Abstract
Histoplasmosis is a serious infectious disease in humans caused by Histoplasma spp. (Onygenales), whose natural reservoirs are thought to be soil enriched with bird and bat guano. The true global burden of histoplasmosis is underestimated and frequently the pulmonary manifestations are misdiagnosed as tuberculosis. Molecular data on epidemiology of Histoplasma are still scarce, even though there is increasing recognition of histoplasmosis in recent years in areas distant from the traditional endemic regions in the Americas. We used multi-locus sequence data from protein coding loci (ADP-ribosylation factor, H antigen precursor, and delta-9 fatty acid desaturase), DNA barcoding (ITS1/2+5.8s), AFLP markers and mating type analysis to determine the genetic diversity, population structure and recognise the existence of different phylogenetic species among 436 isolates of Histoplasma obtained globally. Our study describes new phylogenetic species and the molecular characteristics of Histoplasma lineages causing outbreaks with a high number of severe outcomes in Northeast Brazil between 2011 and 2015. Genetic diversity levels provide evidence for recombination, common ancestry and clustering of Brazilian isolates at different geographic scales with the emergence of LAm C, a new genotype assigned to a separate population cluster in Northeast Brazil that exhibited low diversity indicative of isolation. The global survey revealed that the high genetic variability among Brazilian isolates along with the presence of divergent cryptic species and/or genotypes may support the hypothesis of Brazil being the center of dispersion of Histoplasma in South America, possibly with the contribution of migratory hosts such as birds and bats. Outside Brazil, the predominant species depends on the region. We confirm that histoplasmosis has significantly broadened its area of occurrence, an important feature of emerging pathogens. From a practical point of view, our data point to the emergence of histoplasmosis caused by a plethora of genotypes, and will enable epidemiological analysis focused on understanding the processes that lead to histoplasmosis. Further, the description of this diversity opens avenues for comparative genomic studies, which will allow progress toward a consensus taxonomy, improve understanding of the presence of hybrids in natural populations of medically relevant fungi, test reproductive barriers and to explore the significance of this variation.
Key words: Emerging pathogens, Epidemiology, Genetic diversity, Histoplasma capsulatum, Histoplasmosis, Population structure
Introduction
Histoplasmosis is a life-threatening systemic infection caused by the fungal pathogen Histoplasma capsulatum. This pathogen was first described in Panama in 1906 by Samuel T. Darling (Darling 1906), followed shortly thereafter by reported infections throughout the Americas (Riley and Watson, 1926, Negroni, 1940, Furcow, 1958) and later in widely scattered places around the world (Rocher, 1950, Ashbee et al., 2008, Antinori, 2014, Baker et al., 2019). The first epidemiological investigations in the mid-1940s revealed that Darling’s disease was more frequent than previously thought (Christie & Peterson 1945). There is an increasing awareness of histoplasmosis in Central and South America in patients with HIV/AIDS, and deaths from histoplasmosis in this region may outnumber deaths from tuberculosis (Pasqualotto and Queiroz-Telles, 2018, Linder and Kauffman, 2019, Nacher et al., 2019). Currently, histoplasmosis is one of the world’s leading systemic infections, with incidence ranging from 0.1 to 1 case per 100 000 inhabitants per year in temperate climates, 10 to 100 cases per 100 000 in the humid tropics, and over 100 cases per 100 000 in high risk groups and during outbreaks (Colombo et al., 2011, Adenis et al., 2018). Although most infections are not outbreak-associated, outbreaks of histoplasmosis linked to a common source do occasionally occur and often involve activities that disrupt soil, especially samples that contains bird or bat droppings (Queiroz-Telles et al. 2017).
Mammals become infected after inhaling Histoplasma propagules from the environment (Deepe 2018), which leads, in the vast majority of human cases, to a primary pulmonary infection that is unapparent, subclinical or completely benign (Eissenberg & Goldman 1991). The remaining patients may develop chronic progressive lung disease, chronic cutaneous or systemic disease, or an acute fulminating, rapidly fatal, systemic infection (Kauffman, 2007, Oladele et al., 2018). The infection is also very common in wild (e.g., marsupials, rodents, armadillos, sloths, bats) and domestic animals (e.g., dogs and cats) in endemic areas (Emmons, 1950, Seyedmousavi et al., 2018). However, mammals appear to be dead-end hosts of Histoplasma since there is no person-to-person or animal-to-person spread.
For over a century histoplasmosis was attributed to a single etiological agent, H. capsulatum (Onygenales). Judging from different clinical manifestations, morphologies and wide-ranging geographical occurrence, three different varieties are historically recognised: (i) H. capsulatum var. capsulatum, which is a broadly dispersed New World human pathogen, responsible for classic histoplasmosis (Eissenberg & Goldman 1991); (ii) H. capsulatum var. duboisii, a human pathogen confined to Central and West Africa, causing African histoplasmosis, whose principal features are skin or bone lesions (Loulergue et al., 2007, Pakasa et al., 2018); and (iii) H. capsulatum var. farciminosum, an Old World animal pathogen, responsible for epizootic lymphangitis in equines (Selim et al. 1985).
Previous studies exploring phylogeography, gene flow and hybridisation processes have shown that the causal agents of histoplasmosis encompass several cryptic species, which are structured according to their geographical origin (Van Dyke et al. 2019). Kasuga and colleagues studied 137 isolates recovered from 25 countries on six continents and were able to recognise at least eight clades within seven phylogenetic species, including: (i) North American class 1 clade (NAm 1); (ii) North American class 2 clade (NAm 2); (iii) Latin American group A clade (LAm A); (iv) Latin American group B clade (LAm B); (v) Australian clade; (vi) Netherlands (Indonesian?) clade; (vii) Eurasian clade and (viii) African clade (Kasuga et al., 1999, Kasuga et al., 2003). Recently, based on the genealogic concordance for phylogenetic species recognition (GCPSR) and population structures methods, six new phylogenetic species were proposed: LAm A1; LAm A2; RJ (which corresponds to the LAm A clade described by Kasuga et al., 2003); LAm B1; LAm B2 (corresponding to LAm B clade, (Kasuga et al. 2003); and BAC1 (a bat-associated species-specific clade) (Teixeira et al. 2016).
A more robust and comprehensive taxonomic scenario was recently proposed for Histoplasma based on whole-genome data from 30 isolates embedded in five of the groups proposed by Kasuga et al. (Kasuga et al. 2003), namely the NAm 1, NAm 2, LAm A, Panama/H81 lineage, and Africa groups. Sepúlveda and colleagues demonstrated the presence of independently evolving lineages, which were elevated to species status and named H. capsulatum sensu stricto (s. str.) when referring to the Panamanian lineage (H81 lineage), Histoplasma mississippiense sp. nov. (formerly known as NAm 1), Histoplasma ohiense sp. nov. (formerly known as NAm 2) and Histoplasma suramericanum sp. nov. (formerly known as LAm A) (Sepúlveda et al. 2017).
Although Histoplasma occurs globally, it appears that the phylogenetic species are not evenly distributed. For example, infections due to H. mississippiense and H. ohiense are more common in the USA, where they occur in syntopy. Also, H. capsulatum s. str. and H. suramericanum coexist in sympatry, since their geographical distributions overlap in central and possibly northwestern Colombia (Sepúlveda et al., 2017, Gomez et al., 2019, Sahaza et al., 2019). Moreover, H. capsulatum var. duboisii, the agent of African histoplasmosis is found predominantly on continental Africa and Madagascar and is argued to be a separate species (Sepúlveda et al., 2017, Valero et al., 2018, Van Dyke et al., 2019). Along with geographical differences, there is differential virulence among phylogenetically related species of Histoplasma (Sepúlveda et al. 2014). Whether these differences are responsible for differences in clinical presentations needs to be confirmed in a large group of patients. Previous studies indicate that H. suramericanum causes an acute pulmonary disease with a pronounced lung pathology that leads to high mortality, whereas H. mississippiense and H. ohiense cause a chronic pulmonary disease (Durkin et al., 2004, Sepúlveda et al., 2017). H. capsulatum var. duboisii was recently associated with AIDS and disseminated disease and to a lesser extent with skin lesions in immunocompetent persons (Oladele et al., 2018, Valero et al., 2018).
Several studies at the molecular level have been conducted to elucidate the genetic diversity and population structure of Histoplasma using molecular markers, ranging from DNA sequencing to DNA fingerprinting (Damasceno et al., 2016, Sahaza et al., 2019). In Brazil, high genetic diversity correlates with the geographical origin (Zancope-Oliveira et al., 2005, Muniz Mde et al., 2010, Almeida et al., 2019). However, the origin of the Brazilian Histoplasma populations and their relationship with the global populations remain unknown, especially considering the recently proposed taxonomic changes. Hence, the aims of the present study were to understand the genetic diversity of Histoplasma isolates in Brazil and explore their evolutionary relationships with H. capsulatum s. str., H. capsulatum var. duboisii, H. suramericanum, H. mississippiense and H. ohiense. We used multi-locus sequence data from protein coding loci (ADP-ribosylation factor, H antigen precursor, and delta-9 fatty acid desaturase), DNA barcoding (ITS1/2+5.8s), AFLP markers and mating type analysis to determine the genetic diversity, population structure and recognise the existence of different phylogenetic species of Histoplasma isolates obtained from Northeast and Southeast Brazil. Our data included a wider global panel of isolates leading us to hypothesise that the high genetic variability among Brazilian isolates along with the presence of divergent cryptic species and/or genotypes support the idea that Brazil is the center of dispersion of Histoplasma in South America, possibly with the contribution of migratory hosts such as birds and bats.
Material and Methods
Fungal strains
Isolates were of clinical (n = 102) or veterinary origin (n = 2), from different geographic regions of Brazil, comprising the main endemic areas of histoplasmosis (Table S1). Clinical isolates of H. capsulatum were recovered from patients with acute pulmonary, chronic pulmonary and disseminated forms of histoplasmosis. Isolates were stored as slant cultures on Sabouraud dextrose agar (Difco Laboratories, Detroit, MI, USA) at room temperature. The total dataset comprised 474 operational taxonomic units (OTUs) (Table S1).
DNA extraction and molecular characterisation
Total DNA was obtained and purified directly from 14 day-old monosporic colonies on Sabouraud slants by following the Fast DNA kit protocol (MP Biomedicals, USA), as previously described (Rodrigues et al. 2014). All isolates were characterised at the species level by PCR using a Histoplasma-specific primer pair (Table 1) targeting the gene encoding the M-antigen, by using primers Msp2F and Msp2R (Guedes et al. 2003) or by using sequence-characterised amplified region (SCAR) markers 1281-1283220 (PCR220) and 1281-1283230 (PCR230), as described before (Frías De León et al. 2012). Reference strains representing the main phylogenetic groups in Histoplasma were included in all experiments (Table S1). Diagnostic values using each primer pair included sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV). The receiver operating characteristic (ROC) curves were prepared and analyzed to determine the sensitivity and specificity of each PCR assay (Msp2F-Msp2R, PCR220, PCR230). The area under the ROC curve (AUC) was calculated to evaluate the diagnostic value of each PCR assay. P-values ≤0.05 were considered statistically significant. To measure the degree of concordance of each PCR assay, we calculated the Kappa statistic and its 95 % confidence interval (CI). Kappa values were interpreted as follows: 0.00–0.20, poor agreement; 0.21–0.40, fair agreement; 0.41–0.60, moderate agreement; 0.61–0.80, good agreement; 0.81–1.00, very good agreement (Altman 1991).
Table 1.
Primers used in this study for generic amplification, sequencing and genotyping.
| Locus/Region | Primer | Primer sequence 5′to 3′ | Annealing temperature (°C) | Orientation | Amplicon (bp) | Reference |
|---|---|---|---|---|---|---|
| ARF | ARF1 | AGAATATGGGGCAAAAAGGA | 65–56∗ | Forward | 470 | (Kasuga et al. 1999) |
| ARF2 | CGCAATTCATCTTCGTTGAG | 65–56∗ | Reverse | (Kasuga et al. 1999) | ||
| H-anti | H-anti3 | CGCAGTCACCTCCATACTATC | 65–56∗ | Forward | 410 | (Kasuga et al. 1999) |
| H-anti4 | GCGCCGACATTAACCC | 65–56∗ | Reverse | (Kasuga et al. 1999) | ||
| OLE | Ole3 | TTTAAACGAAGCCCCCACGG | 65–56∗ | Forward | 424 | (Kasuga et al. 1999) |
| Ole4 | CACCACCTCCAACAGCAGCA | 65–56∗ | Reverse | (Kasuga et al. 1999) | ||
| ITS | ITS5 | GGAAGTAAAAGTCGTAACAAGG | 52 | Forward | 630 | (White et al. 1990) |
| ITS4 | TCCTCCGCTTATTGATATGC | 52 | Reverse | (White et al. 1990) | ||
| MAT 1-1 | MAT1-1S | CGTGGTTAGTTACGGAGGCA | 60 | Forward | 440 | (Bubnick & Smulian 2007) |
| MAT1-1AS | TGAGGATGCGAGTGATGGGA | 60 | Reverse | (Bubnick & Smulian 2007) | ||
| MAT 1-2 | MAT1-2S | ACACAGTAGCCCAACCTCTC | 60 | Forward | 528 | (Bubnick & Smulian 2007) |
| MAT1-2AS | TCGACAATCCCATCCAATACCG | 60 | Reverse | (Bubnick & Smulian 2007) | ||
| M-anti | Msp2F | CGGGCCGCGTTTAACAGCGCC | 55 | Forward | 279 | (Guedes et al. 2003) |
| Msp2R | ACCAGCGGCCATAAGGACGTC | 55 | Reverse | (Guedes et al. 2003) | ||
| SCAR marker | 1281-1283220F | CATTGTTGGAGGAACCTGCT | 55 | Forward | 220 | (Frías De León et al. 2012) |
| 1281-1283220R | GAGCTGCAGGATGTTTGTTG | 55 | Reverse | (Frías De León et al. 2012) | ||
| SCAR marker | 1281-1283230F | GGAGCCATGACGTTAAATGG | 55 | Forward | 230 | (Frías De León et al. 2012) |
| 1281-1283230R | TATTGCCAATGGGTTTGTCA | 55 | Reverse | (Frías De León et al. 2012) |
arf: ADP-ribosylation factor; H-anti: H antigen precursor; ole: Delta-9 fatty acid desaturase; ITS: Internal transcribed spacers + rRNA genes; MAT: Mating types; M-anti: M antigen precursor; Scar marker: Sequence-Characterised Amplified Region. ∗Touchdown PCR
Multi-locus sequence analysis (MLSA)
For PCR amplification of specific regions, the partial protein-coding genes used in this study were ADP-ribosylation factor (arf), H-antigen precursor (H-anti), and delta-9 fatty acid desaturase (ole). The reactions were performed as previously described (Kasuga et al. 1999) (Table 1). H. capsulatum ITS regions were directly amplified from genomic DNA with primers ITS5 and ITS4 (White et al. 1990) and used as barcoding markers (Irinyi et al. 2015). PCRs were performed in a final volume of 25 μL, including 12.5 μL PCR Master Mix buffer (2×), which consisted of 3 mM MgCl2, 400 mM each dNTP, and 50 U/mL of Taq Polymerase (Promega Corporation, Madison, WI, USA); 9.5 μL of water, 1 μL each of forward and reverse primers (10 pmol/μL; Integrated DNA Technologies, Coralville, IA, USA), and 1 μL of target DNA [100 ng/μL]. The sequencing reactions were carried out with the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA), and the sequencing products were determined using an ABI3730xL Genetic Analyzer 48-well capillary sequencer (Applied Biosystems). The sequences generated in both orientations were assembled into single sequences via CAP3 implemented in the BioEdit software (Hall 1999). Sequences were aligned with MAFFT v.7 (Katoh & Standley 2013) and the retrieved alignments were manually edited to avoid mispaired bases. All sequences were deposited online with GenBank (accession numbers: MK893472–MK893880; Table S1).
Characterisation of the mating type idiomorphs
PCR primers designed to specifically amplify either the MAT1-1 or the MAT1-2 region were used to determine the mating types, as described before (Bubnick & Smulian 2007). Approximately 50 ng of genomic DNA was used for PCR with two sets of oligonucleotide primers: MAT1-1S (5-CGT GGT TAG TTA CGG AGG CA-3) and MAT1-1AS (5-TGA GGA TGC GAG TGA TGG GA-3), which amplify a 440 bp fragment from the α box region of the MAT1-1 idiomorph, and MAT1-2S (5-ACA CAG TAG CCC AAC CTC TC-3) and MAT1-2AS (5-TCG ACA ATC CCA TCC AAT ACC G-3), which amplify a 528 bp fragment from the HMG domain gene, present in the MAT1-2 idiomorph (Bubnick & Smulian 2007). PCRs were performed with PCR Master Mix buffer (Promega) as described above under the following conditions: 4 min at 94 °C; followed by 35 cycles of 1 min at 94 °C, 1 min at 60 °C, and 1 min at 72 °C; and a final step of 5 min at 72 °C. Samples were visualised on agarose gels as described above.
Phylogenetic reconstruction
Multi-locus sequence analysis (MLSA) was performed by sequencing PCR-amplified gene fragments. A subset of 104 H. capsulatum strains was selected for MLSA (Kasuga et al. 2003). The oligonucleotide primer sequences used for MLSA in this study are listed in Table 1. Genetic relationships were investigated by phylogenetic analysis using the neighbor joining (NJ), maximum likelihood (ML), and maximum parsimony (MP) methods. Phylogenetic trees were constructed with MEGA7 (Kumar et al. 2016). Evolutionary distances were computed using the Kimura 2-parameter distance (Kimura 1980), and the robustness of branches was assessed by bootstrap analysis of 1 000 replicates (Felsenstein 1985).
Amplified fragment length polymorphism
The amplified fragment length polymorphism (AFLP) analysis was conducted according to a modified version of the protocol (Vos et al. 1995) described previously (Najafzadeh et al. 2011). The restriction enzymes used were EcoRI (selective primer: 5′-GAC TGC GTA CCA ATT CNN-3′) and MseI (selective primer: 5′-GAT GAG TCC TGA GTA ANN-3′). Selective amplification was carried out with the fluorescent-labeled (FAM) primer pair EcoRI-AC and MseI-CT. The AFLP products were separated with an ABI3100 Genetic Analyzer, along with the GeneScan 500 ROX internal lane size standard (Applied Biosystems). Selection of the amplified restriction products was automated, and only strong and high-quality fragments were considered. The size of the AFLP fragments was determined with the BioNumerics v7.6 software package (Applied Maths). Binary AFLP matrices were created from the presence (1) or absence (0) at probable fragment positions. Pairwise, genetic distances were expressed as the complement of the Dice coefficient (Dice 1945). Dendrograms were produced according to the unweighted pair-group mean arithmetic method (UPGMA). The cophenetic correlation and standard deviation were used to express the consistency of a given cluster, by calculating the correlation between the dendrogram-derived similarities and the matrix similarities. To evaluate the performance of primer pair EcoRI-AC and MseI-CT, the following polymorphism indices for dominant markers were calculated: polymorphic information content (PIC) (Botstein et al. 1980), expected heterozygosity (H) (Liu 1998), effective multiplex ratio (E) (Powell et al. 1996), arithmetic mean heterozygosity (Havp) (Powell et al. 1996), marker index (MI) (Powell et al., 1996, Varshney et al., 2007), discriminating power (D) (Tessier et al. 1999), and resolving power (Rp) (Prevost & Wilkinson 1999).
Dimensioning analysis
Principal component analysis (PCA) and multi-dimensional scaling (MDS) were used as alternative grouping methods, producing three-dimensional plots in which the entries were spread according to their relatedness. Dimensioning techniques were executed with AFLP data after conversion into a band matching table. Automated band matching was performed on all fingerprint entries within the comparison, considering a minimum profiling of 5 %, with the optimization and position tolerances for selecting bands set to 0.10 %. Default settings were applied for PCA and MDS in BioNumerics v7.6, subtracting the average for characters. In addition, the Self-Organizing Map (SOM), a popular artificial neural network algorithm in the unsupervised learning category, was used to classify AFLP entries in a two-dimensional space (map) according to their likeliness (Kohonen 2001). The Kohonen map size was set to 100 (i.e., the number of nodes of the neural network in each direction). All Figs were exported and treated using Corel Draw X8.
Genetic population analysis
The nucleotide (π) as well as the haplotype (Hd) diversities (Nei 1987) were estimated using DnaSP version 6 (Rozas et al. 2017). Sites containing gaps and missing data were not considered in the analysis. Haplotype network analysis was constructed using the median-joining method (Bandelt et al. 1999), implemented in NETWORK v4.6.1.0 (Fluxus Technology, Suffolk, UK). In addition, recombination possibilities were investigated using the neighbor-net method (Bryant & Moulton 2004), which in the presence of recombination, leads to reticulated relationships, described by the uncorrected-P distance or by the split decomposition method (Bandelt & Dress 1992) both implemented in SplitsTree v.4b06 (Huson & Bryant 2006). Additional measures of recombination were estimated using the PHI-test (P < 0.05, demonstrating significant evidence of recombination). Analysis of AFLP data in STRUCTURE (v2.3.4) (Pritchard et al. 2000) was performed using the admixture model, allowing alpha to be inferred and assuming correlated allele frequencies, using a burn-in period of 10 000 Markov chain Monte Carlo (MCMC) replications followed by 10 000 sampling replications, with 20 independent runs performed for K values one to twelve. Analysis of three-gene multi locus concatenated sequences was performed using the linked model (specifying linkage within individual loci) with a burn-in of 20 000 MCMC replications (after initial 500 admixture replications), followed by 100 000 MCMC sampling replications, with 20 independent runs performed for K values one to fifteen. The optimal numbers of burn-in and sampling replications were determined by plotting alpha and Ln-likelihood from extended runs for convergence of the Markov chains. All data were analyzed using the method of Evanno and colleagues as implemented in StructureHARVESTER(v0.6.94) (Evanno et al., 2005, Earl and vonHoldt, 2012) to determine the optimal number of clusters (K). Consensus population distributions were obtained with CLUMPP (v1.1.2) (Jakobsson & Rosenberg 2007), using the full search for the AFLP data, and the greedy algorithm (1 000 000 random repeats) for the multi-locus gene data. Final plots were generated using ggplot2 (Gómez-Rubio 2017) in R (Team 2014). In addition, to assess the existence of topological congruence between dendrograms (AFLP and MLSA) and the associated confidence level, we calculated the congruence index (Icong), as described by de Vienne and colleagues (de Vienne et al. 2007), based on maximum agreement subtrees (MAST).
Results
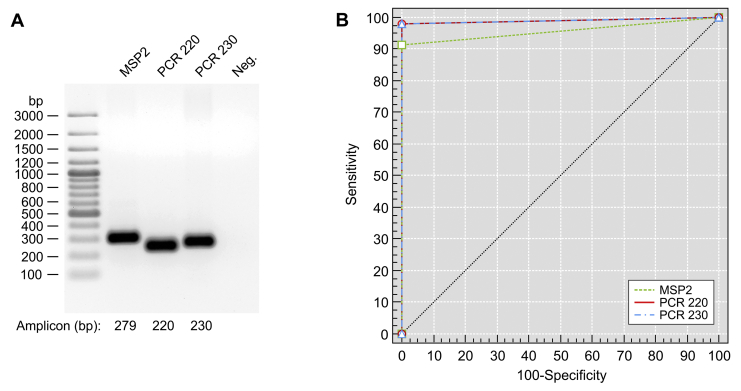
To confirm the identity of our 104 Brazilian isolates as Histoplasma by PCR, we initially chose three Histoplasma-specific primer pairs targeting the gene encoding the M-antigen (Guedes et al. 2003) and two SCAR markers (Frías De León et al. 2012) (Fig. 1A). Among the available strains, 95 (91.3 %), 102 (98.1 %) and 102 (98.1 %) samples were positive for Msp2, PCR220 and PCR230 respectively (see Table S1). As a control, positive amplifications for all 104 strains using ITS5 and ITS4 primers targeting the rDNA suggested the absence of PCR inhibitors for isolates that failed to amplify using Msp2 or SCARs primers. The sensitivity levels of Msp2, PCR220 and PCR230 were 91.4 %, 98.1 % and 98.1 %, respectively, and the specificity was 100 % for each assay. Positive predictive values were 100 % (95 % CI 96.2–100) for Msp2 and 100 % (95 % CI 96.4–100) for SCARs markers; and negative predictive values were 62.5 % (95 % CI 40.6–81.7) for Msp2 and 88.3 % (95 % CI 63.6–98.5) for SCARs markers (Table 2). The concordance of results by the Kappa measure of agreement was 0.67782 (95 % CI: 0.50300–0.85264; good agreement) between Msp2 and each of the SCARs markers, and 0.93137 (95 % CI: 0.83718–1; very good agreement) between SCARs markers. PCR-based identification was assumed to have powerful diagnostic value with high AUC for Msp2 0.957 (95 % CI: 0.903–0.986; P < 0.0001) and SCARs markers 0.990 (95 % CI: 0.952–1; P < 0.0001), indicating a low number of false-negatives and no false-positives (Fig. 1B, Table 2). Therefore, these markers were able to recognise several genotypes in the H. capsulatum clade.
Fig. 1.
Molecular identification of Histoplasma capsulatum by conventional PCR. A. Successful amplification with specific primer sets and Histoplasma capsulatum isolate CEMM 05-2-035 (=H2) as a template. Lane 1, 100 bp DNA ladder (Fermentas, USA) for size determinations; Lane 2, Msp2F-Msp2R primer pair amplification (279 bp); Lane 3, 1281-1283220 primer pair amplification (PCR 220; 220 bp); Lane 4, 1281-1283230 primer pair amplification (PCR 230; 230 bp); Lane 5, negative control. Further information about isolates used and amplification success can be found in Table S1. B. Receiver operating characteristic (ROC) curves for primer pairs Msp2F-Msp2R (MSP2), 1281-1283220 (PCR 220) and 1281-1283230 (PCR 230) based on 104 specimens. Despite high genetic diversity across H. capsulatum clusters, primer pairs usually employed in diagnosis successfully identified the new genetic groups recognised in this study. In PCR220 and PCR230, lines are superimposed, indicating equivalent accuracy. The classification variable was a dichotomous variable that indicated the diagnosis (0 = negative, 1 = positive).
Table 2.
Comparison of sensitivity, specificity, positive and negative predictive values, and area under the ROC curve among different diagnostic markers used to identify Histoplasma spp. cultures (n = 104).
| Diagnostic markers | Sen (%) | Sp (%) | PPV (%) | PPV 95 % CI |
NPV (%) | NPV 95 % CI |
AUC | AUC 95 % CI |
AUC P-value |
|---|---|---|---|---|---|---|---|---|---|
| M-anti | 91.4 | 100 | 100 | 96.2–100 | 62.5 | 40.61–81.7 | 0.957 ± 0.0139 | 0.903–0.986 | <0.0001 |
| PCR220 | 98.1 | 100 | 100 | 96.4–100 | 88.3 | 63.6–98.5 | 0.990 ± 0.00677 | 0.952–1 | <0.0001 |
| PCR230 | 98.1 | 100 | 100 | 96.4–100 | 88.3 | 63.6–98.5 | 0.990 ± 0.00677 | 0.952–1 | <0.0001 |
Sen, sensitivity; Sp, specificity; PPV, positive predictive value; NPV, negative predictive value; AUC, area under the curve.
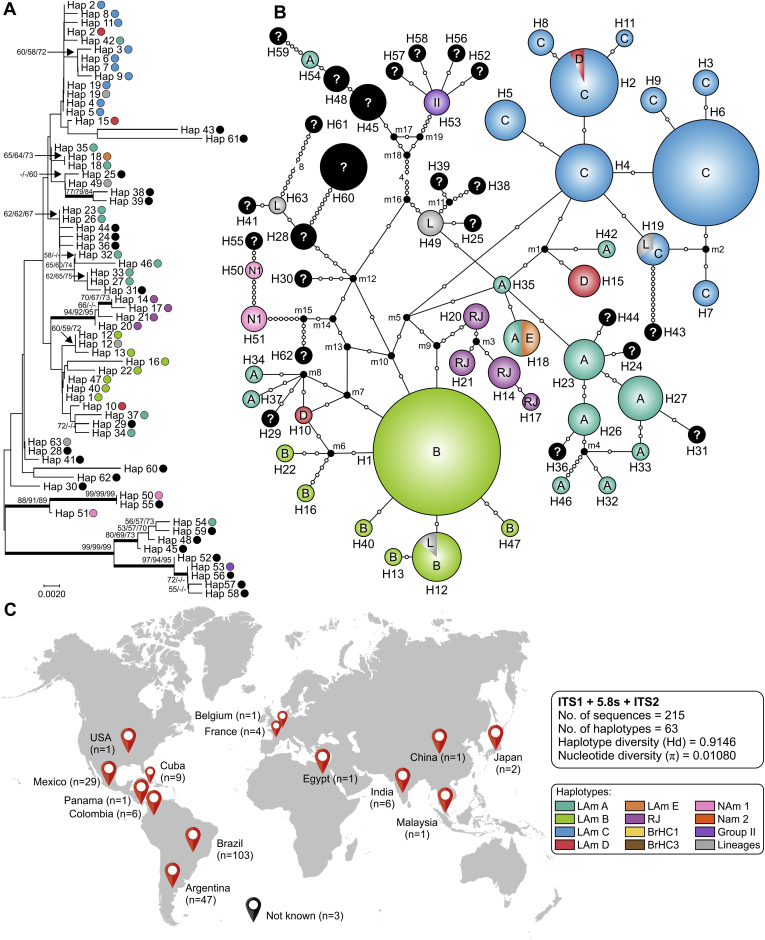
To determine the validity of the rDNA internal transcribed spacer (ITS) region as a marker for diagnostics of human-pathogenic Histoplasma species, we selected 215 ITS sequences (103 generated in this study and 112 reference sequences retrieved from GenBank) for phylogenetic analysis. From these entries, only a few sequences were long enough (>500 bp including ITS1+5.8S+ITS2) to be used for reliable alignment. We covered an epidemiologically diverse panel of clinical isolates with a global distribution including the main endemic areas in the Americas (Argentina, Brazil, Colombia, Cuba, Mexico, Panama and USA), as well as isolates that originated from Belgium, China, Egypt, France, Germany, India, Japan and Malaysia (Fig. 2C). Aligned ITS sequences were 582 bp long, including 497 invariable characters, 81 variable characters, 51 parsimony informative (8.76 %), and 30 singletons. Positions containing gaps and missing data were eliminated. Phylogenetic relationships among 215 sequences representing main genetic groups were inferred based on the Tamura 3-parameter (Gamma) substitution model (Table 3). As the trees obtained from the NJ, ML and MP analyses were identical in their topologies, only the NJ tree with bootstrap support values is presented here (Fig. 2). Using ITS as a single barcode, we were able to identify the H. capsulatum complex, but we failed to distinguish among cryptic species or new genotypes, as phylogenetic species were hitchhiking through different clades (e.g., LAm A isolates, recently named H. suramericanum).
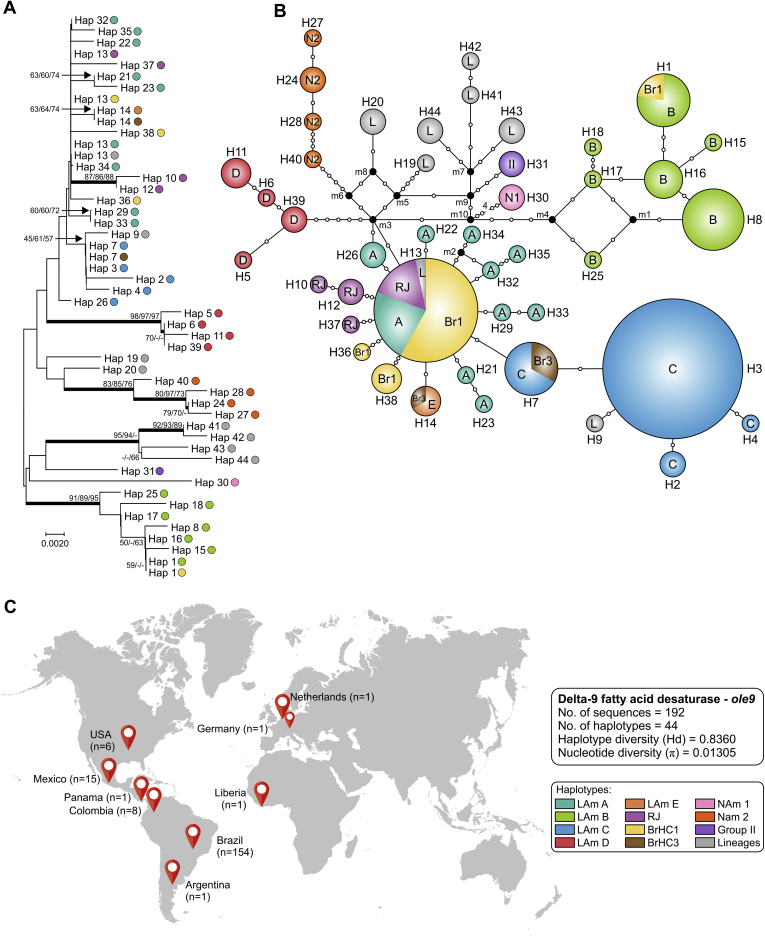
Fig. 2.
Phylogeny, haplotype and structure among Histoplasma capsulatum genotypes. A. Phylogenetic relationships, as inferred from neighbor joining analysis of the ITS sequences (ITS1+5.8S+ITS2; n = 215 OTU), covering the main haplotypes of H. capsulatum. Numbers above the tree branches are the bootstrap values for NJ, ML and MP methods. The branches with bootstrap support higher than 70 % are indicated in bold. B. Median-joining haplotype network of H. capsulatum isolates, covering all the ITS haplotypes found in this study. The size of the circumference is proportional to the haplotype frequency. The haplotypes are color coded according to the genetic group to which they were assigned. Mutational steps are represented by white dots. The black dots (median vectors) represent unsampled or extinct haplotypes in the population. C. Distribution patterns of H. capsulatum ITS sequences used in this study. Note that clade naming follows the original appearance of the isolates in the literature (Kasuga et al., 2003, Taylor et al., 2005, Muniz Mde et al., 2010). NJ, neighbor joining; ML, maximum likelihood; MP, maximum parsimony. Further information about isolate source and GenBank accession number can be found in Table S1.
Table 3.
Amplification success, phylogenetic data and the substitution models used in the phylogenetic and haplotype analysis, per locus.
| Locus/Region | ARF | H-anti | OLE | ITS | MLSA | |
|---|---|---|---|---|---|---|
| Amplification success (%) | 97.11 | 99.03 | 98.07 | 99.03 | – | |
| Histoplasma isolates | This study (n) | 101 | 103 | 102 | 103 | 101 |
| GenBank (n) | 116 | 92 | 91 | 116 | 77 | |
| Total (n) | 217 | 195 | 193 | 219 | 178 | |
| Best model | K2+G | K2+G | K2 | T92+G | K2+G+I | |
| MP–consistency index | 0,814815 | 0,704918 | 0,750000 | 0,661765 | 0,500000 | |
| MP–retention index | 0,960317 | 0,920000 | 0,921986 | 0,910156 | 0,901961 | |
| MP–composite index (for all sites) | 0,837200 | 0,749278 | 0,786761 | 0,714515 | 0,572341 | |
| MP–Tree length | 78 | 97 | 75 | 107 | 301 | |
| MP–No. of tress | 9 | 2 | 1 | 5 | 2 | |
| N. of sequences | 211 | 188 | 192 | 215 | 179 | |
| N. of sites | 453 | 385 | 399 | 582 | 1237 | |
| Total number of sites (excluding sites with gaps / missing data): | 430 | 369 | 390 | 550 | 1192 | |
| Conserved characters | 392 | 309 | 336 | 497 | 1053 | |
| Variable characters | 60 | 71 | 62 | 81 | 176 | |
| Parsimony-informative characters | 40 | 38 | 48 | 51 | 98 | |
| Singletons | 20 | 33 | 14 | 30 | 78 | |
| Nucleotide diversity (pi) | 0,01397 | 0,01665 | 0,01305 | 0,01080 | 0,01346 | |
| Number of haplotypes (h) | 55 | 52 | 44 | 63 | 88 | |
| Haplotype diversity (Hd) | 0,9101 | 0,9165 | 0,8360 | 0,9146 | 0,9746 | |
| Recombination phi test (Φ) (P-value) | No (P = 0.3754) | No (P = 0.1068) | No (P = 0.6897) | No (P = 0.3) | Yes (P = 1.539e-7) | |
ARF: ADP-ribosylation factor; H-anti: H antigen precursor; OLE: Delta-9 fatty acid desaturase; ITS: Internal transcribed spacers + rRNA genes; MLSA: Multi-locus sequence analysis; MP: maximum parsimony.
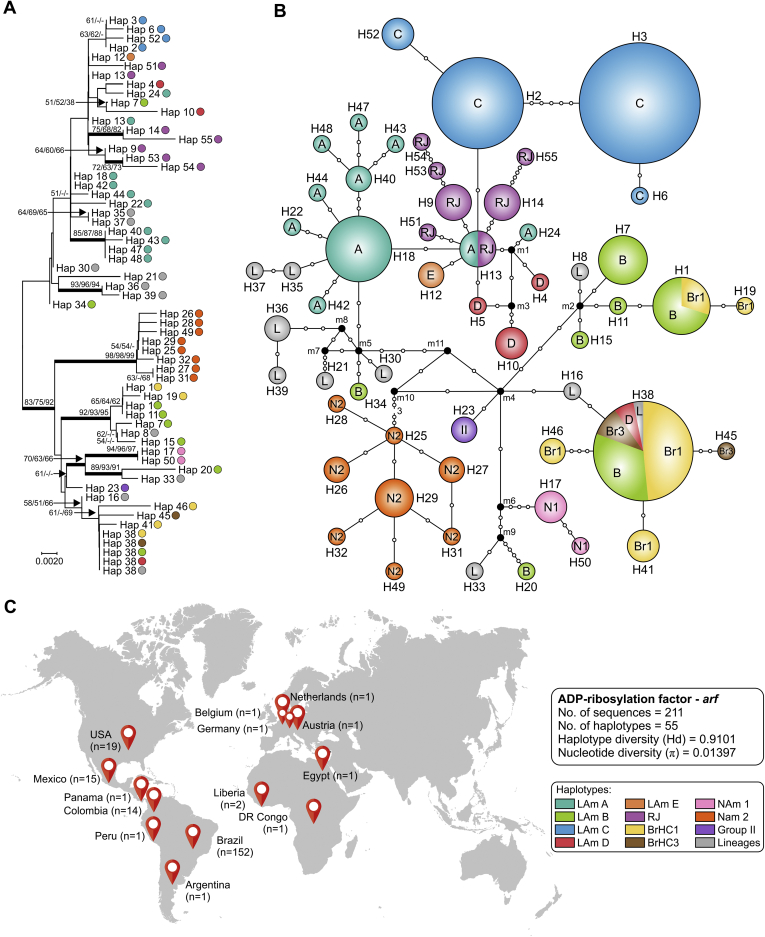
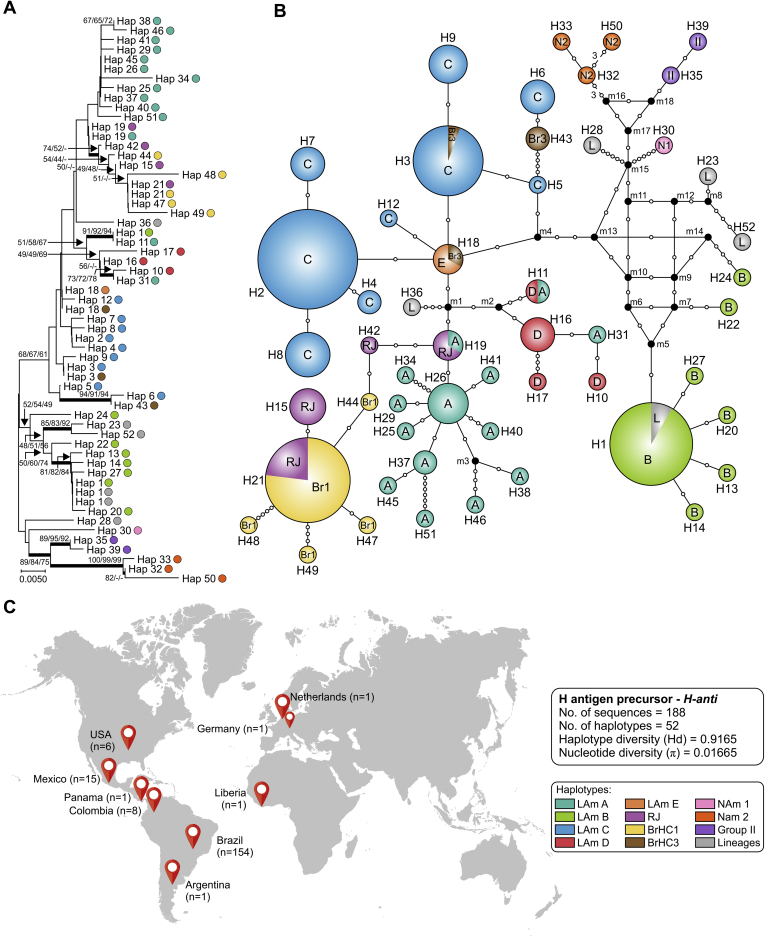
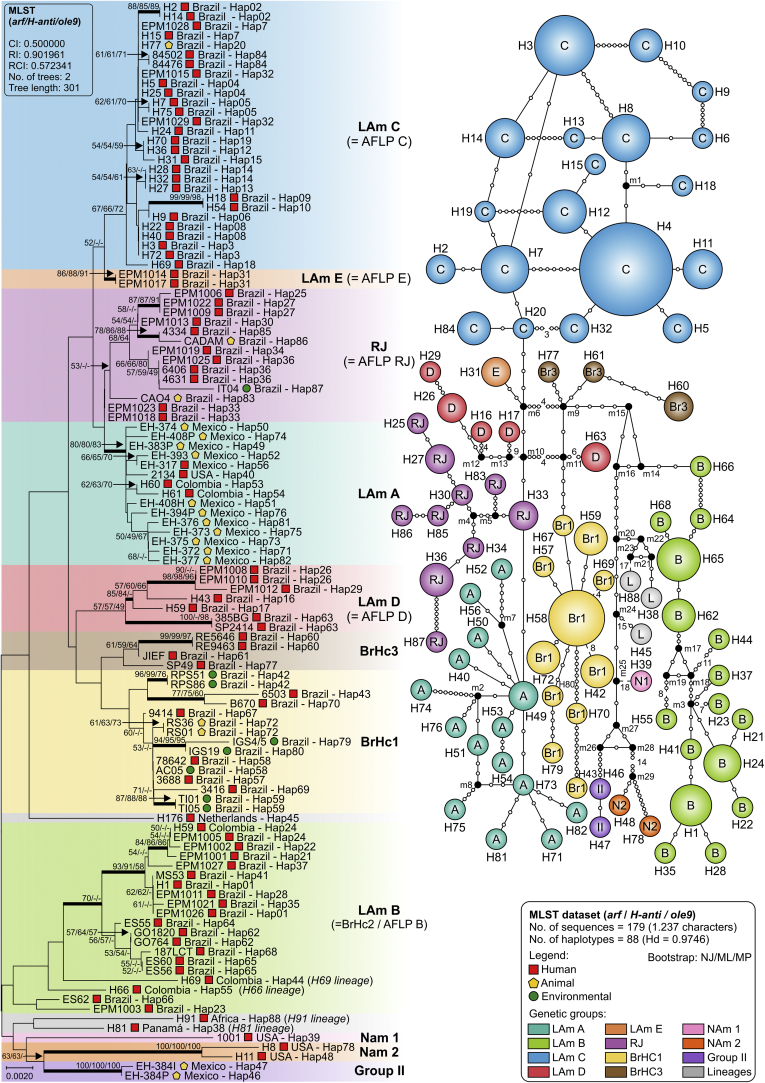
After a survey in the GenBank to recover protein-coding sequences from Histoplasma representing globally distributed isolates, we obtained arf (n = 217), H-anti (n = 195), and ole (n = 193) alignments of 453, 385, and 399 positions, of which 60 (13.2 %), 71 (18.4 %) and 62 (15.5 %) were variable, respectively. Using MEGA7, models with the lowest Bayesian information criterion (BIC) scores were considered to describe the best substitution model and K2+G was selected for arf and H-anti, K2 for ole, and K2+G+I for the concatenated 3-loci dataset (n = 101). These models were used in the NJ and ML analyses (Table 3). Widespread topology differences among arf (Fig. 3), H-anti (Fig. 4), and ole (Fig. 5) were observed, indicating that each independent dataset does not contain enough polymorphisms to resolve all relationships. On the other hand, MLSA greatly improved tree resolution, as the accumulation of nucleotide substitutions produced long branches revealing several cryptic clusters (Fig. 6), here named LAm C-E. Interestingly, genetic clusters often correlate with geographic origin of the isolates, as highlighted by the existence of LAm C, an emerging clade recognised here as causing histoplasmosis in HIV-infected patients who live in Northeast Brazil (Ceará state). In addition, two samples of veterinarian origin (H77 = CEMM 03-3-055 and H78 = CEMM 03-6-059; LAm C) were found as the etiological agent of feline histoplasmosis.
Fig. 3.
Phylogeny, haplotype and structure among Histoplasma capsulatum genotypes. A. Phylogenetic relationships, as inferred from neighbor joining analysis of the ADP-ribosylation factor sequences (arf; n = 211 OTU), covering the main haplotypes of H. capsulatum. Numbers above the tree branches are the bootstrap values for NJ, ML and MP methods. The branches with bootstrap support higher than 70 % are indicated in bold. B. Median-joining haplotype network of H. capsulatum isolates, covering all the arf haplotypes found in this study. The size of the circumference is proportional to the haplotype frequency. The haplotypes are color coded according to the genetic group to which they were assigned. Mutational steps are represented by white dots. The black dots (median vectors) represent unsampled or extinct haplotypes in the population. C. Distribution patterns of H. capsulatum arf sequences used in this study. Note that clade naming follows the original appearance of the isolates in the literature (Kasuga et al., 2003, Taylor et al., 2005, Muniz Mde et al., 2010). NJ, neighbor joining; ML, maximum likelihood; MP, maximum parsimony. Further information about isolate source and GenBank accession number can be found in Table S1.
Fig. 4.
Phylogeny, haplotype and structure among Histoplasma capsulatum genotypes. A. Phylogenetic relationships, as inferred from neighbor joining analysis of the H antigen precursor sequences (H-anti; n = 188 OTU), covering the main haplotypes of H. capsulatum. Numbers above the tree branches are the bootstrap values for NJ, ML and MP methods. The branches with bootstrap support higher than 70 % are indicated in bold. B. Median-joining haplotype network of H. capsulatum isolates, covering all the H-anti haplotypes found in this study. The size of the circumference is proportional to the haplotype frequency. The haplotypes are color coded according to the genetic group to which they were assigned. Mutational steps are represented by white dots. The black dots (median vectors) represent unsampled or extinct haplotypes in the population. C. Distribution patterns of H. capsulatum H-anti sequences used in this study. Note that clade naming follows the original appearance of the isolates in the literature (Kasuga et al., 2003, Taylor et al., 2005, Muniz Mde et al., 2010). NJ, neighbor joining; ML, maximum likelihood; MP, maximum parsimony. Further information about isolate source and GenBank accession number can be found in Table S1.
Fig. 5.
Phylogeny, haplotype and structure among Histoplasma capsulatum genotypes. A. Phylogenetic relationships, as inferred from neighbor joining analysis of the delta-9 fatty acid desaturase sequences (ole; n = 192 OTU), covering the main haplotypes of H. capsulatum. Numbers above the tree branches are the bootstrap values for NJ, ML and MP methods. The branches with bootstrap support higher than 70 % are indicated in bold. B. Median-joining haplotype network of H. capsulatum isolates, covering all the ole haplotypes found in this study. The size of the circumference is proportional to the haplotype frequency. The haplotypes are color coded according to the genetic group to which they were assigned. Mutational steps are represented by white dots. The black dots (median vectors) represent unsampled or extinct haplotypes in the population. C. Distribution patterns of H. capsulatum ole sequences used in this study. Note that clade naming follows the original appearance of the isolates in the literature (Kasuga et al., 2003, Taylor et al., 2005, Muniz Mde et al., 2010). NJ, neighbor joining; ML, maximum likelihood; MP, maximum parsimony. Further information about isolate source and GenBank accession number can be found in Table S1.
Fig. 6.
Phylogeny and haplotype among Histoplasma capsulatum genotypes. A. Phylogenetic relationships, as inferred from neighbor joining analysis of concatenated ADP-ribosylation factor, H-antigen precursor and delta-9 fatty acid desaturase sequences (n = 179 OTU), covering the main genetic groups of H. capsulatum. Numbers above the tree branches are the bootstrap values for NJ, ML and MP methods. The branches with bootstrap support higher than 70 % are indicated in bold. B. Median-joining haplotype network of H. capsulatum isolates. The size of the circumference is proportional to the haplotype frequency. The haplotypes are color coded according to the genetic group to which they were assigned. Mutational steps are represented by white dots. The black dots (median vectors) represent unsampled or extinct haplotypes in the population. Note that clade naming follows the original appearance of the isolates in the literature (Kasuga et al., 2003, Taylor et al., 2005, Muniz Mde et al., 2010). ML, maximum likelihood; MP, maximum parsimony; NJ, neighbor joining. Further information about isolate source and GenBank accession number can be found in Table S1.
For each locus (ITS, arf, ole or H-anti) and for the combined dataset (arf, ole and H-anti), median-joining haplotype networks were built (Fig. 2, Fig. 3, Fig. 4, Fig. 5, Fig. 6B), and the Brazilian isolates were analyzed together with the global Histoplasma isolates, displaying an overall star-like topology. The network topology clearly reflects the genetic differences among the different clusters observed in MLSA. A low number of median vectors were noted in all networks, especially among protein-coding loci. It often predicts the existence of unsampled or extinct haplotypes, supporting the success of the sampling approach adopted here. Judging from the newly detected group LAm C, there is emergence of a single high frequency haplotype at the center of the network (e.g., Hap 6 in Fig. 2; Hap 3 in Fig. 3, Fig. 5; Hap 2 in Fig. 4 and Hap 4 in Fig. 6), surrounded by low frequency derived haplotypes, a pattern typically indicative of recent population expansion or a recent selective sweep (Sarma et al. 2012). Nucleotide and haplotype diversities were high for all markers, with a range from π = 0.01080 to 0.01665 and Hd = 0.83 to 0.91; and the ITS dataset with 219 sequences showed the largest number of haplotypes (n = 63), whereas H-anti (Hd = 0.9165) showed higher values of haplotype diversity, followed closely by ITS (Hd = 0.9146) (Table 3). Haplotype networks constructed for different loci of the Brazilian isolates showed ancestral relationships with different cryptic lineages identified from the South American population. Brazilian haplotypes classified as LAm C always showed closer relationship to other sympatric genetic groups and the nearest taxa were LAm E and RJ, followed by LAm A, a clade with isolates of human and animal origin recovered from Brazil, Colombia, Mexico and USA (Fig. 7).
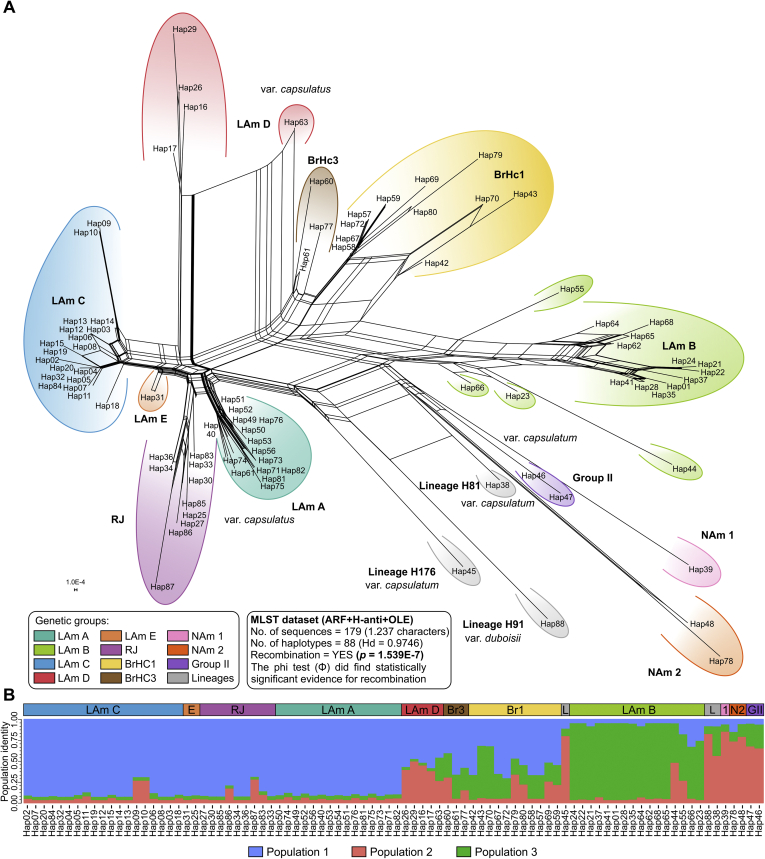
Fig. 7.
A. The neighbor network using the uncorrected p-distance among a core set of Histoplasma genotypes based on 1 237 nucleotide positions derived from the ADP-ribosylation factor, H-antigen precursor and delta-9 fatty acid desaturase loci (n = 179 OTU). Sets of parallel edges (reticulations) in the networks indicate locations of incongruence and potential recombination. Recombination within H. capsulatum genotypes was also supported by the PHI-test (Φ) (P = 1.539E-7). B. Bayesian cluster analyses with STRUCTURE of 88 H. capsulatum haplotypes based on MLSA dataset. Each vertical bar represents one individual MLSA haplotype and its probabilities of being assigned to clusters. Note that clade naming follows the original appearance of the isolates in the literature (Kasuga et al., 2003, Taylor et al., 2005, Muniz Mde et al., 2010). Further information about isolate source can be found in Table S1.
To provide a more accurate representation of genetic groups’ relationships, a phylogenetic network was constructed based on the concatenated alignment of the three coding regions by the split-decomposition method, which revealed clear evidence of phylogenic incongruence and potential recombination (Fig. 7A). Remarkably, large conflicting signals were detected, supporting recombination events at some point in evolutionary history. Not surprisingly, the PHI-test strongly supported the occurrence of recombination (P = 1.539e-7). STRUCTURE analyses using the MLSA dataset revealed three genetic clusters (k = 3) among 179 Histoplasma isolates (Fig. 7B). Within each country, the genetic composition of the individual clusters was very heterogeneous, ranging from isolates presenting high membership, especially in LAm C (Northeast Brazil), as well as strains with admixed ancestry, as in LAm D (Northeast, Midwest and Southeast Brazil). Judging from a threshold value used of 0.8 to identify single clusters, all groups presented admixed ancestry (Fig. 7B). Therefore, admixture was identified within the Brazilian populations (Northeast and Southeast), further suggesting that recombination is present among the loci assessed (Fig. 7B).
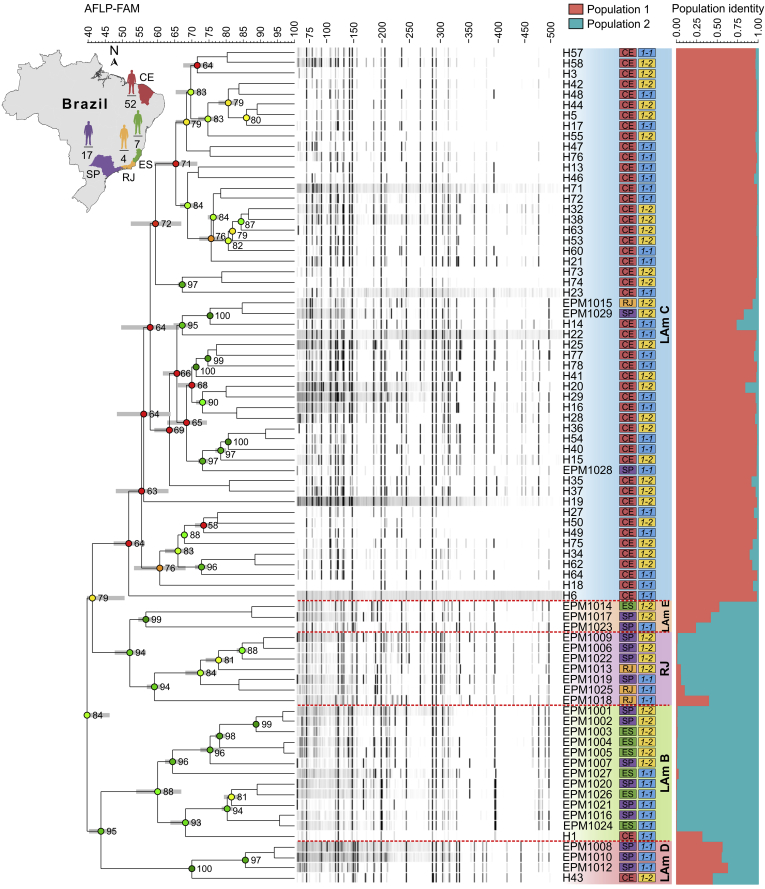
From a subset of 80 Brazilian isolates, listed in Fig. 8, we obtained 75 variable AFLP markers that could be scored unambiguously as present or absent in each Histoplasma isolate (Fig. 8). At a cutoff level of 53 %, the isolates grouped into five clearly separated groups (LAm B–E and RJ; global cophenetic correlation = 0.84; Shannon-Wiener diversity index = 1.05; Simpson's diversity = 52.97 %). The largest group, corresponding to LAm C (n = 53, 66.2 %; cophenetic correlation = 0.64), reveals the emerging genotypes mainly found in Northeast Brazil. Regarding the distribution of the 75 polymorphic markers, we detected the largest number of bands (scored bands = 70; PIC = 0.4711) and the presence of 64 markers in at least two isolates clustered in group LAm C, indicating the importance of this group for genetic diversity of Histoplasma in Northeast Brazil. Remaining isolates corresponded to the genetic clusters LAm B (n = 13, 16.2 %; cophenetic correlation = 0.88; PIC = 0.3164), LAm D (n = 4, 5 %; cophenetic correlation = 1.0; PIC = 0.2347), LAm E (n = 3, 3.7 %; cophenetic correlation = 0.99; PIC = 0.2392) and RJ (n = 7, 8.7 %; cophenetic correlation = 0.94; PIC = 0.2880). We used the allele frequencies to calculate polymorphism indices for dominant markers including the polymorphic information content (PIC = 0.3671), expected heterozygosity (H = 0.4845), and arithmetic mean heterozygosity (Havp = 0.0001) to reveal that there was remarkable variability among the genetic groups. The informativeness of AFLP markers was also estimated using effective multiplex ratio (E = 44.1125), marker index (MI = 0.0036), discriminating power (D = 0.6541), and resolving power (Rp = 30.0750). A strong and positive linear relationship was observed between PIC and RP values of EcoRI-AC/MseI-CT supporting its ability to distinguish genotypes (Pearson correlation = 0.96, r2 = 0.93, P = 0.0079). A summary of AFLP marker attributes calculated for selective primers EcoRI-AC/MseI-CT of Histoplasma species are given in Table 4.
Fig. 8.
The UPGMA dendrogram based on amplified fragment length polymorphism (AFLP) fingerprint, generated with a total of four selective bases (EcoRI-AC/MseI-CT) for 80 Histoplasma capsulatum originated from Brazil. AFLP results show a significant grouping according to geographical origin (Northeast and Southeast Brazil) and MLSA clusters, revealing five differentiated AFLP groups (LAm B, C, D, E and RJ). The dendrogram shows cophenetic correlation values (circles) for a given clade and its standard deviation (gray bar). Patterns of mating-type idiomorphs’ distribution across AFLP groups are shown. Fragments between 50 and 500 bp are shown. For pairwise, genetic distances calculation, Dice coefficient was used. The cophenetic correlation of the dendrogram is 0.84. Bayesian cluster analyses with STRUCTURE (k = 2) of 80 H. capsulatum samples based on AFLP. Each vertical bar represents one individual and its probabilities of being assigned to clusters. Further information about isolate source can be found in Table S1. CE, Ceará (Northeast); ES, Espírito Santo; RJ, Rio de Janeiro; SP, São Paulo (Southeast); 1-1, MAT1-1 mating type idiomorph; 1-2, MAT1-2 mating type idiomorph. Note that clade naming follows the original appearance of the isolates in the literature (Kasuga et al., 2003, Taylor et al., 2005, Muniz Mde et al., 2010).
Table 4.
Summary of polymorphism statistics calculated for selective primers EcoRI-AC/MseI-CT of Histoplasma species.
| Genetic group | Scored bands | H | PIC | E | Havp | MI | D | Rp |
|---|---|---|---|---|---|---|---|---|
| LAm B (n = 13) | 59 | 0.3941 | 0.3164 | 43.0769 | 0.0005 | 0.0221 | 0.4672 | 18.1538 |
| LAm C (n = 53) | 70 | 0.4711 | 0.3601 | 43.4151 | 0.0001 | 0.0055 | 0.6154 | 26.0755 |
| LAm D (n = 4) | 54 | 0.2716 | 0.2347 | 45.2500 | 0.0013 | 0.0569 | 0.2984 | 9.5000 |
| LAm E (n = 3) | 54 | 0.2778 | 0.2392 | 45.0000 | 0.0017 | 0.0772 | 0.3064 | 12.0000 |
| RJ (n = 7) | 66 | 0.3489 | 0.2880 | 51.1429 | 0.0008 | 0.0386 | 0.3999 | 19.1429 |
| Overall (n = 80) | 75 | 0.4845 | 0.3671 | 44.1125 | 0.0001 | 0.0036 | 0.6541 | 30.0750 |
D = discriminating power; E = effective multiplex ratio; H = expected heterozygosity; Havp = mean heterozygosity; MI = marker index; PIC = polymorphism information content; Rp = resolving power.
We found strong correlation among genetic diversity, population structure and their geographical origin. STRUCTURE analyses based on AFLP data revealed two populations, one from the Northeast (Population 1 = LAm C, mainly derived from Fortaleza, Ceará) and one from the Southeast (Population 2 = LAm B, LAm D, LAm E and RJ, originated from endemic areas in the states of Rio de Janeiro, São Paulo and Espírito Santo). In addition, the population structure reflects shared AFLP markers among individuals embedded in cluster LAm E, which shared 54 markers (84.3 %) with LAm C and 27 markers with LAm D. Therefore, our data support LAm D (E = 45.2500; Havp = 0.0013) and LAm E (E = 45.0000; Havp = 0.0017) as hybrid groups in Histoplasma. On the other hand, a low degree of shared markers was observed among the LAm C and the clusters LAm B (E = 43.0769; Havp = 0.0005) and RJ (E = 51.1429; Havp = 0.0008), suggesting longstanding separation, as recently suggested for other cryptic species in Histoplasma (Teixeira et al., 2016, Sepúlveda et al., 2017).
Our AFLP data support the view that cryptic speciation is a common phenomenon in Histoplasma and that cryptic species can be recognised using fingerprint markers. Furthermore, this study shows that AFLP data are a valuable supplement to DNA sequence data in that they may detect a finer level of genetic variation. In this scenario, the topologies of MLSA trees and the AFLP dendrogram showed strict correspondence, with all clinical clades being recognised using both markers (P = 2.28 e-05; Icong = 1.52) (de Vienne et al. 2007). However, single loci analysis based on protein coding regions or ITS region as a barcoding marker failed to recognise cryptic species in separate analyses.
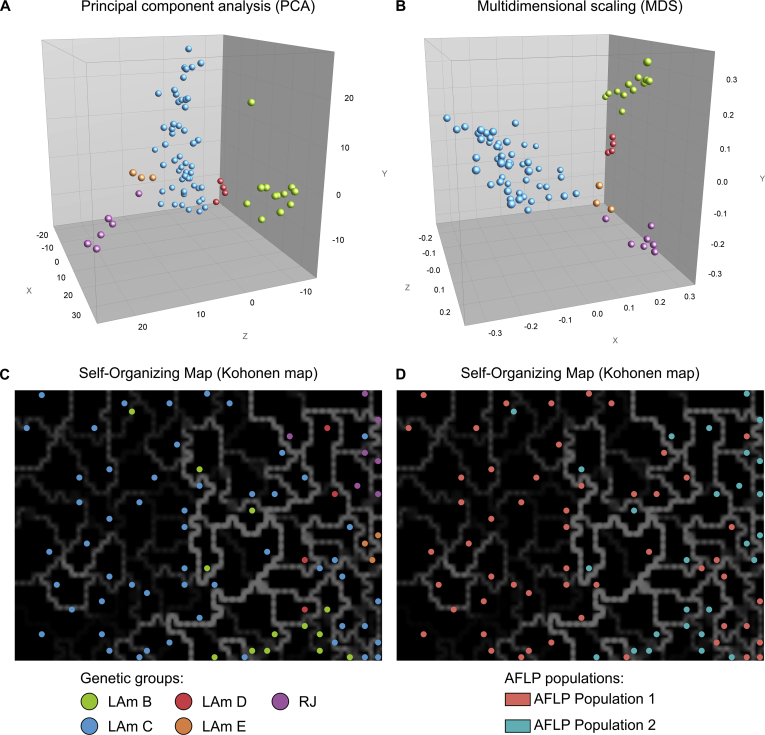
Analysis of AFLP data through PCA (Fig. 9A) and MDS (Fig. 9B) corroborates the clusters recognised by MLSA analysis. PCA revealed that the first three principal components described 35.9 % of the variation in the AFLP data (Fig. 9). Self-organizing maps based on AFLP fingerprints were used and are plotted according to genetic clusters (Fig. 9C) or AFLP populations (Fig. 9D). Unlike PCA, the distance between entries in the SOMs is not in proportion to the taxonomic distance between the entries. Rather, SOMs contain areas of high distance and areas of high similarity (Felix et al. 2015). In Fig. 9C,D, the lighter and thicker the line (white, gray) between black blocks, the more distant are those samples contained in the black block, from the adjacent black block. The models were organized into a meaningful two-dimensional order in which similar Histoplasma genotypes are closer to each other in the grid than the more dissimilar ones. Closely related LAm C genotypes are bounded by thin faint white/gray lines (e.g., isolates H44, H46, H48 and H49 from LAm C, Population 1; all belonging to MLSA Hap 04), implying high similarity of the strains, supporting the emergence of a group with low genetic diversity among HIV patients (Fig. 9C). On the other hand, genetically distant samples are separated by lighter and thicker lines (white, gray) between black blocks (e.g., isolates belonging to population 1 versus population 2; Fig. 9D).
Fig. 9.
The distribution of the studied AFLP fingerprints (EcoRI-AC/MseI-CT) of 80 Histoplasma capsulatum originated from Brazil, using principal component analysis (PCA), multi-dimensional scaling (MDS) and self-organizing mapping (SOM). The dimensioning analyses were performed using BioNumerics v7.6 to determine the consistency of the differentiation of the populations defined by the cluster analysis. A and B show the PCA and MDS of AFLP data with the first three principal components describing the greatest variation plotted on the X (15.4 %), Y (12 %), and Z (8.6 %) axes. The SOM revealed that Northeast isolates (n = 50; Ceará) are embedded in areas of high similarity mostly bounded by thin faint white/gray lines according to the genetic group (C) or AFLP population (D), confirming the low genetic diversity in MLSA data. The lighter and thicker the line (white, gray) between black blocks, the more distant are those samples contained in the black block, from the adjacent black block. Isolates were color coded according to their genetic groups (A, B and C) or AFLP population (D).
A mating type-specific PCR assay was used to amplify either the MAT1-1 or the MAT1-2 region among 104 H. capsulatum isolates. The MAT1-1 region was observed in 54 isolates, while the MAT1-2 region was observed among 49 isolates. The only exception was observed for isolate EPM1011 (LAm B, São Paulo, Brazil), which failed to amplify both MAT alleles despite positive amplification using ITS5 and ITS4 primers, therefore excluding the possibility of PCR inhibitors. The distribution of each sexual idiomorph within a geographical region (Northeast and Southeast) and molecular types (LAm B-E, RJ) are presented in Table 5. The distributions of MAT1-1 or MAT1-2 idiomorph were not significantly skewed (1:1 ratio) within Brazilian states or molecular types, as evidenced by Chi-square tests, supporting the presence of random mating within each population.
Table 5.
Distribution of mating type alleles as determined by PCR with mating type allele-specific primers in Histoplasma capsulatum isolates.
| Origin/Group | No. of isolates | No. of isolates by mating type |
Chi-square value | P-value | |
|---|---|---|---|---|---|
| MAT 1-1 (%) | MAT 1-2 (%) | ||||
| Histoplasma (Northeast) | 75 | 40 (53,33) | 35 (46,66) | 0.333 | 0.5637 |
| Histoplasma (Southeast) | 291 | 14 (50,00) | 14 (50,00) | 0 | 1.0000 |
| Histoplasma (Brazil) | 1041 | 54 (52,42) | 49 (47,57) | 0.243 | 0.6222 |
| LAm B | 141 | 7 (53,84) | 6 (46,15) | 0.077 | 0.7815 |
| LAm C | 74 | 40 (54,05) | 34 (45,94) | 0.486 | 0.4855 |
| LAm D | 5 | 3 (60,00) | 2 (40,00) | 0.200 | 0.6547 |
| LAm E | 2 | 0 (0,00) | 2 (100,00) | - | - |
| RJ | 8 | 4 (50,00) | 4 (50,00) | 0 | 1.0000 |
One isolate (EPM1011; Brazil - Southeast, LAm B) failed to amplify a MAT gene product.
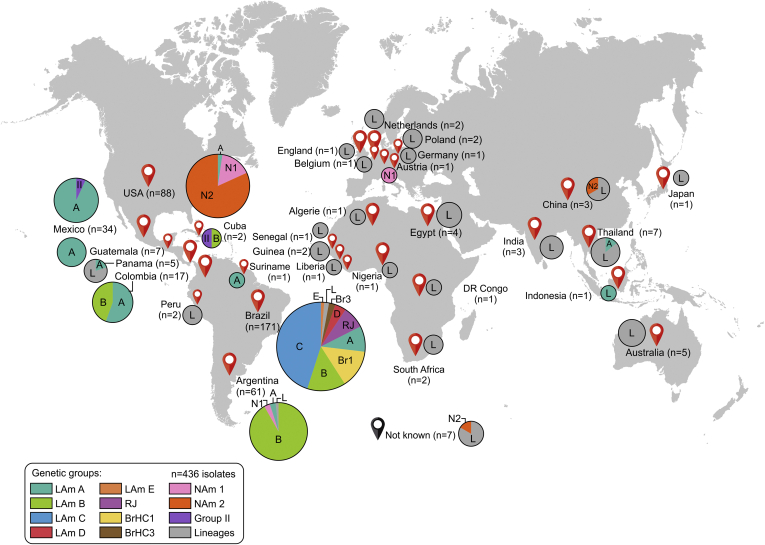
Although classically histoplasmosis is described as an endemic disease in the Americas, our data call attention to global distribution of the disease related to different genotypes of Histoplasma (Fig. 10). Brazil harbors nine highly diverse genetic groups of Histoplasma, and here we describe the recent emergence of LAm C among HIV patients living in Fortaleza, Ceará. Outside Brazil, the predominant genetic group depends on the region. Although most of the cases reported in the literature correspond to the case series observed in the USA (Benedict and Mody, 2016, Benedict et al., 2019), more specifically in Mississippi and Ohio where H. mississippiense and H. ohiense are endemic, no taxonomic diversity is observed (Fig. 10).
Fig. 10.
Global distribution patterns of 436 Histoplasma spp. isolates based on DNA sequencing. The sizes of circumferences are roughly proportional to the numbers of strains included. Codes reported within the pies denote genetic groups. Further information about isolate source and GenBank accession number can be found in Table S1.
Discussion
The history of human histoplasmosis can be separated into four periods. The first began in 1906 when Darling described a rare and often fatal disease (Darling 1906). Then during the second period (1940s), most of the sero-epidemiological studies were conducted, employing the histoplasmin skin test, which revealed that Darling’s disease was very common and widely distributed, causing pulmonary infection ranging from benign to fatal cases (Christie and Peterson, 1945, Joseph Wheat, 2003, Antinori, 2014). We are now at the end of the third period, where molecular data provide a unique opportunity to recognise and identify phylogenetic species and genetic populations (Kasuga et al., 1999, Kasuga et al., 2003, Sepúlveda et al., 2017). A well-corroborated phylogenetic classification has now begun to emerge for Histoplasma. The fourth period will involve comparative genomic studies, based on whole-genome sequencing data, for which purpose a global sampling strategy will be necessary (Sepúlveda et al. 2017). A few examples of other taxonomic distinctions of closely related clades in the Onygenales include Blastomyces (Brown et al. 2013), Coccidioides (Fisher et al. 2002), Emergomyces (Dukik et al. 2017), Emmonsia (Jiang et al. 2018) and Paracoccidioides (Turissini et al. 2017).
Our study describes new phylogenetic species and the molecular characteristics of Histoplasma lineages causing outbreaks with a high number of severe outcomes in Northeast Brazil between 2011 and 2015. A noteworthy difference was observed for the distribution of the emerging genotype LAm C and the affected host population. For instance, H. capsulatum LAm C is the major lineage causing histoplasmosis in HIV patients in Northeast Brazil (Ceará), while it shows low genetic diversity, as revealed by the haplotype networks and Kohonen self-organizing maps. Interestingly, this group first appeared in Rio de Janeiro in 1998 (subclade BrHc1C = isolates 84476, 84502, and 84564), but with low frequency (Muniz Mde et al. 2010). Moreover, LAm C has not been reported outside Brazil, as observed from sequences deposited in the GenBank, and subsequent re-analysis has revealed a low degree of homology. Similarly, H. capsulatum LAm D, E and RJ were recovered only from patients in the Southeast region of Brazil. To some extent, our study confirms previous molecular studies that demonstrated high genetic variability in H. capsulatum and structured populations with limited geographic distribution in Brazil (Zancope-Oliveira et al., 2005, Muniz Mde et al., 2010, Brilhante et al., 2012). Muniz and co-workers (Muniz Mde et al. 2010) grouped 51 isolates into three major clusters using several DNA fingerprinting methods, which correlates with their geographical origins. BrHc1, with subclades BrHc1A–D, was predominantly found in Rio de Janeiro, followed by Mato Grosso do Sul and São Paulo. Clade BrHc2, a more widespread group spanning an area of over 2 000 km, was detected in Espírito Santo, Rio Grande do Sul, and central Brazil. BrHc3 was found in Pernambuco and Ceará. MLSA revealed that clade BrHc3, including isolates RE5646 and RE9463 from Pernambuco (Northeast Brazil) or BrHc1 (from Rio de Janeiro) are genetically distant taxa from LAm C, supporting the novelty of the emergence described here. This agrees with data from a study reported previously by Teixeira and colleagues (Teixeira et al. 2016), who used the MLSA scheme proposed by Kasuga to investigate the global phylogenetic relationship.
From an epidemiological perspective, we present here the largest survey to date (n = 436 isolates; Fig. 10). Our phylogenetic analysis reveals that histoplasmosis is wider spread than previously assumed, indicating that etiological agents can be divided into several phylogenetically distinct groups (Teixeira et al. 2016). Our findings go beyond the genetic diversity previously reported, which strengthens the importance of more epidemiological investigations in Brazil. This is supported by the high incidence of histoplasmosis in Brazil, with 2.19 cases per 1 000 hospital admissions (Giacomazzi et al. 2016), especially among HIV patients (Almeida et al., 2019, Falci et al., 2019). The geographic center of origin should contain the highest allelic and genotypic diversities, which was confirmed here by MLSA and AFLP analysis, supporting the hypothesis that Brazil is the center of origin of Histoplasma spp. in Latin America, not only due to genetic diversity, but also driven by recombination and evidence of sexual reproduction in these populations. Remarkably, in some pathogenic fungi such as Aspergillus and Cryptococcus, abundant evidence of sexual recombination has also been used as a signature for an ancestral population (Hagen et al., 2013, Ashu et al., 2017).
Recent global estimates are of annual incidence of ∼100 000 cases of disseminated histoplasmosis (Bongomin et al. 2017). However, in many countries the annual incidence of histoplasmosis is not known (Denning 2016), and little or no genetic information has been reported in studies of the agents of histoplasmosis around the world. In Latin America, for example, histoplasmosis is one of the most common opportunistic infections among people living with HIV, and approximately 30 % of HIV/AIDS patients diagnosed with histoplasmosis die from it, especially among patients who did not receive highly active antiretroviral therapy (HAART) (Colombo et al. 2011). Moreover, the pulmonary manifestations may be misdiagnosed as tuberculosis (Linder & Kauffman 2019). Here we provide strong evidence based on molecular epidemiology that there is enormous genetic diversity of Histoplasma in Brazil, a situation that deserves to be explored in terms of virulence traits, antifungal susceptibility and clinical outcomes.
Histoplasma capsulatum and allied species cause devastating infection in immunocompromised patients, which is difficult to diagnose rapidly enough to save the patient’s life (Nacher et al. 2018). When new genotypes were detected in Brazil, we wondered whether the existing molecular assays would be capable of detecting such diversity. Surprisingly, we obtained satisfactory results for detection of Histoplasma DNA using Msp2, PCR220 and PCR230 assays, which provided a generic identification as Histoplasma spp. Molecular techniques can detect Histoplasma spp. even in cases with negative serology (e.g., immunoblotting or double immunodiffusion) negative direct mycological examination (potassium hydroxide wet mount slides) or negative cultures, potentially enabling early diagnosis (Dantas et al. 2018).
DNA barcoding indicated that H. capsulatum s. str., H. mississippiense, H. ohiense and H. suramericanum (Sepúlveda et al. 2017), the classic and new phylogenetic cryptic groups (Kasuga et al. 2003) and the historical taxonomic varieties (var. capsulatum, var. farciminosum, and var. duboisii) could not be entirely distinguishable by fixed mutations in the ITS1+5.8S+ITS2 in the rDNA operon. Like the PCR-based diagnosis evaluated here, ITS sequencing may provide a generic diagnosis as previously reported by other groups (Estrada-Barcenas et al., 2014, Landaburu et al., 2014, Irinyi et al., 2015) and may supplement traditional morphology-based taxonomy. Indeed, the definitive diagnosis of histoplasmosis can be difficult when studying tissue sections, so the ITS-based identification can play an important role because it will differentiate Histoplasma from other morphologically similar agents such as the small variant of Blastomyces, or the capsule-deficient cryptococci (Guimarães et al., 2006, Guarner and Brandt, 2011). The lack of resolution of ITS has also been reported for other taxa and prevents its universal application (Kiss 2012). For some fungi, the second largest subunit of ribosomal polymerase II (RPB2) and another recently introduced marker, mini-chromosome maintenance protein (MCM7), were utilised with success (Stielow et al. 2015). This reinforces the urgent need to search for alternative reliable barcode markers to improve resolution and unambiguously identify classic and new Histoplasma phylogenetic species during routine diagnosis. Therefore, the availability of a “gold standard” phylogeny will make it possible to evaluate the informativeness of other DNA fingerprinting systems for phylogenetic analysis and species recognition. As an alternative, we introduced AFLP genotyping to assess the genetic diversity of Brazilian Histoplasma isolates down to the species level. Our results using AFLP markers matched the major genetic group phylogeny assigned by arf, ole and H-anti gene polymorphisms.
AFLP and MLSA indicated negligible gene flow among LAm D, LAm E and RJ, as well between this group ('population 2') and LAm C ('population 1'). Hybrid speciation and introgressive hybridisation occurs more readily between allopatric species and has been suggested to be an important evolutionary force that generates diversity by the recombination of genetic material among divergent lineages. In Histoplasma, whole genomic sequencing has been used to demonstrate the occurrence of recent admixture (Sepúlveda et al. 2017). Extensive backcrossing between hybrid and the parental species can sometimes lead to introgression if foreign genetic material is integrated into the genomes of either parent (Baack and Rieseberg, 2007, Fogelqvist et al., 2015). Such important mechanisms whereby new species may emerge can increase fitness and virulence traits and lead to the development of reproductive isolation, a phenomenon often demonstrated among pathogenic fungi (Stukenbrock 2016). However, introgressed alleles are present in low frequencies in Histoplasma, supporting the view that they are deleterious (Maxwell et al. 2018), which may contribute to reproductive barriers.
Neighbor-net, PHI-test and structure analysis supports a recombination history among Histoplasma clades, as well as admixture in population structure with clear evidence of hybrids between clusters. This could have influenced the evolution, radiation, and potentially the emergence of new genotypes of Histoplasma in Northeast Brazil. Alternatively, conflicting phylogenetic signals may reflect maintenance of ancestral polymorphism, whereby incomplete lineage sorting leads to ambiguous phylogenetic relationships even when genetic isolation occurs instantly for all genes (Retchless & Lawrence 2010). The use of comparative genomics is necessary to disentangle these competing hypotheses.
In fungi, both sexual (e.g., mating) and asexual origins (e.g., fusion of cells or hyphae) are evident in the emerging list of apparent fungal hybrids (Schardl and Craven, 2003, Kohn, 2005). Therefore, we assessed the distribution and frequencies of the two mating types in Brazilian populations of Histoplasma in an attempt to understand sexual reproduction modes. Since H. capsulatum and related species are heterothallic, and our analyses showed equal frequencies of mating types idiomorphs (1:1), we assume that sexual reproduction is a common event among Brazilian Histoplasma isolates. In H. capsulatum s.l., some studies have revealed that mating type idiomorphs are equally distributed in the soil but deviate in clinical samples, with the majority being MAT1-1 (Kwon-Chung et al., 1974, Kwon-Chung et al., 1984) or MAT 1-2 (Rodríguez-Arellanes et al. 2013), which are usually associated with the geographical origin of the isolates (Muniz et al. 2014). Nevertheless, skewed MAT loci distribution has been demonstrated for H. capsulatum var. duboisii in Africa (Valero et al. 2018) which may be related to paucity of sexual reproduction and/or strong selection for pleiotropic effects of a mating type allele (Nieuwenhuis & James 2016) or a phenomenon of small populations (Valero et al. 2018). Therefore, it is tempting to hypothesise that different Histoplasma spp. may have developed a mixed mode of reproduction, ranging from rare to frequent sex, depending on the species and the population characteristics. However, this hypothesis of species-specific reproductive modes in Histoplasma should be further investigated in a bigger number of strains.
Notably, the mating type locus has previously been associated with differential virulence in medically relevant fungi (Xu et al. 2017). This holds true for sexual pathogens such as Cryptococcus neoformans (Kwon-Chung et al. 1992), Aspergillus fumigatus (Cheema & Christians 2011), and Mucor irregularis (Xu et al. 2017). However, mating type appears to have no influence on the virulence process of H. capsulatum s.l. because no significant difference was found between strains MAT1-1 and MAT1-2 in a murine model of infection (Kwon-Chung & Hill 1981). A similar conclusion was drawn for the animal-borne Sporothrix brasiliensis, where both MAT1-1 and MAT1-2 strains were observed to be highly virulent (Della Terra et al. 2017), but deviated in MAT frequency in genetic population studies (Teixeira et al. 2015).
Using a large number of isolates from geographically and ecologically diverse regions, our study allowed us to address several fundamental questions about the epidemiology of Histoplasma. Our findings go beyond those previously reported by others, since we were able to recognise new phylogenetic lineages. Not surprisingly, we identified high genetic diversity and a structured population of Histoplasma in Brazil, matching the different geographic sources. Our findings significantly broaden the area of histoplasmosis occurrence, an important feature of emerging pathogens. Therefore, the correct diagnosis will be fundamental to elevate the epidemiological status in one of the most important systemic mycoses of the Americas. Moreover, with the description of new genotypes, we highlight that ITS is not sufficient for routine distinction of classic and new Histoplasma species or taxonomic varieties, although it can provide a genus-level identification, which is crucial to guide appropriate clinical therapy. Species-specific reproductive modes in Histoplasma ranging from rare to frequent sex may drive dissimilar genetic diversity in this widely distributed pathogen, a hypothesis that should be further investigated in a substantial number of globally sampled strains. From a practical point of view, our data point to the emergence of histoplasmosis caused by a plethora of genotypes and will allow the future proposal of public policies to contain the spread of histoplasmosis.
Limitations of our study include the number of samples associated with clinical status and that MLSA was based only on a few genes. Although those markers have been well characterised by other groups (Kasuga et al., 1999, Kasuga et al., 2003), we believe that phylogenomics can greatly improve recognition of species, speciation events and evolution of Histoplasma (Sepúlveda et al. 2017). With the description of such diversity, we open avenues for future comparative genomic studies, which will hopefully lead to a consensus taxonomy, improve understanding of the presence of hybrids in natural populations of medically relevant fungi, test reproductive barriers and explore the significance of these variations.
Acknowledgments
AMR and ZPC acknowledge the financial support of the São Paulo Research Foundation (FAPESP 2017/27265-5 and FAPESP 2018/21460-3), the Coordination for the Improvement of Higher Education Personnel (CAPES), and the National Council for Scientific and Technological Development (CNPq 552161/2011-0). These agencies had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
Peer review under responsibility of Westerdijk Fungal Biodiversity Institute.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.simyco.2020.02.001.
Contributor Information
A.M. Rodrigues, Email: amrodrigues.amr@gmail.com.
Z.P. de Camargo, Email: zpcamargo1@gmail.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Adenis A.A., Valdes A., Cropet C. Burden of HIV-associated histoplasmosis compared with tuberculosis in Latin America: a modelling study. Lancet Infectious Diseases. 2018;18:1150–1159. doi: 10.1016/S1473-3099(18)30354-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida M.A., Almeida-Silva F., Guimaraes A.J. The occurrence of histoplasmosis in Brazil: A systematic review. International Journal of Infectious Diseases. 2019;86:147–156. doi: 10.1016/j.ijid.2019.07.009. [DOI] [PubMed] [Google Scholar]
- Altman D.G. Chapman and Hall; London: 1991. Practical statistics for medical research; p. 624. [Google Scholar]
- Antinori S. Histoplasma capsulatum: more widespread than previously thought. American Journal of Tropical Medicine and Hygiene. 2014;90:982–983. doi: 10.4269/ajtmh.14-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashbee H.R., Evans E.G.V., Viviani M.A. Histoplasmosis in Europe: report on an epidemiological survey from the European Confederation of Medical Mycology Working Group. Medical Mycology. 2008;46:57–65. doi: 10.1080/13693780701591481. [DOI] [PubMed] [Google Scholar]
- Ashu E.E., Hagen F., Chowdhary A. Global population genetic analysis of Aspergillus fumigatus. mSphere. 2017;2 doi: 10.1128/mSphere.00019-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baack E.J., Rieseberg L.H. A genomic view of introgression and hybrid speciation. Current Opinion in Genetics and Development. 2007;17:513–518. doi: 10.1016/j.gde.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker J., Setianingrum F., Wahyuningsih R. Mapping histoplasmosis in South East Asia – implications for diagnosis in AIDS. Emerging Microbes and Infections. 2019;8:1139–1145. doi: 10.1080/22221751.2019.1644539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H.-J., Dress A.W.M. Split decomposition: a new and useful approach to phylogenetic analysis of distance data. Molecular Phylogenetics and Evolution. 1992;1:242–252. doi: 10.1016/1055-7903(92)90021-8. [DOI] [PubMed] [Google Scholar]
- Bandelt H.J., Forster P., Röhl A. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Benedict K., Beer K.D., Jackson B.R. Histoplasmosis-related healthcare use, diagnosis, and treatment in a commercially insured population, United States. Clinical Infectious Diseases. 2020;70:1003–1010. doi: 10.1093/cid/ciz324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict K., Mody R.K. Epidemiology of histoplasmosis outbreaks, United States, 1938-2013. Emerging Infectious Diseases. 2016;22 doi: 10.3201/eid2203.151117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongomin F., Gago S., Oladele R. Global and multi-national prevalence of fungal diseases—estimate precision. Journal of Fungi. 2017;3:57. doi: 10.3390/jof3040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D., White R.L., Skolnick M. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. American Journal of Human Genetics. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- Brilhante R.S., Ribeiro J.F., Lima R.A. Evaluation of the genetic diversity of Histoplasma capsulatum var. capsulatum isolates from north-eastern Brazil. Journal of Medical Microbiology. 2012;61:1688–1695. doi: 10.1099/jmm.0.044073-0. [DOI] [PubMed] [Google Scholar]
- Brown E.M., McTaggart L.R., Zhang S.X. Phylogenetic analysis reveals a cryptic species Blastomyces gilchristii, sp. nov. within the human pathogenic fungus Blastomyces dermatitidis. PloS One. 2013;8 doi: 10.1371/journal.pone.0059237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D., Moulton V. Neighbor-Net: an agglomerative method for the construction of phylogenetic networks. Molecular Biology and Evolution. 2004;21:255–265. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- Bubnick M., Smulian A.G. The MAT1 locus of Histoplasma capsulatum is responsive in a mating type-specific manner. Eukaryotic Cell. 2007;6:616–621. doi: 10.1128/EC.00020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheema M.S., Christians J.K. Virulence in an insect model differs between mating types in Aspergillus fumigatus. Medical Mycology. 2011;49:202–207. doi: 10.3109/13693786.2010.512301. [DOI] [PubMed] [Google Scholar]
- Christie A., Peterson J.C. Pulmonary calcification in negative reactors to tuberculin. American Journal of Public Health and the Nation’s Health. 1945;35:1131–1147. doi: 10.2105/ajph.35.11.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo A.L., Tobon A., Restrepo A. Epidemiology of endemic systemic fungal infections in Latin America. Medical Mycology. 2011;49:785–798. doi: 10.3109/13693786.2011.577821. [DOI] [PubMed] [Google Scholar]
- Damasceno L.S., Leitao T.M., Taylor M.L. The use of genetic markers in the molecular epidemiology of histoplasmosis: a systematic review. European Journal of Clinical Microbiology and Infectious Diseases. 2016;35:19–27. doi: 10.1007/s10096-015-2508-5. [DOI] [PubMed] [Google Scholar]
- Dantas K.C., Freitas RSd, da Silva M.V. Comparison of diagnostic methods to detect Histoplasma capsulatum in serum and blood samples from AIDS patients. PLoS One. 2018;13 doi: 10.1371/journal.pone.0190408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling S.T. A protozoön general infection producing pseudotubercles in the lungs and focal necroses in the liver, spleen and lymphnodes. JAMA. 1906;XLVI:1283–1285. [Google Scholar]
- de Vienne D.M., Giraud T., Martin O.C. A congruence index for testing topological similarity between trees. Bioinformatics. 2007;23:3119–3124. doi: 10.1093/bioinformatics/btm500. [DOI] [PubMed] [Google Scholar]
- Deepe G.S., Jr. Outbreaks of histoplasmosis: the spores set sail. PLoS Pathogens. 2018;14 doi: 10.1371/journal.ppat.1007213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Terra P.P., Rodrigues A.M., Fernandes G.F. Exploring virulence and immunogenicity in the emerging pathogen Sporothrix brasiliensis. PLoS Neglected Tropical Diseases. 2017;11 doi: 10.1371/journal.pntd.0005903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning D.W. Minimizing fungal disease deaths will allow the UNAIDS target of reducing annual AIDS deaths below 500 000 by 2020 to be realized. Philosophical Transactions of the Royal Society of London B Biological Sciences. 2016;371 doi: 10.1098/rstb.2015.0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dice L.R. Measures of the amount of ecologic association between species. Ecology. 1945;26:297–302. [Google Scholar]
- Dukik K., Muñoz J.F., Jiang Y. Novel taxa of thermally dimorphic systemic pathogens in the Ajellomycetaceae (Onygenales) Mycoses. 2017;60:296–309. doi: 10.1111/myc.12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin M.M., Connolly P.A., Karimi K. Pathogenic differences between North American and Latin American strains of Histoplasma capsulatum var. capsulatum in experimentally infected mice. Journal of Clinical Microbiology. 2004;42:4370–4373. doi: 10.1128/JCM.42.9.4370-4373.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl D.A., vonHoldt B.M. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources. 2012;4:359–361. [Google Scholar]
- Eissenberg L.G., Goldman W.E. Histoplasma variation and adaptive strategies for parasitism: new perspectives on histoplasmosis. Clinical Microbiology Reviews. 1991;4:411–421. doi: 10.1128/cmr.4.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmons C.W. Histoplasmosis: animal reservoirs and other sources in nature of pathogenic fungus, Histoplasma. American Journal of Public Health and the Nation’s Health. 1950;40:436–440. doi: 10.2105/ajph.40.4.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada-Barcenas D.A., Vite-Garin T., Navarro-Barranco H. Genetic diversity of Histoplasma and Sporothrix complexes based on sequences of their ITS1-5.8S-ITS2 regions from the BOLD System. Revista Iberoamericana De Micologia. 2014;31:90–94. doi: 10.1016/j.riam.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Evanno G., Regnaut S., Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Falci D.R., Monteiro A.A., Braz Caurio C.F. Histoplasmosis, an underdiagnosed disease affecting people living with HIV/AIDS in Brazil: results of a multicenter prospective cohort study using both classical mycology tests and histoplasma urine antigen detection. Open Forum Infectious Diseases. 2019;6:ofz073. doi: 10.1093/ofid/ofz073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix B., Roussel S., Pot B. Harmonization of PFGE profile analysis by using bioinformatics tools: example of the Listeria monocytogenes European Union Reference Laboratory network. In: Jordan K., Dalmasso M., editors. Methods in Molecular Biology. Humana Press; New York: 2015. pp. 9–28. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Evolution confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fisher M.C., Koenig G.L., White T.J. Molecular and phenotypic description of Coccidioides posadasii sp. nov., previously recognized as the non-California population of Coccidioides immitis. Mycologia. 2002;94:73–84. [PubMed] [Google Scholar]
- Fogelqvist J., Verkhozina A.V., Katyshev A.I. Genetic and morphological evidence for introgression between three species of willows. BMC Evolutionary Biology. 2015;15:193. doi: 10.1186/s12862-015-0461-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frías De León M.G., Arenas López G., Taylor M.L. Development of specific sequence-characterized amplified region markers for detecting Histoplasma capsulatum in clinical and environmental samples. Journal of Clinical Microbiology. 2012;50:673–679. doi: 10.1128/JCM.05271-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furcow M.L. Recent studies on the epidemiology of histoplasmosis. Annals of the New York Academy of Sciences. 1958;72:129–163. doi: 10.1111/j.1749-6632.1958.tb44226.x. [DOI] [PubMed] [Google Scholar]
- Giacomazzi J., Baethgen L., Carneiro L.C. The burden of serious human fungal infections in Brazil. Mycoses. 2016;59:145–150. doi: 10.1111/myc.12427. [DOI] [PubMed] [Google Scholar]
- Gómez-Rubio V. Ggplot2: elegant graphics for data analysis. Journal of Statistical Software. 2017;2:77. Vol 1, Book Review. [Google Scholar]
- Gomez L.F., Arango M., McEwen J.G. Molecular epidemiology of Colombian Histoplasma capsulatum isolates obtained from human and chicken manure samples. Heliyon. 2019;5 doi: 10.1016/j.heliyon.2019.e02084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarner J., Brandt M.E. Histopathologic diagnosis of fungal infections in the 21st century. Clinical Microbiology Reviews. 2011;24:247–280. doi: 10.1128/CMR.00053-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedes HLdM., Guimarães A.J., Muniz MdM. PCR assay for identification of Histoplasma capsulatum based on the nucleotide sequence of the M antigen. Journal of Clinical Microbiology. 2003;41:535–539. doi: 10.1128/JCM.41.2.535-539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimarães A.J., Nosanchuk J.D., Zancopé-Oliveira R.M. Diagnosis of histoplasmosis. Brazilian Journal of Microbiology. 2006;37:1–13. doi: 10.1590/S1517-83822006000100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen F., Ceresini P.C., Polacheck I. Ancient dispersal of the human fungal pathogen Cryptococcus gattii from the Amazon rainforest. PLoS One. 2013;8 doi: 10.1371/journal.pone.0071148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Huson D.H., Bryant D. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Irinyi L., Serena C., Garcia-Hermoso D. International Society of Human and Animal Mycology (ISHAM)-ITS reference DNA barcoding database--the quality controlled standard tool for routine identification of human and animal pathogenic fungi. Medical Mycology. 2015;53:313–337. doi: 10.1093/mmy/myv008. [DOI] [PubMed] [Google Scholar]
- Jakobsson M., Rosenberg N.A. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Dukik K., Muñoz J.F. Phylogeny, ecology and taxonomy of systemic pathogens and their relatives in Ajellomycetaceae (Onygenales): Blastomyces, Emergomyces, Emmonsia, Emmonsiellopsis. Fungal Diversity. 2018;90:245–291. [Google Scholar]
- Joseph Wheat L. Current diagnosis of histoplasmosis. Trends in Microbiology. 2003;11:488–494. doi: 10.1016/j.tim.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Kasuga T., Taylor J.W., White T.J. Phylogenetic relationships of varieties and geographical groups of the human pathogenic fungus Histoplasma capsulatum Darling. Journal of Clinical Microbiology. 1999;37:653–663. doi: 10.1128/jcm.37.3.653-663.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga T., White T.J., Koenig G. Phylogeography of the fungal pathogen Histoplasma capsulatum. Molecular Ecology. 2003;12:3383–3401. doi: 10.1046/j.1365-294x.2003.01995.x. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman C.A. Histoplasmosis: a clinical and laboratory update. Clinical Microbiology Reviews. 2007;20:115–132. doi: 10.1128/CMR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kiss L. Limits of nuclear ribosomal DNA internal transcribed spacer (ITS) sequences as species barcodes for fungi. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E1811. doi: 10.1073/pnas.1207143109. author reply E1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn L.M. Mechanisms of fungal speciation. Annual Review of Phytopathology. 2005;43:279–308. doi: 10.1146/annurev.phyto.43.040204.135958. [DOI] [PubMed] [Google Scholar]
- Kohonen T. Springer Series in Information Sciences; 2001. Self-organizing maps; p. 502. [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung K.J., Bartlett M.S., Wheat L.J. Distribution of the two mating types among Histoplasma capsulatum isolates obtained from an urban histoplasmosis outbreak. Sabouraudia. 1984;22:155–157. [PubMed] [Google Scholar]
- Kwon-Chung K.J., Edman J.C., Wickes B.L. Genetic association of mating types and virulence in Cryptococcus neoformans. Infection and Immunity. 1992;60:602–605. doi: 10.1128/iai.60.2.602-605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung J.K., Hill W.B. Virulence of the two mating types of Emmonsiella capsulata and the mating experiments with Emmonsiella capsulata var. duboisii. In: Vroey D., Vanbreuseghem R., editors. Sexuality and pathogenicity of fungi. Masson; Paris, France: 1981. pp. 48–56. [Google Scholar]
- Kwon-Chung K.J., Weeks R.J., Larsh H.W. Studies on Emmonsiella capsulata (Histoplasma capsulatum). II. Distribution of the two mating types in 13 endemic states of the United States. American Journal of Epidemiology. 1974;99:44–49. doi: 10.1093/oxfordjournals.aje.a121583. [DOI] [PubMed] [Google Scholar]
- Landaburu F., Cuestas M.L., Rubio A. Genetic diversity of Histoplasma capsulatum strains isolated from Argentina based on nucleotide sequence variations in the internal transcribed spacer regions of rDNA. Mycoses. 2014;57:299–306. doi: 10.1111/myc.12159. [DOI] [PubMed] [Google Scholar]
- Linder K.A., Kauffman C.A. Histoplasmosis: Epidemiology, diagnosis, and clinical manifestations. Current Fungal Infection Reports. 2019:1–9. [Google Scholar]
- Liu B.H. CRC Press; Boca Raton: 1998. Statistical genomics: linkage, mapping, and QTL analysis. [Google Scholar]
- Loulergue P., Bastides F., Baudouin V. Literature review and case histories of Histoplasma capsulatum var. duboisii infections in HIV-infected patients. Emerging Infectious Diseases. 2007;13:1647–1652. doi: 10.3201/eid1311.070665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell C.S., Sepulveda V.E., Turissini D.A. Recent admixture between species of the fungal pathogen Histoplasma. Evolution Letters. 2018;2:210–220. doi: 10.1002/evl3.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniz Mde M., Morais E.S.T.P., Meyer W. Comparison of different DNA-based methods for molecular typing of Histoplasma capsulatum. Applied and Environmental Microbiology. 2010;76:4438–4447. doi: 10.1128/AEM.02004-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniz M.M., Sousa C.N., Evangelista Oliveira M.M. Sexual variability in Histoplasma capsulatum and its possible distribution: what is going on? Revista Iberoamericana De Micologia. 2014;31:7–10. doi: 10.1016/j.riam.2013.10.002. [DOI] [PubMed] [Google Scholar]
- Nacher M., Blanchet D., Bongomin F. Histoplasma capsulatum antigen detection tests as an essential diagnostic tool for patients with advanced HIV disease in low and middle income countries: a systematic review of diagnostic accuracy studies. PLoS Neglected Tropical Diseases. 2018;12 doi: 10.1371/journal.pntd.0006802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacher M., Leitao S.T., Gómez L.B. The fight against HIV-associated disseminated histoplasmosis in the Americas: unfolding the different stories of four centers. Journal of Fungi. 2019;5 doi: 10.3390/jof5020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafzadeh M.J., Sun J., Vicente V.A. Molecular epidemiology of Fonsecaea species. Emerging Infectious Diseases. 2011;17:464–469. doi: 10.3201/1703.100555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negroni P. A mycological study of the first South American case of histoplasmosis. Revista del Instituto Bacteriologico. 1940;9:239–294. [Google Scholar]
- Nei M. Columbia University Press; New York: 1987. Molecular evolutionary genetics; p. 512. [Google Scholar]
- Nieuwenhuis B.P.S., James T.Y. The frequency of sex in fungi. Philosophical Transactions of the Royal Society of London B Biological Sciences. 2016;371:20150540. doi: 10.1098/rstb.2015.0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oladele R.O., Ayanlowo O.O., Richardson M.D. Histoplasmosis in Africa: an emerging or a neglected disease? PLoS Neglected Tropical Diseases. 2018;12 doi: 10.1371/journal.pntd.0006046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakasa N., Biber A., Nsiangana S. African histoplasmosis in HIV-negative patients, kimpese, Democratic Republic of the Congo. Emerging Infectious Diseases. 2018;24:2068–2070. doi: 10.3201/eid2411.180236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualotto A.C., Queiroz-Telles F. Histoplasmosis dethrones tuberculosis in Latin America. Lancet Infectious Diseases. 2018;18:1058–1060. doi: 10.1016/S1473-3099(18)30373-6. [DOI] [PubMed] [Google Scholar]
- Powell W., Morgante M., Andre C. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Molecular Breeding. 1996;2:225–238. [Google Scholar]
- Prevost A., Wilkinson M.J. A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theoretical and Applied Genetics. 1999;98:107–112. [Google Scholar]
- Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queiroz-Telles F., Fahal A.H., Falci D.R. Neglected endemic mycoses. Lancet Infectious Diseases. 2017;17:e367–e377. doi: 10.1016/S1473-3099(17)30306-7. [DOI] [PubMed] [Google Scholar]
- Retchless A.C., Lawrence J.G. Phylogenetic incongruence arising from fragmented speciation in enteric bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11453–11458. doi: 10.1073/pnas.1001291107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley W.A., Watson C.J. Histoplasmosis of Darling. American Journal of Tropical Medicine and Hygiene. 1926;s1-6:271–282. [Google Scholar]
- Rocher G. Research on the existence of histoplasmic and coccidioidal infections in France. Journal Français de Medecine et Chirurgie Thoraciques. 1950;4:293–297. [PubMed] [Google Scholar]
- Rodrigues A.M., de Hoog G.S., Zhang Y. Emerging sporotrichosis is driven by clonal and recombinant Sporothrix species. Emerging Microbes and Infections. 2014;3:e32. doi: 10.1038/emi.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Arellanes G., Nascimento de Sousa C., de Medeiros-Muniz M. Frequency and genetic diversity of the MAT1 locus of Histoplasma capsulatum isolates in Mexico and Brazil. Eukaryotic Cell. 2013;12:1033–1038. doi: 10.1128/EC.00012-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J., Ferrer-Mata A., Sánchez-DelBarrio J.C. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- Sahaza J.H., Duarte-Escalante E., Canteros C. Analyses of the genetic diversity and population structures of Histoplasma capsulatum clinical isolates from Mexico, Guatemala, Colombia and Argentina, using a randomly amplified polymorphic DNA-PCR assay. Epidemiology and Infection. 2019;147 doi: 10.1017/S0950268819000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma D.K., Prakash A., O'Loughlin S.M. Genetic population structure of the malaria vector Anopheles baimaii in north-east India using mitochondrial DNA. Malaria Journal. 2012;11:76. doi: 10.1186/1475-2875-11-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardl C.L., Craven K.D. Interspecific hybridization in plant-associated fungi and oomycetes: a review. Molecular Ecology. 2003;12:2861–2873. doi: 10.1046/j.1365-294x.2003.01965.x. [DOI] [PubMed] [Google Scholar]
- Selim S.A., Soliman R., Osman K. Studies on histoplasmosis farciminosi (epizootic lymphangitis) in Egypt. European Journal of Epidemiology. 1985;1:84–89. doi: 10.1007/BF00141797. [DOI] [PubMed] [Google Scholar]
- Sepúlveda V.E., Márquez R., Turissini D.A. Genome sequences reveal cryptic speciation in the human pathogen Histoplasma capsulatum. MBio. 2017;8 doi: 10.1128/mBio.01339-17. e01339-01317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepúlveda V.E., Williams C.L., Goldman W.E. Comparison of phylogenetically distinct Histoplasma strains reveals evolutionarily divergent virulence strategies. MBio. 2014;5 doi: 10.1128/mBio.01376-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedmousavi S., Bosco SdMG., de Hoog S. Fungal infections in animals: a patchwork of different situations. Medical Mycology. 2018;56:S165–S187. doi: 10.1093/mmy/myx104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stielow J.B., Lévesque C.A., Seifert K.A. One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia. 2015;35:242–263. doi: 10.3767/003158515X689135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenbrock E.H. The role of hybridization in the evolution and emergence of new fungal plant pathogens. Phytopathology. 2016;106:104–112. doi: 10.1094/PHYTO-08-15-0184-RVW. [DOI] [PubMed] [Google Scholar]
- Taylor M.L., Chavez-Tapia C.B., Rojas-Martinez A. Geographical distribution of genetic polymorphism of the pathogen Histoplasma capsulatum isolated from infected bats, captured in a central zone of Mexico. FEMS Immunology and Medical Microbiology. 2005;45:451–458. doi: 10.1016/j.femsim.2005.05.019. [DOI] [PubMed] [Google Scholar]
- Team R.C. R Foundation for Statistical Computing; Vienna, Austria: 2014. R: A Language and Environment for Statistical Computing.http://wwwR-projectorg/ [Google Scholar]
- Teixeira MdM., Patané J.S.L., Taylor M.L. Worldwide phylogenetic distributions and population dynamics of the genus Histoplasma. PLoS Neglected Tropical Diseases. 2016;10 doi: 10.1371/journal.pntd.0004732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MdM., Rodrigues A.M., Tsui C.K.M. Asexual propagation of a virulent clone complex in human and feline outbreak of sporotrichosis. Eukaryotic Cell. 2015;14:158–169. doi: 10.1128/EC.00153-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier C., David J., This P. Optimization of the choice of molecular markers for varietal identification in Vitis vinifera L. Theoretical and Applied Genetics. 1999;98:171–177. [Google Scholar]
- Turissini D.A., Gomez O.M., Teixeira M.M. Species boundaries in the human pathogen Paracoccidioides. Fungal Genetics and Biology. 2017;106:9–25. doi: 10.1016/j.fgb.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valero C., Gago S., Monteiro M.C. African histoplasmosis: new clinical and microbiological insights. Medical Mycology. 2018;56:51–59. doi: 10.1093/mmy/myx020. [DOI] [PubMed] [Google Scholar]
- Van Dyke M.C.C., Teixeira M.M., Barker B.M. Fantastic yeasts and where to find them: the hidden diversity of dimorphic fungal pathogens. Current Opinion in Microbiology. 2019;52:55–63. doi: 10.1016/j.mib.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney R.K., Chabane K., Hendre P.S. Comparative assessment of EST-SSR, EST-SNP and AFLP markers for evaluation of genetic diversity and conservation of genetic resources using wild, cultivated and elite barleys. Plant Science. 2007;173:638–649. [Google Scholar]
- Vos P., Hogers R., Bleeker M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T.J., Bruns T., Lee S. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M., Gelfand D., Shinsky J., White T., editors. PCR Protocols: A Guide to Methods and Applications. Academic Press; New York: 1990. pp. 315–322. [Google Scholar]
- Xu W., Liang G., Peng J. The influence of the mating type on virulence of Mucor irregularis. Scientific Reports. 2017;7:10629. doi: 10.1038/s41598-017-10954-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zancope-Oliveira R.M., Morais e Silva Tavares P., Muniz M.M. Genetic diversity of Histoplasma capsulatum strains in Brazil. FEMS Immunology and Medical Microbiology. 2005;45:443–449. doi: 10.1016/j.femsim.2005.05.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.