Abstract
Objectives:
To develop and validate a radiomics model for preoperative identification of lymph node metastasis (LNM) in patients with early-stage cervical squamous cell carcinoma (CSCC).
Methods:
Total of 190 eligible patients were randomly divided into training (n = 100) and validation (n = 90) cohorts. Handcrafted features and deep-learning features were extracted from T2W fat suppression images. The minimum redundancy maximum relevance algorithm and LASSO regression with 10-fold cross-validation were used for key features selection. A radiomics model that incorporated the handcrafted-signature, deep-signature, and squamous cell carcinoma antigen (SCC-Ag) levels was developed by logistic regression. The model performance was assessed and validated with respect to its calibration, discrimination, and clinical usefulness.
Results:
Three handcrafted features and three deep-learning features were selected and used to build handcrafted- and deep-signature. The model, which incorporated the handcrafted-signature, deep-signature, and SCC-Ag, showed satisfactory calibration and discrimination in the training cohort (AUC: 0.852, 95% CI: 0.761–0.943) and the validation cohort (AUC: 0.815, 95% CI: 0.711–0.919). Decision curve analysis indicated the clinical usefulness of the radiomics model. The radiomics model yielded greater AUCs than either the radiomics signature (AUC = 0.806 and 0.779, respectively) or the SCC-Ag (AUC = 0.735 and 0.688, respectively) alone in both the training and validation cohorts.
Conclusion:
The presented radiomics model can be used for preoperative identification of LNM in patients with early-stage CSCC. Its performance outperforms that of SCC-Ag level analysis alone.
Advances in knowledge:
A radiomics model incorporated radiomics signature and SCC-Ag levels demonstrated good performance in identifying LNM in patients with early-stage CSCC.
Introduction
Cervical carcinoma is the fourth most frequently diagnosed cancer and the fourth leading cause of cancer death in females.1 Early-stage cervical carcinoma (between International Federation of Gynecology and Obstetrics (FIGO) stages Ib and IIa) can be cured by surgery or radiotherapy, which have similar rates of effectiveness.2,3 Surgery is widely chosen, because it removes the primary disease, preserves fertility and ovarian function, and permits better targeting of adjuvant therapy through accurate surgical staging.3,4 Lymph node metastasis (LNM) is the most important independent risk factor for recurrence and survival, therefore, pelvic and/or para-aortic lymphadenectomy is recommended to be performed simultaneously with the surgical removal of the primary tumor.3 But, only 15 to 25% patients with early-stage cervical cancer had LNM,5 which means a significant portion of patients undergo lymphadenectomy unnecessarily and suffer from procedure-related morbidity. Moreover, many scholars believe that, for cervical cancer with LNM, chemoradiation should be the preferred therapeutic method.3,6 Therefore, accurate identification of LNM in patients with early-stage cervical cancer is crucial for making appropriate treatment decisions.
Magnetic resonance imaging (MRI) is recommended as an adjunct to clinical examination. It is highly sensitive and specific for depicting important prognostic factors such as tumor size, parametrial and pelvic side-wall invasion, and adjacent organ invasion, but it is unable to accurately identify LNM, especially for small metastatic lymph nodes.7,8 Several studies have reported that radiomics analysis of the primary cancer lesions has higher sensitivity and specificity in predicting LNM.9–13 A number of studies found that radiomics analysis of MRIs can enable superior prediction of LNM in patients with early-stage cervical cancer, with area under curves (AUCs) of 0.753 to 0.895.14,15 Most of the studies included all cervical cancers, regardless of individual histopathological type. Additionally, there were also no clinical risk factors included in final analyses. However, several studies of other cancer types have found that prediction efficiency is superior after embedding the radiomics and clinical factors in the predictive model.9,13,16
Squamous cell carcinoma (SCC) is the predominant histological subtype of cervical cancer and accounts for more than 80% of cervical cancer incidences.3 A number of studies have found that the level of serum squamous cell carcinoma antigen (SCC-Ag) in the preoperative phase may be a useful marker for predicting LNM of cervical squamous cell carcinoma (CSCC), especially in its early stage.17–19
To the best of our knowledge, there has been no study incorporating radiomics and SCC-Ag levels in the LNM predictive model for patients with early-stage CSCC. Therefore, the objective of this study was to develop and validate a radiomics model that incorporated the radiomics signature and SCC-Ag levels for the preoperative prediction of LNM in patients with early-stage CSCC.
Methods and materials
Patients
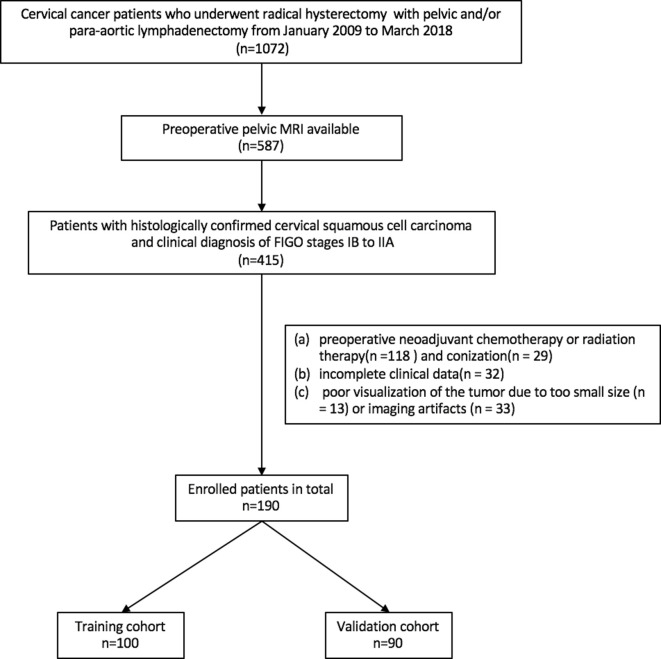
An institutional review board approval (GDREC2018521H(R1)) was obtained for this retrospective study, and the informed consent requirement was waived. Patients who underwent radical hysterectomy with pelvic and/or para-aortic lymphadenectomy between January 2009 and March 2018 were identified from the institutional database. The inclusion criteria were as follows: (a) histologically confirmed CSCC; (b) a clinical diagnosis between FIGO (2009) stages IB to IIA; and (c) a pelvic MRI test performed less than 3 weeks before surgical resection. Exclusion criteria were: (a) preoperative neoadjuvant chemotherapy or radiation therapy (n = 118) and conization (n = 29); (b) incomplete clinical data (n = 32); or (c) poor visualization of the tumor due to small size (less than 3 slices of ROI, n=13)or imaging artifacts (n = 33). A total of 190 consecutive patients who met the criteria were randomly divided into a training cohort (n = 100, mean age, 49.92 years ± 9.56; range, 22 to 73 years) and a validation cohort (n = 90, mean age, 49.91 years ± 9.23; range, 31 to 74 years). Figure 1 presents the patient recruitment pathway, along with the inclusion and exclusion criteria.
Figure 1.
Patient selection flowchart
Baseline clinical data (age, preoperative FIGO stage, levels of squamous cell carcinoma antigen(SCC-Ag), and menstrual status, as well as pregnancy, parturition, and abortion numbers) and histopathological data (histological subtype and lymph node (LN) status) were derived from medical records. Laboratory analysis of SCC-Ag was done via routine blood tests within 1 week before surgery. The threshold value for SCC-Ag levels was <2.35 µ g/L and ≥2.35 µ g/L, according to the study done by Xu et al, which found that 2.35 µ g/L was the optimum cutoff of SCC-Ag levels for predicting LNM of CSCC.17 Maximum diameter of tumor was obtained and defined as the longest diameter measured on both sagittal and transversal T2W images.
MRI acquisition and segmentation
The magnetic resonance (MR) images were obtained on a 3.0 T MR images (Signa Excite HD 3.0, GE Healthcare, Milwaukee, WI) or a 1.5 T MR imager (Achieva 1.5T, Philips Healthcare, Best, Netherlands), equipped with an 8-channel body array coil. Sagittal fat-suppressed T2W (T2-FS) sequence was utilized for the following analysis. The scanning parameters were as follows: 1) GE scanner: TR/TE = 4000/140 ms, field of view (FOV) = 280×280 mm2, matrix = 320×192, slice thickness = 4 mm, slice gap = 0.4 mm; and 2) Philips scanner: TR/TE = 3700/90 ms, FOV = 260×204 mm2, matrix = 260×162, slice thickness = 4 mm, slice gap = 0.4 mm.
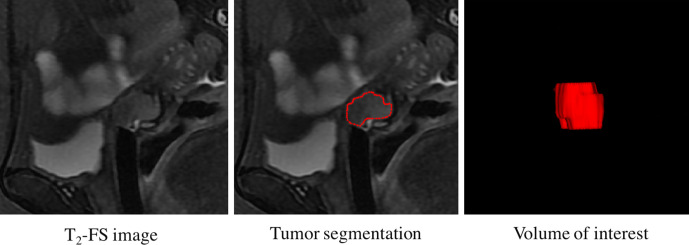
Sagittal T2-FS images in the format of Digital Imaging and Communications in Medicine were retrieved from the Picture Archiving and Communication System (PACS, Carestream Health) for tumor segmentation. We used the ITK-SNAP open-source software (V.3.6.0, www.itk-snap.org) for three-dimensional manual segmentation. The region of interest (ROI) was manually delineated on the sagittal T2-FS images along the border of each tumor slice (Figure 2). To evaluate the intra- and interobserver reproducibility of the radiomics analysis, we initially randomly chose 30 cases for segmentation. ROI segmentation was performed by two radiologists with 9 (reader 1) and 13 (reader 2) years’ experience in abdominal MR interpretation, respectively. To assess intraobserver reproducibility, reader one then repeated the same procedure two weeks later. Features with intraclass correlation coefficient (ICC) larger than 0.75 were considered as having good agreements. The ROIs of the remaining cases were outlined by reader 1.
Figure 2.
Tumor segmentation flowchart
Radiomics feature extraction
Both the deep learning features and hand-crafted features were extracted based on the T2-FS images. T2-FS images of each patient were normalized by minimum-maximum normalization in order to convert all pixels into a range of [1, 50] integral intensities.20,21 The hand-crafted feature extraction was implemented with an in-house Matlab code (MATLAB 2013a, MathWorks, Natick, MA). A total of 8715 features were extracted, including 14 first-order statistics features, 14 size and shape-based features, 63 texture features and 8624 wavelet features (Supplementary Material 1). The deep learning features were extracted from the final features layer of the model Convolutional Neural Networks-fast (CNN-F),22 pre-trained on ImageNet.23 Table 1 shows the CNN architectures. The single-channel slice containing the largest ROI was resized into 3-channel image, from which the bounding box of the ROI was cropped out as the input of the deep model. Ultimately, a total of 4096 deep learning features were extracted.
Table 1.
CNN architectures
| Arch. | conv1 | conv2 | conv3 | conv4 | conv5 | full6 | full7 | full8 |
|---|---|---|---|---|---|---|---|---|
| CNN-F | 64 × 11×11 stride = 4 pad = 0, LRN, ×2 pool |
256 × 5×5 stride = 1 pad = 2, LRN, ×2 pool |
256 × 3×3 stride = 1 pad = 1, |
256 × 3×3 stride = 1 pad = 1, |
256 × 3×3 stride = 1 pad = 1, ×2 pool |
4096 drop-out |
4096 drop-out |
1000 soft-max |
Note: The CNN model has five convolution layers and three fully connected layers. For the convolution layers, the first row descripts the number and the sizes of convolution filters as “num x size x size”; the second row descripts the convolution stride; the third row descripts the spatial padding; the last indicates the max-pooling down-sampling factor. LRN indicates local response normalization.24 For the full-connected layers, the first row descripts the dimensionality of the output. Full6 and full7 are regularized with dropout,25 while the last layer acts as a multi way soft-max classifier.
Radiomics feature selection
The model was built based on the deep learning and handcrafted features. Interclass correlation coefficients were applied to perform a reproducibility assessment of both features. The features with correlation coefficients > 0.75 were chosen for further analysis.
The following methods and processes were used for the selection of key handcrafted features and deep learning features, respectively. Firstly, prognostic value of each feature was test using the concordance statistics (C-statistics), a generalization of the area under the receiver operating characteristic (ROC) curve (AUC). Features with C-statistics above 0.560 were considered as predictive features. Secondly, the minimum redundancy maximum relevance (mRMR) algorithm was applied to further select the features that could best characterize the relation with the label while ensuring that the features were mutually dissimilar from each other. Thirdly, the lasso algorithm was used to select the most prognostic features in training cohort through 10-fold cross-validation. Feature selection was performed only on the training cohort in order to maintain the independence between the training cohort and validation cohort, and validation cohort was untouched.
Development of the radiomics model
After feature selection, a handcrafted- and deep-signature was developed with selected key features. Then, the radiomics scores of handcrafted- and deep-signature were calculated.
A multivariate logistic regression method was used to analyze the candidate variables (SCC-Ag and radiomics scores of the handcrafted signature and the deep signature) affected by LNM. Backward step-wise selection was used with a significance level of 0.05 for variable retention. Finally, radiomics model was developed.
Assessment predictive performance of the radiomics model
Calibration curves were performed to assess the calibration of the radiomics model, accompanied with the Hosmer-Lemeshow test. To quantify the discrimination performance of the radiomics model, AUCs were calculated in training and validation cohort. A nomogram was presented as an intuitive tool for clinicians to assess the risk of LNM in patients with early-stage CSCC. Decision curve analysis was also conducted to determine the clinical impact of the radiomics model. What’s more, ROC analyses were executed to compare the discriminatory efficacy of the radiomics model to those of the radiomics signature and the SCC-Ag alone.
Statistical analysis
Statistical analyses were implemented with SPSS 21 and R software (v.3.4.0, https://www.r-project.org). An independent samples t-test or Mann-Whitney U-test, where appropriate, were used to evaluate the differences in continuous variables (age and numbers of pregnancies, parturitions, and abortions) between the LNM (+) and LNM (-) groups in the training and validation cohorts, while the chi-squared test was used to assess the differences in categorical variables (menstrual status, FIGO stage, and SCC-Ag) between the LNM (+) and LNM (-) groups. The same statistical analysis was applied to compare the differences between the two cohorts regarding the LNM (+) and LNM (-) groups separately. A P-value below 0.05 was considered as statistically significant using two-sided testing.
Results
Clinical characteristics
Patient characteristics in the training and validation cohorts were given in Table 2. There were no significant differences in the clinical and histopathological characteristics (age, preoperative FIGO stage, menstrual status, and numbers of pregnancies, parturitions, and abortions, as well as SCC-Ag levels and LN status) between the two cohorts. LNM positivity was 23 and 28% in the training and validation cohorts, respectively. In the characteristic analysis, age and SCC-Ag levels were found significantly different between the LNM (+) and LNM (-) groups in both the training and validation cohorts.
Table 2.
Characteristics of Patients in the Training and Validation Cohorts
| Training cohort | Validation cohort | P# | |||||
|---|---|---|---|---|---|---|---|
| Characteristic | LNM(+) | LNM(-) | P | LNM(+) | LNM(-) | P | 0.449 |
| (23, 23%) | (77, 77%) | (25, 28%) | (65, 72%) | ||||
| Age (years, mean ± SD) | 46.17 ± 8.72 | 51.04 ± 9.57 | 0.031 | 46.20 ± 8.55 | 51.34 ± 9.15 | 0.017 | 0.995 |
| Menstrual status (N. %) | 0.148 | 0.055 | 0.544 | ||||
| Premenopausal | 17 (17%) | 44 (44%) | 19 (21%) | 30 (33%) | |||
| Postmenopausal | 6 (6%) | 33 (33%) | 6 (7%) | 35 (39%) | |||
| Pregnancy no. (mean ± SD) | 3.96 ± 1.92 | 4.36 ± 1.99 | 0.31 | 3.92 ± 1.68 | 3.83 ± 1.95 | 0.67 | 0.104 |
| Parturition no. (mean ± SD) | 2.96 ± 1.61 | 3.25 ± 1.65 | 0.448 | 2.56 ± 1.39 | 2.75 ± 1.32 | 0.65 | 0.063 |
| Abortion no. (mean ± SD) | 0.96 ± 1.30 | 1.12 ± 1.28 | 0.485 | 1.24 ± 1.20 | 1.12 ± 1.51 | 0.355 | 0.962 |
| FIGO Stage (N. %) | 0.394 | 0.299 | 0.727 | ||||
| ⅠB | 16 (16%) | 46 (46%) | 14 (16%) | 44 (49%) | |||
| ⅡA | 7 (7%) | 31 (31%) | 11 (12%) | 21 (23%) | |||
| SCC-Ag (N. %) | <0.001 | <0.001 | 0.987 | ||||
| ≤2.35 | 8 (8%) | 63 (63%) | 11 (12%) | 53 (59%) | |||
| >2.35 | 15 (15%) | 14 (14%) | 14 (16%) | 12 (13%) | |||
| Maximum diameter of tumor (mm, mean ± SD) | 33.80 ± 8.42 | 27.52 ± 10.42 | 0.01 | 33.94 ± 9.31 | 25.56 ± 11.11 | 0.001 | 0.492 |
| score_hc (mean ± SD) | 0.38 ± 0.19 | 0.19 ± 0.16 | <0.001 | 0.32 ± 0.18 | 0.18 ± 0.16 | 0.001 | 0.702 |
| score_dl (mean ± SD) | 0.33 ± 0.20 | 0.20 ± 0.13 | 0.006 | 0.27 ± 0.19 | 0.15 ± 0.11 | 0.004 | 0.053 |
LNM, lymph node metastasis; SCC-Ag, squamous cell carcinoma antigen; score_hc, radiomics score of handcrafted signature; score_dl, radiomics score of deep signature.
Note: P value was derived from the univariable association analyses between each of the clinical characteristics and lymph node status. P# represented the difference of each clinical characteristic between the training and validation cohorts.
Feature selection and radiomics signature building
In total, 8715 handcrafted features and 4096 deep learning features were extracted for each patient. Through the feature selection, three key handcrafted features and three key deep learning features were selected. A handcrafted signature and a deep signature were building by a logistic regression using the three key handcrafted features and three key deep learning features, respectively. The performance of handcrafted- and deep signature in training and validation cohort were showed in Table 3. Radiomics scores of handcrafted signature and deep signature can be calculated in training and validation cohort by the following formula:
Table 3.
performance of handcrafted signature, deep signature and clinical factor
| Training cohort | Validation cohort | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| cutoff | AUC | ACC | SEN | SPE | AUC | ACC | SEN | SPE | |
| SCC-Ag | 0.500 | 0.735 | 0.780 | 0.652 | 0.818 | 0.688 | 0.744 | 0.560 | 0.815 |
| Handcrafted signature | −0.855 | 0.794 | 0.790 | 0.739 | 0.805 | 0.725 | 0.700 | 0.440 | 0.800 |
| Deep signature | −1.246 | 0.735 | 0.720 | 0.739 | 0.714 | 0.718 | 0.767 | 0.560 | 0.846 |
SCC-Ag, squamous cell carcinoma antigen;AUC, area under curve; ACC, accuracy; SEN, sensitivity; SPE, specificity.
for radiomics score of handcrafted signature.
for radiomics score of deep signature.
In the training and validation cohort, handcrafted signature and deep signature showed statistically significant differences between LNM (+) and LNM (-) (Table 2).
Development and validation of the radiomics model
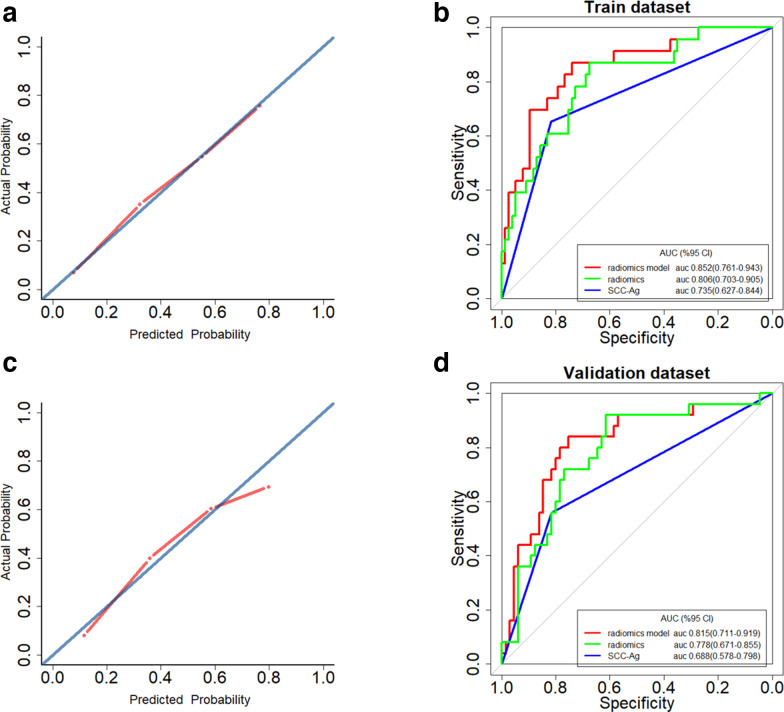
SCC-Ag, radiomics scores of the handcrafted signature and deep signature were independent predictors of LNM in patients with early-stage CSCC, indicated by multivariate logistic regression analysis (Table 4). A radiomics model incorporating these three predictors was generated in the training cohort. In the training cohort, good calibration could be depicted from the calibration curve of the radiomics model (Figure 3A), with a non-significant p-value of 0.753 yielded in the Hosmer–Lemeshow test. Besides, the radiomics model shown good discrimination performance in the training cohort (Figure 3B), with an AUC of 0.852 (95% CI: 0.761–0.943). In the validation cohort, the radiomics model showed similar calibration and discrimination performance (Figure 3C and D), with a non-significant Hosmer–Lemeshow test statistic (p = 0.055) and an AUC of 0.815 (95% CI: 0.711–0.919).
Table 4.
Multivariate logistic regression analysis for LNM in the training cohort
| Intercept and variable | Multivariate logistic regression | |
|---|---|---|
| OR (95% CI) | P-value | |
| intercept | 0.021 (0.004–0.077) | <0.001 |
| SCC-Ag (<2.35 µ g/L and ≥2.35 µ g/L) | 5.400 (1.709–18.144) | 0.005 |
| score_hc | 36.967 (1.207–1772.010) | 0.049 |
| score_dl | 42.940 (1.651–1331.957) | 0.025 |
OR, Odds Ratio; 95% CI, 95% confidence interval; SCC-Ag, squamous cell carcinoma antigen; LNM, lymph node metastasis;score_hc, radiomics score of handcrafted signature; score_dl, radiomics score of deep signature.
Figure 3.
Calibration curves of the radiomics model in the training cohort (A) and validation cohort (C). Calibration curves depicted the calibration of the radiomics model in terms of agreement between the predicted risk of lymph node metastases (LNM) and observed outcomes of LNM. The diagonal blue line represented a perfect prediction by an ideal model, and the dotted red line represented the predictive performance of the model. Closer fit of the red line to the diagonal blue line represented a better prediction. Receiver operating characteristic (ROC) curves to discriminate LNM (+) from LNM (-) for the radiomics model, and the radiomics signature and SCC-Ag alone in the training cohort (B) and the validation cohort (D). The radiomics model achieved stronger discriminatory ability than either the radiomics signature or SCC-Ag alone in discriminating LNM (+) from LNM (-) both in the training cohort and in the validation cohort.
In addition, as shown in Figure 3B and D, the radiomics model yielded the greatest AUCs, which indicated that the model achieved better predictive efficacy than either the radiomics signature or the SCC-Ag alone. Thermodynamics signature alone achieved AUCs of 0.806 (95%CI: 0.703–0.905) in the training cohort and 0.779 (95%CI: 0.671–0.885) in the validation cohort. The AUCs of the SCC-Ag alone in the training cohort and validation cohort were 0.735 (95%CI: 0.627–0.844) and 0.688 (95%CI: 0.578–0.798), respectively.
Clinical usefulness of the radiomics model
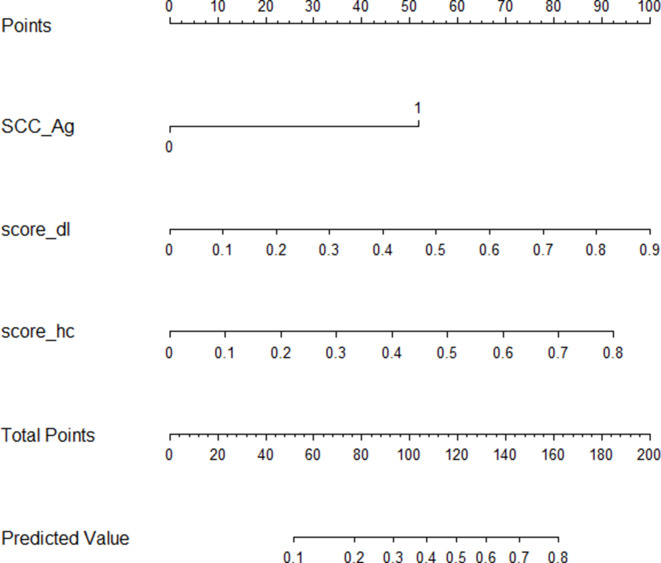
To provide a visualization tool for clinicians in assessing the risk of LNM in early-stage CSCC patients, the radiomics nomogram was developed by radiomics model (Figure 4). The decision curve analysis (DCA) for the radiomics model compared with that of SCC-Ag level analysis is presented in Figure 5.The DCA indicated that, across the majority of the range of risk thresholds, the radiomics model had the highest net benefit compared with the results of either the radiomics signature or the SCC-Ag alone and simple strategies such as the treat-all strategy or the treat-none scheme strategy.
Figure 4.
Radiomics model nomogram. The radiomics nomogram incorporating radiomics signature and SCC-Ag was generated in the training cohort. Abbreviations: SCC-Ag, squamous cell carcinoma antigen; score_hc, radiomics score of handcrafted signature; score_dl, radiomics score of deep signature.
Figure 5.
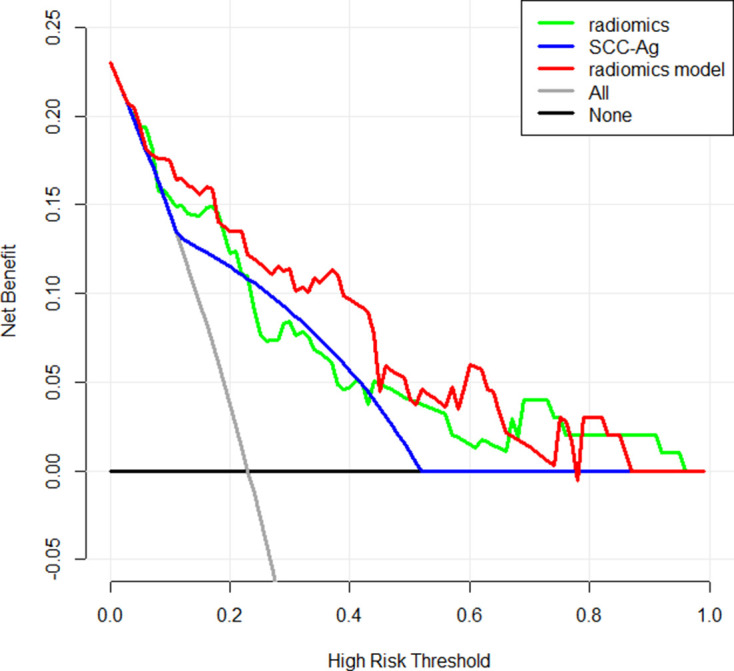
Decision curve analysis for the radiomics model compared with the radiomics signature and SCC-Ag alone. The x-axis represented the threshold probability and the y-axis measured the net benefit. The red line represented the radiomics model. The blue line represented the SCC-Ag. The green line represented the assumption that all patients had lymph node metastases (LNM). The black line represented the hypothesis that no patients had LNM. Across the majority of the range of risk thresholds, the radiomics model had the highest net benefit compared with that of either the radiomics signature or the SCC-Ag alone, and simple strategies such as the treat-all strategy or the treat-none strategy.
Discussion
In the present study, we developed and validated a nomogram for the preoperative diagnosis of LNM in patients with early-stage CSCC. The radiomics nomogram, which incorporated the radiomics signature based on T2-FS images and SCC-Ag levels, exhibited favorable discrimination of LN status with an AUC as high as 0.852 in the training cohort, and demonstrated similar discrimination in the validation cohort (AUC = 0.815). The comparable discrimination implied that the nomogram that incorporated the radiomics signature and SCC-Ag levels was robust in quantifying an individual’s risk of LNM.
LN status is one of the most important factors affecting clinical decision-making between either surgery or radical chemoradiotherapy. Many attempts have been made to improve the precision of LN status evaluation before operating. Several previous studies found that radiomics analysis of MRIs, with AUCs of 0.753 to 0.895, can enable superior prediction of LNM in patients with early-stage cervical cancer.14,15,26 A number of studies also have reported that elevated levels of SCC-Ag in early-stage cervical cancer were significantly associated with LNM, suggesting that SCC-Ag may be a useful marker for predicting LNM of CSCC.17,19,27 In our study, the AUCs of the SCC-Ag level alone in the training cohort and validation cohort were 0.735 and 0.688, respectively, which is similar to previous study (with AUC of 0.763).17SCC-Ag is present in the normal squamous epithelium and is produced in large amounts by malignant proliferative squamous epithelium. It is an important tumor marker for CSCC and widely used for the diagnosis of CSCC, therapeutic effect evaluation, and follow-up monitoring.28–31 Unlike previous studies, our study incorporated the radiomics signature and SCC-Ag levels to develop a radiomics model for preoperative prediction of LNM in patients with early-stage CSCC. The results suggested that the radiomics model achieved greater predictive efficacy than either the radiomics signature or the SCC-Ag levelalone.
Most radiomics features are sensitive to image acquisition and reconstruction parameters, which affects its reproducibility and validity.32 Because of the inconsistent scanning sequences and parameters, and the low number of cases in our study, we only took the role of radiomics features based on T2-FS into account in detecting LNM. However, the predictive efficacy is satisfactory in our study, which is close to the results of the study done by Wu et al (with AUCs of 0.895 and 0.847 in the training and validation cohorts, respectively).15 T2-weighted imaging (T2WI) is the basic gynecologic pelvic MR imaging sequence, and the pathological processes of the uterus are best identified on T2WI.33–35 Compared to other MR imaging sequences, we can accurately delineate the local extent of a tumor and depict extra uterine tumor spread on T2WI. Most of the studies based on MR used T2 or T2-FS weighted sequence as the basic sequence for radiomics studies, including the studies of cervical cancer.14,15,36 In Wu’s study, radiomics model based on T2WI was more sensitivity than other models in predicting LNM in cervical cancer.15A study on soft tissue sarcoma found that radiomics model based on T2-FS showed the best reproducibility among the model based on contrast-enhanced T1W fat-saturated sequence and the combined model.37 Of course, we believe that the discriminative performance of the radiomics model that combine the radiomics signature based on T2WI, diffusion-weighted imaging (DWI), and dynamic contrast-enhanced (DCE) imaging with SCC-Ag level analysis may be improved as well, which is worth future exploration.
Compared with two previous studies regarding using radiomics to detect LNM in patients with early-stage cervical cancer,14,15 our study has three main advantages. Firstly, our study is focused on squamous cell carcinoma, which is the most pathological subtype of cervical cancer and can be learned by biopsy. Secondly, our study verified that SCC-Ag was a useful marker for predicting LNM of CSCC. The radiomics model that incorporated radiomics and SCC-Ag levels achieved greater predictive efficacy than SCC-Ag level analysis alone. Thirdly, in our study, deep learning features were extracted and utilized with hand-crafted features for model building. The deep learning features and handcrafted features were combined through stacking algorithm, which build the final model with the predictions of base learner. Stacking algorithm can achieve a better result by a compromise between bias and variance.38
There are also several limitations of our study. First, it is a single-centered retrospective study and the sample size of patients recruited is relatively small. A multicentred study with a larger sample size will be able to confirm the discriminative efficacy of the radiomics model we propose in the present study. Second, we only extracted radiomics features from T2-FS images. Functional MR imaging has proved valuable for tissue characterization.39 For example, DWI can provide information about water mobility, tissue cellularity and the integrity of cellular membranes. DCE can be used to evaluate tissue perfusion and capillary permeability. Therefore, future studies are essential for exploring the discriminative performance of the radiomics model based on the incorporation of conventional and functional MRI and SCC-Ag analysis.
In conclusion, our study constructed a radiomics nomogram that incorporated both the radiomics signature based on a hand-crafted features model and a deep learning features model and SCC-Ag levels, and this nomogram can be conveniently applied in the preoperative prediction of LNM in patients with early-stage CSCC.
Footnotes
Conflicts of interest: This work was supported by the National Key Research and Development Plan of China [grant number2017YFC1309100], National Natural Scientific Foundation of China [grant numbers 81701782, 81771912], Science and Technology Planning Project of Guangdong Province [grant numbers2017B020227012, 2015B010131011] and the Medical Research Foundation of Guangdong Province [grant numberA2019476].
The authors Lifen Yan and Huasheng Yao contributed equally to the work.
The authors Jinglei Li, Zaiyi Liu and Changhong Liang contributed equally to the work.
Contributor Information
Lifen Yan, Email: sarahyanlifen@126.com.
Huasheng Yao, Email: 245150249@qq.com.
Ruichun Long, Email: longruichun0301@163.com.
Lei Wu, Email: wl858998458@163.com.
Haotian Xia, Email: 1563939760@qq.com.
Jinglei Li, Email: lijinglei80@126.com.
Zaiyi Liu, Email: zyliu@163.com.
Changhong Liang, Email: liangchanghong@gdph.org.cn.
REFERENCES
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68: 394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Landoni F, Maneo A, Colombo A, Placa F, Milani R, Perego P, et al. Randomised study of radical surgery versus radiotherapy for stage IB-IIA cervical cancer. Lancet 1997; 350: 535–40. doi: 10.1016/S0140-6736(97)02250-2 [DOI] [PubMed] [Google Scholar]
- 3.NCCN Clinical Practice Guidelines in oncology: cervical cancer.(2019.V5). Available from Available from: https://www.nccn.org/professionals/physician_gls/pdf/cervical.pdf.
- 4.Kim D-Y, Shim S-H, Kim S-O, Lee S-W, Park J-Y, Suh D-S, et al. Preoperative nomogram for the identification of lymph node metastasis in early cervical cancer. Br J Cancer 2014; 110: 34–41. doi: 10.1038/bjc.2013.718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Querleu D, Leblanc E, Cartron G, Narducci F, Ferron G, Martel P. Audit of preoperative and early complications of laparoscopic lymph node dissection in 1000 gynecologic cancer patients. Am J Obstet Gynecol 2006; 195: 1287–92. doi: 10.1016/j.ajog.2006.03.043 [DOI] [PubMed] [Google Scholar]
- 6.Baalbergen A, Veenstra Y, Stalpers L. Primary surgery versus primary radiotherapy with or without chemotherapy for early adenocarcinoma of the uterine cervix. Cochrane Database Syst Rev 2013;: CD006248.CD006248. doi: 10.1002/14651858.CD006248.pub3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi HJ, Ju W, Myung SK, Kim Y. Diagnostic performance of computer tomography, magnetic resonance imaging, and positron emission tomography or positron emission tomography/computer tomography for detection of metastatic lymph nodes in patients with cervical cancer: meta-analysis. Cancer Sci 2010; 101: 1471–9. doi: 10.1111/j.1349-7006.2010.01532.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selman TJ, Mann C, Zamora J, Appleyard T-L, Khan K. Diagnostic accuracy of tests for lymph node status in primary cervical cancer: a systematic review and meta-analysis. CMAJ 2008; 178: 855–62. doi: 10.1503/cmaj.071124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang Y-Q, Liang C-H, He L, Tian J, Liang C-S, Chen X, et al. Development and validation of a Radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol 2016; 34: 2157–64. doi: 10.1200/JCO.2015.65.9128 [DOI] [PubMed] [Google Scholar]
- 10.Tan X, Ma Z, Yan L, Ye W, Liu Z, Liang C. Radiomics nomogram outperforms size criteria in discriminating lymph node metastasis in resectable esophageal squamous cell carcinoma. Eur Radiol 2019; 29: 392–400. doi: 10.1007/s00330-018-5581-1 [DOI] [PubMed] [Google Scholar]
- 11.Wu S, Zheng J, Li Y, Yu H, Shi S, Xie W, et al. A Radiomics nomogram for the preoperative prediction of lymph node metastasis in bladder cancer. Clin Cancer Res 2017; 23: 6904–11. doi: 10.1158/1078-0432.CCR-17-1510 [DOI] [PubMed] [Google Scholar]
- 12.Yang X, Pan X, Liu H, Gao D, He J, Liang W, et al. A new approach to predict lymph node metastasis in solid lung adenocarcinoma: a radiomics nomogram. J Thorac Dis 2018; 10(Suppl 7): S807–19. doi: 10.21037/jtd.2018.03.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu L, Yang P, Liang W, Liu W, Wang W, Luo C, et al. A radiomics approach based on support vector machine using Mr images for preoperative lymph node status evaluation in intrahepatic cholangiocarcinoma. Theranostics 2019; 9: 5374–85. doi: 10.7150/thno.34149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kan Y, Dong D, Zhang Y, Jiang W, Zhao N, Han L, et al. Radiomic signature as a predictive factor for lymph node metastasis in early-stage cervical cancer. J Magn Reson Imaging 2019; 49: 304–10. doi: 10.1002/jmri.26209 [DOI] [PubMed] [Google Scholar]
- 15.Wu Q, Wang S, Chen X, Wang Y, Dong L, Liu Z, et al. Radiomics analysis of magnetic resonance imaging improves diagnostic performance of lymph node metastasis in patients with cervical cancer. Radiother Oncol 2019; 138: 141–8. doi: 10.1016/j.radonc.2019.04.035 [DOI] [PubMed] [Google Scholar]
- 16.Wu M, Tan H, Gao F, Hai J, Ning P, Chen J, et al. Predicting the grade of hepatocellular carcinoma based on non-contrast-enhanced MRI radiomics signature. Eur Radiol 2019; 29: 2802–11. doi: 10.1007/s00330-018-5787-2 [DOI] [PubMed] [Google Scholar]
- 17.Xu D, Wang D, Wang S, Tian Y, Long Z, Ren X. Correlation between squamous cell carcinoma antigen level and the clinicopathological features of early-stage cervical squamous cell carcinoma and the predictive value of squamous cell carcinoma antigen combined with computed tomography scan for lymph node metastasis. Int J Gynecol Cancer 2017; 27: 1935–42. doi: 10.1097/IGC.0000000000001112 [DOI] [PubMed] [Google Scholar]
- 18.Gadducci A, Tana R, Cosio S, Genazzani AR. The serum assay of tumour markers in the prognostic evaluation, treatment monitoring and follow-up of patients with cervical cancer: a review of the literature. Crit Rev Oncol Hematol 2008; 66: 10–20. doi: 10.1016/j.critrevonc.2007.09.002 [DOI] [PubMed] [Google Scholar]
- 19.Duk JM, Groenier KH, de Bruijn HW, Hollema H, ten Hoor KA, van der Zee AG, et al. Pretreatment serum squamous cell carcinoma antigen: a newly identified prognostic factor in early-stage cervical carcinoma. J Clin Oncol 1996; 14: 111–8. doi: 10.1200/JCO.1996.14.1.111 [DOI] [PubMed] [Google Scholar]
- 20.Liang C, Cheng Z, Huang Y, He L, Chen X, Ma Z, et al. An MRI-based Radiomics classifier for preoperative prediction of Ki-67 status in breast cancer. Acad Radiol 2018; 25: 1111–7. doi: 10.1016/j.acra.2018.01.006 [DOI] [PubMed] [Google Scholar]
- 21.Sasikala V, LakshmiPrabha V. A comparative study on the Swarm intel- ligence based feature selection approaches for fake and real fingerprint classification. 2015 International Conference on Soft-Computing and Net- works Security 2015;: 1–8. [Google Scholar]
- 22.Chatfield K, Simonyan K, Vedaldi A, AJapa Z. Return of the devil in the details: Delving deep into convolutional nets. 2014;.
- 23.Deng J, Dong W, Socher R, Li L-J LK. Fei-Fei L: Imagenet: a large-scale hierarchical image database. IEEE conference on computer vision and pattern recognition In: 2009;: ; 200920-25 June 2009; Miami, Florida, USA. Ieee. [Google Scholar]
- 24.Krizhevsky A, Sutskever I, Hinton G. ImageNet classification with deep Convolutional neural networks. NIPS Curran Associates Inc 2012; 25. [Google Scholar]
- 25.Hinton GE, Srivastava N, Krizhevsky A, Sutskever I. Salakhutdinov Rr: improving neural networks by preventing co-adaptation of feature detectors. Computer Science 2012; 3: 212–23. [Google Scholar]
- 26.Xiao M, Ma F, Li Y, Li Y, Li M, Zhang G, et al. Multiparametric MRI-based Radiomics nomogram for predicting lymph node metastasis in early-stage cervical cancer. J Magn Reson Imaging 2020; 52: 885–96. doi: 10.1002/jmri.27101 [DOI] [PubMed] [Google Scholar]
- 27.Zhou Z, Li W, Zhang F, Hu K. The value of squamous cell carcinoma antigen (SCCA) to determine the lymph nodal metastasis in cervical cancer: a meta-analysis and literature review. PLoS One 2017; 12: e0186165. doi: 10.1371/journal.pone.0186165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang S, Nam B-H, Park J-Y, Seo S-S, Ryu S-Y, Kim JW, et al. Risk assessment tool for distant recurrence after platinum-based concurrent chemoradiation in patients with locally advanced cervical cancer: a Korean gynecologic Oncology Group study. J Clin Oncol 2012; 30: 2369–74. doi: 10.1200/JCO.2011.37.5923 [DOI] [PubMed] [Google Scholar]
- 29.Reesink-Peters N, van der Velden J, Ten Hoor KA, Boezen HM, de Vries EGE, Schilthuis MS, et al. Preoperative serum squamous cell carcinoma antigen levels in clinical decision making for patients with early-stage cervical cancer. J Clin Oncol 2005; 23: 1455–62. doi: 10.1200/JCO.2005.02.123 [DOI] [PubMed] [Google Scholar]
- 30.Markovina S, Wang S, Henke LE, Luke CJ, Pak SC, DeWees T, et al. Serum squamous cell carcinoma antigen as an early indicator of response during therapy of cervical cancer. Br J Cancer 2018; 118: 72–8. doi: 10.1038/bjc.2017.390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Charakorn C, Thadanipon K, Chaijindaratana S, Rattanasiri S, Numthavaj P, Thakkinstian A. The association between serum squamous cell carcinoma antigen and recurrence and survival of patients with cervical squamous cell carcinoma: a systematic review and meta-analysis. Gynecol Oncol 2018; 150: 190–200. doi: 10.1016/j.ygyno.2018.03.056 [DOI] [PubMed] [Google Scholar]
- 32.Schick U, Lucia F, Dissaux G, Visvikis D, Badic B, Masson I, et al. MRI-derived radiomics: methodology and clinical applications in the field of pelvic oncology. Br J Radiol 2019; 92: 20190105. doi: 10.1259/bjr.20190105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Freeman SJ, Aly AM, Kataoka MY, Addley HC, Reinhold C, Sala E. The revised FIGO staging system for uterine malignancies: implications for MR imaging. Radiographics 2012; 32: 1805–27. doi: 10.1148/rg.326125519 [DOI] [PubMed] [Google Scholar]
- 34.Balleyguier C, Sala E, Da Cunha T, Bergman A, Brkljacic B, Danza F, et al. Staging of uterine cervical cancer with MRI: guidelines of the European Society of urogenital radiology. Eur Radiol 2011; 21: 1102–10. doi: 10.1007/s00330-010-1998-x [DOI] [PubMed] [Google Scholar]
- 35.Zhang W, Zhang J, Yang J, Xue H, Cao D, Huang H, et al. The role of magnetic resonance imaging in pretreatment evaluation of early-stage cervical cancer. Int J Gynecol Cancer 2014; 24: 1292–8. doi: 10.1097/IGC.0000000000000169 [DOI] [PubMed] [Google Scholar]
- 36.Wu Q, Shi D, Dou S, Shi L, Liu M, Dong L, et al. Radiomics analysis of multiparametric MRI Evaluates the pathological features of cervical squamous cell carcinoma. J Magn Reson Imaging 2019; 49: 1141–8. doi: 10.1002/jmri.26301 [DOI] [PubMed] [Google Scholar]
- 37.Peeken JC, Spraker MB, Knebel C, Dapper H, Pfeiffer D, Devecka M, et al. Tumor grading of soft tissue sarcomas using MRI-based radiomics. EBioMedicine 2019; 48: 332–40. doi: 10.1016/j.ebiom.2019.08.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mostafaei S, Abdollahi H, Kazempour Dehkordi S, Shiri I, Razzaghdoust A, Zoljalali Moghaddam SH, et al. Ct imaging markers to improve radiation toxicity prediction in prostate cancer radiotherapy by stacking regression algorithm. Radiol Med 2020; 125: 87–97. doi: 10.1007/s11547-019-01082-0 [DOI] [PubMed] [Google Scholar]
- 39.Jalaguier-Coudray A, Villard-Mahjoub R, Delouche A, Delarbre B, Lambaudie E, Houvenaeghel G, et al. Value of dynamic contrast-enhanced and diffusion-weighted MR imaging in the detection of pathologic complete response in cervical cancer after neoadjuvant therapy: a retrospective observational study. Radiology 2017; 284: 432–42. doi: 10.1148/radiol.2017161299 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.