In recent years, Lake Baikal has undergone significant changes in the composition of coastal communities associated with the increasing anthropogenic influence and global climate changes. In this context, we carried out metagenomic sequencing of the DNA viral community of an integral near-bottom water sample from the littoral zone of the lake.
ABSTRACT
In recent years, Lake Baikal has undergone significant changes in the composition of coastal communities associated with the increasing anthropogenic influence and global climate changes. In this context, we carried out metagenomic sequencing of the DNA viral community of an integral near-bottom water sample from the littoral zone of the lake.
ANNOUNCEMENT
Lake Baikal is the world’s deepest, oldest, and largest by volume freshwater lake (1). It has unique environmental conditions and a great biological diversity of mainly endemic flora and fauna (2). Since 2011, the first signs of eutrophication have been observed in the coastal zone of the lake, including changes in the composition and vertical zonation of benthic algae, the presence of unfavorable microbiological indicators, disease and mass mortality of sponges, and others (3–9). Viruses are an important component of the aquatic biota; therefore, the study of viral diversity is useful for assessing their role and dynamics in processes occurring in the coastal zone of the lake.
A shotgun metagenomic study of the DNA viral community, including sample preparation, sequencing, and bioinformatic analysis, was mainly performed as described in our previous studies (10, 11). Briefly, 10-liter water samples were collected in the southern basin of Lake Baikal (near Bolshiye Koty, Russia, 51.9023 N, 105.1028 E) at depths of 10, 12, and 15 m in May 2018 using lightweight diving equipment and a bathometer. The samples were filtered through 0.2-μm nitrocellulose filters (Sartorius) and pooled. The virus-like particles were concentrated with a tangential flow filtration system and the Vivaspin-20 ultrafiltration device (30 kDa; Sartorius) and treated with DNase I and RNase A (Thermo Fisher Scientific). Viral DNA was extracted with a ZR viral DNA kit (Zymo Research).
The preparation and sequencing of DNA libraries were performed in the Center of Shared Scientific Equipment “Persistence of Microorganisms” (ICIS UB RAS, Russia). The paired-end libraries were prepared using a NEBNext Ultra II FS DNA library prep kit for Illumina (New England Biolabs) according to the manufacturer’s protocol. Sequencing of the libraries was conducted on a MiSeq genome sequencer using a MiSeq reagent kit v. 3 (2 × 300 cycles; Illumina).
The primary processing (quality control and trimming) of the virome data set was performed using the R package ShortReads v. 1.48.0 (12); reads with average quality less than 20 and length less than 200 bp were removed. Taxonomic identification of viral sequences was performed using the BLASTn v. 2.5.0 algorithm (13) against the NCBI RefSeq viral complete genome database (14) as described before (11). The BLASTn v. 2.5.0 parameters used were as follows: cost to open a gap, 2; cost to extend a gap, 1; word size for word finder algorithm, 12; penalty for a nucleotide mismatch, 1; and reward for a nucleotide match, 1. The sequence reads were considered “identified” if they had a relative in the reference database with an E value of ≤105 and a bit score of ≥50. For the functional annotation of viral sequences, we used the local BLASTx v. 2.5.0 application (13) and the Clusters of Orthologous Groups (COG) database (15). The BLASTx v. 2.5.0 parameters used were as follows: cost to open a gap, 6; cost to extend a gap, 2; word size for word finder algorithm, 6; E value, ≤105; and bit score, ≥50.
The raw data contained 5,329,629 paired sequence reads (or 10,659,258 single reads). After quality processing, we obtained 8,449,571 single reads; of these, 187,941 sequences (2.2% of the data set) were identified as viral, belonging to 26 families of DNA viruses (78.06% of reads), unclassified to the range of family (21.8%), and RNA viruses (0.14%). Thirteen families were the most numerous and together accounted for more than 77% of affiliated virome sequences (Fig. 1). In total, we revealed a high viral diversity in the samples of near-bottom water of Lake Baikal (1,219 virotypes and 22 functional categories of proteins and enzymes).
FIG 1.
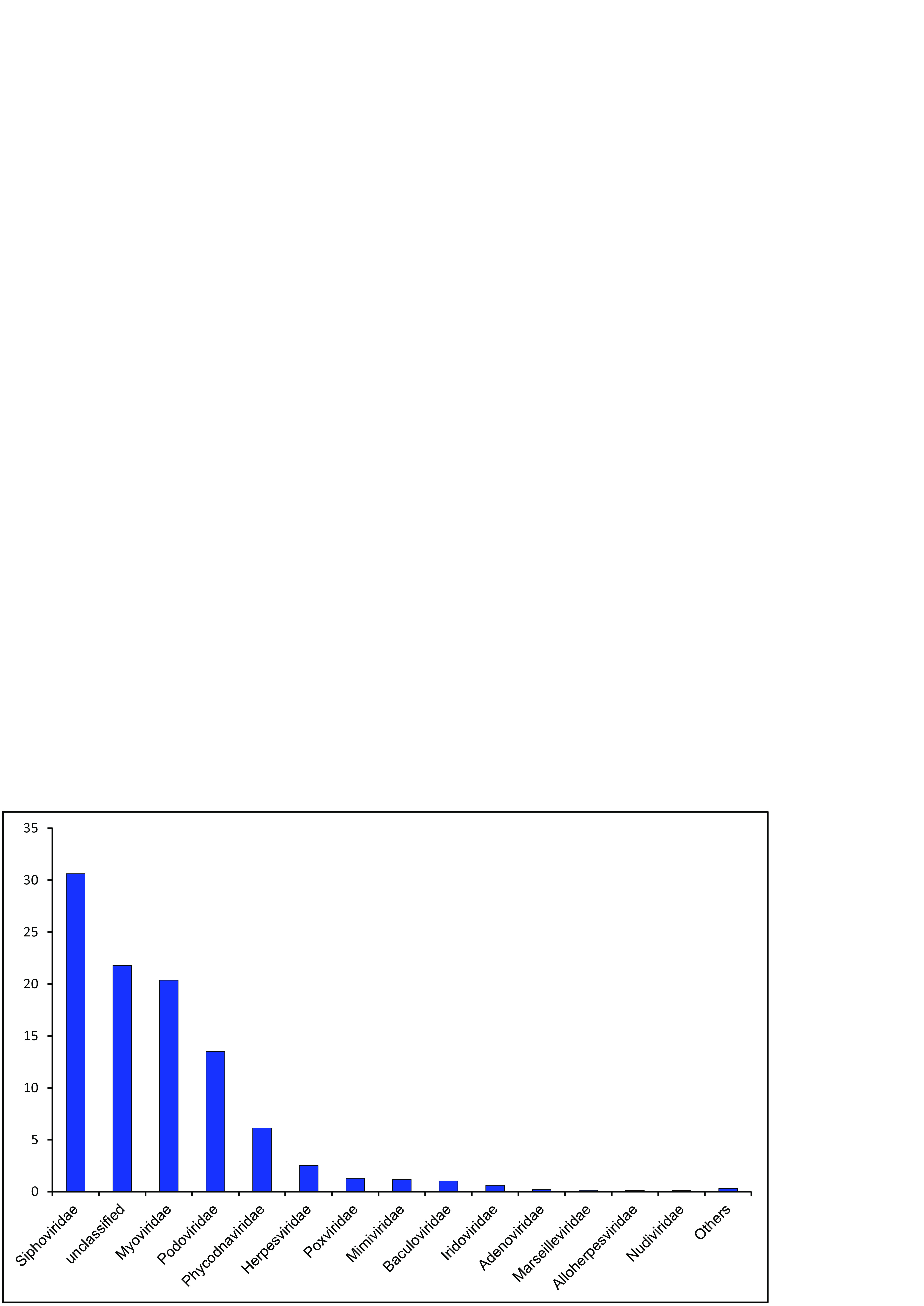
Proportion (percentage) of hits to known viral families in the Lake Baikal virome data set.
Our metagenomic research allowed us to describe the taxonomic and functional composition of viral communities in near-bottom coastal waters of Lake Baikal, which will make it possible to assess the role of viruses in the benthic community.
Data availability.
The raw sequence data have been deposited in the NCBI SRA repository via BioProject PRJNA577390 (BioSample SAMN16330433).
ACKNOWLEDGMENTS
This work was supported by the Russian Science Foundation (RSF project no. 19-14-00088). Sample collection was carried out within the framework of the Siberian Branch of the Russian Academy of Sciences basic budget funding from project no. 0345-2019-0002 (АААА-А16-116122110066-1).
The bioinformatics data analysis was performed in part on the equipment of the Irkutsk Supercomputer Center of the Siberian Branch of the Russian Academy of Sciences (HPC-cluster “Akademik V.M. Matrosov”).
REFERENCES
- 1.Galaziy GI. (ed) 1993. Atlas of Lake Baikal. Roskartografiya, Moscow, Russia. [Google Scholar]
- 2.Kozhova OM, Izmesteva LR. 1998. Lake Baikal: evolution and biodiversity. Backhuys Publishers, Leiden, The Netherlands. [Google Scholar]
- 3.Kravtsova LS, Izhboldina LA, Khanaev IV, Pomazkina GV, Domysheva VM, Kravchenko OS, Grachev MA. 2012. Disturbances of the vertical zoning of green algae in the coastal part of the Listvennichnyi gulf of Lake Baikal. Dokl Biol Sci 447:350–352. doi: 10.1134/S0012496612060026. [DOI] [PubMed] [Google Scholar]
- 4.Kravtsova LS, Izhboldina LA, Khanaev IV, Pomazkina GV, Rodionova EV, Domysheva VM, Sakirko MV, Tomberg IV, Kostornova TY, Kravchenko OS, Kupchinsky AB. 2014. Nearshore benthic blooms of filamentous green algae in Lake Baikal. J Great Lakes Res 40:441–448. doi: 10.1016/j.jglr.2014.02.019. [DOI] [Google Scholar]
- 5.Timoshkin OA, Samsonov DP, Yamamuro M, Moore MV, Belykh OI, Malnik VV, Sakirko MV, Shirokaya AA, Bondarenko NA, Domysheva VM, Fedorova GA, Kochetkov AI, Kuzmin AV, Lukhnev AG, Medvezhonkova OV, Nepokrytykh AV, Pasynkova EM, Poberezhnaya AE, Potapskaya NV, Rozhkova NA, Sheveleva NG, Tikhonova IV, Timoshkina EM, Tomberg IV, Volkova EA, Zaitseva EP, Zvereva YM, Kupchinsky AB, Bukshuk NA. 2016. Rapid ecological change in the coastal zone of Lake Baikal (East Siberia): is the site of the world’s greatest freshwater biodiversity in danger? J Great Lakes Res 42:487–497. doi: 10.1016/j.jglr.2016.02.011. [DOI] [Google Scholar]
- 6.Khodzher TV, Domysheva VM, Sorokovikova LM, Tomberg IV, Sakirko MV. 2018. Hydrochemical studies in Lake Baikal: history and nowadays. Limnol Freshw Biol 1:2–9. doi: 10.31951/2658-3518-2018-A-1-2. [DOI] [Google Scholar]
- 7.Khanaev IV, Kravtsova LS, Maikova OO, Bukshuk NA, Sakirko MV, Kulakova NV, Butina TV, Nebesnykh IA, Belikov SI. 2018. Current state of the sponge fauna (Porifera: Lubomirskiidae) of Lake Baikal: sponge disease and the problem of conservation of diversity. J Great Lakes Res 44:77–85. doi: 10.1016/j.jglr.2017.10.004. [DOI] [Google Scholar]
- 8.Bondarenko NA, Logacheva NF. 2017. Structural changes in phytoplankton of the littoral zone of Lake Baikal. Hydrobiol J 53:16–24. doi: 10.1615/HydrobJ.v53.i2.20. [DOI] [Google Scholar]
- 9.Bondarenko NA, Ozersky T, Obolkina LA, Tikhonova IV, Sorokovikova EG, Sakirko MV, Potapov SA, Blinov VV, Zhdanov AA, Belykh OI. 2019. Recent changes in the spring microplankton of Lake Baikal, Russia. Limnologica 75:19–29. doi: 10.1016/j.limno.2019.01.002. [DOI] [Google Scholar]
- 10.Butina TV, Bukin YS, Krasnopeev AS, Belykh OI, Tupikin AE, Kabilov MR, Sakirko MV, Belikov SI. 2019. Estimate of the diversity of viral and bacterial assemblage in the coastal water of Lake Baikal. FEMS Microbiol Lett 366:fnz094. doi: 10.1093/femsle/fnz094. [DOI] [PubMed] [Google Scholar]
- 11.Butina TV, Khanaev IV, Kravtsova LS, Maikova OO, Bukin YS. 2020. Metavirome datasets from two endemic Baikal sponges Baikalospongia bacillifera. Data Brief 29:105260. doi: 10.1016/j.dib.2020.105260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgan M, Anders S, Lawrence M, Aboyoun P, Pagès H, Gentleman R. 2009. ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics 25:2607–2608. doi: 10.1093/bioinformatics/btp450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 14.Pruitt KD, Tatusova T, Maglott DR. 2005. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tatusov RL, Galperin MY, Natale DA, Koonin EV. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw sequence data have been deposited in the NCBI SRA repository via BioProject PRJNA577390 (BioSample SAMN16330433).


