Methicillin-resistant Staphylococcus aureus sequence type 72 (ST72) is prevalent in South Korea and has shown resistance to multiple antimicrobials. ST72 isolates display different levels of resistance to the antistaphylococcal lysostaphin. Draft genome sequencing of ST72 human isolates exhibiting lysostaphin resistance or susceptibility was performed to better understand the mechanism of lysostaphin resistance using subtractive genomics.
ABSTRACT
Methicillin-resistant Staphylococcus aureus sequence type 72 (ST72) is prevalent in South Korea and has shown resistance to multiple antimicrobials. ST72 isolates display different levels of resistance to the antistaphylococcal lysostaphin. Draft genome sequencing of ST72 human isolates exhibiting lysostaphin resistance or susceptibility was performed to better understand the mechanism of lysostaphin resistance using subtractive genomics.
ANNOUNCEMENT
Community-associated methicillin-resistant Staphylococcus aureus (MRSA) sequence type 72 (ST72) has emerged as a nosocomial infection that causes pneumonia, endocarditis, and skin and soft tissue infections (SSTIs) in South Korea (1, 2). Due to the multidrug resistance of community-associated ST72, we evaluated the efficacy of lysostaphin as an effective antistaphylococcal bacteriocin that specifically eradicates S. aureus by cleaving the pentaglycine bridge in the cell wall (3, 4).
Human ST72 isolates (K07-561 and K07-204) used in the study were originally isolated by the Asia Pacific Foundation for Infectious Diseases, in South Korea (2). The different levels of susceptibility/resistance of K07-561 and K07-204 to lysostaphin were evaluated (4). The isolate K07-561 showed lysostaphin susceptibility, while K07-204 displayed an ∼1,000 times higher level of lysostaphin resistance upon treatment with 2 U of lysostaphin for 5 min (Fig. 1). These human ST72 isolates, K07-204 (lysostaphin resistant) and K07-561 (lysostaphin susceptible), represent an attractive model for elucidating the unknown mechanism of antibiotic and enzybiotic resistance. Therefore, we selected the isolates K07-204 and K07-561 as representative strains for genome sequencing to understand the genetic differences between the resistant and sensitive phenotypes.
FIG 1.
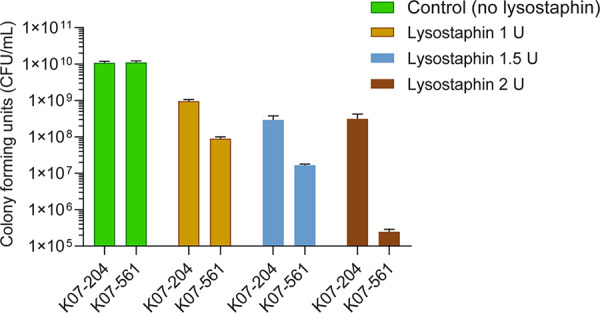
Responses of S. aureus ST72 strains to lysostaphin. Differential responses of K07-204 and K07-561 at various concentrations of lysostaphin are shown.
A single colony was inoculated in 10 ml of tryptic soy broth (TSB) under orbital shaking culture conditions (200 rpm) at 37°C for 16 h. The cells were harvested by centrifugation. Genomic DNA from K07-204 and K07-561 was isolated by the phenol-chloroform extraction method (5). Briefly, the high-molecular-weight genomic DNA was sheared by using a Covaris S2 ultrasonicator system to obtain 350-bp DNA fragments. Libraries were prepared with the TruSeq Nano DNA sample preparation kit (Illumina, Inc., San Diego, CA, USA) by following the manufacturer’s protocol. Products were quantified using a Bioanalyzer 2100 system (Agilent, Santa Clara, CA, USA). Genomes were sequenced with a 150-bp paired-end Illumina NovaSeq 6000 platform. The quality control results were generated using FastQC software (v.0.10.1). Trimming of low-quality bases with a score below Q20 was performed using Sickle (v.1.3.3) (6), resulting in 21,289,096 and 28,904,426 surviving read pairs for K07-204 and K07-561, respectively. The genomes were assembled de novo using IDBA-UD (v.1.1.1) (7). The genome assembly of K07-204 and K07-561 yielded 66 and 58 contigs, respectively. The predicted lengths of the de novo-assembled genomes of K07-204 and K07-561 were found to be 2,310,534 bp and 2,149,433 bp, respectively. The GC contents of K07-204 and K07-561 were 32.92% and 32.88%, respectively. Contigs were annotated using the Prokaryotic Genome Annotation Pipeline (PGAP) (8). The K07-204 genome contained 2,168 coding genes, 40 tRNA sequences, and 1 rRNA sequence. K07-561 contained 2,040 coding genes and 21 tRNA sequences in its genome. The available genome sequences and their subtractive genomics in future studies will not only decode the novel genetic basis of the mechanisms of resistance to antibiotics and lysostaphin but also broaden the therapeutic interventions used against MRSA.
Data availability.
The draft genome sequencing data for K07-204 and K07-561 are available in the GenBank database under the accession numbers JACSIU000000000.1 and JACORE000000000.1, respectively. Raw sequencing reads for K07-204 and K07-561 were deposited in the Sequence Read Archive (SRA) under BioProject/BioSample accession numbers PRJNA637991/SAMN15163378 and PRJNA637996/SAMN15163770, respectively.
ACKNOWLEDGMENT
This study was supported by National Research Foundation of Korea grants to A.K.C. (grant 2020R1I1A1A01070635) and K.K.K. (grant 2017M3A9E4078553).
REFERENCES
- 1.Joo EJ, Chung DR, Kim SH, Baek JY, Lee NY, Cho SY, Ha YE, Kang CI, Peck KR, Song JH. 2017. Emergence of community-genotype methicillin-resistant Staphylococcus aureus in Korean hospitals: clinical characteristics of nosocomial infections by community-genotype strain. Infect Chemother 49:109–116. doi: 10.3947/ic.2017.49.2.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ko KS, Lim SK, Jung SC, Yoon JM, Choi JY, Song JH. 2011. Sequence type 72 meticillin-resistant Staphylococcus aureus isolates from humans, raw meat and soil in South Korea. J Med Microbiol 60:442–445. doi: 10.1099/jmm.0.026484-0. [DOI] [PubMed] [Google Scholar]
- 3.Schindler CA, Schuhardt VT. 1964. Lysostaphin: a new bacteriolytic agent for the Staphylococcus. Proc Natl Acad Sci U S A 51:414–421. doi: 10.1073/pnas.51.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim JH, Chaurasia AK, Batool N, Ko KS, Kim KK. 2019. Alternative enzyme protection assay to overcome the drawbacks of the gentamicin protection assay for measuring entry and intracellular survival of staphylococci. Infect Immun 87:e00119-19. doi: 10.1128/IAI.00119-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ausubel FM, Brent B, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (ed). 1997. Short protocols in molecular biology: a compendium of methods from Current Protocols in Molecular Biology, 3rd ed. Wiley, New York, NY. [Google Scholar]
- 6.Joshi NA, Fass JN. 2011. Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files. https://github.com/najoshi/sickle.
- 7.Peng Y, Leung HC, Yiu SM, Chin FY. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 8.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The draft genome sequencing data for K07-204 and K07-561 are available in the GenBank database under the accession numbers JACSIU000000000.1 and JACORE000000000.1, respectively. Raw sequencing reads for K07-204 and K07-561 were deposited in the Sequence Read Archive (SRA) under BioProject/BioSample accession numbers PRJNA637991/SAMN15163378 and PRJNA637996/SAMN15163770, respectively.


