Abstract
As one of the bicyclic metabolic pathways of one-carbon metabolism, methionine metabolism is the pivot linking the folate cycle to the transsulfuration pathway. In addition to being a precursor for glutathione synthesis, and the principal methyl donor for nucleic acid, phospholipid, histone, biogenic amine, and protein methylation, methionine metabolites can participate in polyamine synthesis. Methionine metabolism disorder can aggravate the damage in the pathological state of a disease. In the occurrence and development of chronic liver diseases (CLDs), changes in various components involved in methionine metabolism can affect the pathological state through various mechanisms. A methionine-deficient diet is commonly used for building CLD models. The conversion of key enzymes of methionine metabolism methionine adenosyltransferase (MAT) 1 A and MAT2A/MAT2B is closely related to fibrosis and hepatocellular carcinoma. In vivo and in vitro experiments have shown that by intervening related enzymes or downstream metabolites to interfere with methionine metabolism, the liver injuries could be reduced. Recently, methionine supplementation has gradually attracted the attention of many clinical researchers. Most researchers agree that adequate methionine supplementation can help reduce liver damage. Retrospective analysis of recently conducted relevant studies is of profound significance. This paper reviews the latest achievements related to methionine metabolism and CLD, from molecular mechanisms to clinical research, and provides some insights into the future direction of basic and clinical research.
Subject terms: Metabolic engineering, Pathogenesis
Introduction
Chronic liver disease (CLD) represents a significant public health concern worldwide.1 Viruses, metabolic dysfunction, autoimmune diseases, and alcoholism are the causes of chronic liver injury, all of which almost causes liver fibrosis.2,3 In addition, CLD is an established risk factor for hepatocellular carcinoma (HCC).4 Metabolic disorders characterize most liver diseases. Patients with CLD often have alterations in glucose,5 trace elements,6–11 lipid,12 and protein metabolism.13–15 The effects of liver diseases on amino acid (AA) metabolism have received widespread attention. Abnormally high concentrations of cysteine, methionine, and aromatic AAs were observed in patients with cirrhosis.16 The serum levels of branched-chain AAs decreased in patients with CLD.13 Nutritional therapy is used to control and manage liver metabolism, and this may improve liver function and have a positive influence on liver diseases.17 A better understanding of nutrient metabolism, in which methionine metabolism plays a vital role, could help identify novel therapeutic targets for preventing CLD.
Methionine, an essential proteogenic AA, is necessary for average growth and development,18 and breakdown of methionine in the small intestine generates free methionine. Subsequently, the free methionine is absorbed and used for protein synthesis or is converted to S-adenosylmethionine (SAM/AdoMet).19 SAM acts as a major methyl and sulfate group donor in numerous biochemical reactions20 and is recommended for the treatment of certain diseases. SAM synthesis is suppressed in CLD, therefore, considerable interest has been focused on utilizing SAM for reducing disease severity (Table 1).19 However, clinical research on methionine supplementation remains insufficient and the results are controversial.21 Here, we offer an in-depth review of the latest achievements related to the physiological and pathophysiological roles of methionine metabolism in liver diseases, from molecular mechanisms to clinical research. We also provide some recommendations for further research.
Table 1.
The main CLDs affected by methionine deficiency, and of any of the enzymes that participate in the transsulfuration pathway
| Chronic liver disease | Adverse consequences of methionine deficiency | References |
|---|---|---|
| Viral hepatitis | Low STAT methylation level, the change of MAT1A/MAT2A and the lower deposition of H3K4me3 on HBV-DNA | 31,57–59 |
| Alcoholic liver disease |
Cystathionine and serum homocysteine elevate. MATα1 level, GSH, folate and vitamin B6, and B12 decrease. Decreased ratio of SAM/SAH directly affects the methylation level, ethanol tampers with multiple enzymes, including MAT, BHMT, and various MTs. The lack of PRMT causes lower PE methylation, which leads to SAM accumulation and sensitivity to oxidative stress. |
73,74,76–78,81–83 |
| Nonalcoholic fatty liver disease | Hepatic Fgf21 mRNA was increased, which is a modulator of energy homeostasis. FFA accumulates and can cause lipotoxicity through JNK1 activation. CD36 level, the PC/PE ratio, and serum homocysteine increase. | 93,96,98–100 |
| Liver fibrosis and cirrhosis | The phosphorylation of MATα2 and MATβ proteins enhanced. The binding of E2F-4 to MAT2A promoter attenuates. SAM/SAH ratio and DNA methylation decrease. | 22,113,117 |
| Hepatocellular carcinoma | GNMT is downregulated, MAT1A expression decreases while MAT2A increases. The activity of ODC increases. High levels of CBS express in HCC, which involve in cell proliferation. The expression of SAHH/AHCY is inhibited. | 48,109,131,138,152 |
The physiological role of methionine metabolism
Methionine metabolism can be divided into the methionine cycle, transsulfuration pathway, and salvage cycle (Fig. 1). First, methionine adenosyltransferases (MATs) catalyze the biosynthesis of SAM from methionine and ATP.18,22 Under the catalysis of methyltransferases (MTs), SAM donates its methyl group for methylation and converts itself to S-adenosyl-homocysteine (SAH).23,24 SAH is then converted by S-adenosyl-homocysteine hydrolase (SAHH/AHCY) to homocysteine (Hcy).25 Hcy promotes glutathione (GSH) synthesis by entering the transsulfuration pathway or is converted back to methionine by methionine synthase (MTR/MS), accordingly completing the methionine cycle. MTR requires the methylated form of vitamin B12 (cobalamin) and uses 5-methyltetrahydrofolate, as a methyl donor for catalysing Hcy remethylation.24 Parallel to this process, betaine homocysteine methyltransferase (BHMT) can catalyze the formation of SAM from the methyl donor betaine.25,26 In the transsulfuration pathway, cystathionine-β-synthase (CBS) catalyzes cystathionine synthesis through the condensation of Hcy and serine. Then, cystathionine-γ-lyase hydrolyzes cystathionine to produce cysteine for GSH synthesis. In addition, cystathionine-γ-lyase and CBS catalyze the production of hydrogen sulfide in these processes.18 Methionine can also be recovered from methylthioadenosine (MTA), a by-product of polyamine (PA) biosynthesis, via the methionine salvage pathway. Furthermore, SAM is decarboxylated by AdoMet decarboxylase to form decarboxylated SAM (dcSAM). After donating of an aminopropyl group for PA synthesis, dcSAM transforms to MTA, which is then converted back to methionine via six enzymatic steps.27–29
Fig. 1.
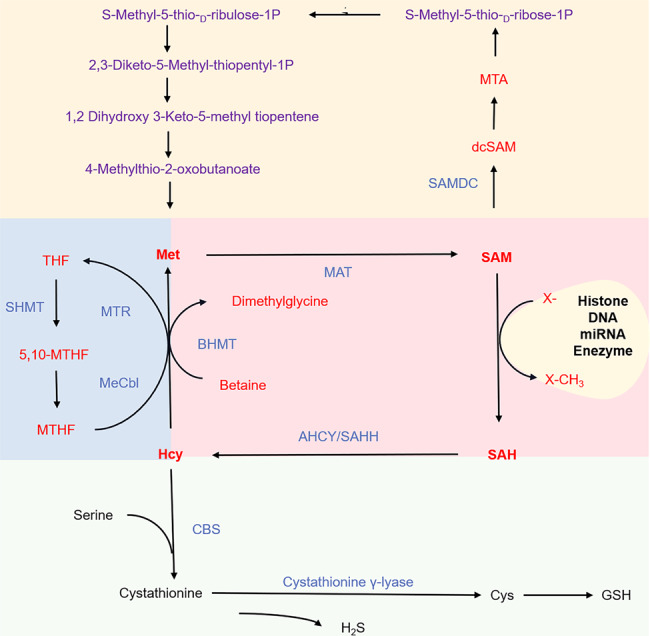
Response of methionine metabolism in the liver. There are four main participants in this pathway, namely methionine, S-adenosylmethionine (SAM), S-adenosyl homocysteine (SAH), and homocysteine (Hcy). Methionine adenosyltransferase (MAT) converts methionine to SAM and then uses a methyl donor catalyzed methyl donor. Another product of these reactions is SAH, which is reduced by S-adenosine homocysteine protease (AHCY/SAHH) to adenosine and Hcy. Methionine metabolism involves the folate cycle, the transsulfuration pathway, and the salvage pathway. AHCY adenosylhomocysteinase, BHMT betaine homocysteine methyltransferase; GSH glutathione; Hcy homocysteine, SAM S-adenosylmethionine, SAH S-adenosyl homocysteine, Met methionine, MTs methyltransferase, CBS cystathionine-β-synthase, Cbl cobalamin, vitamin B12, MeCbl methylcobalamin, MTA 5′‐methylthioadenosine, dcSAM decarboxylated SAM, MTHFR methylenetetrahydrofolate reductase, SHMT serine hydroxymethyltransferase
SAM is the principal methyl donor for the methylation of phospholipids, nucleic acids, and biogenic amines.19 It is likewise the second most common enzymatic cofactor after ATP.30 Methionine concentration is closely related to the ratio of SAM to SAH, which affects many methylation reactions, including histone methylation.31,32 Alterations in the methylation status contribute to many pathophysiological conditions, including cancer, obesity, and ageing.33,34 Methionine is among the major targets of reactive oxygen species (ROS).35–37 Low methionine concentration can also lead to cell morphological changes and cell proliferation.31 Maladjustment of methionine metabolism, which plays a crucial role in cellular physiology, occurs in sundry diseases.18 Liver lesions are closely related to profound alterations in methionine metabolism.38 Copeland et al. linked the alterations in the methionine cycle and liver injury for the first time.39 A systematic review reported that SAM improves some liver biochemical parameters and symptoms in patients with intrahepatic cholestasis,40 which is a feature of several CLDs.41 Furthermore, in recent years, N6-methyladenosine (m6A) modifications have been proven to be related to liver injuries. Deregulation of m6A regulators in host hepatocytes may contribute to the development of viral hepatitis.42 Methionine metabolism is closely related to m6A methylation.42,43 A study revealed a mechanism of homeostatic regulation of SAM synthesis in mammalian cells that involves dynamic m6A modifications in the MAT2A 3′ UTR.44 After methionine depletion, splicing of the MAT2A-retained intron is rapidly induced.45
Methionine metabolism is closely related to various metabolic pathways (Fig. 2). The folate cycle, coupled with the methionine cycle, constitutes a double ring metabolic pathway. All such bicyclic pathways are collectively referred to as one-carbon metabolism.46,47 Methionine metabolism is the pivot linking the folate cycle to the transsulfuration pathway. The intermediate Hcy, which is a sulfur-containing, nonprotein, toxic AA,48 connects the transsulfuration pathway with the methionine cycle. Hcy clearance is essential for genetic protection.25 As the first enzyme in the transsulfuration pathway, CBS has a SAM regulatory site and mutation at this site results in homocystinuria,30 which is closely associated with cancer.48 GSH synthesis also links the transsulfuration pathway with glutamine metabolism, which upregulated by many oncogenic insults and mutations.49 The gene for 5-methylthioadenosine phosphorylase (MTAP), a key enzyme of the methionine salvage pathway, is frequently deleted in human cancers.50,51 Inhibition of protein arginine N-methyltransferase 5 (PRMT5) has recently emerged as a potential therapy against MTAP-deficient cancers.52 MTA inhibits PRMT5 by competing with SAM for binding to the catalytic site.50 Methionine restriction (MR) is sufficient for eliminating MTA accumulation to levels found in MTAP-expressing cells.53 Besides, methionine metabolism is closely related to PA metabolism. An increased in PA levels caused by ornithine decarboxylase (ODC) activation may lead to the pro- or anti-inflammatory roles of PAs.29 Thus, methionine metabolism plays an important role in various biological metabolisms.
Fig. 2.
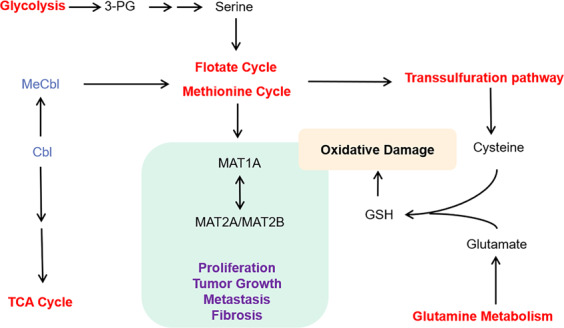
Cross talk between methionine metabolism and the other metabolism. Glycolysis produces ATP and 3-phosphoglycerate (3-PG), which are used in serine synthesis and folate cycle. ATP can be used to transform methionine into SAM. Cobalamin (Cbl) is closely related to TCA, while the methylcobalamin (MeCbl) is related to the folate cycle and methionine cycle. Glutamic acid produced by glutamine metabolism can be used in the synthesis of GSH. 3-PG 3-phosphoglycerate, Cbl cobalamin, vitamin B12, MeCbl methylcobalamin, TCA cycle tricarboxylic acid cycle
Methionine metabolism and CLDs
Role of methionine metabolism in viral hepatitis
Hepatitis B virus (HBV)54 and hepatitis C virus (HCV) cause chronic liver injury.55 Significant ethnic differences are observed in viral infection rates.55 HCV infection can downregulate protein arginine N-methyltransferase 1 (PRMT1) through protein phosphatase 2A (PP2A), while PRMT1 can catalyze STAT-1 methylation on Arg 31.56 STAT-1 is a transcription factor that participates in viral signaling responses and responses to interferon (IFN) signalling activation.57 Duong et al.58,59 demonstrated that downregulation of IFN-sensitive gene expression by interference with STAT-1 methylation can promote interaction with the protein inhibitor of activated STATs (PIAS). After SAM administration, the antiviral effect of IFN was enhanced.59 Feld et al. also showed that adding SAM to peginterferon (PEG-IFN) and ribavirin improves the kinetics of the early antiviral response.60 Sonia Amelia Lozano-Sepulveda et al. suggested that SAM can diminish HCV expression in cells partly by modulating antioxidant enzymes, synthesizing GSH, and switching the MAT1A/MAT2A turnover.57 The MAT1A/MAT2A ratio is relevant to the survival of patients with HCC.61 This transformation may be conducive to the transition of viral hepatitis to liver cancer. SAM can balance the MAT1A/MAT2A ratio, which may reduce or even prevent the further development of the disease. Furthermore, in a HCV study involving ethanol feeding, betaine treatment attenuated the damage caused by ethanol metabolism to STAT-1 methylation.62 However, in a phase II randomized controlled trial, SAM neither reduced liver damage in patients with HCV cirrhosis nor improved the liver function.21 Considering the pharmacokinetics of SAM, the high chemical reactivity of the methyl group of SAM and its spontaneous decomposition may lead to adverse effects. Thus, the efficacy and safety of SAM against HCV need to be investigated in further studies.63
Current therapies for chronic hepatitis B (CHB) mainly use PEG-IFN and orally administered nucleotide analogs.55 Deposition of H3K4me3 on HBV-DNA was reduced in HBV early antigen negative stage samples from CHB patients, which correlated with the levels of viral transcripts,64 while the H3K4me3 signature is modulated in response to a decrease in SAM and SAH levels.31 Liu et al. demonstrated that the X protein of HBV enhances the binding of transcription factors NF-κB and cAMP-response element-binding protein to the promoter of the MAT2A gene, and thus regulates its expression, which is essential in HBV-mediated HCC progression.65 Guo et al. showed that SAM concentrations are related to the severity of HBV-related liver disease.66 HBV inhibited STAT-1 methylation dramatically. Combined with IFN-α, SAM treatment effectively improved STAT-1 methylation and attenuated STAT-1 - PIAS1 binding.67 In establishing a practical cure for chronic HBV infection, the significance of T cells has been confirmed.68 Sinclair et al. showed that a steady supply of methionine is required for T cells to remain activated, and T cell activation increases the demand for methionine.69 Bing et al. found that the combination therapy of glucocorticoids, SAM, and IFN-α is possibly useful for CHB patients.70 HBV CpG methylation is closely linked to hepatocarcinogenesis. DNA methylation also plays a major role by silencing tumor suppressors in HCV-infected patients with HCC. Between HBV and HCV samples, in terms of both expression and methylation levels, researchers found that the greatest differences were three genes: human leukocyte antigen, STAT-1, and 2′5′oligoadenylate synthetase 2. The in-depth study of methylation differential genes will help elucidate diverse mechanisms of HBV and HCV pathogenesis, and further benefit antiviral therapies.71 Considering the important role of the methionine cycle in methylation, the relationship between the methionine cycle and HBV and HCV at the molecular level needs further exploration (Fig. 3). The role of methionine metabolism in virus invasion needs additional research.
Fig. 3.
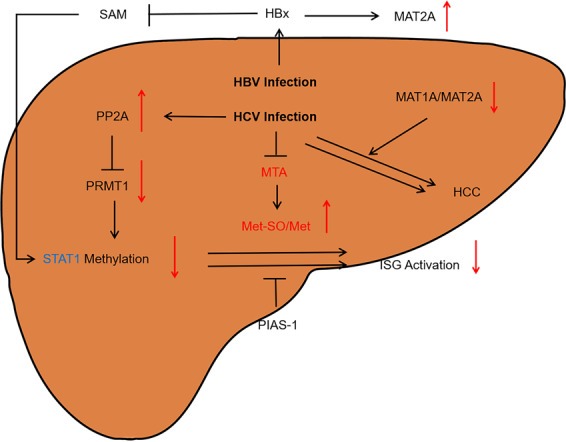
The metabolism of methionine in viral infections of hepatitis B and C. The hepatic polyamine synthesis and transsulfuration pathway activities are impaired in virus infection. Methylthioadenosine (MTA) is a sulfur-containing adenine nucleoside produced from SAM during the synthesis of polyamines, including spermine and spermidine. The level of MTA significantly decreases during the late stage of HCV infection in cells. Moreover, Met is particularly susceptible to elevated ROS levels. Upon reacting to ROS, protein-bound Met is readily oxidized to form methionine sulfoxide (Met-SO). The increased Met-SO level and Met-SO/Met ratio indicate increased oxidative stress subsequent to decrease liver function. Furthermore, the STAT-1 genes showed significant difference between HBV and HCV. Met methionine, Met-SO methionine sulfoxide, HBx the X protein of HBV, ISG interferon-stimulated gene, PP2A protein phosphatase 2A, PRMT1 protein arginine methyltransferase 1, PIAS protein inhibitor of activated STATs
Role of methionine metabolism in alcoholic liver disease
Prolonged exposure to ethanol causes sustained and noticeable liver damage,72 from steatosis to alcoholic steatohepatitis to fibrosis, even cirrhosis.23 Alteration in methionine metabolism plays a vital role in the development of alcoholic liver injury.23,73 Long-term ethanol consumption results in increased Hcy and SAH levels. The increase in SAH levels is sufficient for sensitizing the liver and hepatocytes to tumor necrosis factor (TNF) cytotoxicity.74 The development of steatosis and the inhibition of proteasome activity, both hallmark signatures of alcohol-induced liver damage, occur as a result of the reduced SAM/SAH ratio.75 The decreased SAM/SAH ratio directly affects the methylation level.74 Ethanol tampers with the function of multiple enzymes, including MAT, BHMT, and various MTs.76,77 Decreased MAT activity is attributable to alcohol-induced oxidative stress and reactive aldehydes, which can inactivate the liver-specific MAT. The lack of metabolic products due to impaired methionine metabolism inhibits remethylation of Hcy, the form of GSH, thus weakening defenses against oxidative stress.23,78 The increase in SAH levels and hypomethylation may severely impact the expression/activity of caspase-8, which correlates with enhanced apoptosis of alcoholic liver disease (ALD).79,80 Furthermore, individuals with alcoholic hepatitis have lowered levels of MATα1, which directly interacts with CYP2E1 and facilitates CYP2E1 methylation at a critical arginine residue.81 Besides, SAM can participate in lipid synthesis through the phosphotidylethanolamine N-methyltransferase pathway, which is essential for forming very-low-density lipoproteins (VLDLs).82 Lack of phosphatidylethanolamine (PE) methylation leads to the SAM accumulation, which results in hypermethylation of histones and the major phosphatase PP2A, dependency on cysteine, and sensitivity to oxidative stress.83
By activating the Nrf2-ARE pathway, methionine availability promotes endogenous antioxidant responses and plays a key role in inhibiting ROS-induced oxidative stress.84 In various experimental models of liver diseases, both SAM and betaine attenuated ethanol-induced liver injury.23,75 SAM supplementation reverses the depletion of SAM and GSH in ethanol-fed animals, and restores the fluidity of the mitochondrial inner-membrane. In addition, it alleviates steatosis and hepatocyte necrosis.19 The marked impact of SAM supplementation is prevention of mitochondrial DNA damage and mitoribosome dissociation.85 SAM attenuates injuries by regulating the metabolism of beneficial cytokines.74 SAM also downregulates potentially toxic pro-inflammatory cytokines.79 Betaine per se does not directly interact with oxidants. In the methionine cycle, it mainly mediates SAM synthesis,86 and thus restores the SAM/SAH ratio, and recovers DNA methylation and gene expression.87 Betaine also alleviates alcohol-induced free fatty acid (FFA) accumulation by correcting an alcohol-induced imbalance in fatty acid (FA) synthesis and oxidation by targeting hepatic sterol regulatory element-binding protein (SREBP)-1c, FA synthase, and peroxisome proliferator-activated receptor γ (PPARγ) coactivator 1α.88 SAM exhibits direct antioxidant activity by scavenging ROS.86 In clinical trials, SAM increases the levels of the cellular antioxidant GSH in patients with ALD, and the survival of patients with less advanced liver cirrhosis improves with SAM.89,90 Prednisolone plus SAM can produce an improved therapeutic response.91 The maladjustment of methionine and SAM metabolism has been well-accepted in ALD, while the effect of this maladjustment on downstream products has not been fully investigated.79 Multiple pieces of evidence have linked ethanol-induced abnormal methionine metabolism to deficiencies of folate, and vitamin B6 and B12, which are key factors in ALD pathogenesis.73 The trial of SAM treatment for ALD is inconclusive. Larger and longer-term clinical trials are needed, and supplementation with other compounds important for methionine metabolism, such as vitamin B6, should be considered in ALD patients. Betaine should also include be investigated as a supplement in large-scale clinical studies.90
Role of methionine metabolism in nonalcoholic fatty liver disease
Nonalcoholic fatty liver disease (NAFLD) is a result of defects in multiple metabolic pathways leading to the accumulation of triglycerides (TGs) in the liver.92 NAFLD frequently progresses to nonalcoholic steatohepatitis (NASH), which is a result of prolonged inflammation and hepatocyte damage, ultimately causing fibrosis, HCC, and even death.93,94 Human NASH is associated with hypomethylation of liver DNA.92 A previous study showed a lower rate of methionine transmethylation in insulin-resistant patients with NASH.95 Gene deletion or the lack of unstable methyl groups in the form of methionine and choline undermines the ability to synthesize SAM, which leads to the development of steatosis and its rapid progress toward NASH.96,97 Treatment with a methionine and choline-deficient diet (MCD) is a routine, and useful method for inducing NASH in rodents.93 Saturated FFA accumulates in MCD-fed mice and can cause lipotoxicity through JNK1 activation, leading to mitochondrial damage and ROS production.98 Notably, MAT1A-KO mice exhibit chronic liver SAM deficiency, and spontaneously develop steatohepatitis and HCC.81 The liver of these mice have high TGs, diglycerides, FAs, and ceramide. CD36 content significantly augments in MAT1A-KO mice.92 Increasing the CD36 level contributes to hepatic TG storage.99 Besides, the levels of Hcy, an intermediary in liver methionine metabolism, are elevated in patients with NAFLD.100 Elevated serum Hcy concentrations are related to the histological severity of NAFLD.101 However, high serum Hcy levels are negatively associated with NASH, and significant fibrosis in patients with NAFLD. The pathophysiological mechanisms between Hcy and NAFLD are multifactorial, and not fully understood.102
SAM is the key methyl donor for phosphatidylcholine (PC) synthesis, which is required for the export of VLDLs from the liver.95 Beyond its role as a methyl donor, SAM can act as a metabolic regulator, controlling processes, such as regeneration, differentiation, and organ sensitivity to different injuries.103 Mitochondrial polarization was restored in MAT1A-KO hepatocytes upon incubation with SAM.92 Adding methionine to a high-cholesterol diet can significantly reduce hepatic steatosis (HS), oxidative stress, and fibrosis caused by high choline alone.104 However, due to the lack of early diagnosis, conducting human research in the initial stage of NASH is impossible. Moreover, Maria del Bas et al. found that selenium and vitamin E deficiency together cause an increase in SAM, and suppress MYC expression in livers of hamsters on a high-fat diet, which accelerated the development of hamster NASH. Therefore, under certain circumstances, increasing hepatic SAM might not be an efficient strategy.103 Similarly, Yao et al. showed that excess SAM is harmful. A high-methionine diet (HMD) caused hyperhomocysteinemia (HHcy) and HS in mice.99 The association between HHcy and NAFLD has been well investigated.104,105 Conversely, reducing or limiting methionine intake has beneficial effects, such as increased energy expenditure, extended lifespan, improved insulin sensitivity, and reduced adiposity.106–108 Dietary MR can substantially lower circulating levels of methionine in SAHH-deficient patients.109 Gao et al. showed that dietary MR can induce particular metabolic profiles rapidly in clinical settings.110 According to some studies, methionine induces hypercholesterolemia by facilitating choline synthesis in the liver. Nevertheless, sitagliptin in conjunction with a high-choline diet exacerbates oxidative damage, leading to symptoms that are similar to those of NASH, whereas the HMD will partially attenuate the negative effects.104 Therefore, whether NAFLD patients need a large amount of SAM supplementation needs to be investigated. Furthermore, methionine and intermediates formed during its metabolism may play a separate or synergistic role, thereby conferring hepatoprotective effects; however, this needs to be explored further.
Role of methionine metabolism in liver fibrosis and cirrhosis
Liver fibrosis is a characteristic of almost all CLDs and remains a crucial determinant of clinical prognosis.1 In CLD, the imbalance between the new deposition of the extracellular matrix and its resorption leads to the development of fibrosis, indicating the liver’ s response to repeated wound healing. Eventually, it may lead to cirrhosis.111 Hepatic fibrosis can be considered bidirectional dynamic development and regression. Activated hepatic stellate cells (HSCs) are considered a key factor in fibrosis pathogenesis.112,113 Two MAT genes, MAT2A and MAT2B, are required for HSC activation. For the first time, Ramani et al. identified that the phosphorylation of MATα2 and MATβ proteins is enhanced during HSC activation. The stability of these proteins favors human HSC trans-differentiation.22 MAT2B affects HSC activation through ERK and PI-3K signalling mechanisms, whereas MAT2A affects the growth of HSCs by influencing the changes in SAM levels.113 In quiescent HSCs, PPARγ is a negative regulator of MAT2A. A transition from the quiescence state to the activation state abolishes this control and permits PPARβ to increase MAT2A transcription.114 Besides, leptin-induced multiple signalling pathways mediate HSC activation.115 MAT2B-KO completely blocks leptin-mediated induction of STAT3 phosphorylation.116 A recent study showed that leptin-induced β-catenin signalling attenuats E2F-4 binding to the MAT2A promoter, thus increasing its activity.117 HSC activation causes a decline in SAM and MTA levels, a decrease in the SAM/SAH ratio, and overall hypomethylation of DNA.113 The decrease in SAM levels is related to lower MAT II activity during activation.118
Ramani et al. revealed that mutations in the phosphorylation sites of Y371/Y374 in MATα2, and T257/Y259 in MATβ inhibit HSC activation.22 Mutation of specific phosphorylation sites may be used as a strategy for treating fibrosis and even liver cirrhosis caused by HSC activation. A previous meta-analysis confirmed the protective effects and safety of SAM for CLD. Animal experiments have shown that depletion of SAM level is related to liver fibrosis.66 In an ethanol-LPS fibrotic liver rat model, SAM addition inhibits oxidative stress and HSC activation, thereby significantly reducing liver damage and fibrosis.119 Pharmacological doses of SAM and MTA can downregulate the expression of MAT genes and interrupt leptin-mediated signalling.116 SAM significantly inhibited type I collagen secretion and increased NF-κB activity. SAM also increased type I collagen polyubiquitination.120 Furthermore, SAM inhibits HSC contraction by interfering with the formation of F-actin stress fibers and phosphorylated myosin light chains.121 SAM act as potent inhibitors of Wnt signalling and Wnt-induced lysosomal extracellular protein digestion.122,123 Wnt/β-catenin signalling is a major therapeutic target for liver fibrosis.124 A recent study found that intracellular SAM concentration is regulated by the TGF-β1/p65/MAT2A signalling pathway, and may be targeted in liver fibrosis treatment.125 Studies regarding SAM and reversion of liver fibrosis are still lacking. Liver sinusoidal endothelial cells (LSECs), which maintain liver homeostasis, as well as HSC and Kupffer cell quiescence, are the main players in resolving fibrosis.126 Exploration of the relationship between methionine metabolism, LSECs, and fibrosis regression may provide unexpected findings.
Role of methionine metabolism in HCC
Globally, HCC is the fourth most common cause of cancer-related death.127,128 Surgery remains the most effective treatment with curative potential, and novel treatments are urgently needed.66 MAT1A is a marker for normal liver differentiation, and the expression of MAT2A and MAT2B increases during rapid liver growth and dedifferentiation.129,130 During the general development of the fetal liver, the originally expressed MAT2A is gradually replaced by MAT1A.116,131 MAT2A and its gene product, MAT IIα (dimer formed from α2 subunits), are overexpressed in various human epithelial tumors. TNF-α upregulates MAT2A via NF-κB and adaptor-related protein complex 1.132 MAT2A overexpression improves the activity of the V-Maf Avian Musculoaponeurotic Fibrosarcoma Oncogene Homolog G (MAFG) promoter. High MAFG expression correlates with tumor progression and reduced survival time.61 Liu et al. revealed that in liver cancer, hypoxia activates MAT2A expression through hypoxia-inducible factor-1α, resulting in increased MAT II enzyme activity and reduced SAM production, which then induces genomic DNA demethylation.133 Tumor cell proliferation is inhibited by histone acetylation, which promotes MAT IIα ubiquitylation and subsequent proteasomal degradation.132 Inhibition of MAT1A expression leads to tumor growth, invasion, and metastasis.130 A recent study showed that upregulation of forkhead box M1 decreases MAT1A, while raises NF-κB expression; thus, forming a feed-forward loop that enhances tumorigenesis.134 Prohibitin 1 (PHB1) and MAT1A positively regulate each other, while PHB1 and MAT1A mutually regulate c-MYC/MAFG/c-MAF.135 Silencing of MAT2B could remarkably inhibit migration and invasion.136 A cross talk between MAT2B and HuR and SIRT1 protein influences the therapeutic effect of resveratrol on liver cancer cells.137 Moreover, glycine N-methyltransferase (GNMT), which is the most abundant liver MT regulating the availability of SAM, is downregulated in HCC. Deletion of the gene for GNMT promotes a shift in metabolism and the transfer of nutrients from glucose formation to utilization of elevated levels of SAM. 138 Furthermore, MCD, as a commonly used model for inducing CLD and even HCC, is closely link to cancer.139–142 Choline depletion affects lipid metabolism and transport.143 Jiang et al. found that choline supplementation increases global DNA methylation and the expression of peroxisomal acyl-coenzyme A oxidase 1, which mediates FA β-oxidation.144 Liu et al. showed that higher choline intake can improve the overall general health.145 In juvenile black seabream, choline supplementation suppressed NF-κB activation and increased the expression of lipolysis pathway genes.146 The increased uptake of choline by HCCs cells promotes phospholipid formation, DNA hypermethylation, and hepatocyte proliferation. Gougelet et al. showed that choline-deficient diet reverses these effects and promotes regression of HCC that overexpress β-catenin in mice.147 Some controversies regarding MR do exist. Limited intake of methionine attenuates steatosis and delays the development of NASH through various signal transduction pathways and effector molecules, including SREBPs, sirtuins, and the growth hormone/insulin-like growth factor-1 axis.148 Dietary MR and cysteine restriction have beneficial effects on circulating biomarkers, including FGF21,149 and MR protects against metabolic diseases and ageing, represses cancer development, and improves cancer therapy.150,151 However, MAT1A-KO mice, characterized by chronic SAM deficiency, exhibit macrovesicular steatosis, mononuclear cell infiltration in periportal areas, and HCC development.38 MR leads to insufficient SAM, leading to MAT1A/MAT2A transition and the overall DNA hypomethylation, decreased DNA reduction, and genomic instability and abnormal signal transduction are related, including c-MYC overexpression, increased PA synthesis, RAS/ERK, PI-3K upregulated/AKT, and LKB1/AMPK axis. The decrease in SAM levels leads to HCC cell proliferations, cell survival, and microvascular formation.96
Reduced SAM levels and dysregulation of MATs are considered potential therapeutic targets for HCC. Early studies have shown that in exogenous SAM-treated rats, ODC activity, and PA synthesis are significantly reduced in preneoplastic liver lesions.152 When mice are treated with SAM or ursodeoxycholic acid, MAFG induction is weakened during bile duct ligation. When a combination of these drugs are administered, MAFG induction is completely blocked.61 When MAT1A expression increases, the LIN28B promoter region becomes highly methylated, increasing the expression of let-7.130 Increasing MAT1A expression seems to be an effective treatment strategy. Overexpressing MAT1A in the Huh7 cell line steadily increased SAM levels and cell apoptosis, decreased cell growth, and decreased the expression of angiogenesis genes. MATα1 also interacts with p53 and DNA damage-regulated gene 1 in hepatoma cells.129 Furthermore, SAM treatment altered the homeostasis of MAT1A and MAT2A by altering the balance of AUF1 and methyl-HuR/HuR, which was first identified to inhibit MAT2A mRNA stability.153 SAM maintains MAT1A expression, but inhibits MAT2A expression, in hepatocytes. Pharmacological doses of SAM and its metabolite MTA promote apoptosis of liver cancer cells, while resisting the apoptosis of normal liver cells;116 thus, SAM is an attractive chemopreventive agent.129 Exogenous SAM can inhibit HCC development by recovering the normal level of SAM.131 However, 24-day intravenous infusion of SAM did not affect the size of the formed tumors, which may be due to the compensatory induction of GNMT, and prevented SAM accumulation.38 A study revealed that tumor-initiating cells become addicted to exogenous methionine because of highly elevated methionine cycle activity. Even transient inhibition of the methionine cycle is sufficient to weaken the tumor-initiating capability.154 Treatment effectiveness of MR is dependent on many factors. More studies are warranted to determine the regulatory effects.155 SAM showed a meaningful clinical value for patients with advanced tumors and improved prognosis; however, the efficacy of SAM treatment needs further exploration in randomized prospective clinical trials.66 In addition, glutamine metabolism is a hot research topic.156 Local glutamine deficiency promotes tumor dedifferentiation by inhibiting histone demethylation.157 Glutamine controls ROS through GSH synthesis.49 Glutamine metabolism relies on the methionine cycle in one-carbon metabolism to exert its anti-ROS function. Compared with glutamine metabolism, methionine metabolism is less studied, and thus is worthy of further research. Meanwhile, targeting multiple metabolic pathways to suppress the tumor growth is the best treatment strategy.156
Conclusion
In conclusion, as an important part of one-carbon metabolism, methionine metabolism is closely related to diverse pathophysiological processes.19 Accumulating preclinical evidence indicates that alterations in the methionine cycle play a pathogenetic role in CLD.38 The switch of MAT1A to MAT2A/MAT2B reduces the levels of SAM, which is an essential factor in fibrosis and liver cancer.129 Preventing or even reversing this transformation will be the direction of future research. Controlling SAM levels precisely for liver injury is important, but SAM regulation is not well understood. A previous study showed that METTL16 is associated with SAM homeostasis.45 Future studies can focus on the relationship between methionine metabolism and m6A. In clinical research, whether methionine supplementation is necessary for CLD remains controversial. According to the existing research, methionine supplementation can be combined with basic clinical drugs; and this may have unexpected results. High-quality, prospective clinical trials are required to prove or refute the benefit of SAM supplementation.19 Moreover, SAM is transported across the intestinal epithelium by a strictly paracellular mechanism in the absence of membrane transporters. As a highly polar molecule, SAM is not likely to penetrate lipid membranes. Several studies have reported that both Caco-2 cells and hepatocytes exhibit very low uptake of SAM.158 In mammalian cells, transport of SAM appears to occur exclusively in brain endothelia, but not in non-pathological cells of the periphery.159 Exogenous SAM does not penetrate the plasma membrane, but equilibrates with a small sucrose-inaccessible compartment on the outer side of this membrane.160 These studies thus have indicated that in the treatment of CLD, the impact of SAM supplementation may not be direct. Studies have shown that exogenous SAM is utilized for phospholipid methylation on the outer surface of the plasma membrane.161 Exogenous SAM-mediated control of DNA methylation and gene expression could be a mechanism of the SAM anti-progression effect.162 The mechanisms of intestinal absorption and hepatic uptake of exogenously administered SAM, and the mechanism of its hepatoprotection remain unknown.158 SAM supplementation reduces CLD severity.40 This molecular mechanism is closely related to the role of SAM participating in methylation reactions to provide methyl groups, entering the transsulfuration pathway to metabolize and synthesize GSH, and participating in the one-carbon cycle. However, not many studies have investigated how exogenous SAM intervenes in intracellular metabolism, and additional studies are warranted.
Supplementary information
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (81870423 and 82073914), the Major Project of the Natural Science Research of Jiangsu Higher Education Institutions (19KJA310005), the Open Project of Jiangsu Key Laboratory for Pharmacology and Safety Evaluation of Chinese Materia Medica (JKLPSE202005, JKLPSE201815, and JKLPSE 201804), the Project of the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and the Postgraduate Research & Practice Innovation Program of Jiangsu Province (SJCX20_0536 and KYCX20_1493).
Author contributions
The construction of the main framework: Z.L., F.Z., and S.Z. Collection of references: B.L., Y.S., S.S., and S.X. References are summarized and organized: Z.L. and F.W. Literature analysis: Z.L., F.Z., Z.Z., J.S., and MH. Important revision of important intellectual content of the manuscript: Z.F., Z.L., and Z.S. Write manuscripts: Z.L. Funding: F.Z. and S.Z. All authors approved the final version of the manuscript.
Competing interests
The authors declare no competing interests.
Contributor Information
Feng Zhang, Email: 300631@njucm.edu.cn.
Shizhong Zheng, Email: szhengnucm@163.com.
Supplementary information
The online version of this article (10.1038/s41392-020-00349-7) contains supplementary material, which is available to authorized users.
References
- 1.Lemoinne S, Friedman SL. New and emerging anti-fibrotic therapeutics entering or already in clinical trials in chronic liver diseases. Curr. Opin. Pharm. 2019;49:60–70. doi: 10.1016/j.coph.2019.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Salarian M, et al. Early detection and staging of chronic liver diseases with a protein MRI contrast agent. Nat. Commun. 2019;10:4777. doi: 10.1038/s41467-019-11984-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Younossi ZM, Marchesini G, Pinto-Cortez H, Petta S. Epidemiology of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis: implications for liver transplantation. Transplantation. 2019;103:22–27. doi: 10.1097/TP.0000000000002484. [DOI] [PubMed] [Google Scholar]
- 4.Patten DA, Shetty S. Chronic liver disease: scavenger hunt for novel therapies. Lancet. 2018;391:104–105. doi: 10.1016/S0140-6736(17)32671-5. [DOI] [PubMed] [Google Scholar]
- 5.Vanni E, et al. Sites and mechanisms of insulin resistance in nonobese, nondiabetic patients with chronic hepatitis C. Hepatology. 2009;50:697–706. doi: 10.1002/hep.23031. [DOI] [PubMed] [Google Scholar]
- 6.Himoto, T. & Masaki, T. Associations between zinc deficiency and metabolic abnormalities in patients with chronic liver disease. Nutrients10, 88–105 (2018). [DOI] [PMC free article] [PubMed]
- 7.Clemente MG, Mandato C, Poeta M, Vajro P. Pediatric non-alcoholic fatty liver disease: recent solutions, unresolved issues, and future research directions. World J. Gastroenterol. 2016;22:8078–8093. doi: 10.3748/wjg.v22.i36.8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sahin M, et al. Changes in liver tissue trace element concentrations during hepatitis B viral infection treatment. Biol. Trace Elem. Res. 2019;188:245–250. doi: 10.1007/s12011-018-1414-y. [DOI] [PubMed] [Google Scholar]
- 9.Nangliya V, et al. Study of trace elements in liver cirrhosis patients and their role in prognosis of disease. Biol. Trace Elem. Res. 2015;165:35–40. doi: 10.1007/s12011-015-0237-3. [DOI] [PubMed] [Google Scholar]
- 10.Gatiatulina ER, et al. Evaluation of tissue metal and trace element content in a rat model of non-alcoholic fatty liver disease using ICP-DRC-MS. J. Trace Elem. Med. Biol. 2017;39:91–99. doi: 10.1016/j.jtemb.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 11.Yu L, et al. Lactobacillus plantarum CCFM639 alleviate trace element imbalance-related oxidative stress in liver and kidney of chronic aluminum exposure mice. Biol. Trace Elem. Res. 2017;176:342–349. doi: 10.1007/s12011-016-0843-8. [DOI] [PubMed] [Google Scholar]
- 12.Gong Z, Tas E, Yakar S, Muzumdar R. Hepatic lipid metabolism and non-alcoholic fatty liver disease in aging. Mol. Cell Endocrinol. 2017;455:115–130. doi: 10.1016/j.mce.2016.12.022. [DOI] [PubMed] [Google Scholar]
- 13.Gaggini, M. et al. Altered metabolic profile and adipocyte insulin resistance mark severe liver fibrosis in patients with chronic liver disease. Int. J. Mol. Sci.20, 6333–6347 (2019). [DOI] [PMC free article] [PubMed]
- 14.Morgan MY, Milsom JP, Sherlock S. Plasma ratio of valine, leucine and isoleucine to phenylalanine and tyrosine in liver disease. Gut. 1978;19:1068–1073. doi: 10.1136/gut.19.11.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaggini M, et al. Altered amino acid concentrations in NAFLD: impact of obesity and insulin resistance. Hepatology. 2018;67:145–158. doi: 10.1002/hep.29465. [DOI] [PubMed] [Google Scholar]
- 16.Morgan MY, Marshall AW, Milsom JP, Sherlock S. Plasma amino-acid patterns in liver disease. Gut. 1982;23:362–370. doi: 10.1136/gut.23.5.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mora SI, Garcia-Roman J, Gomez-Nanez I, Garcia-Roman R. Chronic liver diseases and the potential use of S-adenosyl-L-methionine as a hepatoprotector. Eur. J. Gastroenterol. Hepatol. 2018;30:893–900. doi: 10.1097/MEG.0000000000001141. [DOI] [PubMed] [Google Scholar]
- 18.Parkhitko AA, Jouandin P, Mohr SE, Perrimon N. Methionine metabolism and methyltransferases in the regulation of aging and lifespan extension across species. Aging Cell. 2019;18:e13034. doi: 10.1111/acel.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anstee QM, Day CP. S-adenosylmethionine (SAMe) therapy in liver disease: a review of current evidence and clinical utility. J. Hepatol. 2012;57:1097–1109. doi: 10.1016/j.jhep.2012.04.041. [DOI] [PubMed] [Google Scholar]
- 20.Wunsch E, et al. Effect of S-adenosyl-L-methionine on liver biochemistry and quality of life in patients with primary biliary cholangitis treated with ursodeoxycholic acid. A prospective, open label pilot study. J. Gastrointestin. Liver Dis. 2018;27:273–279. doi: 10.15403/jgld.2014.1121.273.icz. [DOI] [PubMed] [Google Scholar]
- 21.Morgan TR, et al. A phase II randomized, controlled trial of S-adenosylmethionine in reducing serum α-fetoprotein in patients with hepatitis C cirrhosis and elevated AFP. Cancer Prev. Res. (Philos.) 2015;8:864–872. doi: 10.1158/1940-6207.CAPR-15-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramani K, Donoyan S, Tomasi ML, Park S. Role of methionine adenosyltransferase alpha2 and beta phosphorylation and stabilization in human hepatic stellate cell trans-differentiation. J. Cell Physiol. 2015;230:1075–1085. doi: 10.1002/jcp.24839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kharbanda KK. Methionine metabolic pathway in alcoholic liver injury. Curr. Opin. Clin. Nutr. Metab. Care. 2013;16:89–95. doi: 10.1097/MCO.0b013e32835a892a. [DOI] [PubMed] [Google Scholar]
- 24.Froese DS, Fowler B, Baumgartner MR. Vitamin B12, folate, and the methionine remethylation cycle-biochemistry, pathways, and regulation. J. Inherit. Metab. Dis. 2019;42:673–685. doi: 10.1002/jimd.12009. [DOI] [PubMed] [Google Scholar]
- 25.Shen W, et al. Homocysteine-methionine cycle is a metabolic sensor system controlling methylation-regulated pathological signaling. Redox Biol. 2020;28:101322. doi: 10.1016/j.redox.2019.101322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez-Miguelsanz J, et al. Betaine homocysteine S-methyltransferase emerges as a new player of the nuclear methionine cycle. Biochim. Biophys. Acta Mol. Cell Res. 2017;1864:1165–1182. doi: 10.1016/j.bbamcr.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 27.Bae DH, Lane DJR, Jansson PJ, Richardson DR. The old and new biochemistry of polyamines. Biochim. Biophys. Acta Gen. Subj. 2018;1862:2053–2068. doi: 10.1016/j.bbagen.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 28.Pegg AE. Functions of polyamines in mammals. J. Biol. Chem. 2016;291:14904–14912. doi: 10.1074/jbc.R116.731661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minois N, Carmona-Gutierrez D, Madeo F. Polyamines in aging and disease. Aging. 2011;3:716–732. doi: 10.18632/aging.100361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ducker GS, Rabinowitz JD. One-carbon metabolism in health and disease. Cell Metab. 2017;25:27–42. doi: 10.1016/j.cmet.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mentch SJ, et al. Histone methylation dynamics and gene regulation occur through the sensing of one-carbon metabolism. Cell Metab. 2015;22:861–873. doi: 10.1016/j.cmet.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu, W. et al. One-carbon metabolism supports S-adenosylmethionine and histone methylation to drive inflammatory macrophages. Mol. cell75, 1147–1160 (2019). [DOI] [PubMed]
- 33.Bergman Y, Cedar H. DNA methylation dynamics in health and disease. Nat. Struct. Mol. Biol. 2013;20:274–281. doi: 10.1038/nsmb.2518. [DOI] [PubMed] [Google Scholar]
- 34.Bogdanović, O. & Lister, R. DNA methylation and the preservation of cell identity. Curr. Opin. Genet. Dev.46, 9–14 (2017). [DOI] [PubMed]
- 35.Lim JM, Kim G, Levine RL. Methionine in proteins: it’s not just for protein initiation anymore. Neurochem. Res. 2019;44:247–257. doi: 10.1007/s11064-017-2460-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Misiti F, Clementi ME, Giardina B. Oxidation of methionine 35 reduces toxicity of the amyloid beta-peptide(1-42) in neuroblastoma cells (IMR-32) via enzyme methionine sulfoxide reductase A expression and function. Neurochem. Int. 2010;56:597–602. doi: 10.1016/j.neuint.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 37.Luo S, Levine RL. Methionine in proteins defends against oxidative stress. FASEB J. 2009;23:464–472. doi: 10.1096/fj.08-118414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pascale, R. M., Peitta, G., Simile, M. M. & Feo, F. Alterations of methionine metabolism as potential targets for the prevention and therapy of hepatocellular carcinoma. Medicina55, 296–320 (2019). [DOI] [PMC free article] [PubMed]
- 39.Copeland DH, Salmon WD. The occurrence of neoplasms in the liver, lungs, and other tissues of rats as a result of prolonged choline deficiency. Am. J. Pathol. 1946;22:1059–1079. [PubMed] [Google Scholar]
- 40.Noureddin M, Sander-Struckmeier S, Mato JM. Early treatment efficacy of S-adenosylmethionine in patients with intrahepatic cholestasis: a systematic review. World J. Hepatol. 2020;12:46–63. doi: 10.4254/wjh.v12.i2.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henkel SA, et al. Expanding etiology of progressive familial intrahepatic cholestasis. World J. Hepatol. 2019;11:450–463. doi: 10.4254/wjh.v11.i5.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen M, Wong C-M. The emerging roles of N6-methyladenosine (m6A) deregulation in liver carcinogenesis. Mol. Cancer. 2020;19:44. doi: 10.1186/s12943-020-01172-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao G, Li H-B, Yin Z, Flavell RA. Recent advances in dynamic m6A RNA modification. Open Biol. 2016;6:160003. doi: 10.1098/rsob.160003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shima H, et al. S-adenosylmethionine synthesis is regulated by selective N(6)-adenosine methylation and mRNA degradation involving METTL16 and YTHDC1. Cell Rep. 2017;21:3354–3363. doi: 10.1016/j.celrep.2017.11.092. [DOI] [PubMed] [Google Scholar]
- 45.Pendleton KE, et al. The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169:824–835 e814. doi: 10.1016/j.cell.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meiser J, et al. Serine one-carbon catabolism with formate overflow. Sci. Adv. 2016;2:e1601273. doi: 10.1126/sciadv.1601273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat. Rev. Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hasan, T. et al. Disturbed homocysteine metabolism is associated with cancer. Exp. Mol. Med.51, 1–13 (2019). [DOI] [PMC free article] [PubMed]
- 49.Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat. Rev. Cancer. 2016;16:619–634. doi: 10.1038/nrc.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mavrakis KJ, et al. Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5. Science. 2016;351:1208–1213. doi: 10.1126/science.aad5944. [DOI] [PubMed] [Google Scholar]
- 51.Marjon K, et al. MTAP deletions in cancer create vulnerability to targeting of the MAT2A/PRMT5/RIOK1 axis. Cell Rep. 2016;15:574–587. doi: 10.1016/j.celrep.2016.03.043. [DOI] [PubMed] [Google Scholar]
- 52.Barbarino, M. et al. PRMT5 silencing selectively affects MTAP-deleted mesothelioma: in vitro evidence of a novel promising approach. J. Cell Mol. Med. 24, 5565–5577 (2020). [DOI] [PMC free article] [PubMed]
- 53.Sanderson SM, Mikhael PG, Ramesh V, Dai Z, Locasale JW. Nutrient availability shapes methionine metabolism in p16/-deleted cells. Sci. Adv. 2019;5:eaav7769. doi: 10.1126/sciadv.aav7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anderson, R. T. et al. Challenges, considerations, and principles to guide trials of combination therapies for chronic hepatitis B virus. Gastroenterology156, 529–533 (2019). [DOI] [PubMed]
- 55.Nguyen, M. H., Wong, G., Gane, E., Kao, J.-H. & Dusheiko, G. Hepatitis B virus: advances in prevention, diagnosis, and therapy. Clin. Microbiol. Rev.33, e00046-19 (2020). [DOI] [PMC free article] [PubMed]
- 56.Ganesan M, et al. Acetaldehyde accelerates HCV-induced impairment of innate immunity by suppressing methylation reactions in liver cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2015;309:G566–G577. doi: 10.1152/ajpgi.00183.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lozano-Sepulveda SA, et al. S-adenosyl-L-methionine modifies antioxidant-enzymes, glutathione-biosynthesis and methionine adenosyltransferases-1/2 in hepatitis C virus-expressing cells. World J. Gastroenterol. 2016;22:3746–3757. doi: 10.3748/wjg.v22.i14.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Duong FH, Christen V, Filipowicz M, Heim MH. S-Adenosylmethionine and betaine correct hepatitis C virus induced inhibition of interferon signaling in vitro. Hepatology. 2006;43:796–806. doi: 10.1002/hep.21116. [DOI] [PubMed] [Google Scholar]
- 59.Duong FHT, Filipowicz M, Tripodi M, La Monica N, Heim MH. Hepatitis C virus inhibits interferon signaling through up-regulation of protein phosphatase 2A. Gastroenterology. 2004;126:263–277. doi: 10.1053/j.gastro.2003.10.076. [DOI] [PubMed] [Google Scholar]
- 60.Feld JJ, et al. S-adenosyl methionine improves early viral responses and interferon-stimulated gene induction in hepatitis C nonresponders. Gastroenterology. 2011;140:830–839. doi: 10.1053/j.gastro.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu T, et al. Mechanisms of MAFG dysregulation in cholestatic liver injury and development of liver cancer. Gastroenterology. 2018;155:557–571 e514. doi: 10.1053/j.gastro.2018.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ganesan M, et al. FAT10 suppression stabilizes oxidized proteins in liver cells: Effects of HCV and ethanol. Exp. Mol. Pathol. 2015;99:506–516. doi: 10.1016/j.yexmp.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 63.Tsikas D, Hanff E, Bollenbach A. -Adenosyl-L-methionine towards hepatitis C virus expression: need to consider -Adenosyl-L-methionine’s chemistry, physiology and pharmacokinetics. World J. Gastroenterol. 2017;23:7343–7346. doi: 10.3748/wjg.v23.i40.7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Flecken, T. et al. Mapping the heterogeneity of histone modifications on hepatitis B virus dna using liver needle biopsies obtained from chronically infected patients. J. Virol.93, e02036-18 (2019). [DOI] [PMC free article] [PubMed]
- 65.Liu Q, et al. The X protein of hepatitis B virus inhibits apoptosis in hepatoma cells through enhancing the methionine adenosyltransferase 2A gene expression and reducing S-adenosylmethionine production. J. Biol. Chem. 2011;286:17168–17180. doi: 10.1074/jbc.M110.167783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guo T, He Y, Ma W, Liu Z, Liu Q. Feasibility and efficacy of S-adenosyl-L-methionine in patients with HBV-related HCC with different BCLC stages. Gastroenterol. Res Pract. 2016;2016:4134053. doi: 10.1155/2016/4134053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li J, et al. Inhibition of STAT1 methylation is involved in the resistance of hepatitis B virus to Interferon alpha. Antivir. Res. 2010;85:463–469. doi: 10.1016/j.antiviral.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 68.Bertoletti A, Tan AT, Koh S. T-cell therapy for chronic viral hepatitis. Cytotherapy. 2017;19:1317–1324. doi: 10.1016/j.jcyt.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 69.Sinclair, L. V. et al. Antigen receptor control of methionine metabolism in T cells. Elife8, e44210 (2019). [DOI] [PMC free article] [PubMed]
- 70.Bing Y, et al. Glucocorticoid-induced S-adenosylmethionine enhances the interferon signaling pathway by restoring STAT1 protein methylation in hepatitis B virus-infected cells. J. Biol. Chem. 2014;289:32639–32655. doi: 10.1074/jbc.M114.589689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun S, et al. A comprehensive genome-wide profiling comparison between HBV and HCV infected hepatocellular carcinoma. BMC Med. Genomics. 2019;12:147. doi: 10.1186/s12920-019-0580-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Osna NA, et al. Prolonged feeding with guanidinoacetate, a methyl group consumer, exacerbates ethanol-induced liver injury. World J. Gastroenterol. 2016;22:8497–8508. doi: 10.3748/wjg.v22.i38.8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Halsted CH, Medici V. Vitamin-dependent methionine metabolism and alcoholic liver disease. Adv. Nutr. 2011;2:421–427. doi: 10.3945/an.111.000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Watson WH, Burke TJ, Doll MA, McClain CJ. S-adenosylhomocysteine inhibits NF-kappaB-mediated gene expression in hepatocytes and confers sensitivity to TNF cytotoxicity. Alcohol Clin. Exp. Res. 2014;38:889–896. doi: 10.1111/acer.12315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ganesan M, et al. Creatine supplementation does not prevent the development of alcoholic steatosis. Alcohol. Clin. Exp. Res. 2016;40:2312–2319. doi: 10.1111/acer.13214. [DOI] [PubMed] [Google Scholar]
- 76.Chen Y, et al. Glutathione and transsulfuration in alcohol-associated tissue injury and carcinogenesis. Adv. Exp. Med. Biol. 2018;1032:37–53. doi: 10.1007/978-3-319-98788-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Auta J, Zhang H, Pandey SC, Guidotti A. Chronic alcohol exposure differentially alters one-carbon metabolism in rat liver and brain. Alcohol Clin. Exp. Res. 2017;41:1105–1111. doi: 10.1111/acer.13382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Halsted CH. B-Vitamin dependent methionine metabolism and alcoholic liver disease. Clin. Chem. Lab Med. 2013;51:457–465. doi: 10.1515/cclm-2012-0308. [DOI] [PubMed] [Google Scholar]
- 79.Barve S, et al. Interactions of cytokines, S-adenosylmethionine, and S-adenosylhomocysteine in alcohol-induced liver disease and immune suppression. J. Gastroenterol. Hepatol. 2006;21:S38–S42. doi: 10.1111/j.1440-1746.2006.04590.x. [DOI] [PubMed] [Google Scholar]
- 80.Hao F, et al. Inhibition of Caspase-8 does not protect from alcohol-induced liver apoptosis but alleviates alcoholic hepatic steatosis in mice. Cell Death Dis. 2017;8:e3152. doi: 10.1038/cddis.2017.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Murray B, et al. Methionine adenosyltransferase alpha1 is targeted to the mitochondrial matrix and interacts with cytochrome P450 2E1 to lower its expression. Hepatology. 2019;70:2018–2034. doi: 10.1002/hep.30762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Choi, S. et al. Methyl-sensing nuclear receptor liver receptor homolog-1 regulates mitochondrial function in mouse hepatocytes. Hepatology71, 1055–1069 (2019). [DOI] [PMC free article] [PubMed]
- 83.Ye C, Sutter BM, Wang Y, Kuang Z, Tu BP. A metabolic function for phospholipid and histone methylation. Mol. Cell. 2017;66:180–193 e188. doi: 10.1016/j.molcel.2017.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang Z, et al. l-Methionine activates Nrf2-ARE pathway to induce endogenous antioxidant activity for depressing ROS-derived oxidative stress in growing rats. J. Sci. Food Agric. 2019;99:4849–4862. doi: 10.1002/jsfa.9757. [DOI] [PubMed] [Google Scholar]
- 85.King AL, et al. The methyl donor S-adenosylmethionine prevents liver hypoxia and dysregulation of mitochondrial bioenergetic function in a rat model of alcohol-induced fatty liver disease. Redox Biol. 2016;9:188–197. doi: 10.1016/j.redox.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bingül İ, et al. Betaine treatment decreased oxidative stress, inflammation, and stellate cell activation in rats with alcoholic liver fibrosis. Environ. Toxicol. Pharm. 2016;45:170–178. doi: 10.1016/j.etap.2016.05.033. [DOI] [PubMed] [Google Scholar]
- 87.Vadigepalli R, Hoek JB. Introduction to the virtual issue alcohol and epigenetic regulation: do the products of alcohol metabolism drive epigenetic control of gene expression in alcohol-related disorders? Alcohol. Clin. Exp. Res. 2018;42:845–848. doi: 10.1111/acer.13630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang W, et al. Betaine attenuates chronic alcohol‑induced fatty liver by broadly regulating hepatic lipid metabolism. Mol. Med. Rep. 2017;16:5225–5234. doi: 10.3892/mmr.2017.7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ghorbani Z, Hajizadeh M, Hekmatdoost A. Dietary supplementation in patients with alcoholic liver disease: a review on current evidence. Hepatobiliary Pancreat. Dis. Int. 2016;15:348–360. doi: 10.1016/S1499-3872(16)60096-6. [DOI] [PubMed] [Google Scholar]
- 90.Singh S, Osna NA, Kharbanda KK. Treatment options for alcoholic and non-alcoholic fatty liver disease: a review. World J. Gastroenterol. 2017;23:6549–6570. doi: 10.3748/wjg.v23.i36.6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tkachenko P, et al. Prednisolone plus S-adenosil-L-methionine in severe alcoholic hepatitis. Hepatol. Int. 2016;10:983–987. doi: 10.1007/s12072-016-9751-4. [DOI] [PubMed] [Google Scholar]
- 92.Alonso C, et al. Metabolomic identification of subtypes of nonalcoholic steatohepatitis. Gastroenterology. 2017;152:1449–1461 e1447. doi: 10.1053/j.gastro.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tanaka N, et al. Role of fibroblast growth factor 21 in the early stage of NASH induced by methionine- and choline-deficient diet. Biochim. Biophys. Acta. 2015;1852:1242–1252. doi: 10.1016/j.bbadis.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim SH, et al. Ezetimibe ameliorates steatohepatitis via AMP activated protein kinase-TFEB-mediated activation of autophagy and NLRP3 inflammasome inhibition. Autophagy. 2017;13:1767–1781. doi: 10.1080/15548627.2017.1356977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kalhan SC, et al. Methionine and protein metabolism in non-alcoholic steatohepatitis: evidence for lower rate of transmethylation of methionine. Clin. Sci. 2011;121:179–189. doi: 10.1042/CS20110060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Frau M, Feo F, Pascale RM. Pleiotropic effects of methionine adenosyltransferases deregulation as determinants of liver cancer progression and prognosis. J. Hepatol. 2013;59:830–841. doi: 10.1016/j.jhep.2013.04.031. [DOI] [PubMed] [Google Scholar]
- 97.Luo J, Li Y-N, Wang F, Zhang W-M, Geng X. S-adenosylmethionine inhibits the growth of cancer cells by reversing the hypomethylation status of c-myc and H-ras in human gastric cancer and colon cancer. Int. J. Biol. Sci. 2010;6:784–795. doi: 10.7150/ijbs.6.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mridha AR, et al. NLRP3 inflammasome blockade reduces liver inflammation and fibrosis in experimental NASH in mice. J. Hepatol. 2017;66:1037–1046. doi: 10.1016/j.jhep.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yao L, et al. Hyperhomocysteinemia activates the aryl hydrocarbon receptor/CD36 pathway to promote hepatic steatosis in mice. Hepatology. 2016;64:92–105. doi: 10.1002/hep.28518. [DOI] [PubMed] [Google Scholar]
- 100.Pastore A, et al. Plasma levels of homocysteine and cysteine increased in pediatric NAFLD and strongly correlated with severity of liver damage. Int. J. Mol. Sci. 2014;15:21202–21214. doi: 10.3390/ijms151121202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lai Z, et al. Association of hepatic global DNA methylation and serum one-carbon metabolites with histological severity in patients with NAFLD. Obesity (Silver Spring) 2020;28:197–205. doi: 10.1002/oby.22667. [DOI] [PubMed] [Google Scholar]
- 102.Xu Y, Guan Y, Yang X, Xia Z, Wu J. Association of serum homocysteine levels with histological severity of NAFLD. J. Gastrointest. Liver Dis. 2020;29:51–58. doi: 10.15403/jgld-529. [DOI] [PubMed] [Google Scholar]
- 103.Maria Del Bas J, et al. Hepatic accumulation of S-adenosylmethionine in hamsters with non-alcoholic fatty liver disease associated with metabolic syndrome under selenium and vitamin E deficiency. Clin. Sci. 2019;133:409–423. doi: 10.1042/CS20171039. [DOI] [PubMed] [Google Scholar]
- 104.Kumar A, et al. High levels of dietary methionine improves sitagliptin-induced hepatotoxicity by attenuating oxidative stress in hypercholesterolemic rats. Nutr. Metab. 2020;17:2. doi: 10.1186/s12986-019-0422-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ai, Y. et al. Homocysteine induces hepatic steatosis involving ER stress response in high methionine diet-fed mice. Nutrients9, 346–356 (2017). [DOI] [PMC free article] [PubMed]
- 106.Forney LA, et al. Sexually dimorphic effects of dietary methionine restriction are dependent on age when the diet is introduced. Obesity. 2020;28:581–589. doi: 10.1002/oby.22721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Stone KP, Wanders D, Orgeron M, Cortez CC, Gettys TW. Mechanisms of increased in vivo insulin sensitivity by dietary methionine restriction in mice. Diabetes. 2014;63:3721–3733. doi: 10.2337/db14-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hasek BE, et al. Dietary methionine restriction enhances metabolic flexibility and increases uncoupled respiration in both fed and fasted states. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010;299:R728–R739. doi: 10.1152/ajpregu.00837.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stender S, et al. Adult-onset liver disease and hepatocellular carcinoma in S-adenosylhomocysteine hydrolase deficiency. Mol. Genet. Metab. 2015;116:269–274. doi: 10.1016/j.ymgme.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gao X, et al. Dietary methionine influences therapy in mouse cancer models and alters human metabolism. Nature. 2019;572:397–401. doi: 10.1038/s41586-019-1437-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Du, K. et al. Hedgehog-YAP signaling pathway regulates glutaminolysis to control activation of hepatic stellate cells. Gastroenterology154, 1465–1489 (2018). [DOI] [PMC free article] [PubMed]
- 112.Lackner C, Tiniakos D. Fibrosis and alcohol-related liver disease. J. Hepatol. 2019;70:294–304. doi: 10.1016/j.jhep.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 113.Ramani K, et al. Changes in the expression of methionine adenosyltransferase genes and S-adenosylmethionine homeostasis during hepatic stellate cell activation. Hepatology. 2010;51:986–995. doi: 10.1002/hep.23411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ramani K, Tomasi ML. Transcriptional regulation of methionine adenosyltransferase 2A by peroxisome proliferator-activated receptors in rat hepatic stellate cells. Hepatology. 2012;55:1942–1953. doi: 10.1002/hep.25594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cao Q, et al. Leptin suppresses microRNA-122 promoter activity by phosphorylation of foxO1 in hepatic stellate cell contributing to leptin promotion of mouse liver fibrosis. Toxicol. Appl Pharm. 2018;339:143–150. doi: 10.1016/j.taap.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 116.Ramani K, et al. Leptin’s mitogenic effect in human liver cancer cells requires induction of both methionine adenosyltransferase 2A and 2beta. Hepatology. 2008;47:521–531. doi: 10.1002/hep.22064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cheng, F. et al. Leptin promotes methionine adenosyltransferase 2A expression in hepatic stellate cells by the downregulation of E2F-4 via the beta-catenin pathway. FASEB J. 34, 5578–5589 (2020). [DOI] [PubMed]
- 118.Martinez-Chantar ML, et al. Methionine adenosyltransferase II beta subunit gene expression provides a proliferative advantage in human hepatoma. Gastroenterology. 2003;124:940–948. doi: 10.1053/gast.2003.50151. [DOI] [PubMed] [Google Scholar]
- 119.Karaa A, Thompson KJ, McKillop IH, Clemens MG, Schrum LW. S-adenosyl-L-methionine attenuates oxidative stress and hepatic stellate cell activation in an ethanol-LPS-induced fibrotic rat model. Shock. 2008;30:197–205. doi: 10.1097/shk.0b013e318160f417. [DOI] [PubMed] [Google Scholar]
- 120.Thompson KJ, et al. S-adenosyl-L-methionine inhibits collagen secretion in hepatic stellate cells via increased ubiquitination. Liver Int. 2011;31:891–901. doi: 10.1111/j.1478-3231.2011.02512.x. [DOI] [PubMed] [Google Scholar]
- 121.Matsui H, Kawada N. Effect of S-adenosyl-L-methionine on the activation, proliferation and contraction of hepatic stellate cells. Eur. J. Pharm. 2005;509:31–36. doi: 10.1016/j.ejphar.2004.12.041. [DOI] [PubMed] [Google Scholar]
- 122.Albrecht LV, Bui MH, De, Robertis EM. Canonical Wnt is inhibited by targeting one-carbon metabolism through methotrexate or methionine deprivation. Proc. Natl Acad. Sci. USA. 2019;116:2987–2995. doi: 10.1073/pnas.1820161116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhou J-Y, et al. Methionine and its hydroxyl analogues improve stem cell activity to eliminate deoxynivalenol-induced intestinal injury by reactivating Wnt/β-catenin signaling. J. Agric. Food Chem. 2019;67:11464–11473. doi: 10.1021/acs.jafc.9b04442. [DOI] [PubMed] [Google Scholar]
- 124.Nishikawa, K., Osawa, Y. & Kimura, K. Wnt/β-catenin signaling as a potential target for the treatment of liver cirrhosis using antifibrotic drugs. Int. J. Mol. Sci.19, 3103–3115 (2018). [DOI] [PMC free article] [PubMed]
- 125.Wang K, et al. TGF-β1/p65/MAT2A pathway regulates liver fibrogenesis via intracellular SAM. EBioMedicine. 2019;42:458–469. doi: 10.1016/j.ebiom.2019.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lafoz, E., Ruart, M., Anton, A., Oncins, A. & Hernandez-Gea, V. The endothelium as a driver of liver fibrosis and regeneration. Cells9, 929–955 (2020). [DOI] [PMC free article] [PubMed]
- 127.Yang JD, et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019;16:589–604. doi: 10.1038/s41575-019-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ho S-Y, et al. Evolution of etiology, presentation, management and prognostic tool in hepatocellular carcinoma. Sci. Rep. 2020;10:3925. doi: 10.1038/s41598-020-61028-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Murray B, Barbier-Torres L, Fan W, Mato JM, Lu SC. Methionine adenosyltransferases in liver cancer. World J. Gastroenterol. 2019;25:4300–4319. doi: 10.3748/wjg.v25.i31.4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lu SC, Mato JM, Espinosa-Diez C, Lamas S. MicroRNA-mediated regulation of glutathione and methionine metabolism and its relevance for liver disease. Free Radic. Biol. Med. 2016;100:66–72. doi: 10.1016/j.freeradbiomed.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lozano-Rosas MG, et al. Diminished S-adenosylmethionine biosynthesis and its metabolism in a model of hepatocellular carcinoma is recuperated by an adenosine derivative. Cancer Biol. Ther. 2020;21:81–94. doi: 10.1080/15384047.2019.1665954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yang HB, et al. Acetylation of MAT IIalpha represses tumour cell growth and is decreased in human hepatocellular cancer. Nat. Commun. 2015;6:6973. doi: 10.1038/ncomms7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liu Q, et al. Hypoxia induces genomic DNA demethylation through the activation of HIF-1alpha and transcriptional upregulation of MAT2A in hepatoma cells. Mol. Cancer Ther. 2011;10:1113–1123. doi: 10.1158/1535-7163.MCT-10-1010. [DOI] [PubMed] [Google Scholar]
- 134.Li, Y. et al. Reciprocal regulation between forkhead Box M1/NF-kB and methionine adenosyltransferase 1A drives liver cancer. Hepatology (2020). 10.1002/hep.31196. [DOI] [PMC free article] [PubMed]
- 135.Fan W, et al. Prohibitin 1 suppresses liver cancer tumorigenesis in mice and human hepatocellular and cholangiocarcinoma cells. Hepatology. 2017;65:1249–1266. doi: 10.1002/hep.28964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wu L, et al. MAT2B mediates invasion and metastasis by regulating EGFR signaling pathway in hepatocellular carcinoma. Clin. Exp. Med. 2019;19:535–546. doi: 10.1007/s10238-019-00579-2. [DOI] [PubMed] [Google Scholar]
- 137.Yang H, et al. Methionine adenosyltransferase 2B, HuR, and sirtuin 1 protein cross-talk impacts on the effect of resveratrol on apoptosis and growth in liver cancer cells. J. Biol. Chem. 2013;288:23161–23170. doi: 10.1074/jbc.M113.487157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hughey CC, et al. Glycine -methyltransferase deletion in mice diverts carbon flux from gluconeogenesis to pathways that utilize excess methionine cycle intermediates. J. Biol. Chem. 2018;293:11944–11954. doi: 10.1074/jbc.RA118.002568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Carril E, et al. Metabolic impact of partial hepatectomy in the non-alcoholic steatohepatitis animal model of methionine-choline deficient diet. J. Pharm. Biomed. Anal. 2020;178:112958. doi: 10.1016/j.jpba.2019.112958. [DOI] [PubMed] [Google Scholar]
- 140.Ye J-Z, et al. Dynamic alterations in the gut microbiota and metabolome during the development of methionine-choline-deficient diet-induced nonalcoholic steatohepatitis. World J. Gastroenterol. 2018;24:2468–2481. doi: 10.3748/wjg.v24.i23.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Toriguchi K, et al. Attenuation of steatohepatitis, fibrosis, and carcinogenesis in mice fed a methionine-choline deficient diet by CCAAT/enhancer-binding protein homologous protein deficiency. J. Gastroenterol. Hepatol. 2014;29:1109–1118. doi: 10.1111/jgh.12481. [DOI] [PubMed] [Google Scholar]
- 142.Rajesh, Y. & Sarkar, D. Molecular mechanisms regulating obesity-associated hepatocellular carcinoma. Cancers12, 1290 (2020). [DOI] [PMC free article] [PubMed]
- 143.Liu Z-Y, et al. Serum choline is associated with hepatocellular carcinoma survival: a prospective cohort study. Nutr. Metab. 2020;17:25. doi: 10.1186/s12986-020-00445-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jiang X, Greenwald E, Jack-Roberts C. Effects of choline on DNA methylation and macronutrient metabolic gene expression in in vitro models of hyperglycemia. Nutr. Metab. Insights. 2016;9:11–17. doi: 10.4137/NMI.S29465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Liu, Z.-Y. et al. Dietary choline, rather than betaine intake, is associated with hepatocellular carcinoma mortality. Food Funct.11, 7866–7877 (2020). [DOI] [PubMed]
- 146.Jin M, et al. Dietary choline supplementation attenuated high-fat diet-induced inflammation through regulation of lipid metabolism and suppression of NFκB activation in juvenile black seabream (Acanthopagrus schlegelii) J. Nutr. Sci. 2019;8:e38. doi: 10.1017/jns.2019.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gougelet A, et al. Hepatocellular carcinomas with mutational activation of beta-catenin require choline and can be detected by positron emission tomography. Gastroenterology. 2019;157:807–822. doi: 10.1053/j.gastro.2019.05.069. [DOI] [PubMed] [Google Scholar]
- 148.Mladenović D, Radosavljević T, Hrnčić D, Rasic-Markovic A, Stanojlović O. The effects of dietary methionine restriction on the function and metabolic reprogramming in the liver and brain - implications for longevity. Rev. Neurosci. 2019;30:581–593. doi: 10.1515/revneuro-2018-0073. [DOI] [PubMed] [Google Scholar]
- 149.Olsen T, et al. Effects of dietary methionine and cysteine restriction on plasma biomarkers, serum fibroblast growth factor 21, and adipose tissue gene expression in women with overweight or obesity: a double-blind randomized controlled pilot study. J. Transl. Med. 2020;18:122. doi: 10.1186/s12967-020-02288-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Xu Q, et al. HNF4α regulates sulfur amino acid metabolism and confers sensitivity to methionine restriction in liver cancer. Nat. Commun. 2020;11:3978. doi: 10.1038/s41467-020-17818-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kitada M, Ogura Y, Monno I, Koya D. The impact of dietary protein intake on longevity and metabolic health. EBioMedicine. 2019;43:632–640. doi: 10.1016/j.ebiom.2019.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pascale RM, Feo CF, Calvisi DF, Feo F. Deregulation of methionine metabolism as determinant of progression and prognosis of hepatocellular carcinoma. Transl. Gastroenterol. Hepatol. 2018;3:36. doi: 10.21037/tgh.2018.06.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Vazquez-Chantada M, et al. HuR/methyl-HuR and AUF1 regulate the MAT expressed during liver proliferation, differentiation, and carcinogenesis. Gastroenterology. 2010;138:1943–1953. doi: 10.1053/j.gastro.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang Z, et al. Methionine is a metabolic dependency of tumor-initiating cells. Nat. Med. 2019;25:825–837. doi: 10.1038/s41591-019-0423-5. [DOI] [PubMed] [Google Scholar]
- 155.Martinez Y, et al. The role of methionine on metabolism, oxidative stress, and diseases. Amino Acids. 2017;49:2091–2098. doi: 10.1007/s00726-017-2494-2. [DOI] [PubMed] [Google Scholar]
- 156.Li T, Le A. Glutamine metabolism in cancer. Adv. Exp. Med. Biol. 2018;1063:13–32. doi: 10.1007/978-3-319-77736-8_2. [DOI] [PubMed] [Google Scholar]
- 157.Pan M, et al. Regional glutamine deficiency in tumours promotes dedifferentiation through inhibition of histone demethylation. Nat. Cell Biol. 2016;18:1090–1101. doi: 10.1038/ncb3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.McMillan JM, Walle UK, Walle T. S-adenosyl-L-methionine: transcellular transport and uptake by Caco-2 cells and hepatocytes. J. Pharm. Pharm. 2005;57:599–605. doi: 10.1211/0022357056082. [DOI] [PubMed] [Google Scholar]
- 159.Chishty M, Reichel A, Abbott NJ, Begley DJ. S-adenosylmethionine is substrate for carrier mediated transport at the blood-brain barrier in vitro. Brain Res. 2002;942:46–50. doi: 10.1016/S0006-8993(02)02654-9. [DOI] [PubMed] [Google Scholar]
- 160.Van Phi L, Söling HD. Methyl group transfer from exogenous S-adenosylmethionine on to plasma-membrane phospholipids without cellular uptake in isolated hepatocytes. Biochem. J. 1982;206:481–487. doi: 10.1042/bj2060481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bontemps F, Van Den Berghe G. Metabolism of exogenous S-adenosylmethionine in isolated rat hepatocyte suspensions: methylation of plasma-membrane phospholipids without intracellular uptake. Biochem. J. 1997;327:383–389. doi: 10.1042/bj3270383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Garcea R, et al. Protooncogene methylation and expression in regenerating liver and preneoplastic liver nodules induced in the rat by diethylnitrosamine: effect of variations of S-adenosylmethionine:S-adenosylhomocysteine ratio. Carcinogenesis. 1989;10:1183–1192. doi: 10.1093/carcin/10.7.1183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


