Abstract
Purpose
To evaluate the noninvasive prenatal testing (NIPT) results of 36,913 cases in Jiangxi province of central China and explore its application value in prenatal screening and diagnosis.
Methods
This retrospective analysis included 36,913 singleton pregnant women who underwent NIPT because of moderate-/high-risk pregnancy or voluntary requirements between January 2017 and December 2019 in our hospital. Chromosomal abnormalities such as trisomies 21, 18, and 13 (T21, T18, T13) and sex chromosome aneuploidies (SCAs) were judged by standard Z-score analysis. Positive NIPT results were confirmed by amniocentesis and karyotyping. Pregnancy outcomes were followed up via telephone interview.
Results
A total of 1.01% (371/36,913) positive cases were detected by NIPT, comprising 137, 46, 31, and 157 cases of T21, T18, T13, and SCAs, respectively. A total of 116 of T21, 27 of T18, 13 of T13, and 51 of SCAs were confirmed to be true positive; all normal cases that had been followed up were verified to be true negative. The NIPT sensitivity in T21, T18, T13, and SCAs was 100.00% individually, whereas the specificity was 99.94% (36,488/36,509), 99.95% (36,579/36,598), 99.95% (36,594/36,612), and 99.72% (36,472/36,574), respectively. Furthermore, the negative predictive values of T21, T18, T13, and SCAs were all 100%, while the positive predictive values were 84.67%, 58.70%, 41.94%, and 33.33%, respectively.
Conclusion
NIPT is highly sensitive and has a low false positive rate in testing clinically significant fetal aneuploidies of general reproductive women. However, this technique cannot substitute for amniocentesis and karyotyping, and detailed genetic counseling is also essential for the high-risk group of NIPT.
Keywords: Noninvasive prenatal testing (NIPT), Chromosomal abnormalities, Prenatal diagnosis, Performance
Introduction
Chromosomal abnormalities, which include trisomy 21 (T21), trisomy 18 (T18), trisomy 13 (T13), and sex chromosome aneuploidies (SCAs), are the main causes of birth defects, especially in pregnancies of numerous older women in China, considering their “two-child policy” [1]. These chromosomal diseases often result in mental retardation and growth or developmental delay, accompanied by severe deformity of facial features, limbs, or other aspects. Unfortunately, no curative treatment is currently available for such birth defects. Therefore, an accurate and effective method that detects fetuses with high chromosomal aneuploidy risk is necessary to reduce birth defects. Serological screening is the traditional method of prenatal screening, but it has a low detection rate, high false positive rate (FPR), and low positive predictive value (PPV) [2]. Other prenatal diagnosis methods, such as amniocentesis, chorionic villus sampling, and umbilical cord blood sampling, are generally accurate, but the high risk of procedure-related miscarriage may limit their clinical application [3, 4].
Since cell-free DNA (cfDNA) was detected in the peripheral blood of pregnant women by Lo YM in 1997 [5], and with the rapid development of high-throughput sequencing technology in recent years, noninvasive prenatal testing (NIPT) has been gradually developed to analyze cfDNA and deduce the presence of fetal chromosomal abnormalities through amplification, high-throughput sequencing technology, and bioinformatics processing [6]. Compared with serological screening, sonographic screening, and other traditional interventional prenatal diagnosis methods, NIPT is noninvasive, has high sensitivity, and can avoid the risk of abortion, infection, or injury caused by invasive interventional operations [7]. Therefore, NIPT is rapidly being employed to detect the fetal chromosome aneuploidies of T21, T18, T13, and SCAs around the world. The American College of Medical Genetics and Genomics (ACMG), along with several other professional associations, has issued statements and guidelines suggesting that NIPT is a screening test to identify pregnancies at risk for common autosomal aneuploidies [8–11]. In addition, a continuously updated meta-analysis of Gil MM showed that screening by analysis of cfDNA in maternal blood (including NIPT) in singleton pregnancies could detect > 99% of fetuses with T21, 98% of fetuses with T18, and 99% of fetuses with T13 at a combined FPR of 0.13%, and the performance of cfDNA testing for T21 in twin pregnancy is similar to that reported in singleton pregnancy [12, 13]. Since NIPT was proposed as a screening test in China in 2011, several studies from multiple centers have reported that it can offer a perfect performance for detecting chromosomal abnormalities of fetus [14–16]. A recent study of 31,515 singleton pregnancies in southeastern China showed a high sensitivity and specificity for T21, T18, T13, and SCAs, and the positive predictive values were high for T21 and T18 and moderate for T13 and SCAs [16]. Therefore, NIPT has been implemented into public healthcare systems as either a first-line test or a supplement to the existing prenatal screening; more and more pregnant women are willing to choose NIPT [17–19].
However, large-scale NIPT performance in the general population of central China remains uninvestigated. In this study, we aimed to retrospectively analyze and follow up 36,913 pregnant women who received NIPT to assess the accuracy and feasibility of NIPT in prenatal testing and find the possible reasons for false-positive and false-negative results.
Materials and methods
Study subjects
Singleton pregnant women who came to Prenatal Diagnosis Center of Jiangxi Maternal and Child Health Hospital between January 2017 and December 2019 were recruited for this study. All the participants had undergone pretest counseling and provided an informed written consent before blood sample collection. Based on maternal age and other risk factors, we categorized subjects into four groups as follows: advanced maternal age (≥ 35 years) with high risk, advanced maternal age with low risk, normal maternal age (< 35 years) with high risk, and normal maternal age with low risk. In this study, “high risk” was defined as a high/moderate risk of Down syndrome, the presence of ultrasound soft markers, or a history of previous Down syndrome pregnancy. In contrast, the absence of any of these risk factors defined “low risk”.
Sample collection and preparation
Approximately 5 mL of peripheral blood was collected from the pregnant women in tubes primed with ethylenediaminetetraacetic acid. Within 96 h after blood collection, plasma was isolated from the blood samples in accordance with the two-step centrifugation protocol [20]. The samples were first centrifuged at 1600g under 4 °C for 10 min. Then, the supernatant was collected and re-centrifuged at 16,000g for 10 min to remove the remaining white blood cells or cell debris. Thereafter, plasma samples were frozen at – 80 °C.
cfDNA extraction and DNA sequencing
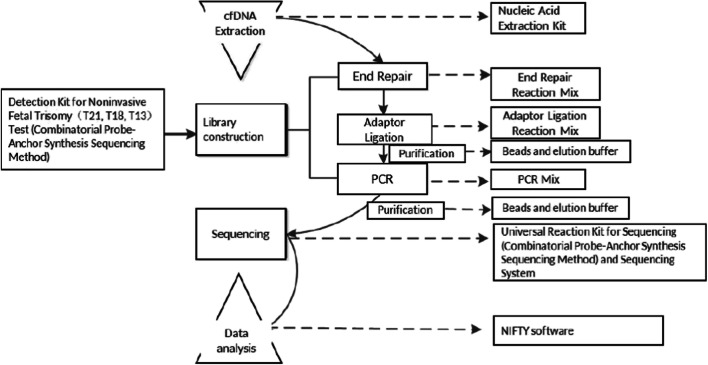
Plasma cfDNA was extracted from the isolated plasma samples according to the manufacturer’s instructions of Nucleic Acid Extraction Kit (Beijing Genomics Institution, BGI, China), and the concentration of cfDNA was measured with a Qubit™ Fluorometer (Thermo Fisher Scientific, USA). Combinatorial probe-anchor synthesis sequencing method was used for DNA sequencing; 96 libraries were sequenced with 45-cycle single-end sequencing on BGISEQ-500 platforms. After the specific adaptor ligation, amplification, and sequencing of cfDNA, we obtained the effective reads of each chromosome by sequencing alignment. Through comparison with cut-off values, fetal chromosomal aneuploidies (T21, T18, and T13) could be identified [21]. Figure 1 illustrates the detection process based on the BGI user manual.
Fig. 1.
The workflow of detection process detection kit for noninvasive fetal trisomy (T21, T18, T13) test (combinatorial probe-anchor synthesis sequencing method) utilizes combinatorial probe-anchor synthesis sequencing method to detect cell-free fetal DNA in maternal plasma samples. After the specific adaptor ligation, amplification, purification, and sequencing of cell-free DNA, the effective reads of each chromosome are obtained through sequencing alignment. Through comparison with cut-off values, fetal chromosomal aneuploidy can be detected.
Prenatal diagnosis and pregnancy follow-up
When the Z-score of chromosomes 21, 18, or 13 of the sample to be detected is ≥ 3, it is considered positive. Every participant received genetic counseling after NIPT screening. Subjects with positive NIPT results were suggested to be verified with invasive testing for prenatal diagnosis. To obtain information about neonatal outcome and newborn growth, we followed up all participants via telephone interviews 1 month after the expected date of delivery.
Data and statistical analysis
To characterize NIPT performance, we calculated the sensitivity, specificity, FPR/false negative rate (FNR), and predictive values through the following formulas. Sensitivity = TP/(TP + FN), specificity = TN/(FP + TN), FNR = FN/all cases, FPR = FP/all cases, PPV = TP/(TP + FP), and negative predictive value (NPV) = TN/(FN + TN). TP, FN, TN, and FP stand for true positive, false negative, true negative, and false positive, respectively. All statistical data were analyzed using the statistical software SPSS version 19.0, difference of positive rate between advanced maternal age and normal age was tested for statistical significance using Chi-square test, and P < 0.05 was considered statistically significant.
Results
Patient characteristics and clinical indication
Between January 2017 and December 2019, 37,006 singleton pregnant women were recruited to the study. The median maternal age was 29 years (range 18 to 54 years), and the median gestational age was 17 + 4 weeks (range 12 to 32 weeks). However, 306 (0.83%) cases required repeat blood sampling because of borderline Z-score or low fetal fraction, and 213 of them obtained effective NIPT results. Ultimately, 36,913 samples that obtained informative results were included in this study while 93 (0.25%) were excluded.
In total, we identified 371 positive NIPT results from the 36,913 pregnancies, yielding an overall abnormal rate of 1.01%. The advanced maternal age (≥ 35 years) with high-risk group had the highest rate (1.88%) of chromosomal abnormalities, whereas the normal age (< 35 years) with low-risk group had the lowest (0.69%). The positive rate was significantly higher in advanced maternal age than in normal age (1.30% vs. 0.90%, P < 0.05) (Table 1).
Table 1.
Detection of fetal aneuploidies in different indications
| Indications | Detected number | Positive number | Positive rate (%) |
|---|---|---|---|
| Advanced maternal age | 9516 | 124 | 1.30 |
| With high risk | 1118 | 21 | 1.88 |
| With low risk | 8398 | 103 | 1.23 |
| Normal maternal age | 27397 | 247 | 0.90 |
| With high risk | 12575 | 145 | 1.15 |
| With low risk | 14822 | 102 | 0.69 |
| Total | 36913 | 371 | 1.01 |
Chromosomal abnormality and prenatal diagnosis
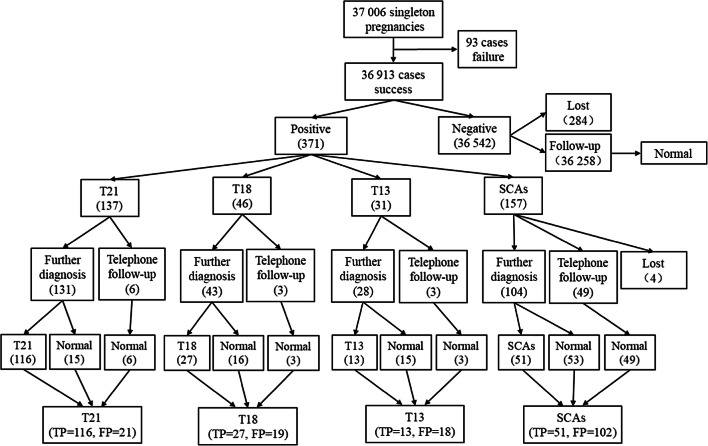
Among the 371 positive cases identified by NIPT, the most common chromosomal abnormality was SCAs (157 cases, 42.32%), followed by T21 (137 cases, 36.93%), T18 (46 cases, 12.40%), and T13 (31 cases, 8.35%). Positive NIPT results were verified by amniocentesis and karyotyping. After genetic counseling, 277 pregnancies underwent prenatal diagnostic testing, and 190 cases were confirmed to be true positive (105, 25, 12, and 48 cases of T21, T18, T13, and SCAs, respectively) (Table 2). Of the 94 pregnancies who refused to take the prenatal diagnostic testing, four cases were lost to follow-up, 29 cases had received karyotyping of their newborns, and 17 cases had chromosome abnormalities (11, 2, 1, and 3 cases of T21, T18, T13, and SCAs, respectively) (Table 3). Figure 2 presents a complete flowchart of NIPT results and follow-up.
Table 2.
Fetal karyotypes of NIPT positives
| Prenatal diagnosis (number) | Fetal karyotypes | Number |
|---|---|---|
| T21 (117) | 47,XN,+21 | 96 |
| 47,XN,+21/46,XN | 6 | |
| 46,XN,rob(13;21),+21 | 2 | |
| 46,XN,rob(14;21),+21 | 1 | |
| 46,XN | 12 | |
| T18 (40) | 47,XN,+18 | 25 |
| 46,XN | 15 | |
| T13 (26) | 47,XN,+13 | 12 |
| 46,XN | 14 | |
| SCAs (94) | 45,X | 5 |
| 47,XXX | 10 | |
| 47,XXY | 17 | |
| 47,XYY | 3 | |
| 45,X/46,XX | 2 | |
| 47,XXX/46,XX | 4 | |
| 46,X,del(X) | 2 | |
| 45,X/46,X,i(X)(q10) | 1 | |
| 46,X,add(X) | 3 | |
| 46,X,psu dic(X)(p11.2) | 1 | |
| 46,XN | 46 |
Table 3.
Follow-up of all NIPT positives
| T21 | T18 | T13 | SCAs | |
|---|---|---|---|---|
| Amniocentesis karyotypes | 117 | 40 | 26 | 94 |
| Abnormal | 105 | 25 | 12 | 48 |
| Normal | 12 | 15 | 14 | 46 |
| Newborn karyotypes | 14 | 3 | 2 | 10 |
| Abnormal | 11 | 2 | 1 | 3 |
| Normal | 3 | 1 | 1 | 7 |
| No karyotyping | 6 | 3 | 3 | 49 |
| Total | 137 | 46 | 31 | 153a |
aFour cases were lost to follow-up
Fig. 2.
Flowchart of NIPT results and follow-up
Performance of NIPT for detecting fetal chromosomal aneuploidies
Based on the results of amniocyte karyotyping and newborn follow-up, 116 out of 137 (84.67%) cases for T21, 27 out of 46 (58.70%) cases for T18, 13 out of 31 (41.94%) cases for T13, and 51 out of 153 (33.33%) cases for SCAs were confirmed to be true positive; all normal cases of NIPT were verified to be true negative, except 284 cases, which had been lost to follow-up. To evaluate the performance of NIPT, we calculated key parameters such as sensitivity, specificity, FPR, FNR, PPV, and NPV. The sensitivity and NPV for T21, T18, T13, and SCAs were all 100%, whereas the FN and FNR were 0. The overall specificity for detecting these four chromosomal anomalies ranged from 99.72 to 99.95%, and the PPV for T21, T18, T13, and SCAs was 84.67%, 58.70%, 41.94%, and 33.33%, respectively (Table 4).
Table 4.
Performance of NIPT for detecting fetal chromosomal aneuploidies
| Chromosome abnormality | TP | FP | TN | FN | Sensitivity (%) | Specificity (%) | FPR (%) | FNR (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| T21 | 116 | 21 | 36,488 | 0 | 100 | 99.94 | 0.06 | 0 | 84.67 | 100 |
| T18 | 27 | 19 | 36,579 | 0 | 100 | 99.95 | 0.05 | 0 | 58.70 | 100 |
| T13 | 13 | 18 | 36,594 | 0 | 100 | 99.95 | 0.05 | 0 | 41.94 | 100 |
| SCAs | 51 | 102 | 36,472 | 0 | 100 | 99.72 | 0.28 | 0 | 33.33 | 100 |
Discussion and conclusion
Chromosomal abnormalities, with an increasing incidence of approximately 1.5 in 100 births in China, constitute one of the major causes of birth defects [22, 23]. A valid and accurate prenatal screening or diagnostic method is necessary for the prenatal diagnosis, in order to reduce the incidence of birth defects and improve the quality of the population. In 2016, the American Society of Medical Genetics suggested that NIPT could be used as a first-line screening method for T21, T18, and T13 in all pregnant women [8]. Since then, NIPT has been widely used for the prenatal screening of T21, T18, T13, and SCAs, and an increasing number of studies have successfully evaluated the performance of NIPT around the world [8–10, 14–16]. However, a large-scale clinical study on the efficacy of NIPT in Jiangxi province of central China is still unavailable. Therefore, we hope that the present study, which includes 36,913 cases, can provide a satisfactory performance report to settle this issue.
This study identified 371 high-risk patients by NIPT, among which 306 received further diagnosis and 207 were confirmed, and 21, 19, 18, and 102 cases of T21, T18, T13, and SCAs respectively, were confirmed to be false positive. Meanwhile, except for 284 cases which were lost to follow-up, no false-negative result was found. Overall, the sensitivity and specificity of NIPT for detecting chromosomal abnormalities were high, reaching 100% and 99.94% for T21, 100% and 99.95% for T18 and T13, and 100% and 99.72% for SCAs, consistent with those reported in previous studies in China [18, 24]. Furthermore, the PPVs of T21, T18, T13, and SCAs were 84.67%, 58.70%, 41.94%, and 33.33%, respectively, indicating a high PPV for T21, a moderate PPV for T18, and a low PPV for T13 and SCAs. Hence, some of the positive cases turned out to be false positive. Although our PPV results are consistent with previous reports worldwide [25–27], several reasons contribute to having false-positive results, including maternal chromosomal abnormality [28, 29], confined placental mosaicism [30, 31], vanishing twin [32], maternal copy number variations [33], fetal fraction [34], and so on. In addition, a short-term telephone follow-up to ascertain whether the newborns are normal or not is not entirely reliable and can also contribute to false-positive results. In our study, 17.5% (65 cases of 371) of pregnant women with positive NIPT results refused to undergo chromosomal diagnosis, excluding 4 cases lost to follow-up, 61 cases were identified to have normal newborns merely by telephone follow-up in 3 months after delivery, which were too early to judge the chromosomal abnormalities, especially for SCAs. Moreover, the lower guanine/cytosine deoxyribonucleotide content of chromosome 13 can cause the sequencing bias and may relate to the lower PPV of T13 [35].
Serological screening for trisomies based on maternal serum-free β-human chorionic gonadotrophin (β-HCG), pregnancy-associated plasma protein-A (PAPP-A), and α-fetoprotein (AFP) levels is still widely used in China. These biochemical markers, combined with maternal age, are usually used for screening for Down syndrome and can achieve a detection rate of 70–80% but a PPV of almost 10% [19]. Compared with NIPT for detection of T21 in our study, the sensitivity and specificity of serological screening seemed unsatisfactory. If all of the samples in our study had been detected by serological screening, more T21 fetuses would have been missed, and more false-positive cases would have been detected, resulting in additional amniocentesis; thus, more pregnancies would likely have been lost after invasive operation. Therefore, NIPT has shown more accuracy than serological screening, and ACMG have proposed NIPT to be offered for pregnant women of different ages.
With the full liberalization of the two-child policy and the popularization of late marriage and late childbirth, the proportion of older pregnant women (≥ 35 years old) in China is increasing. Older pregnant women are vulnerable to the influence of internal and external environment, ovarian function degeneration, and egg aging. Consequently, they are at an increased risk of chromosomal variation in the embryonic stage [36, 37]. Thus, pregnancy at an advanced age has become an independent risk factor for chromosomal aneuploidy formation [38]. According to the latest guideline about the management of prenatal diagnosis technology in China, primiparous women beyond 35 years old should undergo an invasive diagnostic procedure, and NIPT should be used modestly. In our study, 9516 pregnant women with an advanced age (≥ 35 years) still selected NIPT as the prenatal screening method, and 124 of them obtained positive NIPT results (1.30%), which were significantly higher than those with a normal maternal age (0.90%), suggesting a statistically significant difference (P < 0.05). Pregnant women with an advanced age can also acquire a satisfactory NIPT performance, indicating that NIPT can be offered as a prenatal screening method for any maternal age, along with the stress brought on by the two-child policy to the prenatal diagnosis system in China. Through NIPT application, almost 72% of older pregnant women were prevented from undergoing invasive tests, thereby leading to a 91% reduction in the number of procedure-related pregnancy loss [39]. Thus, adequate genetic counseling is necessary for the screening of older pregnant women, and NIPT can be used as the main screening method to detect and diagnose fetal chromosomal abnormalities early and effectively reduce the birth of children with defects.
However, this study has several limitations that should be clarified. Although no false-negative result was found in our study, 284 negative cases of NIPT were lost to follow-up. Additionally, observing false-negative results merely by the appearance of newborns is difficult. Although the International Society for Prenatal Diagnosis recommends NIPT as the first-line prenatal screening test for all pregnant women, most of our subjects still selected NIPT as the second-line screening test, main reason may be because of its high cost. Nevertheless, the market price of NIPT has already reduced to nearly 1800RMB (US$266) in our city; it is still higher than serological screening (approximately $31), but clearly less expensive than amniocentesis karyotype (approximately $960). Thus, more and more parents will choose NIPT as their first-line prenatal screening test in the future. In addition, when the content of cfDNA is below 4.0%, NIPT may fail or result in false-positive and false-negative results [40]. Our study initially examined 37,006 single fetal pregnancies, and 93 of them failed to obtain effective results because the fetal concentration was below 4.0%, with a failure rate of 0.25%, which is lower than that reported in previous studies [41–43]. Therefore, combining invasive prenatal diagnosis is necessary to avoid false positives. Overall, with the increase in the quantity of test specimens and the information analysis method of optimizing of large-scale genome sequencing technology applied in fetal chromosomal aneuploidy, NIPT will have a wide prospect of clinical application.
In summary, NIPT has great advantages in accuracy, specificity, and acceptance of pregnant women, especially in T21 screening; thus, it can effectively avoid birth defect occurrence and improve the quality of the birth population. Notably, NIPT is a prenatal screening method but not a substitute for prenatal diagnosis. Establishing a perfect genetic counseling system is crucial, and clinicians should correctly understand the advantages and limitations of NIPT technology in testing before and after detailed genetic counseling for pregnant women. In clinical application, this study, which focuses on the individual situation of pregnant women, fully informs the detection range and limitations of NIPT in pregnant women according to the need of clinical application. However, further research is required to determine how to integrate the data of NIPT prenatal screening in various regions into the national gene bank to improve the accuracy of NIPT prenatal screening and reduce the testing cost.
Authors’ contributions
Wan Lu and Yan-Qiu Liu conceived and designed research. Wan Lu and Ting Huang conducted and performed experiments. Xin-Rong Wang, Ji-Hui Zhou, Hui-Zhen Yuan, and Yan Yang contributed to track the objects of research. Ting-Ting Huang and Dan-Ping Liu analyzed data. Wan Lu wrote the manuscript. All authors read and approved the manuscript.
Data availability
Completely transparent.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Code availability
Not applicable.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Teng X, Shane MI, Pan S. The changing situation about maternal age, risk factors and pregnancy outcomes after the two-child policy: a retrospective cohort study. Ann Palliat Med. 2020;9(3):824-34. doi:10.21037/apm.2020.04.27. [DOI] [PubMed]
- 2.Carlson LM, Vora NL. Prenatal diagnosis: screening and diagnostic tools. Obstet Gynecol Clin North Am. 2017;44(2):245–256. doi: 10.1016/j.ogc.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Enzensberger C, Pulvermacher C, Degenhardt J, Kawacki A, Germer U, Gembruch U, Krapp M, Weichert J, Axt-Fliedner R. Fetal loss rate and associated risk factors after amniocentesis, chorionic villus sampling and fetal blood sampling. Ultraschall Med. 2012;33(7):E75–EE9. doi: 10.1055/s-0031-1299388. [DOI] [PubMed] [Google Scholar]
- 4.Cheng Y, Leung WC, Leung TY, Choy KW, Chiu R, Lo TK, Kwok KY, Sahota DS. Women's preference for non-invasive prenatal DNA testing versus chromosomal microarray after screening for Down syndrome: a prospective study. BJOG. 2018;125(4):451–459. doi: 10.1111/1471-0528.15022. [DOI] [PubMed] [Google Scholar]
- 5.Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350(9076):485–487. doi: 10.1016/s0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 6.Lo YM. Noninvasive prenatal detection of fetal chromosomal aneuploidies by maternal plasma nucleic acid analysis: a review of the current state of the art. BJOG. 2009;116(2):152–157. doi: 10.1111/j.1471-0528.2008.02010.x. [DOI] [PubMed] [Google Scholar]
- 7.Vossaert L, Wang Q, Salman R, McCombs AK, Patel V, Qu C, et al. Validation studies for single circulating trophoblast genetic testing as a form of noninvasive prenatal diagnosis. Am J Hum Genet. 2019;105(6):1262–1273. doi: 10.1016/j.ajhg.2019.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gregg AR, Skotko BG, Benkendorf JL, Monaghan KG, Bajaj K, Best RG, et al. Noninvasive prenatal screening for fetal aneuploidy, 2016 update: a position statement of the American College of Medical Genetics and Genomics. Genet Med. 2016;18(10):1056–1065. doi: 10.1038/gim.2016.97. [DOI] [PubMed] [Google Scholar]
- 9.Manotaya S, Xu H, Uerpairojkit B, Chen F, Charoenvidhya D, Liu H, Petcharaburanin N, Liu Y, Tang S, Wang X, Dansakul S, Thomsopa T, Gao Y, Zhang H, Xu H, Jiang H. Clinical experience from Thailand: noninvasive prenatal testing as screening tests for trisomies 21, 18 and 13 in 4736 pregnancies. Prenat Diagn. 2016;36(3):224–231. doi: 10.1002/pd.4775. [DOI] [PubMed] [Google Scholar]
- 10.Oepkes D, Page-Christiaens GC, Bax CJ, Bekker MN, Bilardo CM, Boon EM, et al. Trial by Dutch laboratories for evaluation of non-invasive prenatal testing. Part I-Clinical Impact. Prenat Diagn. 2016;36(12):1083–1090. doi: 10.1002/pd.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehrich M, Tynan J, Mazloom A, Almasri E, McCullough R, Boomer T, Grosu D, Chibuk J. Genome-wide cfDNA screening: clinical laboratory experience with the first 10,000 cases. Genet Med. 2017;19(12):1332–1337. doi: 10.1038/gim.2017.56. [DOI] [PubMed] [Google Scholar]
- 12.Gil MM, Accurti V, Santacruz B, Plana MN, Nicolaides KH. Analysis of cell-free DNA in maternal blood in screening for aneuploidies: updated meta-analysis. Ultrasound Obstet Gynecol. 2017;50(3):302–314. doi: 10.1002/uog.17484. [DOI] [PubMed] [Google Scholar]
- 13.Gil MM, Galeva S, Jani J, Konstantinidou L, Akolekar R, Plana MN, Nicolaides KH. Screening for trisomies by cfDNA testing of maternal blood in twin pregnancy: update of The Fetal Medicine Foundation results and meta-analysis. Ultrasound Obstet Gynecol. 2019;53(6):734–742. doi: 10.1002/uog.20284. [DOI] [PubMed] [Google Scholar]
- 14.Hu H, Wang L, Wu J, Zhou P, Fu J, Sun J, Cai W, Liu H, Yang Y. Noninvasive prenatal testing for chromosome aneuploidies and subchromosomal microdeletions/microduplications in a cohort of 8141 single pregnancies. Hum Genomics. 2019;13(1):14. doi: 10.1186/s40246-019-0198-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian C, Deng T, Zhu X, Gong C, Zhao Y, Wei Y, Li R, Xu X, He M, Zhang Z, Cheng J, BenWillem M, Qiao J. Evidence of compliance with and effectiveness of guidelines for noninvasive prenatal testing in China: a retrospective study of 189,809 cases. Sci China Life Sci. 2020;63(3):319–328. doi: 10.1007/s11427-019-9600-0. [DOI] [PubMed] [Google Scholar]
- 16.Xu L, Huang H, Lin N, Wang Y, He D, Zhang M, Chen M, Chen L, Lin Y. Non-invasive cell-free fetal DNA testing for aneuploidy: multicenter study of 31 515 singleton pregnancies in southeastern China. Ultrasound Obstet Gynecol. 2020;55(2):242–247. doi: 10.1002/uog.20416. [DOI] [PubMed] [Google Scholar]
- 17.McCullough RM, Almasri EA, Guan X, Geis JA, Hicks SC, Mazloom AR, et al. Non-invasive prenatal chromosomal aneuploidy testing--clinical experience: 100,000 clinical samples. PloS One. 2014;9(10):e109173. doi: 10.1371/journal.pone.0109173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu H, Liu H, Peng C, Deng T, Fu X, Chung C, Zhang E, Lu C, Zhang K, Liang Z, Yang Y. Clinical Experience of Non-Invasive Prenatal Chromosomal Aneuploidy Testing in 190,277 Patient Samples. Curr Mol Med. 2016;16(8):759–766. doi: 10.2174/1566524016666161013142335. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Liu H, He Y, Xu W, Ma Q, He Y, Lei W, Chen G, He Z, Huang J, Liu J, Liu Y, Huang Q, Yu F. Clinical performance of non-invasive prenatal served as a first-tier screening test for trisomy 21, 18, 13 and sex chromosome aneuploidy in a pilot city in China. Hum Genomics. 2020;14(1):21. doi: 10.1186/s40246-020-00268-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dan S, Wang W, Ren J, Li Y, Hu H, Xu Z, Lau TK, Xie J, Zhao W, Huang H, Xie J, Sun L, Zhang X, Wang W, Liao S, Qiang R, Cao J, Zhang Q, Zhou Y, Zhu H, Zhong M, Guo Y, Lin L, Gao Z, Yao H, Zhang H, Zhao L, Jiang F, Chen F, Jiang H, Li S, Li Y, Wang J, Wang J, Duan T, Su Y, Zhang X. Clinical application of massively parallel sequencing-based prenatal noninvasive fetal trisomy test for trisomies 21 and 18 in 11,105 pregnancies with mixed risk factors. Prenat Diagn. 2012;32(13):1225–1232. doi: 10.1002/pd.4002. [DOI] [PubMed] [Google Scholar]
- 21.Chen S, Lau TK, Zhang C, Xu C, Xu Z, Hu P, Xu J, Huang H, Pan L, Jiang F, Chen F, Pan X, Xie W, Liu P, Li X, Zhang L, Li S, Li Y, Xu X, Wang W, Wang J, Jiang H, Zhang X. A method for noninvasive detection of fetal large deletions/duplications by low coverage massively parallel sequencing. Prenat Diagn. 2013;33(6):584–590. doi: 10.1002/pd.4110. [DOI] [PubMed] [Google Scholar]
- 22.Dai L, Zhu J, Liang J, Wang YP, Wang H, Mao M. Birth defects surveillance in China. World J Pediatr. 2011;7(4):302–310. doi: 10.1007/s12519-011-0326-0. [DOI] [PubMed] [Google Scholar]
- 23.Zhu Y, Lu S, Bian X, Wang H, Zhu B, Wang H, Xu Z, Xu L, Yan W, Zeng Y, Chen Z, Tang S, Shen G, Miao Z. A multicenter study of fetal chromosomal abnormalities in Chinese women of advanced maternal age. Taiwan J Obstet Gynecol. 2016;55(3):379–384. doi: 10.1016/j.tjog.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y, Yu Q, Mao X, Lei W, He M, Lu W. Noninvasive prenatal testing for chromosome aneuploidies and subchromosomal microdeletions/microduplications in a cohort of 42,910 single pregnancies with different clinical features. Hum Genomics. 2019;13(1):60. doi: 10.1186/s40246-019-0250-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamada T, Sekizawa A, Fujii Y, Hirose T, Samura O, Suzumori N, et al. Maternal age-specific risk for trisomy 21 based on the clinical performance of NIPT and empirically derived NIPT age-specific positive and negative predictive values in Japan. J Hum Genet. 2018;63(10):1035–1040. doi: 10.1038/s10038-018-0453-8. [DOI] [PubMed] [Google Scholar]
- 26.Che H, Villela D, Dimitriadou E, Melotte C, Brison N, Neofytou M, van den Bogaert K, Tsuiko O, Devriendt K, Legius E, Esteki MZ, Voet T, Vermeesch JR. Noninvasive prenatal diagnosis by genome-wide haplotyping of cell-free plasma DNA. Genet Med. 2020;22(5):962–973. doi: 10.1038/s41436-019-0748-y. [DOI] [PubMed] [Google Scholar]
- 27.Zhu X, Chen M, Wang H, Guo Y, Chau MHK, Yan H, et al. Clinical utility of expanded noninvasive prenatal screening and chromosomal microarray analysis in high risk pregnancies. Ultrasound Obstet Gynecol. 2020. 10.1002/uog.22021 [Online ahead of print]. [DOI] [PubMed]
- 28.Yao H, Zhang L, Zhang H, Jiang F, Hu H, Chen F, Jiang H, Mu F, Zhao L, Liang Z, Wang W. Noninvasive prenatal genetic testing for fetal aneuploidy detects maternal trisomy X. Prenat Diagn. 2012;32(11):1114–1116. doi: 10.1002/pd.3946. [DOI] [PubMed] [Google Scholar]
- 29.Cheung SW, Patel A, Leung TY. Accurate description of DNA-based noninvasive prenatal screening. N Engl J Med. 2015;372(17):1675–1677. doi: 10.1056/NEJMc1412222. [DOI] [PubMed] [Google Scholar]
- 30.Hall AL, Drendel HM, Verbrugge JL, Reese AM, Schumacher KL, Griffith CB, Weaver DD, Abernathy MP, Litton CG, Vance GH. Positive cell-free fetal DNA testing for trisomy 13 reveals confined placental mosaicism. Genet Med. 2013;15(9):729–732. doi: 10.1038/gim.2013.26. [DOI] [PubMed] [Google Scholar]
- 31.Grati FR, Ferreira J, Benn P, Izzi C, Verdi F, Vercellotti E, Dalpiaz C, D’Ajello P, Filippi E, Volpe N, Malvestiti F, Maggi F, Simoni G, Frusca T, Cirelli G, Bracalente G, Re AL, Surico D, Ghi T, Prefumo F. Outcomes in pregnancies with a confined placental mosaicism and implications for prenatal screening using cell-free DNA. Genet Med. 2020;22(2):309–316. doi: 10.1038/s41436-019-0630-y. [DOI] [PubMed] [Google Scholar]
- 32.Grömminger S, Yagmur E, Erkan S, Nagy S, Schöck U, Bonnet J, Smerdka P, Ehrich M, Wegner RD, Hofmann W, Stumm M. Fetal aneuploidy detection by cell-free DNA sequencing for multiple pregnancies and quality issues with vanishing twins. J Clin Med. 2014;3(3):679–692. doi: 10.3390/jcm3030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snyder MW, Simmons LE, Kitzman JO, Coe BP, Henson JM, Daza RM, et al. Copy-number variation and false positive prenatal aneuploidy screening results. N Engl J Med. 2015;372(17):1639–1645. doi: 10.1056/NEJMoa1408408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Canick JA, Palomaki GE, Kloza EM, Lambert-Messerlian GM, Haddow JE. The impact of maternal plasma DNA fetal fraction on next generation sequencing tests for common fetal aneuploidies. Prenat Diagn. 2013;33(7):667–674. doi: 10.1002/pd.4126. [DOI] [PubMed] [Google Scholar]
- 35.Benn P. Non-invasive prenatal testing using cell free DNA in maternal plasma: recent developments and future prospects. J Clin Med. 2014;3(2):537–565. doi: 10.3390/jcm3020537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richardson MC, Guo M, Fauser BC, Macklon NS. Environmental and developmental origins of ovarian reserve. Hum Reprod Update. 2014;20(3):353–369. doi: 10.1093/humupd/dmt057. [DOI] [PubMed] [Google Scholar]
- 37.Ge ZJ, Schatten H, Zhang CL, Sun QY. Oocyte ageing and epigenetics. Reproduction. 2015;149(3):R103–R114. doi: 10.1530/rep-14-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crawford NM, Steiner AZ. Age-related infertility. Obstet Gynecol Clin North Am. 2015;42(1):15–25. doi: 10.1016/j.ogc.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 39.Taylor-Phillips S, Freeman K, Geppert J, Agbebiyi A, Uthman OA, Madan J, Clarke A, Quenby S, Clarke A. Accuracy of non-invasive prenatal testing using cell-free DNA for detection of Down, Edwards and Patau syndromes: a systematic review and meta-analysis. BMJ Open. 2016;6(1):e010002. doi: 10.1136/bmjopen-2015-010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzumori N, Sekizawa A, Takeda E, Samura O, Sasaki A, Akaishi R, Wada S, Hamanoue H, Hirahara F, Kuriki H, Sawai H, Nakamura H, Yamada T, Miura K, Masuzaki H, Yamashita T, Kamei Y, Namba A, Murotsuki J, Tanemoto T, Fukushima A, Haino K, Tairaku S, Matsubara K, Maeda K, Kaji T, Ogawa M, Osada H, Nishizawa H, Okamoto Y, Kanagawa T, Kakigano A, Endo M, Kitagawa M, Ogawa M, Izumi S, Katagiri Y, Takeshita N, Kasai Y, Naruse K, Neki R, Masuyama H, Hyodo M, Kawano Y, Ohba T, Ichizuka K, Nagamatsu T, Watanabe A, Nishikawa N, Hamajima N, Shirato N, Yotsumoto J, Nishiyama M, Koide K, Hirose T, Sago H. Classification of factors involved in nonreportable results of noninvasive prenatal testing (NIPT) and prediction of success rate of second NIPT. Prenat Diagn. 2019;39(2):100–106. doi: 10.1002/pd.5408. [DOI] [PubMed] [Google Scholar]
- 41.Taneja PA, Snyder HL, de Feo E, Kruglyak KM, Halks-Miller M, Curnow KJ, Bhatt S. Noninvasive prenatal testing in the general obstetric population: clinical performance and counseling considerations in over 85 000 cases. Prenat Diagn. 2016;36(3):237–243. doi: 10.1002/pd.4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benn P, Valenti E, Shah S, Martin K, Demko Z. Factors associated with informative redraw after an initial no result in noninvasive prenatal testing. Obstet Gynecol. 2018;132(2):428–435. doi: 10.1097/aog.0000000000002728. [DOI] [PubMed] [Google Scholar]
- 43.Qiao L, Mao J, Liu M, Liu Y, Song X, Tang H, et al. Experimental factors are associated with fetal fraction in size selection noninvasive prenatal testing. Am J Transl Res. 2019;11(10):6370–6381. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Completely transparent.