Abstract
The taxonomy of honey bee A. mellifera contains a lot of issues due to the specificity of population structure, features of biology and resolutions of honey bee subspecies discrimination methods. There are a lot of transition zones between ranges of subspecies which led to the gradual changes of characteristics among neighbor subspecies. The modern taxonomic pattern of honey bee Apis mellifera is given in this paper. Thirty-three distinct honey bee subspecies are distributed across all Africa (11 subspecies), Western Asia and the Middle East (9 subspecies), and Europe (13 subspecies). All honey bee subspecies are subdivided into 5 evolutionary lineages: lineage A (10 subspecies) and its sublineage Z (3 subspecies), lineage M (3 subspecies), lineage C (10 subspecies), lineage O (3 subspecies), lineage Y (1 subspecies), lineage C or O (3 subspecies).
Keywords: Apis mellifera, Honey bee, Subspecies, Taxonomy
1. Introduction
The honey bee species Apis mellifera Linnaeus, 1758 is distributed in a wide range with various climatic conditions and is subdivided into numerous subspecies Ruttner, 1988, Sheppard et al., 1997, Engel, 1999). Different taxonomic revisions described 20–30 subspecies for the A. mellifera (Buttel-Reepen, 1906, Enderlein, 1906, Skorikov, 1929a, Skorikov, 1929b, Maa, 1953, Ruttner, 1988, Engel, 1999). The taxonomy of honey bee A. mellifera is contradictory and remains a number of errors and issues. Some honey bee subspecies described only by morphometry without any molecular proves (Maa, 1953, Ruttner, 1988, Arias and Sheppard, 1996, Franck et al., 2000, Smith, 2002, Kandemir et al., 2011). The subspecies composition of the evolutionary lineages A (+Z), M, C, O, Y has not yet been resolved and need in comprehensive molecular evidence (Smith, 1991, Smith et al., 1997; Garnery et al., 1992; Kandemir and Kence, 1995; Alburaki et al., 2011, Alburaki et al., 2013).
Since the honey bee subspecies are described as geographic races the confusion in their geographical localization and genes distribution are observed in Africa (Ruttner, 1992, Hepburn and Radloff, 1998, Dukku, 2016), in Europe (Ruttner, 1988, Cornuet and Garnery, 1991, Smith, 1991, Franck et al., 1998), and in Near East (Franck et al., 2000, Kandemir et al., 2006; Alburaki et al., 2011, Alburaki et al., 2013). The clarification of all confusions in honey bee Apis mellifera taxonomy is the main aim of this paper. Here we described all new, old, synonymic, and misspelled taxonomic names of honey bees A. mellifera.
2. The approaches to identify the taxonomic affiliation of the honey bee A. Mellifera
The methods of identification of the taxonomic affiliation of honey bee A. mellifera based on morphometry are subdivided into two groups: 1. the body parts measurement; 2. the wing shapes analysis. For discrimination of honey bee subspecies, the classical morphometry uses the measurement of 36 characters of body parts (Ruttner, 1988). Other popular morphometrical approaches for identification of the taxonomic affiliation of honey bee colonies based on analysis wing shapes. There are three different wing morphometry based approaches for discrimination of honey bee subspecies: classical wing morphometry (DuPraw, 1965), the DAWINO (Discriminant Analysis with Numerical Output) (www.beedol.cz) and geometric morphometry (Bookstein, 1991). Classical wing morphometry captures variation in wing shape by calculating 11 angles between 18 junctions in the wing venation, which constitute a subset of a suite of 17 angles. The DAWINO method consists of the full set of DuPraw’s angles, supplemented by 7 linear measurements, 5 indices, and one area. The geometric morphometry uses DuPraw’s coordinates of points called landmarks which is analyzed by multivariate statistical methods (Miguel et al., 2011).
The allozyme based method of identifying the taxonomic affiliation of honey bees based on the variability of isoenzymes. In honey bees six enzymes are polymorphic: malate dehydrogenase MDH1 (EC 1.1.1.37) (7 alleles) (Smith and Glenn, 1995); malik-enzyme ME (EC 1.1.1.40) (4 alleles) (Sheppard and Berlocher, 1984); esterase EST-3 (EC 3.1.1) (8 alleles) (Ivanova et al., 2010); alkaline phosphatase ALP (EC 3.1.3.1) (3 alleles) (Ivanova et al., 2010); phosphoglucomutase PGM (EC 5.4.2.2) (5 alleles) (Ivanova et al., 2010); hexokinase HK (EC 2.7.1.1) (5 alleles) (Del Lama et al., 1990).
The mitochondrial DNA (mtDNA) based methods of identification the taxonomic affiliation of honey bees based on its nucleotide polymorphism. There are three main directions of mtDNA based discrimination of honey bees: 1. Restriction Fragment Length Polymorphisms by restriction enzymes (RFLP) (Smith and Brown, 1988, Smith et al., 1989, Smith, 1991; Garnery et al., 1992) and RFLP of fragments amplified by PCR (PCR-RFLP) (Hall and Smith, 1991, Arias and Sheppard, 2005, Stevanovic et al., 2010, Ivanova et al., 2010); 2. Polymorphism of PCR products with specific (Arias and Sheppard, 2005) or random primers – Random amplified polymorphic DNA (RAPD) amplification (Hunt and Page, 1994, Suazo et al., 1998, Tunca and Kence, 2011); 3. Sequencing of the mtDNA with the quest of single nucleotide polymorphisms (SNPs) (Arias and Sheppard, 1996, Arias and Sheppard, 2005, Franck et al., 1998, Franck et al., 2000, Franck et al., 2001, Franck et al., 2001, Collet et al., 2006, Shaibi et al., 2009, Pinto et al., 2012, Pinto et al., 2014, Ilyasov et al., 2016, Ilyasov et al., 2019). The study of mtDNA was based on the analysis of complete or partial mitochondrial genome or intergenic sequences IGS. The most polymorphic IGS is the region located between genes COX1 and COX2 of mtDNA (Hall and Smith, 1991, Crozier et al., 1991, Arias and Sheppard, 1996, Jensen et al., 2005, Il'yasov et al., 2007, Ilyasov et al., 2011, Ilyasov et al., 2016, Ilyasov et al., 2019, Cánovas et al., 2008, Muli et al., 2014).
The nuclear DNA (nDNA) based methods of identification of the taxonomic affiliation of honey bees focused on its nucleotide polymorphism. The polymorphism of nDNA can be detected by three main approaches: 1. Polymorphism of PCR products with specific (Arias and Sheppard, 2005, Il’yasov et al., 2015, Kent et al., 2011, Kent et al., 2012) or random RAPD primers amplification (Hunt and Page, 1994, Waldschmidt et al., 2002, Kumar and Khan, 2014); 3. Sequencing of the mtDNA with the quest of single nucleotide polymorphisms (SNPs) (Chapman et al., 2015, Chapman et al., 2016, Harpur et al., 2014, Harpur et al., 2015, Henriques et al., 2018, Il’yasov et al., 2015, Ilyasov et al., 2016, Pinto et al., 2014, Wallberg et al., 2014, Wallberg et al., 2019, Whitfield et al., 2006); 4. Whole-genome sequencing by NGS methods (Weinstock et al., 2007, Whitfield et al., 2006, Wallberg et al., 2014, Wallberg et al., 2019). The most polymorphic nDNA loci are microsatellite repeats – Short Tandem Repeats (STR) that are detected by PCR with specific primers (De La Rúa et al., 2001, Estoup et al., 1995, Franck et al., 2000, Franck et al., 2001, Garnery et al., 1998, Il’yasov et al., 2015, Ilyasov et al., 2016, Miguel et al., 2011, Solignac et al., 2003).
3. The current taxonomic pattern of the honey bee A. Mellifera
The taxonomy of honey bee Apis mellifera Linnaeus, 1758 (synonym: Apis mellifica Linnaeus, 1761, Apis gregaria Geoffroy, 1762, Apis cerifera Scopoli, 1770, Apis daurica Fischer von Waldheim, 1843) has a lot of unsolved issues. The number of honey bee subspecies and their subdivision on evolutionary lineages is not well clear yet (Smith, 1991, Smith et al., 1997; Garnery et al., 1992; Kandemir and Kence, 1995; Alburaki et al., 2011, Alburaki et al., 2013).
The subspecies of honey bee A. mellifera were divided into three lineages (A, M, C) by morphometry firstly (about 20 subspecies) (Ruttner et al., 1978, Moritz et al., 1986, Smith and Brown, 1988, Smith, 1991; Garnery et al., 1992; Estoup et al., 1995), into four lineages (A, M, C, O) by morphometry and molecular data later (about 25 subspecies) (Ruttner, 1988, Smith and Brown, 1988, Cornuet et al., 1988, Lebdigrissa et al., 1991, Cornuet and Garnery, 1991, Smith, 1991, Estoup et al., 1995, Arias and Sheppard, 1996, Franck et al., 1998, Palmer et al., 2000) and five lineages (A (+sublineage Z), M, C, O, Y) by molecular data finally (about 30 subspecies) (Franck et al., 2001; Alburaki et al., 2011, Alburaki et al., 2013, Meixner et al., 2013).
The greatest problem in the discrimination of subspecies was the presence of transition zones between the area of subspecies which led to gradient changes in morphometric and molecular features. Transition is observed between 8 subspecies (A. m. adansonii – A. m. jemenitica, A. m. sahariensis – A. m. intermissa, A. m. intermissa – A. m. major, A. m. intermissa – A. m. scutellata, A. m. capensis – A. m. unicolor) in Africa (Ruttner, 1992, Hepburn and Radloff, 1998, Dukku, 2016); between 3 subspecies (A. m. intermissa – A. m. iberiensis, A. m. iberiensis – A. m. mellifera) in Europe (Ruttner, 1988, Cornuet and Garnery, 1991, Smith, 1991, Franck et al., 1998); between 4 subspecies (A. m. meda – A. m. anatoliaca, A. m. anatoliaca – A. m. syriaca, A. m. syriaca – A. m. lamarckii) in Near East (Franck et al., 2000, Kandemir et al., 2006; Alburaki et al., 2011, Alburaki et al., 2013).
The honey bee subspecies can be affiliated variously by different methods. The subspecies A. m. adansonii, A. m. monticola, A. m. scutellata, A. m. capensis and A. m. unicolor cannot to be discriminated by morphometry (Ruttner et al., 1978), by mtDNA (Garnery et al., 1995; Sheppard et al., 1996), but can be discriminated by other morphometry (Ruttner, 1988), by allozyme (Meixner et al., 2000), by mtDNA (Franck et al., 2001, Kasangaki et al., 2017, Techer et al., 2017). The subspecies A. m. jemenitica, A. m. adansonii cannot be discriminated by morphometry (Radloff et al., 1998) but can be discriminated by other morphometry (Ruttner, 1988, Dukku, 2016), by mtDNA (Franck et al., 2001). The subspecies A. m. carpathica, A. m. carnica, A. m. macedonica cannot be discriminated by morphometry (Radloff et al., 1998) but can be discriminated by other morphometry (Foti et al., 1965, Engel, 1999, Cauia et al., 2008, Mărghitas et al., 2008, Teleky et al., 2009), by mtDNA (Mărghitas et al., 2009, Bouga et al., 2011, Coroian et al., 2014, Syromyatnikov et al., 2018; Ilyasov et al., 2018). The subspecies A. m. rodopica, A. m. macedonica cannot to be discriminated by morphometry (Engel, 1999), by allozyme (Bouga et al., 2005, Kandemir et al., 2005, Ivanova et al., 2010), but can be discriminated by other morphometry (Foti et al., 1965, Engel, 1999, Cauia et al., 2008, Mărghitas et al., 2008, Teleky et al., 2009), by mtDNA (Lazarov, 1936, Tzonev, 1960, Velichkov, 1970, Bouga et al., 2011), by microsatellites (Nikolova, 2011, Uzunov et al., 2014), by mtDNA (Bouga et al., 2005, Martimianakis et al., 2011; Meixner et al., 2013; Radoslavov et al., 2017).
The honey bee subspecies can be variously assigned to the evolutionary lineages by different methods. The subspecies A. m. syriaca, A. m. anatoliaca, A. m. meda assigned to the O lineage by morphometry (Ruttner, 1988), by mtDNA (Arias and Sheppard, 1996), but assigned to the A lineage (Z sublineage) by mtDNA (Arias and Sheppard, 1996, Franck et al., 2000, Bouga et al., 2005; Alburaki et al., 2011, Alburaki et al., 2013). The subspecies A. m. caucasia assigned to the O lineage by morphometry (Ruttner, 1988, Adl et al., 2007, Meixner et al., 2007, Kandemir et al., 2011), but assigned to the C lineage by mtDNA (Franck et al., 2000; Alburaki et al., 2011, Alburaki et al., 2013), by allozyme (Smith, 1991; Garnery et al., 1992; Kandemir and Kence, 1995, Ivanova et al., 2011), by mtDNA (Cornuet and Garnery, 1991, Koulianos and Crozier, 1997, Smith et al., 1997, Garnery et al., 1998, Palmer et al., 2000, Franck et al., 2000, Smith, 2002, Özdïl et al., 2009, Mărghitas et al., 2009; Ilyasov et al., 2018). The subspecies A. m. cypria, A. m. adami assigned to the O lineage by morphometry (Ruttner, 1988) but assigned to the C lineage by mtDNA and allozyme (Bouga et al., 2005). The subspecies A. m. lamarckii assigned to the A lineage by morphometry (Ruttner, 1988) but assigned to the O lineage by mtDNA (Arias and Sheppard, 1996). The subspecies A. m. jemenitica assigned to the A lineage by morphometry (Ruttner, 1988) but assigned to the Y lineage by mtDNA (El-Niweiri and Moritz, 2008; Alburaki et al., 2011, Alburaki et al., 2013, Coulibaly et al., 2019).
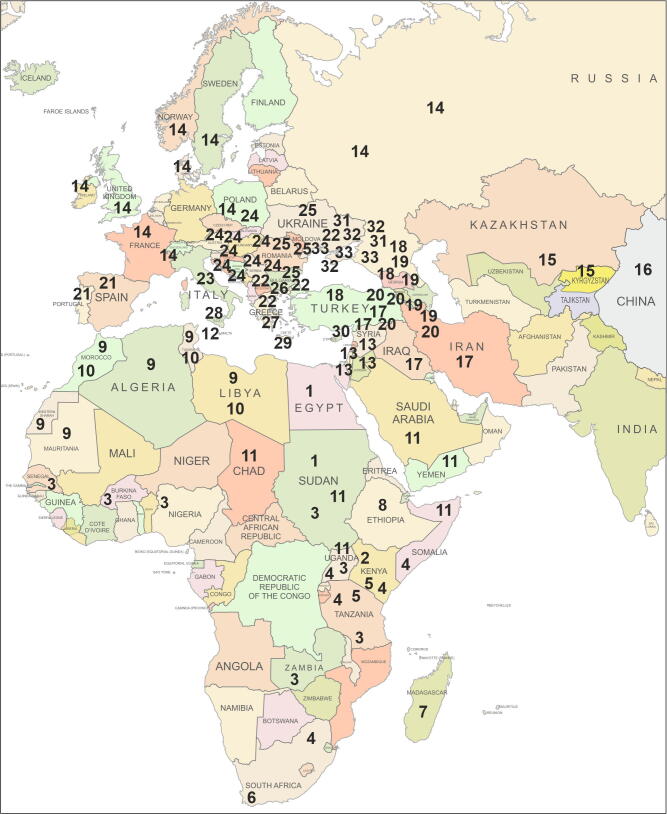
In the current review of all available literature, the existence of 33 subspecies of honey bee A. mellifera is stated. These 33 subspecies subdivided into five evolutionary lineages: lineage A − 10 subspecies, lineage A (sublineage Z) – 3 subspecies, lineage M − 3 subspecies, lineage C − 10 subspecies, lineage O − 3 subspecies, lineage Y − 1 subspecies, undefined lineage C or O − 3 subspecies. Geographically these subspecies distributed in three regions: Africa inhabited by 11 subspecies, Western Asia and the Middle East inhabited by 9 subspecies, Europe inhabited by 13 subspecies (Table 1). The spatial distribution of 33 subspecies of honey bee A. mellifera was shown on geographic map (Fig. 1).
Table 1.
The current taxonomy of the honey bee Apis mellifera.
| No | Subspecies name | Common Name | Lineage | Distribution |
|---|---|---|---|---|
| Africa (11 subspecies) | ||||
| Apis mellifera lamarckii Cockerell 1906 | The Egyptian honey bee | O | Egypt, Sudan | |
| Apis mellifera litorea Smith 1961 | The East African coastal honey bee | A | Kenya | |
| Apis mellifera adansonii Latreille 1804 | The West African honey bee | A | Nigeria, Burkina Faso, Uganda, Tanzania, Zambia, Senegal, Sudan | |
| Apis mellifera scutellata Lepeletier de Saint Fargeau, 1836 | The African honey bee | A | Kenya, Tanzania, Uganda, Republic of South Africa, Somalia | |
| Apis mellifera monticola Smith 1961 | The East African Mountain honey bee | A | Mountains of Kenya, Tanzania | |
| Apis mellifera capensis Escholtz 1822 | The Cape honey bee | A | Cape region in the Republic of South Africa | |
| Apis mellifera unicolor Latreille 1804 | The Madagascar honey bee | A | Madagascar | |
| Apis mellifera simensisMeixner et al., 2011 | The Ethiopian honey bee | A | Ethiopia | |
| Apis mellifera sahariensis Baldensperger 1932 | The Saharan honey bee | A | Morocco, Algeria, Tunisia, Libya, Mauritania, Western Sahara | |
| Apis mellifera intermissa Maa 1953 (synonym: A. m. major Ruttner et al., 1978) (Kerr, 1992; Hepburn and Radloff, 1998) | The Tellian honey bee | A | Morocco, Libya, Tunisia | |
| Apis mellifera jemenitica Ruttner 1976 (synonyms: Apis mellifera yemenitica Ruttner, 1988, Apis mellifera nubica Ruttner, 1976, Apis mellifera sudanensis Ruttner, 1988, Apis mellifera bandasii Radloff and Hepburn, 1997, and Apis mellifera woyi-gambellAmssalu et al., 2004) (Hepburn and Radloff, 1998; Amssalu et al., 2004; Meixner et al., 2011) | The Arabian honey bee | Y | Arabian Peninsula, Chad, Saudi Arabia, Somalia, Sudan, Uganda, Yemen | |
| Western Asia and the Middle East (9 subspecies) | ||||
| Apis mellifera ruttneriSheppard et al., 1997 | The Maltese honey bee | A | Malta | |
| Apis mellifera syriaca Skorikov 1929b | The Syrian honey bee | A (Z) | Syria, Israel, Lebanon, Palestine, Jordan | |
| Apis mellifera mellifera Linnaeus 1758 (synonyms: Apis mellifica germanica Pollmann, 1879, Apis mellifica nigrita Lucas, 1882, Apis mellifica mellifica lehzeni Buttel-Reepen, 1906, Apis mellifica mellifica silvarum Goetze, 1964) | The European dark honey bee | M | France, United Kingdom, Switzerland, European part of Russia, Poland, Denmark, Norway, Sweden, Ireland | |
| Apis mellifera pomonella Sheppard and Meixner 2003 | The Tian Shan honey bee | O | Tian Shan mountains of Kazakhstan, Kyrgyzstan | |
| Apis mellifera sinisxinyuan Chen et al., 2016 | The Xinyuan honey bee | M | Uygur Autonomous Region of China | |
| Apis mellifera meda Skorikov 1929b | The Persian honey bee | A (Z) | Iran, Iraq, Syria, Turkey | |
| Apis mellifera caucasia Pollmann 1889 | The Caucasian honey bee | C | South Russia, Turkey, Georgia | |
| Apis mellifera remipes Gerstäcker1862 (synonym: Apis mellifera armeniaca Skorikov 1929b) (Hepburn and Radloff, 1998) | The Armenian honey bee | O | South Russia, Armenia, Iran, Georgia | |
| Apis mellifera anatoliaca Maa 1953 | The Anatolian honey bee | A (Z) | Iran, Armenia, Syria, Turkey | |
| Europe (13 subspecies) | ||||
| Apis mellifera iberiensis Engel 1999 (new name for Apis mellifera iberica Ruttner 1988, preoccupied, nec Skorikov 1929) | The Spanish honey bee | M | Spain, Portugal | |
| Apis mellifera macedonica Ruttner, 1988 | The Macedonian honey bee | C | Bulgaria, Greece, Macedonia, Ukraine | |
| Apis mellifera ligustica Spinola 1806 | The Italian honey bee | C | Italia | |
| Apis mellifera carnica Pollman 1879 (synonyms: Apis mellifica hymettea Pollmann 1879, Apis mellifera carniolica Koschevnikov 1900, Apis mellifica banatica Grozdanic 1926, Apis mellifera banata Skorikov 1929b) (Hepburn and Radloff, 1998) | The Carniolan honey bee | C | Slovenia, Bulgaria, Poland, Austria, Croatia, Bosnia and Herzegovina, Serbia, Hungary, Romania | |
| Apis mellifera carpathica Foti 1965 | The Carpathian honey bee | C | Ukraine, Bulgaria, Romania, Moldova | |
| Apis mellifera rodopica Petrov 1991 | The Bulgarian honey bee | C | Bulgaria | |
| Apis mellifera cecropia Kiesenweiter 1860 | The Greek honey bee | C | Greece | |
| Apis mellifera siciliana Dalla Torre 1896 (synonym: Apis mellifera sicula Montagano 1911) (Hepburn and Radloff, 1998) | The Sicilian honey bee | C | Sicily | |
| Apis mellifera adami Ruttner, 1975 | The Cretan honey bee | C | Crete | |
| Apis mellifera cypria Pollman 1879 | The Cyprus honey bee | C | Cyprus | |
| Apis mellifera artemisia Engel 1999 (new name for preoccupied: Apis mellifera acervorum Skorikov 1929b) | The Russian steppe honey bee | C or O | South Russia, Ukraine | |
| Apis mellifera sossimai Engel 1999 (new name for preoccupied: Apis mellifera cerifera Gerstäcker 1862) | The Ukrainian honey bee | C or O | Crimea, South Russia, Ukraine | |
| Apis mellifera taurica Alpatov 1935 | The Crimean Honey Bee | C or O | Crimea, South Russia, Ukraine | |
Fig. 1.
The spatial distribution of 33 subspecies of honey bee A. mellifera on geographic map. The numbers correspond to numbers of subspecies in the table.
4. Conclusions
The current taxonomic pattern of the honey bee A. mellifera is described. Thirty-three distinct honey bee subspecies are presented in Africa (11 subspecies), Western Asia and the Middle East (9 subspecies), Europe (13 subspecies), which subdivided into 5 evolutionary lineages: lineage A (10 subspecies), sublineage Z of the lineage A (3 subspecies), lineage M (3 subspecies), lineage C (10 subspecies), lineage O (3 subspecies), lineage Y (1 subspecies), lineage C or O (3 subspecies). The taxonomy of honey bee A. mellifera has a lot of issues due to the specificity of population structure and resolution of honey bee subspecies discrimination methods. Transition zones between ranges of honey bee subspecies led to the gradual changes of all characteristics between neighbors. There is constant gene flow between most of the subspecies due to the lack of natural isolation mechanisms. Some isolated populations remained on mountains and islands, like populations of the dark European honey bees on the Ural Mountains (Russia), Ireland (Republic of Ireland and Northern Ireland), and Læsø Island (Denmark). Leading by professor Nikolenko A.G. under the Grant RFBR No. 19-54-70002 e-Asia_t we are studying the taxonomy of the sisterly honey bee species Apis cerana based on molecular tools, and our further paper could be "A revision of subspecies structure of eastern honey bee Apis cerana".
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by postdoctoral fellowships from Incheon National University to RI (2017-2019).
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Rustem A. Ilyasov, Email: apismell@hotmail.com.
Jun-ichi Takahashi, Email: jit@cc.kyoto-su.ac.jp.
Hyung Wook Kwon, Email: hwkwon@inu.ac.kr.
References
- Adl M.B.F., Gencer H.V., Firatli C., Bahreini R. Morphometric characterization of Iranian (Apis mellifera meda), Central Anatolian (Apis mellifera anatoliaca) and Caucasian (Apis mellifera caucasica) honey bee populations. J. Apic. Res. 2007;46(4):225–231. doi: 10.3896/IBRA.1.46.4.03. [DOI] [Google Scholar]
- Alburaki M., Moulin S., Legout H.E.E., Alburaki A., Garnery L. Mitochondrial structure of eastern honeybee populations from Syria, Lebanon and Iraq. Apidologie. 2011;42(5):628–641. doi: 10.1007/s13592-011-0062-4. [DOI] [Google Scholar]
- Alburaki M., Bertrand B., Legout H., Moulin S., Alburaki A., Sheppard W.S. A fifth major genetic group among honeybees revealed in Syria. BMC Genet. 2013;14:1–10. doi: 10.1186/1471-2156-14-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias M.C., Sheppard W.S. Molecular phylogenetics of honey bee subspecies (Apis mellifera L.) inferred from mitochondrial DNA sequence. Mol. Phylogenet. Evol. 1996;5(3):557–566. doi: 10.1006/mpev.1996.0050. [DOI] [PubMed] [Google Scholar]
- Arias M.C., Sheppard W.S. Phylogenetic relationships of honey bees (Hymenoptera: Apinae: Apini) inferred from nuclear and mitochondrial DNA sequence data. Mol. Phylogenet. Evol. 2005;37(1):25–35. doi: 10.1016/j.ympev.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Amssalu B., Nuru A., Radloff S.E., Hepburn H.R. Multivariate morphometric analysis of honeybees (Apis mellifera) in the Ethiopian region. Apidologie. 2004;35(1):71–81. doi: 10.1051/apido:2003066. [DOI] [Google Scholar]
- Bookstein F.L. Cambridge University Press; New York, USA: 1991. Morphometric Tools for Landmark Data, Geometry and Biology. [Google Scholar]
- Bouga M., Alaux C., Bienkowska M., Büchler R., Carreck N.L., Cauia E. A review of methods for discrimination of honey bee populations as applied to European beekeeping. J. Apic. Res. 2011;50:51–84. doi: 10.3896/IBRA.1.50.1.06. [DOI] [Google Scholar]
- Bouga M., Harizanis P.C., Kilias G., Alahiotis S. Genetic divergence and phylogenetic relationships of honey bee Apis mellifera (Hymenoptera: Apidae) populations from Greece and Cyprus using PCR-RFLP analysis of three mtDNA segments. Apidologie. 2005;36(3):335–344. doi: 10.1051/apido:2005021. [DOI] [Google Scholar]
- Buttel-Reepen H.V. Beiträge zur systematik, biologie, sowie zur geschichtlichen und geographischen verbreitung der honigbiene (Apis mellifica L.), ihrer varietaten und der iiberigen apis-arten. Mitteilungen aus dem zoologischen museum in Berlin. Apistica. 1906;3:117–201. [Google Scholar]
- Cánovas F., Serrano J., Galián J., De La Rúa P. Geographical patterns of mitochondrial DNA variation in Apis mellifera iberiensis (Hymenoptera: Apidae) J. Zool. Syst. Evol. Res. 2008;46(1):24–30. doi: 10.1111/j.1439-0469.2007.00435.x. [DOI] [Google Scholar]
- Cauia E., Usurelu D., Magdalena L.M., Cimponeriu D., Apostol P., Siceanu A. Preliminary researches regarding the genetic and morphometric characterization of honeybees (A. mellifera L.) from Romania. Sci. Pap. Anim. Sci. Biotechnol. 2008;41(2):278–286. [Google Scholar]
- Chapman N.C., Harpur B.A., Lim J., Rinderer T.E., Allsopp M.H., Zayed A. A SNP test to identify Africanized honeybees via proportion of African ancestry. Mol. Ecol. Resour. 2015;15(6):1346–1355. doi: 10.1111/1755-0998.12411. [DOI] [PubMed] [Google Scholar]
- Chapman N.C., Harpur B.A., Lim J., Rinderer T.E., Allsopp M.H., Zayed A. Hybrid origins of Australian honeybees (Apis mellifera) Apidologie. 2016;47(1):26–34. doi: 10.1007/s13592-015-0371-0. [DOI] [Google Scholar]
- Collet T., Ferreira K.M., Arias M.C., Soares A.E.E., Del Lama M.A. Genetic structure of Africanized honey bee populations (Apis mellifera L.) from Brazil and Uruguay viewed through mitochondrial DNA COI-COII patterns. Heredity. 2006;97:329–335. doi: 10.1038/sj.hdy.6800875. [DOI] [PubMed] [Google Scholar]
- Cornuet J.-M., Daoudi A., Mohssine E.H., Fresnaye J. Etude biometrique de colonies d'abeilles marocaines. Apidologie. 1988;19:355–366. [Google Scholar]
- Cornuet J.-M., Garnery L. Mitochondrial DNA variability in honeybees and its phylogeographic implications. Apidologie. 1991;22(6):627–642. doi: 10.1051/apido:19910606. [DOI] [Google Scholar]
- Coroian C.O., Munoz I., Schluns E.A., Paniti-Teleky O.R., Erler S., Furdui E.M. Climate rather than geography separates two European honeybee subspecies. Mol. Ecol. 2014;23(9):2353–2361. doi: 10.1111/mec.12731. [DOI] [PubMed] [Google Scholar]
- Coulibaly, K.a.S., Majeed, M.Z., Chen, C., Yeo, K., Shi, W., Ma, C.S., 2019. Insights into the maternal ancestry of Côte d’Ivoire honeybees using the intergenic region COI-COII. Insects 10 (4). https://doi.org/10.3390/Insects10040090. [DOI] [PMC free article] [PubMed]
- Crozier Y.C., Koulianos S., Crozier R.H. An improved test for Africanized honey bee mitochondrial DNA. Experientia. 1991;47:968–969. doi: 10.1007/BF01929894. [DOI] [PubMed] [Google Scholar]
- De La Rúa P., Galian J., Serrano J., Moritz R.F.A. Molecular characterization and population structure of the honeybees from the Balearic Islands (Spain) Apidologie. 2001;32(5):417–427. doi: 10.1051/apido:2001141. [DOI] [Google Scholar]
- Del Lama M.A., Lobo J.A., Soares A.E.E., Del Lama S.N. Genetic differentiation estimated by isozymic analysis of Africanized honey bee populations from Brazil and from Central America. Apidologie. 1990;21:271–280. doi: 10.1051/apido:19900401. [DOI] [Google Scholar]
- Dukku U.H. Evaluation of morphometric characters of honeybee (Apis mellifera L.) populations in the Lake Chad Basin in Central Africa. Adv. Entomol. 2016;04(02):75–89. doi: 10.7287/peerj.preprints.1680v1. [DOI] [Google Scholar]
- Dupraw E.J. The recognition and handling of honeybee specimens in non-Linnean taxonomy. J. Apic. Res. 1965;4:71–84. doi: 10.1080/00218839.1965.11100107. [DOI] [Google Scholar]
- El-Niweiri M.a.A., Moritz R.F.A. Mitochondrial discrimination of honeybees (Apis mellifera) of Sudan. Apidologie. 2008;39(5):566–573. doi: 10.1051/apido:2008039. [DOI] [Google Scholar]
- Enderlein G. New honeybees and contribution to the distribution of the genus Apis. Stettiner Entomologische Zeitung. 1906;67:331–334. [Google Scholar]
- Engel M.S. The taxonomy of recent and fossil honey bees (Hymenoptera, Apidae, Apis) J. Hymenoptera Res. 1999;8(2):165–196. doi: 10.1007/978-1-4614-4960-7_18. [DOI] [Google Scholar]
- Estoup A., Garnery L., Solignac M., Cornuet J.-M. Microsatellite variation in honey bee (Apis mellifera L.) populations: hierarchical genetic structure and test of the infinite allele and stepwise mutation models. Genetics. 1995;140:679–695. doi: 10.1080/00218839.1999.11100990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foti, N., Lungu, M., Pelimon, C., Barac, I., Copaitici, M., Marza, E., 1965. Researches on morphological characteristics and biological features of the bee population in Romania. In: Proceedings of the Proceedings of XXth Jubiliar International Congress of Beekeeping Apimondia, Bucharest, Romania, pp. 171–176.
- Franck P., Garnery L., Loiseau A., Oldroyd B.P., Hepburn H.R., Solignac M. Genetic diversity of the honeybee in Africa: microsatellite and mitochondrial data. Heredity. 2001;86(4):420–430. doi: 10.1046/j.1365-2540.2001.00842.x. [DOI] [PubMed] [Google Scholar]
- Franck P., Garnery L., Solignac M., Cornuet J.-M. The origin of west European subspecies of honeybees (Apis mellifera): new insights from microsatellite and mitochondrial data. Evolution. 1998;52(4):1119–1134. doi: 10.2307/2411242. [DOI] [PubMed] [Google Scholar]
- Franck P., Garnery L., Solignac M., Cornuet J.-M. Molecular confirmation of a fourth lineage in honeybees from the Near East. Apidologie. 2000;31:167–180. doi: 10.1051/apido:2000114. [DOI] [Google Scholar]
- Garnery L., Franck P., Baudry E., Vautrin D., Cornuet J.M., Solignac M. Genetic diversity of the west European honey bee (Apis mellifera mellifera and A. m. iberica). II. Microsatellite loci. Genet. Sel. Evol. 1998;30(Suppl 1):49–74. doi: 10.1186/1297-9686-30-s1-s49. [DOI] [Google Scholar]
- Garnery L., Cornuet J.M., Solignac M. Evolutionary history of the honey bee Apis mellifera inferred from mitochondrial DNA analysis. Mol. Ecol. 1992;1(3):145–154. doi: 10.1111/j.1365-294X.1992.tb00170.x. [DOI] [PubMed] [Google Scholar]
- Garnery L., Mosshine E.H., Oldroyd B.P., Cornuet J.-M. Mitochondrial DNA variation in Moroccan and Spanish honey bee populations. Mol. Ecol. 1995;4:465–472. doi: 10.1111/j.1365-294X.1995.tb00240.x. [DOI] [Google Scholar]
- Hall H.G., Smith D.R. Distinguishing African and European honey bee matrilines using amplified mitochondrial DNA. PNAS. 1991;88:4548–4552. doi: 10.1073/pnas.88.10.4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpur B.A., Chapman N.C., Krimus L., Maciukiewicz P., Sandhu V., Sood K. Assessing patterns of admixture and ancestry in Canadian honey bees. Insectes Soc. 2015;62(4):479–489. doi: 10.1007/s00040-015-0427-1. [DOI] [Google Scholar]
- Harpur B.A., Kent C.F., Molodtsova D., Lebon J.M.D., Alqarni A.S., Owayss A.A. Population genomics of the honey bee reveals strong signatures of positive selection on worker traits. Proc. Natl. Acad. Sci. USA. 2014;111(7):2614–2619. doi: 10.1073/pnas.1315506111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepburn, H.R., Radloff, S.E., 1998. Honeybees of Africa Springer Berlin Heidelberg. https://doi.org/10.1007/978-3-662-03604-4.
- Henriques D., Wallberg A., Chavez-Galarza J., Johnston J.S., Webster M.T., Pinto M.A. Whole genome SNP-associated signatures of local adaptation in honeybees of the Iberian Peninsula. Sci. Rep. 2018;8(1):11145. doi: 10.1038/s41598-018-29469-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt G.J., Page R.E.J. Linkage analysis of sex determination in the honey bee (Apis mellifera) Mol. Gen. Genet. 1994;244(5):512–518. doi: 10.1007/BF00583902. [DOI] [PubMed] [Google Scholar]
- Ilyasov R.A., Kutuev I.A., Petukhov A.V., Poskryakov A.V., Nikolenko A.G. Phylogenetic relationships of dark European honeybees Apis mellifera mellifera L. from the Russian Ural and West European populations. J. Apicult. Sci. 2011;55(1):67–76. [Google Scholar]
- Ilyasov R.A., Park J., Takahashi J., Kwon H.W. Phylogenetic uniqueness of honeybee Apis cerana from the Korean peninsula inferred from the mitochondrial, nuclear, and morphological data. J. Apicul. Sci. 2018;62(2):189–214. doi: 10.2478/JAS-2018-0018. [DOI] [Google Scholar]
- Ilyasov R.A., Nikolenko A.G., Tuktarov V.R., Goto K., Takahashi J.I., Kwon H.W. Comparative analysis of mitochondrial genomes of the honey bee subspecies A. m. caucasica and A. m. carpathica and refinement of their evolutionary lineages. J. Apic. Res. 2019;58(4):567–579. doi: 10.1080/00218839.2019.1622320. [DOI] [Google Scholar]
- Il'yasov R.A., Petukhov A.V., Poskryakov A.V., Nikolenko A.G. Local honeybee (Apis mellifera mellifera L.) populations in the Urals. Russian J. Genet. 2007;43(6):709–711. [PubMed] [Google Scholar]
- Il’yasov R.A., Poskryakov A.V., Petukhov A.V., Nikolenko A.G. Genetic differentiation of local populations of the dark European bee Apis mellifera mellifera L. in the Urals. Russian J. Genet. 2015;51(7):677–682. doi: 10.1134/S1022795415070042. [DOI] [PubMed] [Google Scholar]
- Ilyasov R.A., Poskryakov A.V., Petukhov A.V., Nikolenko A.G. New approach to the mitotype classification in black honeybee Apis mellifera mellifera and Iberian honeybee Apis mellifera iberiensis. Russian J. Genet. 2016;52:281–291. doi: 10.1134/S1022795416020058. [DOI] [PubMed] [Google Scholar]
- Ivanova E., Bienkowska M., Petrov P. Allozyme polymorphism and phylogenetic relationships in Apis mellifera subspecies selectively reared in Poland and Bulgaria. Folia Biol. 2011;59:3–4. doi: 10.3409/fb59_3-4.121-126. [DOI] [PubMed] [Google Scholar]
- Ivanova E., Staykova T., Petrov P. Allozyme variability in populations of local Bulgarian honey bee. Biotechnol. Biotechnol. Equip. 2010;24(2):371–374. doi: 10.1080/13102818.2010.10817868. [DOI] [Google Scholar]
- Jensen A.B., Palmer K.A., Boomsma J.J., Pedersen B.V. Varying degrees of Apis mellifera ligustica introgression in protected populations of the black honeybee, Apis mellifera mellifera, in northwest Europe. Mol. Ecol. 2005;14(1):93–106. doi: 10.1111/j.1365-294X.2004.02399.x. [DOI] [PubMed] [Google Scholar]
- Kandemir I., Kence A. Allozyme variability in a central Anatolian honeybee (Apis mellifera L.) population. Apidologie. 1995;26:503–510. doi: 10.1051/apido:19950607. [DOI] [Google Scholar]
- Kandemir I., Kence M., Kence A. Morphometric and electrophoretic variation in different honeybee (Apis mellifera L.) populations. Turkish J. Vet. Anim. Sci. 2005;29(3):885–890. [Google Scholar]
- Kandemir I., Kence M., Sheppard W.S., Kence A. Mitochondrial DNA variation in honey bee (Apis mellifera L.) populations from Turkey. J. Apic. Res. 2006;45(1):33–38. doi: 10.1080/00218839.2006.11101310. [DOI] [Google Scholar]
- Kandemir I., Ozkan A., Fuchs S. Reevaluation of honeybee (Apis mellifera) microtaxonomy: a geometric morphometric approach. Apidologie. 2011;42(5):618–627. doi: 10.1007/s13592-011-0063-3. [DOI] [Google Scholar]
- Kasangaki P., Nyamasyo G., Ndegwa P., Kajobe R., Angiro C., Kato A. Mitochondrial DNA (mtDNA) markers reveal low genetic variation and the presence of two honey bee races in Uganda's agro-ecological zones. J. Apic. Res. 2017;56(2):112–121. doi: 10.1080/00218839.2017.1287997. [DOI] [Google Scholar]
- Kent C.F., Issa A., Bunting A.C., Zayed A. Adaptive evolution of a key gene affecting queen and worker traits in the honey bee, Apis mellifera. Mol. Ecol. 2011;20(24):5226–5235. doi: 10.1111/j.1365-294X.2011.05299.x. [DOI] [PubMed] [Google Scholar]
- Kent C.F., Minaei S., Harpur B.A., Zayed A. Recombination is associated with the evolution of genome structure and worker behavior in honey bees. Proc. Natl. Acad. Sci. USA. 2012;109(44):18012–18017. doi: 10.1073/pnas.1208094109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr W.E. Abelhas Africanas su introduction y expansion en el continente Americano; Subspecies y ecotipas Africanos. Industrial Apiculture. 1992;13:12–21. [Google Scholar]
- Koulianos S., Crozier R.H. Mitochondrial sequence characterisation of Australian commercial and feral honeybee strains, Apis mellifera L. (Hymenoptera: Apidae), in the context of the species worldwide. Aust. J. Entomol. 1997;36:359–364. doi: 10.1111/j.1440-6055.1997.tb01486.x. [DOI] [Google Scholar]
- Kumar Y., Khan M.S. Genetic variability of European honey bee, Apis mellifera in mid hills, plains and tarai region of India. Afr. J. Biotechnol. 2014;13(8):916–925. doi: 10.5897/AJB2013.13142. [DOI] [Google Scholar]
- Lazarov A. Brief contribution for the study of local Bulgarian bee. Works Bulgarian Natural. Soc. 1936;6:156–158. [Google Scholar]
- Lebdigrissa K., Msadda K., Cornuet J.-M., Fresnaye J. Phylogenetic relationships between the Tunisian honeybee Apis mellifera intermissa and neighbouring African and west Mediterranean honeybee breeds. Landbouwtijdschrift. 1991;44:1231–1238. [Google Scholar]
- Maa T.C. An inquiry into the systematics of the tribus Apidini or honeybees (Hymenoptera) Treubia. 1953;21:525–640. [Google Scholar]
- Mărghitas, L.A., Dezmirean, D., Teleky, O.R., Furdui, E., Moise, A., Stan, L., et al., 2009. Biodiversity testing of Transylvanian honeybee populations using mtDNA markers. Bull. Univ. Agric. Sci. Veterinary Med. Anim. Sci. Biotechnol. 66 (1–2), 402–406. https://doi.org/10.15835/buasvmcn-asb:66:1-2:3391.
- Mărghitas L.A., Teleky O., Dezmirean D., Rodica M., Bojan C., Coroian C. Morphometric differences between honey bees Apis mellifera carpatica populations from Transylvanian area. Lucrări stiintifice Zootehnie si Biotehnologii. 2008;41(2):309–315. [Google Scholar]
- Martimianakis S., Klossa-Kilia E., Bouga M., Kilias G. Phylogenetic relationships of Greek Apis mellifera subspecies based on sequencing of mtDNA segments (COI and ND5) J. Apic. Res. 2011;50(1):42–50. doi: 10.3896/IBRA.1.50.1.05. [DOI] [Google Scholar]
- Meixner M.D., Arias M.C., Sheppard W.S. Mitochondrial DNA polymorphisms in honey bee subspecies from Kenya. Apidologie. 2000;31:181–190. [Google Scholar]
- Meixner M.D., Leta M.A., Koeniger N., Fuchs S. The honey bees of Ethiopia represent a new subspecies of Apis mellifera - Apis mellifera simensis n. ssp. Apidologie. 2011;42(3):425–437. doi: 10.1007/s13592-011-0007-y. [DOI] [Google Scholar]
- Meixner M.D., Pinto M.A., Bouga M., Kryger P., Ivanova E., Fuchs S. Standard methods for characterising subspecies and ecotypes of Apis mellifera. J. Apic. Res. 2013;52(4):1–28. doi: 10.3896/IBRA.1.52.4.05. [DOI] [Google Scholar]
- Meixner M.D., Worobik M., Wilde J., Fuchs S., Koeniger N. Apis mellifera mellifera in eastern Europe - morphometric variation and determination of its range limits. Apidologie. 2007;38:191–197. doi: 10.1051/apido:2006068. [DOI] [Google Scholar]
- Miguel I., Baylac M., Iriondo M., Manzano C., Garnery L., Estonba A. Both geometric morphometric and microsatellite data consistently support the differentiation of the Apis mellifera M evolutionary branch. Apidologie. 2011;42(2):150–161. doi: 10.1051/apido/2010048. [DOI] [Google Scholar]
- Moritz R.F.A., Hawkins C.F., Crozier R.H., Mackinlay A.G. A mitochondrial DNA polymorphism in honeybees (Apis mellifera L.) Experientia. 1986;42(3):322–324. doi: 10.1007/BF01942522. [DOI] [Google Scholar]
- Muli E., Patch H., Frazier M., Frazier J., Torto B., Baumgarten T. Evaluation of the distribution and impacts of parasites, pathogens, and pesticides on honey bee (Apis mellifera) populations in East Africa. PLoS ONE. 2014;9(4) doi: 10.1371/journal.pone.0094459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolova S. Genetic variability of local Bulgarian honey bees Apis mellifera macedonica (rodopica) based on microsatellite DNA analysis. J. Apicult. Sci. 2011;55(2):117–129. [Google Scholar]
- Özdïl F., Yildiz M.A., Hall H.G. Molecular characterization of Turkish honey bee populations (Apis mellifera) inferred from mitochondrial DNA RFLP and sequence results. Apidologie. 2009;40(5):570–576. doi: 10.1051/apido/2009032. [DOI] [Google Scholar]
- Palmer M.R., Smith D.R., Kaftanoglu O. Turkish honeybees: genetic variation and evidence of a fourth lineage of Apis mellifera mtDNA. J. Hered. 2000;91:42–46. doi: 10.1093/jhered/91.1.42. [DOI] [PubMed] [Google Scholar]
- Pinto M.A., Henriques D., Chávez-Galarza J., Kryger P., Garnery L., Van Der Zee R. Genetic integrity of the dark European honey bee (Apis mellifera mellifera) from protected populations: a genome-wide assessment using SNPs and mtDNA sequence data. J. Apic. Res. 2014;53:269–278. doi: 10.3896/IBRA.1.53.2.08. [DOI] [Google Scholar]
- Pinto M.A., Munoz I., Chavez-Galarza J., De La Rúa P. The Atlantic side of the Iberian Peninsula: A hot-spot of novel African honey bee maternal diversity. Apidologie. 2012;43:663–673. doi: 10.1007/s13592-012-0141-1. [DOI] [Google Scholar]
- Radloff S.E., Hepburn H.R., Fuchs S. Ecological and morphological differentiation of the honeybees. Apis mellifera Linnaeus (Hymenoptera: Apidae). of West Africa. Afr. Entomol. 1998;6(1):17–23. [Google Scholar]
- Radoslavov G., Hristov P., Shumkova R., Mitkov I., Sirakova D., Bouga M. A specific genetic marker for the discrimination of native Bulgarian honey bees (Apis mellifera rodopica): duplication of COI gene fragment. J. Apic. Res. 2017;56(3):196–202. doi: 10.1080/00218839.2017.1307713. [DOI] [Google Scholar]
- Ruttner, F., 1988. Biogeography and taxonomy of honeybees. Springer-Verlag Berlin Heidelberg GmbH, 288. https://doi.org/10.1016/0169-5347(89)90176-6.
- Ruttner, F., 1992. Naturgeschichte der Honigbienen. 455.
- Ruttner F., Tassencourt L., Louveaux J. Biometrical statistical analysis of the geographic variability of Apis mellifera L. Apidologie. 1978;9:363–382. doi: 10.1051/apido:19780408. [DOI] [Google Scholar]
- Shaibi T., Fuchs S., Moritz R.F.A. Morphological study of honeybees (Apis mellifera) from Libya. Apidologie. 2009;40:97–105. doi: 10.1051/apido/2008068. [DOI] [Google Scholar]
- Sheppard W.S., Arias M.C., Grech A., Meixner M.D. Apis mellifera ruttneri, a new honey bee subspecies from Malta. Apidologie. 1997;28:287–293. doi: 10.1051/apido:19970505. [DOI] [Google Scholar]
- Sheppard W.S., Berlocher S.H. Enzyme polymorphism in Apis mellifera from Norway. J. Apic. Res. 1984;23:64–69. doi: 10.1080/00218839.1984.11100610. [DOI] [Google Scholar]
- Sheppard, W.S., Rinderer, T.E., Meixner, M.D., Yoo, H.R., Stelzer, J.A., Al., E., 1996. Hinfl variation in mitochondrial DNA of old world honey bee subspecies. J. Heredity 87, 35–40. https://doi.org/10.1093/oxfordjournals.jhered.a022950.
- Skorikov A.S. Beiträge zur Kenntnis der Kenntnis der kaukasischen Honigbienenrassen. Rep. Bureau Appl. Entomol. (Russia) 1929;4:1–59. [Google Scholar]
- Skorikov A.S. Eine neue Basis fur eine Revision der Gattung Apis L. Rep. Bureau Appl. Entomol. (Russia) 1929;4:249–270. [Google Scholar]
- Smith D.R. Mitochondrial DNA and honeybee biogeography. In: Smith D.R., editor. Diversity in the genus Apis. Westview Press; Boulder, CO: 1991. pp. 131–176. [Google Scholar]
- Smith D.R. Genetic diversity in Turkish honey bees. Uludağ Arıcılık Dergisi. 2002;2:10–17. doi: 10.1080/09064700600641681. [DOI] [Google Scholar]
- Smith D.R., Brown W.M. Polymorphisms in mitochondrial DNA of European and Africanized honey bees (Apis mellifera) Experientia. 1988;44:257–260. doi: 10.1007/BF01941730. [DOI] [PubMed] [Google Scholar]
- Smith D.R., Crespi B.J., Bookstein F.L. Fluctuating asymmetry in the honey bee, Apis mellifera: effects of ploidy and hybridization. J. Evol. Biol. 1997;10(4):551–574. doi: 10.1007/s000360050041. [DOI] [Google Scholar]
- Smith D.R., Glenn T.C. Allozyme polymorphisms in Spanish honeybees (Apis mellifera iberica) J. Hered. 1995;86(1):12–16. doi: 10.1093/oxfordjournals.jhered.a111518. [DOI] [PubMed] [Google Scholar]
- Smith D.R., Taylor O.R., Brown W.M. Neotropical Africanized honey bees have African mitochondrial DNA. Nature. 1989;339:213–215. doi: 10.1038/339213a0. [DOI] [PubMed] [Google Scholar]
- Solignac M., Vautrin D., Loiseau A., Mougel F., Baudry E., Estoup A. Five hundred and fifty microsatellite markers for the study of the honeybee (Apis mellifera L.) genome. Mol. Ecol. Notes. 2003;3:307–311. doi: 10.1046/j.1471-8286.2003.00436.x. [DOI] [Google Scholar]
- Stevanovic J., Stanimirovic Z., Radakovic M., Kovacevic S.R. Biogeographic study of the honey bee (Apis mellifera L.) from Serbia, Bosnia and Herzegovina and Republic of Macedonia Based on mitochondrial DNA analyses. Russian J. Genet. 2010;46(5):603–609. doi: 10.1134/S1022795410050145. [DOI] [PubMed] [Google Scholar]
- Suazo A., Mctiernan R., Hall H.G. Differences between African and European honey bees (Apis mellifera L.) in random amplified polymorphic DNA (RAPD) J. Hered. 1998;89(1):32–36. doi: 10.1093/jhered/89.1.32. [DOI] [Google Scholar]
- Syromyatnikov M.Y., Borodachev A.V., Kokina A.V., Popov V.N. A molecular method for the identification of honey bee subspecies used by beekeepers in Russia. Insects. 2018;9:E10. doi: 10.3390/insects9010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Techer M.A., Clémencet J., Simiand C., Turpin P., Garnery L., Reynaud B. Genetic diversity and differentiation among insular honey bee populations in the southwest Indian Ocean likely reflect old geographical isolation and modern introductions. PLoS ONE. 2017;12(12) doi: 10.1371/journal.pone.0189234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleky, O.R., Bojan, C., Moise, A., Coroian, C., Dezmirean, D., Mărghitaș, L.A., 2009. Ecotypes differentiation within honeybee (Apis mellifera carpatica) from Transylvania. Bulletin of University of Agricultural Sciences and Veterinary Medicine. Anim. Sci. Biotechnol. 64, 1–6. https://doi.org/http://dx.doi.org/10.15835/buasvmcn-asb:64:1-2:2256.
- Tunca R.I., Kence M. Genetic diversity of honey bee (Apis mellifera L.: hymenoptera: Apidae) populations in Turkey revealed by RAPD markers. Afr. J. Agric. Res. 2011;6:6217–6225. doi: 10.5897/AJAR10.386. [DOI] [Google Scholar]
- Tzonev I. Biometric investigations on the A. mellifera (methods, season dynamics and family changeability) News Student Scient. Soc. 1960:157–191. [Google Scholar]
- Uzunov A., Meixner M.D., Kiprijanovska H., Andonov S., Gregorc A., Ivanova E. Genetic structure of apis mellifera macedonica in the Balkan peninsula based on microsatellite DNA polymorphism. J. Apic. Res. 2014;53(2):288–295. doi: 10.3896/IBRA.1.53.2.10. [DOI] [Google Scholar]
- Velichkov V. Honey bee races in Bulgaria. Beekeeping. 1970;10:7–11. [Google Scholar]
- Waldschmidt A.M., Marco-Junior P., Barros E.G., Campos L.A. Genetic analysis of Melipona quadrifasciata Lep. (Hymenoptera: Apidae, Meliponinae) with RAPD markers. Braz. J. Biol. 2002;62(4B):923–928. doi: 10.1590/s1519-69842002000500022. [DOI] [PubMed] [Google Scholar]
- Wallberg A., Bunikis I., Pettersson O.V., Mosbech M.-B., Childers A.K., Evans J.D. A hybrid de novo genome assembly of the honeybee, Apis mellifera, with chromosome-length scaffolds. BMC Genomics. 2019;20(1):275. doi: 10.1186/s12864-019-5642-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallberg A., Han F., Wellhagen G., Dahle B., Kawata M., Haddad N. A worldwide survey of genome sequence variation provides insight into the evolutionary history of the honeybee Apis mellifera. Nat. Genet. 2014;46(10):1081–1088. doi: 10.1038/ng.3077. [DOI] [PubMed] [Google Scholar]
- Weinstock G.M., Robinson G.E., Gibbs R.A., Worley K.C., Evans J.D., Maleszka R. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2007;443(7114):931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield C.W., Ben-Shahar Y., Brillet C., Leoncini I., Crauser D., Leconte Y. Genomic dissection of behavioral maturation in the honey bee. Proc. Natl. Acad. Sci. 2006;103(44):16068–16075. doi: 10.1073/pnas.0606909103. [DOI] [PMC free article] [PubMed] [Google Scholar]