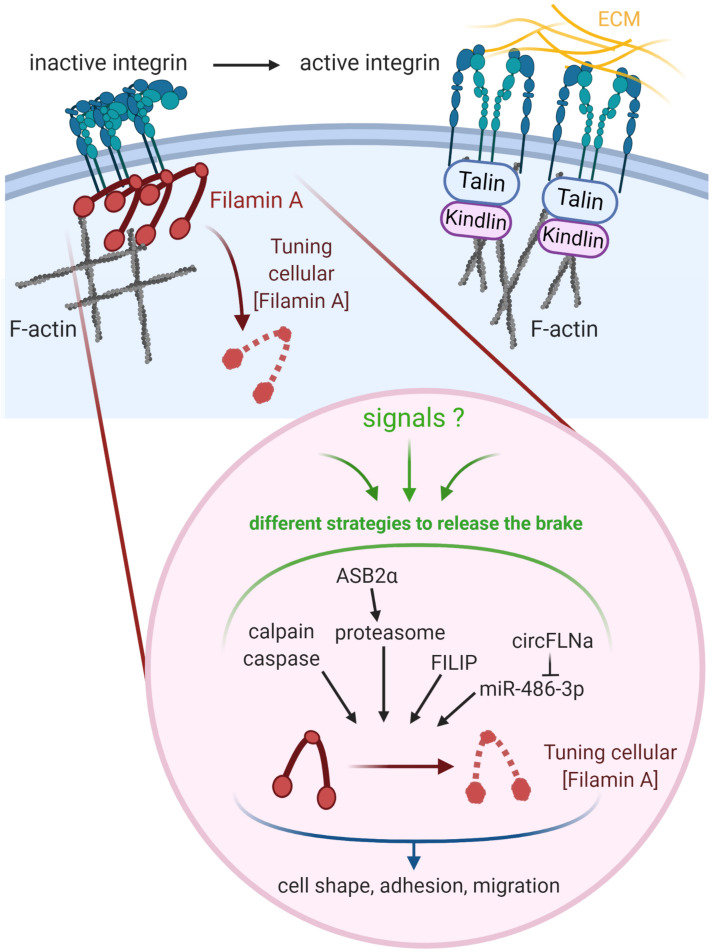
Tuning the cellular concentration of Filamin A represents another level of regulation expected to impact integrin activation, although this has not been formally demonstrated yet (Figure 2). Filamins are regulated by proteolysis, which provides an irreversible regulatory mechanism for processes requiring Filamin removal. Filamin A is cleaved by calpain and caspase at the two hinge regions, producing a 170 kDa protein encompassing the ABD and Filamin A repeats 1 to 15 and a 110 kDa protein that is further cleaved to generate a 90 kDa fragment containing repeats 16 to 23 (Figure 1) (Gorlin et al., 1990; Browne et al., 2000). Filamins A and B are also regulated by proteasomal degradation which represents a fast, reversible, localized and selective regulatory mechanism that allows cells to acutely adapt or fine-tune cellular processes. Surprisingly, only few proteins linked to cytoskeleton dynamics, cell adhesion and migration have been shown to be regulated by this proteolysis pathway in non-muscle cells (Schaefer et al., 2012). Of interest, control of the cellular concentration of Filamins A through ubiquitin-mediated protein degradation represents a seminal example of proteasomal degradation of an actin-binding and -crosslinking protein. Using several molecular and cellular biology approaches, we and others demonstrated that the ASB2α E3 ubiquitin ligase (E3) triggers ubiquitylation and proteasomal degradation of Filamins A and B (Heuze et al., 2008; Burande et al., 2009; Lamsoul et al., 2013; Razinia et al., 2013; Sakane et al., 2013; Spinner et al., 2015; Metais et al., 2018). ASB2α is the specificity subunit of a multimeric E3 of the Cullin 5-RING Ligase family involved in the recruitment of proteins to be ubiquitylated (Lamsoul et al., 2016). By degrading Filamins A and B, ASB2α regulates cell spreading, adhesion and cell migration (Heuze et al., 2008; Baldassarre et al., 2009; Lamsoul et al., 2011, 2013; Spinner et al., 2015). Furthermore, our recent results support a model of cardiac cell differentiation that relies on a key role for ASB2α in remodeling the actin cytoskeleton through induced-degradation of Filamin A (Metais et al., 2018). Indeed, the timely controlled removal of Filamin A ensures critical functions in differentiating cardiac muscle cells suggesting that Filamin A degradation is necessary to modify the actin cytoskeleton organization and properties in order to build the sarcomere, and thus for heartbeats. In addition, the Filamin A interacting protein (FILIP) interacts with Filamin A and induces its degradation with impacts on the mode of neuron migration (Nagano et al., 2004; Sato and Nagano, 2005). More recently, Filamin A expression was shown to be regulated by a microRNA (miR) and a circular RNA (circRNA). Indeed, miR-486-3p can bind Filamin A 3′UTR thereby reducing Filamin A expression while circFLNA sponges miR-486-3p resulting in increased Filamin A expression (Wang et al., 2019). Important questions remain: Does Filamin A degradation directly translate into increased integrin activation? What is the biological relevance of variable Filamin A levels in different cell subtypes or at discrete stages of cell differentiation? Why cells have evolved so many different mechanisms to regulate Filamin A activity and to up-regulate or down-regulate Filamin A concentration?
FIGURE 2.
Tuning the cellular concentration of Filamin A represents a pivotal mechanism to regulate integrin-dependent adhesion and migration. Regulation of integrins involves both integrin inhibitors (e.g., Filamin A) and integrin activators (e.g., Talin, Kindlin). Filamin A protein is regulated through cleavages by calpain and caspase, ASB2α-mediated degradation by the proteasome, and interaction with FILIP. Filamin A mRNA is down-regulated by miR-486-3p which is itself sponged by circFLNA. Created with BioRender.com.
Filamin A in Cell Adhesion and Migration
The first evidence for a role of Filamin A in cell motility was provided in 1992 (Cunningham et al., 1992). Indeed, at the cellular level, Filamin A deficiency in a human melanoma cell line promotes plasma membrane blebbing and causes loss of motility. The role of Filamin A in migration was further supported by the finding that nonsense mutations in the Filamin A gene are associated with the neuronal migration disorder periventricular heterotopia (Fox et al., 1998). However, the role of Filamin A in cell motility is more complex. By providing a physical link between integrins and the actin cytoskeleton and by negatively regulating integrins, Filamin A exerts key roles in regulating positively or negatively cell adhesion and migration according to cell types and/or conditions. Furthermore, Filamin A binding partners may vary according to cell types and/or in response to microenvironment cues such as extracellular matrix components, chemokines or shear flow. This is likely to influence cell adhesion and migration. While the loss of Filamins A or B alone has no effect on cell motility, loss of both filamins following knockdown/knockout or ASB2α-mediated degradation have highlighted the role of filamins in different aspects of cell motility (Heuze et al., 2008; Baldassarre et al., 2009; Lynch et al., 2011; Lamsoul et al., 2013; Spinner et al., 2015). It is tempting to speculate that the functions of Filamin A in many cell types may have been missed in assays using Filamin A single knockout/knockdown cells because of compensation by Filamin B (Sheen et al., 2002; Baldassarre et al., 2009). Filamin-depleted cells exhibit impaired cell spreading (Heuze et al., 2008; Kim et al., 2008; Baldassarre et al., 2009; Lynch et al., 2011). In addition, increased adhesion of Filamin A-depleted neutrophils has been described (Sun et al., 2013). Furthermore, Filamin A knockdown or ASB2α-mediated Filamin A degradation enhances adhesion of myeloid leukemia cells to fibronectin (Lamsoul et al., 2011). In contrast, Roth et al. found that Filamin A was dispensable for adhesion of differentiated HL-60 cells (Roth et al., 2017). However, Filamin A depleted primary murine neutrophils display increased spreading on and higher adhesion in shear-free conditions to β2 integrin ligands, indicating that Filamin A is a negative regulator of β2 integrin adhesion in neutrophils (Uotila et al., 2017). Although Filamin A negatively regulates β2 integrin adhesion in Jurkat T cells, its absence leads to a reduction of primary T cell adhesion to integrin ligands under conditions of shear flow and to a reduced trafficking into lymph nodes and sites of inflammation (Moser et al., 2009; Savinko et al., 2018). Interestingly, Filamin A and vimentin can cooperate to regulate integrin-mediated cell spreading and cell adhesion (Kim et al., 2010a, b).
Filamins A and B depleted cells exhibit impaired initiation of migration of fibrosarcoma HT1080 cells (Baldassarre et al., 2009). Filamin A silencing increases cell adhesion and decreased migration of the bronchial carcinoid H727 cells (Vitali et al., 2017). In contrast, silencing of Filamin A inhibits Snail-induced adhesion and increases migration of colon adenocarcinoma HT29 cells (Wieczorek et al., 2017). In accordance with these results, Filamin A is required to mediate SST2 effects on adhesion and migration of the pancreatic endocrine QGP1 cells (Vitali et al., 2016). Filamin A also positively regulates directional migration of bone osteosarcoma U-2 OS cells and mouse embryonic fibroblasts by suppressing Rac 1 activity downstream of β1 integrins (Jacquemet et al., 2013). Knockdown of Filamins A and B in fibrosarcoma cells was also shown to augment matrix metalloproteinase activity increasing their invasive potential (Baldassarre et al., 2012). These results are in agreement with the fact that ASB2α regulates immature dendritic cell migration by promoting extracellular matrix proteolysis (Lamsoul et al., 2013). Conversely, Filamin A stabilizes podosomes in macrophages and is required for their mesenchymal but not for their amoeboid migration (Guiet et al., 2012). In addition, in the absence of Filamin A, macrophages display impaired migration associated with reduced atherosclerosis in mice (Bandaru et al., 2019). Several evidences indicate that Filamin A regulates the intracellular trafficking of β1 integrins (Meyer et al., 1998; Kim et al., 2010b). This is likely to affect β1 integrin-dependent processes. On the basis of these scattered observations, it is clear that we still miss today a unified view of the roles of Filamin A in cell adhesion and migration.
Concluding Remarks
As reviewed here, the timely proteolysis and/or removal of Filamin A have emerged as pivotal mechanisms to regulate its cellular concentration and integrin-dependent adhesion and migration. When integrating the knowledge gained about the function of Filamin A beyond its integrin regulation role, one is tempted to speculate that this key protein at the interface between multiple receptors, signaling pathways and the actin cytoskeleton exerts different and specific cellular functions in response to a wide-range of environmental cues. As exemplified by the wide spectrum of developmental malformations and diseases caused by mutations in its gene, Filamin A indeed stands out as a major molecular player in different biological processes. In this context, it will be particularly interesting to further investigate how the multiple mechanisms able to adjust Filamin A concentration and activity contribute to its function in different cellular and physiological settings.
Author Contributions
All authors contributed to both the review conceptualization and the writing process.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors apologize to their colleagues whose work could not be cited due to space constraints.
Footnotes
Funding. Research in our lab related to the subject of this review was supported by INSERM, CNRS, University of Toulouse and grants from the Fondation ARC pour la Recherche sur le Cancer, the Comité Midi-Pyrénées de la Ligue contre le Cancer and Sanofi iAwards Europe (to PGL) and the CNRS IRP program (grant “SysTact” to LD).
References
- Arjonen A., Alanko J., Veltel S., Ivaska J. (2012). Distinct recycling of active and inactive beta1 integrins. Traffic 13 610–625. 10.1111/j.1600-0854.2012.01327.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldassarre M., Razinia Z., Brahme N. N., Buccione R., Calderwood D. A. (2012). Filamin A controls matrix metalloproteinase activity and regulates cell invasion in human fibrosarcoma cells. J. Cell Sci 125 3858–3869. 10.1242/jcs.104018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldassarre M., Razinia Z., Burande C. F., Lamsoul I., Lutz P. G., Calderwood D. A. (2009). Filamins regulate cell spreading and initiation of cell migration. PLoS One 4:e7830. 10.1371/journal.pone.0007830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandaru S., Ala C., Salimi R., Akula M. K., Ekstrand M., Devarakonda S., et al. (2019). Targeting filamin a reduces macrophage activity and atherosclerosis. Circulation 140 67–79. 10.1161/circulationaha.119.039697 [DOI] [PubMed] [Google Scholar]
- Bellanger J. M., Astier C., Sardet C., Ohta Y., Stossel T. P., Debant A. (2000). The Rac1- and RhoG-specific GEF domain of Trio targets filamin to remodel cytoskeletal actin. Nat. Cell Biol. 2 888–892. 10.1038/35046533 [DOI] [PubMed] [Google Scholar]
- Berrou E., Adam F., Lebret M., Planche V., Fergelot P., Issertial O., et al. (2017). Gain-of-function mutation in filamin a potentiates platelet integrin alphaIIbbeta3 activation. Arterioscler. Thromb. Vasc. Biol. 37 1087–1097. 10.1161/atvbaha.117.309337 [DOI] [PubMed] [Google Scholar]
- Bouvard D., Pouwels J., De Franceschi N., Ivaska J. (2013). Integrin inactivators: balancing cellular functions in vitro and in vivo. Nat. Rev. Mol. Cell Biol. 14 430–442. 10.1038/nrm3599 [DOI] [PubMed] [Google Scholar]
- Bouvard D., Vignoud L., Dupe-Manet S., Abed N., Fournier H. N., Vincent-Monegat C., et al. (2003). Disruption of focal adhesions by integrin cytoplasmic domain-associated protein-1 alpha. J. Biol. Chem. 278 6567–6574. 10.1074/jbc.m211258200 [DOI] [PubMed] [Google Scholar]
- Browne K. A., Johnstone R. W., Jans D. A., Trapani J. A. (2000). Filamin (280-kDa actin-binding protein) is a caspase substrate and is also cleaved directly by the cytotoxic T lymphocyte protease granzyme B during apoptosis. J. Biol. Chem. 275 39262–39266. 10.1074/jbc.c000622200 [DOI] [PubMed] [Google Scholar]
- Burande C. F., Heuze M. L., Lamsoul I., Monsarrat B., Uttenweiler-Joseph S., Lutz P. G. (2009). A label-free quantitative proteomics strategy to identify E3 ubiquitin ligase substrates targeted to proteasome degradation. Mol. Cell Proteomics 8 1719–1727. 10.1074/mcp.m800410-mcp200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood D. A., Huttenlocher A., Kiosses W. B., Rose D. M., Woodside D. G., Schwartz M. A., et al. (2001). Increased filamin binding to beta-integrin cytoplasmic domains inhibits cell migration. Nat. Cell Biol. 3 1060–1068. 10.1038/ncb1201-1060 [DOI] [PubMed] [Google Scholar]
- Chantaravisoot N., Wongkongkathep P., Loo J. A., Mischel P. S., Tamanoi F. (2015). Significance of filamin A in mTORC2 function in glioblastoma. Mol. Cancer 14:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Stracher A. (1989). In situ phosphorylation of platelet actin-binding protein by cAMP-dependent protein kinase stabilizes it against proteolysis by calpain. J. Biol. Chem. 264 14282–14289. [PubMed] [Google Scholar]
- Clark A. R., Sawyer G. M., Robertson S. P., Sutherland-Smith A. J. (2009). Skeletal dysplasias due to filamin A mutations result from a gain-of-function mechanism distinct from allelic neurological disorders. Hum. Mol. Genet. 18 4791–4800. 10.1093/hmg/ddp442 [DOI] [PubMed] [Google Scholar]
- Cunningham C. C., Gorlin J. B., Kwiatkowski D. J., Hartwig J. H., Janmey P. A., Byers H. R., et al. (1992). Actin-binding protein requirement for cortical stability and efficient locomotion. Science 255, 325–327. 10.1126/science.1549777 [DOI] [PubMed] [Google Scholar]
- Dalkilic I., Schienda J., Thompson T. G., Kunkel L. M. (2006). Loss of FilaminC (FLNc) results in severe defects in myogenesis and myotube structure. Mol. Cell Biol. 26 6522–6534. 10.1128/mcb.00243-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das M., Ithychanda S. S., Qin J., Plow E. F. (2011). Migfilin and filamin as regulators of integrin activation in endothelial cells and neutrophils. PLoS One 6:e26355. 10.1371/journal.pone.0026355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donada A., Balayn N., Sliwa D., Lordier L., Ceglia V., Baschieri F., et al. (2019). Disrupted filamin A/alphaIIbbeta3 interaction induces macrothrombocytopenia by increasing RhoA activity. Blood 133 1778–1788. 10.1182/blood-2018-07-861427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff R. M., Tay V., Hackman P., Ravenscroft G., McLean C., Kennedy P., et al. (2011). Mutations in the N-terminal actin-binding domain of filamin C cause a distal myopathy. Am. J. Hum. Genet. 88 729–740. 10.1016/j.ajhg.2011.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlicher A. J., Nakamura F., Hartwig J. H., Weitz D. A., Stossel T. P. (2011). Mechanical strain in actin networks regulates FilGAP and integrin binding to filamin A. Nature 478 260–263. 10.1038/nature10430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esue O., Tseng Y., Wirtz D. (2009). Alpha-actinin and filamin cooperatively enhance the stiffness of actin filament networks. PLoS One 4:e4411. 10.1371/journal.pone.0004411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrington-Rock C., Firestein M. H., Bicknell L. S., Superti-Furga A., Bacino C. A., Cormier-Daire V., et al. (2006). Mutations in two regions of FLNB result in atelosteogenesis I and III. Hum. Mutat. 27 705–710. 10.1002/humu.20348 [DOI] [PubMed] [Google Scholar]
- Farrington-Rock C., Kirilova V., Dillard-Telm L., Borowsky A. D., Chalk S., Rock M. J., et al. (2008). Disruption of the Flnb gene in mice phenocopies the human disease spondylocarpotarsal synostosis syndrome. Hum. Mol. Genet. 17 631–641. 10.1093/hmg/ddm188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Chen M. H., Moskowitz I. P., Mendonza A. M., Vidali L., Nakamura F., et al. (2006). Filamin A (FLNA) is required for cell-cell contact in vascular development and cardiac morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 103 19836–19841. 10.1073/pnas.0609628104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J. W., Lamperti, Eksioglu Y. Z., Hong S. E., Feng Y., Graham D. A., et al. (1998). Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron 21 1315–1325. 10.1016/s0896-6273(00)80651-0 [DOI] [PubMed] [Google Scholar]
- Gardel M. L., Nakamura F., Hartwig J. H., Crocker J. C., Stossel T. P., Weitz D. A. (2006). Prestressed F-actin networks cross-linked by hinged filamins replicate mechanical properties of cells. Proc. Natl. Acad. Sci. U.S.A. 103 1762–1767. 10.1073/pnas.0504777103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetsch S. C., Martin C. M., Embree L. J., Garry D. J. (2005). Myogenic progenitor cells express filamin C in developing and regenerating skeletal muscle. Stem Cells Dev. 14 181–187. 10.1089/scd.2005.14.181 [DOI] [PubMed] [Google Scholar]
- Gorlin J. B., Yamin R., Egan S., Stewart M., Stossel T. P., Kwiatkowski D. J., et al. (1990). Human endothelial actin-binding protein (ABP-280, nonmuscle filamin): a molecular leaf spring. J. Cell Biol. 111 1089–1105. 10.1083/jcb.111.3.1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiet R., Verollet C., Lamsoul I., Cougoule C., Poincloux R., Labrousse A., et al. (2012). Macrophage mesenchymal migration requires podosome stabilization by filamin A. J. Biol. Chem. 287 13051–13062. 10.1074/jbc.m111.307124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer A., Rider L., Oladimeji P., Cook L., Li Q., Mattingly R. R., et al. (2013). Tyrosyl phosphorylated PAK1 regulates breast cancer cell motility in response to prolactin through filamin A. Mol. Endocrinol. 27 455–465. 10.1210/me.2012-1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart A. W., Morgan J. E., Schneider J., West K., McKie L., Bhattacharya S., et al. (2006). Cardiac malformations and midline skeletal defects in mice lacking filamin A. Hum. Mol. Genet. 15 2457–2467. 10.1093/hmg/ddl168 [DOI] [PubMed] [Google Scholar]
- Heikkinen O. K., Ruskamo S., Konarev P. V., Svergun D. I., Iivanainen T., Heikkinen S. M., et al. (2009). Atomic structures of two novel immunoglobulin-like domain pairs in the actin cross-linking protein filamin. J. Biol. Chem. 284 25450–25458. 10.1074/jbc.m109.019661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuze M. L., Lamsoul I., Baldassarre M., Lad Y., Leveque S., Razinia Z., et al. (2008). ASB2 targets filamins A and B to proteasomal degradation. Blood 112 5130–5140. 10.1182/blood-2007-12-128744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsmann S., Rintanen N., Sethi R., Brown N. H., Ylanne J. (2016). Evidence for the mechanosensor function of filamin in tissue development. Sci. Rep. 6:32798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries J. D., Chastney M. R., Askari J. A., Humphries M. J. (2019). Signal transduction via integrin adhesion complexes. Curr. Opin. Cell Biol. 56 14–21. 10.1016/j.ceb.2018.08.004 [DOI] [PubMed] [Google Scholar]
- Ithychanda S. S., Das M., Ma Y. Q., Ding K., Wang X., Gupta S., et al. (2009a). Migfilin, a molecular switch in regulation of integrin activation. J. Biol. Chem. 284 4713–4722. 10.1074/jbc.m807719200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ithychanda S. S., Hsu D., Li H., Yan L., Liu D., Das M., et al. (2009b). Identification and characterization of multiple similar ligand-binding repeats in filamin: implication on filamin-mediated receptor clustering and cross-talk. J. Biol. Chem. 284 35113–35121. 10.1074/jbc.m109.060954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto D. V., Huehn A., Simon B., Huet-Calderwood C., Baldassarre M., Sindelar C. V., et al. (2018). Structural basis of the filamin A actin-binding domain interaction with F-actin. Nat. Struct. Mol. Biol. 25 918–927. 10.1038/s41594-018-0128-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemet G., Morgan M. R., Byron A., Humphries J. D., Choi C. K., Chen C. S., et al. (2013). Rac1 is deactivated at integrin activation sites through an IQGAP1-filamin-A-RacGAP1 pathway. J. Cell Sci. 126 4121–4135. 10.1242/jcs.121988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay D., Garcia E. J., de la Luz Ibarra M. (2004). In situ determination of a PKA phosphorylation site in the C-terminal region of filamin. Mol. Cell. Biochem. 260 49–53. 10.1023/b:mcbi.0000026052.76418.55 [DOI] [PubMed] [Google Scholar]
- Kasza K. E., Koenderink G. H., Lin Y. C., Broedersz C. P., Messner W., Nakamura F., et al. (2009). Nonlinear elasticity of stiff biopolymers connected by flexible linkers. Phys. Rev. E Stat. Nonlin. Soft. Matter. Phys. 79:041928. [DOI] [PubMed] [Google Scholar]
- Kawamoto S., Hidaka H. (1984). Ca2+-activated, phospholipid-dependent protein kinase catalyzes the phosphorylation of actin-binding proteins. Biochem. Biophys. Res. Commun. 118 736–742. 10.1016/0006-291x(84)91456-6 [DOI] [PubMed] [Google Scholar]
- Kechagia J. Z., Ivaska J., Roca-Cusachs P. (2019). Integrins as biomechanical sensors of the microenvironment. Nat. Rev. Mol. Cell Biol. 20 457–473. 10.1038/s41580-019-0134-2 [DOI] [PubMed] [Google Scholar]
- Kesner B. A., Milgram S. L., Temple B. R., Dokholyan N. V. (2010). Isoform divergence of the filamin family of proteins. Mol. Biol. Evol. 27 283–295. 10.1093/molbev/msp236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiema T., Lad Y., Jiang P., Oxley C. L., Baldassarre M., Wegener K. L., et al. (2006). The molecular basis of filamin binding to integrins and competition with talin. Mol. Cell 21 337–347. 10.1016/j.molcel.2006.01.011 [DOI] [PubMed] [Google Scholar]
- Kim C., Ye F., Ginsberg M. H. (2011). Regulation of integrin activation. Annu. Rev. Cell Dev. Biol. 27 321–345. [DOI] [PubMed] [Google Scholar]
- Kim H., Nakamura F., Lee W., Hong C., Perez-Sala D., McCulloch C. A. (2010a). Regulation of cell adhesion to collagen via beta1 integrins is dependent on interactions of filamin A with vimentin and protein kinase C epsilon. Exp. Cell Res. 316 1829–1844. 10.1016/j.yexcr.2010.02.007 [DOI] [PubMed] [Google Scholar]
- Kim H., Nakamura F., Lee W., Shifrin Y., Arora P., McCulloch C. A. (2010b). Filamin A is required for vimentin-mediated cell adhesion and spreading. Am. J. Physiol. Cell Physiol. 298 C221–C236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Sengupta A., Glogauer M., McCulloch C. A. (2008). Filamin A regulates cell spreading and survival via beta1 integrins. Exp. Cell Res. 314 834–846. 10.1016/j.yexcr.2007.11.022 [DOI] [PubMed] [Google Scholar]
- Kircher P., Hermanns C., Nossek M., Drexler M. K., Grosse R., Fischer M., et al. (2015). Filamin A interacts with the coactivator MKL1 to promote the activity of the transcription factor SRF and cell migration. Sci. Signal. 8:ra112. 10.1126/scisignal.aad2959 [DOI] [PubMed] [Google Scholar]
- Krakow D., Robertson S. P., King L. M., Morgan T., Sebald E. T., Bertolotto C., et al. (2004). Mutations in the gene encoding filamin B disrupt vertebral segmentation, joint formation and skeletogenesis. Nat. Genet. 36 405–410. 10.1038/ng1319 [DOI] [PubMed] [Google Scholar]
- Kumar A., Shutova M. S., Tanaka K., Iwamoto D. V., Calderwood D. A., Svitkina T. M., et al. (2019). Filamin A mediates isotropic distribution of applied force across the actin network. J. Cell Biol. 218 2481–2491. 10.1083/jcb.201901086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lad Y., Jiang P., Ruskamo S., Harburger D. S., Ylanne J., Campbell I. D., et al. (2008). Structural basis of the migfilin-filamin interaction and competition with integrin beta tails. J. Biol. Chem. 283 35154–35163. 10.1074/jbc.m802592200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lad Y., Kiema T., Jiang P., Pentikainen O. T., Coles C. H., Campbell I. D., et al. (2007). Structure of three tandem filamin domains reveals auto-inhibition of ligand binding. EMBO J. 26 3993–4004. 10.1038/sj.emboj.7601827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamsoul I., Burande C. F., Razinia Z., Houles T. C., Menoret D., Baldassarre M., et al. (2011). Functional and structural insights into ASB2α, a novel regulator of integrin-dependent adhesion of hematopoietic cells. J. Biol. Chem. 286 30571–30581. 10.1074/jbc.m111.220921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamsoul I., Metais A., Gouot E., Heuze M. L., Lennon-Dumenil A. M., Moog-Lutz C., et al. (2013). ASB2alpha regulates migration of immature dendritic cells. Blood 122 533–541. 10.1182/blood-2012-11-466649 [DOI] [PubMed] [Google Scholar]
- Lamsoul I., Uttenweiler-Joseph S., Moog-Lutz C., Lutz P. G. (2016). Cullin 5-RING E3 ubiquitin ligases, new therapeutic targets? Biochimie 122 339–347. 10.1016/j.biochi.2015.08.003 [DOI] [PubMed] [Google Scholar]
- Legate K. R., Fassler R. (2009). Mechanisms that regulate adaptor binding to beta-integrin cytoplasmic tails. J. Cell Sci. 122 187–198. 10.1242/jcs.041624 [DOI] [PubMed] [Google Scholar]
- Li C., Yu S., Nakamura F., Pentikainen O. T., Singh N., Yin S., et al. (2010). Pro-prion binds filamin A, facilitating its interaction with integrin beta1, and contributes to melanomagenesis. J. Biol. Chem. 285 30328–30339. 10.1074/jbc.m110.147413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Lu Y., Stemmer P. M., Chen F. (2015). Filamin A phosphorylation by Akt promotes cell migration in response to arsenic. Oncotarget 6 12009–12019. 10.18632/oncotarget.3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light S., Sagit R., Ithychanda S. S., Qin J., Elofsson A. (2012). The evolution of filamin-a protein domain repeat perspective. J. Struct. Biol. 179 289–298. 10.1016/j.jsb.2012.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Das M., Yang J., Ithychanda S. S., Yakubenko V. P., Plow E. F., et al. (2015). Structural mechanism of integrin inactivation by filamin. Nat. Struct. Mol. Biol. 22 383–389. 10.1038/nsmb.2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy C. J., Sim K. S., Yong E. L. (2003). Filamin-A fragment localizes to the nucleus to regulate androgen receptor and coactivator functions. Proc. Natl. Acad. Sci. U.S.A. 100 4562–4567. 10.1073/pnas.0736237100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Lian G., Lenkinski R., De Grand A., Vaid R. R., Bryce T., et al. (2007). Filamin B mutations cause chondrocyte defects in skeletal development. Hum. Mol. Genet. 16 1661–1675. 10.1093/hmg/ddm114 [DOI] [PubMed] [Google Scholar]
- Lynch C. D., Gauthier N. C., Biais N., Lazar A. M., Roca-Cusachs P., Yu C. H., et al. (2011). Filamin depletion blocks endoplasmic spreading and destabilizes force-bearing adhesions. Mol. Biol. Cell 22 1263–1273. 10.1091/mbc.e10-08-0661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai A., Muharram G., Barrow-McGee R., Baghirov H., Rantala J., Kermorgant S., et al. (2014). Distinct c-Met activation mechanisms induce cell rounding or invasion through pathways involving integrins, RhoA and HIP1. J. Cell Sci. 127 1938–1952. 10.1242/jcs.140657 [DOI] [PubMed] [Google Scholar]
- McGrath M. J., Binge L. C., Sriratana A., Wang H., Robinson P. A., Pook D., et al. (2013). Regulation of the transcriptional coactivator FHL2 licenses activation of the androgen receptor in castrate-resistant prostate cancer. Cancer Res. 73 5066–5079. 10.1158/0008-5472.can-12-4520 [DOI] [PubMed] [Google Scholar]
- Metais A., Lamsoul I., Melet A., Uttenweiler-Joseph S., Poincloux R., Stefanovic S., et al. (2018). Asb2alpha-Filamin A axis is essential for actin cytoskeleton remodeling during heart development. Circ. Res. 122:e34. [DOI] [PubMed] [Google Scholar]
- Meyer S. C., Sanan D. A., Fox J. E. (1998). Role of actin-binding protein in insertion of adhesion receptors into the membrane. J. Biol. Chem. 273 3013–3020. 10.1074/jbc.273.5.3013 [DOI] [PubMed] [Google Scholar]
- Moser M., Bauer M., Schmid S., Ruppert R., Schmidt S., Sixt M., et al. (2009). Kindlin-3 is required for beta2 integrin-mediated leukocyte adhesion to endothelial cells. Nat. Med. 15 300–305. 10.1038/nm.1921 [DOI] [PubMed] [Google Scholar]
- Moser M., Nieswandt B., Ussar S., Pozgajova M., Fassler R. (2008). Kindlin-3 is essential for integrin activation and platelet aggregation. Nat. Med. 14 325–330. 10.1038/nm1722 [DOI] [PubMed] [Google Scholar]
- Nagano T., Morikubo S., Sato M. (2004). Filamin A and FILIP (Filamin A-Interacting Protein) regulate cell polarity and motility in neocortical subventricular and intermediate zones during radial migration. J. Neurosci. 24 9648–9657. 10.1523/jneurosci.2363-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura F., Heikkinen O., Pentikainen O. T., Osborn T. M., Kasza K. E., Weitz D. A., et al. (2009). Molecular basis of filamin A-FilGAP interaction and its impairment in congenital disorders associated with filamin A mutations. PLoS One 4:e4928. 10.1371/journal.pone.0004928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura F., Osborn T. M., Hartemink C. A., Hartwig J. H., Stossel T. P. (2007). Structural basis of filamin A functions. J. Cell Biol. 179 1011–1025. 10.1083/jcb.200707073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura F., Pudas R., Heikkinen O., Permi P., Kilpelainen I., Munday A. D., et al. (2006). The structure of the GPIb-filamin A complex. Blood 107 1925–1932. 10.1182/blood-2005-10-3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura F., Song M., Hartwig J. H., Stossel T. P. (2014). Documentation and localization of force-mediated filamin A domain perturbations in moving cells. Nat. Commun. 5:4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta Y., Hartwig J. H. (1996). Phosphorylation of actin-binding protein 280 by growth factors is mediated by p90 ribosomal protein S6 kinase. J. Biol. Chem. 271 11858–11864. 10.1074/jbc.271.20.11858 [DOI] [PubMed] [Google Scholar]
- Ohta Y., Hartwig J. H., Stossel T. P. (2006). FilGAP, a Rho- and ROCK-regulated GAP for Rac binds filamin A to control actin remodelling. Nat. Cell Biol. 8 803–814. 10.1038/ncb1437 [DOI] [PubMed] [Google Scholar]
- Ohta Y., Suzuki N., Nakamura S., Hartwig J. H., Stossel T. P. (1999). The small GTPase RalA targets filamin to induce filopodia. Proc. Natl. Acad. Sci. U.S.A. 96 2122–2128. 10.1073/pnas.96.5.2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rantala J. K., Pouwels J., Pellinen T., Veltel S., Laasola P., Mattila E., et al. (2011). SHARPIN is an endogenous inhibitor of beta1-integrin activation. Nat. Cell Biol. 13 1315–1324. 10.1038/ncb2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razinia Z., Baldassarre M., Bouaouina M., Lamsoul I., Lutz P. G., Calderwood D. A. (2011). The E3 ubiquitin ligase specificity subunit ASB2alpha targets filamins for proteasomal degradation by interacting with the filamin actin-binding domain. J. Cell Sci. 124 2631–2641. 10.1242/jcs.084343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razinia Z., Baldassarre M., Cantelli G., Calderwood D. A. (2013). ASB2alpha, an E3 ubiquitin ligase specificity subunit, regulates cell spreading and triggers proteasomal degradation of filamins by targeting the filamin calponin homology 1 domain. J. Biol. Chem. 288 32093–30105. 10.1074/jbc.m113.496604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razinia Z., Makela T., Ylanne J., Calderwood D. A. (2012). Filamins in mechanosensing and signaling. Annu. Rev. Biophys. 41 227–246. 10.1146/annurev-biophys-050511-102252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson S. P., Twigg S. R., Sutherland-Smith A. J., Biancalana V., Gorlin R. J., Horn D., et al. (2003). Localized mutations in the gene encoding the cytoskeletal protein filamin A cause diverse malformations in humans. Nat. Genet. 33 487–491. 10.1038/ng1119 [DOI] [PubMed] [Google Scholar]
- Roth H., Samereier M., Begandt D., Pick R., Salvermoser M., Brechtefeld D., et al. (2017). Filamin A promotes efficient migration and phagocytosis of neutrophil-like HL-60 cells. Eur. J. Cell Biol. 96 553–566. 10.1016/j.ejcb.2017.05.004 [DOI] [PubMed] [Google Scholar]
- Sakane A., Alamir Mahmoud, Abdallah A., Nakano K., Honda K., Kitamura T., et al. (2013). Junctional Rab13-binding protein (JRAB) regulates cell spreading via filamins. Genes Cells 18 810–822. 10.1111/gtc.12078 [DOI] [PubMed] [Google Scholar]
- Sasaki A., Masuda Y., Ohta Y., Ikeda K., Watanabe K. (2001). Filamin associates with Smads and regulates transforming growth factor-beta signaling. J. Biol. Chem. 276 17871–17877. 10.1074/jbc.m008422200 [DOI] [PubMed] [Google Scholar]
- Sato M., Nagano T. (2005). Involvement of filamin A and filamin A-interacting protein (FILIP) in controlling the start and cell shape of radially migrating cortical neurons. Anat. Sci. Int. 80 19–29. 10.1111/j.1447-073x.2005.00101.x [DOI] [PubMed] [Google Scholar]
- Sato T., Ishii J., Ota Y., Sasaki E., Shibagaki Y., Hattori S. (2016). Mammalian target of rapamycin (mTOR) complex 2 regulates filamin A-dependent focal adhesion dynamics and cell migration. Genes Cells 21 579–593. 10.1111/gtc.12366 [DOI] [PubMed] [Google Scholar]
- Savinko T., Guenther C., Uotila L. M., Llort Asens M., Yao S., Tojkander S., et al. (2018). Filamin A is required for optimal T cell integrin-mediated force transmission, flow adhesion, and T cell trafficking. J. Immunol. 200 3109–3116. 10.4049/jimmunol.1700913 [DOI] [PubMed] [Google Scholar]
- Savoy R. M., Chen L., Siddiqui S., Melgoza F. U., Durbin-Johnson B., Drake C., et al. (2015). Transcription of Nrdp1 by the androgen receptor is regulated by nuclear filamin A in prostate cancer. Endocr. Relat. Cancer 22 369–386. 10.1530/erc-15-0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A., Nethe M., Hordijk P. L. (2012). Ubiquitin links to cytoskeletal dynamics, cell adhesion and migration. Biochem. J. 442 13–25. 10.1042/bj20111815 [DOI] [PubMed] [Google Scholar]
- Schmoller K. M., Lieleg O., Bausch A. R. (2009). Structural and viscoelastic properties of actin/filamin networks: cross-linked versus bundled networks. Biophys. J. 97 83–89. 10.1016/j.bpj.2009.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma C. P., Ezzell R. M., Arnaout M. A. (1995). Direct interaction of filamin (ABP-280) with the beta 2-integrin subunit CD18. J. Immunol. 154 3461–3470. [PubMed] [Google Scholar]
- Sheen V. L., Feng Y., Graham D., Takafuta T., Shapiro S. S., Walsh C. A. (2002). Filamin A and Filamin B are co-expressed within neurons during periods of neuronal migration and can physically interact. Hum. Mol. Genet. 11 2845–2854. 10.1093/hmg/11.23.2845 [DOI] [PubMed] [Google Scholar]
- Smith L., Page R. C., Xu Z., Kohli E., Litman P., Nix J. C., et al. (2010). Biochemical basis of the interaction between cystic fibrosis transmembrane conductance regulator and immunoglobulin-like repeats of filamin. J. Biol. Chem. 285 17166–17176. 10.1074/jbc.m109.080911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinner C. A., Uttenweiler-Joseph S., Metais A., Stella A., Burlet-Schiltz O., Moog-Lutz C., et al. (2015). Substrates of the ASB2alpha E3 ubiquitin ligase in dendritic cells. Sci. Rep. 5:16269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Forster C., Nakamura F., Glogauer M. (2013). Filamin-A regulates neutrophil uropod retraction through RhoA during chemotaxis. PLoS One 8:e79009. 10.1371/journal.pone.0079009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Costell M., Fassler R. (2019). Integrin activation by talin, kindlin and mechanical forces. Nat. Cell Biol. 21 25–31. 10.1038/s41556-018-0234-9 [DOI] [PubMed] [Google Scholar]
- Tadokoro S., Shattil S. J., Eto K., Tai V., Liddington R. C., de Pereda J. M., et al. (2003). Talin binding to integrin beta tails: a final common step in integrin activation. Science 302 103–106. 10.1126/science.1086652 [DOI] [PubMed] [Google Scholar]
- Takala H., Nurminen E., Nurmi S. M., Aatonen M., Strandin T., Takatalo M., et al. (2008). Beta2 integrin phosphorylation on Thr758 acts as a molecular switch to regulate 14-3-3 and filamin binding. Blood 112 1853–1862. 10.1182/blood-2007-12-127795 [DOI] [PubMed] [Google Scholar]
- Tirupula K. C., Ithychanda S. S., Mohan M. L., Naga Prasad S. V., Qin J., Karnik S. S. (2015). G protein-coupled receptors directly bind filamin A with high affinity and promote filamin phosphorylation. Biochemistry 54 6673–6683. 10.1021/acs.biochem.5b00975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng Y., An K. M., Esue O., Wirtz D. (2004). The bimodal role of filamin in controlling the architecture and mechanics of F-actin networks. J. Biol. Chem. 279 1819–1826. 10.1074/jbc.m306090200 [DOI] [PubMed] [Google Scholar]
- Tu Y., Wu S., Shi X., Chen K., Wu C. (2003). Migfilin and Mig-2 link focal adhesions to filamin and the actin cytoskeleton and function in cell shape modulation. Cell 113 37–47. 10.1016/s0092-8674(03)00163-6 [DOI] [PubMed] [Google Scholar]
- Ueda K., Ohta Y., Hosoya H. (2003). The carboxy-terminal pleckstrin homology domain of ROCK interacts with filamin-A. Biochem. Biophys. Res. Commun. 301 886–890. 10.1016/s0006-291x(03)00048-2 [DOI] [PubMed] [Google Scholar]
- Uotila L. M., Guenther C., Savinko T., Lehti T. A., Fagerholm S. C. (2017). Filamin A regulates neutrophil adhesion, production of reactive oxygen species, and neutrophil extracellular trap release. J. Immunol. 199 3644–3653. 10.4049/jimmunol.1700087 [DOI] [PubMed] [Google Scholar]
- Vadlamudi R. K., Li F., Adam L., Nguyen D., Ohta Y., Stossel T. P., et al. (2002). Filamin is essential in actin cytoskeletal assembly mediated by p21-activated kinase 1. Nat. Cell Biol. 4 681–690. 10.1038/ncb838 [DOI] [PubMed] [Google Scholar]
- van der Flier A., Sonnenberg A. (2001). Structural and functional aspects of filamins. Biochim. Biophys. Acta 1538 99–117. 10.1016/s0167-4889(01)00072-6 [DOI] [PubMed] [Google Scholar]
- van der Ven P. F., Obermann W. M., Lemke B., Gautel M., Weber K., Furst D. O. (2000). Characterization of muscle filamin isoforms suggests a possible role of gamma-filamin/ABP-L in sarcomeric Z-disc formation. Cell Motil. Cytoskeleton 45 149–162. [DOI] [PubMed] [Google Scholar]
- Velkova A., Carvalho M. A., Johnson J. O., Tavtigian S. V., Monteiro A. N. (2010). Identification of Filamin A as a BRCA1-interacting protein required for efficient DNA repair. Cell Cycle 9 1421–1433. 10.4161/cc.9.7.11256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vial D., McKeown-Longo P. J. (2012). Epidermal growth factor (EGF) regulates alpha5beta1 integrin activation state in human cancer cell lines through the p90RSK-dependent phosphorylation of filamin A. J. Biol. Chem. 287 40371–40380. 10.1074/jbc.m112.389577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali E., Boemi I., Rosso L., Cambiaghi V., Novellis P., Mantovani G., et al. (2017). FLNA is implicated in pulmonary neuroendocrine tumors aggressiveness and progression. Oncotarget 8 77330–77340. 10.18632/oncotarget.20473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali E., Cambiaghi V., Zerbi A., Carnaghi C., Colombo P., Peverelli E., et al. (2016). Filamin-A is required to mediate SST2 effects in pancreatic neuroendocrine tumours. Endocr. Relat. Cancer 23 181–190. 10.1530/erc-15-0358 [DOI] [PubMed] [Google Scholar]
- Vorgerd M., van der Ven P. F., Bruchertseifer V., Lowe T., Kley R. A., Schroder R., et al. (2005). A mutation in the dimerization domain of filamin c causes a novel type of autosomal dominant myofibrillar myopathy. Am. J. Hum. Genet. 77 297–304. 10.1086/431959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade E. M., Halliday B. J., Jenkins Z. A., O’Neill A. C., Robertson S. P. (2020). The X-linked filaminopathies: synergistic insights from clinical and molecular analysis. Hum. Mutat. 41 865–883. 10.1002/humu.24002 [DOI] [PubMed] [Google Scholar]
- Waldt N., Seifert A., Demiray Y. E., Devroe E., Turk B. E., Reichardt P., et al. (2018). Filamin A phosphorylation at serine 2152 by the serine/threonine kinase Ndr2 controls TCR-induced LFA-1 activation in T Cells. Front. Immunol. 9:2852. 10.3389/fimmu.2018.02852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Nakamura F. (2019a). Identification of Filamin A mechanobinding partner I: smoothelin specifically interacts with the Filamin A mechanosensitive domain 21. Biochemistry 58 4726–4736. 10.1021/acs.biochem.9b00100 [DOI] [PubMed] [Google Scholar]
- Wang J., Nakamura F. (2019b). Identification of Filamin A mechanobinding partner II: fimbacin is a novel actin cross-linking and Filamin A binding protein. Biochemistry 58 4737–4743. 10.1021/acs.biochem.9b00101 [DOI] [PubMed] [Google Scholar]
- Wang J. X., Liu Y., Jia X. J., Liu S. X., Dong J. H., Ren X. M., et al. (2019). Upregulation of circFLNA contributes to laryngeal squamous cell carcinoma migration by circFLNA-miR-486-3p-FLNA axis. Cancer Cell Int. 19:196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener K. L., Partridge A. W., Han J., Pickford A. R., Liddington R. C., Ginsberg M. H., et al. (2007). Structural basis of integrin activation by talin. Cell 128 171–182. 10.1016/j.cell.2006.10.048 [DOI] [PubMed] [Google Scholar]
- Wieczorek K., Wiktorska M., Sacewicz-Hofman I., Boncela J., Lewinski A., Kowalska M. A., et al. (2017). Filamin A upregulation correlates with Snail-induced epithelial to mesenchymal transition (EMT) and cell adhesion but its inhibition increases the migration of colon adenocarcinoma HT29 cells. Exp. Cell Res. 359 163–170. 10.1016/j.yexcr.2017.07.035 [DOI] [PubMed] [Google Scholar]
- Woo M. S., Ohta Y., Rabinovitz I., Stossel T. P., Blenis J. (2004). Ribosomal S6 kinase (RSK) regulates phosphorylation of filamin A on an important regulatory site. Mol. Cell Biol. 24 3025–3035. 10.1128/mcb.24.7.3025-3035.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamak A., Hu D., Mittal N., Buikema J. W., Ditta S., Lutz P. G., et al. (2020). Loss of Asb2 impairs cardiomyocyte differentiation and leads to congenital double outlet right ventricle. iScience 23:100959 10.1016/j.isci.2020.100959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida N., Ogata T., Tanabe K., Li S., Nakazato M., Kohu K., et al. (2005). Filamin A-bound PEBP2beta/CBFbeta is retained in the cytoplasm and prevented from functioning as a partner of the Runx1 transcription factor. Mol. Cell Biol. 25 1003–1012. 10.1128/mcb.25.3.1003-1012.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J., Wang Q., Lu H., Brenneman M., Fan F., Shen Z. (2009). The cytoskeleton protein filamin-A is required for an efficient recombinational DNA double strand break repair. Cancer Res. 69 7978–7985. 10.1158/0008-5472.can-09-2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Tian F., Sandzen J., Cao R., Flaberg E., Szekely L., et al. (2007). Filamin B deficiency in mice results in skeletal malformations and impaired microvascular development. Proc. Natl. Acad. Sci. U.S.A. 104 3919–3924. 10.1073/pnas.0608360104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Chen Z., Zhang L., Zhu M., Tan C., Zhou X., et al. (2020). Loss of Filamin C is catastrophic for heart function. Circulation 141 869–871. 10.1161/circulationaha.119.044061 [DOI] [PMC free article] [PubMed] [Google Scholar]