Abstract
Garlic (Allium sativum L.), is a predominant spice, which is used as an herbal medicine and flavoring agent, since ancient times. It has a rich source of various secondary metabolites such as flavonoids, terpenoids and alkaloids, which have various pharmacological properties. Garlic is used in the treatment of various ailments such as cancer, diabetes and cardiovascular diseases. The present study aims to explore the plausible mechanisms of the selected phytocompounds as potential inhibitors against the known drug targets of non-small-cell lung cancer (NSCLC). The phytocompounds of garlic were identified by gas chromatography-mass spectrometry (GC–MS) technique. Subsequently, the identified phytocompounds were subjected to molecular docking to predict the binding with the drug targets, epidermal growth factor receptor (EGFR), human epidermal growth factor receptor 2 (HER2), echinoderm microtubule-associated protein-like 4-anaplastic lymphoma kinase (EML4-ALK) and group IIa secretory phospholipase A2 (sPLA2-IIA). Molecular dynamics is used to predict the stability of the identified phytocompounds against NSCLC drug targets by refining the intermolecular interactions formed between them. Among the 12 phytocompounds of garlic, three compounds[1,4-dimethyl-7-(1-methylethyl)-2-azulenyl]phenylmethanone, 2,4-bis(1-phenylethyl)-phenol and 4,5–2 h-oxazole-5-one,4-[3,5-di-t-butyl-4-methoxyphenyl] methylene-2-phenyl were identified as potential inhibitors, which might be suitable for targeting the different clinical forms of EGFR and dual inhibition of the studied drug targets to combat NSCLC. The result of this study suggest that these identified phytocompounds from garlic would serve as promising leads for the development of lead molecules to design new multi-targeting drugs to address the different clinical forms of NSCLC.
Keywords: Docking, Garlic, Inhibitors, Lung cancer, Molecular dynamics simulations
1. Introduction
The most common cause of neoplasia-related deaths that occur worldwide is lung cancer (Bankovic et al., 2010, Al-Dhabi et al., 2015). Lung cancer, is a malignant tumour characterized by abnormal cell growth in the tissue lining of the lung and is categorized into Non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC) depending upon its cell type. NSCLC signifies 80% of all lung cancers, with adenocarcinoma accounting for 40% of all cases (Rom et al., 2000). Mutator phenotype and induction of genomic instability are the key events in the early stages of cancer. The comprehensive molecular investigation of NSCLC has made it possible to identify the foremost genes that are critical in carcinogenesis process.The significant genes reported are epidermal growth factor receptor (EGFR), human epidermal growth factor receptor (HER), echinoderm microtubule-associated protein-like 4 anaplastic lymphoma kinase (EML4-ALK) and phospholipase A2 group IIA (PLA2G2A) (Daga et al., 2015, Valsalam et al., 2019a). Among these genes, 10–40% of the NSCLC patients show active mutation in EGFR gene. The EGFR family of gene belongs to receptor tyrosine kinases and has four known members: EGFR or HER1/ErbB1, HER2/ErbB2, HER3/ErbB3, and HER4/ErbB4 (Wieduwilt and Moasser, 2008, Valsalam et al., 2019a). Among these, EGFR and HER2 are the key receptors that possess an intracellular protein kinase domain with tyrosine kinase activity, which are expressed in lung cancer. The amplification or overexpression of both genes has been connected in the clinical course of several cancers, including NSCLC.40% of Asian and 10–15% of the Caucasian patient’s exhibit EGFR mutations. Mutations, duplications, deletions and insertions in EGFR exon 18 have been commonly reported (Elango et al., 2017, Galli et al., 2018). HER2, a member of the ErbB tyrosine kinase family is the preferred binding partner of the ErbB receptors and HER2/EGFR heterodimer has a potent signal transduction when compared to EGFR homodimers. The oncogenic driver mutations in the HER2 gene are found as insertion mutations in exon 20 (Kramarski et al., 1998). A fusion type protein tyrosine kinase found in 4–5% NSCLC patients is EML4-ALK. This is the most recently identified oncogene and arises due to the inversion on the short arm of chromosome 2. Inversion is seen in the exons 1–13 of EML4 and exons 20–29 of ALK. This results in the formation of a chimeric protein EML4-ALK (Rikova et al., 2007). Phospholipase A2 (PLA2) belongs to phospholipase family, which initiates downstream generation of cancer promoting eicosanoids by releasing arachidonic acid from cell membranes. Secretory phospholipase A2 (sPLA2-IIA) has been identified as a possible biomarker in some cancers, including lung, prostate, and esophageal cancer (Yu et al., 2012). sPLA2-IIA plays a key role in preventing the development of gastric cancer and also the overexpression of sPLA2-IIA promote in vitro cancer cell aggressiveness and in vivo cancer growth in lung cancer cells (Ganesan et al., 2013).
Tyrosine kinase inhibitors have become the mainstay in the treatment involving these genes in NSCLC. Gefitinib, Lapatinib, Erlotinib (EGFR and HER2 tryrosine kinase inhibtors), Crizotinib, Ceritinib, Alectinib (EML4-ALK tyrosine kinase inhibitor) and Varespladib (sPLA2 inhibitor) have been attractive drugs for treating patients with NSCLC (Zhang et al., 2012). These small molecules also showed remarkable results in binding effectively to mutant oncogenes until the development of acquired drug resistance in patients. Reason behind this activity is due to the steric interference led by the mutations in the oncogenes and the characteristics of the inhibitor binding. Apart from drug resistance, patients also suffer from treatment-related adverse effects such as rashes and diarrhoea (Stella et al., 2012). Hence, a search for a safe and reliable treatment of NSCLC is much required. Though the therapeutic outlooks of lung cancer have endured intense changes over the last decade, primarily due to the advantages experienced through targeted therapy, yet traditional compounds obtained from nature are considered safe and effective. Secondary metabolites and small molecules from plants have been widely accepted as lead agents in the treatment of several cancers (Banaganapalli et al., 2013). Compounds obtained from spices and vegetables consumed as a part of the diet are of immense interest in recent years. Garlic, ginger, pepper, cardamom, Curcumin etc. are a part of the daily consumed food as well as traditional medicine (Zheng et al., 2016, Elango et al., 2016a, Elango et al., 2016b, Fowsiya et al., 2016). A wide range of these active phytocompounds are being explored through economic computational methods and laboratory trails of these agents support their clinical use in treating lung cancer (Glorybai et al., 2015, Haritha et al., 2016, Helan et al., 2016, Ilavenil et al., 2017). Several clinical studies have demonstrated that the phytocompounds of garlic have a great role in the prevention of cancer (Park et al., 2016a, Park et al., 2016b, Park et al., 2017, Surendra et al., 2016a). Research findings revealed the correlation between excess garlic intake and reduction in risks of the pancreas, colon, stomach, esophagus and breast cancers (Fleischauer and Arab, 2001, Al-Dhabi and Arasu, 2016, Barathikannan et al., 2016, Cuong et al., 2017). Epidemiological studies conducted in the Chinese population indicated the correlation between raw garlic consumption and its protection against lung cancer (Myneni et al., 2016). Recent research findings explored a novel constituent, N-Benzyl-N-methyl-dodecan-1-amine (BMDA) from garlic bulbs. It has the potential to inhibit cell migration in A549 lung cancer cells (Kaowinn et al., 2018, Valsalam et al., 2019a). In this context, bioactive metabolic compounds were identified from a garlic cultivar grown at the hills in the Western Ghats using chromatographic technique and were scrutinised using a computational approach in this study against protein targets to identify their potency in inhibiting the oncogenes involved in NSCLC.
2. Materials and methods
2.1. Collection of plant materials and extraction
Garlic bulbs were collected from Attuvampatti, Kodaikanal, India. The bulbs were sliced, dried in air and grinded to obtain garlic powder. The fresh garlic powder was subjected to Soxhlet extraction using ethanol as a solvent for 48 h at the respective boiling point of the solvent (Lin et al., 1999). Whatman No.1 filter paper is used to filter the extract and then concentrated in vacuum at 40 °C using a rotary evaporator. The extract was preserved at 4 °C for further analysis.
2.2. Identification of the phytocompounds by GC–MS analysis
The GC–MS analysis of phytocompounds from the ethanol extract of garlic was done using Perkin Elmer Clarus 680 gas chromatography-mass spectrometer with a fused silica column, packed with Elite-5MS (5% biphenyl, 95% dimethylpolysiloxane, 30 m × 0.25 mm ID × 250 μm df).Spectroscopic detection by GC–MS involved an electron ionization system which utilized high-energy electrons (70 eV) while helium was used as the carrier gas at a constant flow of 1 ml/min. The initial temperature was set at 60 °C with an increasing rate of 3 °C/min and holding time of about 10 min. Finally, the temperature was increased to 300 °C at 10 °C/min.
The injector temperature was set at 260 °C during the chromatographic run. The injection of 1 μL of the extract was performed in splitless mode. The phytocompounds present in the ethanol extract of garlic was determined and compounds are identified by retention time based on the database of National Institute Standard and Technology (NIST).
2.3. Molecular docking studies
The identified phytocompounds of garlic from GC–MS analysis was subjected as ligands to perform molecular docking studies. Molecular docking study was executed with AutoDock v4.2.3in order to predict the binding and structure of the intermolecular complex between the drug targets and the identified phytocompounds (Morris et al., 1998). The three-dimensional structures of key drug targets, EGFR, HER2, EML4-ALK and sPLA2-IIA were retrieved from the RCSB protein data bank (PDB) of high resolution and co-crystallized with respective inhibitors, which was tabulated in Table 1.
Table 1.
Selected drug targets with their structural information.
| Drug Target | PDB Id | Resolution (Å) | Clinical forms |
|---|---|---|---|
| EGFR | 1 M17 | 1.55 | Wild type |
| 5HG8 | 1.42 | T790M-mutant | |
| 5XGN | 3.00 | T790M/C797S-containing mutant | |
| 4LRM | 3.53 | Exon 20 insertion mutant | |
| HER2 | 3PP0 | 2.25 | Wild type |
| ALK | 4Z55 | 1.55 | Wild type, 1 mutation |
| sPLA2-IIA | 5G3N | 3.53 | Wild type |
The main focus of the study is to inhibit EGFR at the different clinical forms of NSCLC. Therefore, the chain A of four EGFR structures from PDB were utilized to mimic the wild type, T790M-containing mutant,T790M/C797S-containing mutant and exon 20 insertion mutants (PDB Id: 1 M17, Resolution: 1.55 Å; PDB Id: 5HG8, Resolution: 1.42 Å; PDB Id: 5XGN, Resolution: 3 Å and PDB Id: 4LRM, Resolution: 3.526 Å respectively) (Stamos et al., 2002, Cheng et al., 2016, Kong et al., 2017, Yasuda et al., 2013). These mutants reflect the different clinical manifestations and pharmacological action of tyrosine kinase inhibitors. In order to explore the possibility of the identified phytocompounds for multi-targeted therapy of NSCLC, the docking calculations were carried out with other proven drug targets with the chain A of HER2, ALK and sPLA2-IIA (PDB Id: 3PP0, Resolution: 2.25 Å, PDB Id: 4Z55, Resolution: 1.55 Å and PDB Id: 5G3N, Resolution: 3.526 Å respectively) (Aertgeerts et al., 2011, Michellys et al., 2016, Giordanetto et al., 2016). Since our interest is to study the binding of identified phytocompounds with the catalytic domain of EML4-ALK, we have utilized the structure of the catalytic domain of ALK for the docking studies, given that the experimental structure of fusion protein of EML4-ALK is not available yet and the catalytic domain is identical in both EML4-ALK and ALK.
2.4. Protein and ligand preparation
The structures of drug targets were preprocessed by removing ligands, heteroatoms and water molecules from their respective PDB crystal structures. The structures were also cleaned in Discovery Studio v4.5 (DS) to model incomplete residues, standardize atom order, correct connectivity/bond orders, and retain one set of alternate conformations and addition of hydrogens (Dassault Systemes BIOVIA). Then, the respective structures of drug targets were processed in AutoDock suite by merging non-polar hydrogens into their respective heavy atoms, followed by addition of Gasteiger-Marsili charges and the atom types were fixed (Morris et al., 2009). The structures of identified phytocompounds were retrieved from PubChem database (Kim et al., 2016) and imported to AutoDockTools as ‘Sybyl mol2′ format. The ligands were processed via ‘Ligand’ menu that fixes the torsion tree, non-polar hydrogens, charges, and atom types.
2.5. Docking calculations
Docking was performed with the drug targets and the identified phytocompounds with the Lamarckian genetic algorithm (LGA) in AutoDock v4.2.3 (Morris et al., 1998). The ligand binding pockets of respective drug targets were identified from the bound co-crystallized inhibitors from PDB structure and subsequently confirmed with the published literature. A grid maps for all atom types of each protein were generated independently by fixing the grid box using AutoGrid utility. The grid spacing was set to 0.375 Å, such a way that it covers the complete ligand-binding pocket. The grid box was centered based on the co-crystallized inhibitors of the respective drug targets. The initial population size was fixed as 300 and 27,000 as the maximum number of generations for 100 independent LGA runs with 25,000,000 as the maximum number of energy evaluations. In each run, the best individual from each generation was propagated to the next generation via ‘elitism’ and the remaining docking parameters were set to default (Saravanan et al., 2012). Root mean square deviation (RMSD) is used to cluster each molecule, by analysing the docked positions and it is used to determine the optimal binding of the identified phytocompounds with their respective drug targets. The phytocompounds were visually inspected for their intermolecular interactions (hydrogen bonds, van der Waals and hydrophobic interactions) with the respective drug targets with the help of DS. The phytocompounds with satisfactory binding with the respective drug targets were investigated further by molecular dynamics simulations.
2.6. Molecular dynamics simulations
Classical MD simulations of each system was carried out with CHARMM27 all-atom force field using GROMACS v5.1.4 program while the topology of the phytocompounds were generated by the SwissParam webserver, which was summarized in Table 2 (Bjelkmar et al., 2010, Zoete et al., 2011). The independent MD simulations were carried out on the intermolecular complex from docking studies, which serve as the initial structure for each system. Each system is neutralized with counter-ions in a dodecahedron box with TIP3P water model followed by energy minimization with a force below 1000 kJ/mol via steepest descent algorithm. The initial equilibration was performed at constant volume (NVT) for 1 ns at a temperature of 300 K, followed by second equilibration for 1 ns at constant pressure (NPT) of 1 bar.
Table 2.
GC–MS profile of ethanol extract of garlic.
| S. No | Compound Name | Short Names | % of Peak Area | Retention time | Molecular formula | Molecular weight |
|---|---|---|---|---|---|---|
| 1. | 2-HYDROXY PROPANAMIDE | Propanamide | 4.06 | 2.85 | C3H7O2N | 89 |
| 2. | P-TERT-BUTYL PHENOL | Butyl-phenol | 11.54 | 12.81 | C10H14O | 150 |
| 3. | 3-HYDROXY-2,2-DIMETHYL-, 3-HYDROXY-2,2-DIMETHYLPROPYL PROPANOIC ACID | Propanoic-acid | 3.35 | 13.42 | C10H20O4 | 204 |
| 4. | 1-OXASPIRO[2.5]OCT-5-ENE, 8,8-DIMETHYL-4-METHYLENE | Oxaspiro | 2.65 | 16.44 | C10H14O | 150 |
| 5. | 4-(1,1-DIMETHYLPROPYL)- PHENOL | Dimethylpropyl | 4.72 | 16.68 | C11H16O | 164 |
| 6. | HEXESTROL | Hexestrol | 2.21 | 16.97 | C18H22O2 | 270 |
| 7. | 4-(1-PHENYLETHYL)- PHENOL | Phenylethyl | 6.22 | 17.18 | C14H14O | 198 |
| 8. | N-HEXADECANOIC ACID | Decanoic-acid | 3.79 | 19.02 | C16H32O2 | 256 |
| 9. | (Z)-14-TRICOSENYL FORMATE | Tricosenyl | 5.19 | 20.44 | C24H46O2 | 366 |
| 10. | [1,4-DIMETHYL-7-(1-METHYLETHYL)-2-AZULENYL]PHENYL METHANONE | Methanone | 9.42 | 23.26 | C22H22O | 302 |
| 11. | 2,4-BIS(1-PHENYLETHYL)- PHENOL | Bis-Phenylethyl | 15.27 | 22.50 | C22H22O | 302 |
| 12. | 4,5-2H-OXAZOLE-5-ONE, 4-[3,5-DI-T-BUTYL-4-METHOXYPHENYL]METHYLENE-2-PHENYL | Oxazole | 31.53 | 26.95 | C25H29O3N | 391 |
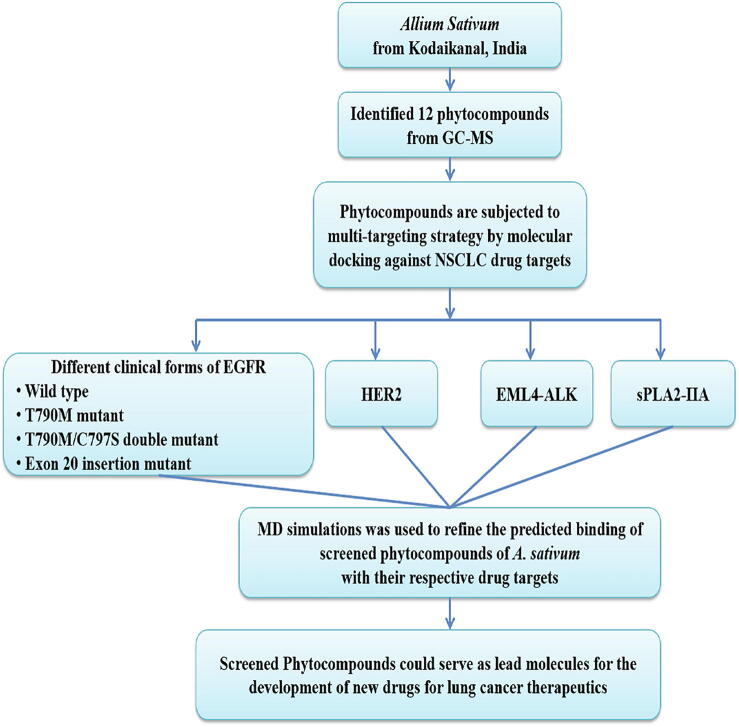
Since we are refining the intermolecular complex from docking studies, the production runs of 20 ns in NPT ensemble at an integration time of 0.2 fs were performed using leapfrog algorithm. LINCS algorithm was used to constrain all bonds while the ionic interactions were calculated using particle mesh Ewald algorithm (Hess, 2008). In total, 20,000 conformations for each system were collected from MD trajectories and subsequently investigated with DS, GROMACS built-in utilities, visual molecular dynamics (VMD) (Humphrey et al., 1996). Further, the intermolecular complexes of the identified phytocompounds with drug targets were manually inspected for satisfactory binding. The research methodology of the current study is depicted in Fig. 1.
Fig. 1.
Methodology of the in silico Molecular Docking analysis.
3. Results and discussion
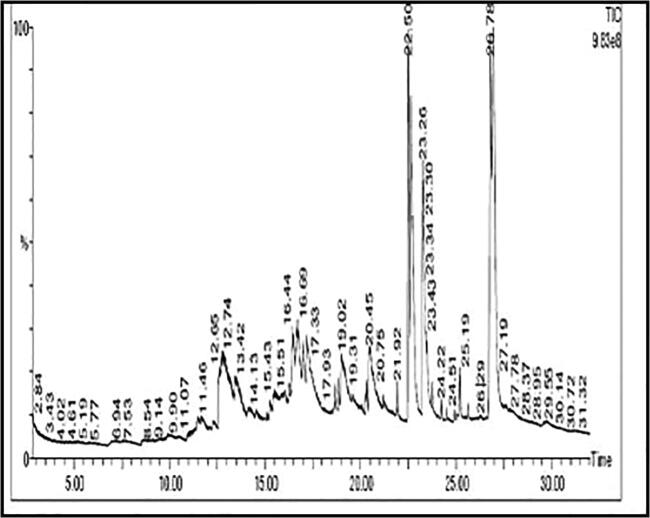
In general, Garlic produces biologically active metabolites with a variety of properties (Surendra et al., 2016. 162:b, Surendra et al., 2016c). Natural compounds as drug targets were necessary before computing their potency against NSCLC oncogenes. Identification of the active compounds present in the ethanolic extract of the garlic bulbs was done with the help of Gas Chromatography and Mass Spectroscopy technique (GC–MS). Based on a vigilant analysis of the chromatogram and MS fragmentation data when compared with the NIST library, we identified 12 compounds (Fig. 2 & Table 2). Numerals indicating the percentage of peak area aided in identifying the levels of the individual compounds in the ethanolic extract. The compound with the highest peak area percentage was 4,5–2 h-oxazole-5-one,4-[3,5-di-t-butyl-4-methoxyphenyl]methylene-2-phenyl accounting for 31.53%. Other compounds that dominated the chromatogram were 2,4-bis(1-phenylethyl)- phenol, p-tert-butyl phenol and1,4-dimethyl-7-(1-methylethyl)-2-azulenyl]phenyl methanone. Verma et al., 2014 has reported that Cadabatrifoliata leaves containing 1,4-dimethyl-7-(1-methylethyl)-2-azulenyl] phenyl Methanone have antimicrobial, antipyretic and anthelmintics activities. Melappa et al., 2017 in a study has reported the use of 2,4-bis(1-phenylethyl)- phenol and p-tert-butyl phenol identified from endophytic fungi as antimitotic and antiproliferative agents. 2,4-bis(1-phenylethyl)-phenol also showed antioxidant property (Kurian et al.,2010). Compounds from the present study were identified for the first time in ethanolic garlic extract and are hardly reported for their anti-proliferative effects in NSCLC oncogenes; therefore we further proceeded in studying their interaction using computational studies.
Fig. 2.
GC–MS chromatogram of ethanol extract of A.sativum.
To assess the suitability of the identified phytocompounds from garlic as potential anticancer agents, the computational studies were carried out on the known drug targets of NSCLC, EGFR, HER2, EML4-ALK, and sPLA-IIA. The docking scores of the studied natural products were comparatively low, owing to their large size, huge contribution of hydrophobic interactions. The phytocompounds were bound well in the desired ligand-binding pocket of wild type and mutants of EGFR via hydrogen bonds along with significant hydrophobic interactions. Molecular docking of the identified phytocompounds with drug target, EGFR on its different clinical forms is given in Table 3. Docking studies revealed that the phytocompounds methanone, bis-phenylethyl, oxazoleone, hexestrol and oxazole had the potential to bind with the different clinical forms of EGFR. These phytocompounds can serve as a starting arsenal to develop lead molecules for EGFR-based therapeutics.
Table 3.
Molecular docking analysis of phytocompounds from garlic with different forms of EGFR.
| S.No. | Inhibitor | 1 M17 |
5HG8 |
5XGN |
4LRM |
||||
|---|---|---|---|---|---|---|---|---|---|
| delG# | Confs$ | delG# | Confs$ | delG# | Confs$ | delG# | Confs$ | ||
| 1 | Methanone | −8.23 | 89 | −8.53 | 38 | −9.45 | 59 | −9.10 | 65 |
| 2 | Bis-Phenylethyl | −8.14 | 61 | −7.06 | 17 | −7.7 | 51 | −8.5 | 44 |
| 3 | Oxazole | −6.94 | 33 | −7.07 | 32 | −7.25 | 67 | −7.78 | 45 |
| 4 | Hexestrol | −7.05 | 17 | −7.7 | 21 | −7.16 | 28 | −6.96 | 55 |
| 5 | Dimethylpropyl | −5.24 | 81 | −5.89 | 93 | −5.3 | 100 | −5.34 | 98 |
| 6 | Oxaspiro | −5.14 | 63 | −5.49 | 89 | −5.28 | 100 | −5.18 | 97 |
| 7 | Butyl-Phenol | −5.07 | 77 | −5.49 | 84 | −4.98 | 100 | −4.93 | 88 |
| 8 | Deconoic-Acid | −4.54 | 23 | −5.42 | 19 | −5.22 | 38 | −5.15 | 19 |
| 9 | Tricosenyl | −4.22 | 14 | −4.46 | 12 | −4.56 | 22 | −4.16 | 9 |
| 10 | Propanamide | −3.94 | 39 | −4.49 | 70 | −4.14 | 62 | −4.11 | 31 |
| 11 | Propanoic-Acid | −4.79 | 30 | −5.79 | 38 | −5.77 | 37 | −4.88 | 35 |
| 12 | Phenylethyl | −6.55 | 98 | −6.92 | 57 | −5.57 | 40 | −6.9 | 87 |
# - Binding energy of the conformation from the largest cluster
$ - number of conformations in the largest cluster of 100 LGA runs.
From the context of different clinical forms of EGFR and the predictions based on the number of conformations in the largest cluster, the binding of phytocompounds were discussed below. Methanone is found to be the best among the studied phytocompounds in terms of docking score. Furthermore, Methanone bound well to EGFR in mutant-clinical forms than its wild type. This makes the compound more attractive as the potential inhibitor against EGFR. The next hit compound is found to be bis-phenylethyl. Bis-Phenylethyl is not binding well in T790M-mutant containing structure. However, it is found to be considerably good in double- and exon-mutant forms of EGFR. A Similar scenario was also observed in the case of Oxazole. Interestingly, hexestrol is the only identified phytocompound, which bound well with the exon-mutant containing structure (4LRM) than the rest of EGFR clinical forms. Apart from the above-mentioned compounds, the phytocompounds dimethylpropyl, oxaspiro, and butyl-phenol showed comparatively less binding with different clinical forms of EGFR that cannot be neglected. These three compounds alone bound with T790M-mutant containing structure (5HG8) effectively. The relatively high conformations in the largest cluster of these compounds reflect that their binding are reliable and their structures are quite planar or ring-containing molecules in nature These molecules could be useful as fragments for the drug design and development for EGFR-based therapeutics. Decanoic-acid, propanoic-acid and tricosenyl, as well as propanamide, are the only compounds which showed poor binding with EGFR structures, which are potential because of their planar/linear and small structure respectively.
To test the suitability of the identified phytocompounds with the other drug targets for NSCLC were investigated via molecular docking. The proteins HER2, ALK and sPLA2-IIA were considered as other drug targets for the possibility of dual inhibition against NSCLC. The observations of the phytocompounds with other drug targets HER2and sPLA2-IIA are similar to that of EGFR except for ALK. Interestingly, the phytocompounds revealed effective binding with HER2 and sPLA2-IIA. The phytocompounds Methanone, Bis-Phenylethyl, Hexestrol, and Oxazole reflects that their binding with ALK is not satisfactory. However, the phytocompounds Dimethylpropyl, Oxaspiro, and Butyl-Phenol showed significant binding with ALK which combinatorial chemists could exploit for drug design and development. As expected, Decanoic-acid and Tricosenyl, as well as Propanamide, are the only compounds which showed poor binding with the drug targets. The interactions of phytocompounds with their respective drug targets were summarized in Table 4.
Table 4.
Molecular docking analysis of phytocompounds with drug targets for dual inhibition against NSCLC.
| S.No. | Inhibitor | ALK |
HER2 |
SPLA-IIA |
|||
|---|---|---|---|---|---|---|---|
| delG# | Confs$ | delG# | Confs$ | delG# | Confs$ | ||
| 1 | Methanone | −7.73 | 33 | −9.65 | 98 | −10.18 | 100 |
| 2 | Bis-Phenylethyl | −7.37 | 28 | −9.89 | 46 | −9.84 | 48 |
| 3 | Oxazole | −6.72 | 27 | −7.92 | 59 | −9.59 | 55 |
| 4 | Hexestrol | −5.86 | 29 | −7.42 | 22 | −7.19 | 72 |
| 5 | Dimethylpropyl | −5.94 | 67 | −6.44 | 81 | −5.79 | 100 |
| 6 | Oxaspiro | −5.61 | 97 | −5.88 | 75 | −5.86 | 100 |
| 7 | Butyl-Phenol | −5.55 | 73 | −6.02 | 94 | −5.48 | 100 |
| 8 | Deconoic-Acid | −5.02 | 39 | −6.13 | 28 | −5.97 | 26 |
| 9 | Tricosenyl | −3.67 | 10 | −7.43 | 11 | −6.21 | 25 |
| 10 | Propanamide | −3.89 | 29 | −3.7 | 45 | −3.79 | 59 |
| 11 | Propanoic-Acid | −6.74 | 94 | −5.74 | 85 | −6.48 | 93 |
| 12 | Phenylethyl | −7.33 | 41 | −7.89 | 91 | −7.2 | 99 |
# - Binding energy of the conformation from the largest cluster
$ - number of conformations in the largest cluster of 100 LGA runs
Molecular dynamics (MD) simulations is a widely used tool to reliably predict and refine the binding of small molecules with proteins (Alonso et al., 2006). It also estimates whether the binding between the small molecules and proteins are stable. The purpose of MD simulations in the current study is to refine the predicted binding of screened phytocompounds of garlic with their respective drug targets from molecular docking studies and to study the inter-molecular interactions and stability of phytocompounds with their respective drug targets thereby the discovery of potential anti-lung cancer lead molecules, the predicted complexes from docking studies were considered as the initial structures for MD simulations.
The present study was on the double and exon mutants and based on the reliability of docking predictions, the inter-molecular interactions of methanone, bis-phenylethyl, and oxazole with their respective structures along with wild type for comparison studies were investigated. The stability of the complexes was inspected with RMSD and radius of gyration. The RMSD values of protein backbone atoms were satisfactorily low with different clinical forms of EGFR while wild type has less deviation. The RMSD analysis reflects that the intermolecular complexes were relatively stable. The analysis of gyration of EGFR structures in complex with the phytocompounds also reflects that the phytocompounds bound with EGFR structures are compact. A similar profile has been observed with the other drug targets, ALK, HER2, and sPLA2-IIA. The overall analysis reflects that the MD simulations are reliable for assessing the interactions of phytocompounds with EGFR structures.
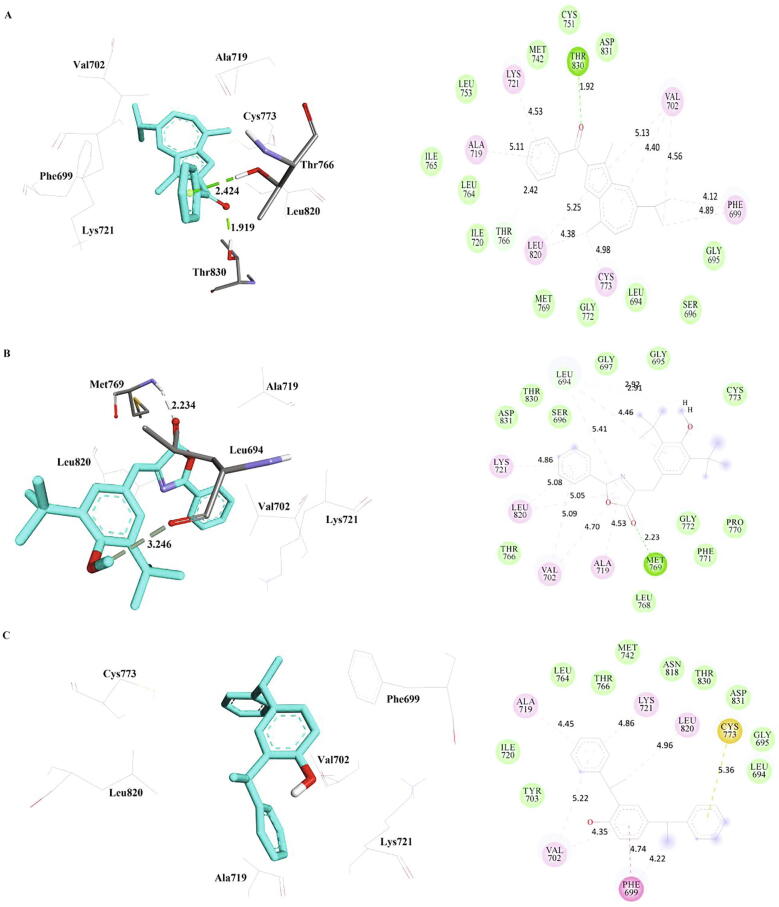
3.1. Interactions of the phytocompounds with wild type EGFR
MD simulations revealed that the phytocompounds methanone, bis-phenylethyl, and oxazole were bound well with the wild type EGFR, which were shown in Table 5 and Fig. 3. The oxygen atom of Methanone is interacting with Thr830 of wild type EGFR (wtEGFR) via hydrogen bond with a distance of 1.92 Å. In addition, the phenyl ring of methanone interacts with Thr766 via Pi-Donor hydrogen bond with a distance of 2.42 Å. In addition, several hydrophobic (Alkyl and Pi-Alkyl) interactions were observed with residues Phe699, Val702, Ala719, Lys721, Cys773, and Leu820 of wtEGFR along with several hydrophobic residues is in close proximity. Further investigations revealed that methanone is firmly interacting with wtEGFR via hydrogen bonds and hydrophobic interactions. Oxazole compound formed a hydrogen bond with Met769 with an oxygen atom of the oxazole ring while the methoxy moiety formed a carbon hydrogen bond with Leu694. In addition, the residues Val702, Ala719, Lys721 and Leu820 involved in hydrophobic (Pi-Alkyl) interactions. However, no hydrogen bond is observed between bis-phenylethyl and wtEGFR, Pi-Sulphur and Pi-Pi T-shaped bonds were observed with residues, Cys773, and Phe699 respectively. In addition, several hydrophobic interactions were observed with residues, Phe699, Val702, Ala719, and Lys721.
Table 5.
The inter-molecular interactions of the phytocompounds, Methanone, Oxazole and Bis-Phenylethyl with wild type EGFR (PDB Id: 1 M17) refined by MD simulations.
| Inhibitor | Hydrogen Bond | Pi-Donor Hydrogen Bond | Carbon Hydrogen Bond | Pi-Sulphur | Pi-Pi T-shaped |
Alkyl | Pi-Alkyl | Vander Waals Interactions | Contact Residues |
|---|---|---|---|---|---|---|---|---|---|
| Methanone | Thr830 | Thr766 | – | – | – | Val702, Cys773, Leu820 | Phe699[2], Val702[2], Ala719, Lys721, Leu820 | – | Leu694,Gly695, Ser696, Ile720, Met742, Cys751, Leu753, Leu764, Ile765, Thr766, Met769, Gly772, Asp831 |
| Oxazole | Met769 | – | Leu694 | – | – | – | Leu694[2], Val702[2], Ala719, Lys721, Leu820[2] | Glu695, Thr766, Leu768, Gly772, Cys773, Asp831 | Ser696, Gly697, Pro770, Phe771, Thr830 |
| Bis-Phenylethyl | – | – | – | Cys773 | Phe699 | Leu820 | Phe699, Val702[2], Ala719, Lys721 | – | Leu694, Gly695, Tyr703, Ile720, Met742, Leu764, Thr766, Asn818, Thr830, Asp831 |
Fig. 3.
The inter-molecular interactions of the phytocompounds, a) Methanone b) Oxazole and c) Bis-Phenylethyl with wild type EGFR (PDB Id: 1 M17) refined by MD simulations.
It is noteworthy to mention that similar binding profile was observed between several FDA approved drugs/small molecules/natural compounds and wtEGFR that leads to chemoprevention. Notably, the residues Phe699, Lys721, Met769, Thr830 and Asp831 of wtEGFR involved in the binding with natural products, which is observed in methanone (Singh and Bast, 2014). In addition, rugosaflavanoid derivatives were bound with the residues Leu694 and Gly772 exhibited anticancer activity, which is also observed with methanone (Puranik et al., 2017). In addition, oxazole interacted with residues Met769, Lys721, Thr830, and Asp831 of wtEGFR, along with residues Phe771, Cys773, Leu694, Leu768, and Gly772 which were reported as key interactions (Singh and Bast, 2014).
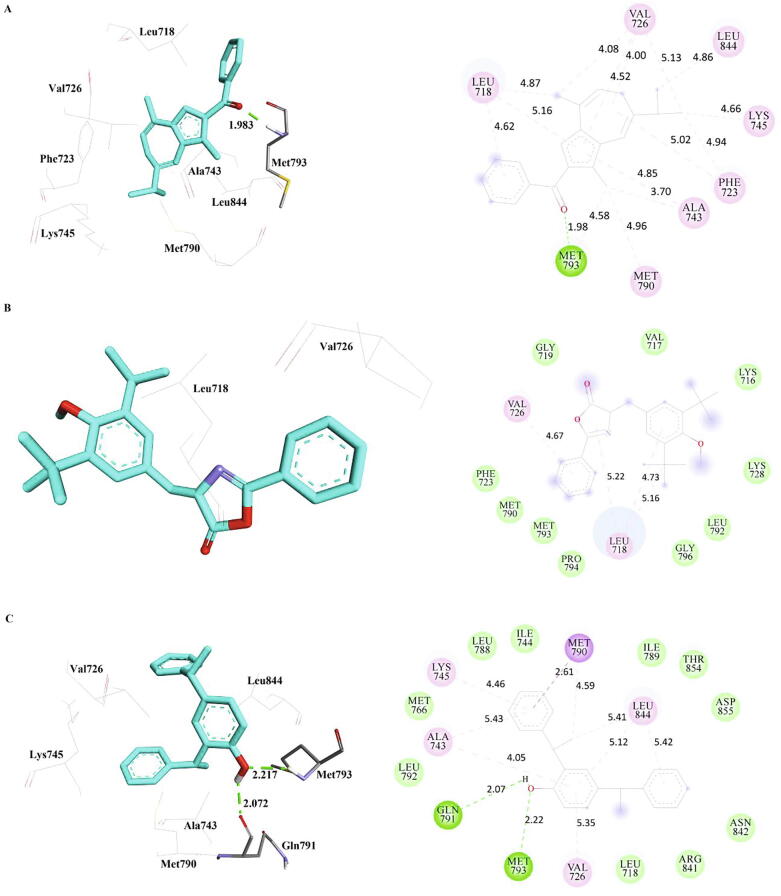
3.2. Interactions of the phytocompounds with double mutant EGFR
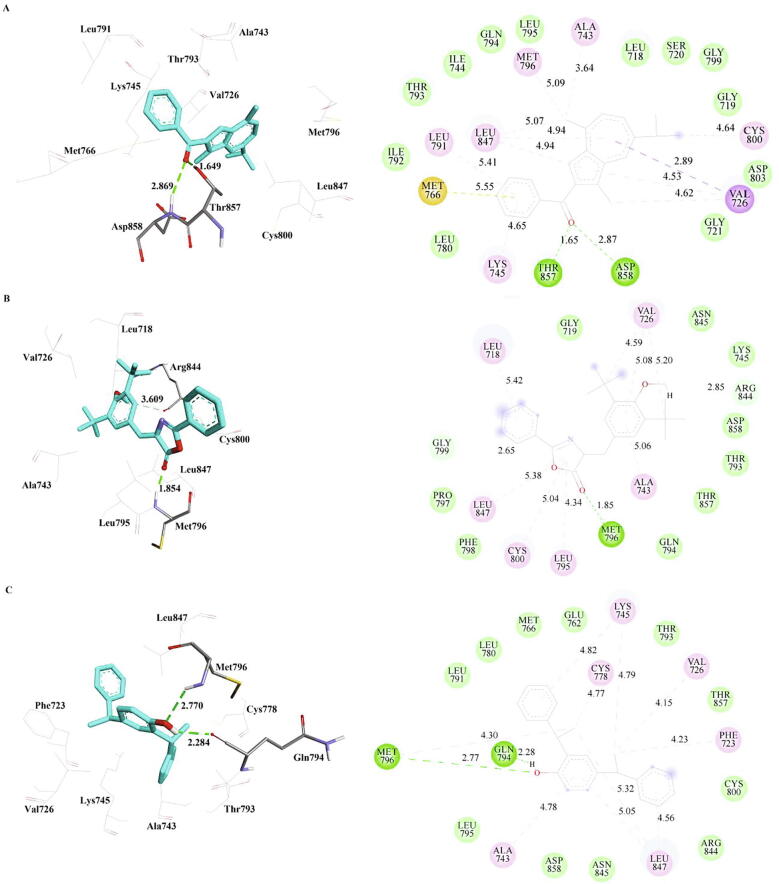
The interactions of the compounds methanone, bis-phenylethyl and oxazole with double mutant EGFR (dmEGFR) were presented in Table 6 and Fig. 4. The hydrogen bond interaction is formed between the oxygen group of methanone with the Met793 of dmEGFR with a distance of 1.98 Å. Alkyl and pi-alkyl interactions were observed with residues Leu718, Val726, Ala743, Lys745, Met790, Met793, and Leu844. These interactions revealed that methanone has better binding with dmEGFR via a hydrogen bond, alkyl and pi-alkyl interactions. Hydrogen bond was not observed in oxazole compound, but it forms alkyl and Pi-alkyl interactions with the residues Leu718 and Val726, while bis-phenylethyl forms hydrogen bond with Gln791 and Met793 along with hydrophobic interactions with Met790 and Leu844 of dmEGFR.
Table 6.
The inter-molecular interactions of the phytocompounds with double mutant EGFR (PDB Id: 5XGN) refined by MD simulations.
| Inhibitor | Hydrogen Bond | Pi-Sigma | Alkyl | Pi-Alkyl | Contact Residues |
|---|---|---|---|---|---|
| Methanone | Met793 | – | Leu718, Val726[3], Ala743, Lys745, Met790, Met793, Leu844 | Leu718[2], Phe723[2], Val726, Ala743 | Gly719, Lys728, Gln791, Leu792, Pro794, Gly796, Thr854 |
| Oxazole | – | – | Leu718 | Leu718[2], Val726 | Lys716, Val717, Gly719, Phe723, Lys728, Met790, Leu792, Met793, Pro794, Gly796 |
| Bis-Phenylethyl | Gln791, Met793 | Met790 | Met790, Leu844 | Val726, Ala743[2], Lys745, Leu844[2] | Leu718, Ile744, Met766, Leu788, Ile789, Leu792, Arg841, Asn842, Thr854, Asp855 |
Fig. 4.
The inter-molecular interactions of the phytocompounds, a) Methanone b) Oxazole and c) Bis-Phenylethyl with double mutant EGFR (PDB Id: 5XGN) refined by MD simulations.
To summarize, the interactions of the phytocompounds methanone, bis-phenylethyl, and oxazole with dmEGFR were satisfactory. Methanone forms interaction with dmEGFR via the key residues such as Val726, Ala743, Lys745, and Met790, whereas Lys745 forms interaction with both bis-phenylethyl and oxazole. These interactions have been observed with the first allosteric inhibitor, EAI045 in combination with the antibody cetuximab that inhibit EGFR kinase effectively (Zhao et al., 2018).
3.3. Interactions of the phytocompounds with exon mutant EGFR
The intermolecular interactions of phytocompounds with exon mutant EGFR (emEGFR) refined by MD simulations indicates that Thr857 and Asp858 forms hydrogen bond with methanone whereas Met796 had a hydrogen bond with oxazole and bis-phenylethyl. In addition, bis-phenylethyl also formed a hydrogen bond with Gln794. Pi-sigma and Pi-sulphur interactions were observed between Val726 and Met766 with methanone. Val726, Ala743, Met796, Cys800, Leu847, Lys745, Leu791, Leu718, Leu795, and Phe723 forms Alkyl and Pi-Alkyl interactions. The inter-molecular interactions of phytocompounds with emEGFR are presented in the Table 7 and Fig. 5. Interaction with residues Leu718, Ala743, and Lys745 were exhibited by pyrimidine-based benzylcarbamate derivatives that inhibit proliferation and signalling pathways of emEGFR and HER2, which was observed in the studies of phytocompounds (Jang et al., 2018). Gefitnib and Imatnib drugs remain as the key therapy for lung cancer treatment as they exhibit tyrosine kinase inhibitor activity of EGFR. The rates of patients who respond to Gefitinib is upto 40% and mostly belong to the Asian ethnic group but approximately 77% patients exhibits mutational changes in EGFR gene as compared to 7% refractory to Gefitinib and belong to NSCLC category (Nand et al., 2016). Drug targets of the present study revealed good binding interaction with the EGFR gene with mutations. Shaik et al., 2019 has reported CUCM-36((1E,4Z,6E)-1-(3,4-Diphenoxyphenyl)-5-hydroxy-7-(4-hydroxy-3 phenoxy phenyl)-1,4,6-heptatrien-3-one) with better affinity towards wild type and mutant forms of EGFR which was similar to our results. Granulatimide, Danorubicin, Penicinoline and Austocystin D, compounds from traditional Chinese medicine were shown to exhibit inhibitory effect towards EGFR mutant genes, which supports the interaction of the compounds in the present study as garlic is one of the important constituent of traditional Chinese medicine.
Table 7.
The molecular interactions between the phytocompounds and exon mutant EGFR (PDB Id: 4LRM) refined by molecular dynamics simulations.
| Inhibitor | Hydrogen Bond | Carbon Hydrogen Bond | Pi-Sigma | Pi-Sulphur | Alkyl | Pi-Alkyl | Contact Residues |
|---|---|---|---|---|---|---|---|
| Methanone | Thr857, Asp858 | – | Val726 | Met766 | Val726, Ala743, Met796, Cys800, Leu847 | Val726, Lys745, Leu791, Leu847[2] | Leu718, Gly719, Ser720, Gly721, Ile744, Leu780, Ile792, Thr793, Gln794, Leu795, Gly799, Asp803 |
| Oxazole | Met796 | Gly799, Arg844 | – | – | Val726[2] | Leu718, Val726, Ala743, Leu795, Cys800, Leu847 | Gly718, Lys745, Thr793, Gln794, Pro797, Phe798, Asn845, Thr857, Asp858 |
| Bis-Phenylethyl | Gln794, Met796 | – | – | – | Val726, Lys745, Cys778, Met796, Leu847 | Phe723, Ala743, Lys745, Leu847 | Glu762, Met766, Leu780, Leu791, Thr793, Leu795, Cys800, Arg844, Asn845, Thr857, Asp858 |
Fig. 5.
The inter-molecular interactions of the phytocompounds, a) Methanone b) Oxazole and c) Bis-Phenylethyl with exon mutant EGFR (PDB Id: 4LRM) refined by MD simulations.
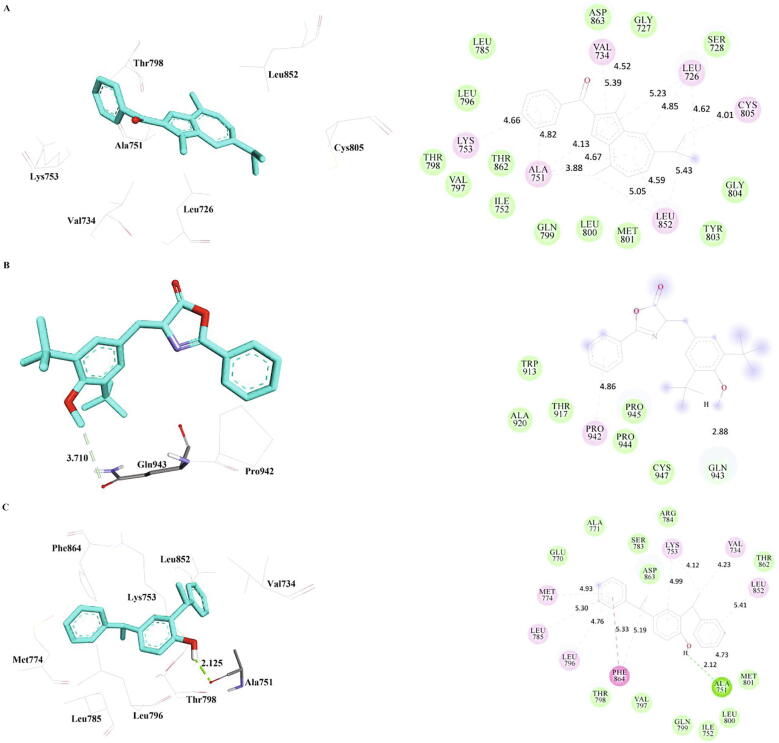
3.4. Interactions of the phytocompounds with HER2
Among the phytocompounds, bis-phenylethyl interacts with the oxygen atom of Ala751 of HER2 via a hydrogen bond with a distance of 2.12 Å, while oxazole forms carbon hydrogen bond with the oxygen atom of Gln943. Furthermore, bis-phenylethyl had Pi-Pi T-shaped interaction with Phe864 of HER2. Various hydrophobic interactions were observed between Leu726, Val734, Ala751, Lys753, Leu852, Cys805, Pro942, Met774, Leu785, Leu796, Phe864 of HER2 and the phytocompounds. It has been reported that Ala751, Lys753, and Leu796 are involved in the interactions between cannabisin F and HER2. Cannabisin F is an acyclic bis-phenylpropane lignanamides found in the fruits of Cannabis sativa, has been reported for its cytotoxic activity against lung cancer and cervical cancer (Hu et al., 2016). The interactions of methanone, oxazole, and bis-phenylethyl with HER2 were presented in Table 8 and Fig. 6. Secondary metabolites from plants show good potency as drugs to target lung cancer. Especially flavonoids have seen to possess good binding interaction with the oncogenes.HER2 does not bind to ligand directly, but can form heterodimers with other receptors of ERBB family (Ricciardi et al., 2014). HER2 activation involves in various number of signal transduction pathways including JAK/STAT and PI3K/AKT, RAS/MAP/MEK pathways. HER2 is considered as a promising therapeutic agent for lung cancer therapy. The overexpression of HER2 is observed in 6–30% of lung cancer cases. An insertion mutation in exon 20 of HER2 activates downstream signalling pathways and induced lung tumorigenesis in animal models. 2–20% of lung cancer patients showed HER2 gene amplification (Pillai et al., 2017).
Table 8.
Inter-molecular interactions of the phytocompounds against HER2 (PDB Id: 3PP0) refined by MD simulations.
| Inhibitor | Hydrogen Bond | Carbon Hydrogen Bond | Pi-Pi T-shaped | Alkyl | Pi-Alkyl | Contact Residues |
|---|---|---|---|---|---|---|
| Methanone | – | – | – | Leu726, Val734, Ala751, Cys805, Leu852[2] | Leu726[2], Val734, Ala751[3], Lys753, Leu852 | Gly727, Ser728, Ile752, Leu785, Leu796, Val797, Thr798, Gln799, Leu800, Met801, Tyr803, Gly804, Thr862, Asp863 |
| Oxazole | – | Gln943 | – | – | Pro942 | Trp913, Thr917, Ala920, Pro944, Pro945, Cys947 |
| Bis-Phenylethyl | Ala751 | – | Phe864 | Val734, Lys753 | Ala751, Lys753, Met774, Leu785, Leu796, Leu852, Phe864 | Ile752, Glu770, Ala771, Ser783, Arg784, Val797, Thr798, Gln799, Leu800, Met801, Thr862, Asp863 |
Fig. 6.
The inter-molecular interactions of the phytocompounds, a) Methanone b) Oxazole and c) Bis-Phenylethyl with exon mutant EGFR (PDB Id: 3PP0) refined by MD simulations.
7-hydroxyflavone, 2-(-4-fluorophenyl)- 4n-chromen-4-one, 3-hydroxyflavone and 8-methylflavone,4-hydroxyflavone,6,3- dimethyl flavone were found to be the best compounds indicating minimum binding energy in examination with HER2 receptor (Singhal et al., 2017)
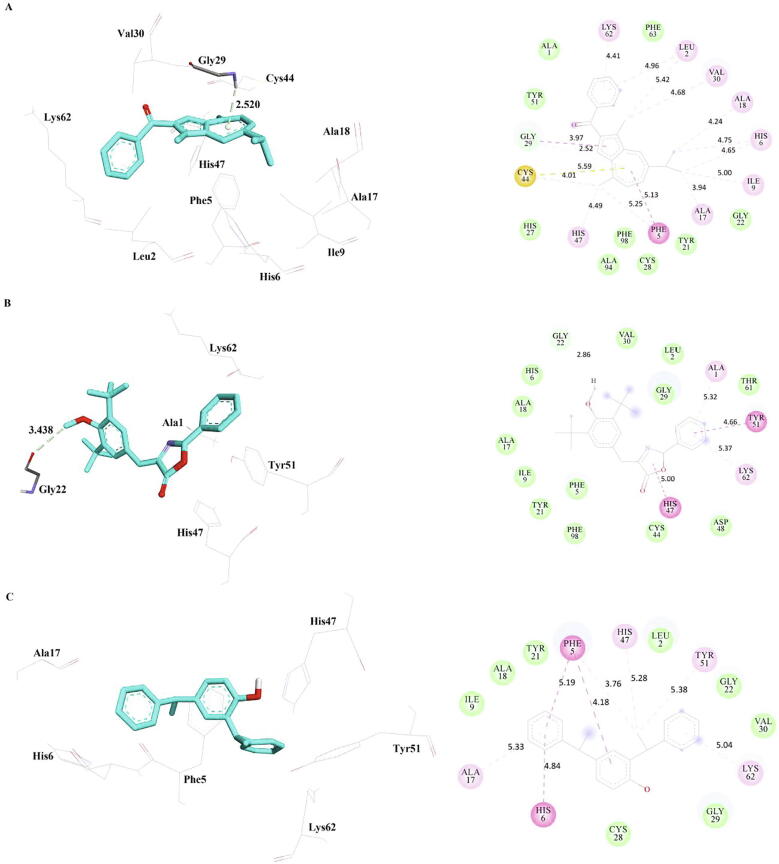
3.5. Interactions of the phytocompounds with sPLA2-IIA
Hydrogen bond was not observed between the phytocompounds and sPLA2-IIA, but Pi-donor hydrogen bond was formed between methanone and Gly29 of sPLA2-IIA, a Carbon hydrogen bond formed between oxazole and Gly22 of sPLA2-IIA. Other than these interactions, various other interactions were observed and it is summarized in Table 9 and interactions were visualized in Fig. 7. Xanthone derivatives have a enormous range of pharmacological properties, such as those involving antimalarial, anti-inflammatory and anticancer properties (Miladiyah et al., 2018). Molecular docking studies of xanthone derivatives against sPLA2-IIA were reported in a research finding, in which Interactions with Gly29, His47, His6 plays an important role. (Chen et al., 2017). These residues are also involved in the interaction of identified phytocompounds with sPLA2-IIA which substantiates the various pharmacological benefits of garlic. Kumar et al., 2012 reported the formation of one hydrogen bond interaction of PLA 2 for the ligands Curcumin, Ellagic acid and Caffeic acid, whereas three hydrogen bond interactions was seen with Catechin. The present study does not report any hydrogen bond formation but the presence of a pi donor bond was seen between methanone and Gly 29 of sPLA-IIA. Research findings on in vitro studies on sPLA2 inhibition demonstrated that it has the capability to decrease proliferation of human lung cancer cells and it also reduces the tumour growth in murine models (Halpern et al., 2018).
Table 9.
The inter-molecular interactions of the phytocompounds with sPLA2-IIA (PDB Id: 5G3N) refined by molecular dynamics simulations.
| Inhibitor | Pi-Donor Hydrogen Bond | Carbon Hydrogen Bond | Pi-Sulphur | Pi-Pi Stacked | Pi-Pi T-shaped | Amide-Pi Stacked | Alkyl | Pi-Alkyl | Contact Residues |
|---|---|---|---|---|---|---|---|---|---|
| Methanone | Gly29 | – | Cys44 | – | Phe5 | Gly29 | Leu2, Ile9, Ala17, Ala18, Val30 | Leu2, Phe5, His6[2], His47, Lys62 | Ala1, Tyr21, Gly22, His27, Cys28, Tyr51, Phe63, Ala94, Phe98 |
| Oxazole | – | Gly22 | – | – | His47, Tyr51 | – | – | Ala1, Lys62 | Leu2, Phe5, His6, Ile9, Ala17, Ala18, Tyr21, Gly29, Val30, Cys44, Asp48, Tyr61, Phe98 |
| Bis-Phenylethyl | – | – | – | Phe5, His6 | Phe5 | – | – | Phe5, Ala17, His47, Tyr51, Lys62 | Leu2, Ile9, Ala18, Tyr21, Gly22, Cys28, Gly29, Val30 |
Fig. 7.
The inter-molecular interactions of the phytocompounds, a) Methanone b) Oxazole and c) Bis-Phenylethyl with sPLA2-IIA (PDB Id: 5G3N) refined by molecular dynamics simulations.
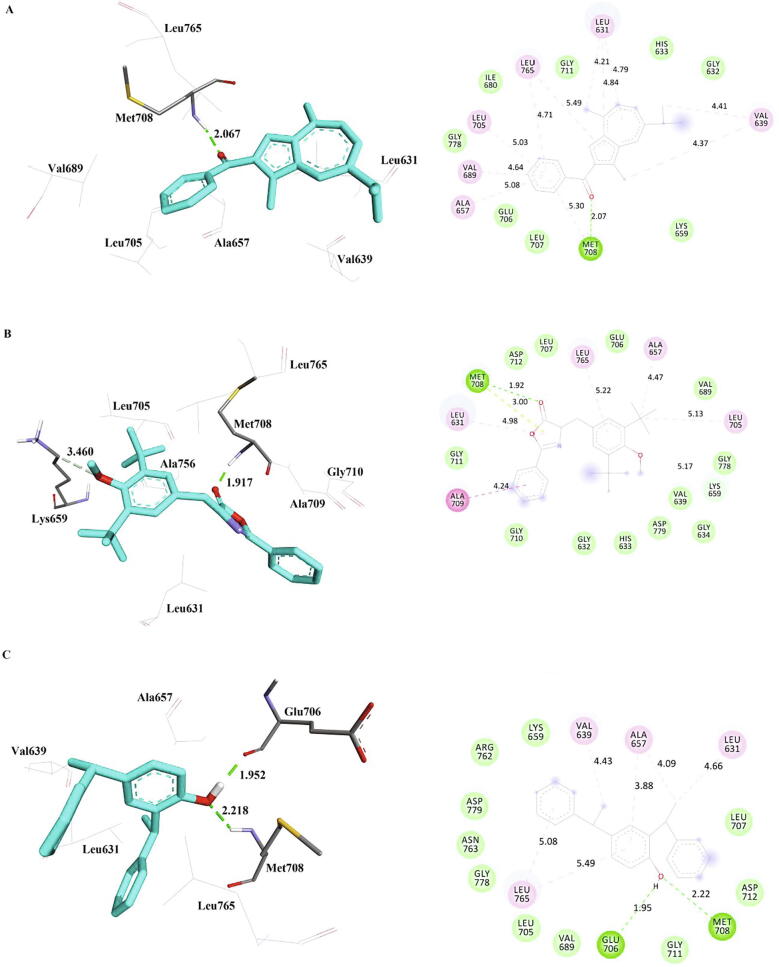
3.6. Interactions of the phytocompounds with EML4-ALK
Hydrogen bond interactions were formed between Met708 of EML4-ALK with all the three phytocompounds with a hydrogen bond distance of 2.07, 1.917, and 2.218 respectively (Fig. 8). This illustrates that Met708 plays a major role in binding affinity of EML4-ALK with the garlic compounds. Additionally, Glu706 of bis-phenylethyl forms a hydrogen bond with a distance of 1.952 with EML4-ALK. Residues like Leu631, Lys659, Ala657, and Leu765 are involved in the alkyl and pi-alkyl interactions (Table 10). These residues also show interactions with 7-azaindole based ALK inhibitors, which showed excellent binding efficiency against wild type EML4-ALK and L1196M mutant enzyme (Gummadi et al., 2013). Matrine, an alkaloid present in Sophora flarescens showed a binding energy of − 6.19 kcal/mol ,Triptolide, a diterpenoid from Tripterygium wilfordii showed a binding energy of − 5.69 kcal/mol, these were related to the binding energies obtained in the present study (Anusha et al., 2019). The phytocompounds can therefore be considered as potent inhibitors of ELM4-ALK.
Fig. 8.
The inter-molecular interactions of the phytocompounds, a) Methanone b) Oxazole and c) Bis-Phenylethyl with EML4-ALK (PDB Id: 4Z55) refined by molecular dynamics simulations.
Table 10.
Inter-molecular interactions of the phytocompounds with EML4-ALK (PDB Id: 4Z55) refined by molecular dynamics simulations.
| Inhibitor | Hydrogen Bond | Carbon Hydrogen Bond | Pi-Lone Pair | Amide-Pi Stacked | Alkyl | Pi-Alkyl | van der Waals Interactions | Contact Residues |
|---|---|---|---|---|---|---|---|---|
| Methanone | Met708 | – | – | – | Leu631, Val639[2] | Leu631[2], Ala657, Val689, Leu705, Met720 Leu765[2] |
– | Gly632, His633, Lys659, Ile680, Glu706, Leu707 Gly711, Gly778 |
| Oxazole | Met708 | Lys659 | Met708 | Ala709 | Ala657, Leu705 | Leu631, Leu765 | Gly710 | Gly632, His633, Gly634 Val639, Val689, Glu706, Leu707, Gly711 Asp712, Gly778 Asp779 |
| Bis-Phenylethyl | Glu706, Met708 | – | – | Leu631, Val639, Ala657 | Ala657 Leu765 [2] |
– | Lys659 Val689, Val689, Leu705 Leu707, Gly711, Asp712, Arg762, Asn763, Gly778 Asp779 |
4. Conclusion
Despite the advances in drug development against cancer, NSCLC still imposes great challenges. EGFR has been an attractive drug target for the treatment of NSCLC, but the evolution of drug resistance, there is an urgent need to discover effective drugs against EGFR-mutants. Therefore, the ongoing combination trials might revolutionize the treatment paradigm for EGFR-mutant NSCLC. Previous research findings suggested that inhibiting the distinct clones of EGFR-mutant NSCLC with EGFR inhibitors along with other drug targets like HER2, EML4-ALK might be the solution. The identified phytocompounds could be the starting arsenal for the development of lead molecules targeting EGFR-mutants and other drug targets. Computational studies revealed that these garlic phytocompounds inhibit wild type EGFR, as well as double mutant and exon mutant. These compounds are also subjected for their dual inhibitory effect against known lung cancer targets of NSCLC. The garlic phytocompounds including methanone, bis-phenylethyl, oxazole could serve as lead molecules for the development of drugs to combat lung cancer.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
Dr. Saravanan Parameswaran, Postdoctoral Fellowship from the BK21Plus Program, the Ministry of Education, Republic of Korea and Bio & Medical Technology Development Program of the National Research Foundation (NRF) & funded by the Korean government (MSIT) (No. NRF-2018M3A9A7057263). The authors extend their appreciation to the Researchers supporting project number (RSP- 2020/173) King Saud University, Riyadh, Saudi Arabia.
Footnotes
Peer review under responsibility of King Saud University.
References
- Aertgeerts K., Skene R., Yano J., Sang B.C., Zou H., Snell G., Ishikawa T., Tanaka T., Miki H., Ohta Y., Sogabe S. Structural analysis of the mechanism of inhibition and allosteric activation of the kinase domain of HER2 protein. J. Biol. Chem. 2011;286:18756–18765. doi: 10.1074/jbc.M110.206193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Dhabi NA, Arasu MV, Rejiniemon TS. 2015. In vitro antibacterial, antifungal, antibiofilm, antioxidant, and anticancer properties of isosteviol isolated from endangered medicinal plant Pittosporum tetraspermum. Evidence-Based Complementary and Alternative Medicine.2015. [DOI] [PMC free article] [PubMed]
- Al-Dhabi NA, Arasu MV. 2016. Quantification of phytochemicals from commercial Spirulina products and their antioxidant activities.. Evidence-Based Complementary and Alternative Medicine. 2016. [DOI] [PMC free article] [PubMed]
- Alonso H., Bliznyuk A.A., Gready J.E. Combining docking and molecular dynamic simulations in drug design. Med. Res. Rev. 2006;26:531–568. doi: 10.1002/med.20067. [DOI] [PubMed] [Google Scholar]
- Anusha D., Sharanya S., Ramya R., David D.C. Anticancer screening of the phytochemicals present in the medicinal plant Vitex Negundo against mutant anaplastic Lymphoma Kinase (ALK) protein: An In –silico Approach. Biomed Pharmacol J. 2019;12:993–1000. doi: 10.13005/bpj/1727. [DOI] [Google Scholar]
- Banaganapalli B., Mulakayala C., Pulaganti M., Mulakayala N., Anuradha C.M., Suresh Kumar C., Shaik N.A., Yousuf Al-Aama J., Gudla D. Experimental and computational studies on newly synthesized resveratrol derivative: a new method for cancer chemoprevention and therapeutics? OMICS. 2013;17:568–583. doi: 10.1089/omi.2013.0014. [DOI] [PubMed] [Google Scholar]
- Bankovic J., Stojsic J., Jovanovic D., Andjelkovic T., Milinkovic V., Ruzdijic S., Tanic N. Identification of genes associated with non-small-cell lung cancer promotion and progression. Lung cancer (Amsterdam, Netherlands) 2010;67:151–159. doi: 10.1016/j.lungcan.2009.04.010. [DOI] [PubMed] [Google Scholar]
- Barathikannan K., Venkatadri B., Khusro A., Al-Dhabi N.A., Agastian P., Arasu M.V., Choi H.S., Kim Y.O. Chemical analysis of Punica granatum fruit peel and its in vitro and in vivo biological properties. BMC Complimnetary and Alternative Medicine. 2016;16:264. doi: 10.1186/s12906-016-1237-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjelkmar P., Larsson P., Cuendet M.A., Hess B., Lindahl E. Implementation of the CHARMM Force Field in GROMACS: Analysis of Protein Stability Effects from Correction Maps, Virtual Interaction Sites, and Water Models. J Chem Theory Comput. 2010;6:459–466. doi: 10.1021/ct900549r. [DOI] [PubMed] [Google Scholar]
- Chen X., Leng J., Rakesh K.P., Darshini N., Shubhavathi T., Vivek H.K., Mallesha N., Qin H.L. Synthesis and molecular docking studies of xanthone attached amino acids as potential antimicrobial and anti-inflammatory agents. Med. Chem. Commun. 2017;8:1706–1719. doi: 10.1039/c7md00209b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Nair S.K., Murray B.W., Almaden C., Bailey S., Baxi S., Behenna D., Cho-Schultz S., Dalvie D., Dinh D.M., Edwards M.P., Feng J.L., Ferre R.A., Gajiwala K.S., Hemkens M.D., Jackson-Fisher A., Jalaie M., Johnson T.O., Kania R.S., Kephart S., Lafontaine J., Lunney B., Liu K.K., Liu Z., Matthews J., Nagata A., Niessen S., Ornelas M.A., Orr S.T., Pairish M., Planken S., Ren S., Richter D., Ryan K., Sach N., Shen H., Smeal T., Solowiej J., Sutton S., Tran K., Tseng E., Vernier W., Walls M., Wang S., Weinrich S.L., Xin S., Xu H., Yin M.J., Zientek M., Zhou R., Kath J.C. Discovery of 1-{(3R,4R)-3-[({5-Chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}prop-2-en-1-one (PF-06459988), a Potent, WT Sparing, Irreversible Inhibitor of T790M-Containing EGFR Mutants. J Med Chem. 2016;59:2005–2024. doi: 10.1021/acs.jmedchem.5b01633. [DOI] [PubMed] [Google Scholar]
- Cuong D.M., Arasu M.V., Jeon J., Park Y.J., Kwon S.-J., Al-Dhabi N.A., Park S.U. Medically important carotenoids from Momordica charantia and their gene expressions in different organs. Saudi Journal of Biological Sciences. 2017;24:1913–1919. doi: 10.1016/j.sjbs.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daga A., Ansari A., Patel S., Mirza S., Rawal R., Umrania V. Current drugs and drug targets in non-small cell lung cancer: limitations and opportunities. Asian Pac J Cancer Prev. 2015;16:4147–4156. doi: 10.7314/apjcp.2015.16.10.4147. [DOI] [PubMed] [Google Scholar]
- Elango G., Roopan S.M., Al-Dhabi N.A., Arasu M.V., Damodharan K.I., Elumalai K. Nanomedicine, and Biotechnology; Artificial Cells: 2017. Cocos nucifera coir-mediated green synthesis of Pd NPs and its investigation against larvae and agricultural pest. [DOI] [PubMed] [Google Scholar]
- Elango G., Roopan S.M., Al-Dhabi N.A., Arasu M.V., Dhamodaran K.I., Elumalai K. Coir mediated instant synthesis of Ni-Pd nanoparticles and its significance over larvicidal, pesticidal and ovicidal activities. Journal of Molecular Liquids. 2016;223:1249–1255. [Google Scholar]
- Elango G., Roopan S.M., Dhamodaran K.I., Elumalai K., Al-Dhabi N.A., Arasu M.V. Spectroscopic investigation of biosynthesized nickel nanoparticles and its larvicidal, pesticidal activities. Journal of Photochemistry & Photobiology, B: Biology. 2016;162:162–167. doi: 10.1016/j.jphotobiol.2016.06.045. [DOI] [PubMed] [Google Scholar]
- Fleischauer A.T., Arab L. Garlic and cancer: a critical review of the epidemiologic literature. J Nutr. 2001;131:1032S–1040S. doi: 10.1093/jn/131.3.1032S. [DOI] [PubMed] [Google Scholar]
- Fowsiya J., Madhumitha G., Al-Dhabi N.A., Arasu M.V. Photocatalytic degradation of Congo red using Carissa edulis extract capped zinc oxide nanoparticles. Journal of Photochemistry & Photobiology, B: Biology. 2016;162:395–401. doi: 10.1016/j.jphotobiol.2016.07.011. [DOI] [PubMed] [Google Scholar]
- Galli G., Corrao G., Imbimbo M., Proto C., Signorelli D., Ganzinelli M., Zilembo N., Vitali M., de Braud F., Garassino M.C., Lo Russo G. Uncommon mutations in epidermal growth factor receptor and response to first and second generation tyrosine kinase inhibitors: A case series and literature review. Lung cancer. 2018;115:135–142. doi: 10.1016/j.lungcan.2017.12.002. [DOI] [PubMed] [Google Scholar]
- Ganesan K., Ivanova T., Wu Y., Rajasegaran V., Wu J., Lee M.H., Yu K., Garrido-Castro A.C., Felip E. HER2 driven non-small cell lung cancer (NSCLC): potential therapeutic approaches. Transl. Lung. Cancer Res. 2013;2:122–127. doi: 10.3978/j.issn.2218-6751.2013.02.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glorybai L, Barathi K.K, Arasu MV, Al-Dhabi NA, Agastian P. 2015. Some biological activities of Epaltes divaricata L. - an in vitro study. Annals of Clinical Microbiology and Antimicrobials. 2015, 14:18. [DOI] [PMC free article] [PubMed]
- Giordanetto F., Pettersen D., Starke I., Nordberg P., Dahlstrom M., Knerr L., Selmi N., Rosengren B., Larsson L.O., Sandmark J., Castaldo M., Dekker N., Karlsson U., Hurt-Camejo E. Discovery of AZD2716: a novel secreted phospholipase A2 (sPLA2) inhibitor for the treatment of coronary artery disease. ACS Med. Chem. Lett. 2016;7:884–889. doi: 10.1021/acsmedchemlett.6b00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gummadi V.R., Rajagopalan S., Looi C.Y., Paydar M., Renukappa G.A., Ainan B.R., Krishnamurthy N.R., Panigrahi S.K., Mahasweta K., Raghuramachandran S., Rajappa M., Ramanathan A., Lakshminarasimhan A., Ramachandra M., Wong P.F., Mustafa M.R., Nanduri S., Hosahalli S. Discovery of 7-azaindole based anaplastic lymphoma kinase (ALK) inhibitors: wild type and mutant (L1196M) active compounds with unique binding mode. Bioorg Med Chem Lett. 2013;23:4911–4918. doi: 10.1016/j.bmcl.2013.06.071. [DOI] [PubMed] [Google Scholar]
- Halpern A.L., Kohtz P.D., Kalatardi S., Meng X., Fullerton D.A., Weyant M.J. Pharmacologic Inhibition of Secretory Phospholipase A2 IIa Reduces Lung Cancer Growth In Vitro and in a Murine Model. J. Am. Coll. Surg. 2018;227:S53. doi: 10.1016/j.jamcollsurg.2018.07.095. [DOI] [Google Scholar]
- Haritha E., Roopan S.M., Madhavi G., Elango G., Al-Dhabi N.A., Arasu M.V. Green chemical approach towards the synthesis of SnO2 NPs in argument with photocatalytic degradation of diazo dye and its kinetic studies. Journal of Photochemistry & Photobiology, B: Biology. 2016;162:441–447. doi: 10.1016/j.jphotobiol.2016.07.010. [DOI] [PubMed] [Google Scholar]
- Helan V., Prince J. J, Naif Abdullah Al-Dhabi, Mariadhas Valan Arasu, A. Ayeshamariam, G. Madhumitha, Selvaraj Mohana Roopan, M. Jayachandran. Neem leaves mediated preparation of NiO nanoparticles and its magnetization, coercivity and antibacterial analysis. Results in Physics. 2016. 6:712–718.
- Hess, B., 2008. P-LINCS: A parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 4, 116-122. https://doi.org/10.1021/ct700200b. https://doi.org/10.1089/omi.2013.0014. [DOI] [PubMed]
- Hu J.B., Dong M.J., Zhang J. A Holistic in silico approach to develop novel inhibitors targeting ErbB1 and ErbB2 Kinases. Trop J Pharm Res. 2016;15:231–239. doi: 10.4314/tjpr.v15i2.3. [DOI] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Ilavenil S., Kim D.H., Srigopalram S., Kuppusamy P., Arasu M.V., Lee K.D., Lee J.C., Song Y.H., Jeong Y.I., Choi K.C. Ferulic acid in Lolium multiflorum inhibits adipogenesis in 3T3-L1 cells and reduced high-fat-diet-induced obesity in Swiss albino mice via regulating p38MAPK and p44/42 signal pathways. Journal of Functional Foods. 2017;37:293–302. [Google Scholar]
- Jang J., Son J., Park E., Kosaka T., Saxon J.A., De Clercq D.J.H., Choi H.G., Tanizaki J., Eck M.J., Jänne P.A., Gray N.S. Discovery of a Highly Potent and Broadly Effective Epidermal Growth Factor Receptor and HER2 Exon 20 Insertion Mutant Inhibitor. Angew. Chem. Int. Ed. Engl. 2018;3:11629–11633. doi: 10.1002/anie.201805187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaowinn S., Kaewpiboon C., Kim J.E., Lee M.R., Hwang D.Y., Choi Y.W., Kim H.W., Park J.K., Song K.M., Lee N.H., Maeng J.S., Chung Y.H. N-Benzyl-N-methyl-dodecan-1-amine, a novel compound from garlic, exerts anti-cancer effects on human A549 lung cancer cells overexpressing cancer upregulated gene (CUG)2. Eur J Pharmacol. 2018;841:19–27. doi: 10.1016/j.ejphar.2018.09.035. [DOI] [PubMed] [Google Scholar]
- Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B.A., Wang J., Yu B., Zhang J., Bryant S.H. PubChem Substance and Compound databases. Nucleic Acids Res. 2016;44:D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L.L., Ma R., Yao M.Y., Yan X.E., Zhu S.J., Zhao P., Yun C.H. Structural pharmacological studies on EGFR T790M/C797S. Biochem. Biophys. Res. Commun. 2017;488:266–272. doi: 10.1016/j.bbrc.2017.04.138. [DOI] [PubMed] [Google Scholar]
- Kumar D.R., Lakshmi P.S., Saravani N., Marimuthu S. In silico Molecular Docking Studies on Porcine Pancreatic Phospholipase A2 against Plant extracts of Phenolic Inhibitors. Int J Res Biomed Biotechnol. 2012;2:8–16. [Google Scholar]
- Kurian G.A., Suryanarayanan S., Raman A., Padikkala J. Antioxidant effects of ethyl acetate extract of Desmodium gangeticum root on myocardial ischemia reperfusion injury in rat hearts. Chin Med. 2010;5:1–7. doi: 10.1186/1749-8546-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Opoku A.R., Geheeb-Keller M., Hutchings A.D., Terblanche S.E., Jäger A.K., van Staden J. Preliminary screening of some traditional zulu medicinal plants for anti-inflammatory and anti-microbial activities. J Ethnopharmacol. 1999;68:267–274. doi: 10.1016/s0378-8741(99)00130-0. [DOI] [PubMed] [Google Scholar]
- Melappa G., Prakash B. In vitro antimitotic, antiproliferative and GC-MS studies on the methanolic extract of endophytic fungi, penicillium species of Tabebuia argentea bur & k. Sch. Farmacia. 2017;5:301–309. [Google Scholar]
- Michellys P.Y., Chen B., Jiang T., Jin Y., Lu W., Marsilje T.H., Pei W., Uno T., Zhu X., Wu B., Nguyen T.N., Bursulaya B., Lee C., Li N., Kim S., Tuntland T., Liu B., Sun F., Steffy A., Hood T. Design and synthesis of novel selective anaplastic lymphoma kinase inhibitors. Bioorg Med Chem Lett. 2016;26:1090–1096. doi: 10.1016/j.bmcl.2015.11.049. [DOI] [PubMed] [Google Scholar]
- Miladiyah I., Jumina J., Haryana S.M., Mustofa M. Biological activity, quantitative structure-activity relationship analysis, and molecular docking of xanthone derivatives as anticancer drugs. Drug Des Devel Ther. 2018;12:149–158. doi: 10.2147/DDDT.S149973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G.M., Goodsell D.S., Halliday R.S., Huey R., Hart W.E., Belew R.K., Olson A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. doi: 10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B. [DOI] [Google Scholar]
- Morris, G.M., Huey, R., Lindstrom, W., Sanner, M.F., Belew, R.K., Goodsell, D.S., Olson, A.J., 2009. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785-2791.https://doi.org/10.1002/jcc.21256. [DOI] [PMC free article] [PubMed]
- Myneni A.A., Chang S.C., Niu R., Liu L., Swanson M.K., Li J., Su J., Giovino G.A., Yu S., Zhang Z.F., Mu L. Raw Garlic Consumption and Lung Cancer in a Chinese Population. Cancer Epidemiol Biomarkers Prev. 2016;25:624–633. doi: 10.1158/1055-9965.EPI-15-0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nand M., Maiti P., Pant R., Kumari M., Chandra S., Pande V. Virtual screening of natural compounds as inhibitors of EGFR 696–1022 T790M associated with non-small cell lung cancer. Bioinformation. 2016;12:311–317. doi: 10.6026/97320630012311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.H., Baskar T.B., Park S.-Y., Kim S.-J., Arasu M.V., Al-Dhabi N.A., Kim J.K., Park S.U. Metabolic profiling and antioxidant assay of metabolites from three radish cultivars (Raphanus sativus) Molecules. 2016;21:157. doi: 10.3390/molecules21020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.J., Baskar T.B., Yeo S.K., Arasu M.V., Al-Dhabi N.A., Lim S.S., Park S.U. Composition of volatile compounds and in vitro antimicrobial activity of nine Mentha spp. SpringerPlus. 2016;5:1628. doi: 10.1186/s40064-016-3283-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.J., Park S.-Y., Arasu M.V., Al-Dhabi N.A., Ahn H.-G., Kim J.K., Park S.U. Accumulation of carotenoids and metabolic profiling in different cultivars of Tagetes flowers. Molecules. 2017;22:313. doi: 10.3390/molecules22020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai R.N., Behera M., Berry L.D., Rossi M.R., Kris M.G., Johnson B.E., Bunn P.A., Ramalingam S.S., Khuri F.R. HER2 mutations in lung adenocarcinomas: A report from the Lung Cancer Mutation Consortium. Cancer. 2017;123:4099–4105. doi: 10.1002/cncr.30869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkas-Kramarski R., Lenferink A.E., Bacus S.S., Lyass L., van de Poll M.L., Klapper L.N., Tzahar E., Sela M., van Zoelen E.J., Yarden Y. The oncogenic ErbB-2/ErbB-3 heterodimer is a surrogate receptor of the epidermal growth factor and betacellulin. Oncogene. 1998;16(10):1249–1258. doi: 10.1038/sj.onc.1201642. [DOI] [PubMed] [Google Scholar]
- Puranik N.V., Srivastava P. First synthesis of rugosaflavonoid and its derivatives and their activity against breast cancer. RSC Advances. 2017;7:33052–33060. doi: 10.1039/c7ra04971d. [DOI] [Google Scholar]
- Ricciardi G.R., Russo A., Franchina T., Ferraro G., Zanghì M., Picone A., Scimone A., Adamo V. NSCLC and HER2: between lights and shadows. J Thorac Oncol. 2014;9:1750–1762. doi: 10.1097/JTO.0000000000000379. [DOI] [PubMed] [Google Scholar]
- Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., Hu Y., Tan Z., Stokes M., Sullivan L., Mitchell J., Wetzel R., Macneill J., Ren J.M., Yuan J., Bakalarski C.E., Villen J., Kornhauser J.M., Smith B., Li D., Zhou X., Gygi S.P., Gu T.L., Polakiewicz R.D., Rush J., Comb M.J. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- Rom W.N., Hay J.G., Lee T.C., Jiang Y., Tchou-Wong K.M. Molecular and genetic aspects of lung cancer. Am. J. Respir. Crit. Care Med. 2000;161:1355–1367. doi: 10.1164/ajrccm.161.4.9908012. [DOI] [PubMed] [Google Scholar]
- Saravanan P., Dubey V.K., Patra S. Potential selective inhibitors against Rv0183 of Mycobacterium tuberculosis targeting host lipid metabolism. Chem. Biol. Drug Des. 2012;79:1056–1062. doi: 10.1111/j.1747-0285.2012.01373x. [DOI] [PubMed] [Google Scholar]
- Shaik N.A., Al-Kreathy H.M., Ajabnoor G.M., Verma P.K., Banaganapalli B. Molecular designing, virtual screening and docking study of novel curcumin analogue as mutation (S769L and K846R) selective inhibitor for EGFR. Saudi J Biol Sci. 2019;26:439–448. doi: 10.1016/j.sjbs.2018.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Bast F. In silico molecular docking study of natural compounds on wild and mutated epidermal growth factor receptor. Med Chem Res. 2014;23:5074–5085. doi: 10.1007/s00044-014-1090-1. [DOI] [Google Scholar]
- Singhal G., Roy A., Bharadvaja N. In-silico study on plant determined flavonoids compounds for the synthetic medications against breast cancer growth. JAHM. 2017;3:116–121. [Google Scholar]
- Stamos J., Sliwkowski M.X., Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J. Biol. Chem. 2002;48:46265–46272. doi: 10.1074/jbc.M207135200. [DOI] [PubMed] [Google Scholar]
- Stella G.M., Luisetti M., Inghilleri S., Cemmi F., Scabini R., Zorzetto M., Pozzi E. Targeting EGFR in non-small-cell lung cancer: lessons, experiences, strategies. Respiratory medicine. 2012;106:173–183. doi: 10.1016/j.rmed.2011.10.015. [DOI] [PubMed] [Google Scholar]
- Surendra T.V., Roopan S.M., Al-Dhabi N.A., Arasu M.V., Sarkar G., Suthindhiran K. Vegetable peel waste for the production of ZnO nanoparticles and its toxicological efficiency, antifungal, hemolytic, and antibacterial activities. Nanoscale Research Letters. 2016;11:546. doi: 10.1186/s11671-016-1750-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surendra T.V., Roopan S.M., Arasu M.V., Al-Dhabi N.A., Rayalu G.M. RSM optimized Moringa oleifera peel extract for green synthesis of M. oleifera capped palladium nanoparticles with antibacterial and hemolytic property. Journal of Photochemistry & Photobiology, B: Biology. 2016. 162::550–557. doi: 10.1016/j.jphotobiol.2016.07.032. [DOI] [PubMed] [Google Scholar]
- Surendra T.V., Roopan S.M., Arasu M.V., Al-Dhabi N.A., Sridharan M. Phenolic compounds in drumstick peel for the evaluation of antibacterial, hemolytic and photocatalytic activities. Journal of Photochemistry & Photobiology, B: Biology. 2016;161:463–471. doi: 10.1016/j.jphotobiol.2016.06.013. [DOI] [PubMed] [Google Scholar]
- Valsalam S., Agastian P., Arasu M.V., Al-Dhabi N.A., Ghilan A.K.M., Kaviyarasu K., Ravindran B., Chang S.W., Arokiyaraj S. Rapid biosynthesis and characterization of silver nanoparticles from the leaf extract of Tropaeolum majus L. and its enhanced in-vitro antibacterial, antifungal, antioxidant and anticancer properties. Journal of Photochemistry & Photobiology, B: Biology. 2019: 191,:65–74. doi: 10.1016/j.jphotobiol.2018.12.010. [DOI] [PubMed] [Google Scholar]
- Verma R., Devre K., Gangrade T. A Review on Phytochemical, Pharmacological, and Pharmacognostical Profile of Cadaba trifoliate. Research and reviews: journal of pharmacognsy and phytochemistry. 2014;2:35–39. [Google Scholar]
- Wieduwilt M.J., Moasser M.M. The epidermal growth factor receptor family: Biology driving targeted therapeutics. Cell. Mol. Life Sci. 2008;65:1566–1584. doi: 10.1007/s00018-008-7440-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda H., Park E., Yun C.H., Sng N.J., Lucena-Araujo A.R., Yeo W.L., Huberman M.S., Cohen D.W., Nakayama S., Ishioka K., Yamaguchi N., Hanna M., Oxnard G.R., Lathan C.S., Moran T., Sequist L.V., Chaft J.E., Riely G.J., Arcila M.E., Soo R.A., Meyerson M., Eck M.J., Kobayashi S.S., Costa D.B. Structural, biochemical, and clinical characterization of epidermal growth factor receptor (EGFR) exon 20 insertion mutations in lung cancer. Sci Transl Med. 2013;5:1–23. doi: 10.1126/scitranslmed.3007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.A., Mauchley D., Li H., Meng X., Nemenoff R.A., Fullerton D.A., Weyant M.J. Knockdown of secretory phospholipase A2 IIa reduces lung cancer growth in vitro and in vivo. J Thorac Cardiovasc Surg. 2012;144:1185–1191. doi: 10.1016/j.jtcvs.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Lee J.C., Lin L., Olivas V., Au V., LaFramboise T., Abdel-Rahman M., Wang X., Levine A.D., Rho J.K., Choi Y.J., Choi C.M., Kim S.W., Jang S.J., Park Y.S., Kim W.S., Lee D.H., Lee J.S., Miller V.A., Arcila M., Ladanyi M., Moonsamy P., Sawyers C., Boggon T.J., Ma P.C., Costa C., Taron M., Rosell R., Halmos B., Bivona T.G. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nature genetics. 2012;44:852–860. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Yao M.Y., Zhu S.J., Chen J.Y., Yun C.H. Crystal structure of EGFR T790M/C797S/V948R in complex with EAI045. Biochem. Biophys. Res. Commun. 2018;502:332–337. doi: 10.1016/j.bbrc.2018.05.154. [DOI] [PubMed] [Google Scholar]
- Zheng J., Zhou Y., Li Y., Xu D.P., Li S., Li H.B. Spices for Prevention and Treatment of Cancers. Nutrients. 2016;8:495. doi: 10.3390/nu8080495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoete V., Cuendet M.A., Grosdidier A., Michielin O. SwissParam: a fast force field generation tool for small organic molecules. J. Comput. Chem. 2011;32:2359–2368. doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]