Abstract
Objective: To explore the effect of smoking on gene expression in human alveolar macrophages and the value of identified key genes in the early diagnosis and prognosis of lung cancers. Methods: We downloaded three data sets (GSE8823, GSE2125, and GSE3212) from the Gene Expression Omnibus (GEO) database, including 31 non-smoking and 33 smoking human alveolar macrophage samples. We identified common differentially expressed genes (DEGs), from which we obtained module genes and hub genes by using STRING and Cytoscape. Then we analyzed the protein-protein interaction (PPI) network of DEGs, hub genes, and module genes and used David online analysis tool to carry out functional enrichment analysis of DEGs and module genes. Results: A total of 85 differentially expressed genes was obtained, including 42 up-regulated genes and 43 down-regulated genes. The Human Protein Atlas and Survival analysis showed that GBP1, ITGAM, CSF1, SPP1, COL1A1, LAMB1 and THBS1 may be closely associated with the carcinogenesis and prognosis of lung cancer. Conclusion: DEGs, module, and hub genes identified in the present study help explain the effects of smoking on human alveolar macrophages and provide candidate targets for diagnosis and treatment of smoking-related lung cancer.
Keywords: Smoking, alveolar macrophages, smoking-related lung cancers, diagnosis, prognosis
Introduction
Cigarette smoke is so harmful that a retrospective study found that secondhand smoke alone killed more than 600,000 people worldwide [1]. Smoking is also a significant risk factor for lung cancer and other lung diseases. It mainly causes damage to the lungs and other organs through the inflammatory response, oxidative stress, genetic changes, cell aging and different pathways [2]. Studies have found that smoking causes changes in alveolar macrophage genes involved in the occurrence and development of lung cancer [3]. For example, tumor-associated macrophages (TAM) can polarize into both M1 and M2 phenotypes. The M1 type is involved in the inflammatory and toxic reactions of tumor cells [4]. Transforming growth factor secreted by M2-type macrophages is associated with tumor progression [5]. In the face of intractable lung cancer, many studies have shown that intervention in the tumor microenvironment from the perspective of alveolar macrophages reduces cancer recurrence and development. In recent years, an anti-tumor strategy centering on macrophages by limiting the infiltration of cancer cells has been suggested as a cancer treatment method [6]. However, the study of the effects of smoking on gene expression in human alveolar macrophages is still immature. Therefore, it is necessary to strengthen the research on the molecular mechanism of the occurrence and development of lung cancer mediated by changes in alveolar macrophages caused by tobacco smoke exposure.
With rapid development in recent years, microarray technology has become more convenient and economical, and it can screen different genes in high throughput for research on the molecular mechanism of related diseases [7]. The limitation of false positives, and the limited number of included samples reduce reliability of the analysis results. Based on microarray technology’s application to generate a large number of data sets, bioinformatics can efficiently utilize the above data to obtain more reliable results [8]. Therefore, we used bioinformatic tools to get three expression profile data sets, including GSE8823, GSE2125, and GSE3212. The DEGs of alveolar macrophages in smoke-exposed and non-smoking individuals were analyzed. Secondly, we screened out the most significant module gene and Hub gene and constructed the protein interaction network diagram of DEGs, module genes, and Hub genes. Then we conducted a functional enrichment analysis of the identified DEGs and module genes. Third, we analyzed the differential expression of Hub genes in lung cancer patients and healthy control groups and further selected 7 Hub genes to explain their value in predicting the prognosis in lung cancer. Finally, we tried to explore the influence of Hub gene mutation on the survival of lung cancer patients. In summary, the differentially identified genes may be therapeutic targets and biomarkers for smoking-related lung disease.
Materials and methods
Microarray data
Data came from GEO (http://www.ncbi.nlm.nih.gov/geo) [9], an online public functional genome data warehouse. We downloaded three gene expression datasets (GSE8823 [10], GSE2125 [11], and GSE3212 [12]). GSE8823 included 11 non-smoking alveolar macrophages samples and 13 smoking alveolar macrophages samples, based on platform GPL570[hg-u133_plus_2] Affymetrix Human Genome U133 Plus 2.0 Array. GSE3212 included five non-smoking alveolar macrophage samples and five smoking alveolar macrophage samples, based on platform GPL80[Hu6800] Affymetrix Human full-length HuGeneFL Array. GSE2125 included 15 non-smoking alveolar macrophages sample and 15 smoking alveolar macrophage samples, based on platform GPL570[hg-u133_plus_2] Affymetrix Human Genome U133 Plus 2.0 Array.
Identification of DEGs
To further analyze the DEGs between smoking alveolar macrophages and non-smoking alveolar macrophages, we used an interactive online analysis tool--GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r). The deletion occurs when one gene corresponds to multiple probes, or one probe does not have a corresponding homologous gene. |logFC (fold change)| ≥1 and adj. p-value <0.01 were considered statistically significant.
PPI network construction, module analysis, and Hub genes selection
To explore the interactions between proteins and further explore the mechanism of disease occurrence or development, we first constructed a PPI network for screened DEGs based on an online protein interaction prediction tool -STRING [13]. Then we imported the resulting data into Cytoscape (version 3.7.2) [14] for analysis. Cytoscape is a biological information platform for visualization of molecular interactions. Third, we use Cytoscape’s plug-in MCODE (version 1.5.1) [15] to filter essential modules in the network diagram. The detailed criteria were as follows: Max depth =100, k-score =2, node score cutoff =0.2, degree cutoff =2, and find clusters in the whole network. Finally, we used cytoHubba [16], a plug-in of Cytoscape, to screen the Hub genes in the network graph, and obtained the top 20 genes with the highest score based on the MCC algorithm.
Functional enrichment analysis of DEGs
The Database for Annotation, Visualization, and Integrated Discovery (DAVID; http://david.ncifcrf.gov) (version 6.8) [17] is an online Database for feature enrichment. We performed GO enrichment analyses on screened DEGs, including biological process (BP), cellular component (CC), molecular function (MF), and KEGG analysis [18]. The visual network diagram presented the GO enrichment analysis results, and Cytoscape plugin, BiNGO (version 3.0.3) [19] drew the network diagram. The setting criteria were as follows: Organism selects Homo sapiens, Ontology file selects GO_full, P<0.05 was considered statistically significant.
Hub genes analysis
First, we conducted KEGG and GO enrichment analysis on Hub genes, and the specific operation was the same as DEGs enrichment analysis. Then the hierarchical clustering of Hub genes is constructed by using UCSC online tools (https://xenabrowser.net/heatmap/). The Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/), UALCAN [20], and The Human Protein Atlas (https://www.proteinatlas.org) were used to analyze the differential expression of Hub genes in lung adenocarcinoma (LUAD), Lung squamous cell carcinoma (LUSC), and standard tissue samples. Among them, immunohistochemical (IHC) results from the Human Protein Atlas database were used to verify the translation level of hub genes. Fourth, we used UALCAN--an online platform for analyzing cancer omics, screening Genes correlated with the Hub Genes in LUSC. Finally, we used Kaplan-Meier plotter to draw the Kaplan-Meier survival graph [21]. Kaplan-Meier plotter is capable of detecting the effect of any gene or gene combination on survival in a variety of tumors. All forms of gene changes such as Missense Mutation, Truncating Mutation, and Amplification of Hub genes in patients with LUSC were analyzed using the cBioPortal database [22]. Furthermore, whether the genetic changes affect the overall and disease-free survival of LUSC patients was further investigated.
Results
Identification of 85 DEGs shared by three gene expression datasets
Three microarray datasets (GSE8823, GSE2125, and GSE3212) were obtained through strict screening, including 1205,974 and 1312 DEGs, respectively. The analysis results were presented by a volcano diagram (Figure 1A-C). There were 85 overlapping genes in the three datasets, including 42 up-regulated genes and 43 down-regulated genes, as shown by the Venn diagram (Figure 1D).
Figure 1.
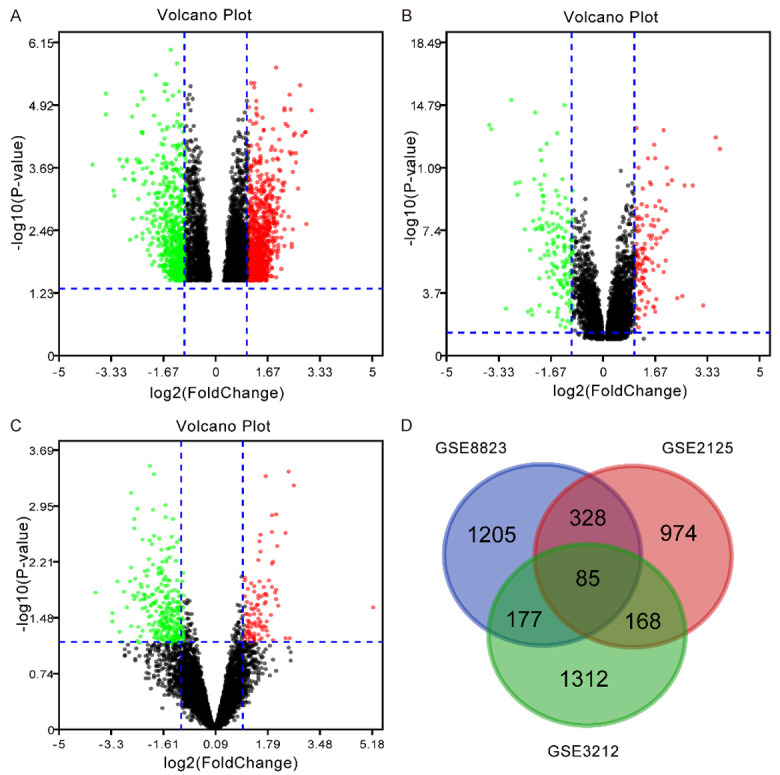
Identification of differentially expressed genes in human alveolar macrophages between smokers and non-smokers. A-C: GEO2R was used to analyze the expression of DEGs in three data sets (GSE8823, GSE2125, and GSE3212 respectively, which are presented as a volcano plots. The red and green dots represent up and down, respectively. D: Venn diagram shows that there are 85 DEGs in the three databases.
PPI network construction, module analysis, and Hub gene selection
First, we constructed the PPI network of DEGs with STRING (Figure 2A). Then, we used Cytoscape’s plug-in MCODE to screen the most significant module genes in the system (Figure 2B). BiNGO, a plug-in of Cytoscape, was used to obtain 20 Hub genes in PPI network (Figure 2C). All Hub genes are shown in Table 1, including gene symbol, full name and primary function.
Figure 2.
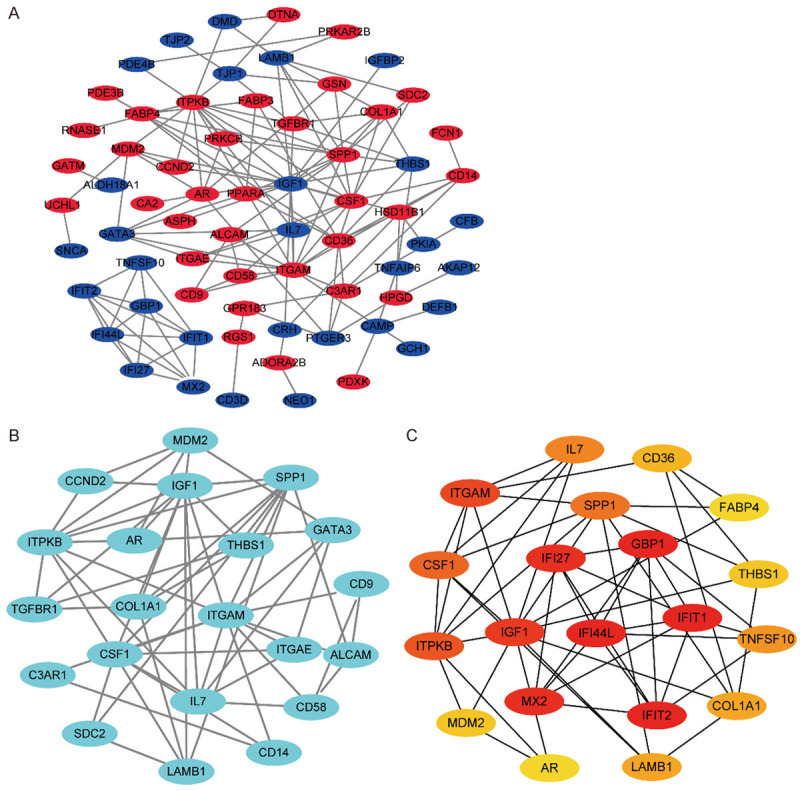
PPI network of DEGs, module genes, and hub genes. A. PPI network of 85 DEGs. Brown and red represent down-regulated and up-regulated genes, respectively. B. PPI network of the most significant module i20 nodes and 58 edges. C. PPI network of 20 hub genes.
Table 1.
Functional roles of 20 hub genes
| No. | Gene symbol | Full name | Function |
|---|---|---|---|
| 1 | GBP1 | Guanylate binding protein 1 | Overexpression of GBP1 promotes the progress of cervical cancer |
| 2 | IFIT2 | Interferon induced protein with tetratricopeptide repeats 2 | Low IFIT2 expression is associated with poor survival in non-small cell lung cancer. |
| 3 | IFIT1 | Interferon induced protein with tetratricopeptide repeats 1 | High expression of IFIT1 in head and neck squamous cell carcinoma is associated with poor prognosis |
| 4 | IFI44L | Interferon induced protein 44 like | IFI44L is associated with metastasis and drug resistance of liver cancer |
| 5 | MX2 | MX dynamin like GTPase 2 | MX2 is a restriction factor of hepatitis B virus replication. |
| 6 | IFI27 | Interferon alpha inducible protein 27 | Down-regulation of the IFI27 gene inhibited the proliferation of oral squamous cell carcinoma |
| 7 | IGF1 | Insulin like growth factor 1 | Association of serum IGF1 and adiponectin are involved in the metastasis of breast cancer. |
| 8 | ITGAM | Integrin subunit alpha M | ITGAM is a risk factor for systemic lupus erythematosus and may also be a protective factor for rheumatoid arthritis |
| 9 | ITPKB | Inositol-trisphosphate 3-kinase B | ITPKB may be associated with lung cancer cell metastasis |
| 10 | CSF1 | Colony stimulating factor 1 | CSF1 is involved in regulating macrophage differentiation and promoting the release of pro-inflammatory factors |
| 11 | SPP1 | Secreted phosphoprotein 1 | SPP1 is closely associated with non-small cell lung cancer, breast cancer, colorectal cancer and other cancers. |
| 12 | IL7 | Interleukin 7 | Il-7 may have anti-tumor effects |
| 13 | TNFSF10 | TNF superfamily member 10 | Viral therapy using TNFSF10 combined with plasminogen k5 may treat cancer |
| 14 | COL1A1 | Collagen type I alpha 1 chain | Down-regulation of COL1A1 expression inhibits the growth and metastasis of breast cancer cells |
| 15 | LAMB1 | Laminin subunit beta 1 | LAMB1 may be a biomarker for colorectal cancer |
| 16 | CD36 | CD36 molecule | CD36 mediates inflammation, molecular adhesion, and apoptosis |
| 17 | MDM2 | MDM2 proto-oncogene | MDM2 inhibitors may have anticancer effects. |
| 18 | THBS1 | Thrombospondin 1 | Activation of THBS1 promotes metastasis of oral squamous cell carcinoma. |
| 19 | AR | Androgen receptor | AR antagonists may be used in the treatment of breast cancer |
| 20 | FABP4 | Fatty acid binding protein 4 | FABP4 inhibitor inhibited oxidative stress in mice with acute lung injury |
Functional enrichment analysis of DEGs and the most significant module
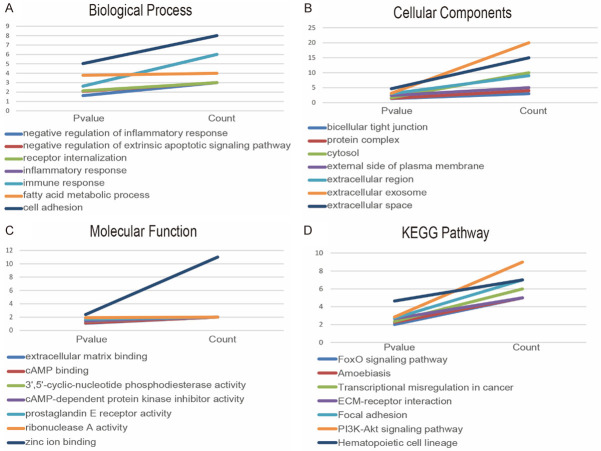
We used the DAVID tool to carry out GO and KEGG enrichment analysis of DEGs. The results of the GO analysis showed that the biological processes (BP) of DEGs were significantly enriched in cell adhesion, fatty acid metabolic process, immune response, inflammatory response, and positive regulation of angiogenesis (Figure 3A). Cellular component (CC) of DEGs is mainly enriched in the cell fraction, plasma membrane part, plasma membrane, extracellular region part, and extracellular region (Figure 3B). The molecular functions (MF) of DEGs were significantly enriched in zinc ion binding, ribonuclease A activity, prostaglandin E receptor activity, 3’,5’-cyclic-amp phosphodiesterase activity, and camp-dependent protein kinase inhibitor activity (Figure 3C). KEGG analysis showed that DEGs mainly concentrated in Hematopoietic cell lineage, Transcriptional misregulation in cancer, pi3k-Akt signaling pathway, focal adhesion, and ECM-receptor interaction (Figure 3D). The results of GO enrichment analysis are presented by the visual network diagram (Figure 3E).
Figure 3.
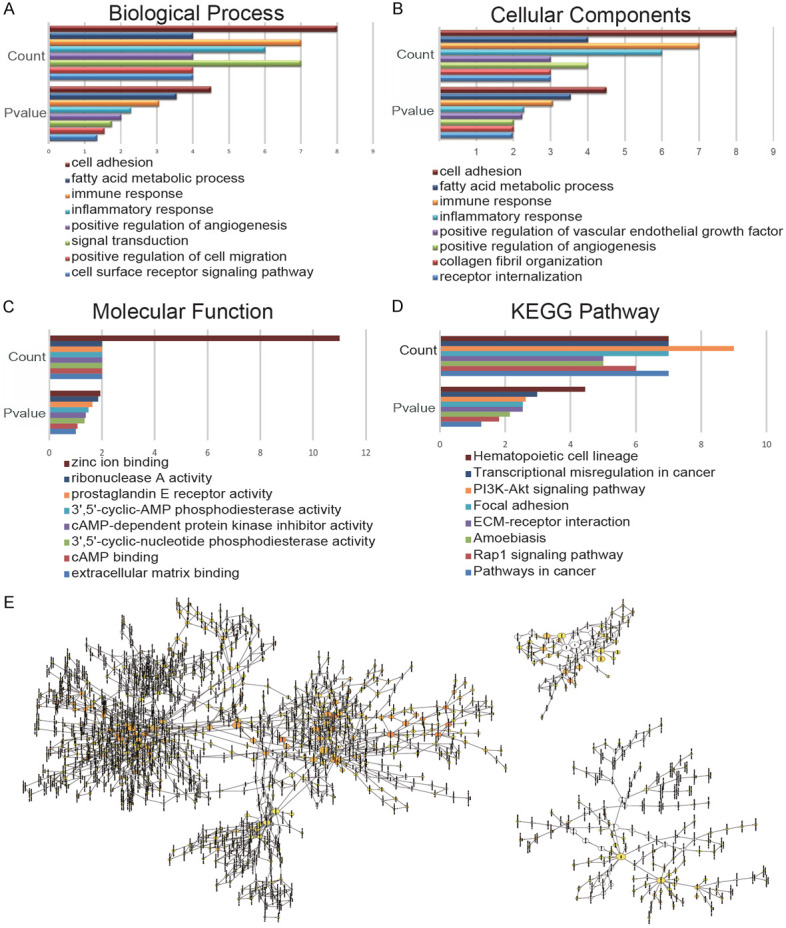
GO and KEGG enrichment analyses of DEGs. Enrichment analysis of BP (A), CC (B), MF (C), and KEGG (D), was carried out by DAVID. (E) The results of GO enrichment analysis are presented as a visual network diagram.
Functional enrichment analysis of the most significant module
DAVID was used for the GO and KEGG enrichment analysis of the most significant module genes. The GO analysis results showed that the BP of module genes were mainly enriched in cell adhesion, fatty acid metabolic process, inflammatory response, immune response, and receptor internalization (Figure 4A). CC changes of module genes are mainly concentrated in extracellular space, extracellular exosome, extracellular region, external side of the plasma membrane, and cytosol (Figure 4B). The variations of MF of module genes were mainly concentrated in zinc ion binding, ribonuclease A activity, prostaglandin E receptor activity, cAMP-dependent protein kinase inhibitor activity, and 3’,5’-cyclic nucleotide phosphodiesterase activity (Figure 4C). KEGG analysis showed that module genes were mainly enriched in Hematopoietic cell lineage, pi3k-Akt signaling pathway, Focal adhesion, ECM-receptor interaction, and Transcriptional misregulation in cancer (Figure 4D).
Figure 4.
GO and KEGG enrichment analyses of the most significant module. Enrichment analysis of BP (A), CC (B), MF (C), and KEGG (D) was carried out by DAVID.
Hub gene analysis
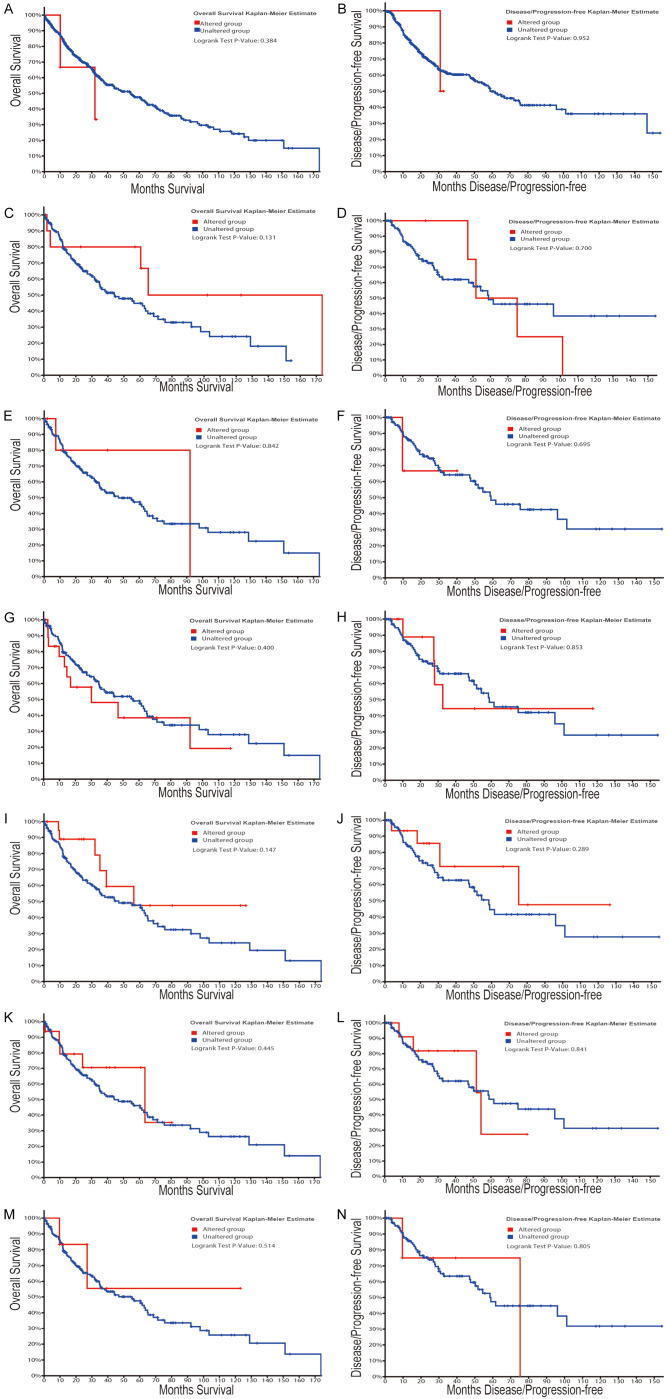
First, we performed hierarchical clustering analysis on the 20 previously screened Hub genes, based on the UCSC online tool. The analysis results showed that there was a difference between the gene expression of the Hub genes in lung cancer tissue samples and standard tissue samples (Figure 5). Then, we used the GEPIA database and UALCAN cancer database to analyze the differential expression of 20 Hub genes in LUAD, LUSC, and standard tissue samples. The results showed that 7 Hub genes were found to be different, including GBP1, COL1A1, SPP1, ITGAM, CSF1, LAMB1, and THBS1. The expression of GBP1 and LAMB1 in LUSC tissues was lower than that in healthy tissues (Figure 6A, 6B, 6K and 6L). Compared with standard tissue samples, the two Hub genes SPP1 and COL1A1 were highly expressed in LUSC patients, and the three Hub genes ITGAM, CSF1 and THBS1 in lung tissues of LUSC patients were down-regulated, with statistically significant differences (P<0.05; Figure 6B-J and 6M-N). Subsequently, IHC staining results obtained from the Human Protein Atlas database were used to verify the high expression of COL1A1 and SPP1 Hub genes, as shown in Figure 7A-L, which showed that the expression levels of these two proteins were higher in LUSC patients than in the healthy control group. We analyzed the top 20 genes most closely related to the seven hub genes in Table 2. Further, we used the KM-plotter to plot the Kaplan-Meier curve of 7 Hub genes. High expression of SPP1 and COL1A1 genes were associated with a better survival rate in lung cancer patients. In contrast, high expression of the other five hub genes SPP1, ITGAM, CSF1, LAMB1, and THBS1 was associated with lower OS, all of which had statistical significance (P<0.05; Figure 8A-G). Finally, we analyzed the gene mutation of 7 Hub genes, and the results were as follows (Figure 9A-G): in 167 cases, 6 percent of GBP1 had gene change: 1 case Truncating Mutation, 1 case, Amplification, and eight instances mRNA High. In 183 cases, 6 percent of the ITGAM gene from genetically altered: 6 cases Missense Mutation, 1 case Amplification, and three instances mRNA High. Among the 200 samples, 3 percent of CSF1 had gene changes: 2. Missense Mutation, 3 Deep Deletion, and one mRNA High. Among 182 cases, 11 percent of SPP1 had gene change: 1 case Missense Mutation, 1 case Amplification, 1 case Deep Deletion, and 17 instances mRNA High. Among the 175 samples, 12 percent of COL1A1 had gene changes. Among 278 samples, 9 percent of LAMB1 had gene changes: 1 Truncating Mutation, 3 Amplification, ten mRNA High, 2 Missense Mutation, and mRNA High mixture. 4 Missense Mutation, 2 Truncating Mutation, 4 Amplification, and six mRNA High. Among the 200 samples, 3 percent of THBS1 had gene changes: 1 case of Missense Mutation, five instances of mRNA High. Mutations in all of the above genes were not associated with overall survival or disease-free survival of LUSC patients (Figure 10A-N).
Figure 5.
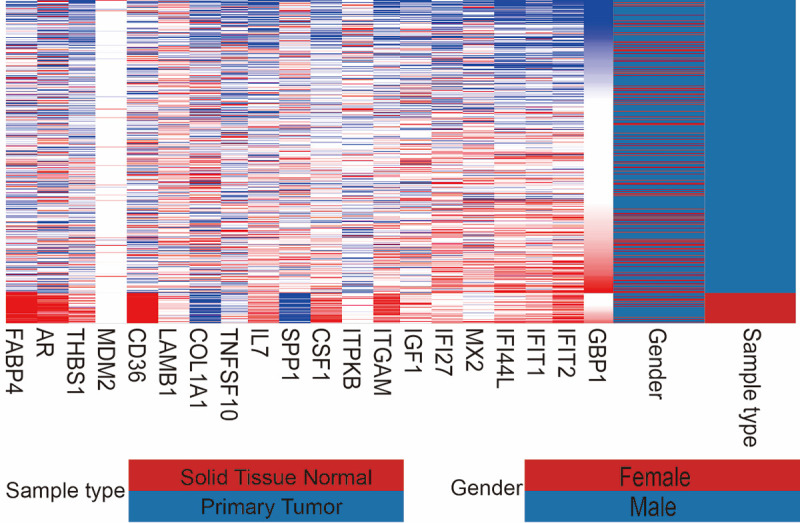
Heatmap of hub genes. Blue is the Primary Tumor specimen, and dark red is Solid Tissue Normal. Red indicates high gene expression, and blue indicates low gene expression.
Figure 6.
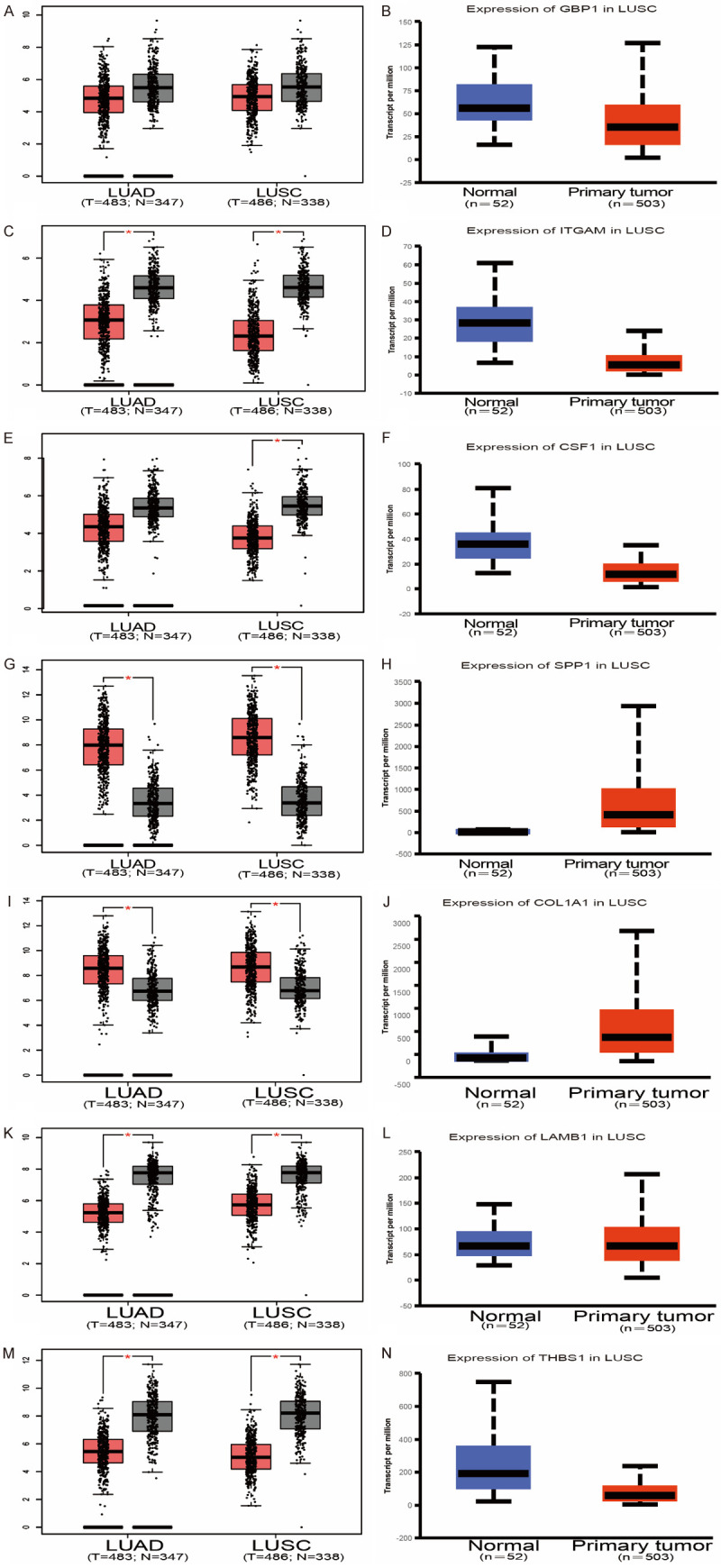
Differential expression of 7 Hub genes in lung cancer and healthy lung tissues. Compared with normal tissues, the expressions of GBP1, ITGAM, CSF1, LAMB1, THBS1 in NSCLC patients were down-regulated, and the expressions of SPP1 and COL1A1 were increased. In addition to GBP1, the difference was statistically significant (*P<0.05). Lung adenocarcinoma (left column); squamous cancer (right column).
Figure 7.
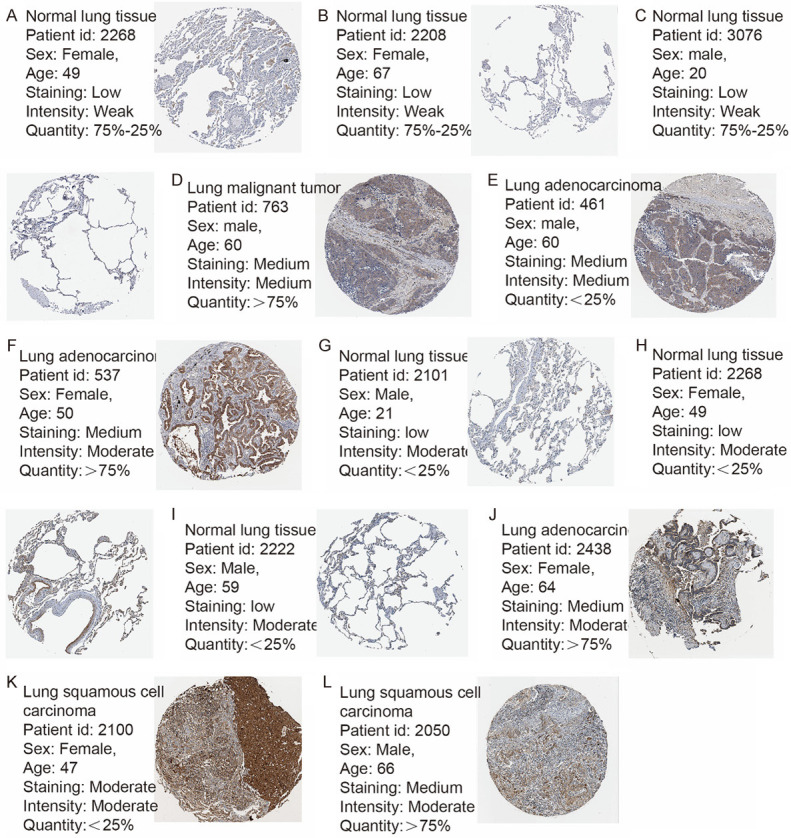
IHC experiment showed that the expression of SPP1 and COL1A1 in NSCLC was higher than that in healthy tissues. (A-C) SPP1 was low in normal tissues, and (D-F) was moderately expressed in COL1A1 tissues. (G-I) COL1A1 expression was low in normal tissues, and (J-L) expression was up-regulated in NSCLC.
Table 2.
The top 20 genes most closely related to the seven hub genes in LUSC
| No. | GBP1 | ITGAM | CSF1 | SPP1 | COL1A1 | LAMB1 | THBS1 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|
|
||||||||
| genes | CC | genes | CC | genes | CC | genes | CC | genes | CC | genes | CC | genes | CC | |
| 1 | GBP2 | 0.77 | SH2B3 | 0.73 | GPR84 | 0.73 | CBR3 | 0.61 | COL5A1 | 0.96 | FSTL1 | 0.61 | BICC1 | 0.70 |
| 2 | BATF2 | 0.75 | NFAM1 | 0.69 | CSF1R | 0.72 | SRXN1 | 0.59 | COL3A1 | 0.95 | LEPRE1 | 0.59 | CRISPLD2 | 0.70 |
| 3 | STAT1 | 0.75 | ITGB2 | 0.68 | CACNA2D4 | 0.68 | GCLM | 0.57 | COL6A3 | 0.92 | FN1 | 0.56 | ITGA1 | 0.67 |
| 4 | TAP1 | 0.74 | DOCK10 | 0.68 | CLEC5A | 0.67 | EPHX1 | 0.56 | COL1A2 | 0.91 | LTBP1 | 0.54 | CTGF | 0.63 |
| 5 | UBE2L6 | 0.73 | LGALS9 | 0.67 | HIC1 | 0.65 | PRDX1 | 0.55 | COL5A2 | 0.91 | LAMC1 | 0.54 | CYR61 | 0.62 |
| 6 | IRF1 | 0.73 | PILRA | 0.66 | CD14 | 0.65 | GSR | 0.55 | PDGFRB | 0.9 | TCF7L1 | 0.53 | SPON1 | 0.62 |
| 7 | GBP5 | 0.73 | SIGLEC9 | 0.66 | RNASE2 | 0.64 | OSGIN1 | 0.54 | ADAMTS2 | 0.89 | CRIM1 | 0.53 | IL1R1 | 0.61 |
| 8 | GBP4 | 0.71 | NCKAP1L | 0.66 | AGAP2 | 0.63 | AKR1B10 | 0.52 | SPARC | 0.88 | LAMA5 | 0.53 | LAMA4 | 0.61 |
| 9 | APOL6 | 0.70 | IL21R | 0.65 | STAB1 | 0.62 | G6PD | 0.51 | NID2 | 0.87 | PXDN | 0.52 | GREM1 | 0.61 |
| 10 | PRF1 | 0.68 | HK3 | 0.65 | ABI3 | 0.62 | NQO1 | 0.51 | BGN | 0.87 | AFAP1 | 0.52 | LRRC32 | 0.61 |
| 11 | LAP3 | 0.68 | FCGR2C | 0.64 | ADAP2 | 0.62 | AKR1C3 | 0.51 | COL11A1 | 0.87 | RBM9 | 0.52 | MRVI1 | 0.60 |
| 12 | PSMB9 | 0.68 | TNFRSF1B | 0.64 | CD300C | 0.62 | MAP2 | 0.49 | COL6A2 | 0.87 | C1QTNF6 | 0.52 | FGF7 | 0.60 |
| 13 | CXCL9 | 0.67 | LILRB4 | 0.64 | ENG | 0.62 | CBR1 | 0.49 | MMP2 | 0.86 | LATS2 | 0.52 | COL6A3 | 0.60 |
| 14 | PSME2 | 0.67 | LRRC25 | 0.64 | LRRC25 | 0.62 | CYP4F3 | 0.49 | LRRC15 | 0.86 | FBLN1 | 0.52 | ACTA2 | 0.60 |
| 15 | CCL4 | 0.67 | SPI1 | 0.63 | TNFRSF1B | 0.61 | CES1 | 0.49 | AEBP1 | 0.86 | ARHGEF17 | 0.51 | ZEB2 | 0.60 |
| 16 | IL18BP | 0.66 | STAT5A | 0.63 | COL8A1 | 0.61 | ME1 | 0.49 | THBS2 | 0.85 | CDK14 | 0.50 | C13orf33 | 0.60 |
| 17 | B2M | 0.65 | CD4 | 0.63 | NLRP3 | 0.61 | TXNRD1 | 0.48 | TIMP2 | 0.85 | GLI3 | 0.50 | NID2 | 0.59 |
| 18 | LAG3 | 0.65 | CD74 | 0.63 | SIGLEC9 | 0.61 | ASPH | 0.48 | ZNF469 | 0.85 | IGDCC4 | 0.50 | CD93 | 0.59 |
| 19 | WARS | 0.65 | HLA-DOA | 0.62 | ITGB7 | 0.60 | AKR1C1 | 0.47 | FNDC1 | 0.84 | KIAA1462 | 0.49 | PMP22 | 0.59 |
| 20 | HLA-E | 0.65 | CLEC5A | 0.62 | SLC7A7 | 0.59 | TDP2 | 0.47 | POSTN | 0.84 | TGFB3 | 0.49 | COL3A1 | 0.59 |
Note: CC correlation coefficient.
Figure 8.
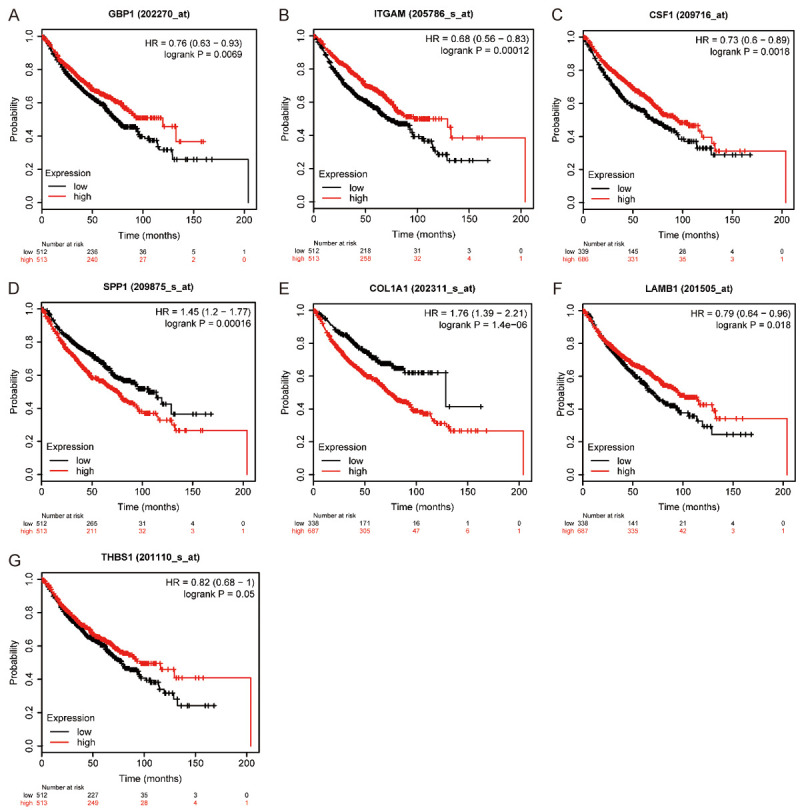
Overall survival analysis of 7 hub genes was conducted using Kaplan-Meier plotter. The down-regulated expressions of GBP1 (A), ITGAM (B), CSF1 (C), LAMB1 (F), and THBS1 (G) in lung cancer patients were associated with poor survival. However, the low expression of SPP1 (D) and COL1A1 (E) in lung cancer patients was associated with a better prognosis (P<0.05).
Figure 9.

Gene alteration of seven hub genes in lung squamous cell carcinoma. The incidences of GBP1 (A), ITGAM (B), CSF1 (C), SPP1 (D), COL1A1 (E), LAMB1 (F), and THBS1 (G) in lung squamous cell carcinoma were 6%, 6%, 3%, 11%, 12%, 9%, and 3%, respectively.
Figure 10.
Correlation analysis between Mutations in 7 hub genes and overall survival or disease-free survival. Mutations in GBP1 (A and B), ITGAM (C and D), CSF1 (E and F), SPP1 (G and H), COL1A1 (I and J), LAMB1 (K and L), and THBS1 (M and N) were not associated with overall survival or disease-free survival of LUSC patients.
Discussion
Cigarette smoke remains a significant risk factor for lung diseases such as COPD and lung cancer. A recent study based on 2770 participants showed that smoking had an odds ratio (OR) of 2.63 (95% CI 1.86-3.73) for COPD [23]. In a systematic review of 12 lung cancer studies conducted by [24], the lung cancer prevalence of passive smokers was RR 1.25 (95% CI 1.10-1.39, P<0.001). In a study by [25], lung cancer mortality in male and female smokers was 4.46 (95% CI 3.10-6.41) and 3.58 (95% CI 2.24-5.73), respectively. The molecular mechanisms of smoking-induced lung cancer are very complex. A growing number of fundamental studies suggest that smoking-induced changes in alveolar macrophages may play an important role. In addition to increasing the number and activation of alveolar macrophages, smoke also induces their secretion of inflammatory mediators, adhesion molecules, and other mediators to increase expression, thus causing lung injury [26]. Therefore, it is urgent to find new candidate genes for smoking-induced lung cancer and screen out the markers with diagnostic and therapeutic potential for lung cancer.
To study the effects of smoking on gene expression in human alveolar macropages, we included a total of three microarray datasets. Finally, 85 DEGs genes were selected, including 42 up-regulated genes and 43 low-expressed genes. We mapped the interaction network of DEGs and module gene and carried out functional enrichment analysis. The changes in BP term of the DEGs enrichment were mainly in the following aspects, including signal transduction, positive regulation of angiogenesis, inflammatory response, immune response, fatty acid metabolic process, and cell adhesion. EGs and module genes include inflammatory response, immune response, fatty acid lean process, and cell ledges. GG pathway analysis revealed that the most significant module was primarily enriched in the following aspects of the FoxO signaling pathway, Amoebiasis, Transcriptional misregulation in cancer, the ECM - receptor interaction, Focal adhesion, pi3k-Akt signaling pathway, and Hematopoietic cell lineage. Parts of the repeated KEGG pathway of the DEGs and the most significant module include Transcriptional misregulation in cancer, pi3k-Akt signaling pathway, Hematopoietic cell lineage, ECM-receptor interaction, and Focal migration. Many studies have confirmed the results of our analysis. Studies have found that cell adhesion inhibitors may play an important therapeutic role in the prevention and treatment of COPD [27]. Inflammatory response and immune response also play an essential role in the occurrence and development of diseases such as COPD and lung cancer [28]. Also, in terms of pathway enrichment, the FoxO signaling pathway is most closely related to the pi3k-Akt signaling pathway [29], especially the FoxOs protein in the FoxO signaling pathway, which plays an essential role in inhibiting tumor cell proliferation and inducing apoptosis. FoxO’s dysfunction is involved in the disease progression of breast cancer, prostate cancer, thymic tumors, and many other tumors [30]. Therefore, an in-depth study of the star molecules in the FoxO signaling pathway may have broad prospects for the discovery and development of anticancer drugs. For lung diseases and some tumors closely related to smoking, the conclusions of these studies are consistent with our results.
We used Cytoscape to screen the critical genes in the DEGs network map, identified the top 20 genes with the strongest correlation, and constructed PPI network, including GBP1, IFIT2, IFIT1, IFI44L, MX2IFI27, IGF1, ITGAM, ITPKB, CSF1, SPP1, IL7, TNFSF10, COL1A1, LAMB1, CD36, MDM2, THBS1, AR, FABP4. Furthermore, we conducted a hierarchical clustering analysis of the above genes, and the results showed that these Hub genes could be clearly distinguished between Primary Tumor and Solid Tissue Normal. Then, we compared the differential expression of 20 Hub genes in healthy tissues and LUAD, LUSC patients in the GEPIA database and screened out seven genes with significant differences, including GBP1, ITGAM, CSF1, SPP1, COL1A1, LAMB1, and THBS1. Then, we verified two genes-SPP1 and COL1A1, which were significantly highly expressed in the samples of patients with lung cancer-in The Human Protein Atlas, and the results further confirmed that the expression levels of SPP1 and COL1A1 in lung cancer tissues were significantly higher than that in healthy tissues.
CSF1 is a growth factor secreted by many types of cells, such as macrophages and cancer cells, which has autocrine and paracrine effects on the CSF1 receptor (CSF1R). Now studies have found that mir-1207-5p activates the expression of CSF1 and is involved in the proliferation, migration, and invasion of lung cancer cells, thus affecting the survival and metastasis of lung cancer patients [31]. In addition to its involvement in the progression of lung cancer, CSF1 is also significantly expressed in breast, ovarian, prostate, and other diseases [32]. Our study found that SPP1 was significantly highly expressed in both LUAD and LUSC patients and that the up-regulation of SPP1 expression in lung cancer patients was closely associated with a low survival rate. Mutations in the SPP1 gene did not affect the overall or disease-free survival of LUSC patients. Our conclusion is consistent with the results of Zhang and Li [33,34]. The latest research also found that SPP1 may be an essential mechanism of afatinib resistance. As we know, afatinib is applied as second-generation EGFR Tyrosine Kinase Inhibitor in the treatment of non-small cell lung cancer, and the high expression of SPP1 can enhance the resistance of afatinib and enhance the invasion ability of lung cancer cells. SPP1 may also be involved in disease progression by mediating macrophage polarization in lung adenocarcinoma [35], and the specific mechanism remains to be further studied. Compared with healthy tissues, LAMB1 expression in lung tissues of patients with pulmonary fibrosis was increased [36]. The up-regulation of LAMB1 expression was associated with the occurrence and development of multiple tumors in colorectal cancer, glioblastoma multiforme (GBM), and others. In our study, it was observed that the expression of THBS1 was down-regulated in lung cancer, and survival analysis found that the high expression of THBS1 was associated with a good prognosis. The above analysis results all suggested that THBS1 plays a protective role in lung tumors, consistent with a prior study [37]. We further analyzed the relationship between THBS1 gene mutation and prognosis in patients with lung cancer and found that disease-free survival rate and overall survival rate of patients with lung cancer were not related to THBS1 gene mutation.
Through literature review, it was found that the correlation between GBP1, ITGAM, COL1A1, and pulmonary diseases has not been widely reported. Our study found that the expression of GBP1 in LUAD and LUSC patients was higher than that in controls, but the difference was not statistically significant. Down-regulation of the GBP1 gene in lung cancer patients was associated with lower survival. Some studies have observed that cytokines induce human ovarian cancer cells to secrete GBP1, and the up-regulation of the GBP1 gene in EOC patients is associated with a good prognosis [38]. However, other studies have shown that GBP1 is involved in cell proliferation. High expression of GBP1 promotes the invasion and migration of glioblastoma cells and predicts a poor prognosis. Moreover, the up-regulation of long non-coding RNA LINC01783 in cervical cancer can regulate the expression of GBP1 and promote disease progression [39]. At present, the research on ITGAM mainly focuses on systemic lupus erythematosus (SLE), and no related research on ITGAM and lung diseases was found. Our bioinformatic analysis showed that ITGAM was protective against lung cancer. COL1A1 is involved in the development of gastric, breast, colon, and pancreatic cancers by promoting the proliferation, invasion, and metastasis of tumor cells. At present, there are still few studies on the relationship between COL1A1 and lung tumors. Oleksiewicz found that the expression of COL1A1 was increased in patients with non-small cell lung cancer [40]. Our research found that COL1A1 was overexpressed in LUAD and LUSC patients and was associated with low survival. Therefore, COL1A1 may be a marker for the diagnosis, treatment, and prognosis of lung cancer.
Conclusion
We used bioinformatic techniques to identify candidate genes for smoking-induced changes in the expression of genes in pulmonary macrophages that are associated with susceptibility to lung disease. The study identified 85 DEGs and 20 Hub genes that may be involved in the development and progression of smoking-related lung cancer. However, the biologic function and clinical diagnostic and prognostic value of these genes in lung cancer need more study.
Data sharing
Figure 10 and Table 2 are available from author on reasonable request. Figure 10: Correlation analysis between survival of the Hub genes and gene alteration. Table 2: The top 20 genes most closely related to the seven hub genes in LUSC.
Acknowledgements
The study was funded by the Tianshan youth program of Xinjiang Uygur autonomous region, China (2018Q090).
Disclosure of conflict of interest
None.
References
- 1.Oberg M, Jaakkola MS, Woodward A, Peruga A, Prüss-Ustün A. Worldwide burden of disease from exposure to second-hand smoke: a retrospective analysis of data from 192 countries. Lancet. 2011;377:139–146. doi: 10.1016/S0140-6736(10)61388-8. [DOI] [PubMed] [Google Scholar]
- 2.Zhou Z, Chen P, Peng H. Are healthy smokers really healthy? Tob Induc Dis. 2016;14:35. doi: 10.1186/s12971-016-0101-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reilly JJ, Chapman HA Jr. Association between alveolar macrophage plasminogen activator activity and indices of lung function in young cigarette smokers. Am Rev Respir Dis. 1988;138:1422–1428. doi: 10.1164/ajrccm/138.6.1422. [DOI] [PubMed] [Google Scholar]
- 4.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 5.Ruffell B, Affara NI, Coussens LM. Differential macrophage programming in the tumor microenvironment. Trends Immunol. 2012;33:119–126. doi: 10.1016/j.it.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pappas K, Papaioannou AI, Kostikas K, Tzanakis N. The role of macrophages in obstructive airways disease: chronic obstructive pulmonary disease and asthma. Cytokine. 2013;64:613–625. doi: 10.1016/j.cyto.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Botling J, Edlund K, Lohr M, Hellwig B, Holmberg L, Lambe M, Berglund A, Ekman S, Bergqvist M, Pontén F, König A, Fernandes O, Karlsson M, Helenius G, Karlsson C, Rahnenführer J, Hengstler JG, Micke P. Biomarker discovery in non-small cell lung cancer: integrating gene expression profiling, meta-analysis, and tissue microarray validation. Clin Cancer Res. 2013;19:194–204. doi: 10.1158/1078-0432.CCR-12-1139. [DOI] [PubMed] [Google Scholar]
- 8.Karim MR, Michel A, Zappa A, Baranov P, Sahay R. Improving data workflow systems with cloud services and use of open data for bioinformatics research. Brief Bioinform. 2018;19:1035–1050. doi: 10.1093/bib/bbx039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kazeros A, Harvey BG, Carolan BJ, Vanni H, Krause A, Crystal RG. Overexpression of apoptotic cell removal receptor MERTK in alveolar macrophages of cigarette smokers. Am J Respir Cell Mol Biol. 2008;39:747–757. doi: 10.1165/rcmb.2007-0306OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woodruff PG, Koth LL, Yang YH, Rodriguez MW, Favoreto S, Dolganov GM, Paquet AC, Erle DJ. A distinctive alveolar macrophage activation state induced by cigarette smoking. Am J Respir Crit Care Med. 2005;172:1383–1392. doi: 10.1164/rccm.200505-686OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heguy A, O’Connor TP, Luettich K, Worgall S, Cieciuch A, Harvey BG, Hackett NR, Crystal RG. Gene expression profiling of human alveolar macrophages of phenotypically normal smokers and nonsmokers reveals a previously unrecognized subset of genes modulated by cigarette smoking. J Mol Med (Berl) 2006;84:318–328. doi: 10.1007/s00109-005-0008-2. [DOI] [PubMed] [Google Scholar]
- 13.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 18.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 20.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin S, Wang X, Zhang X, Guo M, Miura M, Xiao Y. Influence of biomass burning on local air pollution in mainland Southeast Asia from 2001 to 2016. Environ Pollut. 2019;254:112949. doi: 10.1016/j.envpol.2019.07.117. [DOI] [PubMed] [Google Scholar]
- 24.Kim AS, Ko HJ, Kwon JH, Lee JM. Exposure to secondhand smoke and risk of cancer in never smokers: a meta-analysis of epidemiologic studies. Int J Environ Res Public Health. 2018;15:1981. doi: 10.3390/ijerph15091981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu BQ, Peto R, Chen ZM, Boreham J, Wu YP, Li JY, Campbell TC, Chen JS. Emerging tobacco hazards in China: 1. Retrospective proportional mortality study of one million deaths. BMJ. 1998;317:1411–1422. doi: 10.1136/bmj.317.7170.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osińska I, Wołosz D, Domagała-Kulawik J. Association between M1 and M2 macrophages in bronchoalveolar lavage fluid and tobacco smoking in patients with sarcoidosis. Pol Arch Med Wewn. 2014;124:359–364. doi: 10.20452/pamw.2339. [DOI] [PubMed] [Google Scholar]
- 27.Vanderslice P, Biediger RJ, Woodside DG, Berens KL, Holland GW, Dixon RA. Development of cell adhesion molecule antagonists as therapeutics for asthma and COPD. Pulm Pharmacol Ther. 2004;17:1–10. doi: 10.1016/j.pupt.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 28.Wills B, Brahmer JR, Naidoo J. Treatment of complications from immune checkpoint inhibition in patients with lung cancer. Curr Treat Options Oncol. 2018;19:46. doi: 10.1007/s11864-018-0562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sunters A, Madureira PA, Pomeranz KM, Aubert M, Brosens JJ, Cook SJ, Burgering BM, Coombes RC, Lam EW. Paclitaxel-induced nuclear translocation of FOXO3a in breast cancer cells is mediated by c-Jun NH2-terminal kinase and Akt. Cancer Res. 2006;66:212–220. doi: 10.1158/0008-5472.CAN-05-1997. [DOI] [PubMed] [Google Scholar]
- 30.Matsuzaki H, Daitoku H, Hatta M, Tanaka K, Fukamizu A. Insulin-induced phosphorylation of FKHR (Foxo1) targets to proteasomal degradation. Proc Natl Acad Sci U S A. 2003;100:11285–11290. doi: 10.1073/pnas.1934283100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dang W, Qin Z, Fan S, Wen Q, Lu Y, Wang J, Zhang X, Wei L, He W, Ye Q, Yan Q, Li G, Ma J. miR-1207-5p suppresses lung cancer growth and metastasis by targeting CSF1. Oncotarget. 2016;7:32421–32432. doi: 10.18632/oncotarget.8718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones CV, Ricardo SD. Macrophages and CSF-1: implications for development and beyond. Organogenesis. 2013;9:249–260. doi: 10.4161/org.25676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang W, Fan J, Chen Q, Lei C, Qiao B, Liu Q. SPP1 and AGER as potential prognostic biomarkers for lung adenocarcinoma. Oncol Lett. 2018;15:7028–7036. doi: 10.3892/ol.2018.8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li S, Yang R, Sun X, Miao S, Lu T, Wang Y, Wo Y, Jiao W. Identification of SPP1 as a promising biomarker to predict clinical outcome of lung adenocarcinoma individuals. Gene. 2018;679:398–404. doi: 10.1016/j.gene.2018.09.030. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Du W, Chen Z, Xiang C. Upregulation of PD-L1 by SPP1 mediates macrophage polarization and facilitates immune escape in lung adenocarcinoma. Exp Cell Res. 2017;359:449–457. doi: 10.1016/j.yexcr.2017.08.028. [DOI] [PubMed] [Google Scholar]
- 36.Ji X, Wu B, Han R, Yang J, Ayaaba E, Wang T, Han L, Ni C. The association of LAMB1 polymorphism and expression changes with the risk of coal workers’ pneumoconiosis. Environ Toxicol. 2017;32:2182–2190. doi: 10.1002/tox.22431. [DOI] [PubMed] [Google Scholar]
- 37.Weng TY, Wang CY, Hung YH, Chen WC, Chen YL, Lai MD. Differential expression pattern of THBS1 and THBS2 in lung cancer: clinical outcome and a systematic-analysis of microarray databases. PLoS One. 2016;11:e0161007. doi: 10.1371/journal.pone.0161007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carbotti G, Petretto A. Cytokine-induced guanylate binding protein 1 (GBP1) release from human ovarian cancer cells. Cancers (Basel) 2020;12:488. doi: 10.3390/cancers12020488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen WJ, Xiong L, Yang L, Yang LJ, Li L, Huang L, Liang XQ, Xue J, Tan BZ. Long non-coding RNA LINC01783 promotes the progression of cervical cancer by sponging miR-199b-5p to mediate GBP1 expression. Cancer Manag Res. 2020;12:363–373. doi: 10.2147/CMAR.S230171. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Oleksiewicz U, Liloglou T, Tasopoulou KM, Daskoulidou N, Gosney JR, Field JK, Xinarianos G. COL1A1, PRPF40A, and UCP2 correlate with hypoxia markers in non-small cell lung cancer. J Cancer Res Clin Oncol. 2017;143:1133–1141. doi: 10.1007/s00432-017-2381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]