Little is known about the functional importance of mRNA localization to centrosomes. Ryder et al. survey RNA localization to centrosomes within Drosophila embryos and show that regulation of centrocortin mRNA by FMRP ensures normal mitosis and embryonic viability.
Abstract
Centrosomes are microtubule-organizing centers required for error-free mitosis and embryonic development. The microtubule-nucleating activity of centrosomes is conferred by the pericentriolar material (PCM), a composite of numerous proteins subject to cell cycle–dependent oscillations in levels and organization. In diverse cell types, mRNAs localize to centrosomes and may contribute to changes in PCM abundance. Here, we investigate the regulation of mRNA localization to centrosomes in the rapidly cycling Drosophila melanogaster embryo. We find that RNA localization to centrosomes is regulated during the cell cycle and developmentally. We identify a novel role for the fragile-X mental retardation protein in the posttranscriptional regulation of a model centrosomal mRNA, centrocortin (cen). Further, mistargeting cen mRNA is sufficient to alter cognate protein localization to centrosomes and impair spindle morphogenesis and genome stability.
Introduction
The centrosome is a multifunctional organelle that serves as the primary microtubule-organizing center of most animal cells and comprises a central pair of centrioles surrounded by a proteinaceous matrix of pericentriolar material (PCM; Conduit et al., 2015). During mitosis, centrosomes help organize the bipolar mitotic spindle and function to ensure the fidelity of cell division. In interphase, centrosomes contribute to cell polarization, intracellular trafficking, and ciliogenesis (Vertii et al., 2016).
Cell cycle–dependent changes in PCM composition contribute to functional changes in centrosome activity. Upon mitotic entry, centrosomes undergo mitotic maturation, a process by which centrosomes augment their microtubule-nucleating capacity through the recruitment of additional PCM (Palazzo et al., 2000). This process is reversed upon mitotic exit by PCM shedding (Magescas et al., 2019; Mittasch et al., 2020). These dynamic oscillations in PCM composition and organization are essential for centrosome function, and their deregulation is associated with developmental disorders, increased genomic instability, and cancer (Conduit et al., 2015; Nigg and Raff, 2009). Nonetheless, the regulation of PCM dynamics remains incompletely understood.
Centrosomes are essential for early Drosophila embryogenesis, which proceeds through 14 rounds of rapid, synchronous, abridged nuclear cycles (NCs) consisting of S and M phases with no intervening gap phases before cellularization (Foe and Alberts, 1983). From NC 10 to 14, the embryo develops as a syncytial blastoderm, wherein thousands of nuclei and their associated centrosome pairs divide just under the embryonic cortex. Nuclear migration and divisions are coordinated by the centrosomes, and mutations in centrosome-associated genes impair spindle morphogenesis, mitotic synchrony, genome stability, and embryonic viability (Glover et al., 1995; Megraw et al., 1999; Sunkel and Glover, 1988). As in many organisms, the early development of the Drosophila embryo proceeds through a period of transcriptional quiescence and is supported by a maternal supply of mRNA and proteins (Vastenhouw et al., 2019). Thus, PCM dynamics apparent in early embryos rely on posttranscriptional mechanisms.
More than a decade ago, a high-throughput screen for mRNAs with distinct subcellular locations in syncytial Drosophila embryos uncovered a subset of mRNAs localizing to spindle poles (Lécuyer et al., 2007). Many of the centrosome-enriched transcripts identified in that screen encode known centrosome regulators, including cyclin B (cyc B) and pericentrin-like protein (plp; Dalby and Glover, 1992; Martinez-Campos et al., 2004; Raff et al., 1990). These findings raise the possibility that RNA localization, translational control, and other posttranscriptional regulatory mechanisms contribute to centrosome activity and/or function. Consistent with this idea, RNA is known to associate with centrosomes in diverse cell types, including early embryos (Drosophila, Xenopus, zebrafish, and mollusk), surf clams, and cultured mammalian cells (Alliegro and Alliegro, 2008; Alliegro et al., 2006; Bergalet et al., 2020; Blower et al., 2007; Lambert and Nagy, 2002; Lécuyer et al., 2007; Raff et al., 1990; Sepulveda et al., 2018). The functional consequences and the mechanisms that regulate centrosome-localized RNA remain little understood, however (Marshall and Rosenbaum, 2000; Ryder and Lerit, 2018).
Here, we report that multiple RNAs dynamically localize to centrosomes in Drosophila early embryos. We show that these RNAs localize in unique patterns, with some forming higher-order granules and others localizing to centrosomes as individual molecules. We further demonstrate that some RNAs localize to centrosomal subdomains, e.g., centrosome flares, which extend from interphase centrosomes and define the PCM scaffold (Lerit et al., 2015; Megraw et al., 2002; Richens et al., 2015). We identify one centrosomal RNA, centrocortin (cen), which forms micrometer-scale granules that localize asymmetrically to centrosomes. We further define the mechanisms underlying cen mRNA granule formation and function. We find that cen mRNA granules include Cen protein and the translational regulator fragile-X mental retardation protein (FMRP), the orthologue of the fragile X syndrome–related RNA-binding protein encoded by the Fmr1 gene. Our data show that FMRP regulates both the localization and steady-state levels of cen RNA and protein. Moreover, we find that reducing cen dosage is sufficient to ameliorate mitotic spindle defects associated with Fmr1 loss. Finally, we show that mislocalization of cen mRNA prevents the localization of Cen protein to distal centrosomes and is associated with disrupted embryonic nuclear divisions.
Results
Quantitative analysis of mRNA distributions to Drosophila centrosomes
A genome-wide screen identified a cohort of mRNAs showing localization near spindle poles (Lécuyer et al., 2007). To quantitatively assess transcript localization to centrosomes, we combined single-molecule FISH (smFISH) with direct visualization of centrosomes. smFISH permits precise subcellular localization of individual RNA molecules, an important feature when determining enrichment at a relatively small target, such as the centrosome (Raj et al., 2008). For this analysis, we focused on NC 13 embryos, as their prolonged interphase facilitates the collection of sufficient samples for quantification (Foe and Alberts, 1983). We used GFP-Centrosomin (GFP-Cnn) expressed under endogenous regulatory elements to label centrosomes (Lerit et al., 2015). Cnn is a core component of the centrosome scaffold required for the organization of the PCM that defines the outer edge of the centrosome (Conduit et al., 2010, 2014; Megraw et al., 1999). Among the candidate RNAs reported to localize near spindle poles (i.e., Bsg25D, cen, cyc B, plp, small ovary [sov], and partner of inscuteable [pins] mRNAs), we selected five for investigation based on prior data implicating their protein products in centrosome regulation and/or cell division: cyc B, cen, plp, sov, and pins (Lécuyer et al., 2007).
To examine patterns of RNA localization, we developed an automated custom image analysis pipeline that calculates the distribution of RNA transcripts relative to the distance from the centrosome (Fig. S1 A; Materials and methods). Briefly, smFISH signals and centrosomes were segmented, and the distances between individual RNA objects and the closest centrosome were measured. This analysis allowed us to calculate the percentage of mRNA overlapping with the centrosome surface (Fig. S1 B, arrowheads).
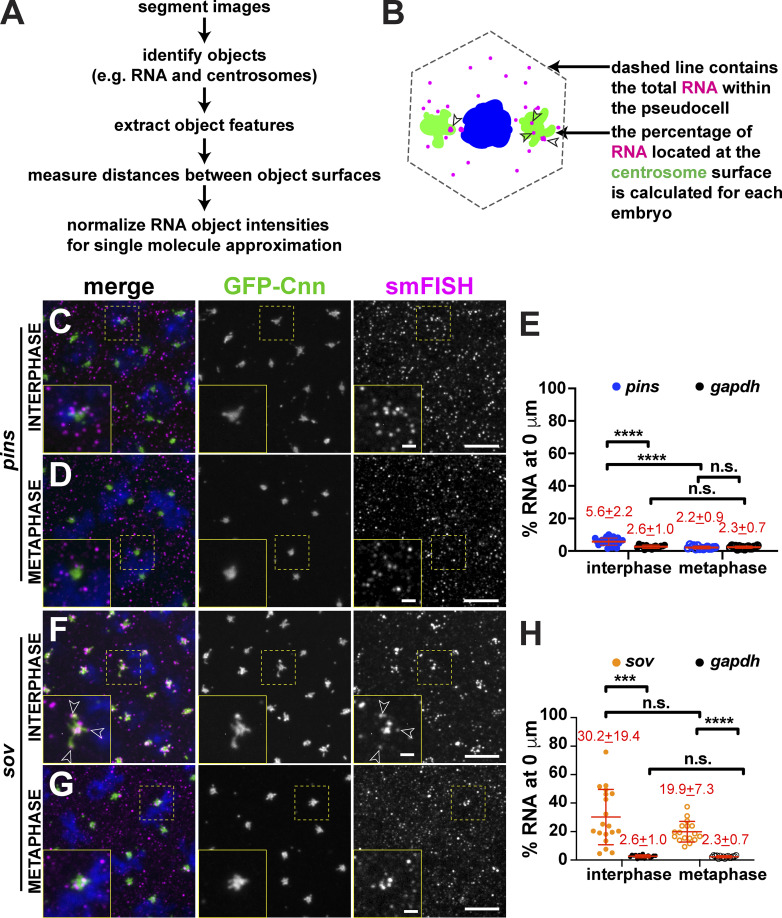
Figure S1.
Determining mRNA enrichment at centrosomes. (A) Workflow used to quantify RNA distributions relative to centrosomes. (B) Cartoon shows total RNA (magenta) within a syncytial Drosophila embryo pseudocell (dashed line). Arrowheads show RNA overlapping with the centrosome (green) surface. (C–H) Maximum-intensity projections and quantification of smFISH for pins or sov mRNAs (magenta) in interphase and metaphase NC 13 embryos expressing GFP-Cnn (green). Boxed regions are enlarged in the insets. Open arrowheads denote association of sov mRNA with centrosome flares. Quantification of the percentage of RNA overlapping with the centrosome surface (0 µm distance) is shown to the right, where each dot represents a single measurement from n = 16 interphase and metaphase (gapdh mRNA), n = 24 interphase and 15 metaphase (pins mRNA), and n = 19 interphase and 17 metaphase (sov mRNA) embryos. Mean ± SD are shown (red text). (C–H) pins (C–E) and sov (F–H). Note that values for gapdh are reproduced from Fig. 1 C to facilitate comparison. Table 1 lists the number of embryos, centrosomes, and RNA objects quantified per condition. ***, P < 0.001; and ****, P < 0.0001 by ANOVA followed by Dunnett’s T3 multiple comparisons test. Scale bars: 5 µm; 1 µm (insets). n.s., not significant.
Table 1. Quantification of RNA localization to centrosomes.
| Genotype | mRNA | NC stage | Cell cycle phase | Embryos (n) | Centrosomes (n) | mRNA objects (n) |
|---|---|---|---|---|---|---|
| GFP-Cnn | cen | NC 10 | Interphase | 20 | 291 | 165,767 |
| GFP-Cnn | cen | NC 10 | Metaphase | 17 | 272 | 95,665 |
| GFP-Cnn | cen | NC 13 | Interphase | 18 | 3,048 | 118,306 |
| GFP-Cnn | cen | NC 13 | Metaphase | 17 | 2,169 | 162,043 |
| GFP-Cnn | cyc B | NC 13 | Interphase | 19 | 3,247 | 177,536 |
| GFP-Cnn | cyc B | NC 13 | Metaphase | 13 | 1,667 | 143,962 |
| GFP-Cnn | gapdh | NC 10 | Interphase | 19 | 298 | 248,645 |
| GFP-Cnn | gapdh | NC 10 | Metaphase | 18 | 298 | 209,717 |
| GFP-Cnn | gapdh | NC 13 | Interphase | 16 | 2,362 | 112,165 |
| GFP-Cnn | gapdh | NC 13 | Metaphase | 16 | 2,096 | 76,762 |
| GFP-Cnn | pins | NC 13 | Interphase | 24 | 3,642 | 117,132 |
| GFP-Cnn | pins | NC 13 | Metaphase | 15 | 1,911 | 74,495 |
| GFP-Cnn | plp | NC 13 | Interphase | 19 | 2,899 | 27,476 |
| GFP-Cnn | plp | NC 13 | Metaphase | 17 | 2,392 | 27,542 |
| GFP-Cnn | sov | NC 13 | Interphase | 19 | 3,689 | 81,150 |
| GFP-Cnn | sov | NC 13 | Metaphase | 17 | 1,960 | 46,055 |
| GFP-γ-Tub | cen | NC 10 | Interphase | 13 | 192 | 122,555 |
| GFP-γ-Tub | cen | NC 10 | Metaphase | 10 | 220 | 86,483 |
| GFP-γ-Tub; Fmr1 | cen | NC 10 | Interphase | 12 | 192 | 100,816 |
| GFP-γ-Tub; Fmr1 | cen | NC 10 | Metaphase | 12 | 205 | 56,786 |
| GFP-γ-Tub | cen | NC 13 | Interphase | 27 | 3,458 | 192,394 |
| GFP-γ-Tub | cen | NC 13 | Metaphase | 12 | 1,228 | 67,961 |
| GFP-γ-Tub; Fmr1 | cen | NC 13 | Interphase | 27 | 3,088 | 142,762 |
| GFP-γ-Tub; Fmr1 | cen | NC 13 | Metaphase | 12 | 1,222 | 46,690 |
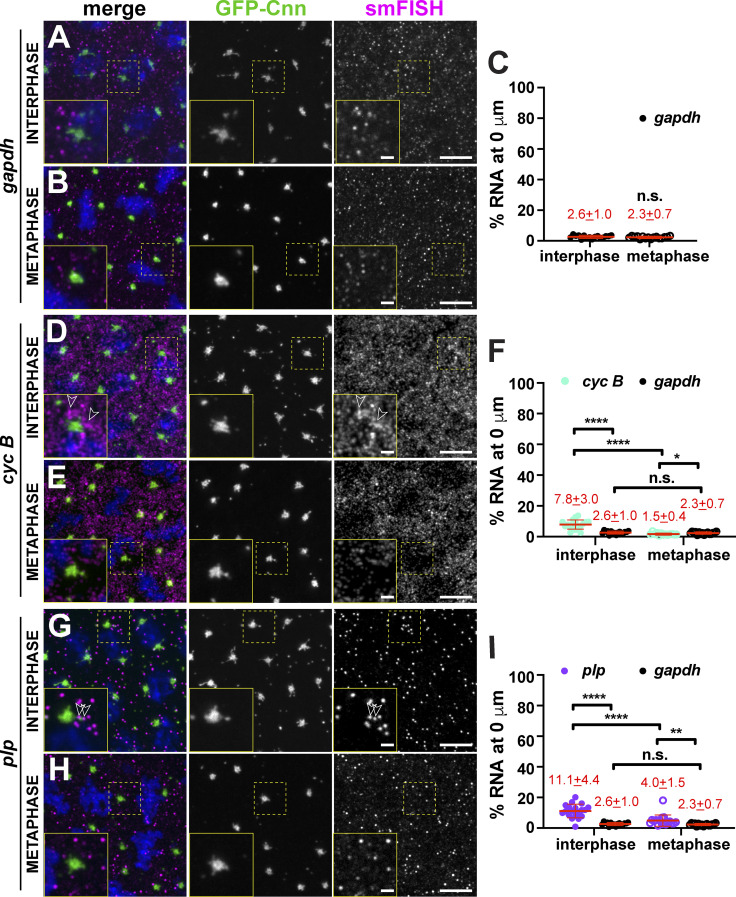
Several prior studies noted an enrichment of cyc B mRNA in the spindle pole region of syncytial Drosophila embryos (Dalby and Glover, 1992; Raff et al., 1990; Vardy and Orr-Weaver, 2007). Therefore, we initially investigated the localization of cyc B relative to a nonlocalizing control RNA, gapdh, to validate our quantitative imaging approach. Consistent with other reports, we observed that cyc B was particularly abundant at the posterior pole (Raff et al., 1990). To standardize measurements of mRNA enriched near somatic centrosomes across samples, we imaged embryos at ∼50% egg length unless otherwise noted. To monitor cell cycle–dependent changes in RNA distribution, centrosome enrichments were calculated during interphase and metaphase. Interphase embryos were selected based on their round nuclei with noncondensed chromosomes and duplicated centrosomes, while metaphase embryos were selected by the presence of a metaphase plate. Throughout the cell cycle, gapdh mRNA appeared dispersed throughout the cytoplasm, and <3% overlapped with centrosomes despite high levels of expression (Fig. 1, A–C; Graveley et al., 2011). By contrast, threefold more cyc B mRNA overlapped with interphase centrosomes (Fig. 1, D–F; P < 0.0001). During metaphase, however, cyc B showed less centrosome enrichment than gapdh, raising the possibility that cyc B mRNA may be actively excluded from mitotic centrosomes (Fig. 1 F; P < 0.05). These findings reveal that cyc B localization to centrosomes is regulated by cell cycle progression. Moreover, these data showcase the utility of our analysis pipeline to quantitatively define RNA localization to centrosomes.
Figure 1.
Quantitative localization of mRNA to centrosomes. Maximum-intensity projections of smFISH (magenta) in NC 13 embryos expressing the centrosome marker GFP-Cnn (green). DAPI labels nuclei blue. Boxed regions enlarged in insets. Open arrowheads mark mRNA at the PCM. Quantification of the percentage of RNA overlapping with the centrosome surface (0 µm distance) is shown to the right, where each dot represents a single measurement from n = 16 interphase and metaphase (gapdh mRNA), 19 interphase and 13 metaphase (cyc B mRNA), and 19 interphase and 17 metaphase (plp mRNA) embryos, respectively. Mean ± SD displayed (red). (A–I) gapdh (A–C), cyc B (D–F), and plp mRNAs (G–I). Note that values for gapdh are reproduced from C for comparison. Table 1 lists the number of embryos, centrosomes, and RNA objects quantified per condition. *, P < 0.05; **, P < 0.01; ****, P < 0.0001 by ANOVA followed by Dunnett’s T3 multiple comparisons test. Scale bars: 5 µm; 1 µm (insets). n.s., not significant.
Multiple mRNAs are enriched at centrosomes in a cell cycle–dependent manner
We next investigated the localization of plp mRNA, as PLP protein cooperates with Cnn to mediate centrosome scaffolding (Lerit et al., 2015; Richens et al., 2015). Recently, orthologous PCNT transcripts were shown to be localized to centrosomes in zebrafish embryos and cultured mammalian cells, specifically during early mitosis (Sepulveda et al., 2018). plp mRNA was significantly enriched at centrosomes throughout the cell cycle, particularly during interphase, when approximately fourfold more plp than gapdh mRNA resides at centrosomes (Fig. 1, G–I; P < 0.0001). Enrichment of plp mRNA is coincident with the formation of interphase centrosome flares containing PLP protein (Lerit et al., 2015), hinting that aspects of plp posttranscriptional regulation may be differentially regulated over the cell cycle.
We similarly analyzed the localization of pins and sov mRNAs to centrosomes. Relative to gapdh, significantly more pins mRNA localized to interphase centrosomes (Fig. S1, C–E). By contrast, sov mRNA was concentrated at centrosomes throughout the cell cycle (∼10-fold more than gapdh; Fig. S1, F–H). As previously noted, sov mRNA tended to localize along centrosome flares (Lécuyer et al., 2007; Fig. S1 F, arrowheads).
Our quantitative analysis underscores transcript-dependent and cell cycle stage–dependent variabilities in centrosome enrichment, suggesting that RNA localization to centrosomes is a regulated process, often favoring localization to interphase centrosomes. Differential mRNA localization over the cell cycle also implies that RNA localization to centrosomes is likely to be a dynamic process.
Dynamic regulation of micrometer-scale cen mRNA granules
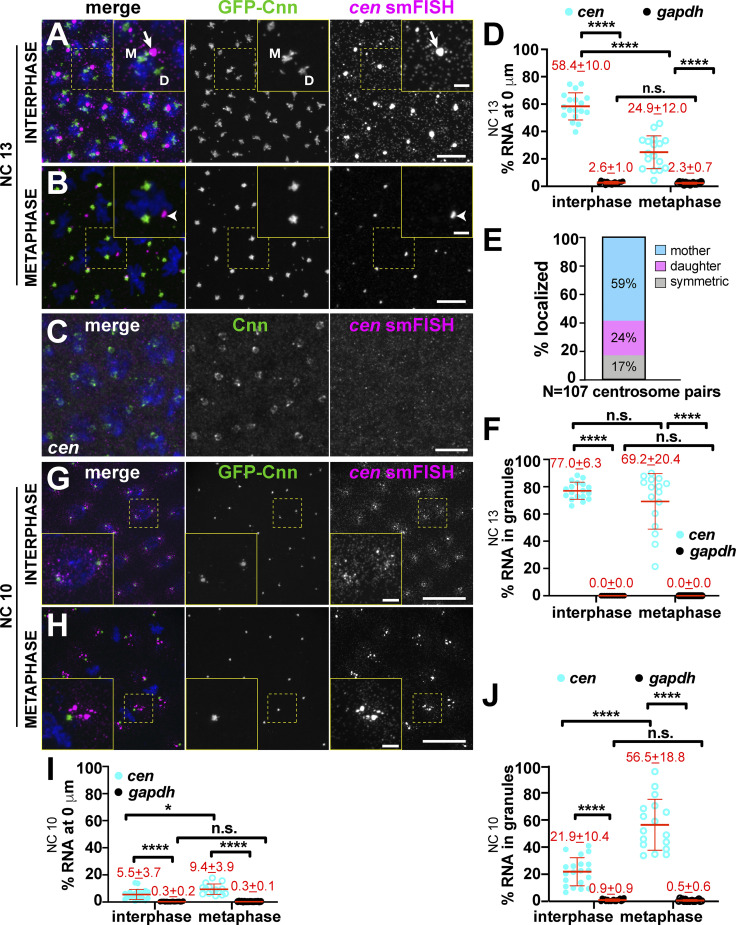
We next investigated the localization of cen mRNA. Cen was previously shown to be required for normal nuclear divisions in Drosophila embryos (Kao and Megraw, 2009). Moreover, cen mRNA was recently localized to embryonic centrosomes (Bergalet et al., 2020). Unlike the other transcripts we investigated, the majority of cen mRNA was enriched at centrosomes during interphase (Fig. 2 A, arrow). cen mRNA displayed a propensity to form higher-order structures, or RNA granules, defined as an overlapping cluster of four or more mRNAs (Little et al., 2015). Throughout NC 13, cen mRNA formed micrometer-scale granules, consistent with recent work (Fig. 2, A and B, arrows and arrowheads; Bergalet et al., 2020). Demonstrating specificity, these signals were not detected in null cen embryos (Fig. 2 C). Quantification revealed that nearly 60% of cen mRNA overlapped with interphase centrosomes (Fig. 2 D). Although this enrichment declined to ∼25% during metaphase, cen mRNA remained significantly enriched at centrosomes relative to gapdh (Fig. 2 D). The majority of cen mRNA resided in granules, which showed a biased localization to the mother centrosome during interphase (Fig. 2, A, E, and F, arrow). During metaphase, cen mRNA granules appeared less tightly associated with centrosomes (Fig. 2, B and F, arrowhead). We conclude that cen mRNA organizes into pericentrosomal granules in a cell cycle–dependent manner, resulting in a bulk enrichment of cen mRNA at centrosomes.
Figure 2.
cen mRNA localization to centrosomes is cell cycle regulated. Maximum-intensity projections of cen smFISH (magenta), where Cnn (green) labels centrosomes. Boxed regions enlarged in insets. (A) Interphase NC 13 embryo showing cen mRNA granules (arrow). Mother (M) and daughter (D) centrosomes noted. (B) Metaphase NC 13 embryo with cen mRNA (arrowhead) displaced from centrosomes. (C) cen mRNA is not detected in cen mutants. (D) Percentage of RNA overlapping with centrosomes in NC 13. Note that values for gapdh are reproduced from Fig. 1 C for comparison. (E) Frequency distribution of cen mRNA granule localization from n = 107 centrosome pairs and n = 5 embryos. (F) Percentage of cen mRNA residing within granules (≥4 overlapping RNA molecules) in NC 13. (G) Interphase NC 10 embryo with cen mRNA symmetrically distributed to centrosomes. (H) Metaphase NC 10 embryo with cen mRNA granules. (I) Percentage of RNA overlapping with centrosomes in NC 10. (J) Percentage of cen mRNA within granules in NC 10. Each dot represents a measurement from a single embryo. Mean ± SD displayed (red) from n = 16 interphase and metaphase (gapdh mRNA), 18 interphase and 17 metaphase (cen mRNA) NC 13; n = 19 interphase and 18 metaphase (gapdh mRNA); and 20 interphase and 17 metaphase (cen mRNA) NC 10 embryos, respectively. Table 1 lists number of objects quantified per condition. *, P < 0.05; ****, P < 0.0001 by ANOVA followed by Dunnett’s T3 multiple comparisons test (D, I, and J) and the Kruskal-Wallis test followed by Dunn’s multiple comparisons test (F). Scale bars: 10 µm; 2.5 µm (insets). n.s., not significant.
The strong enrichment of cen mRNA within pericentrosomal granules prompted us to investigate the timing of their formation. In interphase NC 10 embryos, cen mRNA predominantly existed in single molecules radiating in a gradient from centrosomes (Fig. 2 G). Entry into mitosis correlated with formation of cen mRNA granules that were closely apposed and symmetrically distributed to the two centrosomes (Fig. 2 H). Throughout NC 10, significantly more cen mRNA than gapdh was found at centrosomes (Fig. 2 I). Similarly, significantly more cen mRNA than gapdh was contained within granules, particularly during metaphase (>100-fold difference; Fig. 2 J). These data suggest that the formation of cen mRNA granules is entrained with the cell cycle and correlates with the initiation of cortical nuclear divisions. Our finding that cen mRNA persists in RNA granules in interphase NC 13 embryos suggests that the capacity for cen mRNA granule formation or maintenance is additionally regulated developmentally.
The cen mRNA granule contains Cen protein and requires the centrosome scaffold
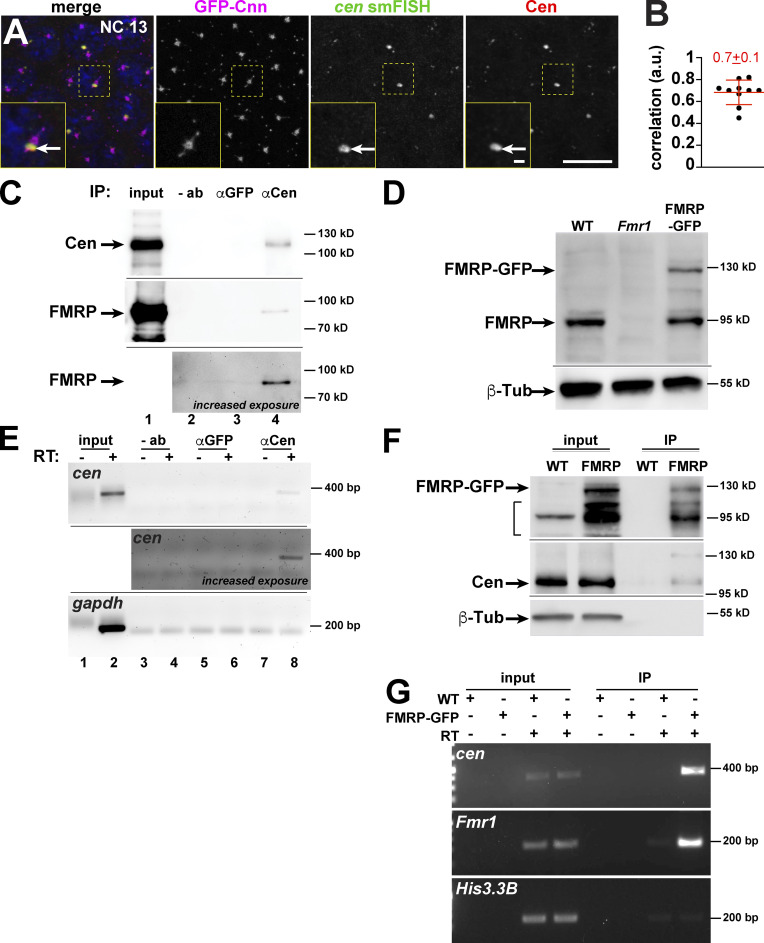
To gain insight into the regulation and function of cen mRNA granules, we first investigated their composition. Recent work uncovered that cen mRNA granules contain Cen protein, and some cen mRNA granules represent sites of local translation (Bergalet et al., 2020). We similarly noted a strong coincidence of cen mRNA and protein at centrosomes, confirming that Cen protein is abundant in cen mRNA granules (Fig. 3, A and B, arrows).
Figure 3.
Composition of the cen mRNA granule. (A) Maximum-intensity projection of a NC 13 embryo expressing GFP-Cnn (magenta) showing colocalization of cen mRNA (green) and protein (red). Boxed region enlarged in insets; arrows, cen mRNA granule. (B) Pearson’s correlation coefficient for cen smFISH and Cen signals. Each dot is a single measurement from n = 10 NC 13 embryos; mean ± SD displayed (red). (C) Blots from Cen immunoprecipitation (IP) using 1–3-h (∼NC 7–14) embryo extracts. Lane 1, 10% input; lane 2, empty beads; lane 3, rabbit anti-GFP antibody; and lane 4, rabbit anti-Cen antibody. Cen pulls down itself (top) and FMRP (middle and bottom). Lower blot shows increased exposure to highlight FMRP, with lane 1 cropped due to oversaturated signal. (D) Blot shows FMRP levels in 0–2-h (up to NC 14) embryos of the indicated genotypes using anti-FMRP antibody. (E) RNA-immunoprecipitation where RT-PCR reactions were run in the presence (+) or absence (–) of reverse transcriptase (RT). Lanes 1 and 2, 10% input; lanes 3 and 4, empty beads; lanes 5 and 6, rabbit anti-GFP antibody; and lanes 7 and 8, rabbit anti-Cen antibody. Middle image shows increased exposure to highlight cen; note that lanes 1 and 2 were cropped owing to oversaturated signal. (F) Blots from FMRP-GFP immunoprecipitation using 0–2-h WT or FMRP-GFP (FMRP) embryonic extracts and GFP-Trap beads probed with rabbit anti-GFP (top), rabbit anti-Cen (middle), and mouse anti-β-Tub antibodies (bottom). GFP pulls out FMRP-GFP and Cen protein. Bracket denotes nonspecific and/or degradation products. (G) RNA-immunoprecipitation from GFP-Trap beads, where Fmr1 (positive control; Ling et al., 2004) and cen mRNAs are pulled down (last lane), while His3.3B mRNA (negative control) is not. Full gels/blots available on FigShare (see Materials and methods). Scale bars: 10 µm; 1 µm (insets). a.u., arbitrary units.
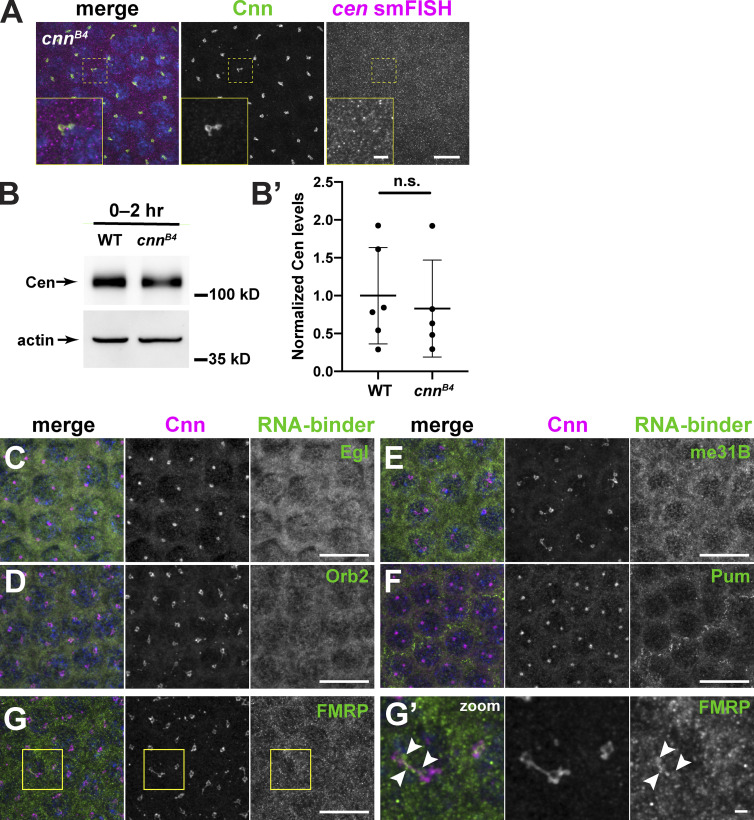
Cen interacts directly with the centrosome scaffold protein Cnn, and a point mutation in Cnn, cnnB4, is sufficient to disrupt Cen–Cnn binding and, consequently, Cen protein localization to centrosomes (Kao and Megraw, 2009). To test whether the centrosome scaffold is required for RNA localization, we examined if cen mRNA localized to pericentrosomal granules in cnnB4 mutants. We found that cen mRNA no longer formed granules in cnnB4 embryos and instead appeared dispersed throughout the cytoplasm (Fig. S2 A). This behavior subsequently allowed us to test if cen mRNA granules were required for Cen translation. We observed no difference in the levels of Cen protein in WT versus cnnB4 mutant 0–2-h embryos (Fig. S2, B and B′). These data suggest that the cen mRNA granule is not required for normal steady-state levels of Cen protein; however, an important caveat is that maternal deposition of Cen may obscure changes resulting from granule loss. Nonetheless, our data support a model in which the centrosome scaffold contributes to the formation and/or localization of the cen mRNA granule, likely via associations between Cen and Cnn.
Figure S2.
cen mRNA granule formation requires the centrosome scaffold. (A) Image shows immunofluorescence for Cnn (green) and cen smFISH (magenta) in an NC 12 cnnB4 embryo. Boxed region is enlarged in inset. Note the absence of large pericentrosomal cen mRNA granules. (B) Immunoblots show Cen protein content in 0–2-h (up to NC 14) WT and cnnB4 lysates. Actin is used as a loading control. (B′) Each dot represents the levels of Cen normalized to the mean relative expression of the actin load control. n.s., not significant (P = 0.672) by unpaired t test from n = 3 independent biological replicates, with n = 2 technical replicates run on the same gel. (C–G′) Images show interphase NC 12 embryos stained for Cnn (magenta) and antibodies for the indicated RNA-binding proteins (RNA-binder, green): Egl (C), Orb2 (D), me31B (E), Pum (F), and FMRP (G). (G′) Inset from G; arrowheads, FMRP overlapping with Cnn. Scale bars: 10 µm; 2 µm (insets).
FMRP associates with cen granules
RNA granules are diverse structures, and RNA-binding proteins are crucial for their formation and function (Singh et al., 2015). Therefore, to provide mechanistic insight into the regulation of the cen mRNA granule, we assayed the centrosomal localization of a few candidate RNA-binding proteins, including maternal expression at 31B (me31B), Pumilio (Pum), Egalitarian (Egl), Orb2, and FMRP (Deshpande et al., 2006; Dienstbier et al., 2009; Gamberi et al., 2006; Fig. S2, C–G′). Among these, FMRP appeared to be cytoplasmic, with a subset of puncta overlapping with centrosomes (Fig. S2 G′, arrowheads). We selected FMRP for further analysis.
To further investigate the composition of cen mRNA granules, we probed for biochemical interactions. We isolated endogenous Cen protein complexes from embryos by immunoprecipitation and detected a specific association with FMRP (Fig. 3 C). Specificity of the FMRP antibody was validated by immunoblot (Fig. 3 D). We similarly coisolated cen mRNA, but not the control gapdh, from Cen pull-downs (Fig. 3 E). The interaction between Cen and FMRP was confirmed by using a construct that expresses FMRP-GFP under endogenous regulatory elements (Sudhakaran et al., 2014) and reversing the direction of immunoprecipitation. Immunoblotting confirmed that FMRP-GFP was not overexpressed relative to endogenous FMRP levels (Fig. 3 D). Using GFP-Trap beads, we isolated FMRP protein and detected a specific association with Cen (Fig. 3 F). FMRP associates with Fmr1 mRNA (Ling et al., 2004). We confirmed this interaction and also detected a specific interaction between FMRP and cen mRNA, but not the control Histone H3.3B (His3.3B; Fig. 3 G). Therefore, the cen mRNA granule represents a ribonucleoprotein (RNP) complex comprising cen mRNA, Cen, FMRP, and likely other constituents. These data also hint that FMRP may mediate aspects of cen mRNA regulation.
FMRP functions as a negative regulator of cen mRNA granule formation and localization to centrosomes
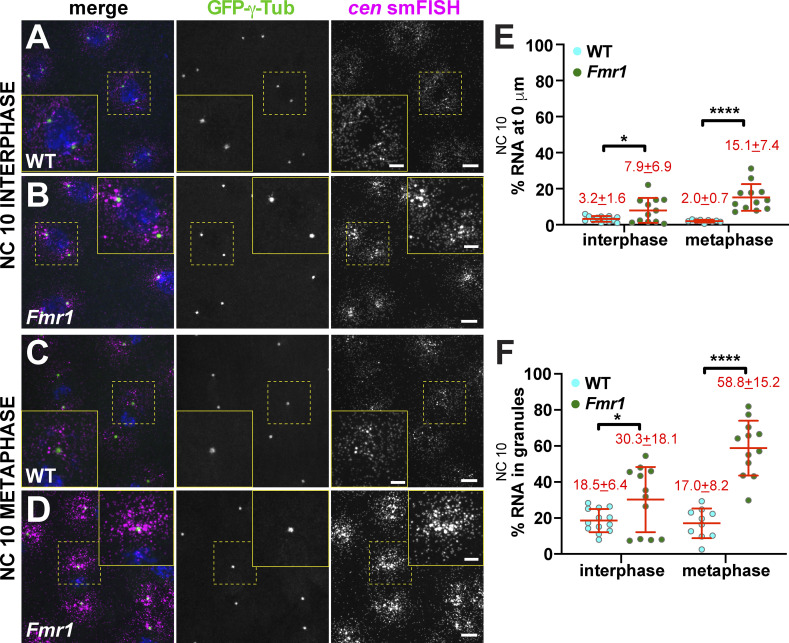
FMRP is a multifunctional RNA-binding protein implicated in RNA localization, stability, and translational regulation (Banerjee et al., 2018). To determine if FMRP contributes to cen regulation, we first compared the localization of cen mRNA in WT versus Fmr1 null mutant embryos expressing the PCM marker GFP-γ-Tubulin (GFP-γ-Tub). In WT NC 10 interphase embryos, cen mRNA was predominantly distributed as single molecules enriched near centrosomes, as we previously noted (Fig. 2 G; Fig. 4 A and inset). In Fmr1 embryos, however, significantly more cen mRNA clustered near centrosomes (Fig. 4, B–D). Quantification confirmed that cen mRNA localization to centrosomes and residence within RNA granules were both significantly higher in Fmr1 embryos relative to WT (Fig. 4, E and F). These data suggest that FMRP regulates cen mRNA granule formation and localization to centrosomes.
Figure 4.
Fmr1 regulates cen mRNA granule formation. Maximum-intensity projections of cen smFISH (magenta) in NC 10 embryos expressing GFP-γ-Tub (green). Boxed regions are enlarged at right (yellow box, zoom). (A) WT NC 10 interphase embryo with cen mRNA at centrosomes. (B) Fmr1 embryo with more granular, pericentrosomal cen mRNA. (C) WT NC 10 metaphase embryo. (D) Fmr1 embryo showing increased cen mRNA at centrosomes. (E) Percentage of cen mRNA overlapping with centrosomes in WT versus Fmr1 embryos. (F) Percentage of cen mRNA within granules. Each dot is a single measurement from n = 13 interphase and 10 metaphase WT and n = 12 interphase and metaphase Fmr1 embryos. Mean ± SD displayed (red). Table 1 lists number of objects quantified per condition. *, P < 0.05; ****, P < 0.0001 by unpaired t test. Scale bars: 10 µm; 2.5 µm (insets).
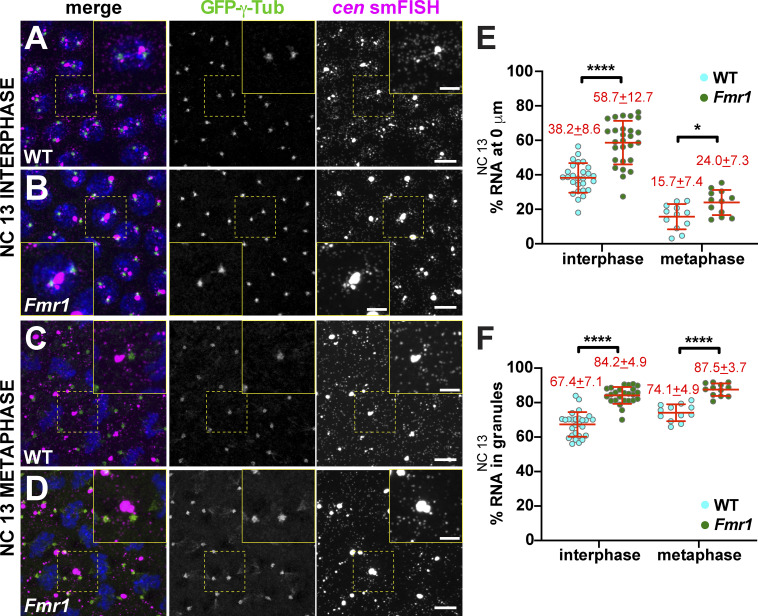
We noted similar trends later in development, during NC 13, when significantly more cen mRNA distributed to pericentrosomal granules in Fmr1 embryos than WT (Fig. 5, A–D). Quantification highlighted a significant enrichment of cen mRNA at centrosomes and within RNA granules in Fmr1 embryos relative to WT, particularly during interphase (Fig. 5, E and F), suggesting that FMRP normally limits cen mRNA localization to centrosomes. In sum, loss of FMRP is associated with more cen mRNA localized to granules, which reside closer to and are more likely to overlap with centrosomes.
Figure 5.
FMRP regulates cen mRNA localization to centrosomes. Maximum-intensity projections of cen smFISH (magenta) in NC 13 embryos expressing GFP-γ-Tub (green). Boxed regions are enlarged at right (yellow box, zoom). (A) WT interphase embryo showing cen mRNA granules. (B) Fmr1 embryo with increased cen mRNA in pericentrosomal granules. (C) WT metaphase embryo with cen mRNA displaced from the centrosome. (D) Fmr1 embryo with cen mRNA at centrosomes. (E) Percentage of cen mRNA overlapping with centrosomes in WT versus Fmr1 embryos. (F) Percentage of cen mRNA within granules. Each dot is a single measurement from n = 27 interphase and n = 12 metaphase WT or Fmr1 NC 13 embryos; mean ± SD displayed (red). Table 1 lists number of objects quantified per condition. *, P < 0.05; ****, P < 0.0001 by unpaired t test. Scale bars: 10 µm; 2.5 µm (insets).
FMRP regulates the abundance of cen mRNA and protein
The early embryo is largely transcriptionally inactive for the first 2 h of development, and most RNAs are maternally endowed (Anderson and Lengyel, 1979). Thus, the enhanced formation of cen mRNA granules in Fmr1 mutants could be attributed to changes in mRNA localization, increased mRNA stability, or both.
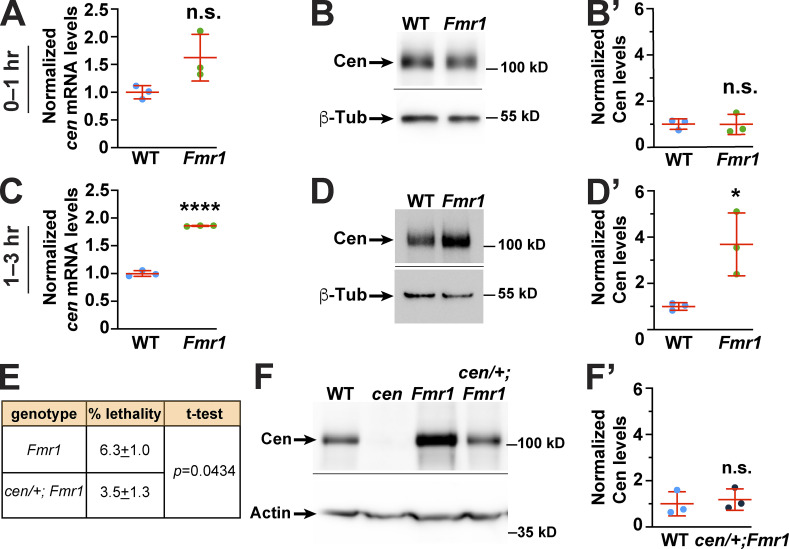
To test if FMRP regulates cen mRNA levels, we examined normalized cen mRNA levels by qPCR. We found no significant change in cen mRNA levels in Fmr1 versus WT 0–1-h embryos, a period encompassing up to NC 7 (P = 0.07 by unpaired t test; Fig. 6 A; Foe et al., 1993). FMRP functions primarily as a translational repressor, and deregulation of FMRP targets in neurons is considered a significant driver of fragile X syndrome pathophysiology (Banerjee et al., 2018; Darnell, 2011). In 0–1-h embryos, total levels of Cen protein were unaffected by loss of Fmr1 (P = 0.9 by unpaired t test; Fig. 6, B and B′). In contrast, within 1–3-h embryos (∼NC 7–14), cen mRNA levels increased 1.8-fold in Fmr1 mutants relative to controls (P < 0.0001 by unpaired t test; Fig. 6 C), and Fmr1 embryos contained significantly more Cen protein than controls (3.7-fold increase relative to WT, P = 0.03 by unpaired t test; Fig. 6, D and D′). Thus, both cen mRNA and protein levels are increased in later-stage Fmr1 embryos. Taken together, these data suggest that FMRP may contribute to cen mRNA turnover and/or translational repression. Although cen mRNA localization and levels may be coupled, such that increased cen mRNA content accounts for augmented cen mRNA localization to centrosomes and translation in Fmr1 mutants, we cannot rule out the possibility that FMRP contributes to multiple aspects of cen mRNA posttranscriptional regulation, either directly or indirectly.
Figure 6.
FMRP regulates levels of cen mRNA and protein levels. (A) Levels of cen RNA were normalized to RP49 as detected by qPCR from 0- to 1-h (up to NC 7) embryos. (B) Blots show Cen protein levels relative to the β-Tub loading control from 0–1-h embryos and quantified in B′. (C) Normalized levels of cen RNA from 1–3-h (∼NC 7–NC 14) embryos. (D) Blots show Cen protein in 1–3-h embryos and quantified in D′. (E) Embryonic lethality rates in Fmr1 versus cen/+; Fmr1 embryos. The mean ± SD is presented from n = 3 biological replicates; P was calculated by unpaired t test. (F) Blots show Cen protein in 1–3-h embryos and quantified in F′. Each dot is a measurement from an independent experiment; mean ± SD are displayed (red). Data in A–D′ and F′ are normalized to the mean relative expression of the controls from n = 3 biological replicates. *, P < 0.05; **** P < 0.0001 by unpaired t test. Full-sized blots available on FigShare (see Materials and methods). n.s., not significant.
cen and FMRP functionally interact to regulate cell division and embryonic viability
FMRP has established roles in mitotic progression. In Drosophila embryos, loss of FMRP results in severe mitotic defects, including improper centrosome separation, loss of mitotic synchrony, and faulty cellularization (Deshpande et al., 2006; Monzo et al., 2006; Papoulas et al., 2010; Sullivan et al., 1993). Using hatch rate analysis as a measure of embryonic viability, we found that while Fmr1 mutants show an average of 6.3% unhatched embryos, cen hemizygosity partially restored viability (P < 0.05 vs. Fmr1 by unpaired t test; Fig. 6 E). Western blot analysis confirmed that cen hemizygosity normalized Cen protein levels in Fmr1 embryos (Fig. 6, F and F′). These data are consistent with a genetic interaction between cen and Fmr1; moreover, they implicate elevated Cen dosage as a driver of Fmr1-mediated embryonic lethality.
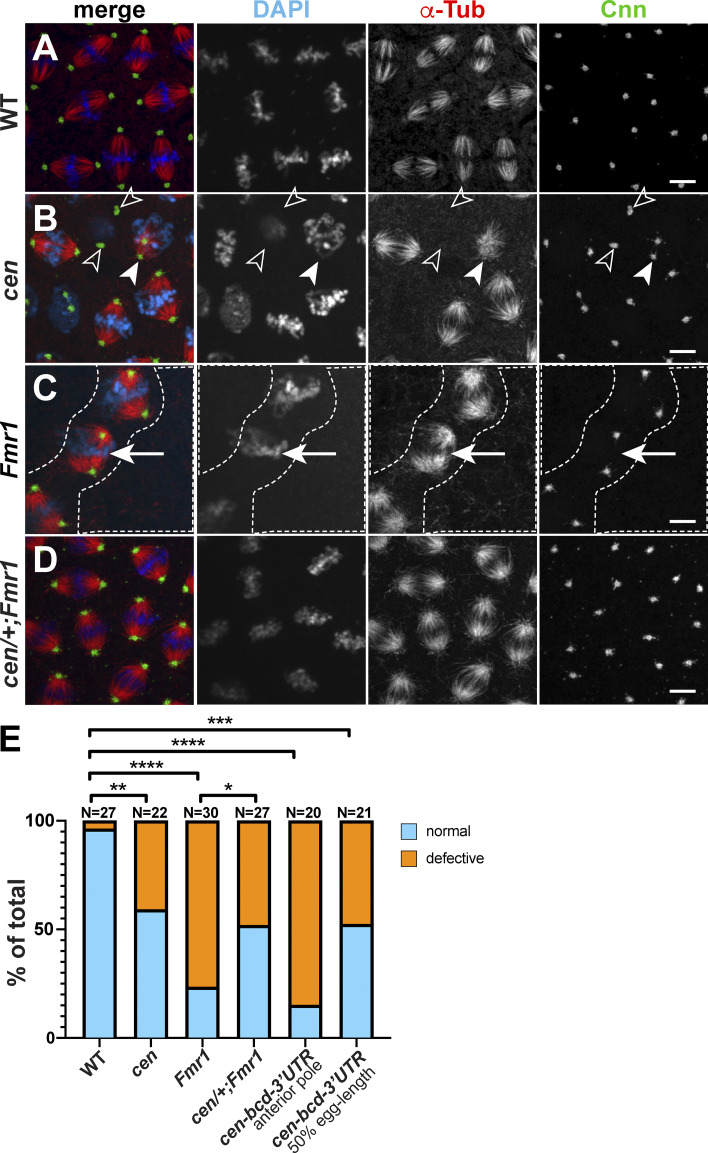
To test if cen genetically modifies the mitotic defects observed in Fmr1 mutant embryos, we tabulated the incidence of abnormal microtubule spindles, including bent, multipolar, monopolar, or fused spindles (Materials and methods). Occasionally, even WT embryos contained aberrant microtubule spindles (3.7%, n = 1/27 embryos; Fig. 7, A and E). However, cen mutant embryos showed increased rates of spindle errors (40.9%, n = 9/22 embryos, P < 0.01 vs. WT; Fig. 7, B and E, arrowheads), in agreement with prior observations (Bergalet et al., 2020; Kao and Megraw, 2009). Similarly, loss of Fmr1 was associated with high rates of spindle defects (76.1%, n = 23/30 embryos, P < 0.0001 cf. WT; Fig. 7, C and E, arrows). Consistent with this result, areas of reduced nuclear density (i.e., nuclear fallout) were noted in Fmr1 embryos (Fig. 7 C, dashed lines). Reducing cen dosage, however, ameliorated the spindle defects in Fmr1 mutants (48.1%, n = 13/27 embryos, P < 0.05; Fig. 7, D and E). Together, these data demonstrate that normal cen dosage is required for spindle morphogenesis and that the up-regulation of cen in Fmr1 embryos contributes to an increased rate of spindle errors and embryonic lethality.
Figure 7.
Cen and FMRP ensure proper mitosis. Maximum-intensity projections of metaphase NC 11 embryos from the indicated genotypes stained for β-Tub to label microtubules (red), Cnn (green), and DAPI (blue). (A) WT show uniform bipolar mitotic spindles. (B) cen embryo with reduced microtubules (open arrowheads) and incomplete centrosome separation (closed arrowheads). (C) Fmr1 embryo with nuclear fallout (dashed lines) and bent spindles (arrows). (D) Hemizygosity for cen partially rescues Fmr1 mutants. (E) Frequency of spindle defects from n = 27 WT, 22 cen, 30 Fmr1, 27 cen/+;Fmr1, 20 cen-bcd-3′UTR (anterior), and 21 cen-bcd-3′UTR (50% egg length) embryos. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by Fisher’s exact test. Scale bars: 5 µm.
Ectopic cen mRNA localization disrupts mitosis
Our data support a model whereby the local concentration of cen mRNA contributes to proper cell cycle progression. To test this model, we engineered a chimeric RNA comprising the cen coding sequence and the bicoid (bcd) 3′UTR, previously shown to be sufficient to mislocalize target RNAs to the anterior pole (Macdonald and Struhl, 1988). For these experiments, we examined embryos from mothers expressing the cen-bcd-3′UTR transgene in the context of the cen null background (hereafter, cen-bcd-3′UTR embryos).
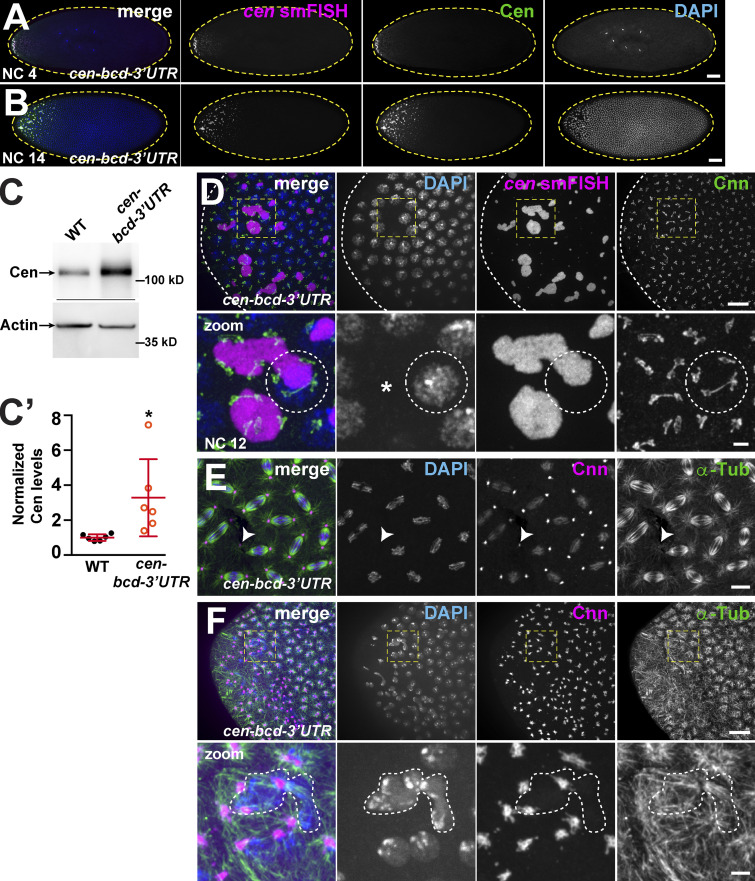
We first confirmed that our transgenic construct successfully mistargeted cen mRNA to the anterior. cen-bcd-3′UTR embryos showed a crescent of cen mRNA and protein at the anterior pole (Fig. 8, A and B). Immunoblotting showed that Cen protein is overexpressed in cen-bcd-3′UTR embryos, similar to Fmr1 mutants (approximately threefold increase vs. WT, P < 0.05 by unpaired t test; Fig. 8, C and C′). Given its restricted localization to the anterior pole, the cen-bcd-3′UTR transgene allowed us to simultaneously test opposing effects of cen dosage. We examined the effect of excess cen by visualizing the anterior pole, and we examined the effect of local cen mRNA depletion by visualizing the embryo midregion.
Figure 8.
Ectopic localization of cen mRNA disrupts nuclear divisions. Maximum-intensity projections of cen-bcd-3′UTR embryos (derived from females expressing a pUASp-cen-bcd-3′UTR transgene and the maternal α-Tub GAL4 driver in the cen null background). (A and B) Embryos labeled with cen smFISH (magenta), DAPI (blue), and Cen (green) with a gradient of cen mRNA and protein (A) and disrupted nuclear spacing at the anterior pole (B). (C) Blots show Cen protein in 1–3-h (∼NC 7–NC 14) embryos and quantified in C′. Cen levels were normalized to the mean WT levels of actin from n = 3 independent biological replicates with n = 2 technical replicates run on the same gel. Mean ± SD is displayed (red). *, P < 0.05 by unpaired t test. (D) NC 12 anterior with large cen RNPs (magenta) decorated by centrosomes (Cnn, green). Dashed circle outlines nucleus and part of a cen RNP with supernumerary centrosomes. (E) NC 12 embryo at ∼50% egg length; arrowhead marks a detached centrosome. (F) NC 12 embryo at anterior pole with disorganized microtubules (α-Tub, green), centrosome position (Cnn, magenta), and dysmorphic nuclei (DAPI; dashed lines). Boxes enlarged below (zoom). Scale bars: 50 µm (A and B); 10 µm (D–F); and 2 µm (insets).
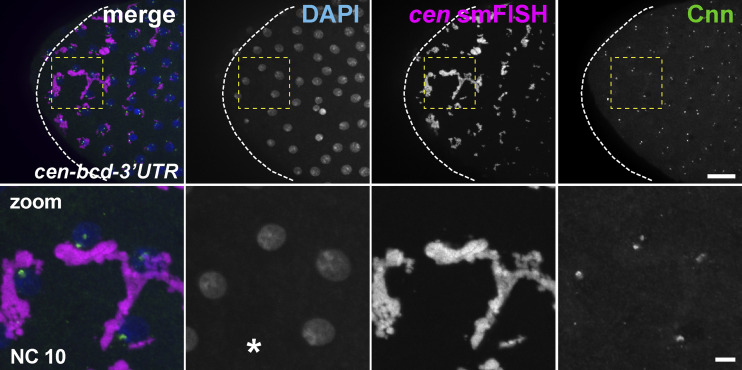
At the anterior of cen-bcd-3′UTR embryos, cen mRNA and protein coalesced into massive RNPs (Fig. 8 D). Consistent with precocious formation of cen mRNA granules, cen-bcd-3′UTR RNPs were also prominent during NC 10 (Fig. S3). Through the use of reporter constructs, it was recently demonstrated that the cen coding sequence is sufficient for centrosome targeting (Bergalet et al., 2020). In agreement with these data, the enlarged cen RNPs observed in cen-bcd-3′UTR embryos associated with centrosomes at the anterior pole (Fig. 8 D, dashed circle). Furthermore, we did not observe cen mRNA or protein localized to more distal centrosomes in cen-bcd-3′UTR embryos, indicating that localization elements within the bcd-3′UTR confine cen mRNA to the anterior pole. These findings further suggest that localization of cen mRNA is necessary and sufficient for Cen protein localization to centrosomes.
Figure S3.
Deregulation of cen mRNA granule formation. (A) Immunoblots show Cen protein content relative to the actin loading control from 1–3-h embryonic extracts and are quantified in A′. Levels of Cen were normalized to the mean WT levels of actin from n = 3 independent biological replicates, each with n = 2 technical replicates run on the same gel. (B) Maximum-intensity projection of an interphase NC 10 embryo for cen mRNA labeled by smFISH (magenta), DAPI (nuclei, blue), and Cnn showing large cen RNPs. Asterisk marks nuclear fallout. Boxed regions enlarged below (zoom). Scale bars: 10 µm; 2 µm (insets).
The restricted localization of cen mRNA and protein to the anterior pole within cen-bcd-3′UTR embryos allowed us to test whether cen mRNA was required locally for error-free mitosis. Examination of mitotic spindles at ∼50% egg length, an area devoid of cen mRNA and protein, revealed an increased rate of microtubule spindle defects (47.6%, n = 10/21 embryos, P < 0.001 vs. WT; Figs. 7 E and 8 E). Together with recent work showing that the native cen 3′UTR recruits I-kappaB kinase ε (IKKε or ik2) mRNA to centrosomes (Bergalet et al., 2020), these data suggest that cen mRNA functions locally to support spindle integrity, perhaps in concert with ik2 mRNA.
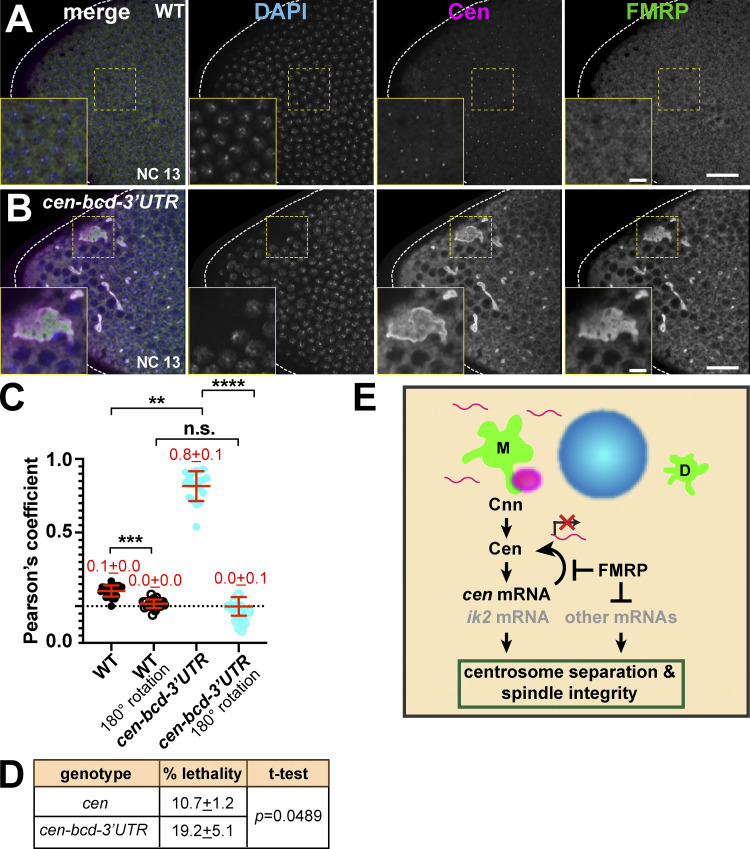
The anterior pole of cen-bcd-3′UTR embryos showed lower nuclear density (i.e., nuclear fallout), dysmorphic nuclei, and mitotic asynchrony, indicative of disrupted nuclear divisions (Fig. 8 F). In accordance with these findings, 85% of cen-bcd-3′UTR embryos displayed spindle defects at the anterior (n = 17/20 embryos, P < 0.0001 vs. WT; Fig. 7 E), as well as nuclei associated with supernumerary centrosomes (Fig. 8 D dashed circle). Notably, the large RNPs that form in cen-bcd-3′UTR embryos were sufficient to recruit FMRP, as evidenced by a significant overlap of Cen and FMRP signals (P < 0.01; Fig. 9, A–C). In addition to overlapping Cen and FMRP signals, we note a concentration of Cen residing at the periphery of the enlarged cen RNPs (Fig. 9 B, insets). Given these phenotypes, we next examined embryonic viability. While cen mutant embryos show an elevated rate of unhatched embryos relative to controls, as previously noted (10.7% unhatched; (Kao and Megraw, 2009), cen-bcd-3′UTR embryos had increased lethality (mean 19.2%; P < 0.05 vs. cen by unpaired t test; Fig. 9 D). We propose a model in which the deregulated balance of Cen levels impairs mitotic spindle organization (Fig. 9 E).
Figure 9.
Deregulation of cen mRNA impairs viability. (A and B) Single optical sections of interphase NC 13 embryos stained for Cen and FMRP; boxes enlarged in inset. (C) Pearson’s coefficient of Cen and FMRP signals; each dot is a measurement from n = 25 WT and n = 19 cen-bcd-3′UTR embryos from two independent experiments. One channel was rotated 180° to test for specificity of colocalization. Mean ± SD is displayed (red). **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by Kruskal-Wallis followed by Dunn’s multiple comparisons test. Scale bars: 20 µm (A and B); 5 µm (insets). n.s., not significant. (D) Embryonic lethality rates in cen versus cen-bcd-3′UTR embryos from three independent experiments. Mean ± SD is shown; P calculated by unpaired t test. (E) Model for FMRP-mediated regulation of cen mRNA. A direct interaction docks Cen to Cnn (green) at the centrosome (Kao and Megraw, 2009). Cen protein interacts with cen mRNA (magenta), which also recruits ik2 mRNA (Bergalet et al., 2020). Our data suggest that cen mRNA localization, organization into granules and levels, and translation are regulated by FMRP. Further, cen mRNA is an important target of FMRP required for spindle integrity and viability.
Discussion
Centrosome-localized RNA has been described in a variety of organismal contexts, and while the conserved feature of mRNA at centrosomes hints at a biological function, the underlying physiological significance has remained unclear (Marshall and Rosenbaum, 2000; Ryder and Lerit, 2018). To address this question, we systematically examined five transcripts shown to enrich near spindle poles to quantitatively define their common and unique localization patterns in Drosophila embryos. We identified subsets of mRNAs showing centrosome enrichment in a cell cycle–regulated and developmentally regulated manner. These nonrandom variances in RNA distributions further imply biological relevance. We tested if RNA localization contributes to normal centrosome functions through in-depth studies with a model transcript, cen mRNA. We identified FMRP as an RNA-binding protein required for regulation of cen RNA localization, organization, and translational control. Further, reducing cen dosage rescued Fmr1-dependent mitotic errors and embryonic lethality. We also directly tested the consequences of mistargeting cen mRNA. Mislocalization of cen mRNA to the anterior abrogated the normal localization of Cen to more distal centrosomes and disrupted spindle organization. Anterior mitotic divisions were also severely disrupted due to the increased local concentration of cen mRNA, which also recruited FMRP. These studies suggest that a normalized local concentration of cen mRNA is essential for normal cell division and genome stability.
Centrosomes as platforms for translational regulation
FMRP is a multifunctional RNA-binding protein with roles in translational repression, activation, RNA localization, and RNA stability (Darnell, 2011; Estes et al., 2008; Greenblatt and Spradling, 2018; Pilaz et al., 2016). In humans, mutations in the gene encoding FMRP, FMR1, are the leading cause of heritable intellectual disability and autism. Although high-throughput studies have identified putative RNA substrates, surprisingly few of these have been validated (Santoro et al., 2012). Our studies demonstrate that cen mRNA is regulated by FMRP, either directly or indirectly, and that titrating cen dosage is sufficient to partially restore embryonic viability in Fmr1 mutants. Consistent with direct regulation of cen mRNA by FMRP, the cen coding sequence contains six putative binding motifs for FMRP, according to RBPmap, an RNA-binding motif predictor (Paz et al., 2014). Moreover, human orthologues of cen, CDR2 and CDR2L, were identified as direct FMRP targets (Ascano et al., 2012). Deregulation of CDR2 and CDR2L is associated with paraneoplastic cerebellar degeneration (Albert et al., 1998; Corradi et al., 1997). Our studies suggest that Drosophila cen may serve as a valuable model to uncover mechanisms underlying FMRP-mediated regulation of CDR2 and CDR2L. Whether FMRP similarly regulates other centrosome-localized mRNAs is an interesting question for future study.
The enhanced recruitment of cen mRNA to heterogeneously sized pericentrosomal granules, coupled with the increased production of Cen protein within Fmr1 mutants, led us to speculate that cen mRNA granules may be sites of local translation, as recently proposed (Bergalet et al., 2020). However, disruption of cen granule formation, as in cnnB4 mutants, does not impair total Cen protein levels. This finding raises the possibility that Cen may be translated at alternate sites or that maternal stores of Cen obscure changes resulting from cen mRNA granule loss. These models are not mutually exclusive, and cen mRNA may be translated at multiple locales. Our data support a model in which centrosomes serve as platforms for translation control, which may be positive or negative depending on the specific transcript and/or cell cycle stage, consistent with the idea that cen mRNA granules are sites of Cen translational regulation (Bergalet et al., 2020).
We show that cen mRNA preferentially localizes to interphase centrosomes; that the centrosome scaffold, Cnn, is required for cen mRNA granule formation and localization; and that FMRP functions as a negative regulator of cen mRNA, limiting cen mRNA stability and translation of Cen protein (Fig. 9 E). We speculate that FMRP represses Cen translation within cen mRNA granules, dampening the local Cen concentration. Consequently, cen mRNA enrichment at centrosomes is exaggerated in Fmr1 mutants. Other factors likely promote Cen translation. Translational repression or derepression may be coupled to cen mRNA granule centrosome proximity, which decreases as embryos enter mitosis. An imbalance of Cen levels at centrosomes, either too little (as in cen mutants) or too much (as in Fmr1 mutants or cen-bcd-3′UTR embryos), impairs centrosome function/spindle integrity and embryonic viability. As the cen 3′UTR recruits ik2 mRNA to centrosomes, the mitotic defects observed following cen perturbation may result from indirect effects via ik2 mRNA (Bergalet et al., 2020). Nonetheless, cen mRNA dosage must be properly regulated for mitotic fidelity.
Differential enrichment of mRNAs on interphase centrosomes
A common trend emerging from our comparative analyses is the greater enrichment of mRNA at interphase versus metaphase centrosomes. One possible explanation is the differential size of interphase centrosomes, which are significantly larger in Drosophila embryos owing to the elaboration of extended centrosome flares, part of the architecture of the centrosome scaffold (Lerit et al., 2015; Megraw et al., 2002; Richens et al., 2015). This pattern contrasts with mammalian centrosomes, which are larger in mitosis (Lawo et al., 2012). According to this size model, a larger centrosome might dock additional RNAs simply because of the increased volume it occupies in the cell. We discount this model based on our finding that a highly expressed control transcript, gapdh, does not enrich at interphase centrosomes. This result also argues against the idea that centrosomes recruit RNA molecules spuriously. Relatively few RNAs localize to centrosomes (Lécuyer et al., 2007; Raff et al., 1990). We show that the localization of centrosome-associated RNA is regulated in space and time.
Why do RNAs localize to interphase centrosomes? Recent work in mammalian cells proposed that some lengthy transcripts may be cotranslationally transported to centrosomes (Chouaib et al., 2020; Sepulveda et al., 2018). This model would account for contemporaneous recruitment and colocalization of centrosome mRNA and proteins and may be pertinent to cen mRNA localization. Of the RNAs overlapping with the centrosome surface, sov was unique in that it appeared to preferentially dock along centrosome flares, localizing to the outer PCM zone. However, we do not detect Sov protein at centrosomes. Instead, Sov resides in the nucleus during interphase and is undetectable after nuclear envelope breakdown (Benner et al., 2019). These findings suggest that Sov is rapidly translocated into the nucleus. Live imaging of RNA transport and nascent protein synthesis is required to rigorously test the dynamics of RNA localization and local translation.
Another model that may account for enrichment of centrosome RNAs at interphase centrosomes is the possibility that RNA contributes to centrosome structure, perhaps by promoting phase transitions (Woodruff et al., 2015, 2017; Zwicker et al., 2014). A common principle of phase transitions is the association of intrinsically disordered proteins with specific RNA molecules to form non–membrane-bound organelles with unique biophysical properties (Berry et al., 2018). Might cen mRNA granules represent phase-separated domains? Congruous with phase separation, Cen protein contains multiple predicted intrinsically disordered domains (Ishida and Kinoshita, 2007). While we cannot rule out the contribution of all centrosomal RNAs, our studies do not suggest that cen mRNA contributes to centrosome structure. Mistargeting cen mRNA to the anterior cortex did not appear to disrupt the organization of distal centrosomes, for example.
Critically, disrupting the PCM scaffold is sufficient to inhibit formation of the cen mRNA granule. We previously showed that the PCM scaffold becomes progressively more structured during the prolonged interphases of later NCs (Lerit et al., 2015). Additionally, the mother centrosome organizes a larger PCM scaffold owing to inherently greater levels of Cnn and PLP (Conduit et al., 2010; Lerit et al., 2015). Collectively, these features may account for the asymmetric localization of cen mRNA to mother centrosomes in late-stage syncytial embryos. These data lead us to conclude that the PCM scaffold organized by Cnn and PLP is upstream of the recruitment and organization of cen mRNA granules (Fig. 9 E).
Many types of RNP granules form within cells, including stress granules, germ granules, P-bodies, etc., which all have unique functions and modes of assembly. The spatial proximity of multiple RNA molecules may facilitate intermolecular RNA interactions subsequently recognized by RNA-binding proteins (Van Treeck and Parker, 2018). The FMRP-containing cen mRNA granule represents one such RNP, and further understanding how it promotes mitotic integrity warrants further investigation. As the early Drosophila embryo is transcriptionally quiescent, posttranscriptional regulatory mechanisms, and especially translational control, are fundamentally important for proper centrosome regulation and function.
Materials and methods
Fly stocks
The following Drosophila strains and transgenic lines were used: y1w1118 (1495; Bloomington Drosophila Stock Center) was used as the WT control unless otherwise noted; PBAC-GFP-Cnn, which expresses Cnn tagged at the N-terminus with EGFP under endogenous regulatory elements (Lerit et al., 2015); Ubi-GFP-γ-Tub23C, which expresses GFP-γ-Tub under the Ubiquitin promotor (Lerit and Rusan, 2013); null cen mutant embryos derived from homozygous cenf04787 animals (18805; Bloomington Drosophila Stock Center; Kao and Megraw, 2009); null Fmr1 mutant embryos derived from Fmr1Δ113M/Fmr13 trans-heterozygotes (Fmr1Δ113M; 67403; Bloomington Drosophila Stock Center; Zhang et al., 2001); Fmr13, gift from T. Jongens, University of Pennsylvania, Philadelphia, PA; Dockendorff et al., 2002); and hypomorphic cnnB4 mutants, a gift from T. Megraw (Florida State University, Tallahassee, FL). The maternal γ-Tub promoter was used to control GAL4 expression (matGAL4; 7063; Bloomington Drosophila Stock Center) to drive expression of pUASp-cen-bcd-3′UTR (this study). FMRP-GFP is a recombineered line expressing FMRP tagged at the C-terminus with GFP under endogenous regulatory elements (gift from M. Ramaswami, Trinity College Dublin, Dublin, Ireland; Sudhakaran et al., 2014). In all experiments, mutant embryos represent progeny derived from mutant mothers to examine maternal effects. Flies were raised on molasses-based Drosophila medium, and crosses were maintained at 25°C in a light- and temperature-controlled chamber.
Construction of transgenic animals
To generate pUASp-cen-bcd-3′UTR, the cen coding sequence was PCR amplified using Phusion high-fidelity DNA polymerase from the cDNA clone LD41224 (Drosophila Genomics Resource Center) using the primers 5′-GCAGGCTCCGCGGCCGCCCCCTTCACCAGGATGGAGGAATCCAATCACGGTTC-3′ and 5′-GAAACTCTCTAACAGCCTCTCATCCAGGTTACTTTTGACGAAACTGATGATGATGACTC-3′. The cen start and stop codons are underlined. The bcd-3′UTR was PCR amplified using Q5 high-fidelity polymerase (M0491S; New England Biolabs) from genomic DNA using the primers 5′-GAGTCATCATCATCAGTTTCGTCAAAAGTAACCTGGATGAGAGGCGTGTTAGAG-3′ and 5′-CTGGGTCGGCGCGCCCACCCTTGTCTAGGTAGTTAGTCACAATTTACCCGAGTAGAGTAG-3′. The cen-bcd-3′UTR fusion was assembled and directionally cloned into the pENTR-D vector (Invitrogen) by Gibson assembly using fivefold molar excess of the bcd-3′UTR. Sequence-verified single-colony clones were shuttled into the destination vector pPW (UASp promoter) using the Gateway cloning system (Invitrogen). Transgenic animals were generated by BestGene (Chino Hills, CA).
Embryonic hatch rate analysis
24-h collections of eggs were collected on yeasted grape juice agar plates, transferred to fresh plates, and aged for 48 h at 25°C. Unhatched embryos were counted from a total of ∼600 embryos, and data presented are mean ± SD from three biological replicates.
Immunofluorescence
Embryos were prepared for immunofluorescence as described in Lerit et al. (2015). Briefly, samples were fixed in PFA, blocked extensively in BBT (PBS supplemented with 0.1% Tween-20 and 0.1% BSA), and incubated overnight at 4°C with primary antibodies diluted in BBT. The next day, samples were further blocked in BBT supplemented with 2% normal goat serum and incubated with secondary antibodies and DAPI for 2 h at room temperature before being mounted in AquaPoly/Mount mounting medium (87001-902; VWR).
The following primary antibodies were used: rabbit anti-Cen (1:500; gift from T. Megraw; Kao and Megraw, 2009), rabbit anti-Cnn (1:3,500; gift from T. Megraw), mouse anti-α-Tub DM1α (1:500; T6199; Sigma-Aldrich), rabbit anti-Egl (1:2,000; gift from R. Lehmann, New York University, New York, NY), rabbit anti-Pum (1:1,000; gift from Martine Simonelig, Institute of Human Genetics, University of Montpellier, Montpellier, France), mouse anti-Orb2 (1:1,000; clone 4G8; Developmental Studies Hybridoma Bank), mouse anti-me31B (1:3,000; gift from A. Nakamura, Kumamoto University, Kumamoto, Japan); and mouse anti-FMRP (1:10; clone 5A11; Developmental Studies Hybridoma Bank). Secondary antibodies and stains were Alexa Fluor 488, 568, or 647 (1:500, Molecular Probes). DAPI was used at 10 ng/ml (Thermo Fisher Scientific).
Detection of RNA by smFISH
smFISH experiments were adapted from manufacturer’s recommended protocols. All steps were performed with RNase-free solutions. Briefly, fixed and rehydrated embryos were washed in PBST (PBS plus 0.1% Tween-20) and washed in wash buffer (WB; 10% formamide and 2× SSC supplemented fresh each experiment with 0.1% Tween-20 and 2 µg/ml nuclease-free BSA [0332-25G; VWR]). Embryos were then incubated with 100 µl of hybridization buffer (HB; 100 mg/ml dextran sulfate and 10% formamide in 2× SSC supplemented fresh each experiment with 0.1% Tween-20, 2 µg/ml nuclease-free BSA, and 10 mM ribonucleoside vanadyl complex (RVC; S1402S; New England Biolabs) for 10–20 min in a 37°C water bath. Stellaris smFISH probes conjugated to Quasar 570 dye (LGC Biosearch Technologies) were designed against the coding region for each gene of interest using the Stellaris RNA FISH probe designer and stored at −20°C as stock solutions of 25 µM in nuclease-free water. Probes are listed in Table S1. After preincubation in HB, embryos were incubated in a 37°C water bath overnight in 25 µl of HB containing a 1:50 dilution of smFISH probe. The next morning, embryos were washed three times for 30 min each in prewarmed WB, stained with DAPI for 1 h at room temperature, washed with PBST, and mounted with Vectashield mounting medium (H-1000; Vector Laboratories). Slides were stored at 4°C and imaged within 1 wk.
For experiments in which immunofluorescence was combined with smFISH, we adapted a protocol from Xu et al. (2015). After overnight incubation with smFISH probes, embryos were washed well in WB, followed by two 10-min washes in 2× SSC–0.1% Tween-20 and four 10-min washes in PBST. Embryos were blocked for 2 h in blocking solution (PBS supplemented with 1 mg/ml nuclease-free BSA, 0.1% Tween-20, and 2 mM RVC, prepared fresh) and incubated overnight in primary antibodies at 4°C. The next day, embryos were washed well in blocking solution, incubated with secondary antibodies and DAPI at room temperature, and washed in PBST before being mounted in Vectashield.
Microscopy
Images were acquired on a Nikon Ti-E system fitted with a Yokogawa CSU-X1 spinning disk head (Yokogawa Corp. of America), Orca Flash 4.0 v2 digital complementary metal–oxide–semiconductor camera (Hamamatsu Corp.), Perfect Focus system (Nikon), and a Nikon LU-N4 solid-state laser launch (15 mW; 405, 488, 561, and 647 nm) using the following Nikon objectives: 100× 1.49-NA Apo total internal reflection fluorescence oil immersion, 40× 1.3-NA Plan Fluor oil immersion, and 20× 0.75-NA Plan Apo. Images were acquired at ambient temperature (∼25°C) using either Vectashield or Aqua-Poly/Mount imaging medium, as described. The microscope was powered through Nikon Elements AR software on a 64-bit HP Z440 workstation (Hewlett-Packard).
Image analysis
For fixed studies, Alexa Fluor 488, 568, or 647 was used. Images were assembled using Fiji (National Institutes of Health; Schindelin et al., 2012), Adobe Photoshop, and Adobe Illustrator software to separate or merge channels, crop regions of interest, generate maximum-intensity projections, and adjust brightness and contrast.
RNA detection and measurements
For quantification of single-molecule RNA distribution relative to centrosomes, single-channel .tif raw images were segmented in three dimensions using code adapted from the Allen Institute for Cell Science Cell Segmenter (Chen et al., 2018). Each segmented image was compared with the original image to validate accurate segmentation. RNA objects of ≥50 pixels in segmented images were identified, and object features were extracted. Extracted features included the raw image total pixel intensity, the object centroid coordinates, and the surface coordinates. Distances were measured from the surface of each RNA object to the surface of the closest centrosome.
For single-molecule normalization, we used a previously described method (Mueller et al., 2013). Single molecules of RNA are objects of 50–100 pixels, as determined by their diffraction-limited 200-nm size. For each RNA probe, we divided the integrated intensity of each RNA object by the averaged integrated intensity of all single-molecule RNAs, allowing an estimate of the number of RNA molecules per object. We then calculated the percentage of total RNA that overlapped with centrosomes. For cen and gapdh mRNAs, we calculated the percentage of RNA in granules, which is the fraction of RNA contained in objects estimated to have ≥4 RNAs. We selected 10 and 4 µm as the upper boundary for the pseudocell radius for NC 10 and NC 13, respectively, based on measuring the centrosome-to-centrosome distances from a set of representative images. A detailed protocol for RNA analysis was recently described (Ryder and Lerit, 2020).
Spindle morphology defects
Mitotic embryos imaged at 40× were examined for the following morphologies: bent spindles, multipolar or fused spindles, acentrosomal spindle poles, and defective centrosome separation. If any spindles within an embryo contained one of these phenotypes, the embryo was considered positive for a spindle morphology defect. Three independent biological replicates were performed for each genotype.
Colocalization analysis
Single optical slices were selected for analysis. For analysis of Cen protein overlap with cen RNA, the entire image was analyzed. For analysis of FMRP overlap with Cen protein in cen-bcd-3′UTR embryos, a 40 × 40-µm region of interest was selected using the Cen protein channel to include Cen protein aggregates. Colocalization was measured on background-subtracted and automatic threshold–masked images using the Coloc 2 plugin for Fiji (Schindelin et al., 2012).
Immunoblotting
Aged embryos were harvested, dechorionated in bleach, flash frozen in liquid nitrogen, and stored at −80°C. 5–10 mg of frozen embryos were lysed with a 1-ml glass dounce homogenizer (Wheaton) in 100 µl lysis buffer (50 mM Hepes, 150 mM NaCl, 2.5 mM MgCl2, 0.1% Triton X-100, and 250 mM sucrose supplemented with 1× EDTA-free protease inhibitor cocktail [04693159001; Roche]), 1 µg/ml Pepstatin A (P5318; Sigma-Aldrich), 1 mM DTT (10197777001; Sigma-Aldrich), and 2 mM RVC). 25 µl of 5× SDS loading dye was added and samples were boiled for 10 min at 95°C then resolved by SDS-PAGE gel and transferred to nitrocellulose membrane by wet transfer. Membranes were blocked for 1 h at room temperature in a 5% dry milk solution diluted in TBST (Tris-based saline with 0.05% Tween-20), washed well with TBST, and incubated overnight at 4°C with primary antibodies. After washing with TBST, membranes were incubated for 1 h in the following secondary antibodies diluted 1:5,000 in TBST, 5% milk: goat anti-mouse HRP (31430; Thermo Fisher Scientific) and goat anti-rabbit HRP (31460, Thermo Fisher Scientific). Membranes were washed well in TBST, and bands were visualized with Clarity ECL substrate (1705061; Bio-Rad) on a Bio-Rad ChemiDoc imaging system.
Densitometry was measured using Fiji software using the region-of-interest measure tool. For each sample, the ratio between the protein of interest and a loading control (e.g., β-Tub) was calculated. The mean relative expression and SD were calculated and normalized to the mean of the biological control. Three independent biological replicates were processed on the same gel.
The following primary antibodies were used: rabbit anti-Cen (1:1,000; gift from T. Megraw), mouse anti-FMRP (1:100; clone 5A11; Developmental Studies Hybridoma Bank); mouse anti-β-Tub (1:1,000; E7; Developmental Studies Hybridoma Bank); and mouse anti-actin (1:1,000; clone JLA20; Developmental Studies Hybridoma Bank).
Immunoprecipitation
∼30 mg of frozen embryos were lysed with a glass dounce in 100 µl lysis buffer (50 mM Hepes, pH 7.4, 150 mM NaCl, 2.5 mM MgCl2, 250 mM sucrose, and 0.1% Triton X-100) supplemented with 1× protease inhibitor cocktail, 1 µg/ml Pepstatin A, 1 mM DTT, 1 U/µl RNase Inhibitor (M0314S; New England Biolabs), and 2 mM RVC. Lysates were cleared by centrifugation, and the supernatant was precleared in 25 µl of washed Protein A/G magnetic agarose beads (88802; Pierce), or blocked magnetic beads (bmp-20; Chromotek) for GFP-Trap of FMRP-GFP, to reduce nonspecific binding. A 0.1 volume of precleared lysates was reserved as input, and the remainder was immunoprecipitated for 2 h at 4°C in rabbit anti-GFP (A-11122; Invitrogen), rabbit anti-Cen, or no antibody as a control and transferred to 25 µl washed Protein A/G magnetic agarose beads for immunoprecipitation for 2 h. GFP-Trap magnetic agarose beads (gtma-10; Chromotek) were used for FMRP-GFP. Beads were then washed well in immunoprecipitation buffer (lysis buffer with 8 U/ml RNase Out and 0.4 mM RVC) and resuspended in 100 µl immunoprecipitation buffer. 50 µl of the beads (20% of volume for GFP-Trap) were analyzed for protein content by SDS-PAGE as described above. RNA was extracted from the other 50 µl of beads (80% of volume for GFP-Trap) using TRI Reagent (T9424; Sigma-Aldrich) and treated with TURBO DNase (AM2238; Thermo Fisher Scientific) before RT-PCR.
cDNA was synthesized from 500 ng of RNA using Superscript IV Reverse Transcriptase (18091050; Thermo Fisher Scientific) according to the manufacturer’s protocol with (RT+) or without (RT–) reverse transcriptase. DNA was amplified by PCR using Phusion High Fidelity DNA Polymerase (M0530L; New England Biolabs). The following primers were used: cen forward, 5′-TAACCGCAGACGGACAAC-3′, and reverse, 5′-GAATGCCCTATGGCTAGAAT-3′; gapdh forward, 5′-CACCCATTCGTCTGTGTTCG-3′, and reverse, 5′-CAACAGTGATTCCCGACCAG-3′; Fmr1 forward, 5′-CATCGTTCGACGGAGTAACA-3′, and reverse, 5′-GGAGCTTGTTGTTGGCTGAT-3′; and His3.3B forward, 5′-CACTCCAACAACTGTCCAGC-3′, and reverse, 5′-GTCCAGCCGACGTTAGATTG-3′.
qPCR
RNA was extracted from ∼5 mg of frozen embryos using TRI Reagent and treated with Ambion Turbo DNase (AM2238; Thermo Fisher Scientific) for 30 min at 37°C, followed by phenol:chloroform extraction. On the same day, RNA concentrations were measured with a spectrophotometer, and cDNA was synthesized from 500 ng of RNA using the iScript kit according to the manufacturer’s protocol (170-8891; Bio-Rad).
qPCR was performed on a Bio-Rad CFX96 real-time system with iTaq Universal SYBR Green Supermix (172–5121; Bio-Rad). Three biological samples were tested in triplicate using 96-well plates (HSP9601; Bio-Rad). cen expression levels were normalized to Ribosomal protein L32 (RP49). The following primers were used: cen forward, 5′-TGAGGATACGACGCTCTGTG-3′, and reverse, 5′-AAAGTACCCCCGGTAACACC-3′, amplicon 78 bp; and RP49 forward, 5′-CATACAGGCCCAAGATCGTG-3′, and reverse, 5′-ACAGCTTAGCATATCGATCCG-3′, amplicon 75 bp.
Statistical analysis
Data were plotted and statistical analysis was performed using Microsoft Excel and GraphPad Prism software. To calculate significance, the distribution was first tested for outliers using the ROUT test with Q = 1%. Identified outliers were excluded from further analysis. Normality was then assessed with a D’Agnostino and Pearson normality test. Data were analyzed by Student’s two-tailed t test, ANOVA, Fisher’s exact test, or the appropriate nonparametric tests and are displayed as mean ± SD. Data for the percentage of RNA overlapping with centrosomes presented in Figs. 1 and 2 were analyzed as one dataset for each developmental stage using the appropriate one-way ANOVA test. Data shown are representative results from at least two independent experiments, as indicated in the figure legends.
Data availability
All code for RNA detection and measurements are available on github at https://github.com/pearlryder/rna-at-centrosomes and https://github.com/pearlryder/cen-at-fmr-null-centrosomes. Full-sized images of all DNA gels and immunoblots are available at FigShare: doi.org/10.6084/m9.figshare.12821564 and doi.org/10.6084/m9.figshare.12821579.
Online supplemental material
Fig. S1 shows the workflow used to analyze mRNA localization. Fig. S2 shows that the centrosome scaffold is required for cen mRNA granule formation. Fig. S3 shows that cen mRNA granules form precociously in cen-bcd-3′UTR embryos. Table S1 lists all smFISH probes used in this study.
Supplementary Material
lists all smFISH probes used in this study.
Acknowledgments
We thank Drs. Liz Gavis, Nasser Rusan, Tim Megraw, Greg Rogers, Ruth Lehmann, Mani Ramaswami, Martine Simonelig, Akira Nakamura, and Tom Jongens for gifts of reagents; Lauren Lym and Jina Lee for technical assistance; and L. Gavis, N. Rusan, and members of our laboratory for constructive feedback.
Stocks obtained from the Bloomington Drosophila Stock Center (National Institutes of Health grant P40OD018537); antibodies from the Developmental Studies Hybridoma Bank, created by the National Institute of Child Health and Human Development of the National Institutes of Health and maintained at the University of Iowa Department of Biology; and reagents from the Drosophila Genomics Resource Center (National Institutes of Health grant 2P40OD010949) were used in this study. This work was supported by National Institutes of Health grants 5K12GM000680 and 1F32GM128407 to P.V. Ryder, National Institutes of Health grants 5K22HL126922 and 1R01GM138544 to D.A. Lerit, and American Heart Association grant 20POST35210023 to J. Fang.
The authors declare no competing financial interests.
Author contributions: P.V. Ryder and D.A. Lerit conceived of the project and designed experiments to visualize and measure centrosome RNA localization. P.V. Ryder performed and analyzed all smFISH, qPCR, immunofluorescence, and Cen blotting experiments and collaborated with J. Fang for Fmr1 mutant data. J. Fang designed and performed FMRP biochemistry. D.A. Lerit analyzed centrosome asymmetry data, generated the cen-bcd-3′UTR transgenic animal, and supervised the project. All authors contributed to manuscript preparation.
References
- Albert, M.L., Darnell J.C., Bender A., Francisco L.M., Bhardwaj N., and Darnell R.B.. 1998. Tumor-specific killer cells in paraneoplastic cerebellar degeneration. Nat. Med. 4:1321–1324. 10.1038/3315 [DOI] [PubMed] [Google Scholar]
- Alliegro, M.C., and Alliegro M.A.. 2008. Centrosomal RNA correlates with intron-poor nuclear genes in Spisula oocytes. Proc. Natl. Acad. Sci. USA. 105:6993–6997. 10.1073/pnas.0802293105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alliegro, M.C., Alliegro M.A., and Palazzo R.E.. 2006. Centrosome-associated RNA in surf clam oocytes. Proc. Natl. Acad. Sci. USA. 103:9034–9038. 10.1073/pnas.0602859103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, K.V., and Lengyel J.A.. 1979. Rates of synthesis of major classes of RNA in Drosophila embryos. Dev. Biol. 70:217–231. 10.1016/0012-1606(79)90018-6 [DOI] [PubMed] [Google Scholar]
- Ascano, M. Jr., Mukherjee N., Bandaru P., Miller J.B., Nusbaum J.D., Corcoran D.L., Langlois C., Munschauer M., Dewell S., Hafner M., et al. 2012. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 492:382–386. 10.1038/nature11737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, A., Ifrim M.F., Valdez A.N., Raj N., and Bassell G.J.. 2018. Aberrant RNA translation in fragile X syndrome: From FMRP mechanisms to emerging therapeutic strategies. Brain Res. 1693(Pt A):24–36. 10.1016/j.brainres.2018.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benner, L., Castro E.A., Whitworth C., Venken K.J.T., Yang H., Fang J., Oliver B., Cook K.R., and Lerit D.A.. 2019. Drosophila Heterochromatin Stabilization Requires the Zinc-Finger Protein Small Ovary. Genetics. 213:877–895. 10.1534/genetics.119.302590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergalet, J., Patel D., Legendre F., Lapointe C., Benoit Bouvrette L.P., Chin A., Blanchette M., Kwon E., and Lecuyer E.. 2020. Inter-dependent Centrosomal Co-localization of the cen and ik2 cis-Natural Antisense mRNAs in Drosophila. Cell Rep. 30:3339–3352.e3336. [DOI] [PubMed] [Google Scholar]
- Berry, J., Brangwynne C.P., and Haataja M.. 2018. Physical principles of intracellular organization via active and passive phase transitions. Rep. Prog. Phys. 81:046601. 10.1088/1361-6633/aaa61e [DOI] [PubMed] [Google Scholar]
- Blower, M.D., Feric E., Weis K., and Heald R.. 2007. Genome-wide analysis demonstrates conserved localization of messenger RNAs to mitotic microtubules. J. Cell Biol. 179:1365–1373. 10.1083/jcb.200705163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Ding L., Viana M.P., Hendershott M.C., Yang R., Mueller I.A., and Rafelski S.M.. 2018. The Allen Cell Structure Segmenter: a new open source toolkit for segmenting 3D intracellular structures in fluorescence microscopy images. bioRxiv. Preprint posted December 8, 2018.
- Chouaib, R., Safieddine A., Pichon X., Imbert A., Kwon O.S., Samacoits A., Traboulsi A.M., Robert M.C., Tsanov N., Coleno E., et al. 2020. A Dual Protein-mRNA Localization Screen Reveals Compartmentalized Translation and Widespread Co-translational RNA Targeting. Dev. Cell. 54:773–791.e5. 10.1016/j.devcel.2020.07.010 [DOI] [PubMed] [Google Scholar]
- Conduit, P.T., Brunk K., Dobbelaere J., Dix C.I., Lucas E.P., and Raff J.W.. 2010. Centrioles regulate centrosome size by controlling the rate of Cnn incorporation into the PCM. Curr. Biol. 20:2178–2186. 10.1016/j.cub.2010.11.011 [DOI] [PubMed] [Google Scholar]
- Conduit, P.T., Feng Z., Richens J.H., Baumbach J., Wainman A., Bakshi S.D., Dobbelaere J., Johnson S., Lea S.M., and Raff J.W.. 2014. The centrosome-specific phosphorylation of Cnn by Polo/Plk1 drives Cnn scaffold assembly and centrosome maturation. Dev. Cell. 28:659–669. 10.1016/j.devcel.2014.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit, P.T., Wainman A., and Raff J.W.. 2015. Centrosome function and assembly in animal cells. Nat. Rev. Mol. Cell Biol. 16:611–624. 10.1038/nrm4062 [DOI] [PubMed] [Google Scholar]
- Corradi, J.P., Yang C., Darnell J.C., Dalmau J., and Darnell R.B.. 1997. A post-transcriptional regulatory mechanism restricts expression of the paraneoplastic cerebellar degeneration antigen cdr2 to immune privileged tissues. J. Neurosci. 17:1406–1415. 10.1523/JNEUROSCI.17-04-01406.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalby, B., and Glover D.M.. 1992. 3′ non-translated sequences in Drosophila cyclin B transcripts direct posterior pole accumulation late in oogenesis and peri-nuclear association in syncytial embryos. Development. 115:989–997. [DOI] [PubMed] [Google Scholar]
- Darnell, J.C. 2011. Defects in translational regulation contributing to human cognitive and behavioral disease. Curr. Opin. Genet. Dev. 21:465–473. 10.1016/j.gde.2011.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande, G., Calhoun G., and Schedl P.. 2006. The drosophila fragile X protein dFMR1 is required during early embryogenesis for pole cell formation and rapid nuclear division cycles. Genetics. 174:1287–1298. 10.1534/genetics.106.062414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dienstbier, M., Boehl F., Li X., and Bullock S.L.. 2009. Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev. 23:1546–1558. 10.1101/gad.531009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dockendorff, T.C., Su H.S., McBride S.M.J., Yang Z., Choi C.H., Siwicki K.K., Sehgal A., and Jongens T.A.. 2002. Drosophila lacking dfmr1 activity show defects in circadian output and fail to maintain courtship interest. Neuron. 34:973–984. 10.1016/S0896-6273(02)00724-9 [DOI] [PubMed] [Google Scholar]
- Estes, P.S., O’Shea M., Clasen S., and Zarnescu D.C.. 2008. Fragile X protein controls the efficacy of mRNA transport in Drosophila neurons. Mol. Cell. Neurosci. 39:170–179. 10.1016/j.mcn.2008.06.012 [DOI] [PubMed] [Google Scholar]
- Foe, V.E., and Alberts B.M.. 1983. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J. Cell Sci. 61:31–70. [DOI] [PubMed] [Google Scholar]
- Foe, V.E., Odell G.M., and Edgar B.A.. 1993. Mitosis and morphogenesis in the Drosophila embryo: Point and counterpoint. ScienceOpen. 149–300.
- Gamberi, C., Johnstone O., and Lasko P.. 2006. Drosophila RNA Binding Proteins. Int. Rev. Cytol. 248:43–139. [DOI] [PubMed] [Google Scholar]
- Glover, D.M., Leibowitz M.H., McLean D.A., and Parry H.. 1995. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell. 81:95–105. 10.1016/0092-8674(95)90374-7 [DOI] [PubMed] [Google Scholar]
- Graveley, B.R., Brooks A.N., Carlson J.W., Duff M.O., Landolin J.M., Yang L., Artieri C.G., van Baren M.J., Boley N., Booth B.W., et al. 2011. The developmental transcriptome of Drosophila melanogaster. Nature. 471:473–479. 10.1038/nature09715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt, E.J., and Spradling A.C.. 2018. Fragile X mental retardation 1 gene enhances the translation of large autism-related proteins. Science. 361:709–712. 10.1126/science.aas9963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida, T., and Kinoshita K.. 2007. PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 35(Web Server):W460–W464. 10.1093/nar/gkm363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao, L.-R., and Megraw T.L.. 2009. Centrocortin cooperates with centrosomin to organize Drosophila embryonic cleavage furrows. Curr. Biol. 19:937–942. 10.1016/j.cub.2009.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert, J.D., and Nagy L.M.. 2002. Asymmetric inheritance of centrosomally localized mRNAs during embryonic cleavages. Nature. 420:682–686. 10.1038/nature01241 [DOI] [PubMed] [Google Scholar]
- Lawo, S., Hasegan M., Gupta G.D., and Pelletier L.. 2012. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat. Cell Biol. 14:1148–1158. 10.1038/ncb2591 [DOI] [PubMed] [Google Scholar]
- Lécuyer, E., Yoshida H., Parthasarathy N., Alm C., Babak T., Cerovina T., Hughes T.R., Tomancak P., and Krause H.M.. 2007. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 131:174–187. 10.1016/j.cell.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Lerit, D.A., and Rusan N.M.. 2013. PLP inhibits the activity of interphase centrosomes to ensure their proper segregation in stem cells. J. Cell Biol. 202:1013–1022. 10.1083/jcb.201303141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerit, D.A., Jordan H.A., Poulton J.S., Fagerstrom C.J., Galletta B.J., Peifer M., and Rusan N.M.. 2015. Interphase centrosome organization by the PLP-Cnn scaffold is required for centrosome function. J. Cell Biol. 210:79–97. 10.1083/jcb.201503117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling, S.C., Fahrner P.S., Greenough W.T., and Gelfand V.I.. 2004. Transport of Drosophila fragile X mental retardation protein-containing ribonucleoprotein granules by kinesin-1 and cytoplasmic dynein. Proc. Natl. Acad. Sci. USA. 101:17428–17433. 10.1073/pnas.0408114101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little, S.C., Sinsimer K.S., Lee J.J., Wieschaus E.F., and Gavis E.R.. 2015. Independent and coordinate trafficking of single Drosophila germ plasm mRNAs. Nat. Cell Biol. 17:558–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald, P.M., and Struhl G.. 1988. cis-acting sequences responsible for anterior localization of bicoid mRNA in Drosophila embryos. Nature. 336:595–598. 10.1038/336595a0 [DOI] [PubMed] [Google Scholar]
- Magescas, J., Zonka J.C., and Feldman J.L.. 2019. A two-step mechanism for the inactivation of microtubule organizing center function at the centrosome. eLife. 8:E47867. 10.7554/eLife.47867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, W.F., and Rosenbaum J.L.. 2000. Are there nucleic acids in the centrosome? Curr. Top. Dev. Biol. 49:187–205. 10.1016/S0070-2153(99)49009-X [DOI] [PubMed] [Google Scholar]
- Martinez-Campos, M., Basto R., Baker J., Kernan M., and Raff J.W.. 2004. The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J. Cell Biol. 165:673–683. 10.1083/jcb.200402130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw, T.L., Kilaru S., Turner F.R., and Kaufman T.C.. 2002. The centrosome is a dynamic structure that ejects PCM flares. J. Cell Sci. 115:4707–4718. 10.1242/jcs.00134 [DOI] [PubMed] [Google Scholar]
- Megraw, T.L., Li K., Kao L.R., and Kaufman T.C.. 1999. The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development. 126:2829–2839. [DOI] [PubMed] [Google Scholar]
- Mittasch, M., Tran V.M., Rios M.U., Fritsch A.W., Enos S.J., Ferreira Gomes B., Bond A., Kreysing M., and Woodruff J.B.. 2020. Regulated changes in material properties underlie centrosome disassembly during mitotic exit. J. Cell Biol. 219:647. 10.1083/jcb.201912036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monzo, K., Papoulas O., Cantin G.T., Wang Y., Yates J.R. III, and Sisson J.C.. 2006. Fragile X mental retardation protein controls trailer hitch expression and cleavage furrow formation in Drosophila embryos. Proc. Natl. Acad. Sci. USA. 103:18160–18165. 10.1073/pnas.0606508103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, F., Senecal A., Tantale K., Marie-Nelly H., Ly N., Collin O., Basyuk E., Bertrand E., Darzacq X., and Zimmer C.. 2013. FISH-quant: automatic counting of transcripts in 3D FISH images. Nat. Methods. 10:277–278. 10.1038/nmeth.2406 [DOI] [PubMed] [Google Scholar]
- Nigg, E.A., and Raff J.W.. 2009. Centrioles, centrosomes, and cilia in health and disease. Cell. 139:663–678. 10.1016/j.cell.2009.10.036 [DOI] [PubMed] [Google Scholar]
- Palazzo, R.E., Vogel J.M., Schnackenberg B.J., Hull D.R., and Wu X.. 2000. Centrosome maturation. Curr. Top. Dev. Biol. 49:449–470. 10.1016/S0070-2153(99)49021-0 [DOI] [PubMed] [Google Scholar]
- Papoulas, O., Monzo K.F., Cantin G.T., Ruse C., Yates J.R. III, Ryu Y.H., and Sisson J.C.. 2010. dFMRP and Caprin, translational regulators of synaptic plasticity, control the cell cycle at the Drosophila mid-blastula transition. Development. 137:4201–4209. 10.1242/dev.055046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz, I., Kosti I., Ares M. Jr., Cline M., and Mandel-Gutfreund Y.. 2014. RBPmap: a web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res. 42(W1):W361–W367. 10.1093/nar/gku406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilaz, L.-J., Lennox A.L., Rouanet J.P., and Silver D.L.. 2016. Dynamic mRNA Transport and Local Translation in Radial Glial Progenitors of the Developing Brain. Curr. Biol. 26:3383–3392. 10.1016/j.cub.2016.10.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff, J.W., Whitfield W.G., and Glover D.M.. 1990. Two distinct mechanisms localise cyclin B transcripts in syncytial Drosophila embryos. Development. 110:1249–1261. [DOI] [PubMed] [Google Scholar]
- Raj, A., van den Bogaard P., Rifkin S.A., van Oudenaarden A., and Tyagi S.. 2008. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods. 5:877–879. 10.1038/nmeth.1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richens, J.H., Barros T.P., Lucas E.P., Peel N., Pinto D.M.S., Wainman A., and Raff J.W.. 2015. The Drosophila Pericentrin-like-protein (PLP) cooperates with Cnn to maintain the integrity of the outer PCM. Biol. Open. 4:1052–1061. 10.1242/bio.012914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder, P.V., and Lerit D.A.. 2020. Quantitative analysis of subcellular distributions with an open-source, object-based tool. Biology Open. 9. . 10.1242/bio.055228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder, P.V., and Lerit D.A.. 2018. RNA localization regulates diverse and dynamic cellular processes. Traffic. 19:496–502. 10.1111/tra.12571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro, M.R., Bray S.M., and Warren S.T.. 2012. Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Annu. Rev. Pathol. 7:219–245. 10.1146/annurev-pathol-011811-132457 [DOI] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulveda, G., Antkowiak M., Brust-Mascher I., Mahe K., Ou T., Castro N.M., Christensen L.N., Cheung L., Jiang X., Yoon D., et al. 2018. Co-translational protein targeting facilitates centrosomal recruitment of PCNT during centrosome maturation in vertebrates. eLife. 7:e34959. 10.7554/eLife.34959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, G., Pratt G., Yeo G.W., and Moore M.J.. 2015. The Clothes Make the mRNA: Past and Present Trends in mRNP Fashion. Annu. Rev. Biochem. 84:325–354. 10.1146/annurev-biochem-080111-092106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhakaran, I.P., Hillebrand J., Dervan A., Das S., Holohan E.E., Hülsmeier J., Sarov M., Parker R., VijayRaghavan K., and Ramaswami M.. 2014. FMRP and Ataxin-2 function together in long-term olfactory habituation and neuronal translational control. Proc. Natl. Acad. Sci. USA. 111:E99–E108. 10.1073/pnas.1309543111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, W., Fogarty P., and Theurkauf W.. 1993. Mutations affecting the cytoskeletal organization of syncytial Drosophila embryos. Development. 118:1245–1254. [DOI] [PubMed] [Google Scholar]
- Sunkel, C.E., and Glover D.M.. 1988. polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J. Cell Sci. 89:25–38. [DOI] [PubMed] [Google Scholar]
- Van Treeck, B., and Parker R.. 2018. Emerging Roles for Intermolecular RNA-RNA Interactions in RNP Assemblies. Cell. 174:791–802. 10.1016/j.cell.2018.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardy, L., and Orr-Weaver T.L.. 2007. The Drosophila PNG kinase complex regulates the translation of cyclin B. Dev. Cell. 12:157–166. 10.1016/j.devcel.2006.10.017 [DOI] [PubMed] [Google Scholar]
- Vastenhouw, N.L., Cao W.X., and Lipshitz H.D.. 2019. The maternal-to-zygotic transition revisited. Development. 146:dev161471. 10.1242/dev.161471 [DOI] [PubMed] [Google Scholar]
- Vertii, A., Hehnly H., and Doxsey S.. 2016. The Centrosome, a Multitalented Renaissance Organelle. Cold Spring Harb. Perspect. Biol. 8:a025049. 10.1101/cshperspect.a025049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff, J.B., Ferreira Gomes B., Widlund P.O., Mahamid J., Honigmann A., and Hyman A.A.. 2017. The Centrosome Is a Selective Condensate that Nucleates Microtubules by Concentrating Tubulin. Cell. 169:1066–1077.e10. 10.1016/j.cell.2017.05.028 [DOI] [PubMed] [Google Scholar]
- Woodruff, J.B., Wueseke O., Viscardi V., Mahamid J., Ochoa S.D., Bunkenborg J., Widlund P.O., Pozniakovsky A., Zanin E., Bahmanyar S., et al. 2015. Centrosomes. Regulated assembly of a supramolecular centrosome scaffold in vitro. Science. 348:808–812. 10.1126/science.aaa3923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H., Sepúlveda L.A., Figard L., Sokac A.M., and Golding I.. 2015. Combining protein and mRNA quantification to decipher transcriptional regulation. Nat. Methods. 12:739–742. 10.1038/nmeth.3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y.Q., Bailey A.M., Matthies H.J., Renden R.B., Smith M.A., Speese S.D., Rubin G.M., and Broadie K.. 2001. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell. 107:591–603. 10.1016/S0092-8674(01)00589-X [DOI] [PubMed] [Google Scholar]
- Zwicker, D., Decker M., Jaensch S., Hyman A.A., and Jülicher F.. 2014. Centrosomes are autocatalytic droplets of pericentriolar material organized by centrioles. Proc. Natl. Acad. Sci. USA. 111:E2636–E2645. 10.1073/pnas.1404855111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists all smFISH probes used in this study.
Data Availability Statement
All code for RNA detection and measurements are available on github at https://github.com/pearlryder/rna-at-centrosomes and https://github.com/pearlryder/cen-at-fmr-null-centrosomes. Full-sized images of all DNA gels and immunoblots are available at FigShare: doi.org/10.6084/m9.figshare.12821564 and doi.org/10.6084/m9.figshare.12821579.