Abstract
Purpose: The study of breast cancer nearly always involves patients close to menopause or older. Therefore, young patients are mostly underrepresented. Our aim in this study was to demonstrate biological differences in breast cancer of young people using as a model available cell lines derived from people with breast cancer younger than 35 years. Methods: Global miRNA expression was analyzed in breast cancer cells from young (HCC1500, HCC1937) and old patients (MCF-7, MDA-MB-231, HCC1806, and MDA-MB-468). In addition, it was compared with same type of results from patients. Results: We observed a differential profile for 155 miRNAs between young and older cell lines. We identified a set of 24 miRNA associated with aggressiveness that were regulating pluripotency of stem cell-related pathways. Combining the miRNA expression data from cell lines and breast cancer patients, 132 miRNAs were differently expressed between young and old samples, most of them previously found in cell lines. MiR-23a-downregulation was also associated with poor survival in young patients. Conclusions: Our results suggest that HCC1500 and HCC1937 cell lines could be suitable cellular models for breast cancer affecting young women. The miR-23a-downregulation could have a potential role as a poor prognosis biomarker in this age group.
Keywords: Breast cancer cell model, breast cancer young women, Cell lines, microRNAs
Introduction
Breast cancer (BC) is the most common malignancy in women worldwide. According to Globocan 2018, BC represents the 22.1% of all cancer diagnosed among women younger than 35 years with a mortality rate of 13.5%. Young age has been described as an independent risk factor for death in BC,1,2 and these patients tend to present more biologically aggressive tumors (basal and Her2-enriched subtypes), which have been associated with a poorer prognosis.3
MicroRNAs (miRNA) have demonstrated involvement in cell development, differentiation, proliferation, and apoptosis and have been linked to many diseases such as cancers.4,5 Previous studies from our group 6 detected a wide range of differentially regulated miRNAs in very young women with BC (<35 years) (BCVY) when compared to tumors diagnosed in older female counterparts or healthy breast tissue, suggesting that the aberrant expression of miRNA genes may be implicated in the aggressiveness of BC in this age group. Moreover, this different expression profile indicates that BCVY appears to be a different biological entity.
BC cell lines are widely used in many aspects of research and particularly as in vitro models in cancer research. Cell lines provide an important experimental tool in cancer research with the major advantage of an infinite supply of a relatively homogeneous cell population, capable of self-replication, which can be widely distributed to facilitate comparative studies.7
Although cell lines have been widely used as cellular models, the number of BC cell lines from young women is quite limited being the vast majority from older patients (>50 years old). The aim of the present study is to analyze miRNA expression differences between two BC cell lines from young women (HCC1500, HCC1937) and four from older counterparts (MCF-7, MDA-MB-231, HCC1806, and MDA-MB-468) in order to observe miRNA expression differences that support previous group results from Peña-Chilet et al.6 In addition, we wanted to examine the validity of HCC1500 and HCC1937 as suitable cellular model for future BCVY studies. To this end, we compared miRNA expression results from cell lines with results from patients previously obtained by Peña-Chilet et al.6
Methods
BC cell lines
The study was performed with three established breast cancer cell lines from the American Type Culture Collection (ATCC, Rockville, MD): HCC1500, HCC1937, and HCC1806. Cell lines were cultured in RPMI 1640 medium supplemented with 1% L-glutamine and 10% fetal bovine serum (GIBCO, New York, NY). The culture conditions were identical in all cell lines: 37 °C and 5% CO2.
RNA isolation
Total RNA from BC cell lines was isolated in passage 4 using High Pure RNA Isolation Kit from Roche following the manufacturer’s protocol. RNA concentration was measured using a NanoDrop ND 2000-UV-vis Spectrophotometer (Thermo Fisher Scientific Inc., Wilmington DE, USA).
RNA integrity determined by RNA integrity number (RIN) value was assessed with the Agilent 2100 Bioanalyzer using RNA 6000 Nano Assay (Agilent Technologies Inc., Santa Clara, CA).
miRNA microarray
Microarray expression profiling was performed using GeneChip® miRNA 2.0 Array (Affymetrix, Santa Clara, CA). We analyzed miRNA expression in two BC cell lines from women younger than 35 years (CLVY): HCC1500 and HCC1937 and the HCC1806 cell line from women older than 45 years (CLO). The chip contains a total of 15 644 probes, in 11 replicates, including 2 210 human miRNA. Hybridization and scanning were performed according to the Affymetrix standard protocol, using Affymetrix Expression Console software.
To compare the miRNA expression profile from CLVY with CLO, we analyzed miRNA expression in additional three CLO (MCF7, MDA-MB-231, and MDA-MB-468) from previous group published studies.8 Expression analysis was performed per triplicate for each BC cell line. Breast cancer cell lines characteristics are included in Table 1. After array normalization by RMA (Robust Multiarray Average) Kernel density estimator showed high right symmetry for all cell samples. Analogously, we appreciated a symmetry in the expression variability within array and among arrays in the box diagrams representation (Supplementary Figure 1A). The symmetry among arrays guarantee that the results from arrays could be comparable to each other.
Table 1.
Breast cancer cell lines characteristics.
| Cell line | Subtype | Receptor expression | Tumor type | Age |
|---|---|---|---|---|
| HCC 1500 | Luminal | EGP2 | IDC | 32 |
| HCC1937 | Basal | EGP2 | IDC | 24 |
| MDA-MB-231 | Basal | EGFR, TGF-β | Carcinoma | 51 |
| MDA-MB-468 | Basal | EGFR, TGF-α | Carcinoma | 51 |
| MCF7 | Luminal | ER, IGFBP | IDC | 69 |
| HCC1806 | Basal | EGP2 | Carcinoma | 60 |
Abbreviations: EGP2: epithelial glycoprotein 2; EGFR: epidermal growth factor receptor; TGF-β/α: transforming growth factor β / α; ER: estrogen receptor; IGFBP: insulin growth factor binding protein; IDC: invasive ductal carcinoma.
Array data processing and analysis
Data quality of *.CEL sample files was analyzed by affy package from R/Bioconductor. All data were normalized by the Robust Multichip Average (RMA) method. We selected 1154 human miRNAs. After filtering the data, in order to determine the differences in expression pattern between CLVY and from CLO, we carried out a differential expression t-test analysis using genefilter package from R Bioconductor. P-values were adjusted for multiple comparisons by Benjamini & Hochberg false discovery rate (FDR). miRNAs with a FDR P-value < .05 were considered statistically significant. Average linkage hierarchical clustering was performed to obtain clusters of data sets, using Heatmap.3 from R Bioconductor. Raw and normalized microarray data will be available from Gene Expression Omnibus (GSE121396).
Validation by RT-qPCR
Validation expression of selected miRNAs was carried out by quantitative real time-PCR (RT-qPCR) in a different set of cellular extracts using new biological triplicates of the six BC cell lines included in the array. We used the TaqMan microRNA Assays (Applied Biosystems by Life Technologies, Carlsbad, CA). Normalization was done with RNU43 snoRNA. The data were managed using the QuantStudio™ Desing & Analysis Software (ThermoFisher). Relative expression was calculated using the comparative Ct method and obtaining the fold-change value (ΔΔCt). The Wilcoxon Rank Sum test for non-normally distributed samples was performed to analyze differences between BC cell lines from different age groups (FDR P-value threshold of .05).
Pathway enrichment analysis and candidate gene searching
DIANA-miRPath pathway enrichment analysis was used to gain insight into global molecular networks and canonical pathways related to differentially expressed miRNAs.9 This software performs an enrichment analysis of multiple miRNA target genes comparing each set of miRNA targets to all known KEGG pathways. Pathways showing a FDR P-value < .05 were considered significantly enriched between classes under comparison.
Comparative miRNA expression study between cell lines and FFPE BC samples
We re-analyzed the hybridization chips from BC FFPE tumor samples previously published 6 (GSE48088) and cell lines in a combined study. There were 12 BC samples from women older than 45 years old and 21 from women younger than 35 years. The combined study was analyzed and normalized following the same steps as in the previously explained BC cell lines study. The Kernel density estimator showed higher similarity among cell lines. Patient samples presented major dispersion in intensity values. However, the mean Log Intensity values were around 1 for all samples (Supplementary Figure 1B). The symmetry among arrays guarantee that the results from cell and patients’ arrays could be comparable to each other and could be analyzed in the same study. We selected miRNAs that were significantly differently expressed between young and old women from both sample types, cell lines, and patients, with a FDR P-value < .05. Significant miRNAs were represented in unsupervised hierarchical clustering and results obtained were compared with previous results from the BC cell lines study.
Relapse free survival (RFS) and overall survival (OS) studies
To test the hypothesis of the potential association of miR-23a, miR-27a, and miR-23a-27a repression with RFS and/or OS, we analyzed the follow-up data of TCGA samples with miRNA expression data. Specifically, TCGA miRNA expression data includes 635 women older than 45-years-old and 23 BCVY samples. We next performed a univariate cox regression study to determine whether repression or overexpression of miR-23a were correlated with underlying clinical conditions in BCVY and BCO. Survival studies were performed using survival R package.
Results
miRNA expression profiling in BC cell lines
After initial pre-processing, the array maintained 2275 probes for human miRNAs. Among them, 1121 were pre-miRNA transcript probes and were removed from the study. Finally, we analyzed differential expression for 1154 miRNAs. Statistical analysis showed that 302 miRNAs were differentially expressed between CLVY (HCC1500 and HCC1937) and their older counterparts (MCF-7, MDA-MB-231, MDA-MB-468, and HCC1806) (P-value < .05) and after adjusting for FDR (FDR < 0.05), 155 miRNAs remained significant. Values obtained are included in Supplementary Table 1.
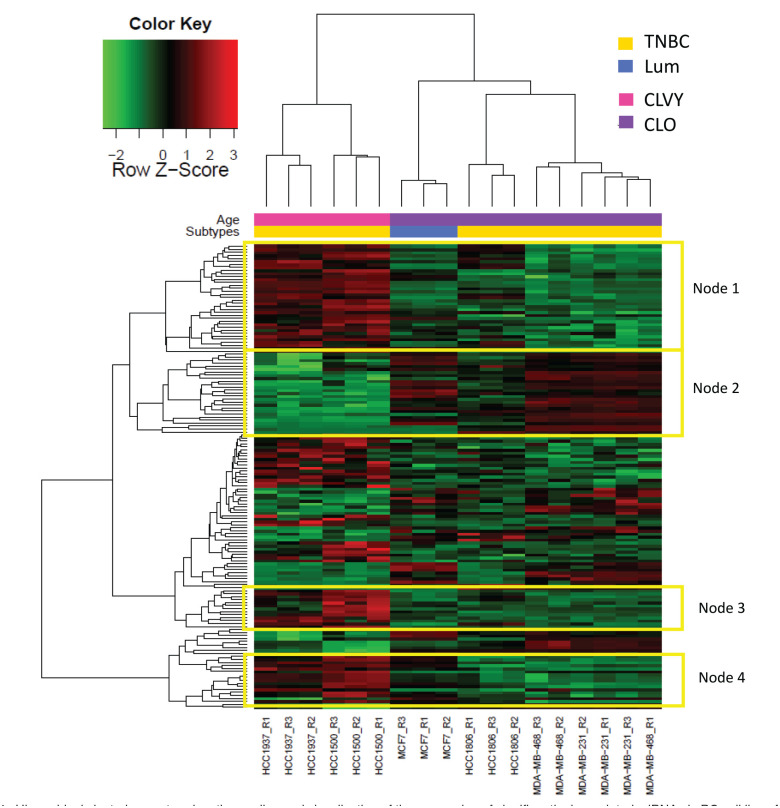
Average linkage hierarchical clustering with data from the 155 significant miRNAs was performed to obtain clusters of data set. Heatmap representation shows two major subgroups, separating CLVY from CLO (Figure 1). Biological triplicates for each BC cell line presented a similar miRNA profile and clustered together in the Heatmap representation, with the exception of one triplicate of MDA-MB-468 cell line. These results show a good reproducibility among biological triplicates. In addition, we found an overall higher miRNA expression in younger cell lines; specifically, 60% of significant miRNAs were overexpressed CLVY (red color in Heatmap representation) and the remaining 40% were repressed in this group.
Figure 1.
Hierarchical clustering centered on the median and visualization of the expression of significantly deregulated miRNAs in BC cell lines from young women. Heatmap representing the expression of 155 miRNAs that were significantly deregulated in breast cancer cell lines from young women. The expression is shown in different intensities, green below the median, red above and black unchanged with respect to the median. Cell lines are represented in pink for women younger than 35 years old and older women are indicated in purple. Molecular subtypes are indicated as triple negative (yellow) and luminal (blue).
TNBC: triple negative breast cancer; Lum: luminal; CLO: breast cancer cell lines from women older than 45 years old; CLVY: breast cancer cell lines from very young women (< 35 years old).
We highlighted 4 sub-nodes that presented homogeneous expression between age groups. Nodes 1, 3, and 4 showed a group of miRNA overexpressed in CLVY that were selected for further pathway enrichment analysis. Interestingly, we identified a group of 24 significant miRNAs that presented a similar expression pattern for cell line HCC1806 and CLVY and they are indicated in node 2. MiRNAs for each sub-node are included in Supplementary Table 2.
Pathway enrichment analysis for BC cell lines
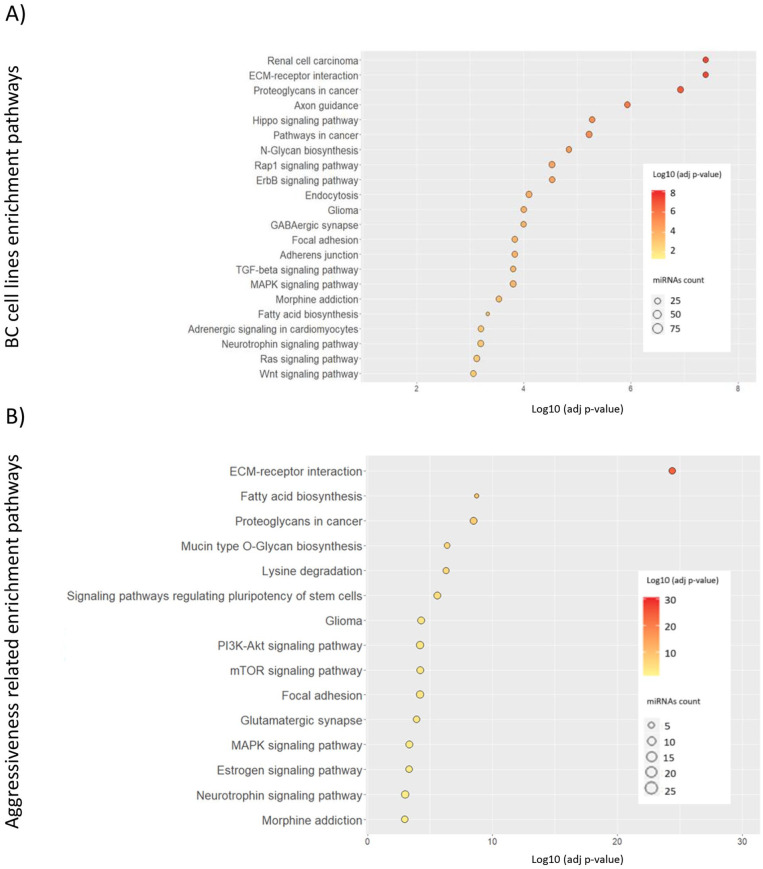
The enrichment study showed 22 pathways that were significantly deregulated (FDR P-value < .05) by the 155 significant miRNAs. We found pathways related to extracellular matrix receptor interaction or proteoglycans. Interestingly, we identified pathways related to the nervous system such as axon guidance, hippo signaling pathways, GABAergic synapse, and neurotrophic signaling. In addition, we identified pathways related to ErbB, Wnt, Ras, PI3 K-Akt or MAPK signaling. Focal adhesion and adherent junctions were also deregulated by significant miRNA found. The list of identified deregulated pathways is represented in Figure 2A.
Figure 2.
Pathways significantly deregulated by miRNA differently expressed in breast cancer cell lines from young women. Enrichment study was performed by DIANA-mirPath and pathways obtained presented a FDR-adjusted P-value < .001. (A) Pathways altered by the 155 significant miRNAs deregulated in breast cancer cell lines from young women and (B) pathways in which the 24 miRNA aggressiveness-related were involved. Dot color indicates a logarithmic transformation of FDR-adjusted P-values (orange for most significant pathways and yellow for lower P-values) and miRNA count indicates the number of miRNAs involved in the represented pathways.
The miRNA expression study showed a group of 24 miRNAs (included in node 2) that present similar expression between CLVY and HCC1806. We sought to analyze the pathways in which these miRNAs were involved. MiRNA enrichment analysis showed about 15 deregulated pathways with a FDR adjusted P-value < .001 (Figure 2B). The most significant pathway, in which 19 of the 24 miRNA studied participated, was related to extracellular matrix receptor interaction. In addition, cancer-related pathways such as cancer-related proteoglycans were observed. Interestingly, we identified relevant signaling pathways in cancer such as mTOR, PI3 K-Akt, ErbB, Ras, Wnt, and MAPK. Focal adhesion pathways were deregulated by the miRNA studied. Furthermore, 23 miRNAs were involved in signaling pathways regulating pluripotency of stem cells.
When studying miRNAs from the selected sub-nodes indicated in Figure 1, KEGG pathway enrichment analysis revealed several pathways overrepresented with a FDR P-value < .05. Most of the pathways observed were deregulated by miRNA from different sub-nodes. However, we identified particular cluster altered pathways. MiRNAs from node 1 participated in cancer metabolism-related pathways such as central carbon or choline. Signaling pathways related with calcium, insulin, MAPK, oxytocin, TFG-beta were also deregulated by miRNA from node 1. MiRNAs from node 3 participated in cell adhesion, N-glycan biosynthesis, and nervous system–related pathways such as hippo signaling. Node 4 showed biological proliferation- or differentiation-related pathways important in cancer development or progression such as AMPK, mTOR, PI3 K-Akt, estrogen, and Wnt signaling pathways. More information about node miRNA enrichment and the FDR P-values associated with each pathway are shown in Supplementary Table 3.
RT-qPCR validation in BC cell lines
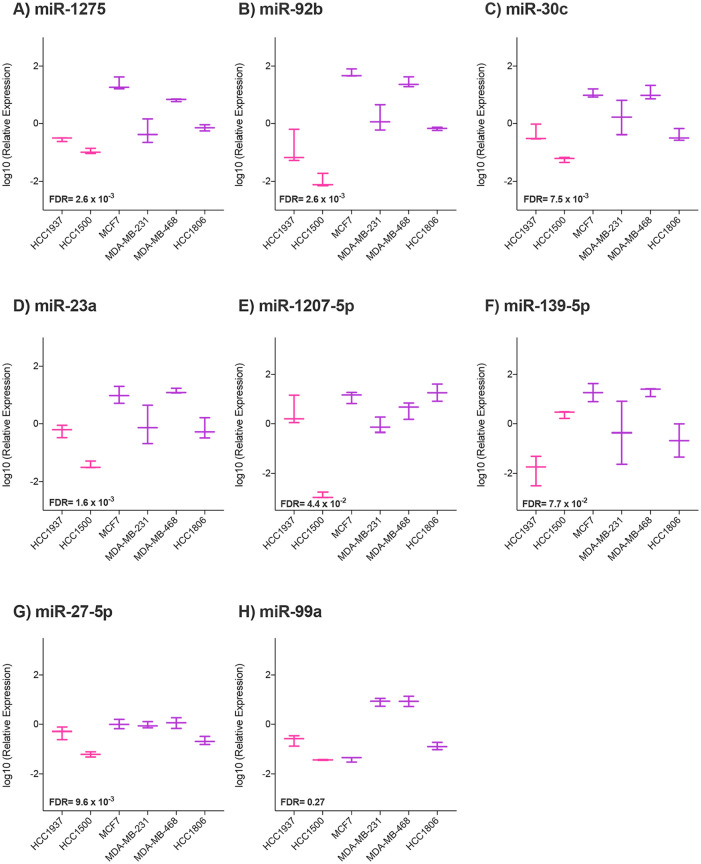
We evaluated the robustness of the expression for eight significant miRNAs obtained in BC cell lines by quantitative real-time PCR (RT-qPCR). We analyzed five repressed miRNAs (hsa-miR-23a, hsa-miR-139-5p, hsa-miR-30c, hsa-miR-27a-5p, and hsa-miR-99a) and 3 overexpressed (hsa-miR-1207-5p, hsa-miR-1275 and hsa-miR-92b). The validation step was performed in a different set of cellular extracts using new biological triplicates of BC cell lines included in the array.
After miRNA amplification by RT-qPCR, the mean Ct for the replicates, the standard deviation and the ΔΔCt was calculated. We remove outlier values and performed the normality Shapiro-Wilk test. All samples tested presented a P-value < .05, so there was evidence that the data were not normally distributed. Afterwards we performed the Wilcoxon rank sum test for non-parametric samples to compare relative expression values for each miRNA between CLVY and CLO.
Results showed significant differences between cell lines from young and older women for 5 of 8 selected miRNAs: miR-1275 (FDR P-value = 2.6 x 10-3), miR-92b (FDR P-value = 2.6 x 10-3), miR-30c (FDR P-value = 7.5 x 10-3), miR-23a (FDR P-value = 1.6 x 10-2), miR-1207-5p (FDR P-value = 4.4 x 10-2), and miR-27a-5p presents a FDR P-value near to the significance (FDR P-value = 9.6 x 10-2). MiR-23a, miR-30c, and miR-27a-5p expression values agreed with downregulation observed in younger cell lines (Figure 3), while the remaining significant miRNAs showed contrary results between RT-qPCR validation and previous array expression data.
Figure 3.
RT-qPCR miRNA expression validation results for breast cancer cell lines. Boxplots represent the sample distribution with the mean for each breast cancer cell line from old and young women. Expression was quantified using the RT-qPCR and calculated by ΔΔCt method. Y axis represents logarithmic transformation of the relative expression. Differences by miRNA between breast cancer cell lines from young (represented in pink color) and older (represented in purple) women were analyzed by Wilcoxon rank sum test: (A) miR-1275, (B) miR-92b, (C) miR-30c, (D) miR-23a, (E) miR-1207-5p, (F) miR-139-5p, (G) miR-27a- 5p, and (H) miR-99a.
Comparison of miRNA expression between BC cell line and FFPE BC samples
In order to evaluate the reproducibility of the miRNA profile observed in cell lines, we performed a comparative study between miRNA expression in BC cell lines and BC FFPE patient tumor samples obtained in previous group studies.6
The differential expression between young and old samples (cell lines and patients) of 1154 human miRNA included in the array were analyzed by t-test. Results showed 305 miRNAs differently expressed in both BC samples from BCVY and CLVY, compared with BC in older patients (BCO) and CLO, with P-value < .05, and 132 of them were adjusted by FDR < 0.05. The 54% of significant miRNAs were upregulated in younger samples and the remaining 46% were repressed in them. The list of significant miRNAs is summarized in Supplementary Table 4.
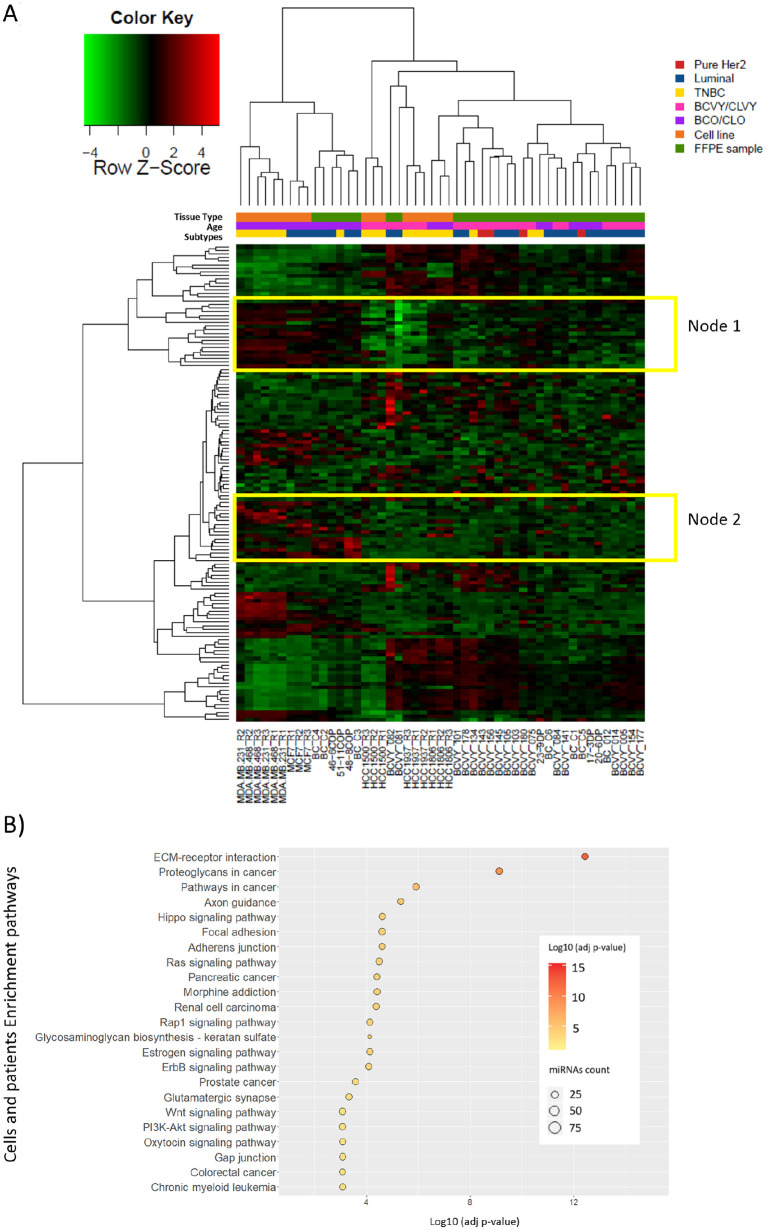
We performed average linkage hierarchical clustering with data from the 132 significant miRNAs to obtain clusters of data set (Figure 4A). With the exception of cell line HCC1806 and a few samples from BCO patients, heatmap representation shows two major subgroups, separating BCVY and CLVY samples from the older counterparts. In agreement with previous observations in BC cell lines, HCC1806 presents a miRNA profile more similar to CLVY and BCVY. Furthermore, in this combined study the expression of cell line HCC1806 is similar to younger samples for all miRNAs studied. Two sub-nodes were selected for further enrichment pathway analysis and miRNAs included are summarized in Supplementary Table 5.
Figure 4.
Hierarchical clustering centered on the median representation and enrichment pathway analysis of the expression of significantly deregulated miRNAs between young and old samples (patients and cell lines). (A) Heatmap representing the expression of 132 miRNAs that were significantly deregulated between young and old samples from BC patient tissue and cell lines. Expression is shown in different intensities, green below the median, red above and black unchanged with respect to the median. Young samples are represented in pink and older are indicated in purple. Molecular subtypes are indicated as Pure Her2 (red), triple negative (yellow) and luminal (blue). Sample precedence is indicated as orange for breast cancer cell lines and green for FFPE patient tissue samples. (B) Pathways significantly deregulated by the 132 miRNA differently expressed in the combined study with breast cancer cell lines and FFPE samples. Enrichment study was performed by DIANA-mirPath and pathways obtained presented a FDR-adjusted P-value < .001. Dot color indicates a logarithmic transformation of FDR-adjusted P-values (orange for most significant pathways and yellow for lower P-values) and miRNA count indicates the number of miRNAs involved in the represented pathways.
TNBC: triple negative breast cancer; Lum: luminal; BCO: breast cancer tumors from women older than 35 years old; BCVY: breast cancer tumors from women younger than 35 years old; CLO: BC cell lines from older patients; CLVY: BC cell lines from very young patients; FFPE: Formalin-fixed, paraffin-embedded.
Next, we studied whether miRNA expression differences observed between BC age groups were related to molecular subtypes. Generalized linear model (GLM) analysis showed that miRNA expression differences that distinguish young from old samples were specific to age and not related to any molecular subtype (P-value = .66). Supporting that, no molecular subtype clusters were observed in Heatmap representation.
Pathway enrichment analysis for BC cell lines and patients
Enrichment pathway analysis with the significant miRNAs that were differently expressed in BC cell lines and patients showed 23 deregulated pathways, most of them previously observed in the miRNA expression in BC cell lines study (Figure 4B). Again, the most significant pathway obtained was related to extracellular matrix (ECM) receptor interaction with an FDR-adjusted P-value = 3.6 x 10-13 in which 74 of the 132 miRNAs studied were involved. Among others, Ras, Rap1, ErbB, PI3 K-Akt, mTOR, and Wnt signaling pathways were significantly deregulated in younger patients. We could also identify nervous system–related pathways as previously observed in cell lines; axon guidance, hippo signaling pathways, and glutamatergic synapse were some of the observed pathways. Pathways related to adherent junctions and focal adhesion were significantly altered by deregulated miRNAs.
We analyzed the enrichment pathways by the two selected sub-nodes. Node 1 showed multiple pathways altered and involved in different functions. We identified deregulation in AMPK, ErbB, mTOR, and prolactin signaling pathways, among others. Most of them were previously observed in the global enrichment study for BC patients and cell lines. Interestingly, node 2 miRNAs participated in circadian rhythm and cardiomyopathies. The list of deregulated pathways by sub-nodes is summarized in Supplementary Table 6 .
Common significant miRNA from BC cell lines and tumor patient samples
We compared results obtained from our BC cell lines study, BC FFPE patients study 6 and combined study with patients and cell lines. Significant miRNA obtained from the three different analysis are represented in Venn diagram (Supplementary Figure 2). Results showed 10 miRNAs that were significantly deregulated in younger samples in the three studies (miR-1207-5p, miR-1275, miR-1973, miR-220b, miR-23a, miR-27b*, miR-3141, miR-3162, miR-4270 and miR-4317). MiR-23a downregulation was validated by RT-qPCR in both young cell lines and BC patient samples in Peña-Chilet.6 This result supports the miR-23a role in BCVY women and its potential biomarker function in this age group.
Clinical importance of miR-23a expression as an independent prognostic factor for relapse-free survival and overall survival in BCVY
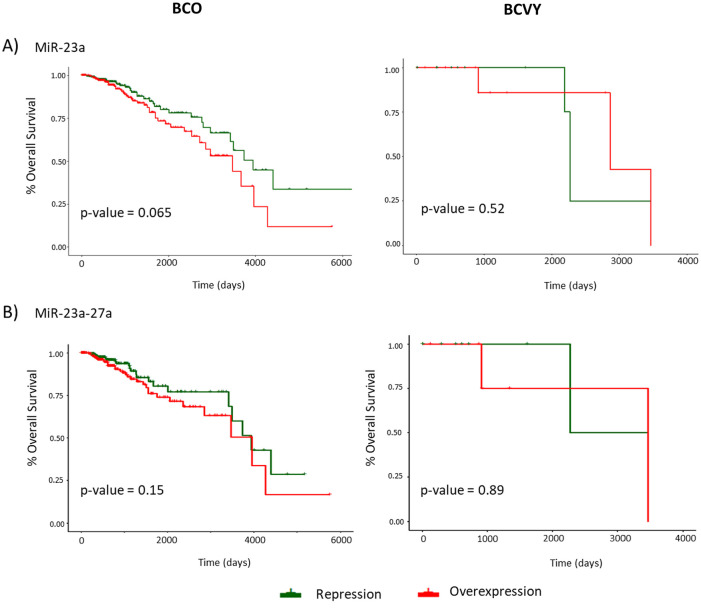
OS study showed no significant correlation between miR-23a and/or miR-27a expression status and relapse, either in BCO or in BCVY women. However, we observed poor survival in BCVY patients with miR-23a repression compared with BCO patients who presented worse prognosis when miR-23a was overexpressed (Figure 5A). MiR-27a did not show the same association with downregulation and survival as previously observed for miR-23a (results not shown). Nevertheless, when both miRNAs (miR-27a and miR23a) were repressed, we observed also poorer survival in BCVY in comparison with BCO, but results were not significant (Figure 5B).
Figure 5.
Representation of relapse-free survival and overall survival curves for miR-23a and miR-23a-27a cluster expression analyzed in breast cancer samples from TCGA. The first column curves represent OS in BCO patients and the second column in BCVY for miR-23A (A) and miR-23a-27a cluster (B) expression. X axis represents follow-up time in days and Y axis indicates the percentage of overall survival. Green curves represent miRNA repression and the red line represents overexpression. P-values were obtained by univariate cox analysis.
Discussion
MiRNAs, a novel class of non-coding RNAs, have emerged as promising diagnostic and therapeutic tools for human cancers because of their critical role in various biological processes during tumor development, such as cell proliferation, differentiation, apoptosis and migration, and subsequent tumorigenesis.10,11 The differential miRNA expression study between the two CLVY (HCC1500 and HCC1937) and CLO (HCC1806, MCF-7, MDA-MB-231 and MDA-MB-468) revealed a group of 155 miRNAs that were significantly differently expressed between cell lines from different age groups. The corresponding cell line replicates cluster together, indicating good reproducibility of miRNA expression profile among biological replicates.
In addition to good reproducibility among replicates, we observed that CLVY cell lines HCC1500 and HCC1937 presented a high similar miRNA expression in comparison with CLO cell lines: MCF7, HCC1806, MDA-MB-231, and MDA-MB-468, as we can appreciated in the heatmap representation where the hierarchical clustering divide clearly CLO from CLVY cell lines. Here, we provide a miRNA profile that distinguish CLVY from CLO which could be highly valuable to understand the mechanisms underlying BC affecting very young women. The TNBC from CLO (MDA-MB-231, MDA-MB-468, and HCC1806) showed a more similar clustering than MCF7 cell line, that was from the luminal molecular subtype. In addition, MCF7 cell line cluster nearer the luminal CLVY, HCC1500, indicating a more similarity in the miRNA expression among molecular subtypes. Previous studies find a similarity among the miRNA expression between molecular subtypes in BC. However, our statistical analysis did not find differences related with the molecular BC subtypes.
We identified a set of 24 miRNAs that presented a similar expression pattern between HCC1806 and CLVY. HCC1806 was established from the tumor of a 60-year-old patient with TNBC 12 with a basaloid features,12-17 that have been related with a worse prognosis than patients with triple-negative tumors.17-19 The enrichment pathway study showed that most of the pathways deregulated by these miRNAs were previously obtained in the BC cell lines study. However, a remarkable deregulation was observed for pluripotency of stem cell–related pathways. BC affecting young women is characterized by more aggressive, more proliferative and larger tumors than in older patients. We hypothesized that the group of 24 miRNAs with similar expression profile between HCC1806 and CLVY might be related to aggressiveness that is a similar feature in BCVY and the previous description of the HCC1806 cell line. However, further studies are needed to address in more depth the potential role of these miRNAs in cancer progression and more aggressive tumor features.
The combined study analyzing miRNA expression data from BC cell lines and patients showed good correlation between young women samples from both cell lines and tumors, and a different miRNA profile between these and samples from older women (cells and patients). Specifically, we identified 132 miRNAs significantly differently expressed between young and older samples and 47 of them were previously observed in the BC cell line study.
Multiple pathways obtained in the enrichment analysis was observed both in the BC cell line study and in the combined study with cell lines and tumor samples. Most of them were related to signaling pathways important in cancer such as mTOR, PI3 K-Akt, ErbB, Ras, Wnt, and MAPK. In addition, we identified deregulated pathways related to the nervous system that have been previously observed in other works on BCVY.20 In agreement with this, previous articles have highlighted the importance of neural elements in the cancer microenvironment which promote cell growth. However, the mechanisms mediating neuronal influences on cancer growth and progression are likely incompletely understood.21
Few other studies report the implication of microRNAs in the etiology of BC in young women,6,20,22 and less information is found in ethnic backgrounds other than Caucasic. Bensen et al,23 2018 reported a SNP association signal on chromosome 17p25.3 located in the primary transcript of miR-3065 in African-American BC cohort. miR-3065 may potentially regulate the expression of the gene BAIAP2. Curiously, BAIAP2 presented a highly differential expression pattern in several BC cell lines, among which there is HCC1937, used in this study. It is well known that BC in African-American women is more aggressive with major proportion of triple negative subtypes and diagnosed at younger ages. Unfortunately, we are not able to detect miR-3065 in our cohort of BCVY, possibly due to the use of miR- expression platform and singularities of our study design.
Cell lines have been used as models for BC research. However, there are few BC cell lines available from young women (<35 years old) and some of them have been eliminated from BC studies due to their uncertain origin, such as cell line MDA-MB-435 (31 years old) which was described as a melanoma cell line 14 years ago and the HBL100 cell line (27 years old) that was discontinued because of the presence of an Y chromosome. The reproducibility and similarity between the tumor sample study and the present BC cell line analysis supports the use of HCC1500 and HCC1937 as a suitable model for the study of BC affecting very young women.
Among the 8 miRNAs included in the validation study in cell lines, we were able to validate the miR-23a, miR-30c and miR-27a-5p was near the significance. Expression results of these miRNAs agree with downregulation in younger cell lines obtained in the miRNA array. The miR-30c has been described as a tumor suppressive miRNA in BC, inhibiting cell migration, invasion and epithelial mesenchymal transition. Here, we observed a significant downregulation of miR-30c in CLVY that could be related with the poor outcome observed in BCVY patients.24
At the same time, the miR-23a downregulation was validated in our BC cell lines study as well as in the patients’ study from Peña-Chilet.6 In addition, miR-27a-5p was also downregulated in CLVY samples and these last two miRNAs are encoded by the same intergenic cluster miR-23a~27a~24-2, located in chromosome 19p13.12.25 Indeed, the mature sequence of miR-23a and miR-27a differ just by one nucleotide. These miRNAs present different expression in several cancers and has diverse effects.26 Analysis of BC tissue-based miRNA panel shows that miR-23a possibly functions as an oncogenic governor and promotes BC progression.27,28 However, miR-23a acts as a negative regulator of oncogene in different cancers such as neprhoblastomas.29 Otherwise, Li et al30 reported that miR-27a could function as a tumor suppressor in BC, suppressing cancer proliferation, migration and tumorigenesis. The downregulation of miR-23a, miR-27a and miR-24-2 was observed in the cell lines miRNA expression study and miR-23a and miR-27a were validated by RT-qPCR. Our results suggest that CLVY present a downregulation of the cluster miR-23a~27a~24-2 which could be a signature in BCVY. However, these results were validated only in miR-23a and miR-27a.
Although we could not obtain significant results, we identified an association between miR-23a-dowregulation and poor survival for young patients. In addition, we also observed poor survival in BCVY samples that presented downregulation of miR-23a and miR-27a whereas older patients showed an inverse association. These results could be indicating a downregulation cluster signature characteristic of BCVY. The sample size for BCVY samples in TCGA is quite limited so further studies with a larger cohort should be addressed in order to examine the downregulation of this miRNA cluster and its association with poor prognostic in BC affecting young women.
Altogether, our results demonstrate the suitability of HCC1500 and HCC1937 as cellular models for future studies of BC affecting the very young. In addition, miR-23a, miR-30c, and miR-27a downregulation was detected and validated in BC cell lines. Although no significant results were reached, we observed an association between miR-23a and miR-23-miR-27a cluster-downregulation and poor survival rates in BCVY. We encourage future studies of cluster miR-23a-miR-27a in larger cohorts of BCVY in order to validate its potential role as a poor prognosis biomarker in this age group.
Supplemental Material
Supplemental material, sj-docx-3-bcb-10.1177_1178223420977845 for miRNA Expression Analysis: Cell Lines HCC1500 and HCC1937 as Models for Breast Cancer in Young Women and the miR-23a as a Poor Prognostic Biomarker by Sara S Oltra, Maria Peña-Chilet, Maria T Martinez, Eduardo Tormo, Juan Miguel Cejalvo, Joan Climent, Pilar Eroles, Ana Lluch and Gloria Ribas in Breast Cancer: Basic and Clinical Research
Supplemental material, sj-docx-4-bcb-10.1177_1178223420977845 for miRNA Expression Analysis: Cell Lines HCC1500 and HCC1937 as Models for Breast Cancer in Young Women and the miR-23a as a Poor Prognostic Biomarker by Sara S Oltra, Maria Peña-Chilet, Maria T Martinez, Eduardo Tormo, Juan Miguel Cejalvo, Joan Climent, Pilar Eroles, Ana Lluch and Gloria Ribas in Breast Cancer: Basic and Clinical Research
Supplemental material, sj-docx-5-bcb-10.1177_1178223420977845 for miRNA Expression Analysis: Cell Lines HCC1500 and HCC1937 as Models for Breast Cancer in Young Women and the miR-23a as a Poor Prognostic Biomarker by Sara S Oltra, Maria Peña-Chilet, Maria T Martinez, Eduardo Tormo, Juan Miguel Cejalvo, Joan Climent, Pilar Eroles, Ana Lluch and Gloria Ribas in Breast Cancer: Basic and Clinical Research
Supplemental material, sj-docx-6-bcb-10.1177_1178223420977845 for miRNA Expression Analysis: Cell Lines HCC1500 and HCC1937 as Models for Breast Cancer in Young Women and the miR-23a as a Poor Prognostic Biomarker by Sara S Oltra, Maria Peña-Chilet, Maria T Martinez, Eduardo Tormo, Juan Miguel Cejalvo, Joan Climent, Pilar Eroles, Ana Lluch and Gloria Ribas in Breast Cancer: Basic and Clinical Research
Supplemental material, sj-docx-7-bcb-10.1177_1178223420977845 for miRNA Expression Analysis: Cell Lines HCC1500 and HCC1937 as Models for Breast Cancer in Young Women and the miR-23a as a Poor Prognostic Biomarker by Sara S Oltra, Maria Peña-Chilet, Maria T Martinez, Eduardo Tormo, Juan Miguel Cejalvo, Joan Climent, Pilar Eroles, Ana Lluch and Gloria Ribas in Breast Cancer: Basic and Clinical Research
Supplemental material, sj-docx-8-bcb-10.1177_1178223420977845 for miRNA Expression Analysis: Cell Lines HCC1500 and HCC1937 as Models for Breast Cancer in Young Women and the miR-23a as a Poor Prognostic Biomarker by Sara S Oltra, Maria Peña-Chilet, Maria T Martinez, Eduardo Tormo, Juan Miguel Cejalvo, Joan Climent, Pilar Eroles, Ana Lluch and Gloria Ribas in Breast Cancer: Basic and Clinical Research
Supplemental material, sj-pdf-1-bcb-10.1177_1178223420977845 for miRNA Expression Analysis: Cell Lines HCC1500 and HCC1937 as Models for Breast Cancer in Young Women and the miR-23a as a Poor Prognostic Biomarker by Sara S Oltra, Maria Peña-Chilet, Maria T Martinez, Eduardo Tormo, Juan Miguel Cejalvo, Joan Climent, Pilar Eroles, Ana Lluch and Gloria Ribas in Breast Cancer: Basic and Clinical Research
Supplemental material, sj-pdf-2-bcb-10.1177_1178223420977845 for miRNA Expression Analysis: Cell Lines HCC1500 and HCC1937 as Models for Breast Cancer in Young Women and the miR-23a as a Poor Prognostic Biomarker by Sara S Oltra, Maria Peña-Chilet, Maria T Martinez, Eduardo Tormo, Juan Miguel Cejalvo, Joan Climent, Pilar Eroles, Ana Lluch and Gloria Ribas in Breast Cancer: Basic and Clinical Research
Acknowledgments
We would like to thank the expert personnel in the Multigenic Analysis Laboratory at the Central Biomedical Research Unit (UCIM) in the University of Valencia and the thorough English revision by an expert colleague.
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by grants from The Ministry of Economy and Competitiveness and the Carlos III Health Institute (PI13/00606) and FEDER. SSO is funded on a FPU pre-doctoral fellowship (FPU13/04976) from MECD, (Spanish Government); GR is funded on a Miguel Servet II contract (CPII14-00013) from the Carlos III Health Institute; MPC is funded by the private patients Foundation LeCado. CIBERONC (CB16/12/00481-CB16/12/00473) is an initiative of the Carlos III Health Institute.
Declaration of Conflicting Interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Availability of Data and Materials: The dataset generated during the current study are available in the Gene Expression Omnibus repository (GSE121396), website: https://www.ncbi.nlm.nih.gov/gds.
Author Contributions: G.R and A.L conceived the study. G.R and M.P supervised the study. G.R and S.O designed all aspects of the research. S.O collected the data and performed computational analysis. P.E, E.T, and J.C collect part of expression data from cell lines. M.P and M.M contributed in the comparative study. M.M and J.M.C contributed in the data acquisition. S.O and G.R wrote the manuscript. All authors revised, read, and approved the final manuscript.
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Livi L, Meattini I, Saieva C, et al. The impact of young age on breast cancer outcome. Euro J Surg Oncol. 2010;36:639-645. [PubMed] [Google Scholar]
- 2. Han W, Kang SY, Korean Breast Cancer Society. Relationship between age at diagnosis and outcome of premenopausal breast cancer: age less than 35 years is a reasonable cut-off for defining young age-onset breast cancer. Breast Cancer Res Treat. 2010;119:193-200. [DOI] [PubMed] [Google Scholar]
- 3. Anders CK, Johnson R, Litton J, Phillips M, Bleyer A. Breast cancer before age 40 years. Semin Oncol. 2009;36:237-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6-7. [DOI] [PubMed] [Google Scholar]
- 5. Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pena-Chilet M, Martinez MT, Perez-Fidalgo JA, et al. MicroRNA profile in very young women with breast cancer. BMC Cancer. 2014;14:529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pandrangi SL, Raju Bagadi SA, Sinha NK, et al. Establishment and characterization of two primary breast cancer cell lines from young Indian breast cancer patients: mutation analysis. Cancer Cell Int. 2014;14:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tormo E, Pineda B, Serna E, et al. MicroRNA profile in response to doxorubicin treatment in breast cancer. J Cell Biochem. 2015;116:2061-2073. [DOI] [PubMed] [Google Scholar]
- 9. Vlachos IS, Zagganas K, Paraskevopoulou MD, et al. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acid Res. 2015;43:W460-W466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hu C, Lv L, Peng J, et al. MicroRNA-375 suppresses esophageal cancer cell growth and invasion by repressing metadherin expression. Oncol Lett. 2017;13:4769-4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang Y, Xu D, Pan J, et al. Dynamic monitoring of circulating microRNAs as a predictive biomarker for the diagnosis and recurrence of papillary thyroid carcinoma. Oncol Lett. 2017;13:4252-4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gazdar AF, Kurvari V, Virmani A, et al. Characterization of paired tumor and non-tumor cell lines established from patients with breast cancer. Int J Cancer. 1998;78:766-774. [DOI] [PubMed] [Google Scholar]
- 13. Fan LZ, Cherian MG. Potential role of p53 on metallothionein induction in human epithelial breast cancer cells. Br J Cancer. 2002;87:1019-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sasa M, Bando Y, Takahashi M, Hirose T, Nagao T. Screening for basal marker expression is necessary for decision of therapeutic strategy for triple-negative breast cancer. Journal of Surgical Oncology. 2008;97:30-34. [DOI] [PubMed] [Google Scholar]
- 15. van de Rijn M, Perou CM, Tibshirani R, et al. Expression of cytokeratins 17 and 5 identifies a group of breast carcinomas with poor clinical outcome. Am J Pathol. 2002;161:1991-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nofech-Mozes S, Trudeau M, Kahn HK, et al. Patterns of recurrence in the basal and non-basal subtypes of triple-negative breast cancers. Breast Cancer Res Treat. 2009;118:131-137. [DOI] [PubMed] [Google Scholar]
- 17. Nielsen TO, Hsu FD, Jensen K, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367-5374. [DOI] [PubMed] [Google Scholar]
- 18. Jacquemier J, Ginestier C, Rougemont J, et al. Protein expression profiling identifies subclasses of breast cancer and predicts prognosis. Cancer Res. 2005;65:767-779. [PubMed] [Google Scholar]
- 19. Rakha EA, Elsheikh SE, Aleskandarany MA, et al. Triple-negative breast cancer: distinguishing between basal and nonbasal subtypes. Clin Cancer Res. 2009;15:2302-2310. [DOI] [PubMed] [Google Scholar]
- 20. Oltra SS, Pena-Chilet M, Vidal-Tomas V, et al. Methylation deregulation of miRNA promoters identifies miR124-2 as a survival biomarker in Breast Cancer in very young women. Sci Rep. 2018;8:14373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Venkatesh H, Monje M. Neuronal activity in ontogeny and oncology. Trends Cancer. 2017;3:89-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hironaka-Mitsuhashi A, Matsuzaki J, Takahashi RU, et al. A tissue microRNA signature that predicts the prognosis of breast cancer in young women. PLoS ONE. 2017;12:e0187638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bensen JT, Graff M, Young KL, et al. A survey of microRNA single nucleotide polymorphisms identifies novel breast cancer susceptibility loci in a case-control, population-based study of African-American women. Breast Cancer Res. 2018;20:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kawaguchi T, Yan L, Qi Q, et al. Overexpression of suppressive microRNAs, miR-30a and miR-200c are associated with improved survival of breast cancer patients. Scientific Reports. 2017;7:15945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang S, He X, Ding J, et al. Upregulation of miR-23a approximately 27a approximately 24 decreases transforming growth factor-beta-induced tumor-suppressive activities in human hepatocellular carcinoma cells. Int J Cancer. 2008;123:972-978. [DOI] [PubMed] [Google Scholar]
- 26. Chhabra R, Dubey R, Saini N. Cooperative and individualistic functions of the microRNAs in the miR-23a~27a~24-2 cluster and its implication in human diseases. Molecular Cancer. 2010;9:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Eissa S, Matboli M, Shehata HH. Breast tissue-based microRNA panel highlights microRNA-23a and selected target genes as putative biomarkers for breast cancer. Transl Res. 2015;165:417-427. [DOI] [PubMed] [Google Scholar]
- 28. Ma F, Li W, Liu C, et al. MiR-23a promotes TGF-beta1-induced EMT and tumor metastasis in breast cancer cells by directly targeting CDH1 and activating Wnt/beta-catenin signaling. Oncotarget. 2017;8:69538-69550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Koller K, Das S, Leuschner I, Korbelius M, Hoefler G, Guertl B. Identification of the transcription factor HOXB4 as a novel target of miR-23a. Genes Chromosomes Cancer. 2013;52:709-715. [DOI] [PubMed] [Google Scholar]
- 30. Li M, Han Y, Zhou H, et al. Transmembrane protein 170B is a novel breast tumorigenesis suppressor gene that inhibits the Wnt/beta-catenin pathway. Cell Death & Disease. 2018;9:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-3-bcb-10.1177_1178223420977845 for miRNA Expression Analysis: Cell Lines HCC1500 and HCC1937 as Models for Breast Cancer in Young Women and the miR-23a as a Poor Prognostic Biomarker by Sara S Oltra, Maria Peña-Chilet, Maria T Martinez, Eduardo Tormo, Juan Miguel Cejalvo, Joan Climent, Pilar Eroles, Ana Lluch and Gloria Ribas in Breast Cancer: Basic and Clinical Research
Supplemental material, sj-docx-4-bcb-10.1177_1178223420977845 for miRNA Expression Analysis: Cell Lines HCC1500 and HCC1937 as Models for Breast Cancer in Young Women and the miR-23a as a Poor Prognostic Biomarker by Sara S Oltra, Maria Peña-Chilet, Maria T Martinez, Eduardo Tormo, Juan Miguel Cejalvo, Joan Climent, Pilar Eroles, Ana Lluch and Gloria Ribas in Breast Cancer: Basic and Clinical Research
Supplemental material, sj-docx-5-bcb-10.1177_1178223420977845 for miRNA Expression Analysis: Cell Lines HCC1500 and HCC1937 as Models for Breast Cancer in Young Women and the miR-23a as a Poor Prognostic Biomarker by Sara S Oltra, Maria Peña-Chilet, Maria T Martinez, Eduardo Tormo, Juan Miguel Cejalvo, Joan Climent, Pilar Eroles, Ana Lluch and Gloria Ribas in Breast Cancer: Basic and Clinical Research
Supplemental material, sj-docx-6-bcb-10.1177_1178223420977845 for miRNA Expression Analysis: Cell Lines HCC1500 and HCC1937 as Models for Breast Cancer in Young Women and the miR-23a as a Poor Prognostic Biomarker by Sara S Oltra, Maria Peña-Chilet, Maria T Martinez, Eduardo Tormo, Juan Miguel Cejalvo, Joan Climent, Pilar Eroles, Ana Lluch and Gloria Ribas in Breast Cancer: Basic and Clinical Research
Supplemental material, sj-docx-7-bcb-10.1177_1178223420977845 for miRNA Expression Analysis: Cell Lines HCC1500 and HCC1937 as Models for Breast Cancer in Young Women and the miR-23a as a Poor Prognostic Biomarker by Sara S Oltra, Maria Peña-Chilet, Maria T Martinez, Eduardo Tormo, Juan Miguel Cejalvo, Joan Climent, Pilar Eroles, Ana Lluch and Gloria Ribas in Breast Cancer: Basic and Clinical Research
Supplemental material, sj-docx-8-bcb-10.1177_1178223420977845 for miRNA Expression Analysis: Cell Lines HCC1500 and HCC1937 as Models for Breast Cancer in Young Women and the miR-23a as a Poor Prognostic Biomarker by Sara S Oltra, Maria Peña-Chilet, Maria T Martinez, Eduardo Tormo, Juan Miguel Cejalvo, Joan Climent, Pilar Eroles, Ana Lluch and Gloria Ribas in Breast Cancer: Basic and Clinical Research
Supplemental material, sj-pdf-1-bcb-10.1177_1178223420977845 for miRNA Expression Analysis: Cell Lines HCC1500 and HCC1937 as Models for Breast Cancer in Young Women and the miR-23a as a Poor Prognostic Biomarker by Sara S Oltra, Maria Peña-Chilet, Maria T Martinez, Eduardo Tormo, Juan Miguel Cejalvo, Joan Climent, Pilar Eroles, Ana Lluch and Gloria Ribas in Breast Cancer: Basic and Clinical Research
Supplemental material, sj-pdf-2-bcb-10.1177_1178223420977845 for miRNA Expression Analysis: Cell Lines HCC1500 and HCC1937 as Models for Breast Cancer in Young Women and the miR-23a as a Poor Prognostic Biomarker by Sara S Oltra, Maria Peña-Chilet, Maria T Martinez, Eduardo Tormo, Juan Miguel Cejalvo, Joan Climent, Pilar Eroles, Ana Lluch and Gloria Ribas in Breast Cancer: Basic and Clinical Research