Benzi and Piatti preview work from Cordeiro and colleagues that describes new insights into the regulation of PLK1 during mitotic checkpoint silencing.
Abstract
Silencing of the spindle assembly checkpoint involves two protein phosphatases, PP1 and PP2A-B56, that are thought to extinguish checkpoint signaling through dephosphorylation of a checkpoint scaffold at kinetochores. In this issue, Cordeiro et al. (2020. J. Cell Biol. https://doi.org/10.1083/jcb.202002020) now show that a critical function of these phosphatases in checkpoint silencing is removal of Polo kinase at kinetochores, which would otherwise autonomously sustain the checkpoint.
The main goal of mitosis is to accurately segregate chromosomes, such that each daughter cell inherits a full complement of genetic information. To accomplish this delicate task, once each chromosome is faithfully duplicated through DNA replication, its identical sister chromatids must attach to spindle microtubules coming from opposite spindle poles through a process known as chromosome biorientation. Kinetochores are proteinaceous assemblies that reside at the centromeric region of chromosomes and are key to this process by capturing spindle microtubules (1). Microtubule capture, however, is inherently error prone, and several cycles of attachment/detachment are often required before chromosomes achieve biorientation. Obviously, chromosome segregation without error correction would be highly detrimental, leading to unbalanced chromosome numbers, referred to as aneuploidies, which are hallmarks of cancer and genetic diseases. Luckily, eukaryotic cells not only possess an error-correction machinery deputed to rectify faulty attachments (2), but they also have a safeguard device, called the spindle assembly checkpoint (SAC), that temporarily halts cells in mitosis to provide them with the necessary time window to fix the errors. SAC signaling fires at unattached kinetochores, which are continuously generated during error correction, and is extinguished once all chromosomes are bioriented, thus resuming mitotic progression and chromosome segregation (3).
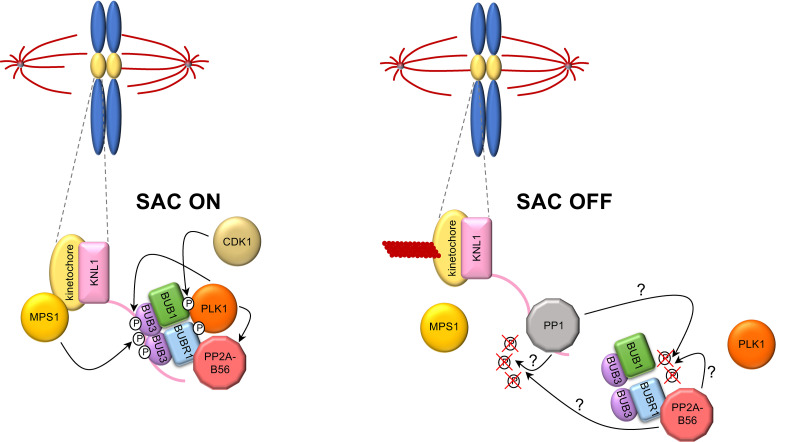
Prevailing models posit that a key trigger of SAC signaling is the phosphorylation of the kinetochore scaffold KNL1 by the SAC kinase MPS1. This creates a phospho-docking site at the MELT repeats (amino acid consensus Met-Glu-Leu-Thr) of KNL1 that recruits the heterotetrameric SAC complex BUB1:BUB3:BUB3:BUBR1 (referred to as BUB complex; Fig. 1), which in turn attracts to the kinetochore other SAC proteins that collectively prevent mitotic progression (3).
Figure 1.
The interplay of SAC kinases and phosphatases at kinetochores. When SAC is activated at an unattached kinetochore (SAC on), MPS1 phosphorylates the kinetochore scaffold KNL1, thereby recruiting the BUB complex. Contextually, CDK1-dependent phosphorylation of BUB1 and BUBR1 generates phospho-docking sites for recruitment of the Polo kinase PLK1, which on one side sustains KNL1 phosphorylation and on the other stimulates BUBR1 binding to PP2A-B56. The latter, in turn, counteracts PLK1 local activity by dislodging PLK1 from the kinetochore. During SAC silencing, local activity of MPS1 is shut off. Additionally, the PP1 phosphatase binds to KNL1 and, together with PP2A-B56, further evicts PLK1 from the kinetochore, possibly through dephosphorylation of its phospho-docking sites in BUB1 and BUBR1. This leads to KNL1 dephosphorylation and displacement of the BUB complex, thus extinguishing SAC signaling (SAC off). Whether PP1 and PP2A-B56, as opposed to other phosphatases, contribute directly to KNL1 dephosphorylation remains an open question.
The protein phosphatases PP1 and PP2A-B56 are recruited to kinetochores through binding to KNL1 and BUBR1, respectively, and are thought to silence the SAC through dephosphorylation of the MELT repeats of KNL1, thus antagonizing MPS1 activity (Fig. 1). Additional mechanisms, such as MPS1 inhibition and stripping of SAC components from kinetochores, have been proposed to contribute to obliterate SAC signaling upon chromosome biorientation (4).
In this issue, compelling evidence from Cordeiro et al. challenges the current view by showing that rather than dephosphorylating KNL1, PP1 and PP2A-B56 actually silence the SAC by down-regulating the activity of Polo kinase (PLK1 in human cells) at kinetochores (5). Polo kinase and MPS1 share a common substrate preference and both can phosphorylate the MELT repeats of KNL1. Additionally, Polo cooperates with MPS1 in SAC signaling in various species, while in organisms where MPS1 is absent, like nematodes, Polo functionally replaces MPS1 (6).
Consistent with previous results (7, 8), Cordeiro et al. show that when kinetochore phosphatases are dampened, PLK1 levels increase at kinetochores through an unknown mechanism, which might involve dephosphorylation of the phosphoepitopes in the Polo-binding motifs generated on the BUB complex by CDK1 (BUB1-pT609 and BUBR1-pT620; 9, 10, 11). This implies that when PP1 and PP2A-B56 are low at kinetochores, PLK1 can amplify SAC signaling through a positive feedback loop by boosting KNL1 phosphorylation independently of Mps1, thereby recruiting the BUB complex and, in turn, increasing amounts of PLK1 (Fig. 1). In agreement with this view, in a sensitized setup where kinetochore phosphatases are crippled along with MPS1, concomitant inhibition of PLK1 is sufficient to bring about KNL1 dephosphorylation and restore SAC signaling. These data led the authors to the provocative conclusion that the primary role of PP1 and PP2A-B56 in SAC silencing is to harness PLK1 activity. This new model is appealing not only because it highlights a novel function for PP1 and PP2A-B56 in SAC silencing, but also because it explains the modest effects that are commonly observed on SAC signaling upon PLK1 inhibition alone. Indeed, kinetochore phosphatases, and primarily PP2A-B56 (12), are already partially active in a SAC-induced mitotic arrest (e.g., upon microtubule depolymerization), as shown here by the increased KNL1 phosphorylation upon their inactivation.
Interestingly, sequence alignment of BUB1 and BUBR1 homologues across the phylogenetic tree reveals that, in metazoans, putative Polo-binding motifs are usually located in the vicinity of hypothetical PP2A-B56–binding motifs, suggesting that they coevolved. The physical proximity of Polo-binding and PP2A-B56–binding motifs in BUB1 and BUBR1 could position the Polo-binding motifs in a favorable arrangement for their PP2A-B56–driven dephosphorylation and, as a consequence, PLK1 clearance from kinetochores (Fig. 1).
The data by Cordeiro et al. represent a paradigm shift in our understanding of SAC silencing for two main reasons. First, consistent with published data (13, 14), PP1 and PP2A-B56 might be involved in this process primarily by inactivating upstream SAC kinases (MPS1 and PLK1), rather than dephosphorylating their substrates. Second, since PLK1 is partially displaced from kinetochores by the above phosphatases already during a SAC arrest, MPS1 inactivation might be the main trigger of SAC silencing. Several mechanisms have been proposed to attenuate MPS1 activity once the SAC is satisfied, such as MPS1 displacement from kinetochores (6) and dephosphorylation of MPS1 in its activation loop (13, 14).
The new model raises a burning question: If PP1 and PP2A do not dephosphorylate KNL1 at MELT repeats, what does? Other phosphatases, whose identity remains elusive, could be involved in KNL1 dephosphorylation. Alternatively, phosphorylated KNL1 might be actively turned over at kinetochores. Nevertheless, at present, the involvement of PP1 and PP2A-B56 in KNL1 dephosphorylation cannot be ruled out, as complete inhibition of kinetochore phosphatases in the experimental setup used here is likely very challenging. Further investigations will be required to solve this central issue.
Another important question that deserves further scrutiny is, how exactly do PP1 and PP2A-B56 inhibit PLK1 activity at kinetochores? Cordeiro et al. propose that they could dephosphorylate the Polo-binding motifs in BUB1/BUBR1. Alternatively, the close proximity of PP2A- and Polo-binding motifs in metazoan BUBR1 homologues could make the association of PLK1 and PP2A with BUBR1 mutually exclusive.
Finally, and most importantly, what is the physiological meaning of the complex interplay between SAC kinases and phosphatases described here? A crucial function of PLK1 bound to the BUB complex in human cells is to stabilize kinetochore-microtubule attachments in prometaphase by recruiting PP2A-B56 through phosphorylation of the PP2A-B56-binding motif in BUBR1 (15). In turn, eviction of PLK1 from kinetochores by PP2A-B56 will have two major outputs: (i) maintain microtubule dynamics at bioriented chromosomes (8) and (ii) stimulate binding of PP1 to KNL1, which primes the system for SAC silencing (16). As soon as MPS1 levels drop at kinetochores and/or other phosphatases intervene to dephosphorylate KNL1, SAC signaling is finally extinguished. The development of fluorescence-based biosensors combined with mathematical modeling will certainly provide in the future further mechanistic insights into such intricate network.
Acknowledgments
The authors apologize to colleagues whose work could not be cited or discussed due to space restrictions.
Work in Piatti's laboratory is supported by the Labex EpiGenMed and the Agence Nationale de la Recherche (ANR-18-CE13-0015-01).
The authors declare no competing financial interests.
References
- 1.Musacchio, A., and Desai A.. 2017. Biology. 10.3390/biology6010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lampson, M.A., and Grishchuk E.L.. 2017. Biology. 10.3390/biology6010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Musacchio, A. 2015. Curr. Biol. 10.1016/j.cub.2015.08.051 [DOI] [Google Scholar]
- 4.Etemad, B., and Kops G.J.. 2016. Curr. Opin. Cell Biol. 10.1016/j.ceb.2016.02.016 [DOI] [PubMed] [Google Scholar]
- 5.Cordeiro, M.H., et al. 2020. J. Cell Biol. 10.1083/jcb.202002020 [DOI] [Google Scholar]
- 6.Pachis, S.T., and Kops G.J.P.L.. 2018. Open Biol. 10.1098/rsob.180109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foley, E.A., et al. 2011. Nat. Cell Biol. 10.1038/ncb2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu, D., et al. 2012. J. Cell Biol. 10.1083/jcb.201205090 [DOI] [Google Scholar]
- 9.Elowe, S., et al. 2007. Genes Dev. 10.1101/gad.436007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qi, W., et al. 2006. Mol. Biol. Cell. 10.1091/mbc.e06-03-0240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong, O.K., and Fang G.. 2007. J. Cell Biol. 10.1083/jcb.200708044 [DOI] [Google Scholar]
- 12.Espert, A., et al. 2014. J. Cell Biol. 10.1083/jcb.201406109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayward, D., et al. 2019. J. Cell Biol. 10.1083/jcb.201905026 [DOI] [Google Scholar]
- 14.Moura, M., et al. 2017. eLife. 10.7554/eLife.25366 [DOI] [Google Scholar]
- 15.Suijkerbuijk, S.J.E., et al. 2012. Dev. Cell. 10.1016/j.devcel.2012.09.005 [DOI] [PubMed] [Google Scholar]
- 16.Liu, D., et al. 2010. J. Cell Biol. 10.1083/jcb.201001006 [DOI] [Google Scholar]