Abstract
The highly contagious nature of Coronavirus disease 2019 (Covid-19) resulted in a global pandemic. Due to the relatively slow and taxing nature of conventional testing for Covid-19, a faster method needs to be in place. The current researches have suggested that visible irregularities found in the chest X-ray of Covid-19 positive patients are indicative of the presence of the disease. Hence, Deep Learning and Image Classification techniques can be employed to learn from these irregularities, and classify accordingly with high accuracy. This research presents an approach to create a classifier model named StackNet-DenVIS which is designed to act as a screening process before conducting the existing swab tests. Using a novel approach, which incorporates Transfer Learning and Stacked Generalization, the model aims to lower the False Negative rate of classification compensating for the 30% False Negative rate of the swab tests. A dataset gathered from multiple reliable sources consisting of 9953 Chest X-rays (868 Covid and 9085 Non-Covid) was used. Also, this research demonstrates handling data imbalance using various techniques involving Generative Adversarial Networks and sampling techniques. The accuracy, sensitivity, and specificity obtained on our proposed model were 95.07%, 99.40% and 94.61% respectively. To the best of our knowledge, the combination of accuracy and false negative rate obtained by this paper outperforms the current implementations. We must also highlight that our proposed architecture also considers other types of viral pneumonia. Given the unprecedented sensitivity of our model we are optimistic it contributes to a better Covid-19 detection.
Keywords: Covid-19, Stacked generalization, Transfer learning, Deep neural networks, Generative adversarial networks, Image segmentation
Introduction
Coronavirus disease 2019 (Covid-19) is defined as illness caused by a novel coronavirus now called severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2; formerly called 2019-nCoV). After the first documented outbreak in Wuhan, China, the spread of Covid-19 virus has increased rapidly worldwide. As of 11 July 2020, 12.4 million cases have been confirmed worldwide. As Covid-19 spreads through contact with contaminated surfaces or infected person, early diagnosis plays an important role in breaking the transmission chain and slowing down the spread of this virus. One of the widely used testing methods is a polymerize chain reaction (PCR) test. Even though PCR tests can determine the presence of the virus very early on, these tests are laborious with several stages at which errors may occur. Moreover, PCR tests have nearly 30 percent false negative rate in the initial week of infection [1], which is useful for confirming the virus but not for clearing the patient as negative for Covid-19. Another testing method is Serologic testing which detects SARS-CoV-2 antibodies in serum or plasma components of blood. However, such antibody tests do not rule out the chance of an active infection.
To make these tests more accurate Chest X-ray (CXR) can be used for the first rough screening process. As CXRs are cheap, less time consuming and effortless they can provide an easy early detection mechanism and thus, can be used to determine the need of a swab test. Moreover, the results of these CXR images can be used to reexamine the negative results of swab tests, reducing the effect of false negative results of the swab tests. The major hurdle in using manual detection from CXR by a radiologist is the insufficient sensitivity in ground-glass opacity (GGO) nodule [2]. This leads to inefficient early detection of the disease using CXR. However, a well-trained deep learning model can differentiate and detect such nodules which can be easily missed by the human eye.
CNNs (convolutional neural networks) show good performance in image classification tasks. Pre-trained CNN architectures can be fine-tuned to achieve high accuracy in image classification tasks, even on a small dataset. However, in some situations, CNNs fall short. In [38] Mateusz Buda et al. demonstrate how CNNs do not perform well in situations where an imbalance exists in the dataset. In the case where one class represents a high majority of the dataset, the CNN classifier leans towards the overrepresented class. This leads to the classifier having a misleading high accuracy. Another case, when the test set is balanced and training set is not balanced; here, the classifier’s decision reflects the prior class probabilities (likelihood of an outcome before new evidence is presented); which leads to low test set accuracy while the classifier’s real discriminative power does not change. Our implementation avoids this situation by using sampling methods (Sect. 2.2.2) before training the CNN models.
In [39] Hosseini et al. demonstrate how CNN do not perform well on some transformed inputs such as negative images, even though they have the same structure and semantics as their normal counterpart. Hosseini et al. found that CNNs have a lower accuracy in identifying/classifying the negative of the image if the CNN is trained on a regular version of the same image. Our implementation avoids this problem as our dataset consists of X-ray images which are not affected by inversion of colors and other such transformations.
CNNs and CXR images have been used previously for detection of various respiratory diseases including COVID-19. Wang and Wong [3] proposed a deep model for COVID19 detection (COVID-Net), which obtained 92.4% accuracy in classifying normal, non-COVID pneumonia, and COVID-19 classes. Apostolopoulos et al. [4] developed the deep learning model for Covid detection with success rates of 98.75% for two classes and 93.48% for three classes.
In this study we have used a combination of four CNN models. CNNs have been used in several computer vision tasks [5]. A significant progress has been recorded in the performance of CNNs in medical studies in the past few years. In this work we aim to propose a system designed to reduce the false negatives by using a combination of four CNN models trained on segmented and non-segmented Chest X-rays of publicly available images. Our proposed architecture is an end-to-end solution for COVID-19 diagnosis which can determine the presence of the disease based on raw Chest X-ray images. The use of Stacked Generalization approach to create the model and evaluating it thoroughly using various metrics and also a unique verification method contribute to the novelty of our approach. Using this technique we were able to achieve a better accuracy and sensitivity of 99.07% and 99.4% respectively.
Related work
Generative Adversarial Networks (GANs) have been proven to handle dataset imbalance by generating synthetic samples of the same. The system implemented in [6] by Abdul Waheed et al. have used ACGAN architecture [7] to generate samples of Covid and Non-Covid X-ray images to handle the lack of image data. The discriminator takes a (112 × 112 × 3) image as in input, where it goes through 5 blocks, each consisting of a convolutional layer, batch normalization layer, activation layer and a dropout layer in the mentioned sequence. The discriminator outputs the probability that the image is real or fake, and the class label it belongs to. The generator takes as input a latent point in space and a class label to output a (112 × 112 × 3) image. In training, discriminator layers are initially set as non-trainable and hence, only the generator gets updated by the discriminator. In addition to their existing dataset, their system generated 1399 synthetic images of Normal X-ray images and 1669 synthetic images of Covid-19 X-ray images. In [8] Khalifa et al. have used a GAN architecture consisting of 5 convolutional layers, 4 ReLU layers, 4 Batch Normalization Layers and 1 Tanh layer in the generator network and 5 convolutional layers, 4 ReLU layers and 3 Batch Normalization layers in the discriminator network. This network resulted in an increase in the dataset by a factor of 10.
Use of deep neural networks in medical diagnosis has been studied for many years and many researchers have successfully achieved great results. Use of CNNs for diagnosis of chest diseases started with a boom after the release of massive Chest X-ray (CXR) dataset by Wang et al. [9] with more than 1 million CXRs. Some researchers have also claimed to outperform radiologists while diagnosing CXR. As an example, Rajpurkar et al. [10] proposed ChexNet a model trained end-to-end on DenseNet-121 [11] pretrained on ImageNet [12] which has outperformed the average F1-Score (0.387) of 4 radiologists with ChexNet F1-Score (0.435). In [13] Shin et al. has investigated the performance of various CNNs for classification of interstitial lung disease. In 2005 Xuanyang et al. [14] focused on the classification of CXR to diagnose SARS where feature mining is done on segmented lungs images and classification by three algorithms was performed. Among the three classification algorithms, Regression trees outperformed shallow neural nets and decision trees. Still, Neural Nets are supposed to be the best methods for pneumonia classification.
In [15] Sethy et al. compared the performance of various CNNs trained using transfer training approach, where models trained on ImageNet were used to initialize the CNNs. The models were used to extract features from the CXRs followed by training SVM on these mined features for Covid classification. While Narin et al. [16] used a transfer learning based approach to train and performed comparative analysis on the results of three CNN models namely Inception ResNet v2, Inception v3 and ResNet50. They used a fivefold cross validation strategy and achieved accuracy of 98% for ResNet50 while performances of Inception ResNet v2 and Inception v3 were 97% and 87% respectively.
Proposed work
False negative (FN) rate which is a crucial metric while determining performance of any model for biomedical image classification, as any particular false case can cause a sense of negligence in one’s mind. Considering the Covid-19 detection, where-in any false detection following no further examination can cause huge impact keeping in mind the novelty of the virus. Various CNNs used for the classification trained using transfer learning method have achieved high accuracies but still the FN rates are comparatively high. The proposed model StackNet-DenVIS has achieved both best accuracy and lowest FN rate. Consideration of the features on which the predictions are made is also important and there too DenVIS performed outstandingly. StackNet-DenVIS is a model built by stacked generalization ensembling of four different CNNs where-in we optimized outputs from CNNs and achieved precise accuracies. We combined multiple Chest X-ray datasets which included Covid-19 Pneumonia images and then handled data imbalance using SMOTE and Tomek Links along with addition of synthetic images with the help of DCGAN. The balanced dataset was then passed through the UNet model where segmentation of lungs was done which in turn helped in making lungs as the only prominent part in the X-ray. Four CNN models DenseNet-121, VGG19 bn, Inception ResNet v2, SE ResNeXt50-32 × 4d were trained on two datasets (1) Non-Segmented Balanced, (2) Segmented Balanced. Best performing models were then selected for ensembling. Figure 1 shows the architecture of the proposed model.
Fig. 1.
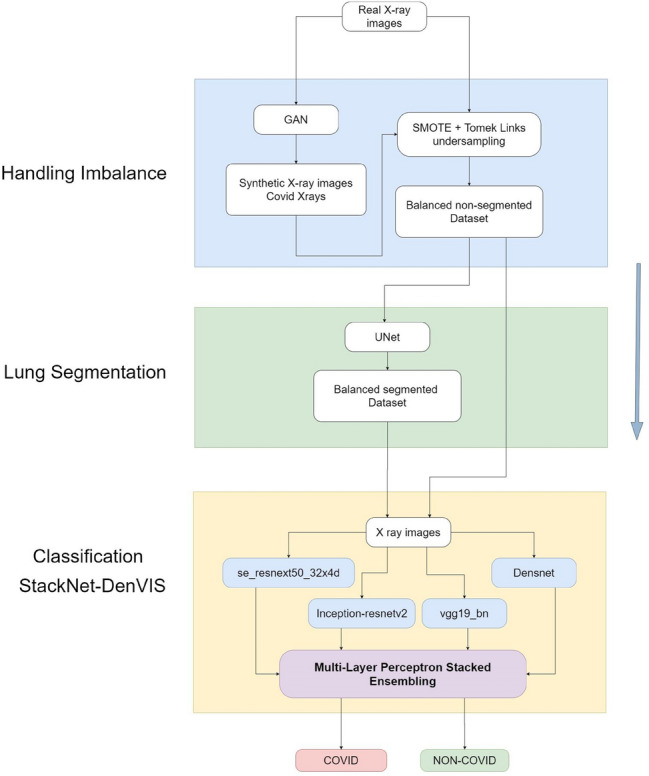
Proposed architecture
The novelty of concatenating two multi-layer perceptron (MLP) models trained on different inputs, helped us in making decisions on both the classes independently detailed in Sect. 2.4.
This reduced the FN rate and also helped in outperforming other models as demonstrated in Sect. 3.2.
Methods
Data preparation
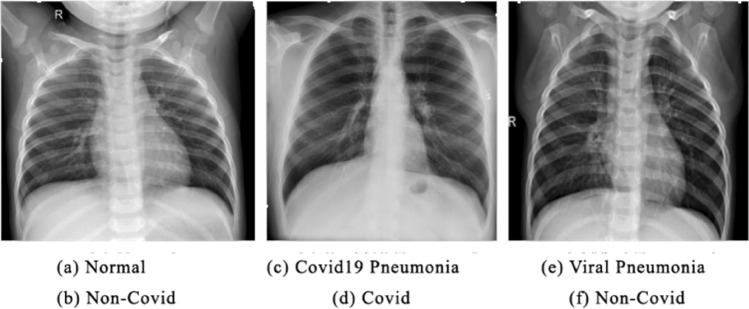
The dataset used to train the models was collected from 4 different sources. Choice of the datasets include (1) Chest X-ray Images [dataset] [17] (2) Covid-19 chest X-ray data [dataset] [3] (3) Covid-19 radiography database [dataset] [18] (4) Covid-19 Chest X-ray dataset [dataset] [19]. These sources were chosen as they were chosen as they were openly accessible. Further, the agglomerated dataset comprises 2 sets, Training and Testing with two labels, Covid and Non-Covid for each set (Fig. 2).
Fig. 2.
a, c and e represent labels of source dataset and b, d, and f represent labels of our dataset
Differentiating the types:
Covid: comprises of images which are labelled as Covid in the source.
Non-Covid: comprises of images labelled as Non-Covid Pneumonia, Healthy and Normal in the source.
Our model classifies the images into two major categories Covid and Non-Covid thus, we labelled all possible types of Pneumonia other than those caused by COVID19 as Non-Covid. Finally, the dataset consists of 700 Covid and 7500 Non-Covid images in the training set while the test set contains 168 Covid and 1596 Non-Covid images.
Preprocessing
Augmenting dataset
CNNs have been proven to be effective in the field of medical imaging in the last few years [5]. However, their efficacy is contingent on the size of the datasets. CNNs have a propensity to overfit on small datasets. This is because the efficiency of generalization is proportional to the size of the dataset. Thus, an acute data imbalance can lead to a biased classifier. After the combination of the datasets mentioned in Sect. 2.1, we get 868 Covid images and 9094 Non-Covid images. It is evident from these statistics that the number of Covid-19 training and testing images are lower than the Non-Covid categories i.e. other viral infections like influenza, H1N1 etc. Figure 3 shows the distribution of Covid and Non-Covid cases for training and testing images. The orange bar represents the Covid images while the green bar represents the Non-Covid images.
Fig. 3.
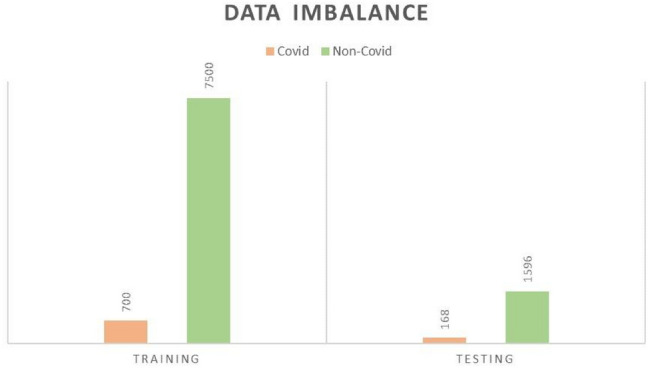
Data imbalance
Data Augmentation is generally used to overcome such data imbalance, in which we artificially extend the training dataset. However, it cannot introduce new unseen data and is limited to operations like flipping, zooming, blurring the images etc. Thus, to introduce unseen data in the training set, we have used Generative Adversarial Networks (GANs). GAN is a generative model with two adversaries; the generator network and the discriminator network. These networks compete against each other to generate new unseen data which is similar to the input distribution. In this research, we have used a version of GAN known as Deep Convolutional GAN (DCGAN).
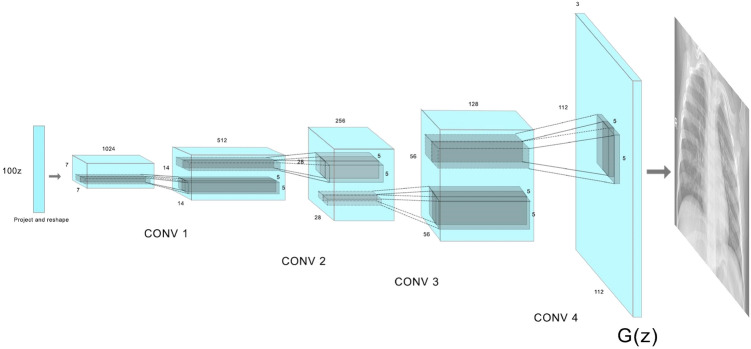
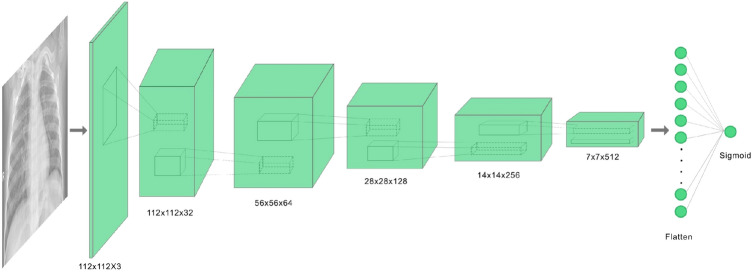
DCGAN is a variant on GAN which uses CNNs. It mainly consists of convolutional layers without any max pooling or fully connected layers. It uses transpose convolution and convolutional strides for up sampling and down sampling respectively. Figures 4 and 5 show the architecture of the generator and discriminator DCGAN used in our research.
Fig. 4.
Generator architecture
Fig. 5.
Discriminator architecture
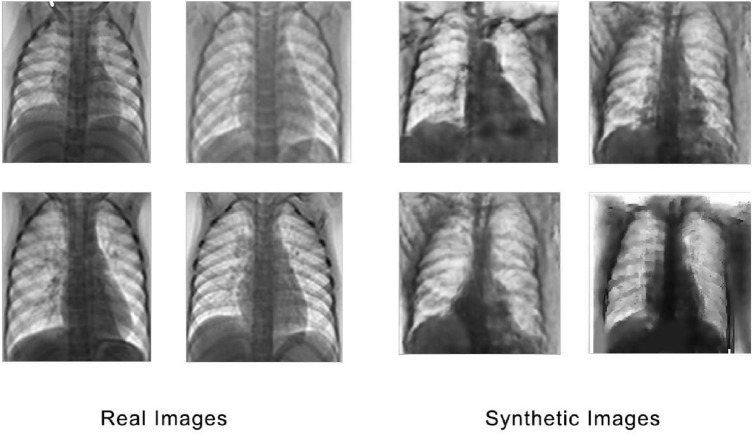
The architecture of the DCGAN is based on the CovidGAN [6] along with a few modifications. The generator takes in a 100-dimensional laten noise vector to produce a 112 × 112 × 3 image. The point in the latent space is interpreted by a dense layer and reshaped to 7 × 7 × 1024. Then using transpose convolution layers the low-resolution image is up sampled to a 112 × 112 × 3 image. The discriminator takes in a 112 × 112 × 3 image and outputs if the generated image is real or fake. Down sampling is performed by a number of convolutional layers, converting the input of 112 × 112 × 3 to 7 × 7 × 512 where each layer has a batch normalization and a dropout layer of probability 0.4. The sigmoid output layer predicts the authenticity of the generated image. The results of the DCGAN are shown in Fig. 6.
Fig. 6.
Comparison of images generated by DCGAN and real images
After combining, relabeling and addition of images, done with the help of GANs, dataset still has an issue of high imbalance where COVID X-rays are in the minority class and considering the use case here i.e. screening the COVID X-rays for Covid-19 detection this issue needs a solution.
SMOTE and Tomek Links Under-Sampling
To overcome the dataset imbalance problem, we have used Synthetic Minority Oversampling Technique (SMOTE) and Tomek Links Under sampling in a pipelined approach as proposed by Gustavo E. A. P. A. Batista [20]. Unlike in simple oversampling, where the same examples of the minority class are copied, SMOTE generates synthetic classes which are close to the feature space of the minority class, thereby increasing the minority class examples. It is a technique based on nearest neighbors judged by Euclidean Distance between data points in feature space. There is a parameter which represents the percentage of Over-Sampling whose value indicates the number of synthetic samples to be created. For each minority instance, k number of nearest neighbors are found such that they also belong to the same class where,
The difference between the feature vector of the considered instance and the feature vectors of the k nearest neighbors are found. So, k number of difference vectors are obtained. The k difference vectors are each multiplied by a random number between 0 and 1 (excluding 0 and 1). Now, the difference vectors, after being multiplied by random numbers, are added to the feature vector of the considered instance (original minority instance) at each iteration.
In this case, the minority class is oversampled with the applied ‘sampling_strategy’ parameter represented as ‘k’ (k = 0.5) i.e. keeping the sampling strategy parameter as 0.5 increases the number of minority class examples by 50%.
Tomek Links [21] is one of the finest techniques for under-sampling in which the Tomek Links are identified from the given samples and according to the stated class that particular sample is removed. It has the effect of making the decision boundary in the training dataset less noisy or ambiguous. A Tomek Link is the distance between two samples from two different classes say x and y such that for any sample z:
Tomek Links basically exist between the nearest neighbors in the feature space and thereby removing the sample of the minority class. In order to apply the Tomek Links configuration, the tomek argument can be passed to the SMOTETomek() class to create an instance of the configuration.
In a pipelined approach the minority Class is oversampled by using SMOTE followed by removing the majority class samples by Tomek Links [22]. The above-mentioned methods are implemented using the imblearn library.
Since the usage of SMOTE [20] and Tomek Links [22] requires the application of feature comparison thus, it is a slow algorithm and the images are required to be stored in the RAM for processing. So to avoid crashing of our system we have used Batch-SMOTE followed by undersampling where-in we divided the combined dataset into four batches containing 2063 images each with Covid images shuffled in such a way that every batch contains at least 125 images.
As shown in Table 1, oversampling the minority class (i.e. ‘Covid’) resulted in at least 900 image samples in each batch. After, undersampling the majority class resulted in matching distribution size for each class in a batch. Finally, a total number of 7490 images with 3745 images in each class were obtained after this process.
Table 1.
Summary of oversampling and undersampling
| Batch | Original | After Oversampling | After Undersampling | Total | |||
|---|---|---|---|---|---|---|---|
| Sr. no | Covid | Non-Covid | Covid | Non-Covid | Covid | Non-Covid | |
| 1 | 200 | 1850 | 925 | 1850 | 925 | 925 | |
| 2 | 250 | 1813 | 907 | 1813 | 907 | 907 | |
| 3 | 175 | 1888 | 944 | 1888 | 944 | 944 | |
| 4 | 125 | 1938 | 969 | 1938 | 969 | 969 | |
| Total | 3745 | 3745 | 7490 | ||||
Lung Segmentation
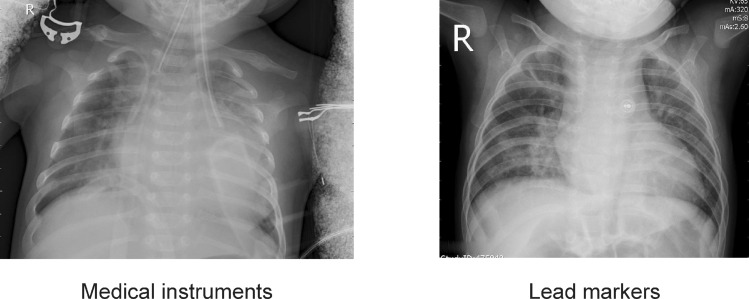
As we train to differentiate between different classes from different datasets, a Neural Network model might learn features specific to the dataset and not specific to the disease. These unwanted features include lead markers and medical instruments or implants the patient might have. Such artifacts can create an intensity variation which might be incorrectly identified by the model as Covid-19 nodule. Moreover, a number of X-rays have the initials of the radiologic technologist. These initials are marked using lead markers. Such lead markers often include the alphabets ‘R’ and ‘L’ indicating the right or left of the patient respectively. These artifacts can influence the model's decision as the model might learn to identify these initials as a feature for classifying the X-ray as positive for Covid-19. As these lead markings are not pertinent to the disease these should be removed. Figure 7 illustrates some of these artefacts.
Fig. 7.
Artefacts in the dataset
Thus, in order to eliminate the unwanted features pertaining to the dataset we perform semantic segmentation of the lungs [23–26]. The goal of semantic segmentation is to generate a high-resolution image same as that of the input but with a label for every pixel. This ensures that only the features relevant to the disease are preserved and it trims away the possible sources of bias such as annotations present on the CXRs and medical instruments attached to the patient etc.
We implement this task of lung segmentation using U-Net [27] trained on Montgomery County Xray Set and Shenzhen Hospital X-ray Set [28]. The U-Net model architecture consists of two major parts; the contracting part with convolutional and max pooling layers and the expansive part consisting of transposed 2D convolutional layers. The model is able to distinguish and localize borders by classifying every pixel making it apposite for the task of segmentation. The segmentation is performed with the obtained mask and then the segmented image is blurred with a pixel radius of 4 to eliminate the sharp edges. Figure 8 shows the Lung segmentation process.
Fig. 8.
Process of lung segmentation
Transfer Learning
Training a CNN model from scratch and achieving best results requires the dataset to be in millions and considering our dataset contains only 7490 images, training a new CNN model from scratch was elusive. Thus, we have used transfer learning as an alternative where-in the usage of pretrained models is suggested.
Rather than training from scratch, in transfer learning, the weights of a particular model are first initialized for a task, utilizing large-scale datasets and then the learnt features are applied over the problem set. Such an approach makes it possible to avoid computational cost, mathematical calculations, hardware limitations and helps in focusing to resolve the determined task.
An important aspect while using transfer learning is the initialization of weights which is done by considering the variations in the source dataset. As ImageNet [12] is widely used as an initializer where the required task to be solved is related to medical images, we have initialized our models with ImageNet weights.
Among the different approaches towards usage of transfer learning, we have used a fine-tuning method which includes training the model on the available dataset with making some of the layers in the model trainable and keeping others non-trainable.
Pretrained Models Used
Considering the false negative rate of State-of-the-art CNNs when working with medical data, we narrowed the experiment to consist of the following choices: DenseNet-121 [11], VGG19 BN [29], Inception ResNet v2 [30] and SE-ResNeXt50-32 × 4d [31]. The objective of selecting the particular models was their performance in the ImageNet Large Scale Visual Recognition Challenge (ILSVRC). In this paper, the technique of discriminative layer training [32] was implemented using the fastai library.
Multi-layer perceptron stacked ensembling
Stacked generalization
Ensembling [33] of different models is a common technique to improve the prediction accuracy. In Deep Learning with Python [34] Francois has stated two basic approaches towards ensembling—weighted and normal averages.
These approaches include averaging the predictions of the models, while weights are added accordingly, to support the most promising model. As the selection of weights is done manually, which creates a gap for manual error, we used the Stacked Generalization [35] method.
In Stacked Generalization the model learns to predict the classes taking the predictions of CNN models as input. This model is also sometimes termed as meta-learner. Stacked Ensembling approach generally outperforms Average Ensembling and Weighted average Ensembling.
Hypothesis
As Covid-19 is a novel disease and highly contagious, even if the patient has a 10% chance of being infected it should be taken into consideration while making predictions. Consider the results in Table 2, for case 1 the WA and NA predicted the probability as 0.23 and 0.47 respectively. As per our hypothesis any probability > 0.2 should be considered as Covid, so the image would be classified as Covid while the Ground Truth says it's Non-Covid, unlike in Stack Ensembling where the same result is stated as Non-Covid with a probability of 0.0046. Table 2 includes only few of the many such cases where Stack Ensembling outperforms other ensembling approaches.
Table 2.
Examples of StackNet-DenVIS vs normal average vs weighted average
| Sr. no | DenseNet | SE_Resnext50-32 × 4d | Inception_resnetv2 | VGG19_bn | Weigted average (WA) |
Normal average (NA) |
StackNet-DenVIS | Ground truth |
|---|---|---|---|---|---|---|---|---|
| 1 | 0.9999 | 0.6783 | 0.1524 | 4.77e−06 | 0.2356 | 0.4756 | 0.0046 | 0 |
| 2 | 0.9993 | 0.0797 | 0.552 | 0.2343 | 0.367 | 0.4663 | 0.9995 | 1 |
| 3 | 0.9999 | 0.2502 | 0.1524 | 0.9999 | 0.6694 | 0.5903 | 0.9995 | 1 |
| 4 | 0.0066 | 0.9999 | 0.2502 | 0.4999 | 0.4631 | 0.4391 | 0.0046 | 0 |
Architecture
The use of Stacked Ensembling not only reduces the variance but also helps in reducing the error rate with significant margin. Unlike in average ensembling, the conditional weights in stacked ensembling are added to the input predictions, thus helping in better generalization. Stacked Ensembling is done in two levels; first the predictions of the CNN models on the training datasets are stored and in the second level Multi-Layer Perceptron (MLP) Model built is trained on the predicted data.
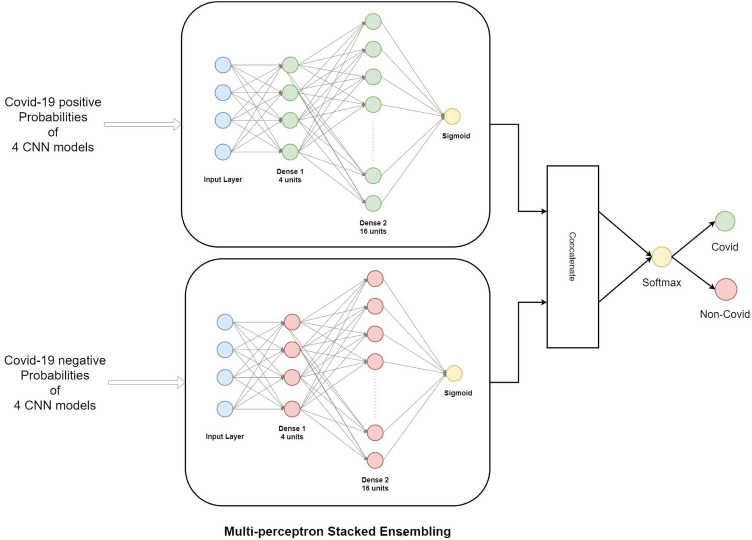
The predicted probabilities by the CNN models are taken as training data rather than taking the class labels. This helps in making the level-2 predictions to be more precise. While the architecture of the MLP Model varies according to the usage, in our case we have used the model as shown in Fig. 9.
Fig. 9.
Architecture of multi-layer perceptron stacked ensembling
Our Stacked model contains two separate MLP models concatenated which then gives two output probabilities. The models are named as ‘C’ and ‘NC’ as they are trained on Covid and Non-Covid probabilities respectively.
Each of the two models contain 1 input, 1 hidden and 1 output layer. The input layer consists of 4 units and a Rectified Linear Unit (ReLU) as activation function. Hidden layer has 16 units and again ReLU activation. The output layer has a single unit with sigmoid activation function. Lastly, after concatenation we have added a final output layer which has two units and softmax function as activation.
This method helps to simultaneously concentrate on variations in both the probabilities without any bias of the other one. Also, consideration of both the probabilities simultaneously helps in accurate prediction while satisfying our hypothesis.
Training the model
General
Since neural networks process inputs using small weight values, inputs with large integer values can disrupt or slow down the learning process. Hence, we have normalized the pixel values so that each pixel has a value between 0 and 1. This is achieved by dividing all pixels values by the largest pixel value; that is 255. This is performed across all channels, regardless of the actual range of pixel values that are present in the image. Furthermore, all the images are resized to 224 × 224 pixels. After these preprocessing steps we had two datasets—segmented and non-segmented. Utilizing these datasets, we trained our models on both of them and took the models with best accuracies for ensembling. For training, we split both the datasets randomly as training 80%, validation 20% and for testing we have a separate dataset with 170 Covid images and 1594 Non-Covid images.
GANs
The GAN model is a composition of a generator and a discriminator where the discriminator is initially set to non-trainable. For each batch of 32 images, the discriminator updates based on the generators output. The model is trained on 868 Covid X-rays for 2000 epochs with Adam optimizer at a learning rate of 0.00002 and momentum of optimizer as 0.5. Implemented using Keras deep learning library on a Tesla K80 GPU with 12 GB RAM, the model takes approximately 12 h to train.
DenseNet
While training DenseNet-121 we replaced the last layer with two fully connected (FC) layers, one with 256 units followed by dropout of 50% to reduce overfitting with activation as ReLu and second with 2 units and activation as softmax. The weights were initialized from a pretrained model on ImageNet. Keeping only the FC block trainable we trained the model using Adam with parameters as standard (1 = 0.9, 2 = 0.999). We trained the model with batches of size 32 for 15 epochs keeping initial learning rate as 0.001.
SE-ResNeXt50-32 × 4d, inception ResNet v2, and VGG19 bn
The training procedure for SE-ResNeXt50-32 × 4d, Inception ResNet v2, and VGG19 bn followed the concept of discriminative layer training [32] and 1-cycle policy [36]. The transfer learning parameters for the mentioned models are as in Table 3. The models were initially trained for 8 epochs on the following hyperparameters: batch size = 64, optimizer function = Adam with parameters (1 = 0.9, 2 = 0.99) and loss function = FlattenedLoss. After the initial 8 epochs, the model is trained for 3 additional epochs with a per cycle maximal learning rate sliced between (1e−6, 1e−4), in addition to the hyperparameters as mentioned. The given procedure was implemented using the fastai library.
Table 3.
The CNN models used and the transfer learning parameters
| Network | Parameter | Value |
|---|---|---|
| SE-ResNeXt50-32 × 4d | Last layers cut | 2 |
| Split at block | 6 | |
| Inception ResNet v2 | Last layers cut | 2 |
| Split at block | 9 | |
| VGG19 bn | Last layers cut | 1 |
| Split at block | 22 | |
| DenseNet-121 | Last layers cut | 1 |
| Split at block | 7 |
Multi-layer perceptron (MLP) stacking model
After training all the CNNs and selecting the best ones a new dataset for the Stacked model was created by predicting Covid and Non-Covid probabilities of the training images from all the selected CNNs. By applying fivefold cross validation on the newly created dataset of predicted values the Stacked model is trained for 200 epochs. We used Adam with standard parameters for training. Using Early Stopping callback, the training is stopped when the generalization error starts to increase.
Metrics
Along with total accuracy, we also compute the Macro-average and the weighted average for each class. Macro average will compute the metric individually for each class and then take average, treating all the classes equally. On the other hand, a weighted average will be computed using true instances of each label. The formulas are as listed below.
where TP is true positives, FP is false positives, and FN is false negatives.
Results
Overall performance
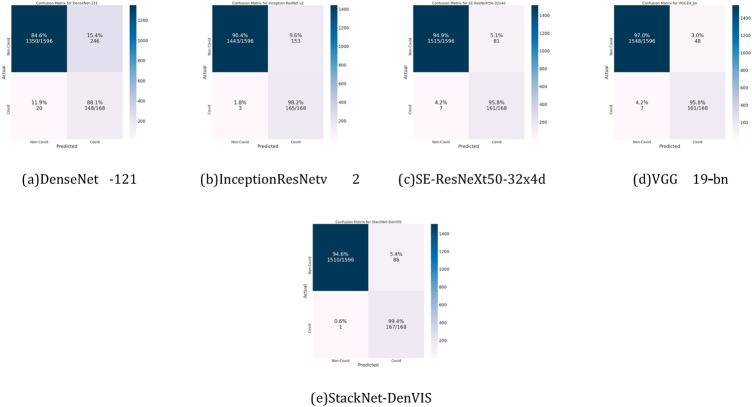
The performance of the proposed implementation was tested on 168 Covid and 1596 Non-Covid images. Parameters like Precision, Recall, F1-Score, Accuracy, Sensitivity, and Specificity were calculated to analyze the performance of the proposed implementation. When operating with medical data, especially highly contagious diseases like COVID-19, the sensitivity metric is more important as compared to the rest. In this case, Sensitivity (or True Positive Rate) denotes how likely an example of COVID-19 positive case is correctly classified as COVID-19 positive. Similarly, when considering an example of a Covid-19 negative case, specificity denotes how likely the example is correctly classified as Covid-19 negative i.e. a ‘Non-Covid’ case. A sensitivity of 99.40% is achieved on our proposed StackNet-DenVIS model. In addition to the given metric, our model achieves a high accuracy of 95.07% and a specificity of 94.61%. Table 4 shows the class-wise performance metrics as achieved on different models used in this implementation. Figure 10 shows confusion matrices for the different models used in this implementation.
Table 4.
Class-wise performance metrics as achieved on different models used in this implementation
| Model | Class | Precision | Recall | F1-Score | Support | Accuracy (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|---|---|
| Covid | 0.6601 | 0.994 | 0.7933 | 168 | ||||
| StackNet-DenVIS | Non-Covid | 0.9993 | 0.9461 | 0.972 | 1596 | 95.07 | 99.4 | 94.61 |
| Macro-average | 0.8297 | 0.9701 | 0.8827 | 1764 | ||||
| Weighted-average | 0.967 | 0.9507 | 0.955 | 1764 | ||||
| Covid | 0.7703 | 0.9583 | 0.9825 | 168 | ||||
| VGG19 bn | Non-Covid | 0.9955 | 0.9699 | 0.8541 | 1596 | 96.88 | 95.83 | 96.99 |
| Macro-average | 0.8829 | 0.9641 | 0.9183 | 1764 | ||||
| Weighted-average | 0.9741 | 0.9688 | 0.9703 | 1764 | ||||
| Covid | 0.6653 | 0.9583 | 0.7854 | 168 | ||||
| SE-ResNeXt50-32 × 4d | Non-Covid | 0.9954 | 0.9492 | 0.9718 | 1596 | 95.01 | 95.83 | 94.92 |
| Macro-average | 0.8303 | 0.9538 | 0.8786 | 1764 | ||||
| Weighted-average | 0.964 | 0.9501 | 0.954 | 1764 | ||||
| Covid | 0.5189 | 0.9821 | 0.679 | 168 | ||||
| Inception ResNet v2 | Non-Covid | 0.9979 | 0.9041 | 0.9487 | 1596 | 91.16 | 98.21 | 90.41 |
| Macro-average | 0.7584 | 0.9431 | 0.8139 | 1764 | ||||
| Weighted-average | 0.9523 | 0.9116 | 0.923 | 1764 | ||||
| Covid | 0.3756 | 0.881 | 0.5267 | 168 | ||||
| DenseNet-121 | Non-Covid | 0.9854 | 0.8459 | 0.9103 | 1596 | 84.92 | 88.1 | 84.59 |
| Macro-average | 0.6805 | 0.8634 | 0.7185 | 1764 | ||||
| Weighted-average | 0.9273 | 0.8492 | 0.8738 | 1764 |
Fig. 10.
Confusion matrices
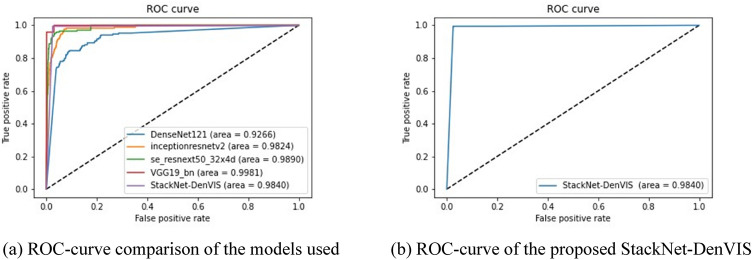
We obtained an AUROC of 98.40% on the proposed StackNet-DenVIS model. The ROC-Curve of our model and the ROC-Curve comparison between the models used is as shown in Fig. 11.
Fig. 11.
ROC curves
Furthermore, we have also explored the inference time for each model. In order to measure inference time in neural networks, we must consider two factors—asynchronous execution and GPU power-saving mode. Asynchronous execution allows the lines of code to execute out of its order. Hence, it is possible that the line of code responsible to stop the recording of time can be executed abruptly and thus, giving the wrong inference time. Moreover, GPU power-saving mode causes the GPU to ‘slow down’ and thus can cause a delay in the actual inference time.
So, in order to avoid the problem caused due to asynchronous execution, we use torch.cuda.synchronize() to perform synchronization and to avoid the delay caused by GPU power-saving mode, we ‘warm-up’ the GPU by initially running inference on a dummy input for 10 iterations. After this step, we perform 300 iterations of running inference on an image; in each iteration we use torch.cuda.Event() with the parameter enable_timing = True to measure inference time in that iteration. After performing this method for our models, we obtain the average inference time for each model as shown in Table 5. Figure 12 shows the graph comparing inference time between each model.
Table 5.
Time consumed by each method on the same image
| Network | Time taken (ms) |
|---|---|
| SE-ResNeXt50-32 × 4d | 40.213 |
| Inception ResNet v2 | 63.982 |
| VGG19 bn | 28.822 |
| DenseNet-121 | 0.191 |
| Ensembled | 215.485 |
Fig. 12.
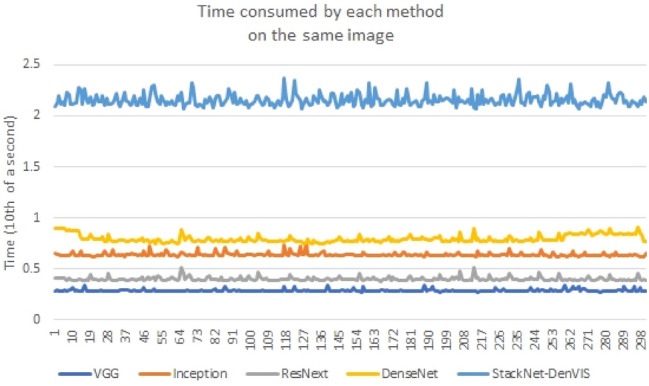
Time consumed by each method on the same image
Feature maps and black box evaluation
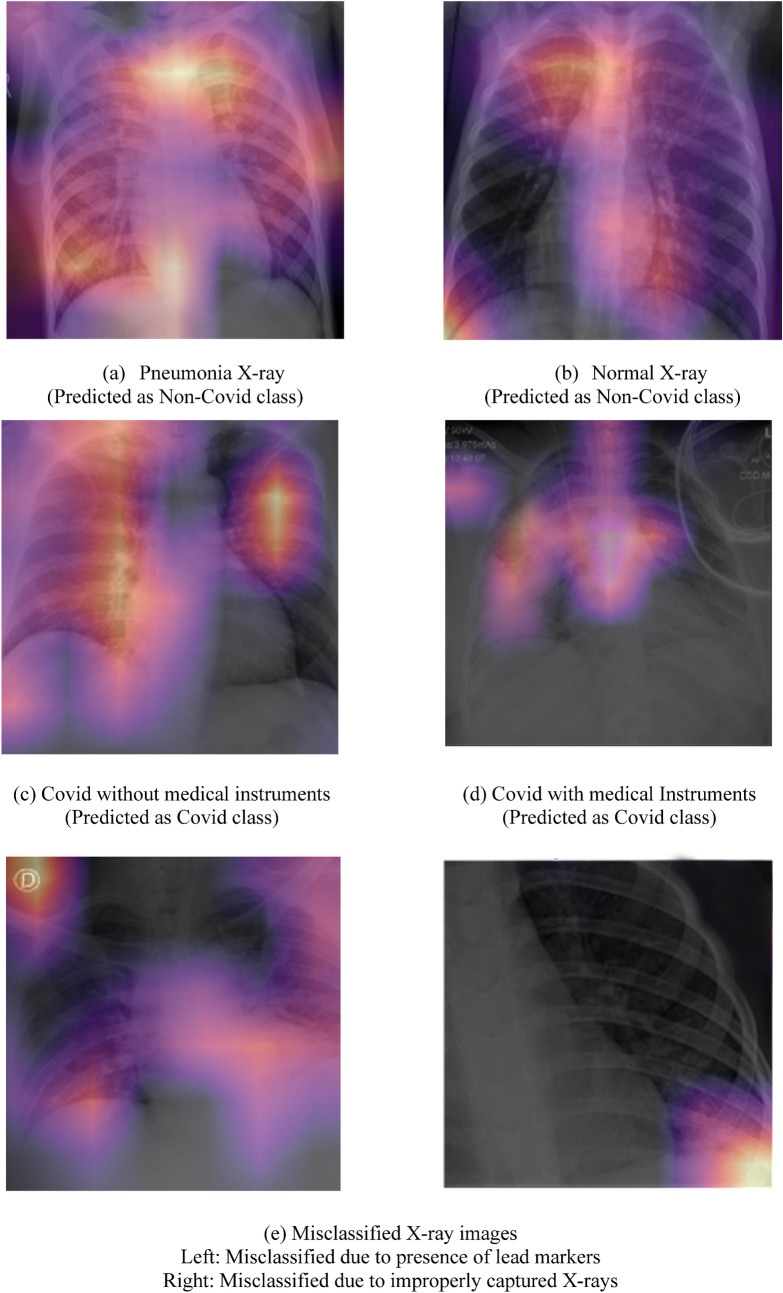
Figure 13 shows the feature map overlays in different scenarios. As we can see, in case of Covid-19 positive patients, the activation region is located inside the lungs, highlighting the Covid-19 nodules inside the lungs. On the other hand, for Normal and Pneumonia X-rays, these nodules are not present and hence, the activation region is away from the lungs. Furthermore, in case of misclassified images certain regions are activated due to hindrance caused in the inference by factors like poor quality of X-ray images or lead markers appearing in the X-ray image and improperly captured X-rays.
Fig. 13.
Features map of various cases
Figure 14a shows another example of a feature map of a Covid positive X-ray. As we can see that the areas of activation are located within the lungs, suggesting the detection of patches in the lungs.
Fig. 14.
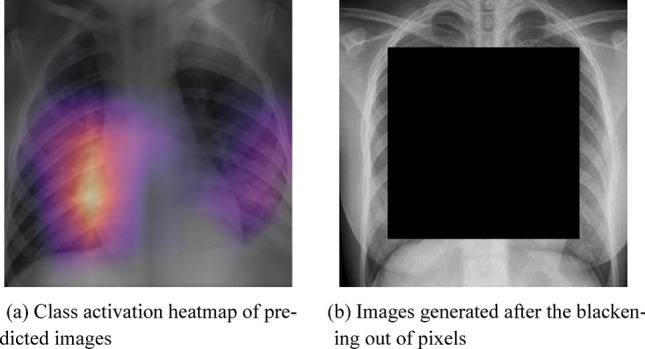
Images of heatmap and result of blackening out center pixels
In order to cross examine the results of the heatmap we conducted an experiment similar to the one suggested by Gianluca Maguolo [37]. In this experiment, we resized a set of 40 testing images and then a square of fixed size in the center of the image was turned black. This covered a majority area of the lungs in the CXRs with a black box. As a result, most of the information pertinent to the disease was removed. Figure 14b shows the images generated after the blackening out of pixels.
Then we tested these images with our trained model. The results were classified into three categories; ‘Covid’ for the images where the model predicts positive for Covid-19. ‘Non-Covid’ for the images where the model predicts negative for Covid-19. And lastly, ‘Ambiguous’ for images with nearly equal probabilities for both ‘Covid’ and ‘Non-Covid’.
We observed that 30 out of 40 images were classified as ambiguous. While only 3 and 7 images were classified as Covid and Non-Covid respectively, indicating that the model learned features correlated to the presence of the disease.
Discussion
Covid-19 X-ray dataset was first uploaded by Cohen [19] which helped many researchers to develop models to diagnose this disease. Majority of the research papers included in the literature conducted their research using the Covid-19 X-rays from this dataset. Sethy and Behera [15] compared different models and various classification methodologies in order to achieve promising results. ResNet50 as a feature extractor and SVM as a classifier gave them an accuracy of 95.83% with 25 Covid-19(+) and 25 Covid-19(−) images. Narin et al. [16] used three CNN models with transfer learning as an approach to training. ResNet50 with an accuracy of 98% outperformed Inception v3 and Inception ResNet v2 with accuracies as 97% and 87% respectively. They used a test set containing 50 Covid-19(+) and 50 Covid-19(−) images. Abdul Waheed et al. [6] proposed the CovidGAN model trained on synthetically augmented Covid-19 X-rays with the help of Generative Adversarial Network and achieved an accuracy of 95%. Sensitivity achieved by this model was 90% on a test data containing 72 Covid-19(+) and 120 Normal. Apostolopoulos [4] compared 5 different CNN models including (VGG19, MobileNet v2, Inception, Xception, Inception ResNet v2) where MobileNet v2 achieved an accuracy of 96.78% and VGG19 achieved an accuracy of 98.75%. Whereas the models were tested for two classes with 224 Covid-19 images and 1204 Non-Covid-19 images. In this research, four deep CNN models ensembled using the Stack Generalization [35] approach called StackNet-DenVIS were used for Covid-19 detection. In the existing works, a single model is used for prediction. Instead of using a single model, we have combined the class probabilities of four CNN models using stacked generalization which serve as a confidence measure for the predictions made. As stacked generalization works by deducing the biases of the generalizers, it can use the set of predictions as a context and conditionally decide to weigh the input predictions differently, potentially resulting in better performance. We obtained an accuracy of 95.07% and a sensitivity of 99.4%. Moreover, the improved sensitivity measure in our research signifies that the Covid-19 positive cases can be detected with better confidence and hence can be used for the screening process. We have also used SMOTE oversampling and Tomek Links undersampling technique in order to get a balanced and less noisy data for training the model. This method is especially useful in this case as the two classes are difficult to distinguish from each other.
In order to evaluate our model thoroughly, we have incorporated evaluation measures other than the standard performance measures. We have included a Black Box Evaluation (Sect. 3.2) to confirm the veracity of our results. This method verifies that the features learned by the model are present in the lungs and hence, is relevant to the disease.
We used a total of 7490 images; 3745 Covid and 3745 Non-Covid images for training our model. The Test set consisted of 168 Covid and 1596 Non-Covid (including images of Normal and other Pneumonia X-rays). We obtained an accuracy of 95.07% and a sensitivity of 99.4%. We obtained superior performance in terms of sensitivity, in comparison to other studies in the literature (Table 6).
Table 6.
Comparison between performance metrics of related works
| Study | Model | Number of cases | Accuracy (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|
| Waheed et al. [6] | CNN with synthetic augmentation |
72 Covid-19 120 Non-Covid-19 |
95.00 | 90.00 | 97.00 |
| Apostolopoulos et al. [4] | MobileNet v2 |
224 Covid-19 (+) 1204 Non-Covid |
96.78 | 98.66 | 96.46 |
| Sethy et al. [15] | ResNet50 plus SVM |
25 Covid-19 (+) 25 Covid-19 (−) |
95.33 | 95.33 | NA |
| Narin et al. [16] | ResNet50 | 50 Covid-19 (+) 50 Covid-19 (−) | 98 | 96 | 100 |
| Proposed network | StackNet-DenVIS |
168 Covid 1596 Non-Covid |
95.07 | 99.40 | 94.61 |
The main limitation of this research pertains to the availability of the dataset. First, the limited number of Covid-19 X-ray images obtained from open source datasets. In this research, although Covid-19 images were oversampled to compensate for the limited availability of images, the resulting quantity was still limited to use for all three stages i.e. training, validation and testing. Secondly, the images generated by GANs used in this research are limited by the available computational power at our disposal i.e. Tesla K80 GPU with 12 GB RAM. Hence, the quality of these generated images can be improved drastically with an advanced GPU and an extended training time. Moreover, the input given to the system is restricted to frontal chest X-rays. In case of lateral chest X-rays the predictions of the model are less accurate due to the shortage of features. In previous researches listed in the literature, common transfer learning techniques were used with few having different classifiers. However, the proposed model is based on the Stack Generalization method. The proposed model can be used as a screening tool for Covid-19. Considering the low cost and low false negative rate as compared to the PCR test, the proposed model can also be used for diagnosis purposes, where the patient has symptoms related to Pneumonia. Keeping in mind the ability of the model to detect Covid-19 nodules in the X-ray, this model will serve as a promising tool during detection procedures. Future work can reduce the computation overhead by increasing the efficiency of the model.
Conclusion
In this research, we proposed a set of models ensembled with a multi-layer perceptron model which is collectively termed as StackNet-DenVIS for the detection of Covid-19 Pneumonia from frontal Chest X-rays. We achieved a sensitivity of 99.4% with a false negative rate of 1 in 168 images for Covid class. While the model was later tested with images modified to censor the lungs with a black box placed on them, which resulted in 30 out of 40 images to be classified as ambiguous. Since this method of evaluating a model on images modified to censor the lungs with a black box placed on them was proposed; to the best of our knowledge, we are the first ones to achieve such results on the given evaluation technique. The proposed model can also be used for timely diagnosis of Covid-19 patients to check the development of pneumonia due to Covid-19. The model can be modified to classify between different types of Pneumonia by training on a balanced dataset and changing the prediction layer to respective number of classes. Since this research includes four computationally expensive training models such as VGG19 bn and DenseNet-121, we intend to make it more robust by working towards the reduction of training time.
Acknowledgements
First and foremost, we would like to acknowledge the healthcare professionals and researchers who are risking their lives to deal with Covid-19 patients. Secondly, we want to acknowledge the various professionals, who helped to collect the relevant data, and their research which made it possible for us to implement our proposed solution, thus contributing to the research and the fight against Covid-19 as well.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Pratik Autee, Email: patautee7@gmail.com.
Sagar Bagwe, Email: sagarbagwe6699@gmail.com.
Vimal Shah, Email: shahvimal1302@gmail.com.
Kriti Srivastava, Email: kriti.srivastava@djsce.ac.in.
References
- 1.Kucirka LM, Lauer SA, Laeyendecker O, Boon D, Lessler J (2020) Variation in false-negative rate of reverse transcriptase polymerase chain reaction-based sars-cov-2 tests by time since exposure. Ann Internal Med [DOI] [PMC free article] [PubMed]
- 2.Zu ZY, Jiang MD, Xu PP, Chen W, Ni QQ, Lu GM, Zhang LJ (2020) Coronavirus disease 2019 (covid-19): a perspective from china. Radiology 200490 [DOI] [PMC free article] [PubMed]
- 3.Wang L, Wong A (2020) Covid-net: a tailored deep convolutional neural network design for detection of covid-19 cases from chest X-ray images. arXiv:2003.09871 [DOI] [PMC free article] [PubMed]
- 4.Apostolopoulos ID, Mpesiana TA (2020) Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med [DOI] [PMC free article] [PubMed]
- 5.Greenspan H, Van Ginneken B, Summers RM. Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique. IEEE Trans Med Imaging. 2016;35(5):1153–1159. doi: 10.1109/TMI.2016.2553401. [DOI] [Google Scholar]
- 6.Waheed A, Goyal M, Gupta D, Khanna A, Al-Turjman F, Pinheiro PR. Covidgan: data augmentation using auxiliary classifier gan for improved covid-19 detection. IEEE Access. 2020;8:91916–91923. doi: 10.1109/ACCESS.2020.2994762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Odena CO, Shlens J (2016) Conditional image synthesis with auxiliary classifier gans. arXiv:1610.09585
- 8.Khalifa NEM, Taha MHN, Hassanien AE, Elghamrawy S (2020) Detection of coronavirus (covid-19) associated pneumonia based on generative adversarial networks and a fine-tuned deep transfer learning model using chest X-ray dataset. arXiv:2004.01184
- 9.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM. Chestx-ray8: hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. IEEE Conf Comput Vision Patt Recogn (CVPR) 2017;2017:3462–3471. [Google Scholar]
- 10.Rajpurkar P, Irvin J, Zhu K, Yang B, Mehta H, Duan T, Ding D, Bagul A, Langlotz C, Shpanskaya K, Lungren MP, Ng AY (2017) Chexnet: radiologist-level pneumonia detection on chest X-rays with deep learning. arXiv:1711.05225
- 11.Huang G, Liu Z, van der Maaten L, Weinberger KQ (2016) Densely connected convolutional networks. arXiv:1608.06993
- 12.Deng J, Dong W, Socher R, Li L. Kai Li, Li Fei-Fei, Imagenet: a large-scale hierarchical image database, in. IEEE Conf Comput Vision Patt Recogn. 2009;2009:248–255. [Google Scholar]
- 13.Shin H, Roth HR, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D, Summers RM. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging. 2016;35(5):1285–1298. doi: 10.1109/TMI.2016.2528162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xuanyang X, Yuchang G, Shouhong W, Xi L (2005) Computer aided detection of sars based on radiographs data mining. In: 2005 IEEE engineering in medicine and biology 27th annual conference, pp 7459–7462 [DOI] [PubMed]
- 15.Sethy P, Santi KB, Kumar P. Detection of coronavirus disease (covid-19) based on deep features and support vector machine. Int J Math Eng Manag Sci. 2020;12:643–651. doi: 10.33889/IJMEMS.2020.5.4.052. [DOI] [Google Scholar]
- 16.Narin A, Kaya C, Pamuk Z (2020) Automatic detection of coronavirus disease (covid-19) using X-ray images and deep convolutional neural networks. arXiv:2003.10849 [DOI] [PMC free article] [PubMed]
- 17.Kermany DS, Zhang K, Goldbaum MH (2018) Labeled optical coherence tomography (oct) and chest X-ray images for classification
- 18.Chowdhury MEH, Rahman T, Khandakar A, Mazhar R, Kadir MA, Mahbub ZB, Islam KR, Khan MS, Iqbal A, Al-Emadi N, Reaz MBI, Islam TI (2020) Can AI help in screening viral and covid-19 pneumonia? arXiv:2003.13145.
- 19.Cohen JP, Morrison P, Dao L, Roth K, Duong TQ, Ghassemi M (2020) Covid-19 image data collection: prospective predictions are the future, arXiv 2006.11988. https://github.com/ieee8023/covid-chestxray-dataset
- 20.Batista GE, Prati RC, Monard MC. A study of the behavior of several methods for balancing machine learning training data. ACM SIGKDD Explorations Newsl. 2004;6(1):20–29. doi: 10.1145/1007730.1007735. [DOI] [Google Scholar]
- 21.Tomek I et al (1976) Two modifications of CNN. In: IEEE transactions on systems, man, and cybernetics SMC-6(11): 769–772. 10.1109/TSMC.1976.4309452
- 22.Batista G, Bazzan A, Monard M-C (2003) Balancing training data for automated annotation of keywords: a case study. pp 10–18
- 23.Tartaglione E, Barbano CA, Berzovini C, Calandri M, Grangetto M (2020) Unveiling covid-19 from chest X-ray with deep learning: a hurdles race with small data. arXiv:2004.05405 [DOI] [PMC free article] [PubMed]
- 24.Candemir S, Jaeger S, Palaniappan K, Musco JP, Singh RK, Xue Z, Karargyris A, Antani S, Thoma G, McDonald CJ. Lung segmentation in chest radiographs using anatomical atlases with nonrigid registration. IEEE Trans Med Imaging. 2013;33(2):577–590. doi: 10.1109/TMI.2013.2290491. [DOI] [PubMed] [Google Scholar]
- 25.Hu S, Hoffman EA, Reinhardt JM. Automatic lung segmentation for accurate quantitation of volumetric X-ray CT images. IEEE Trans Med Imaging. 2001;20(6):490–498. doi: 10.1109/42.929615. [DOI] [PubMed] [Google Scholar]
- 26.Mansoor A, Bagci U, Xu Z, Foster B, Olivier KN, Elinoff JM, Suffredini AF, Udupa JK, Mollura DJ. A generic approach to pathological lung segmentation. IEEE Trans Med Imaging. 2014;33(12):2293–2310. doi: 10.1109/TMI.2014.2337057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention, Springer, pp 234–241
- 28.Jaeger S, Candemir S, Antani S. Wang Y-XJ, Lu P-X, Thoma G (2014) Two public chest X-ray datasets for computer-aided screening of pulmonary diseases. Quant Imaging Med Surg. 2014;4(6):475–477. doi: 10.3978/j.issn.2223-4292.2014.11.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simon M, Rodner E, Denzler J (2016) Imagenet pre-trained models with batch normalization. arXiv:1612.01452
- 30.Szegedy SI, Vanhoucke V, Alemi A (2016) Inception-v4, inception-resnet and the impact of residual connections on learning. arXiv:1602.07261
- 31.Hu J, Shen L, Albanie S, Sun G, Wu E (2017) Squeeze-and-excitation networks. arXiv:1709.01507 [DOI] [PubMed]
- 32.Jin X, Chen Y, Dong J, Feng J, Yan S (2016) Collaborative layer-wise discriminative learning in deep neural networks. arXiv:1607.05440
- 33.Hansen LK, Salamon P. Neural network ensembles. IEEE Trans Pattern Anal Mach Intell. 1990;12(10):993–1001. doi: 10.1109/34.58871. [DOI] [Google Scholar]
- 34.Chollet F (2017) Deep learning with Python, Manning
- 35.Wolpert H. Stacked generalization. Neural Networks. 1992;5(2):241–259. doi: 10.1016/S0893-6080(05)80023-1. [DOI] [Google Scholar]
- 36.Smith LN (2018) A disciplined approach to neural network hyper-parameters: part 1—learning rate, batch size, momentum, and weight decay. arXiv:1803.09820
- 37.Maguolo G, Nanni L (2020) A critic evaluation of methods for covid-19 automatic detection from X-ray images. arXiv:2004.12823 [DOI] [PMC free article] [PubMed]
- 38.Buda M, Atsuto M, Mazurowski MA. A systematic study of the class imbalance problem in convolutional neural networks. Neural Netw. 2018;106:249–259. doi: 10.1016/j.neunet.2018.07.011. [DOI] [PubMed] [Google Scholar]
- 39.Hosseini H, Xiao B, Jaiswal M, Poovendran R (2017) On the limitation of convolutional neural networks in recognizing negative images. arXiv:1703.06857v2