Abstract
Vibrio anguillarum is a Gram-negative marine pathogen causative agent of vibriosis in a wide range of hosts, including invertebrates and teleosts. Lumpfish (Cyclopterus lumpus), a native fish of the North Atlantic Ocean, is utilized as cleaner fish to control sea lice (Lepeophtheirus salmonis) infestations in the Atlantic salmon (Salmo salar) aquaculture industry. V. anguillarum is one of the most frequent bacterial pathogens affecting lumpfish. Here, we described the phenotype and genomic characteristics of V. anguillarum strain J360 isolated from infected cultured lumpfish in Newfoundland, Canada. Koch’s postulates determined in naïve lumpfish showed lethal acute vibriosis in lumpfish. The V. anguillarum J360 genome was shown to be composed of two chromosomes and two plasmids with a total genome size of 4.56 Mb with 44.85% G + C content. Phylogenetic and comparative analyses showed that V. anguillarum J360 is closely related to V. anguillarum strain VIB43, isolated in Scotland, with a 99.8% genome identity. Differences in the genomic organization were identified and associated with insertion sequence elements (ISs). Additionally, V. anguillarum J360 does not possess a pJM1-like plasmid, typically present in virulent isolates from the Pacific Ocean, suggesting that acquisition of this extrachromosomal element and the virulence of V. anguillarum J360 or other Atlantic isolates could increase.
Keywords: Vibrio anguillarum, lumpfish, genomics, insertion elements, evolution
1. Introduction
Vibrio spp. is naturally ubiquitous in aquatic and marine environments [1]. Some members of this genus can cause infections in humans after exposition to contaminated water, such as Vibrio cholerae, the causative agent of cholera, and after consumption of raw contaminated seafood, such as V. parahaemolitycus, V. alginolyticus, and V. vulnificus [1,2,3]. Other members of Vibrio spp. such as V. splendidus, V. nereis, V. harveyi, V. damsela, V. tubiashi, and V. anguillarum are pathogens of aquatic organisms including cultured fish species [4,5]. V. anguillarum is a Gram-negative marine pathogen, which causes vibriosis in a wide range of cultured and wild invertebrates and teleosts hosts, but is also present in brackish and fresh water [6,7,8].
The lumpfish (Cyclopterus lumpus), a native fish of the North Atlantic Ocean, is utilized as a cleaner fish to control sea lice (Lepeophtheirus salmonis) infestations in the Atlantic salmon (Salmo salar) industry [9,10,11]. The lumpfish performs well in cold environments, removing nearly 90% of the sea lice at sea-cages (with a feeding rate of 0.3 sea-lice per day) [9,12,13]. Lumpfish have been reported to be up to 64% more efficacious with respect to sea lice removal when compared to other wrasse species utilized as cleaner fish species, like ballan (Labrus bergylta), corkwing (Crenilabrus melops), rock cook (Centrolabrus exoletus), and goldsinny (Ctenolabrus rupestris) [10,12]. In addition, these wrasses (not including lumpfish) exhibit a reduced activity during winter, because they enter into a hypometabolic state (similar to hibernation) [13,14], and this eventually decreases sea lice removal efficiency [15,16].
V. anguillarum is one of the most frequent pathogens affecting lumpfish in sea-cages [9,10]. In lumpfish, V. anguillarum causes hemorrhagic septicemia, which is characterized by skin lesions, gill hemorrhages, and bacterial aggregations in lymphoid organs [10,17,18]. In Atlantic Canada, V. anguillarum is frequent [19], but it has not been reported in lumpfish.
Currently, 23 serotypes of V. anguillarum have been described, and serotypes O1, O2, and O3 are the most frequent serotypes causing outbreaks in teleosts [18,19]. The V. anguillarum genome consists of two chromosomes, and a large virulent plasmid is present in some isolates [8]. Several virulent strains of V. anguillarum from different geographic locations and fish species have been sequenced [20], including V. anguillarum M3 [7], V. anguillarum NB10 [6], and V. anguillarum 775 [8]. V. anguillarum is more frequently reported in warm-water fish, and several of these genomes have been sequenced, assembled, and annotated, and although virulent strains of V. anguillarum have been isolated from cold water environments, only a few of these isolates have been described [20].
In this study, we describe the complete genome of a V. anguillarum strain isolated from an outbreak in cultured lumpfish in Newfoundland, Canada, and compared its genome to other known V. anguillarum strains. We determined that V. anguillarum J360 produces acute vibriosis in lumpfish, does not harbor virulent plasmids, is closely related to V. anguillarum strains isolated from the Atlantic coasts, and is distantly related to V. anguillarum strains isolated from the Pacific coasts. V. anguillarum J360 also showed high similarity to a V. anguillarum isolated from sea bass (Dicentrarchus labrax) in Scotland. Comparative analysis suggested that insertion sequence elements play a key role in V. anguillarum evolution.
2. Material and Methods
2.1. Bacterial Culture Conditions
A single colony of V. anguillarum J360 was grown routinely in 3 mL of Trypticase soy broth (TSB, Difco, Franklin Lakes, NJ, USA) supplemented with up to 2% NaCl at 15 °C in a 16 mm-diameter glass tube and placed in a roller for 24 h. When required, the culture medium was supplemented with 100 µM of FeCl3, 100 µM 2,2-dipyridyl, or 1.5% bacto-agar (Difco). Chrome Azurol S (CAS) plates were used for the siderophores secretion assay [21]. Blood agar plates (0.5% salmon blood) were used to evaluate hemolytic activity. Bacterial cells were harvested at mid-log phase, at an optical density (O.D. 600 nm) of ≈0.7 (≈4.1 × 108 CFU/mL), and washed three times with phosphate-buffered saline (PBS; 136 mM NaCl, 2.7 mM KCl, 10.1 mM Na2HPO4, 1.5 mM KH2PO4 (pH 7.2)) [22] at 4200× g for 10 min at room temperature.
2.2. V. anguillarum Isolation
V. anguillarum strain J360 was isolated from the head kidney of infected cultured lumpfish in Newfoundland, Canada. Fish with classic vibriosis symptoms were netted and immediately euthanized with an overdose of MS222 (400 mg/L) (Syndel Laboratories, Nanaimo, BC, Canada). Tissue samples (e.g., head kidney, liver, and spleen) were collected and placed into sterile homogenized bags (Nasco whirl-pak®, Fort Atkinson, WI, USA). The infected tissues were weighed and homogenized in PBS up to a final volume of 1 mL. One hundred microliters of the homogenized tissue suspension were plated onto Trypticase soy agar (TSA) plates and incubated at 15 °C for 48 h. Isolated colonies were selected and purified for further analysis. Bacteria stock was preserved at −80 °C in 10% glycerol and 1% peptone solution.
2.3. Matrix-Assisted Laser Desorption/Ionization—Time-of-Flight (MALDI-TOF) Mass Spectrometry and Serotypification Analysis
Serotypification and MALDI-TOF were conducted commercially at the University of Prince Edward Island, Canada. The MALDI Biotyper RTC was conducted according to the MALDI Biotyper 3.1 user manual and parameter settings as previously published [23].
2.4. Biochemical, Enzymatic, and Physiological Characterization
V. angillarum J360 growth curves were conducted in triplicates at 15 °C, 28 °C, and under rich and iron-limited conditions according to established protocols [24]. Briefly, a single colony of V. angillarum J360 was inoculated in 3 mL of TSB and incubated in a roller for 24 h at 15 °C. An inoculum of 300 μL of cells at mid-log phase (OD600 ≈ 0.7) was added to 30 mL of fresh TSB into 250 mL flasks and incubated for 48 h at 15 °C and 28 °C with aeration (180 rpm) in an orbital shaker. Bacterial growth was monitored spectrophotometrically until OD600 ≈ 2.0 ± 0.3 (≈8 × 108 CFU/mL) using a Genesys 10 UV spectrophotometer (Thermo Spectronic, Thermo Fischer Scientific, USA). Growth curves under iron-limited conditions were determined using three different 2,2-dipyridyl concentrations (100, 150, 250 µM). Controls consisted of nonsupplemented TSB. The doubling time was estimated using the optical density (OD) values, g = b − B, where b is the OD value at the end of the time interval and B is the OD value at the beginning of the time interval.
The biochemical profile of V. anguillarum J360 was characterized using API20E, API20NE, and APY ZYM systems (BioMerieux, Marcy-l’Etoile, France) according to the manufacturer’s instructions. The strips were incubated at 15 °C for 48 h and the results were analyzed using APIweb (BioMerieux). V. anguillarum J360 growth was also tested at different temperatures (4 °C, 15 °C, 28 °C, 37 °C) and different concentrations of NaCl (0%, 0.5%, 2%). Motility, hemolysin on TSA (5% salmon blood), siderophore synthesis, catalase activity, and oxidase activity were evaluated using standard methods [25]. The antibiogram of V. anguillarum J360 was determined for tetracycline (10 mg/mL), oxytetracycline (30 mg/mL), ampicillin (10 mg/mL), sulfamethoxazole (STX) (25 mg/mL), chloramphenicol (30 mg/mL), colistin sulphate (10 mg/mL), and oxalinic acid (2 mg/mL) using standard methods [25,26].
2.5. Siderophores Synthesis
V. anguillarum J360 was grown under conditions previously described. Bacterial cells were harvested at mid-log phase, at an optical density (OD600) of ≈0.7 (≈4.1 × 108 CFU/mL), washed three times with phosphate-buffered saline (PBS; 136 mM NaCl, 2.7 mM KCl, 10.1 mM Na2HPO4, 1.5 mM KH2PO4 (pH 7.2)) [22] at 4200× g for 10 min at room temperature, and resuspended in 1 mL of PBS. An inoculum of 300 µL of the bacterial suspension was added to 3 mL TSB medium and TSB medium supplemented with 100 µM of FeCl3 or 100 µM of 2,2-dipyridyl. V. anguillarum J360 was grown at 15 °C for 24 h with aeriation. Following the incubation period, the cells were harvested at mid-log phase, washed twice with PBS, and resuspended in 100 µL of PBS. Five microliters of the concentrated bacterial pellet was inoculated onto CAS agar plates [21] and incubated at 15 °C and 28 °C for 48 h.
2.6. Fish Holding
Fish were produced and maintained at the Dr. Joe Brown Aquatic Research Building (JBARB), Memorial University of Newfoundland (MUN), under the animal protocols #18-1-JS and #18-03-JS. Lumpfish were acclimated to ≈8–10 °C in 500 L tanks supplied with 95–110% air-saturated and UV-treated filtered flow-through seawater, and an ambient photoperiod. Stocking density was maintained at 6.6 kg/m3. The fish were fed daily by automated feeders a commercial diet (Skretting—Europa; 15 crude protein (55%), crude fat (15%), crude fiber (1.5%), calcium (3%), phosphorus (2%), sodium (1%), vitamin A (5000 IU/kg), vitamin D (3000 IU/kg), and vitamin E (200 IU/kg))) at a rate of 0.5% of their body weight per day.
2.7. Infection Assay in Lumpfish
Naive lumpfish (weight ≈ 55 g) were transferred from the JBARB to the AQ3 biocontainment Cold-Ocean Deep-Sea Research Facility (CDRF) for infection assays. Fish were separated into three 500 L tanks containing 60 fish per dose and acclimated for 2 weeks under previously described conditions. The infection procedures were conducted according to established protocols [27]. Briefly, fish were anesthetized with 40 mg of MS222 (Syndel Laboratories, BC, Canada) per liter of sea water and intraperitoneally infected with 100 µL of 106, or 107 CFU per dose of V. anguillarum J360. The Control group (n = 60) was mock-injected with PBS. Mortality was monitored daily until 30 days post-infection. Samples of liver, spleen, and head kidney were taken from moribund fish to re-isolate the pathogen.
2.8. DNA Extraction and Sequencing
V. anguillarum J360 was grown under conditions previously described. Bacterial cells were harvested at mid-log phase, at an optical density OD600 of ≈0.7 (≈4.1 × 108 CFU/mL) and washed three times with PBS. DNA extraction was conducted using a Wizard DNA extraction High Molecular Weight Kit (Promega, Madison, WI, USA). The DNA integrity and purity were evaluated by gel electrophoresis (agarose gel 0.8%) [28] and spectrophotometry (Genova-Nano Spectrophotometer, Jenway, UK). Libraries and sequencing were conducted commercially at Genome Quebec (Montreal, QC, Canada) using PacBio RS II and Miseq Illumina sequencers.
2.9. Genome Assembly, Annotation, and Mapping
PacBio reads were assembled at Genome Quebec using Celera Assembler (August 2013 version). Annotation was completed using the Rapid Annotation Subsystem Technology pipeline (RAST) (http://rast.nmpdr.org/) [29]. The two V. anguillarum J360 chromosomes and large plasmid were submitted to the National Center for Biotechnology Information (NCBI) and re-annotated using the NCBI Prokaryotic Genome Annotation Pipeline.
To detect small plasmids, the Illumina reads were trimmed using CLC Genomics Workbench v20.0 (CLC Bio) and examined for quality using FastQC version 12 [30]. High-quality Illumina reads were assembled using the CLC Genomics Workbench de novo tool and aligned to the reference V. anguillarum J360 chromosomes and large plasmid using the genome finishing module tools with default parameters. Illumina sequences that did not align with the chromosomes or large plasmid were analyzed and annotated using the previously described methods. The V. anguillarum J360 whole genome was mapped by using DNA plotter software [31].
2.10. Whole Genome Comparison and Phylogeny Analysis
The genomes utilized are listed in Table 1. Whole genomes were aligned to calculate the average nucleotide identity (ANI) using the CLC Genomic workbench v20 (CLC Bio) whole genome analysis tool with default parameters (Min. initial seed length = 15; Allow mismatches = yes; Min. alignment block = 100). A minimum similarity (0.8) and a minimum length (0.8) were used as parameters for CDS identity. A comparative heat map was constructed using the heat map tool with default parameters (Euclidean distance method and complete cluster linkages). Phylogenetic analysis was performed in two different software, CLC Genomic workbench v20.0 and MEGA X [32], with the same parameters for robustness comparison purposes, using the extracted alignment from the ANI analysis. Evolutionary history was calculated using the neighbor-joining method [33] with a bootstrap consensus of 500 replicates, and evolutionary distance was computed using the Jukes–Cantor method [34]. Photobacterium damselae 91–197 (AP018045/6) chromosomes were utilized as outgroups [35]. Whole genome dot plots between closely related V. anguillarum strains were constructed using the CLC Genomic workbench v20.0, a whole genome analysis tool to visualize and further analyze genomic differences. Comparative alignment analysis represents homologous regions, translocations, and inversions within the two strain genomes for chromosome-I and for chromosome-II. Homologous regions were identified as locally colinear blocks (LCBs), which represent conserved regions that do not present rearrangements, and genomic gaps (GGs) were identified as unmatched regions. Analysis was performed using the progressive Mauve v20150206 [36].
Table 1.
Geographic origin and hosts species of Vibrio anguillarum isolates.
| Species and Serotype | Geographic Origin, Host Species | Genome Size (bp) | Accession Numbers | References |
|---|---|---|---|---|
| V. anguillarum 775/O1 | USA, Pacific coast/Oncorhynchus kisutch | 4,117,056 | CP002284/5 | [8] |
| V. anguillarum M3/O1 | China, Shandong/Paralichthys olivaceus | 4,117,885 | CP006699/700 | [7] |
| V. anguillarum NB10/O1 | Sweden, Baltic Sea/Oncorhynchus mykiss | 4,373,835 | LK021130/29 | [6] |
| V. anguillarum VIB12/O2 | Greece/Dicentrarchus labrax | 4,897,690 | CP023310/11 | [20] |
| V. anguillarum VIB43/O1 | Scotland, UK/Dicentrarchus labrax | 4,407,865 | CP023054/5 | [20] |
| V. anguillarum CNEVA/O3 | France/Dicentrarchus labrax | 4,256,429 | CP022103/4 | [20] |
| V. anguillarum MHK3/O1 | China/Flounder | 4,015,925 | CP022468/9 | - |
| V. anguillarum 425 | China/Sea bass | 4,373,373 | CP020533/4 | - |
| V. anguillarum 87-9-116/O1 | Finland/Salmo salar | 4,338,125 | CP021980/1 | [20] |
| V. anguillarum JLL237/O1 | Denmark/Oncorhynchus mykiss | 4,286,989 | CP022101/2 | [20] |
| V. anguillarum ATCC-68554/O1 | USA, Pacific coast/Oncorhynchus kisutch | 4,141,906 | CP023209/8 | [20] |
| V. anguillarum 90-11-286 | Denmark/Farm-water sample | 4,342,224 | CP011460/1 | [37] |
| V. anguillarum S3/O1 | Denmark/Oncorhynchus mykiss | 4,272,973 | CP022099/100 | [20] |
| V. campbelli ATCC 25920 | USA, Hawaii/Seawater isolate | 5,178,103 | CP015863/4 | [38] |
| V. fluvialis ATCC 33809 | Bangladesh/Homo sapiens | 4,827,733 | CP014034/5 | [39] |
| V. parahaemolyticus ATCC 17802 | Japan/Seawater isolate | 5,152,461 | CP014046/7 | [40] |
| V. tasmaniensis LGP32 | France/Crassostrea gigas | 4,974,818 | FM954972/3 | [41] |
| Photobacterium damselae 91-197 | USA/Hybrid Striped Bass (Morone sp.) | 4,293,175 | AP018045/6 | [35] |
2.11. Multi-Locus Sequence Analysis of V. anguillarum Housekeeping Genes
Multi-locus analysis (MLSA) was used to infer the phylogeny history of V. anguillarum strains using reference genes including 16S ribosomal RNA subunit (rrn), cell-division protein (ftsZ), glyceraldehyde-3-phosphate dehydrogenase (gapA), gyrase beta subunit (gyrB), rod shape-determining protein (mreB), uridine monophosphate (UMP) kinase or uridylate kinase (pyrH), recombinase A (recA), RNA polymerase alpha subunit (rpoA), and topoisomerase I (topA) gene sequences. Only genes from complete genomes were considered for the MLSA. Sequences were aligned using CLC Genomic workbench v20.0 (CLC Bio). Concatenation of locus sequences was made using Sequence Matrix software v1.7.8 [42]. Phylogenetic analysis was performed using the two software mentioned above with the same parameters. Evolutionary history was estimated using the neighbor-joining method [33] with a bootstrap consensus of 500 replicates, and evolutionary distance was computed using the Jukes–Cantor method [34]. The gene loci and accession numbers are listed in Table S1.
2.12. Genomic Islands Analysis
Detection of genomic islands (GIs) was conducted using IslandViewer v.4 pipeline (https://www.pathogenomics.sfu.ca/islandviewer/browse/), which integrate IslandPath-DIMOB, SIGH-HMM, and IslandPick analysis tools into a single analysis [43]. Analysis was performed for the chromosomes and the plasmids.
2.13. Comparative Analysis of V. aguillarum J360 Large Plasmid
A genomic comparison of virulent and nonvirulent plasmids between V. anguillaurm species was performed using the whole genome alignment tool of CLC Genomics workbench v20.0 with default parameters. Plasmids used in this analysis were: p292-VIB12 (CP023312); p15-VIB43 (CP023056); pVaM3 (CP006701); pJM1 (AY312585); p67vangNB10 (LK021128); and p65-ATCC 68554 (CP023210). Plasmids were aligned to calculate the ANI. A comparative heat map was constructed using the heat map tool with default parameters (Euclidean distance method and complete cluster linkages).
2.14. Statistical Analysis
Fish survival percentages were transformed to arc-sin (). One-way ANOVA was used to determine significance, followed by Tukey’s post-hoc test, to determine significant differences (p < 0.05). All statistical analyses were performed using GraphPad Prism 7 (GraphPad Software, California, CA, USA).
2.15. Ethics Statement
All animal protocols required for this research were approved by the Institutional Animal Care Committee and the Biosafety Committee at Memorial University of Newfoundland (MUN). Animal assays were conducted under protocols #16-92-KG, #18-01-JS #18-03-JS, and biohazard license L-01, approved on 26-05-2020.
3. Results
3.1. Phenotypic, Biochemical, and Enzymatic Characterization
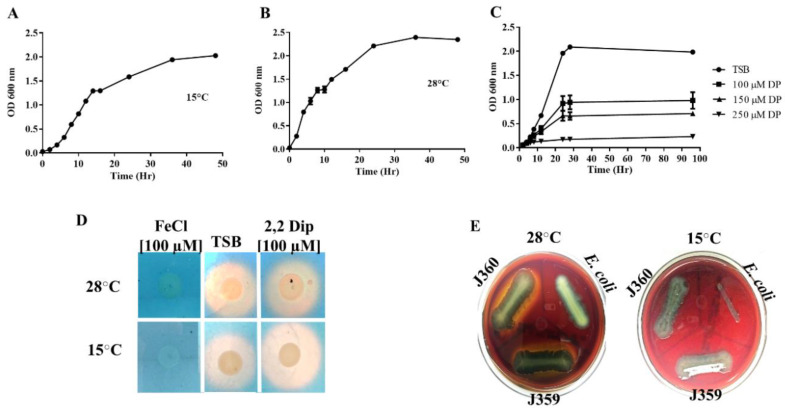
V. anguillarum strain J360 was capable of growing in Tryptic soy broth (TSB) and Luria Bertani (LB) medium up to 30 °C (Table 2). The V. anguillarum J360 doubling time in TSB supplemented with 2% NaCl at 15 °C was 2 h (Figure 1A) and 1 h at 28 °C (Figure 1B). V. anguillarum J360 did not grow at 37 °C, in TCBS selective media at 15 and 28 °C, or in the absence of NaCl. V. anguillarum J360 was shown to be motile and capable of synthesizing type I fimbria, oxidase, and catalase (Table 2). The antibiogram analysis showed that V. anguillarum J360 is ampicillin-resistant and susceptible to tetracycline, oxytetracycline, sulfamethoxazole, chloramphenicol, colistin sulphate, oxalinic acid, and O-129 (Table 2).
Table 2.
Phenotypic characteristics of V. anguillarum J360.
| Characteristic | Vibrio anguillarum J360 |
|---|---|
| Growth at: | |
| 4 °C | + |
| 15 °C | + |
| 28 °C | + |
| 37 °C | − |
| LB NaCl 0% | − |
| LB NaCl 0.5% | + |
| LB NaCl 2% | + |
| Plate Count Agar 50% seawater | + |
| TCBS | − |
| Motility | + |
| Fimbria Type I | + |
| Siderophores synthesis | + |
| Catalase | + |
| Oxidase | + |
| Antibiogram: | Halo diameter (mm) |
| Tetracycline (10 mg/mL) | 31 (Susceptible) |
| Oxytetracycline (30 mg/mL) | 34 (Susceptible) |
| Ampicillin (10 mg/mL) | 0 (Resistant) |
| Sulfamethoxazole (25 mg/mL) | 33 (Susceptible) |
| Chloramphenicol (30 mg/mL) | 35 (Susceptible) |
| Colistin sulphate (10 mg/mL) | 15 (Susceptible) |
| Oxalinic acid (2 mg/mL) | 39 (Susceptible) |
| O-129 | 39 (susceptible) |
Figure 1.
Bacterial growth and physiological characteristics of V. anguillarum J360. (A) V. anguillarum J360 growth in Trypticase soy broth (TSB) at 15 °C for 48 h. Independent triplicates were utilized. (B) V. anguillarum J360 growth in TSB at 28 °C for 48 h. Independent triplicates were utilized. (C) V. anguillarum J360 growth under iron-limited conditions (TSB supplemented with 100, 150, and 250 µM of 2,2-dipyridyl). (E) V. anguillarum J360 hemolysin activity assay on Sheep blood agar plates incubated at 15 °C and 28 °C for 48 h. (D) Siderophore synthesis on chrome azurol S (CAS) agar plates from V. anguillarum J360 grown under iron-enriched conditions (TSB supplemented with 100 µM of FeCl3), standard culture conditions (TSB), and iron-limited conditions (TSB supplemented with 100 µM of 2,2-dipyridyl). Yellow reaction around the V. anguillarum J360 colony indicated positive for siderophore synthesis.
V. anguillarum J360 growth under iron-limited conditions was evaluated at 15 °C on TSB media with different 2,2-dipyridyl concentrations (100, 150, 250 µM). V. anguillarum J360 was able to grow in the presence of high concentrations of 2,2-dipyridyl. The doubling time at 100 µM of 2,2-dipyridyl was 3 h (Figure 1C); however, it was increased to 4 h (OD600 ≈ 0.7) and 5 h (OD600 ≈ 0.2) at 150 and 250 µM of 2,2-dipyridyl, respectively (Figure 1C). No siderophores secretion was observed in the cells grown under iron-enriched conditions. Additionally, there was no significant differences in the size of the halo for siderophores secretion between nonsupplemented TSB and supplemented TSB with 100 µM of 2,2-dipyridyl at 28 °C (Figure 1D). Nonetheless, a small difference in the halo increased diameter was observed for siderophores secretion under iron-limited conditions at 15 °C (Figure 1D). Hemolytic activity was evaluated on blood agar plates at 28 °C and 15 °C. V. anguillarum J360 hemolytic activity was observed only at 28 °C (Figure 1E).
The biochemical and enzymatic profiles indicate that V. anguillarum J360 is able to synthetize alkaline phosphatase, esterase (C4), esterase lipase (C8), lipase (C14), leucine, valine and cysteine arylamidase, and acid phosphatase (Table S2). V. anguillarum J360 reduces nitrates and glucose; produces indole from tryptophan; produces urease, β-galactosidase, arginine hydrolase, esculinase and gelatinase; and is able to utilize arabinose, mannose, mannitol, N-acetyl-glucosamine, and maltose (Tables S3 and S4). The API20NE profile indicated that the isolate was V. fluvialis, with 99.7% probability (Table S3).
3.2. MALDI-TOF and Agglutination Analysis
The MALDI-TOF mass spectrometry score for V. anguillarum was 1.96, indicating that there was a low confidence for identification. The V. anguillarum agglutination test was positive for the O2 serotype and negative for the O1 serotype.
3.3. Infection Assay in Specific Pathogen-Free Lumpfish
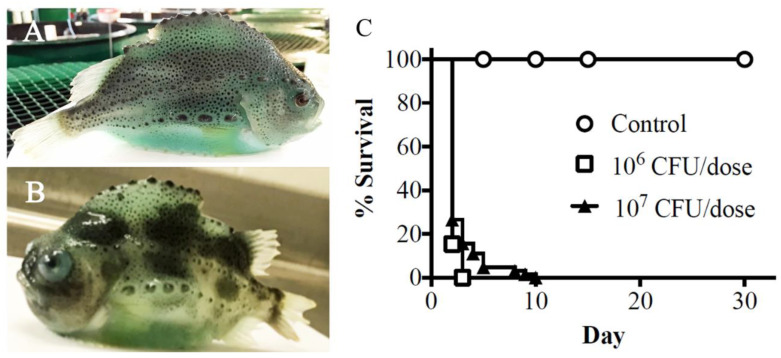
Naïve cultured lumpfish (≈55 g) were intraperitoneally (ip) infected with V. anguillarum J360 to evaluate its virulence (Figure 2A,B). Two groups of 60 lumpfish were injected with 1 × 106 and 1 × 107 CFU/dose, respectively. The control group was mock-infected with PBS, and mortality was monitored until 30 days post-infection (dpi). Mortality began at 2 dpi and reached 100% in both doses at 10 dpi (Figure 2C). Vibriosis clinical signs were observed at 5 dpi, including hemorrhage over the lateral lines, dorsal and/or caudal fins, ventral sucker, vent, mouth, and the operculum. Additionally, infected fish exhibited exophthalmia (Figure 2B).
Figure 2.
Pathogenicity and virulence of V. anguillarum J360 in lumpfish (C. lumpus). (A) Healthy lumpfish cultured at Dr. Joe Brown Aquatic Research Building (JBARB). (B) Lumpfish-infected V. anguillarum J360 (5 days post-infection at 10 °C). (C) Survival of lumpfish intraperitoneally infected with V. anguillarum J360.
3.4. V. anguillarum J360 Genome Sequencing and Annotation
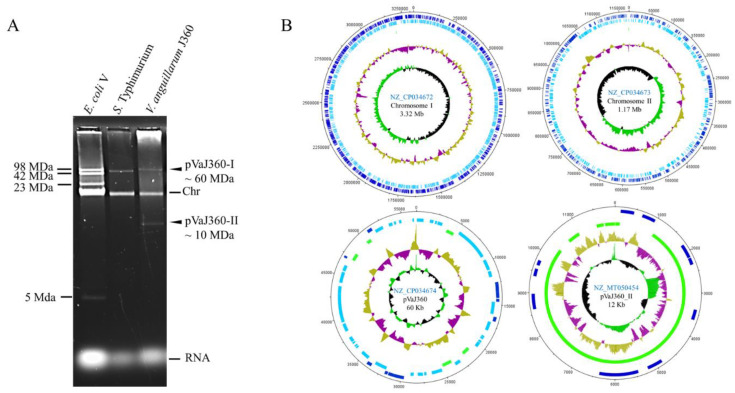
V. anguillarum genomic DNA sequenced by PacBio resulted in five contiguous sequences (contigs). The larger contigs corresponded to circularized chromosome-I (3,320,860 bp), chromosome-II (1,171,281 bp), and a large plasmid pVaJ360-I (56,630 bp) with coverage assemblies of 211, 167, and 36 times, respectively. The plasmid profile of V. anguillarum J360 indicated that there was also a small plasmid (Figure 3A). Using Illumina reads, we were able to assemble pVaJ306-II (11,995 bp) with a coverage of 226 times. The V. anguillarum J360 genome was submitted to NCBI under the BioProject (PRJNA485045) and BioSample (SAMN09781303). The complete genome of V. angullarum J360 possesses two chromosomes [chromosome-I (NZ_CP034672) and chromosome-II (NZ_CP034673)], a large plasmid pVaJ360-I (NZ_CP034674), and a small plasmid pVaJ360-II (MT050454), and has an estimated total length of 4.55 Mb and a G + C content of 44.6% (Figure 3B, Table 3). RAST pipeline annotation predicted a total of 441 subsystems and 3149 coding sequences (CDS) for chromosome-I; a total of 88 subsystems and 1143 CDSs for chromosome-II; a total of 2 subsystems and 96 CDSs for the large plasmid pVaJ360-I; and a total of 24 CDS in a single subsystem and 1 total RNAs sequence for the small plasmid pVaJ360-II (Table 4). The NCBI Prokaryote Genome Annotation pipeline showed a total of 4371 genes predicted, a total of 10 (5S), 9 (16S), and 9 (23S) rRNAs, 105 tRNAs, and 4 non-coding RNAs(ncRNAs) for the whole genome (Table 5).
Figure 3.
V. anguillarum J360 genome visualization. (A) V. anguillarum J360 plasmid profile in 0.5% agarose using nucleic acids alkaline extraction; E. coli V and S. Typhimurium UK-1 (χ3761) were used as markers; Chr: Chromosomal band. (B) Genome map of V. anguillarum strain J360. Chromosome I, chromosome II, large plasmid pVaJ360-I (~60 MDa), and small plasmid pVaJ360-II (~10 MDa) were mapped using DNA plotter. Blue bars represent forward genes; light blue bars represent reverse genes; green bars represent pseudogenes and miscellaneous features. Gold (high) and violet (low) represent G + C content percent; green (high G + C) and black (low G + C) represent skew.
Table 3.
V. anguillarum J360 genome summary.
| Labels | Size (Mb) | Topology | RefSeq ID | INSDC Identifier |
|---|---|---|---|---|
| Chromosome-I | 3.32 | Circular | NZ_CP034672 | CP034672 |
| Chromosome-II | 1.17 | Circular | NZ_CP034673 | CP034673 |
| Plasmid pVaJ360-I | 0.06 | Circular | NZ_CP034674 | CP034674 |
| Plasmid pVaJ360-II | 0.012 | Circular | NZ_MT050454 | MT050454 |
Table 4.
Rapid annotation subsystem technology (RAST) V. anguillarum J360 annotation summary.
| Chromosome-I | Chromosome-II | Plasmid pVaJ360-I | Plasmid pVasJ360-II | |
|---|---|---|---|---|
| Genome size (bp) | 3,320,860 | 1,172,081 | 56,630 | 11,995 |
| G+C content (%) | 44.6 | 44.1 | 43.7 | 47 |
| Number of subsystems | 441 | 88 | 2 | 1 |
| Number of coding sequences | 3149 | 1143 | 96 | 24 |
| Number of RNAs | 129 | 5 | - | 1 |
Table 5.
NCBI prokaryotic Genome Annotation pipeline V. anguillarum J360 genome annotation summary.
| Attribute | Data Provided |
|---|---|
| Annotation pipeline | NCBI |
| Annotation method | Best placed reference protein set; GeneMarks v4.6 |
| Genes (total) | 4371 |
| CDSs (total) | 4234 |
| Genes (coding) | 3966 |
| Genes (RNA) | 137 |
| rRNAs | 10, 9, 9 (5S, 16S, 23S) |
| Complete rRNAs | 10, 9, 9 (5S, 16S, 23S) |
| tRNAs | 105 |
| ncRNAs | 4 |
| Pseudogenes (total) | 268 |
| Pseudogenes (ambiguous residues) | 0 of 268 |
| Pseudogenes (frameshifted) | 105 of 268 |
| Pseudogenes (incomplete) | 163 of 268 |
| Pseudogenes (internal stop) | 45 of 268 |
| Pseudogenes (multiple problems) | 41 of 268 |
3.5. Whole Genome Alignment, Phylogeny, and Synteny
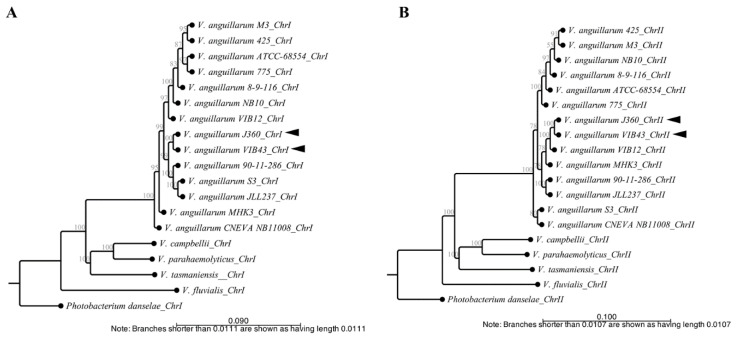
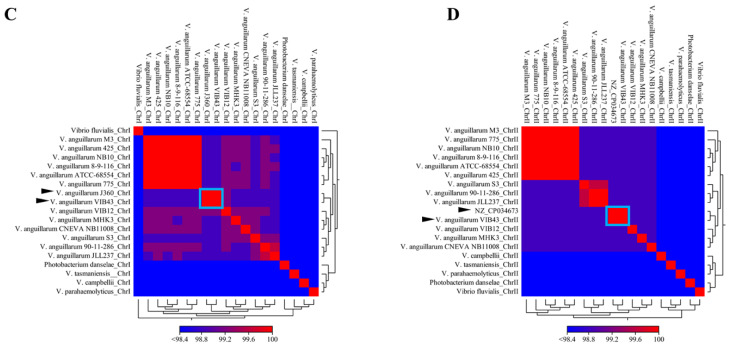
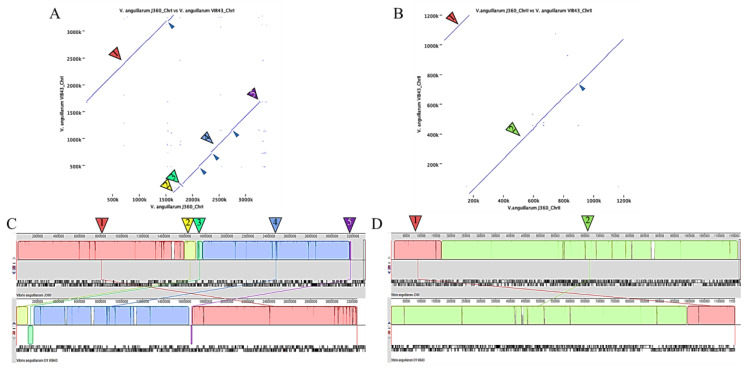
Phylogenetic analysis of Vibrio spp. was performed using CLC Bio with only complete genome sequences (Table 1). Phylogenetic analysis of chromosome-I showed that there are three clusters with three or more strains, whereas V. tasmaniensis, V. parahaemolyticus, and V. campbellii clustered together as one and V. fluvialis clustered separately. By contrast, V. anguillarum species were represented by two clusters plus V. anguillarum strains CNEVA, MHK3, VIB12, NB10, and 8-9-116 that clustered separately. V. anguillarum J360 was closely related to V. anguillarum VIB43 isolated from Scotland, UK (Figure 4A, Table 1). Phylogenetic analysis of chromosome-II indicated four clusters, whereas non-V. anguillarum species clustered together, and V. fluvialis clustered separately (Figure 4B). Similar to chromosome-I, V. anguillarum J360 chromosome-II was closely related to V. anguillarum VIB43 (Figure 4B). The same results were observed in the phylogenetic analysis using MEGA X for chromosome-I (Figure S1A) and chromosome-II (Figure S1B). The heat map indicated that there was a high identity between V. anguillarum J360 and V. anguillarum VIB43 alignments of chromosome-I (Figure 4C) and chromosome-II (Figure 4D), respectively. The ANI analysis for the whole genome alignment indicates a 99.95% for chromosome-I (Figure S2A) and 99.93% for chromosome-II of genome identity (Figure S2B) that support previous observed results. The dot plot visualization matches between V. anguillarum J360, and the closest related strain, V. anguillarum VIB43, showed that there was a high similarity within the genome. However, two inversion events and genomic gaps (GGs) were identified (Figure 5A,B). The whole genome alignment identified several locally collinear blocks (LCBs), described as conserved segments free from genomic rearrangements [36]. The comparative alignment analysis of each chromosome showed five LCBs in chromosome-I (Figure 5C) and two LCBs in chromosome-II (Figure 5D). Additionally, the LCBs identified in both chromosomes are conserved, and in agreement with the reversion events and GGs identified in the dot plot analysis (Figure 5).
Figure 4.
Phylogenetic and comparative genomic analysis of V. anguillarum J360. (A) Phylogenetic history of V. anguillarum J360 chromosome-I. (B) Phylogenetic history of chromosome-II. Evolutionary history was inferred using the neighbor-joining method, with a bootstrap consensus of 500 replicates for taxa analysis. Evolutionary distance was computed using the Jukes–Cantor method. All ambiguous positions were removed for each sequence pair (pairwise deletion option). (C) Heat map visualization of aligned sequences identity for V. anguillarum J360 chromosome-I. (D) Heat map visualization of aligned sequences identity for V. anguillarum J360 chromosome-II. Whole genome alignments and the phylogenetic analysis involved 18 Vibrio sp. listed in Table 1. Photobacterium damselae 91–197 as an outgroup. Analysis was performed using CLC workbench v.20 (CLC Bio). Black arrows represent V. anguillarum J360 genome and V. anguillarum VIB43 as the closest related strain. Light blue square represents the percentage of identity between strains.
Figure 5.
Comparative genome synteny between V. anguillarum J360 and V. anguillarum VIB43. (A) Dot plot analysis for chromosome-I. (B) Dot plot analysis for chromosome-II. Numerated arrows represent homologous regions and solid arrows represent inversions. Dot plots were computed using CLC workbench v.20. (C) Homologous regions identified as locally colinear blocks (LCBs) (numerated arrows) of chromosome-I. LCB-1 (red); LCB-2 (yellow); LCB-3 (spring green); LC-4 (light blue); LCB-5 (magenta). (D) Homologous regions identified as locally colinear blocks (LCBs) (numerated arrows) of chromosome-II. LCB-1 (orange); LCB-2 (green). Small dark-blue arrows indicate genomic gaps or unmatched regions (GGs).
3.6. Multi-Locus Sequence Analysis (MLSA) and Phylogeny
We also utilized MLSA to contrast these results with the whole genome analyses. MLSA was computed using the nine housekeeping genes listed in Table S1. Gene sequences were aligned, concatenated, and analyzed. The phylogenetic analysis performed in CLC Bio showed that there were five clusters with at least two strains, whereas V. anguillarum J360 clustered alone. This analysis indicated that V. anguillarum J360 is closely related to V. anguillarum NB10, which clustered with V. campbelli and V. tasmaniensis, and is distantly related to V. anguillarum 775 and V. anguillarum M3 (Figure S3A). The same results were observed using MEGA X software (Figure S3B).
3.7. Distribution of Genes Associated with Pathogenesis and Environmental Adaptation in V. anguillarum J360
Gene distribution within the V. anguillarum J360 chromosomes was determined for virulence and environmental adaptation-related genes (Table 6). Genes associated with iron homeostasis were identified, including ferrous and ferric transport, and regulatory mechanisms. In chromosome-I genes like ferric iron uptake transcriptional regulator (fur), tonB1, tonB2, and feoABC uptake systems were identified. Hemolysis activity-related genes like heme transport (DYL72_00770, DYL72_02835), TonB-dependent hemoglobin receptors (DYL72_17445, DYL72_00730, DYL72_20920), and heme transporters (CcmB and CcmD) were distributed in both chromosomes. Furthermore, five hemolysin-encoding genes (DYL72_01800, DYL72_07805, DYL72_12295, DYL72_17765) were found distributed in both chromosomes, including a thermolabile hemolysin (DYL72_17760) in chromosome-II.
Table 6.
Predicted genes associated with subsystems of pathogenesis and environmental adaption.
| Gene Subsystem Category and Name | Presence/Absence of Gene in V. anguillarum J360 | GenBank Accession N° | |
|---|---|---|---|
| Chromosome-I | Chromosome-II | ||
| Iron transport and regulation | |||
| iron-regulated protein A | X | DYL72_00705 | |
| tonB2, exbD2 | X | DYL72_00755, DYL72_00745 | |
| tonB1, exbB, exbD1 | X | DYL72_00260, DYL72_00265, DYL72_00270 | |
| fur | X | DYL72_03070 | |
| iron ABC transport permease | X | DYL72_00175, DYL72_00765 | |
| Heme transport | |||
| Ton-B-dependent hemoglobin receptor | X | X | DYL72_17445, DYL72_00730, DYL72_20920 |
| hutXZ | X | DYL72_00735, DYL72_00740 | |
| heme ABC transporter protein | X | DYL72_00770, DYL72_02835 | |
| heme exporter protein ccmBD | X | DYL72_02830, DYL72_02840 | |
| Ferrous and ferric transport | |||
| ferric ABC transporter | X | DYL72_00180 | |
| feoABC | X | DYL72_02945, DYL72_02940, DYL72_02935 | |
| Ferrichrome | |||
| fhuACBD | X | DYL72_06770, DYL72_10590 | |
| Hemolysins | |||
| hemolysin genes | X | X | DYL72_01800, DYL72_07805, DYL72_12295, DYL72_17765 |
| thermolabile hemolysin | X | DYL72_17760 | |
| Toxins-associated genes | |||
| toxins and pseudogenes | X | DYL72_00035, DYL72_00045, DYL72_14975, DYL72_14985 | |
| rtxA, hipA | X | DYL72_01180, DYL72_03440 | |
| ampC | X | DYL72_17850 | |
| type II toxin–antitoxin system RelBE/ParDE/DinJ family | X | DYL72_18560, DYL72_18565, DYL72_18715, DYL72_18750, DYL72_19140, DYL72_19250, DYL72_19960 | |
| type II toxin–antitoxin system prevent-host-death family antitoxin | X | DYL72_18805 | |
| Txe/YoeB family addiction module | X | DYL72_18810 | |
| type II toxin–antitoxin system Phd/YefM family antitoxin | X | DYL72_19255 | |
| type II toxin–antitoxin system YafQ family toxin | X | DYL72_19965 | |
| toxin–antitoxin system subunit antitoxin | X | DYL72_20370 | |
| Metalloproteases | |||
| CPBP family intramembrane metalloprotease | X | DYL72_00295 | |
| pmbA, tldD, ftsH | X | DYL72_05300, DYL72_09550, DYL72_10735 | |
| SprT family zinc-dependent metalloprotease | X | DYL72_09830 | |
| M6 family metalloprotease domain-containing protein | X | DYL72_17780 | |
| Secreted enzymes | |||
| phospholipase gene | X | DYL72_16230 | |
| lipase | X | DYL72_17465, DYL72_18050, DYL72_20375 | |
| Motility and Chemotaxis | |||
| fliRQPONMLKJIHGFE, fliSTD, fliL | X | DYL72_03140-DYL72_03205, DYL72_03225-DYL72_03235, DYL72_08330 | |
| motYA, motB | X | DYL72_12660, DYL72_00275, DYL72_12090 | |
| flhFAB | X | DYL72_02900, DYL72_02905, DYL72_03135, | |
| flag, flaCA | X | DYL72_03240, DYL72_03670, DYL72_03675 | |
| flagellin | X | DYL72_03245, DYL72_03250, DYL72_03255 | |
| flgLKJIHGFEDCB, flgAMNP | X | DYL72_03685-DYL72_03735, DYL72_03750-DYL72_03765 | |
| flagellar basal-body protein | X | DYL72_03775 | |
| pomA | X | DYL72_12085 | |
| flagellar brake protein | X | DYL72_17355, DYL72_20195 | |
| methyl-accepting chemotaxis protein | X (17) | X (16) | |
| chemotaxis response regulator protein-glutamate methylesterase | X | X | DYL72_00595, DYL72_02870, DYL72_16590, |
| cheD, cheW, cheA, cheV, cheY, cheX, cheC | X | X | DYL72_00600, DYL72_00610, DYL72_00620, DYL72_01045, DYL72_02885, DYL72_09235, DYL72_20035 |
| cheV2, cheW2, cheW3, cheA2, cheV3, cheD2, cheW4, cheW5, cheA3, cheV4 | X | X | DYL72_02435, DYL72_02855, DYL72_02860, DYL72_02875, DYL72_03745, DYL72_16595, DYL72_16610, DYL72_16615, DYL72_16620, DYL72_21345 |
| chemotaxis protein | X | X | DYL72_00635, DYL72_16275, DYL72_16460 |
| chemotaxis protein methyltransferase | X | DYL72_03740 | |
| Type IV pilus | |||
| pilQ, pilW, pilM, pilT, pilB | X | DYL72_05800, DYL72_01100, DYL72_05820, DYL72_09795, DYL72_10350 | |
| Type VI secretion system (T6SS) | |||
| tssI, tssBCEFG, tssJ, tssK tssH, tssAM | X | DYL72_16415, DYL72_17000—DYL72_17020, DYL72_17030, DYL72_17045, DYL72_17055, DYL72_17070, DYL72_17075, | |
| tssI2, tssM2, tssKJHFE2, tssC2, tssI3, tssI4, tssI5, tssI6 | X | X | DYL72_17085, DYL72_21115, DYL72_00885, DYL72_00895, DYL72_00900, DYL72_00930, DYL72_00940, DYL72_00945, DYL72_00955, DYL72_00960, DYL72_00975, DYL72_02685, DYL72_10015 |
| tagHO | X | DYL72_17025, DYL72_17065 | |
| Hcp family type VI secretion system effector | X | X | DYL72_16420, DYL72_18695, DYL72_21120, DYL72_02690, DYL72_10010 |
| type VI secretion system tube protein Hcp | X | DYL72_00965 | |
| DotU family type IV/VI secretion system protein | X | X | DYL72_17050, DYL72_00890 |
| type VI secretion protein | X | X | DYL72_17080, DYL72_00950, DYL72_00970, DYL72_00985 |
| type VI secretion system PAAR protein | X | X | DYL72_21095, DYL72_02650, DYL72_02660, DYL72_10045, |
| type VI secretion protein VasB-1 | X | DYL72_00935 | |
| Quorum sensing | |||
| quorum-sensing autoinducer synthase | X | DYL72_17995 | |
| Regulators | |||
| transcriptional regulator LuxR | X | X | DYL72_06050, DYL72_21285 |
| transcriptional regulator LysR | X | DYL72_03060, DYL72_09990 | |
| transcriptional regulator XRE | X | DYL72_10570 | |
| cysB | X | DYL72_13470 | |
| nhaR | X | DYL72_10935 | |
| hfq | X | DYL72_09115 | |
Toxin–antitoxin-associated genes were also identified. We found that V. anguillarum J360 possesses five different type II toxin–antitoxin system protein families, such as RelBE/ParDE/DinJ, Txe/YoeB, PhD/YefM, YafQ, and a prevent-host-death system located in chromosome-II. In addition, a toxin precursor gene (rtxA), a serine/threonine-protein kinase gene (hipA), and four toxin genes (DYL72_00035, DYL72_00045, DYL72_14975, DYL72_14985) were found in chromosome-I, and an antibiotic-resistance gene (ampC) was found in chromosome-II.
Metalloprotease coding genes were found in both chromosomes, including a CPBP family intermembrane metalloprotease (DYL72_00295) and a sprT family zinc-dependent metalloprotease (DYL72_09830) gene, and three metalloprotease genes (pmbA, tldD, and ftsH) are present in chromosome-I. Additionally, a M6 family domain that possesses metallopeptidase activity was found in chromosome-I. Motility genes were found in chromosome-I, except for flagellar motor brake proteins that were found in chromosome-II. Chemotaxis genes were found on both chromosomes, including 17 genes in chromosome-I and 16 genes in chromosome-II that encode for methyl-accepting chemotaxis proteins. Four copies of cheV and five copies of cheW were also found, and these are involved in phosphorylation-dependent excitation and methylation-dependent adaptation, respectively. We identified five type IV pilus-associated genes in chromosome-I, and type VI secretion system-related genes in both chromosomes, including six copies of tssI, and two operons encoding tssBCEFG and tssKJHFE2. Two secretion system families were identified, including the Hcp type VI secretion system family effector and DotU family type IV and type VI secretion system-related proteins. A single quorum-sensing-associated gene was found in chromosome-II, which is a quorum-sensing autoinducer synthase. Transcriptional regulators were found in chromosome-I, such as lysR, cysB, nhaR, and hfq, and luxR was present in both chromosomes.
3.8. Genomic Islands (GIs)
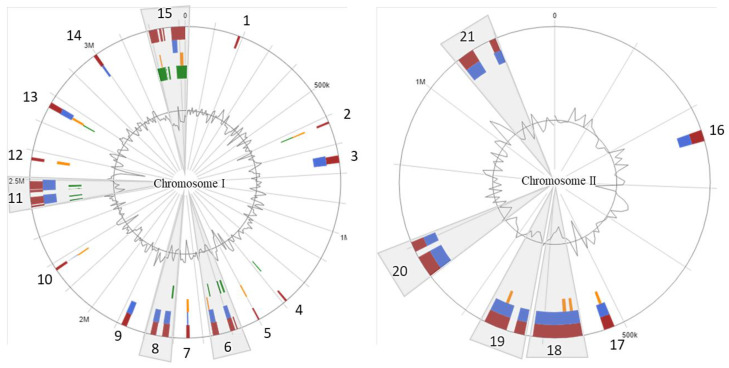
Twenty-one putative GIs were identified within the chromosomes; 15 GIs in chromosome-I and 6 GIs in chromosome-II, respectively (Figure 6; Supplementary file 1 and 2). The GIs size ranged from 5 to 73.4 kb with a total of 724 genes. The largest genomic island (GI15) (Figure 6) consisted of 147 genes (predicted by at least one of the three software used, see Method section), and was flanked by two site-specific integrase genes and a zinc ribbon-domain protein.
Figure 6.
V. anguillarum J360 genomic islands (GIs). Genomic islands (GIs) detected in chromosome-I; genomic islands (GIs) detected in chromosome-II. Red bars represent GIs detected using 3 different packages; blue bars represent GIs detected with the SIGI-HMM package; orange bars represent GIs detected with the IslandPath-DIMOB package; green bars represent GIs detected with the IslandPick package.
Genes encoding for integrases, porins, transposases, and iron transport were found among the GIs. Genes such as phosphonate C-P lyases, associated with the cleavage of carbon-phosphorus compounds for organic phosphorus reservoir in marine bacteria [44], zinc ribbon domain proteins, AbrB/MazE/SpoVT DNA-binding domains, a ParA domain, for a GCN5-related N-acetyltransferase (GNAT) associated with regulatory post-transcriptional acetylation [45], two site-specific integrases, and a MasF transcriptional regulator related to toxin/anti-toxin systems (Supplementary file 1) were identified in these GIs. GI5 is the smallest genomic island identified in V. anguillarum J360 (Figure 6), which possesses five unique genes that encode for acetyltransferase, acyltransferase, formyltransferase, asparagine synthase, and one hypothetical protein (Supplementary File 1).
3.9. V. anguillarum Large Plasmids Analysis
The plasmid profile indicates that V. anguillarum J360 harbors one large plasmid and one small plasmid (Figure 3A). The large plasmid pVaJ360-I (~60 kb) has genes that encode for integrases, DNA-binding proteins, peptidases, site-specific integrases, resolvase, mobile elements, and pro-phages. Comparative analysis showed that pVaJ360-I is not related to V. anguillarum virulent plasmids such as pJM1 (strain 775); p65 (strain ATCC-68554); pJM1-like plasmid, p67 (strain NB10); and p15 (strain VIB43) (Figure S5A). The ANI analysis of these plasmids demonstrated that pVaJ360-I does not have a percentage of identity with pJM1 or pJM1-like plasmids (Figure S5B), suggesting that pVaJ360 is not a virulent plasmid.
The small plasmid pVaJ360-II (~12 kb) has only 10 CDSs that encode for hypothetical proteins, transposases, mobile elements, a transcriptional regulator LysR, a tRNA Glu, and additionally 14 miscellaneous features. BLASTn analysis indicates that this plasmid has no similarities with the pJM1 or pJM1-like plasmids described above.
4. Discussion
Lumpfish across hatcheries and deployment sites frequently show signs of systemic bacterial infection, including skin lesions, gill hemorrhages, and bacterial aggregations in lymphoid organs (i.e., spleen, liver, head kidney) [10]. In the United Kingdom, Iceland, and Norway, several bacterial outbreaks have been reported in lumpfish hatcheries and at cage sites, and the most frequent pathogen detected is V. anguillarum [46]. Thus, it is not surprising that this pathogen was found to be present in Atlantic Canada.
V. anguillarum serotypes O1, O2, and O3 are the most prevalent strains among the 23 serotypes currently described [18,19]. Agglutination assays indicated that V. anguillarum J360 is O2, similar to other V. anguillarum strains isolated from lumpfish infections in the North Atlantic [46]. The biochemical profile obtained using API20NE showed that V. anguillarum J360 was able to reduce sugars, urea, and produce indole, suggesting a 99% possibility for V. fluvialis (Table S3). Although the biochemical profile did not identify V. anguillarum J360, its phenotypic characterization is consistent with other V. anguillarum isolates [25,26], except that V. anguillarum J360 was positive for urease (Table S4). This result is coincident with the presence of the urease encoding gene in chromosome-II (DYL2_19555). In addition, V. anguillarum J360 was not able to grow in TCBS-selective media, suggesting susceptibility to bile salts or a poor adaptation to the culture medium (Table 2) [47].
V. anguillarum J360 showed a thermo-inducible α-hemolysin activity at 28 °C, but no hemolytic activity was observed at temperatures below 15 °C (Figure 1E). Hemolytic activity is an important virulence factor for V. anguillarum [48]. For instance, severe hemorrhages are a typical clinical sign of V. anguillarum infection in fish, including lumpfish [46]. Coincidently, in the current study, V. anguillarum J360 was shown to be highly virulent in lumpfish, and infected lumpfish displayed severe hemorrhagic symptoms at 5 dpi (Figure 2B), similar to other strains described in Marco-Lopez et al. 2013. Koch’s postulates for V. anguillarum J360 showed that lumpfish infected with 1 × 106 and 1 × 107 CFU/dose reached 100% mortality within 10 dpi at 10 °C (Figure 2C). In addition, V. anguillarum was re-isolated from the spleen, liver, and head-kidney, confirming Koch’s postulates. The original V. anguillarum outbreak in cultured lumpfish and the infection assays in the current study showed similar clinical signs (Figure 2B). V. anguillarum hemolytic activity was evident during infection. However, the lumpfish is a cold-water fish typically cultured between 6 and 12 °C [49]. These results contradicted with the V. anguillarum thermo-inducible hemolytic activity at 28 °C (Figure 1E). Perhaps, internal fish conditions (e.g., innate and adaptive immunity) triggered V. anguillarum hemolytic activity. These results suggest that its regulatory mechanisms need further analysis.
V. anguillarum J360 possesses two chromosomes, a large plasmid and a small plasmid (Figure 3 and Table 4). Vibrio spp. and V. anguillarum genomes selected for phylogenetic and comparative genomics analysis possess two chromosomes and one large plasmid (Table 1). Typically, serotype O1 harbors a virulent plasmid, called pJM1 or pJM1-like, and serotypes O2 and O3 strains possess a nonvirulent large plasmid [50,51] or they do not harbor large plasmids [20,37]. V. anguillarum J360 is a O2 serotype that does not harbor a virulence plasmid (Figure S5), suggesting that this strain could increase its virulence if a virulence plasmid is acquired.
The total genome size of V. anguillarum J360 is 4,561,566 bp (Table 4), which is larger than the currently available V. anguillarum genomes (Table 1). This may suggest that V. anguillarum J360 may have acquired genetic material through horizontal gene transfer and/or adapted to its lumpfish host or to environmental conditions in Atlantic Canada.
Phylogenetic distance based on the whole genome alignment analysis of chromosome-I and chromosome-II showed that V. anguillarum J360 is closely related to V. anguillarum VIB43, and distantly related to V. anguillarum VIB12 (Figure 4A,B). Interestingly, V. anguillarum VIB43 and VIB12 were isolated from the same host species, sea bass (Dicentrarchus labrax), but from different locations. V. anguillarum VIB43 was isolated in Scotland and V. anguillarum VIB12 was isolated in the Mediterranean Sea [20]. The ANI analysis between V. anguillarum J360 and V. anguillarum VIB43 showed a 99.93% identity for chromosome-I and 99.95% for chromosome-II (Figure 4C,D and Figure S2A,B), which suggest that these two strains share a common ancestor.
Additionally, the whole genome phylogenetic analysis showed that V. anguillarum J360 and VIB43 are not closely related to V. anguillarum 775, M3, and NB10, which is contradictory to previous MLSA studies that indicated that V. anguillarum VIB43 and VIB12 are closely related to those V. anguillarum strains [20,50]. The MLSA computed the phylogenetic stress based on concatenated conserved sequences [51,52]. In this study, we used nine conserved housekeeping genes, including 16S rRNA, ftsZ, gapA, gyrB, mreB, pyrH, recA, rpoA, and topA (Table S1). In contrast to the whole genome phylogenetic analysis, the MLSA showed that V. anguillarum J360 clusters alone, and the closest related strain is V. anguillarum NB10 isolated from rainbow trout (Oncorhynchus mykiss) in the Gulf of Bothnia, Sweden [6], which is distantly related to V. anguillarum 775 and M3 strains (Figure S3, Table 1). V. anguillarum M3 was isolated from Japanese flounder (Paralichthys olivaceus) in Shandong, China, and classified as closely related to V. anguillarum 775 isolated from Coho salmon (Oncorhynchus kisutch) in the Pacific coast of USA [7,8]. By contrast, MLSA phylogenetic analysis based only on the 16S rRNA gene showed that V. anguillarum NB10 is closely related to V. anguillarum M3 [6].
In contrast to the MLSA analysis, the whole genome phylogenetic analysis of V. anguillarum strains is in concordance with the geographic origin of the strain isolation. For instance, according to the whole genome phylogenetic analysis, V. anguillarum J360 and V. anguillarum VIB43, both isolated in the North Atlantic Ocean, are highly related, and closely related to V. anguillarum strains 90-11-286, S3, and JLL237 isolated in Finland (Table 1; Figure 4). Actually, these geographic locations are a natural habitat for lumpfish populations [53].
In contrast to the whole genome phylogenetic analysis, the MLSA uses protein-coding genes, which evolved at a slow but constant rate, and it could have better resolution, especially at the species level [51,52]. However, the selection and number of coding genes, and alignment method, are variable for MLSA. We found that the phylogenetic analysis using whole genomes is more reliable than MLSA, and the robustness of our analyses showed to be consistent with two different software. In addition, whole genome analyses allow homologous regions, deletion, translocation, and inversion events to be identified.
Genome alignment and synteny analysis between V. anguillarum J360 and V. anguillarum VIB43 showed a high similarity within the chromosome sequences, but inversions and unmatched regions were also observed (Figure 5A,B). This suggests that homologous recombination events play an important role in V. anguillarum evolution, perhaps influenced by insertion sequence (IS) elements such as chromosomal integrons or “super integrons” (SIs) describing Vibrio spp. and several Gram-negative species [20]. We determined that there are five LCBs in chromosome-I (Figure 5C) and two LCBs in chromosome-II (Figure 5D) shared between V. anguillarum J360 and VIB43. Further analysis revealed that all the LCBs present in chromosome-I have small inversion events (Figure 5A,C). In addition, we found that LCBs-1 and -4 have genome gaps (GGs) or unmatching regions in both strains (Figure 5A and Figure S4A). The GGs identified in LCB-1 of V. anguillarum J360 chromosome-I are not present in V. anguillarum VIB43 LCB-1 (Figure S4A). These identified GGs possess several genes that encode for IS families transposases (IS66, ISL3, IS3, IS5) and site-specific integrases previously described in the V. anguillarum VIB43 genome, and with high similarity to V. anguillarum NB10, 775, and ATCC-6855 genomes [6,20]. We found that the unique GGs in LCB-1 of V. anguillarum J360 possess genes related to iron uptake and iron homeostasis (Figure S4A), suggesting that these genes could be acquired by horizontal gene transfer.
In V. anguillarum J360 chromosome-II, two GGs were identified in LCB-1 and LCB-2 (Figure S4B), and both GGs have an IS630-like element belonging to the ISVa15 transposase family. This IS630-like element is not present in V. anguillarum VIB43 LCBs. According to the description of Holm et al. (2018), ISVa3–ISVa20 are new insertion sequence (IS) elements in the V. anguillarum genomic repertory that are responsible for the divergency within strains. This suggests that V. anguillarum J360 and V. anguillarum VIB43 could be derived from a common ancestor and adapted to local environmental conditions and host species.
Pathogenesis-associated genes were found in both chromosomes, but chromosome-I harbors most of the virulence genes and their respective transcriptional regulators (Table 6). No virulence-associated genes were found in the large plasmid pVaJ360-I or in the small plasmid pVaJ360-II. The V. anguillarum virulence plasmid pJM1 possesses intrinsic virulence genes associated with iron uptake, like anguibactin biosynthesis (angA-angE, vabA-E) and anguibactin transport (fatA-fatD) [8,54,55]. By contrast, all the V. anguillarum J360 iron homeostasis-related genes are in its chromosomes. For instance, genes related to ferric-anguiobactin and siderophore uptake (e.g., exbB and exbD2, respectively) are present in chromosome-I. Comparative genomic analyses showed that the large virulent plasmids pJM1, P67-NB10, and p65-ATCC have high similarity (Figure S5A,B). However, the large plasmid pVaJ360-I and the small plasmid pVaJ360-II of V. anguillarum J360 do not present similarity (Figure S5A) or identity (Figure S5B) with other reported plasmid sequences, nor possess virulence-associated genes. This suggests that V. anguillarum J360 does not harbor plasmids previously described in V. anguillarum, including serotype O2 strains [50].
Hemolysins are important virulence factors for V. anguillarum species, and contribute to its attachment, tissue colonization, and iron homeostasis, thus increasing its pathogenicity [56,57]. V. anguillarum J360 has four hemolysin genes, and these are consistent with the hemorrhagic clinical signs observed in lumpfish during the infection assays (Figure 2B). In addition, a thermolabile hemolysin gene is present in chromosome-II, which can be related to the thermo-inducible hemolytic phenotype of V. anguillarum J360 (Figure 1E).
The resistance of V. anguillarum J360 to ampicillin (Table 2) relates to the presence of a class C beta-lactamase (ampC)-encoding gene in chromosome-II. Metalloproteases such as pmbA, tldD, and ftsH genes, which are associated with carbon storage, hydrolysis of peptide bonds, and virulence, were also identified.
V. anguillarum J360 does not possess some of the virulence genes present in V. anguillarum strains isolated from the Pacific coasts, including the metalloproteases empA and prtV [8,58]. Nonetheless, V. anguillarum J360 harbors a M6 family metalloprotease (DYL72_17780) similar to the prtV gene, associated to gelatinase activity (Table S3).
In addition, genes that encoded for secreted enzymes such as phospholipase and lipases were found in chromosome-II, which correlates with the lipase (C14)-positive phenotype observed in the enzymatic profile (Table S3). V. anguillarum J360 possesses several genes associated with flagella and motility, such as the operons fliRQPONMLKJIHGFE (DYL72_03140-DYL72_03205), flgLKJIHGFEDCB (DYL72_03685-DYL72_03735), and motYBA (DYL72_12660, DYL72_00275, DYL72_12090) located in chromosome-I, which is consistent with the mot+ phenotype of V. anguillarum J360 (Table 2).
Virulence factors present in other V. anguillarum genomes (e.g., V. anguillarum 775 and V. anguillarum M3) were not identified in the V. anguillarum J360 genome. These include mannose-sensitive hemagglutinin type 4 pilus (MHSA) [8,54], vstA-vstH genes for a Type VI secretion system [55], and virA-virB genes related to lipopolysaccharides synthesis [58].
In concordance with the locations of the virulence factors, transcriptional regulators such as luxR, lysR, cysB, nhaR, and hfq were mostly located in chromosome-I, and an additional copy of luxR was found in chromosome-II. LuxR belongs to a transcriptional activators family, that together with an N-(3-oxodecanoyl)-L-homoserine lactone (ODHL), mediates the signal transduction mechanisms of quorum-sensing genes such as luxICDABE operon [59]. The duplication of luxR in V. anguillarum J360 suggests that quorum-sensing plays an important role in the biology of this strain.
Genomic Islands (GIs) have been identified in V. anguillarum species, for instance, V. anguillarum NB10 possesses 29 GIs [6], V. anguillarum 775 possesses 10 GIs [8], and V. anguillarum J360 has 21 GIs (Figure 6). We found that GI-19 in chromosome-I (2,994,299..3,011,196 nt) of V. anguillarum NB10 [6] has some homologous regions with V. anguillarum J360 GI-14 in chromosome-I (2,988,473..3,000,617 nt). In addition, we determined that GI-25 (546,066..578,220 nt) and GI-26 (639,559..674,888 nt) in chromosome-II of V. anguillarum NB10 [6] share similar genetic context with GI-18 (552,330..612,508 nt) and GI-19 (622,682..673,456 nt) in chromosome-II of V. anguillarum J360, respectively. However, the low similarity between GIs of V. anguillarum J360 and V. anguillarum NB10 suggests a relatively distant relationship, consistent with the whole genome phylogenetic analysis (Figure 4).
In contrast, 19 GIs were identified in V. anguillarum VIB43 (Figure S6, Supplementary files 3 and 4), which showed high similarities with the GIs founded in V. anguillarum J360. For instances, GI-1, GI-4, and GI-12 present in V. anguillarum VIB43 chromosome-I (Figure S6) possess several homologous regions with GI-8, GI-10, and GI-3 of V. anguillarum J360 chromosome-I (Figure 6), respectively (Supplementary files 1 and 3). In addition, GI-6 shared homologous regions with GIs 11 and 12; however, all these regions are flanked by IS66 or IS66-like family transposases (ISVa9, ISVa15, ISVa11), suggesting that these regions are hot spots for recombination events (Supplementary files 1 and 3). GI-12 of V. anguillarum VIB43 and GI-3 of V. anguillarum J360 are highly conserved (Supplementary 1 and 3). Similar results were observed in chromosome-II. GI-15 and GI-16 of V. anguillarum VIB43 (Figure S6) showed several homologous regions with GI-16 and GI-17 of V. anguillarum J360 (Figure 6), respectively (Supplementary files 2 and 4). GI-17 of V. anguillarum VIB43 has several homologous regions with GIs-18 and GI-19 of V. anguillarum J360 (Supplementary files 2 and 4). The homologous regions between GIs-17 of V. anguillarum VIB43, and GI-18 and GI-19 of V. anguillarum J360 encode for several virulence factors like lipocalin, Hcp tube protein (T6SS), interferase toxin, damage-inducible protein J, secretion proteins, and toxins. These homologous regions are flanked by several IS elements (Supplementary files 2 and 4), which indicates that these three GIs are genomic pathogenic islands, perhaps acquired through horizontal gene transference [60]. These results support the hypothesis that these IS elements (IS66, ISL3, IS3, IS5)6 are responsible for the genomic gaps (GGs) and genomic rearrangements previously mentioned, as well as support the 0.05–8% of genomic differences observed at the identity analyses.
5. Conclusions
In this study, the complete genome of V. anguillarum J360 serotype O2 isolated from infected cultured lumpfish in Newfoundland, Canada was reported. V. anguillarum J360 has a larger genome size (4,549,571 bp) as compared to other available V. anguillarum genomes. The V. anguillarum J360 genome has genes related to antibiotic resistance, hemolysin activity, gelatinase, and lipases that play a major role in virulence. V. anguillarum J360 was shown to be closely related to the V. anguillarum VIB43 strain isolated in Scotland, UK, from sea bass. Comparative genomics revealed that five LCBs are shared between V. anguillarum J360 and V. anguillarum VIB43 chromosome-I, and two LCBs are shared in chromosome-II. Twenty-one genomic islands (GIs) were identified within the chromosomes of V. anguillarum J360. Similar GIs identified in V. anguillarum J360 were found in V. anguillarum VIB43 chromosomes, and this is consistent with the whole genome phylogenetic analysis. V. anguillarum J360 has virulence-associated genes in both chromosomes but does not harbor a virulent plasmid.
Acknowledgments
We thank the staff at the Cold-Ocean and Deep-Sea Research Facility (CDRF) and at the Joe Brown Aquaculture Research Building for expert technical assistance with the fish assays.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/11/1666/s1, Figure S1: Evolutionary taxa relationship of V. anguillarum chromosome-I and chromosome-II, Figure S2: Average nucleotide identity (ANI) comparison of V. anguillarum whole genome alignment, Figure S3: Phylogenetic analysis of V. anguillarum using Multi Locus Sequence Analysis (MLA), Figure S4: Comparative whole genome alignment closeup between V. anguillarum J360 and V. anguillarum VIB43, Figure S5: Comparative analysis V. anguillarum J360 large plasmid pVaJ360-I, Figure S6: V. anguillarum VIB43 genomic islands (GIs), Table S1: Enzymatic profile of V. anguillarum J360 using the API ZYM system, Table S2: Enzymatic profile of V. anguillarum J360 using API20NE, Table S3: Enzymatic profile of V. anguillarum J360 using API20E, Table S4. Housekeeping genes used for MLSA. Supplementary file S1: Genomic islands V. anguillarum J360 chromosome-1; Supplementary file S2: Genomic islands V. anguillarum J360 chromosome-2; Supplementary file S3: Genomic islands V. anguillarum VIB43 chromosome-1; Supplementary file S4: Genomic islands V. anguillarum VIB43 chromosome-2.
Author Contributions
Conceptualization: J.S. and I.V.; methodology: J.S., I.V., T.C., S.C., H.G., J.M., N.O. and D.B.; investigation: J.S., I.V., T.C., S.C., H.G., N.O., J.M., D.B. and J.D.W.; writing—original draft: I.V. and J.S.; resources: J.S., N.O., D.B. and J.D.W.; writing—review and editing: I.V., J.S., J.D.W., T.C., S.C., H.G., J.M., N.O., D.B., J.D.W. and J.S.; visualization: I.V. and J.S.; supervision: J.S. and J.D.W.; funding acquisition: J.S., J.D.W. and D.B. All authors have read and agreed to the published version of the manuscript.
Funding
Research funding was provided by the Ocean Frontier Institute, through an award from the Canada First Research Excellence Fund (sub-module J3), the Government of Newfoundland and Labrador, Department of Fisheries and Land Resources, and NSERC-Discovery grant (RGPIN-2018-05942).
Conflicts of Interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pruzzo C., Huq A., Colwell R., Donelli G. Pathogenic Vibrio Species in the Marine and Estuarine Environment. Springe; Boston, MA, USA: 2005. pp. 217–252. [Google Scholar]
- 2.Baker-Austin C., Oliver J., Alam M., Ali A., Waldor M., Qadri F., Martinez-Urtaza J. Vibrio spp. infections. Nat. Rev. Dis. Primers. 2018;4:8. doi: 10.1038/s41572-018-0005-8. [DOI] [PubMed] [Google Scholar]
- 3.Baker-Austin C., Stockley L., Rangdale R., Martinez-Urtaza J. Environmental occurrence and clinical impact of Vibrio vulnificus and Vibrio parahaemolyticus: A European perspective. Environ. Microbiol. Rep. 2010;2:7–18. doi: 10.1111/j.1758-2229.2009.00096.x. [DOI] [PubMed] [Google Scholar]
- 4.Gram L., Melchiorsen J., Spanggaard B., Huber I., Nielsen T. Inhibition of Vibrio anguillarum by Pseudomonas fluorescens AH2, a possible probiotic treatment of fish. Appl. Environ. Microbiol. 1999;65:969–973. doi: 10.1128/AEM.65.3.969-973.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frans I., Michiels C., Bossier P., Willems K., Lievens B., Rediers H. Vibrio anguillarum as a fish pathogen: Virulence factors, diagnosis and prevention. J. Fish Dis. 2011;34:643–661. doi: 10.1111/j.1365-2761.2011.01279.x. [DOI] [PubMed] [Google Scholar]
- 6.Holm K., Nilsson K., Hjerde E., Willassen N., Milton D. Complete genome sequence of Vibrio anguillarum strain NB10, a virulent isolate from the Gulf of Bothnia. Stand. Genom. Sci. 2015;10:60. doi: 10.1186/s40793-015-0060-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li G., Mo Z., Li J., Xiao P., Hao B. Complete Genome Sequence of Vibrio anguillarum M3, a Serotype O1 Strain Isolated from Japanese Flounder in China. Genome Announc. 2013;1:e00769-13. doi: 10.1128/genomeA.00769-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naka H., Dias G., Thompson C., Dubay C., Thompson F., Crosa J. Complete genome sequence of the marine fish pathogen Vibrio anguillarum harboring the pJM1 virulence plasmid and genomic comparison with other virulent strains of V. anguillarum and V. ordalii. Infect. Immun. 2011;79:2889–2900. doi: 10.1128/IAI.05138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brooker A., Papadopoulou A., Gutierrez C., Rey S., Davie A., Migaud H. Sustainable production and use of cleaner fish for the biological control of sea lice: Recent advances and current challenges. Vet. Rec. 2018;183:383. doi: 10.1136/vr.104966. [DOI] [PubMed] [Google Scholar]
- 10.Powell A., Treasurer C., Pooley A., Keay R., Imsland A., Garcia de Leaniz C. Use of lumpfish for sea-lice control in salmon farming:challenges and opportunities. Rev. Aqua. 2017;10:1–20. doi: 10.1111/raq.12194. [DOI] [Google Scholar]
- 11.Imsland A., Reynolds P., Eliassen G., Hangstad A., Foss A., Vikingstad E., Elvegård T. The use of lumpfish (Cyclopterus lumpus L.) to control sea lice (Lepeophtheirus salmonis Krøyer) infestations in intensively farmed Atlantic salmon (Salmo salar L.) Aquaculture. 2014;424:18–23. doi: 10.1016/j.aquaculture.2013.12.033. [DOI] [Google Scholar]
- 12.McEwan G., Groner M., Cohen A., Imsland A., Revie C. Modelling sea lice control by lumpfish on Atlantic salmon farms: Interactions with mate limitation, temperature and treatment rules. Dis. Aqua. Org. 2019;133:69–82. doi: 10.3354/dao03329. [DOI] [PubMed] [Google Scholar]
- 13.Sayer M., Davenport J. Hypometabolism in torpid goldsinny wrasse subjected to rapid reductions in seawater temperature. J. Fish Biol. 1996;49:64–75. doi: 10.1111/j.1095-8649.1996.tb00005.x. [DOI] [Google Scholar]
- 14.Costa I., Driedzic W., Gamperl A. Metabolic and cardiac responses of cunner Tautogolabrus adspersus to seasonal and acute changes in temperature. Phy. Biochem. Zool. 2013;86:233–244. doi: 10.1086/669538. [DOI] [PubMed] [Google Scholar]
- 15.Deady S., Varian S., Fives J. The use of cleaner-fish to control sea lice on two Irish salmon (Salmo salar) farms with particular reference to wrasse behaviour in salmon cages. Aquaculture. 1995;131:73–90. doi: 10.1016/0044-8486(94)00331-H. [DOI] [Google Scholar]
- 16.Blanco Gonzalez E., de Boer F. The development of the Norwegian wrasse fishery and the use of wrasses as cleaner fish in the salmon aquaculture industry. Fish. Sci. 2017;83:661–670. doi: 10.1007/s12562-017-1110-4. [DOI] [Google Scholar]
- 17.Sørensen U., Larsen J. Serotyping of Vibrio anguillarum. Appl. Environ. Microbiol. 1986;51:593–597. doi: 10.1128/AEM.51.3.593-597.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pedersen K., Grisez L., van Houdt R., Tiainen T., Ollevier F., Larsen J. Extended serotyping scheme for Vibrio anguillarum with the definition and characterization of seven provisional O-serogroups. Curr. Microbiol. 1999;38:183–189. doi: 10.1007/PL00006784. [DOI] [PubMed] [Google Scholar]
- 19.Aquaculture, Department of Aquaculture, editor. Fisheries and Land Resources: Newfoundland and Labrador Aquatic Animal Reportable and Notifiable Disease. Fisheries and Aquaculture; Saint John, NL, Canada: 2020. [(accessed on 26 October 2020)]. p. 2. Available online: https://www.gov.nl.ca/ffa/files/Newfoundland-and-Labrador-Aquatic-Animal-Reportable-and-Notifiable-Diseases-September-2020.pdf. [Google Scholar]
- 20.Holm K., Baekkedal C., Soderberg J., Haugen P. Complete genome sequences of seven Vibrio anguillarum strains as derived from PacBio sequencing. Genome Biol. Evol. 2018;10:1127–1131. doi: 10.1093/gbe/evy074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Louden B., Haarmann D., Lynne A. Use of Blue Agar CAS Assay for Siderophore detection. J. Microbiol. Biol. Edu. 2011;12:51–53. doi: 10.1128/jmbe.v12i1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wood E. Molecular Cloning. A Laboratory Manual. Biochem. Educ. 1983;11:82. doi: 10.1016/0307-4412(83)90068-7. [DOI] [Google Scholar]
- 23.Cameron M., Barkema H., De Buck J., De Vliegher S., Chaffer M., Lewis J., Keefe G. Identification of bovine-associated coagulase-negative staphylococci by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry using a direct transfer protocol. J. Dairy Sci. 2017;100:2137–2147. doi: 10.3168/jds.2016-12020. [DOI] [PubMed] [Google Scholar]
- 24.Connors E., Soto-Dávila M., Hossain A., Vasquez I., Gnanagobal H., Santander J. Identification and validation of reliable Aeromonas salmonicida subspecies salmonicida reference genes for differential gene expression analyses. Infect. Genet. Evol. 2019;73:314–321. doi: 10.1016/j.meegid.2019.05.011. [DOI] [PubMed] [Google Scholar]
- 25.Myhr E., Larsen J., Lillehaug A., Gudding R., Heum M., Håstein T. Characterization of Vibrio anguillarum and closely related species isolated from farmed fish in Norway. Appl. Environ. Microbiol. 1991;57:2750–2757. doi: 10.1128/AEM.57.9.2750-2757.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramasamy P., Gunasekaran D. Assessment of antibiotic sensitivity and pathogenicity of Vibrio spp. and Aeromonas spp. from aquaculture enviroment. Moj Ecol. Environ. Sci. 2018;3:128–136. doi: 10.15406/mojes.2018.03.00077. [DOI] [Google Scholar]
- 27.Chakraborty S., Cao T., Hossain A., Gnanagobal H., Vasquez I., Boyce D., Santander J. Vibrogen-2 vaccine trial in lumpfish (Cyclopterus lumpus) against Vibrio anguillarum. J. Fish Dis. 2019;42:1057–1064. doi: 10.1111/jfd.13010. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J., Russell W. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Press; New York, NY, USA: 2001. [Google Scholar]
- 29.Aziz R., Bartels D., Best A., DeJongh M., Disz T., Edwards R., Formsma K., Gerdes S., Glass E., Kubal M., et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andrews S. FastQC: A Quality Control Tool for High Throughput Sequence Data. [(accessed on 26 October 2020)];2010 Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 31.Carver T., Thomson N., Bleasby A., Berriman M., Parkhill J. DNAPlotter: Circular and linear interactive genome visualization. Bioinformatics. 2009;25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saitou N., Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Microbiol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 34.Jukes T., Cantor C. Evolution of Protein Molecules. In: Munro H., editor. Mammalian Protein Metabolism. Academic Press; Cambridge, MA, USA: 1969. pp. 21–132. [Google Scholar]
- 35.Teru Y., Hikima J., Kono T., Sakai M., Takano T., Hawke J., Takeyama H., Aoki T. Whole-genome sequence of Photobacterium damselae subsp. piscicida strain 91–197, isolated from hybrid striped Bass (Morone sp.) in the United States. Genome Announc. 2017;5:e00600-17. doi: 10.1128/genomeA.00600-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Darling A., Mau B., Blattner F., Perna N. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rasmussen B., Grotkjær T., Alvise P., Yin G., Zhang F., Bunk B., Spröer C., Bentzon-Tilia M., Gram L. Vibrio anguillarum is genetically and phenotypically unaffected by Long-Term continuous exposure to the antibacterial compound tropodithietic acid. Appl. Environ. Microbiol. 2016;82:4802. doi: 10.1128/AEM.01047-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gomez-Gil B., Soto-Rodríguez S., García-Gasca A., Roque A., Vazquez-Juarez R., Thompson F., Swings J. Molecular identification of Vibrio harveyi-related isolates associated with diseased aquatic organisms. Microbiology. 2004;150:1769–1777. doi: 10.1099/mic.0.26797-0. [DOI] [PubMed] [Google Scholar]
- 39.Lee J., Shread P., Furniss A., Bryant T. Taxonomy and description of Vibrio fluvialis sp. nov. (synonym group F vibrios, group EF6) J. Appl. Bacteriol. 1981;50:73–94. doi: 10.1111/j.1365-2672.1981.tb00873.x. [DOI] [PubMed] [Google Scholar]
- 40.Fujino T., Miwatani T., Yasuda J., Kondo M., Takeda Y., Akita Y., Kotera K., Okada M., Nishimune H., Shimizu Y., et al. Taxonomic studies on the bacterial strains isolated from cases of “shirasu” food-poisoning (Pasteurella parahaemolytica) and related microorganisms. Biken J. 1965;8:63–71. [PubMed] [Google Scholar]
- 41.Le Roux F., Zouine M., Chakroun N., Binesse J., Saulnier D., Bouchier C., Zidane N., Ma L., Rusniok C., Lajus A., et al. Genome sequence of Vibrio splendidus: An abundant planctonic marine species with a large genotypic diversity. Environ. Microbiol. 2009;11:1959–1970. doi: 10.1111/j.1462-2920.2009.01918.x. [DOI] [PubMed] [Google Scholar]
- 42.Vaidya G., Lohman D.J., Meier R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 2011;27:171–180. doi: 10.1111/j.1096-0031.2010.00329.x. [DOI] [PubMed] [Google Scholar]
- 43.Bertelli C., Hoad G., Winsor G., Brinkman F., Laird M., Lau B., Williams K. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017;45:W30–W35. doi: 10.1093/nar/gkx343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Villarreal-Chiu J., Quinn J., McGrath J. The genes and enzymes of phosphonate metabolism by bacteria, and their distribution in the marine environment. Front. Microbiol. 2012;3:19. doi: 10.3389/fmicb.2012.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Favrot L., Blanchard J., Vergnolle O. Bacterial GCN5-Related N-Acetyltransferases: From resistance to regulation. Biochemistry. 2016;55:989–1002. doi: 10.1021/acs.biochem.5b01269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marcos-Lopez M., Donald K., Stagg H., McCarthy U. Clinical Vibrio anguillarum infection in lumpsucker Cyclopterus lumpus in Scotland. Vet. Rec. 2013;173:319. doi: 10.1136/vr.101763. [DOI] [PubMed] [Google Scholar]
- 47.Tagliavia M., Salamone M., Bennici C., Quatrini P., Cuttitta A. A modified culture medium for improved isolation of marine vibrios. Microbiol. Open. 2019;8:e00835. doi: 10.1002/mbo3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li L., Mou X., Nelson D. HlyU Is a Positive Regulator of hemolysin expression in Vibrio anguillarum. J. Bacteriol. 2011;193:4779. doi: 10.1128/JB.01033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boyce D., Ang K., Prickett R. Cunner and lumpfish as cleaner fish species in Canada. In: Treasurers J.W., editor. Cleaner fish biology and Aquaculture Applications. 5m Publishing; Sheffield, UK: 2018. [Google Scholar]
- 50.Castillo D., Alvise P., Xu R., Zhang F., Middelboe M., Gram L. Comparative genome analyses of Vibrio anguillarum strains reveal a link with pathogenicity traits. mSystems. 2017;2:e00001-17. doi: 10.1128/mSystems.00001-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steinum T., Karatas S., Martinussen N., Meirelles P., Thompson F., Colquhoun D. Multilocus sequence analysis of close relatives Vibrio anguillarum and Vibrio ordalii. Appl. Environ. Microbiol. 2016;82:5496–5504. doi: 10.1128/AEM.00620-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Glaeser S., Kämpfer P. Multilocus sequence analysis (MLSA) in prokaryotic taxonomy. Syst. Appl. Microbiol. 2015;38:237–245. doi: 10.1016/j.syapm.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 53.Whittaker B., Consuegra S., Garcia de Leaniz C. Genetic and phenotypic differentiation of lumpfish (Cyclopterus lumpus) across the North Atlantic: Implications for conservation and aquaculture. PeerJ. 2018;6:e5974. doi: 10.7717/peerj.5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marsh J., Taylor R. Genetic and transcriptional analyses of the Vibrio cholerae mannose-sensitive hemagglutinin type 4 pilus gene locus. J. Bacteriol. 1999;181:1110–1117. doi: 10.1128/JB.181.4.1110-1117.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weber B., Hasic M., Chen C., Wai S., Milton D. Type VI secretion modulates quorum sensing and stress response in Vibrio anguillarum. Environ. Microbiol. 2009;11:3018–3028. doi: 10.1111/j.1462-2920.2009.02005.x. [DOI] [PubMed] [Google Scholar]
- 56.Hirono I., Masuda T., Aoki T. Cloning and detection of the hemolysin gene of Vibrio anguillarum. Microb. Pathogens. 1996;21:173–182. doi: 10.1006/mpat.1996.0052. [DOI] [PubMed] [Google Scholar]
- 57.Rock J., Nelson D. Identification and characterization of a hemolysin gene cluster in Vibrio anguillarum. Infect. Immun. 2006;74:2777–2786. doi: 10.1128/IAI.74.5.2777-2786.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Naka H., Lopez C., Crosa J. Role of the pJM1 plasmid-encoded transport proteins FatB, C and D in ferric anguibactin uptake in the fish pathogen Vibrio anguillarum. Environ. Microbiol. Rep. 2010;2:104–111. doi: 10.1111/j.1758-2229.2009.00110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Milton D., Hardman A., Camara M., Chhabra S., Bycroft B., Stewart G., Williams P. Quorum sensing in Vibrio anguillarum: Characterization of the vanI/vanR locus and identification of the autoinducer N-(3-oxodecanoyl)-L-homoserine lactone. J. Bacteriol. 1997;179:3004–3012. doi: 10.1128/JB.179.9.3004-3012.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Busschaert P., Frans I., Crauwels S., Zhu B., Willems K., Bossier P., Michiels C., Verstrepen K., Lievens B., Rediers H. Comparative genome sequencing to assess the genetic diversity and virulence attributes of 15 Vibrio anguillarum isolates. J. Fish Dis. 2015;38:795–807. doi: 10.1111/jfd.12290. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.