Abstract
Background
Colorectal cancer (CRC) is a major cause of cancer deaths worldwide. Accumulating evidence suggests a potentially important role of colorectal infection with Fusobacterium nucleatum (F. nucleatum) in colorectal carcinogenesis. We conducted a systematic review, including both a qualitative synthesis and a meta-analysis, to synthesize the evidence from the epidemiological literature on the association between F. nucleatum detection in the colon/rectum and CRC.
Methods
A systematic literature search of Ovid MEDLINE(R), Embase, Web of Science Core Collection, EBM Reviews—Cochrane Database of Systematic Reviews, and CINAHL Plus with Full Text was conducted using earliest inclusive dates up to 4 October 2020. Eligible studies were original, comparative observational studies that reported results on colorectal F. nucleatum detection and CRC. Two independent reviewers extracted the relevant information. Odds ratio (OR) estimates were pooled across studies using the random effects model. Newcastle-Ottawa scale was used to critically appraise study quality.
Results
Twenty-four studies were included in the systematic review, of which 12 were included in the meta-analysis. Studies investigated F. nucleatum in feces, colorectal tissue samples, or both. In most studies included in the systematic review, the load of F. nucleatum was higher, on average, in specimens from CRC patients than in those from CRC-free controls. Meta-analysis showed a positive association between F. nucleatum detection in colorectal specimens and CRC (OR = 8.3; 95% confidence interval (95% CI) 5.2 to 13.0).
Conclusions
The results of this systematic review suggest that F. nucleatum in the colon/rectum is associated with CRC.
Systematic review registration
This systematic review protocol has been registered with the International Prospective Register of Systematic Reviews (PROSPERO) on July 10, 2018 (registration number CRD42018095866).
Supplementary Information
The online version contains supplementary material available at 10.1186/s13643-020-01526-z.
Keywords: Colorectal cancer, Fusobacterium nucleatum, Systematic review, Meta-analysis
Background
Colorectal cancer (CRC) is a significant burden on global public health: it is the fourth and third most commonly diagnosed cancer in men and women [1], respectively, with more than a million new cases per year worldwide [2]. It is also the fourth leading cause of death from cancer in the world [1]. While some CRC cases are attributed to inheritance and inflammatory bowel disease, about 80% of them are sporadic [3]. Thus, identification of etiological factors is essential for efforts to reduce the morbidity and mortality from CRC.
Over the years, epidemiological studies have identified a number of CRC risk factors, such as diet, cigarette smoking, obesity, physical inactivity, diabetes, and certain genetic polymorphisms [4–11]. Furthermore, the role of some bacteria in colon carcinogenesis seems quite plausible [12, 13]. In 2011, Sears and Pardoll suggested that bacteria are the main drivers of the intestinal mucosa immune response and subsequent changes in the function and genetics of epithelial cells, which support oncogenic transformation [14]. These ideas have rapidly gained credibility due to important discoveries on the role of gut microbial dysbiosis and specifically of the bacterium Fusobacterium nucleatum (F. nucleatum) in colorectal carcinogenesis [15–25]. F. nucleatum is one of the dominant species of 500 or more organisms that coexist in the oral cavity [26] and the most prevalent oral species in extra-oral infections [27, 28]. Two virulence factors have been identified for F. nucleatum: an adhesin FadA and a self-transporting protein Fap2 [28]. On the one hand, FadA allows F. nucleatum to invade human epithelial cells, activate β-catenin signaling, induce expression of the oncogenic gene, and promote the growth of colorectal tumor cells [24, 25, 29–32]. On the other hand, the protein Fap2 inhibits the activity of immune cells and thus potentiates the progression of CCR [32, 33]. This suggests that F. nucleatum may participate in the colorectal tumor process and thus be a pro-oncogenic bacterium. In a murine model of CRC (APC +/−), the introduction of F. nucleatum increased tumor multiplicity and the selective recruitment of myeloid cells infiltrating tumors, thereby promoting tumor progression [18]. F. nucleatum also stimulates the recruitment of tumor-infiltrating immune cells, which generate an inflammatory microenvironment conducive to the progression of colorectal neoplasia [18]. Mouse tumors (APC +/−) exposed to F. nucleatum have a pro-inflammatory expression, similar to that observed in human colorectal tumors positive for F. nucleatum [18].
Over the last decade, many subsequent studies have reported an overabundance of F. nucleatum in colorectal tissues and stools from subjects diagnosed with CRC compared with CRC-free “controls.” The literature on this topic has been growing rapidly but has not yet been reviewed. We therefore conducted a systematic review and a meta-analysis to review the available literature on the association between F. nucleatum infection in the colon and CRC.
Methods
This systematic review protocol has been registered with the International Prospective Register of Systematic Reviews (PROSPERO) on 10 July 2018 (registration number CRD42018095866). The protocol for this systematic review was published previously [34]. This systematic review follows the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines as well as the Meta-analysis of Observational Studies in Epidemiology (MOOSE) guidelines (see Additional file 1 for PRISMA checklist).
Search strategy
Literature search covered the following databases: all Ovid MEDLINE(R), Embase, Web of Science Core Collection, EBM Reviews—Cochrane Database of Systematic Reviews, and CINAHL Plus with Full Text. A comprehensive search from each database’s earliest inclusive dates (1946 for Ovid Medline, 1974 for Embase, 1945 for Web of Science, and 2008 for EBM Reviews) to 31 December 2018 was first conducted. Specific details regarding the search strategies appear in Table 1. The electronic literature search was complemented by hand-searching the list of references in the identified publications.
Table 1.
Initial search strategy
| Database and search dates | Search #1 | Search #2 | Search #3 | Search #4 | Search #5 |
|---|---|---|---|---|---|
| All Ovid MEDLINE (R). January 1, 1946, to December 31, 2018 | exp Colonic Polyps/ or exp Colorectal Neoplasms/ | ((colon$ or colorect$ or rect$ or sigmoid) adj5 (polyp$ or adeno$ or cancer$ or carcinoma$ or malignan$ or metastas$ or neoplas$ or oncolog$ or tumo$)).tw. | #1 OR #2 | Fusobacterium nucleatum/ or exp Fusobacterium Infections/ or nucleatum.tw. | #3 AND #4 |
| Embase. January 1, 1974, to December 31, 2018 | exp Colon Polyp/ or exp Colorectal Tumor/ | ((colon$ or colorect$ or rect$ or sigmoid) adj5 (polyp$ or adeno$ or cancer$ or carcinoma$ or malignan$ or metastas$ or neoplas$ or oncolog$ or tumo$)).tw. | #1 OR #2 | Fusobacterium nucleatum/ or exp Fusobacterium Infection/ or nucleatum.tw. | #3 AND #4 |
| CINAHL Plus with Full Text. January 1, 1981, to December 31, 2018 | TX ((colon* or colorect* or rect* or sigmoid) N5 (polyp* or adeno* or cancer* or carcinoma* or malignan* or metastas* or neoplas*or oncolog* or tumo*)) | (MH “Colonic Polyps”) OR (MH “Colorectal Neoplasms+”) | #1 OR #2 | (MH “Fusobacterium Infections+”) OR TX nucleatum | #3 AND #4 |
| Web of Science Core Collection. January 1, 1945, to December 31, 2018 | TOPIC: (((colon* or colorect* or rect* or sigmoid) NEAR/5 (polyp* or adeno* or cancer* or carcinoma* or malignan* or metastas* or neoplas* or oncolog* or tumo*))) | TOPIC: (nucleatum) | #1 AND #2 | ||
| EBM Reviews—Cochrane Database of Systematic Reviews. January 1, 2005, to December 31, 2018 | ((colon$ or colorect$ or rect$ or sigmoid) adj5 (polyp$ or adeno$ or cancer$ or carcinoma$ or malignan$ or metastas$ or neoplas$ or oncolog$ or tumo$)).tw. | nucleatum.tw. | #1 AND #2 |
An update of the literature search was then carried out on 4 October 2020, in order to identify additional human studies that were published in French and English, since the initial search.
Inclusion and exclusion criteria
Eligible studies were original, comparative observational studies that reported results on colorectal F. nucleatum infection in at least two groups: individuals diagnosed with CRC and colorectal-adenoma- and CRC-free subjects (in this article, this population will be referred to as “controls”). No demographic or geographic limitations were applied. Only studies published in English or French were included. Colorectal F. nucleatum had to be investigated either in feces or in biopsies from tumors in CRC patients and from healthy colorectal mucosa in “controls.” Ascertainment of F. nucleatum infection had to be based on microbiological detection and/or quantification tests such as any polymerase chain reaction (PCR) technique, sequencing, or microscopy visualization (e.g., fluorescence in situ hybridization technique (FISH)). Also, Fusobacterium had to be investigated at the species level, and the data had to be available for the particular species of F. nucleatum. Studies reporting data on genus or phylum levels only were thus excluded. The data on the CRC status, the outcome of interest, had to be based on laboratory-confirmed diagnosis (and thus had to be ascertained via a cancer registry or medical records).
Study selection
Two independent reviewers (AIJ and CL) performed the study selection process based on title and abstract. Retained studies were then full-text screened by the same reviewers independently to verify the inclusion criteria. Any disagreement was resolved by discussion. If consensus could not be reached, a third reviewer determined the eligibility and approved the final list of retained studies.
Quality assessment and data extraction
We used the Newcastle-Ottawa Scale (NOS) to assess the quality of the included observational studies. The scale includes three domains: selection (4 items), comparability (1 item), and exposure (3 items). A study can be awarded a maximum of one star for each numbered item within the selection and exposure categories. A maximum of two stars can be given for comparability. Study quality was then classified as poor, fair, or good, according to the Agency for Healthcare Research and Quality (AHRQ) thresholds for converting NOS scores, described as follows: (i) good quality = 3 or 4 stars in selection domain AND 1 or 2 stars in comparability domain AND 2 or 3 stars in outcome/exposure domain; (ii) fair quality = 2 stars in selection domain AND 1 or 2 stars in comparability domain AND 2 or 3 stars in outcome/exposure domain; and (iii) poor quality = 0 or 1 star in selection domain OR 0 stars in comparability domain OR 0 or 1 star in outcome/exposure domain [35–38].
The data from each study were independently extracted by two independent reviewers (AIJ and CL, AIJ and HS) and then reciprocally verified. Disagreements of two reviewers were resolved by discussion. If consensus could not be reached, a third reviewer was consulted. The following information from each article was extracted: authors’ names, country, year of publication, aim of the study, study design, study population, sample size (number of cases and number of controls), study participants’ characteristics, identification sources, criteria of matching (if any), inclusion and exclusion criteria (including any restriction of last antibiotic consumption, precursors of cancer, or inflammatory bowel disease (IBD)), localization of the tumor (colon or rectal or colorectal tumors), type of collected specimens (stools or biopsies or both), exposure definition (frequency of presence of F. nucleatum in specimens, relative abundance, or relative quantification of bacteria load), the technique used to detect and quantify the bacterium load, and main results and the accompanying results of statistical tests. For studies comparing more groups with controls and CRC patients (e.g., adenoma or IBD patients), only data on CRC patients and controls was extracted. Similarly, when any included study performed a second validation bacterial analysis on the same participants or on a subsample, only the results of the first technique were extracted. (See supplementary material)
Statistical analysis
We used two approaches for data synthesis, a narrative and a quantitative synthesis using a meta-analysis. The descriptive synthesis was conducted according to the Centre for Reviews and Dissemination and included text and tables to summarize the findings.
To perform a meta-analysis, we included studies reporting any measure of association between F. nucleatum and CRC, or reporting proportions or numbers of F. nucleatum-positive samples in CRC cases and controls that allowed us to calculate estimates of odds ratios along with the corresponding 95% confidence intervals (CI). Then, a pooled OR estimate and its corresponding 95% CI were calculated. The data was pooled using a random effects model [39]. Heterogeneity across studies was tested using Cochran’s Q and the I2 statistic, and potential publication bias was investigated by visual inspection of funnel plots and Egger’s regression asymmetry test.
A subgroup meta-analysis using a random effects model was subsequently performed in order to investigate the change in F. nucleatum association to CRC by population area, type of colorectal specimen, and microbiological test, as well as to verify the effect of including participants with history of IBD or a recent antibiotic use in the included studies. Comprehensive Meta-Analysis Version 3 was used to conduct the meta-analysis.
Results
Search results
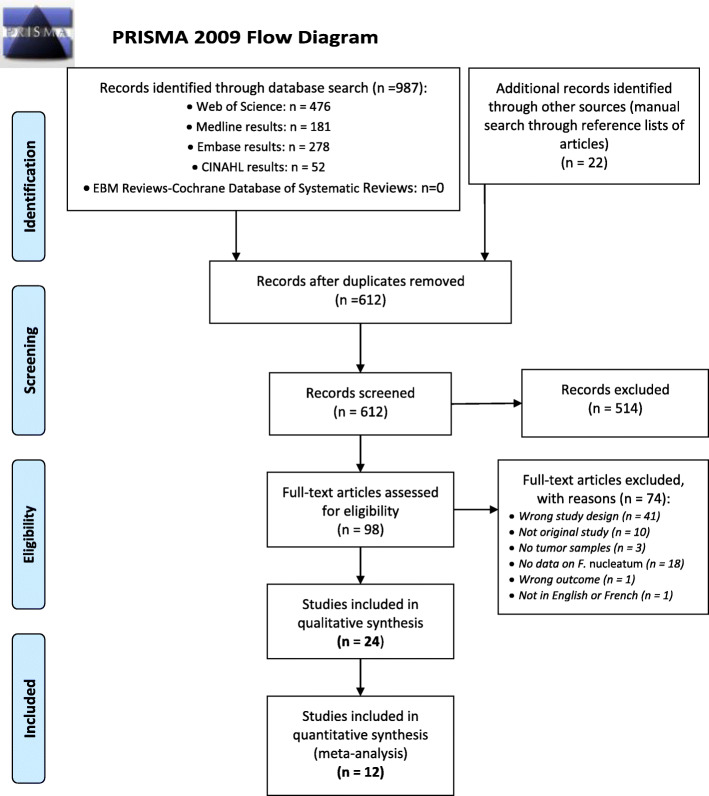
Initial search (31 December 2018) and its recent update (4 October 2020) retrieved 987 records from databases and 22 additional records through manual search of relevant reviews. After removal of 397 duplicates, 612 articles were screened based on titles and abstracts, which resulted in 514 excluded articles. Ninety-eight full-text publications were assessed for eligibility. Of these, 74 were excluded for not meeting inclusion criteria. Finally, 24 studies [40–63] were included in the systematic review, of which 12 were included in the meta-analysis. Figure 1 shows the study flow diagram.
Fig. 1.
Flow diagram for selection of studies included in the systematic review and meta-analysis
Characteristics of studies included in the systematic review
The characteristics of the 24 studies included in the systematic review are summarized in Table 2. Studies were published between 2012 and 2020 and were mostly conducted in Asia: twelve in China, one in Japan, and two in Iran. Six studies were conducted in Europe (Germany, Spain, Italy, Ireland, Norway, and Sweden), and three in the Americas (two in USA and one in Brazil). Studies were designed as “non-nested” case-control (n = 21), cross sectional (n = 1), or nested case-control (n = 2), with poor or fair quality assessment of 14 studies, according to the AHRQ scale. Studies failed often in fulfilling selection of controls and non-response rate items.
Table 2.
Characteristics of studies included in the systematic review
| First author (year) | Country | Study design | No. of CRC /C | Age, mean (SD) | Exclusion of subjects with history of antibiotic use (period) | Exclusion of subjects with history of IBD diagnosis | Specimen type | Specimen collection time | Detection method | Study quality based on NOS scores | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CRC | C | ||||||||||
| Amitay (2017) [40] | Germany | NCC | 46/231 | 66.9 (-) | 62.1 (-) | No | Yes | Feces | Before colonoscopy | 16S rRNA gene analysis | Poor |
| Yu (2015) [41] | China | CC | 42/52 | --- | --- | Yes (last 6 months) | Yes | Feces and biopsy c |
Feces: non-reported Biopsies: left tissues |
454 FLX pyrosequencing and PCR | Poor |
| Zhang (2018) [42] | China | CC | 130/130 | 60.5 (9.8) | 58.6 (8.9) | Yes (last 6 months) | Yes | Feces | Before colonoscopy | 16S rDNA sequencing and PCR | Good |
| Flanagan (2014) [43] | Ireland | CC | 7/25 | --- | --- | No | Yes | Feces | --- | qPCR | Poor |
| Fukugaiti (2015) [44] | Brazil | CC | 7/10 | 65.4 (1.1) | 54.8 (1.3) | Yes (unspecified) | No | Feces | Before colonoscopy | qPCR | Poor |
| Suehiro (2017) [45] | Japan | CC | 158/60 | 69 (-) | 32 (-) | No | No | Feces | Prior to bowel preparation | dd-PCR | Poor |
| Rezasoltani (2018) [46] | Iran | CC | 20/31 | 60.9 (13.5) | 59.8 (17) | Yes (last 6 months) | Yes | Feces | Before colonoscopy | qPCR | Poor |
| Eklof (2017) [47] | Sweden | NCC | 39/65 | 34 (-) | 34 (-) | No | Yes (for C only) | Feces | Prior to bowel preparation | qPCR | Good |
| Guo (2018) [48] | China | CC | 215/156 | --- | --- | Yes (last 3 months) | Yes | Feces | Time of diagnosis, before resection | qPCR | Poor |
| Liang (2017) [49] | |||||||||||
| Cohort I | China | CC | 170/200 | 67.2 (11.6) | 59.3 (5.8) | Yes (last 3 months) | No | Feces | Before or one month after colonoscopy | qPCR | Good |
| Cohort II | China | 33/36 | 63.4 (9.6) | 53.2 (12.2) | |||||||
| Mira-Pascual (2015) [50] | Spain | CC | 7/9 | 71.1 (10.1) | 52.6 (15.2) | No | No | Feces and biopsyd | ≥ 1 week before colonoscopy | qPCR | Good |
| Repass (2018) [51] | USA | CC | 40/40 | --- | --- | No | No | Biopsy | During surgery | qPCR | Fair |
| Russo (2017) [52] | Italy | CC | 10/10 | --- | --- | Yes (last 3 months) | No | Feces | 1 day before surgery for CRC and unspecified for C | qPCR | Poor |
| Vogtmann (2016) [53] | USA | CC | 52/52 | 61.8 (-) | 61.2 (-) | No | No | Feces | Prior to surgery or other treatment | WGSS | Good |
| Wang (2016) [54] | China | CC | 10/10 | 54.8 (-) | 54 (-) | No | No | Feces | --- | PCR | Good |
| Wang (2012) [55] | China | CC | 46/56 | 60a | 49a | Yes (last 3 months) | Yes (for C only) | Feces | Before surgery for CRC patients and unspecified for C | qPCR | Poor |
| Wong (2017) [56] | China | CC | 104/102 | 66.9 (10.1) | 57.1 (5.8) | Yes (last 3 months) | Yes | Feces | Prior to bowel preparation for colonoscopy | qPCR | Poor |
| Wu (2013) [57] | China | CC | 19/20 | 58.3 (8.7) | 53.2 (5.4) | Yes (last 3 months) | Yes (for C only) | Feces | --- | qPCR | Good |
| Xie (2017) [58] | China | CC | 327/242 | 63.5 (10.2) | 60.1 (8.4) | Yes (last month) | Nob | Feces | Prior to bowel preparation for surgery or endoscopy | qPCR | Good |
| Yu (2016) [59] | China | CC | 93/20 | 59.25(-) | --- | No | No | Biopsy | Colonoscopy or surgery | FISH | Poor |
| Yu (2017) [60] | China | CC | 74/54 | 63 (50.7) | 67 (34.9) | Yes (last 3 months) | Yes | Feces | Before or mostly after colonoscopy | Metagenomic sequencing | Poor |
| Tunsjo (2019) [61] | Norway | CC | 23/22 | 70 (-) | 57 (-) | No | No | Feces and biopsy e | Prior to bowel preparation or 1 week after colonoscopy | qPCR | Poor |
| Liu (2020) [62] | China | CC | 53/45 | 52.4 (18.8) | 53.7 (16.7) | Yes (last month) | Yes | Feces | Prior to bowel preparation for colonoscopy | qPCR | Good |
| Kashani (2020) [63] | Iran | CS | 35/45 | --- | --- | No | No | Biopsy | Colonoscopy | PCR | Satisfactoryf |
No. number, CRC colorectal cancer cases, C controls (colorectal-adenoma- and CRC-free subjects), IBD inflammatory bowel disease, NCC nested case-control, CC case-control, CS cross sectional, qPCR quantitative polymerase chain reaction, NOS Newcastle-Ottawa scale, WGSS whole genome shotgun sequencing, dd-PCR droplet digital PCR, FISH fluorescent in situ hybridization
--- Not reported
aMedian
bControls were those with normal or chronic inflamed colorectal mucosa
cBiopsies were taken from only 31/42 CRC and 37/52 C
dBiopsies were taken from 7/7 CRC and 5/9 C
eBiopsies were taken from 21/23 CRC and 11/22 C
fIn NOS adapted for CS studies, the study quality is based on the total score and rated as follow: very good (9–10 points), good (7–8 points), satisfactory (5–6 points), and unsatisfactory (0 to 4 points)
In seven studies, cases and controls were matched for two to four variables, including age, gender, body mass index, ethnicity, and the time period of sample collection [42, 47, 50, 51, 53, 54, 57, 62]. There were 13 studies that excluded subjects with reported antibiotic use in the last month, or in the last 3 months or 6 months, and 11 studies that excluded patients previously diagnosed with IBD.
The majority of studies investigated F. nucleatum in feces only (18 studies), while three studies analyzed biopsies only, and another three studies analyzed both types of specimens. Quantitative PCR was the most used bacteria-detection technique followed by sequencing techniques, while only one study used FISH technique.
In most of the included studies, feces were collected before colonoscopy or surgery, except for the study by Yu et al. [60] where feces were collected more after colonoscopy than before. Tunsjo et al. [61] also reported collecting feces either before colonoscopy or 1 week after. In four studies, no information was provided about the timing of specimen collection [43, 54, 57].
Comparison of Fusobacterium nucleatum load in colorectal specimens between colorectal cancer cases and controls
As shown in Table 3, F. nucleatum quantification (load) in colorectal specimens was reported by 18 studies [41–50, 52, 53, 55, 56, 58, 60–62] including one study with two independent cohorts [49]. Bacteria quantification was mostly performed in stool specimens, except for Yu et al. [41] who quantified the bacteria in both feces and biopsies. Vogtmann et al. [53], Wang et al. [55], and Zhang et al. [42] reported the relative abundance (RA) of the bacteria as a percentage, corresponding to the contribution of F. nucleatum to the total bacteria present in specimens [42, 53, 55]. Their results confirmed that F. nucleatum does not naturally contribute to a healthy gut microbiome (RA varied from 0.001 to 0.003% in controls). When investigated in CRC case specimens, F. nucleatum was significantly more abundant than in controls, but still in very small proportions (RA varied from 0.061 to 0.17%). While only four studies reported results of absolute quantification of F. nucleatum, either as copy number or bacteria counts, the majority of studies performed relative quantification (RQ) of F. nucleatum to the total bacteria present in specimens based on the ΔΔCq method. These RQ studies reported a significantly higher F. nucleatum load in colorectal specimens of CRC patients compared to controls, except for two studies [50, 52]. Fold change in F. nucleatum from controls to CRC cases was estimated by three studies with very different values: 132-fold according to Wong et al. [56], 66-fold according to Tunsjo et al. [61], and 5.2-fold reported by Xie et al. [58].
Table 3.
Studies comparing F. nucleatum load in colorectal specimens of colorectal cancer cases and controls
| First author (year) | Specimen type (No. of specimens CRC/C) | F. nucleatum quantification measures reported | Statistics reported | F. nucleatum load in CRC compared to C | ||
|---|---|---|---|---|---|---|
| C | CRC | p | ||||
| Relative abundance | ||||||
| Zhang (2018) [42] | Feces (130/130) | Relative contribution to total bacteria (%) | Mean | 0.001 | 0.17 | < 0.001 |
| Vogtmann (2016) [53] | Feces (52/53) | Relative contribution to total bacteria (%) | Mean | 0.003 | 0.08 | 0.043 |
| Wang (2012) [55] | Feces (46/56) | Relative contribution to total bacteria (%) | Mean | 0.002 | 0.061 | 0.005 |
| Absolute quantification | ||||||
| Yu (2017) [60] | Feces (74/54) | Bacteria counts | Rank mean |
45.09 IMG 40.32 MLG 54.62 OTU |
78.66 IMG 82.14 MLG 71.71 OTU |
< 0.001 |
| Suehiro (2017) [45] | Feces (158/60) | Absolute copy number of F. nucleatum | Median (min–max) | 17.5 (0–5793) | 317 (0–17,343) | < 0.0001 |
| Fukugaiti (2015) [44] | Feces (7/10) | Log no. copies of F. nucleatum/gram | Mean ± SD (min-max) | 4.0 ± 1.5 (1.0–6.4) | 6.2 ± 1.5 (3.5–8.0) | 0.01 |
| Mira-Pascual (2015) [50] | Feces (6/10) | Log no. gene copies of F. nucleatum/mg | Median (IQR) | 4.16 (3.47–4.85) | 4.70 (3.85–5.15) | --- |
| Rezasoltani (2018) [46] | Feces (20/31) | Cq value (Quantity of F. nucleatum = 10 ( )) | Mean (SD) | 29.16 (3.31) | 17.74 (3.59) | < 0.05 |
| Liu (2020) [62] | Feces (53/45) | Log 10 copies of F. nucleatum ∕ gram | Mean (SD) | ≈3.5 (1) | ≈6 (1) | < 0.01 |
| Relative quantification | ||||||
| Russo (2017) [52] | Feces (10/10) | Ratio: | Mean | ≈2.25 | ≈1.8 | > 0.05 |
| Yu (2015) [41] | Feces (42/52) | ΔCq value | Median [IQR] | ≈26 [2] | ≈19 [2] | < 0.001 |
| Biopsy (31/37) | ≈27 [2] | ≈18 [3] | < 0.001 | |||
| Flanagan (2014) [43] | Feces (7 /25) | 2−ΔCq value | Median | 2−21 | 2−15 | 0.02 |
| Liang (2017) [49] Coh I | Feces (170/200) | 2−ΔCq value | Median | ≈2−17 | ≈2−5 | < 0.0001 |
| Liang (2017) [49] Coh II | Feces (33/36) | 2−ΔCq value | Median | ≈2−16 | ≈2−13 | 0.012 |
| Eklof (2017) [47] | Feces (39/66) | Log 2−ΔCq | Median [IQR] | ≈− 10 [2] | ≈− 7 [5] | < 0.001 |
| Guo (2018) [48] | Feces (215/156) | Log 2−ΔCq | Mean | ≈−4 | ≈− 3 | < 0.0001 |
| Wong (2017) [56] | Feces (104/102) | Log 2−ΔCq (fold change from HC to CRC) | Mean | ≈−7 | ≈−5 (132) | < 0.001 |
| Xie (2017) [58] | Feces (327/242) | Log2 RQ, based on ΔCq (Fold change from HC to CRC) | Mean | ≈−20 | ≈− 16 for early stage and −18 for advanced stage (5.12) |
0.006 < 0.001 |
| Tunsjo (2019) [61] | Feces (23/22) | 2−ΔCq value (fold change from HC to CRC) | Median | ≈2−7 |
≈2−5 (66) |
0.0073 |
CRC colorectal cancer cases, C controls (colorectal-adenoma- and CRC-free CRC and C subjects), SD standard deviation, Coh cohort, OTUs operational taxonomic units, MLGs metagenomic linkage groups, IMG integrated microbial genome, Cq quantification cycle in qPCR, ΔCq the average Cq value of F. nucleatum—the average Cq value of total bacteria (or reference gene), min minimum, max maximum
---Not reported
≈A value read from a graph in the study material
Comparison of frequency of presence of Fusobacterium nucleatum in colorectal specimens between colorectal cancer cases and controls
Rather than absolute or relative quantification of F. nucleatum load, some studies compared the frequency of presence of the bacteria in colorectal specimens between controls and CRC patients, as shown in Table 4. Some studies [40, 44, 49–51, 53, 54, 57, 60, 61, 63] reported the frequency of presence of the bacterium in colorectal specimens when the bacterium was simply detected (by PCR, sequencing, or FISH techniques), while other studies [45, 47, 58] reported the frequency of presence of the bacterium when its load level was above a specific cutoff value. The cutoff values were typically set to the values that served to achieve the highest discrimination between CRC patients and controls in terms of Youden index. Tunsjo et al. [61] set a cutoff value for detecting F. nucleatum in feces, but not in biopsies. As shown in Table 4, the cutoff value was not reported in one study [58] and varied between the three others: 260 copies of F. nucleatum by Suehiro et al. [45] and a 2−ΔCq of 0.00026 (2−12) for both Eklof et al. [47] and Tunsjo et al. [61].
Table 4.
Frequency of presence of F. nucleatum in CRC and controls’ specimens and odds ratio for the association between F. nucleatum and colorectal cancer
| First author (year) | Specimen type (no. of specimens CRC/C) | Definition of specimen positive to F. nucleatum | Prevalence of F. nucleatum (+) | OR [95% CI] | Adjusted OR [95% CI] | ||
|---|---|---|---|---|---|---|---|
| C (%) | CRC (%) | p | |||||
| Amitay (2017) [40] | Feces (46/231) | F. nucleatum is detected | 20 | 51 | < 0.001 | 4.16 [2.15–8.07] | --- |
| Fukugaiti (2015) [44] | Feces (7/10) | F. nucleatum is detected | 90 | 100 | --- | 2.37 [0.08–66.88] | --- |
| Liang (2017) [49] Coh I | Feces (170/200) | F. nucleatum is detected | 72 | 98.2 | < 0.0001 | 21.22 [6.57–68.5] | --- |
| Mira-Pascual (2015) [50] | Feces (7/9) | F. nucleatum is detected | 22.2 | 85 | --- | 19.86 [1.47–268.17] | --- |
| Biopsy (7/5) | F. nucleatum is detected | 0 | 26,6 | --- | 4.6 [0.18–121.18] | --- | |
| Vogtmann (2016) [53] | Feces (52/52) | F. nucleatum is detected | 26.9 | 63.5 | 0.0002 | 4.72 [2.05–10.85] | --- |
| Wang (2016) [54] | Feces (10/10) | F. nucleatum is detected | 10 | 60 | --- | 13.5 [1.2–152.21] | --- |
| Wu (2013) [57] | Feces (19/20) | F. nucleatum is detected | 0 | 68.4 | --- | 85.15 [4.42–1638.85] | --- |
| Yu (2017) [60] | Feces (74/54) | F. nucleatum is detected | 3.7 | 52.7 | 7.53E-08 | 29 [6.57–128] | --- |
| Kashani (2020) [63] | Biopsy (35/45) | F. nucleatum is detected | 24 | 68 | 0.0001 | 6.74 [2.5–18.07] | --- |
| Repass (2018) [51] | Biopsy (40/40) | The product is amplified in the qPCR reaction | 5 | 40 | --- | 12.67 [2.67–60.05] | --- |
| Yu (2016) [59] | Biopsy (93/20) | Average number of bacteria per field ≥ 5, visualized by FISH technique | Invasive 20, in biofilms 10 | Invasive 65.9a, in biofilms 48.4a | < 0.05 | 7.73 [2.38–25.07] e | --- |
| Tunsjo (2019) [61] | Feces (23/22) | F. nucleatum detected with ΔCq values < 12 | 0 | 35 | --- | 24.9 [1.33–463.72] | --- |
| Biopsy (21/11) | F. nucleatum is detected | 18 | 52 | --- | 4.93 [0.85–28.67] | --- | |
| Suehiro (2017) [45] | Feces (158/60) | F. nucleatum detected at a higher level based on cutoff value (> 260 copies )b | 10 | 54 | --- | 10.57 [4.3–25.98] | --- |
| Eklof (2017) [47] | Feces (39/65) | F. nucleatum detected at a higher level based on cutoff value (2−ΔCq > 0.00026c≈2−12 )c | 24.3 | 69.2 | --- | 7 [2.89–16.96] | --- |
| Xie (2017) [58] | Feces (327/242) | F. nucleatum detected at a higher level based on cutoff value (value is not reported) d | --- | --- | --- | 4.31 [2.96–6.28] | 4.28 f [2.27–8.09] |
no. number, CRC colorectal cancer cases, C controls (colorectal-adenoma- and CRC-free subjects), OR odds ratio
---Not reported
aThe value was calculated as the average of reported prevalence in proximal and distal separately
b260 copies of F. nucleatum was the best cutoff point to discriminate between HC and CRC in receiver operating characteristic (ROC) analysis resulting in estimated sensitivity of 54% and estimated specificity of 90%, and the area under the ROC curve was 0.75
cThe cutoff value of 0.00026 (= 2−ΔCq = 2−12) gave the most reliable analysis for detecting cancer in the study patients, with estimated specificity of 76.9%
dROC curve was used to evaluate the diagnostic value of bacterial candidates in distinguishing CRC and controls and to determine the best cutoff values that maximized the Youden index
eOR was calculated based on prevalence of invasive F. nucleatum (in tissues) as its detection was higher than within biofilms
fOR adjusted for age, gender, FIT test, two additional bacteria markers, history of diabetes, and high blood pressure
F. nucleatum was commonly detected in all specimens across studies, except for Mira-Pascual et al. and Wu et al., who did not detect the bacterium in controls’ biopsies and controls’ feces respectively [50, 57]. The frequency of specimens positive to F. nucleatum was higher among CRC patients than controls in all the studies. The study by Yu et al. was the only one to use FISH technique, which allowed for quantifying the bacteria within tissues (called invasive F. nucleatum) and in the biofilm separately. Their results showed a higher frequency of presence of F. nucleatum in tissues than in biofilm.
Association between Fusobacterium nucleatum and colorectal cancer
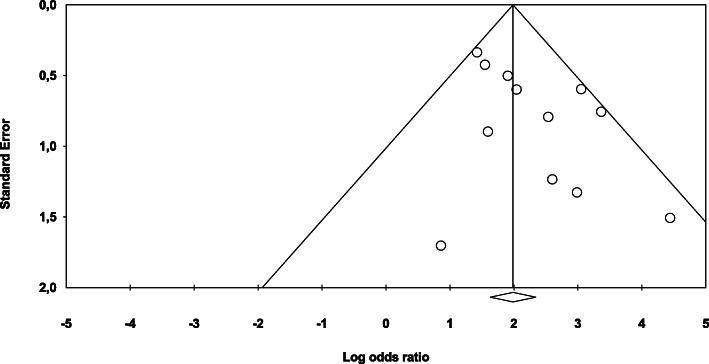
To perform a meta-analysis on the association between F. nucleatum and CRC, we pooled data from 12 studies [40, 44, 49–51, 53, 54, 57, 59–61, 63] that operationally defined the presence of F. nucleatum in terms of the detection of the bacterium in colorectal specimens, with no use of a cutoff value that optimizes distinction between cases and controls, as described above. As shown in Fig. 2, the overall pooled OR and the corresponding 95% CI estimated in a random effects model show a positive association between F. nucleatum detection in colorectal specimens and CRC (OR = 8.3; 95% confidence interval (95% CI) 5.2 to 13.0), with moderate heterogeneity (I2 = 26.32%, p value for heterogeneity = 0.18). Funnel plot for investigating publication bias is presented in Fig. 3. Visual inspection of the funnel plot does not suggest an evident publication bias, which was also confirmed by Egger’s regression test (p = 0.053). Adjusted pooled OR estimate could not be calculated due to non-availability of adjusted OR estimates from the reports of the individual studies.
Fig. 2.
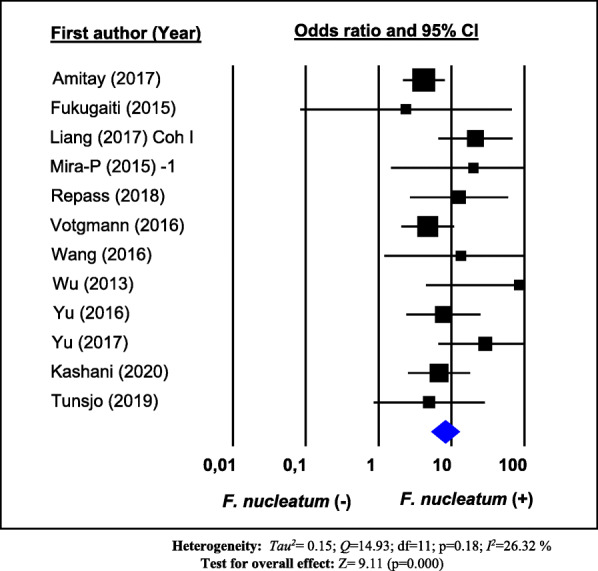
Forest plot of the association between F. nucleatum in colorectal specimens and colorectal cancer. Results of a random-effects meta-analysis of 12 observational studies
Fig. 3.
Funnel plot of the natural logarithm-transformed odds ratio estimates, by the corresponding standard error. Circles, studies in the meta-analysis
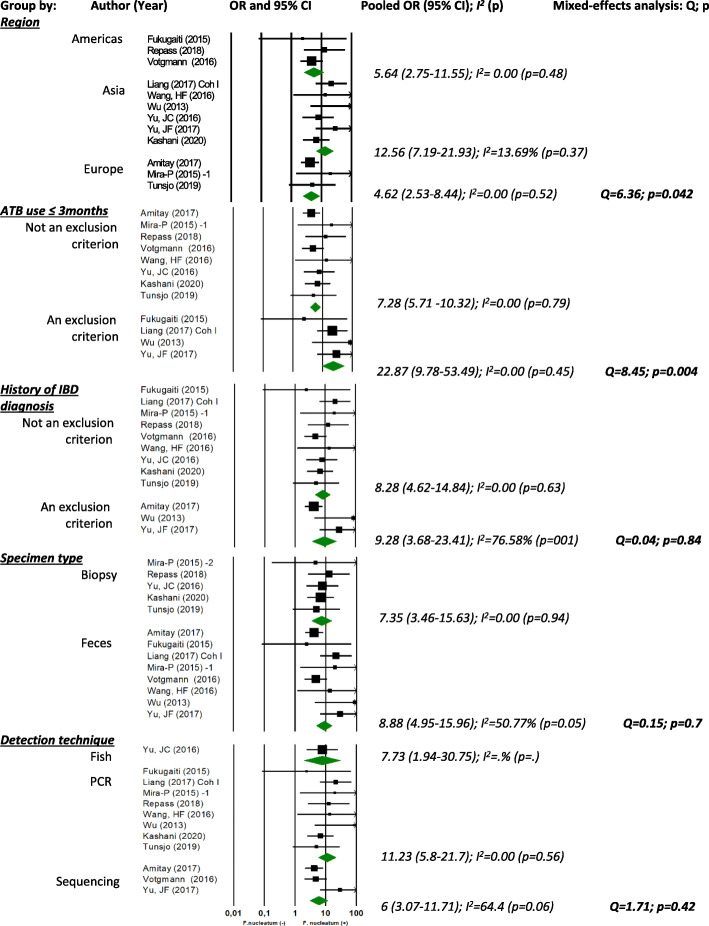
Subgroup meta-analysis shows a stronger association between F. nucleatum and CRC in Asiatic populations, compared to European and American populations, as well as in studies excluding subjects with reported antibiotic use in the last 3 months, compared with studies that did not exclude these subjects (Fig. 4). However, the association was not statistically significantly different by specimen type (stools vs. biopsies), bacterial detection technique (FISH vs. qPCR vs. sequencing), or previous IBD diagnosis as exclusion criteria for study participation.
Fig. 4.
Forest plots for subgroup meta-analysis. ATB, antibiotic; IBD, inflammatory bowel disease; OR, odds ratio
Discussion
This systematic review summarizes results from 24 observational studies that compared the prevalence of the presence of F. nucleatum and/or the mean/median of F. nucleatum load in colorectal specimens, among cases of CRC and controls. Studies used mainly two ways to compare CRC patients and controls in regard to colorectal infection by F. nucleatum: (1) bacterium load expressed by RA (a percentage expressing the relative contribution of F. nucleatum to total bacteria), absolute quantification (bacteria count), or more often by RQ (2−ΔCq value by qPCR technique, with ΔCq = (the average Cq value of F. nucleatum − the average Cq value of total bacteria or reference gene)); and (2) frequency of the presence of F. nucleatum in colorectal specimens.
It is true that RA and RQ are both relative measures of the bacterium load, but many studies used one or the other term to express the same thing, which can be confusing. Thus, in this systematic review, we tried to differentiate between the two terms, RA and RQ, and represent results accordingly. This showed that RA was used less commonly than RQ, even if RA also allows better comparison between healthy and altered microbiome composition, since dysbiosis is, by definition, the loss in representation of different bacterial phyla within the whole bacterial composition of the microbiome.
We also mention some issues with publishing data of F. nucleatum RQ in individual studies. Most of the time, RQ was extracted from papers’ supplementary tables or graphs that were often poorly annotated. Also, even if individual studies based their RQ on a common ΔΔCq method, values were reported differently across studies. Thus, we encourage researchers to standardize the way to report RQ data.
Overall, results of absolute and relative quantification of F. nucleatum were higher in CRC cases compared to controls across most studies. Only three studies reported the fold change of F. nucleatum load from controls to CRC cases, but one was much larger than the others: 5.12-fold by Xie et al. [58], 66-fold by Tunsjo et al. [61], and 132-fold by Wong et al. [56]. When comparing these studies, Xie et al. [58] included subjects with chronic inflamed colorectal mucosa within their control group, while Wong et al. [56] excluded subjects with IBD; Tunsjo et al. [61], for their part, did not report excluding IBD patients from participation. Presence of IBD patients among controls in the study of Xie et al. [58] could probably have blurred the difference in F. nucleatum load between their CRC and controls. In this regard, we mention a study by Strauss et al. who isolated Fusobacterium spp. from 63.6% of patients with gastrointestinal disease compared to 26.5% of healthy controls (P = 0.01), with F. nucleatum representing 69% of recovered Fusobacterium spp. in their IBD patients [64].
Our meta-analysis included 12 studies based on a total of 1098 cases and 1069 controls. Only crude pooled OR could be calculated, and it shows an association between the presence of F. nucleatum in feces or colorectal mucosa and CRC.
All included studies reported results of F. nucleatum detection in specimens collected just before colonoscopy or surgery. Furthermore, the estimated OR for the association between F. nucleatum and CRC was not adjusted for potential confounders. Thus, causal explanation of the “positive” empirical association (as quantified by the pooled-OR estimate) is not warranted, in our view. However, the involvement of F. nucleatum in early CRC carcinogenesis stages has been suggested by other studies that identified the bacterium in precancerous lesions. Its RA was reported to be higher in adenomas than in healthy tissues and lower in adenomas than in carcinomas, reflecting a gradual enrichment of the colon with F. nucleatum in parallel to the adenoma-carcinoma sequence [65–67]. The level of F. nucleatum also seems to increase with advancing stages of dysplasia [43].
Subgroup meta-analysis suggested (even if weakly) that the F. nucleatum–CRC association (if it does exist) may be stronger in Asian populations than in American or European ones. This finding seems to be in line with the results of a recent meta-analysis by Huang et al. [68] on the diagnostic value of fecal F. nucleatum in screening CRC, which had a better performance in Asians. The apparent dependence of the association between F. nucleatum and CRC on population area may be explained by lifestyle differences between populations and/or by diversity in human gut microbiomes at the population level. Nishijima et al. analyzed gut microbiomes of Japanese individuals by comparing metagenomic data obtained from 106 Japanese subjects with those from 11 other nations. They found that gut microbiome of the Japanese is considerably different from those of other populations and cannot be explained by diet alone [69].
We also found that the estimated association between F. nucleatum and CRC was stronger in the subgroup of studies that excluded subjects with recent antibiotic use, compared with the subgroup of studies that did not. This can be explained by a possible bias due to introducing subjects with microbiomes altered by recent antibiotic use.
Some studies failed in reporting critical information, such as time of specimen collection, which was not reported by four studies [41, 43, 54, 57]. Also, in one study [60], feces were collected most frequently after colonoscopy. However, colonic microbiota has been shown to be disturbed by the bowel cleansing protocol and takes about 2 weeks to recover to its original composition depending on the cleansing protocol [70].
F. nucleatum is a very heterogeneous species of the Fusobacteria phylum and has been classified into four to five subspecies: animalis, nucleatum, polymorphum, vincentii/fusiforme. F. nucleatum, subsp. nucleatum, is mainly isolated in periodontal pathological sites, while F. nucleatum subsp. vincentii/fusiforme is often isolated from healthy sites as normal flora. F. nucleatum subsp. animalis and polymorphum are associated with complications of pregnancy, and F. nucleatum subsp. animalis is associated with inflammatory bowel disease [71]. xIn our systematic review, data about subspecies of F. nucleatum was only reported by the study of Amitay et al. [40], in which the four subspecies were identified: ssp. nucleatum, animalis, vincentii, and polymorphum. In a study by Ye et al., five F. nucleatum subspecies were identified in clinical CRC specimens, with ssp. animalis being the most common one [72]. Komiya et al. examined whether identical strains of F. nucleatum could be isolated from colorectal and saliva specimens from the same patient. Saliva and colorectal specimens were analyzed from 14 CRC patients by qPCR, of which 40% exhibited identical strains of F. nucleatum in their colorectal and saliva specimens [73].
The oral cavity can serve as a reservoir for the systemic dissemination of pathogenic bacteria and their toxins, leading to infections and inflammations in distant bodily sites. Several oral species were identified in infections at extraoral sites. Han et al. [28] suggested a spread of oral infection due to transient bacteremia leading to bacterial colonization in extraoral sites, systemic damage by toxins free of oral pathogens, and systemic inflammation caused by soluble antigens of oral pathogens. F. nucleatum is one of the most dominant species of the oral microbiota [26]. It often aggregates with other oral bacteria and plays an essential role in the formation of dental plaque, acting as a bridge between early colonizing bacteria (Gram-positive bacteria) and late colonizing bacteria (Gram-negative bacteria) [74]. Such a mechanism resembles the proposed driver-passenger model in explaining how bacteria in the intestinal microbiota could be involved in carcinogenesis. The first step consists of colonization of the intestine by pathogenic bacteria known as “drivers” with pro-inflammatory and pro-carcinogenic potential (B. fragilis and E. coli in particular). The tumor progression would then cause a modification in the tumor microenvironment, allowing colonization by opportunistic bacteria known as “passengers” (F. nucleatum and Streptococcus gallolyticus in particular), promoting further development of the tumor [75, 76]. The bacterial “drivers” and “passengers” would thus have distinct temporal roles in the pathogenesis of CRC [75]. This model implies that there is not a single bacterium that would alone be incriminated in the occurrence and development of CRC, but rather a bacterial community whose taxonomic composition continues to change throughout the tumorigenic process, thereby allowing specific bacteria to play their role in tumor transformation, according to their virulence and other properties. Moreover, some believe that the oral bacterium F. nucleatum plays a role in the development of CRC within a bacterial community or biofilm, rather than as an individual pathogen [77]. Warren et al. analyzed the bacterial composition of 130 colorectal tumors and their surrounding healthy tissues, and confirmed the over-representation of Fusobacterium, but in the simultaneous presence of two other commensal oral bacteria, Leptotrichia and Campylobacter, in individual tumors [22].
Conclusion
The results of this systematic review and meta-analysis suggest that the F. nucleatum in feces or colorectal mucosa is associated with CRC. Future clinical and epidemiological studies should address the potential role of F. nucleatum in the etiology of CRC. Further, the bacterium should be investigated in the colon at the subspecies level to assess the oral origin of colorectal infection with F. nucleatum.
Supplementary Information
Additional file 1. PRISMA 2009 checklist.
Additional file 2. Supplementary material.
Acknowledgements
The authors gratefully acknowledge the help of Natalie Clairoux and Patrice Dupont (librarians at Bibliothèque de la Santé, Université de Montréal) for helping with the search strategy.
Abbreviations
- CRC
Colorectal cancer
- F. nucleatum
Fusobacterium nucleatum
- 95% CI
95% confidence interval
- OR
Odds ratio
- PRISMA
Preferred Reporting Items for Systematic Reviews and Meta-analyses
- MOOSE
Meta-analysis of Observational Studies in Epidemiology
- NOS
Newcastle-Ottawa scale
- AHRQ
Agency for Healthcare Research and Quality
- RA
Relative abundance
- RQ
Relative quantification
- IBD
Inflammatory bowel disease
- PCR
Polymerase chain reaction
- qPCR
Quantitative polymerase chain reaction
- FISH
Fluorescence in situ hybridization
Authors’ contributions
AIJ carried out the study design, study screening, data extraction, quality appraisal, and data analysis, and drafted the manuscript. IK carried out the study design, advised on all methodological issues, and critically revised the manuscript. CL participated in the study design, participated in the study screening and data extraction, and revised the manuscript. HS participated in data extraction and manuscript drafting and revised the manuscript. EE carried out study design, advised on all methodological issues, and critically revised the manuscript. All authors approved the final version of this manuscript.
Funding
This work was supported by a doctoral scholarship from the Fonds de Recherche du Québec-Santé and an award from Études Supérieures et Postdoctorales of Université de Montréal (to: Amal Idrissi Janati).
Competing interest
None declared
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Arnold M, Sierra MS, Laversanne M, et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66(4):683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 2.Potter JDLN. Genetics of colorectal cancer. New York: Springer Science + Business Media; 2009. [Google Scholar]
- 3.Müller MF, Ibrahim AEK, Arends MJ. Molecular pathological classification of colorectal cancer. Virchows Archiv. 2016;469(2):125–134. doi: 10.1007/s00428-016-1956-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cross AJ, et al. A large prospective study of meat consumption and colorectal cancer risk: an investigation of potential mechanisms underlying this association. Cancer Research. 2010;70(6):2406–2414. doi: 10.1158/0008-5472.CAN-09-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larsson SC, Wolk A. Meat consumption and risk of colorectal cancer: a meta-analysis of prospective studies. International Journal of Cancer. 2006;119(11):2657–2664. doi: 10.1002/ijc.22170. [DOI] [PubMed] [Google Scholar]
- 6.Liang PS, Chen T-Y, Giovannucci E. Cigarette smoking and colorectal cancer incidence and mortality: systematic review and meta-analysis. International Journal of Cancer. 2009;124(10):2406–2415. doi: 10.1002/ijc.24191. [DOI] [PubMed] [Google Scholar]
- 7.Ning Y, Wang L, Giovannucci EL. A quantitative analysis of body mass index and colorectal cancer: findings from 56 observational studies. Obes Rev. 2010;11(1):19–30. doi: 10.1111/j.1467-789X.2009.00613.x. [DOI] [PubMed] [Google Scholar]
- 8.Robsahm TE, Aagnes B, Hjartaker A, et al. Body mass index, physical activity, and colorectal cancer by anatomical subsites: a systematic review and meta-analysis of cohort studies. Eur J Cancer Prev. 2013;22(6):492–505. doi: 10.1097/CEJ.0b013e328360f434. [DOI] [PubMed] [Google Scholar]
- 9.Deng L, Gui Z, Zhao L, Wang J, Shen L. Diabetes mellitus and the incidence of colorectal cancer: an updated systematic review and meta-analysis. Dig Dis Sci. 2012;57(6):1576–1585. doi: 10.1007/s10620-012-2055-1. [DOI] [PubMed] [Google Scholar]
- 10.Cyr A, Dunnagan TA, Haynes G. Efficacy of the health belief model for predicting intention to pursue genetic testing for colorectal cancer. J Genet Couns. 2010;19(2):174–186. doi: 10.1007/s10897-009-9271-7. [DOI] [PubMed] [Google Scholar]
- 11.Cho E, Smith-Warner SA, Ritz J, et al. Alcohol intake and colorectal cancer: a pooled analysis of 8 cohort studies. Ann Intern Med. 2004;140(8):603–613. doi: 10.7326/0003-4819-140-8-200404200-00007. [DOI] [PubMed] [Google Scholar]
- 12.Ray K. Colorectal cancer: fusobacterium nucleatum found in colon cancer tissue--could an infection cause colorectal cancer? Nature Reviews Gastroenterology & Hepatology. 2011;8(12):662. doi: 10.1038/nrgastro.2011.208. [DOI] [PubMed] [Google Scholar]
- 13.Collins D, Hogan AM, Winter DC. Microbial and viral pathogens in colorectal cancer. Lancet Oncol. 2011;12(5):504–512. doi: 10.1016/S1470-2045(10)70186-8. [DOI] [PubMed] [Google Scholar]
- 14.Sears CL, Pardoll DM. Perspective: alpha-bugs, their microbial partners, and the link to colon cancer. J Infect Dis. 2011;203(3):306–311. doi: 10.1093/jinfdis/jiq061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mima K, Nishihara R, Qian ZR, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65(12):1973–1980. doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y, Weng W, Peng J, et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating Toll-like receptor 4 signaling to nuclear factor-kappaB, and up-regulating expression of MicroRNA-21. Gastroenterology. 2017;152(4):851–66.e24. doi: 10.1053/j.gastro.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bashir A, Miskeen AY, Bhat A, Fazili KM, Ganai BA. Fusobacterium nucleatum: an emerging bug in colorectal tumorigenesis. European Journal of Cancer Prevention. 2015;24(5):373–385. doi: 10.1097/CEJ.0000000000000116. [DOI] [PubMed] [Google Scholar]
- 18.Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host & Microbe. 2013;14(2):207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22(2):292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allen-Vercoe E, Jobin C. Fusobacterium and Enterobacteriaceae: important players for CRC? Immunol Lett. 2014;162(2 Pt A):54–61. doi: 10.1016/j.imlet.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allen-Vercoe E, Strauss J, Chadee K. Fusobacterium nucleatum: an emerging gut pathogen? Gut Microbes. 2011;2(5):294–298. doi: 10.4161/gmic.2.5.18603. [DOI] [PubMed] [Google Scholar]
- 22.Warren RL, Freeman DJ, Pleasance S, et al. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome. 2013;1(1):16. doi: 10.1186/2049-2618-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Research. 2012;22(2):299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rubinstein MR, Wang X, Liu W, et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14(2):195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito M, Kanno S, Nosho K, et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int J Cancer. 2015;137(6):1258–1268. doi: 10.1002/ijc.29488. [DOI] [PubMed] [Google Scholar]
- 26.Kapatral V, Anderson I, Ivanova N, et al. Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586. J Bacteriol. 2002;184(7):2005–2018. doi: 10.1128/JB.184.7.2005-2018.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han YW. Fusobacterium nucleatum interaction with host cells- Oral microbial communities: genomic inquiry and interspecies communication. 2011. [Google Scholar]
- 28.Han YW, Wang X. Mobile microbiome: oral bacteria in extra-oral infections and inflammation. J Dent Res. 2013;92(6):485–491. doi: 10.1177/0022034513487559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kostic ADC E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, El-Omar EM, Brenner D, Fuchs CS, Meyerson M, Garrett WS. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han YW, Ikegami A, Rajanna C, et al. Identification and characterization of a novel adhesin unique to oral fusobacteria. J Bacteriol. 2005;187(15):5330–5340. doi: 10.1128/JB.187.15.5330-5340.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fardini Y, Wang X, Temoin S, et al. Fusobacterium nucleatum adhesin FadA binds vascular endothelial cadherin and alters endothelial integrity. Mol Microbiol. 2011;82(6):1468–1480. doi: 10.1111/j.1365-2958.2011.07905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gur C, Ibrahim Y, Isaacson B, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42(2):344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shang FM, Liu HL. Fusobacterium nucleatum and colorectal cancer: a review. World Journal of Gastrointestinal Oncology. 2018;10(3):71–81. doi: 10.4251/wjgo.v10.i3.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Idrissi Janati A, Karp I, Sabri H, Emami E. Is a fusobacterium nucleatum infection in the colon a risk factor for colorectal cancer?: a systematic review and meta-analysis protocol. Syst Rev. 2019;8(1):114. doi: 10.1186/s13643-019-1031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh MK J, Singh S, Thumburu KK, Jaiswal N, Chauhan A, Agarwal A, Paul N, Sagwal S. Filtering the information overload for better decisions. Abstracts of the 23rd Cochrane Colloquium; 2015 3-7 Oct. Vienna: Wiley; 2015. Comparison of Newcastle Ottawa scale (NOS) and Agency for Health Research and Quality (AHRQ) as risk of bias assessment tools for cohort studies. [Google Scholar]
- 36.Katsigianni M, Karageorgiou V, Lambrinoudaki I, Siristatidis C. Maternal polycystic ovarian syndrome in autism spectrum disorder: a systematic review and meta-analysis. Mol Psychiatry. 2019;24(12):1787–1797. doi: 10.1038/s41380-019-0398-0. [DOI] [PubMed] [Google Scholar]
- 37.Perletti G, Monti E, Magri V, et al. The association between prostatitis and prostate cancer. Systematic review and meta-analysis. Arch Ital Urol Androl. 2017;89(4):259–265. doi: 10.4081/aiua.2017.4.259. [DOI] [PubMed] [Google Scholar]
- 38.Pillinger T, Osimo EF, Brugger S, et al. A meta-analysis of immune parameters, variability, and assessment of modal distribution in psychosis and test of the immune subgroup hypothesis. Schizophr Bull. 2019;45(5):1120–1133. doi: 10.1093/schbul/sby160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borenstein M, Hedges LV, Higgins JPT, H RR. Introduction to meta-analysis. UK: Wiley, Ltd ed; 2009. [Google Scholar]
- 40.Amitay ELW S, Vital M, Pieper DH, Hofler D, Gierse IJ, Butt J, Balavarca Y, Cuk K, Brenner H. Fusobacterium and colorectal cancer: causal factor or passenger? Results from a large colorectal cancer screening study. Carcinogenesis. 2017;38(8):781–788. doi: 10.1093/carcin/bgx053. [DOI] [PubMed] [Google Scholar]
- 41.Yu YN, Yu TC, Zhao HJ, et al. Berberine may rescue Fusobacterium nucleatum-induced colorectal tumorigenesis by modulating the tumor microenvironment. Oncotarget. 2015;6(31):32013–32026. doi: 10.18632/oncotarget.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang YZY X, Yu ED, Wang N, Cai QC, Shuai Q, Yan FH, Jiang LF, Wang HX, Liu JX, Chen Y, Li ZS, Jiang QW. Changes in gut microbiota and plasma inflammatory factors across the stages of colorectal tumorigenesis: a case-control study. Bmc Microbiol. 2018;18(1):10. doi: 10.1186/s12866-018-1150-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flanagan LS J, Ebert M, Soucek P, Kunicka T, Liska V, Bruha J, Neary P, Dezeeuw N, Tommasino M, Jenab M, Prehn JH, Hughes DJ. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis. 2014;33(8):1381–1390. doi: 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- 44.Fukugaiti MHI A, Fern; es, M. R.; Ribeiro, U.; Nakano, V.; Avila-Campos, M. J High occurrence of Fusobacterium nucleatum and Clostridium difficile in the intestinal microbiota of colorectal carcinoma patients. Brazil J Microbiol. 2015;46(4):1135–1140. doi: 10.1590/S1517-838246420140665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suehiro YS K, Nishioka M, Hashimoto S, Takami T, Higaki S, Shindo Y, Hazama S, Oka M, Nagano H, Sakaida I, Yamasaki T. Highly sensitive stool DNA testing of Fusobacterium nucleatum as a marker for detection of colorectal tumours in a Japanese population. Ann Clin Biochem. 2017;54(1):86–91. doi: 10.1177/0004563216643970. [DOI] [PubMed] [Google Scholar]
- 46.Rezasoltani SS M, Asadzadeh Aghdaei H, Nazemalhosseini Mojarad E, Dabiri H, Akhavan Sepahi A, Modarressi MH, Feizabadi MM, Zali MR. Applying simple linear combination, multiple logistic and factor analysis methods for candidate fecal bacteria as novel biomarkers for early detection of adenomatous polyps and colon cancer. J Microbiol Method. 2018;155:82–88. doi: 10.1016/j.mimet.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 47.Eklof VL-B A, Zingmark C, Edin S, Larsson P, Karling P, Alexeyev O, Rutegard J, Wikberg ML, Palmqvist R. Cancer-associated fecal microbial markers in colorectal cancer detection. Int J Cancer. 2017;141(12):2528–2536. doi: 10.1002/ijc.31011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo SL L, Xu B, Li M, Zeng Q, Xiao H, Xue Y, Wu Y, Wang Y, Liu W, Zhang G. A simple and novel fecal biomarker for colorectal cancer: ratio of Fusobacterium nucleatum to probiotics populations, based on their antagonistic effect. Clin Chem. 2018;64(9):1327–1337. doi: 10.1373/clinchem.2018.289728. [DOI] [PubMed] [Google Scholar]
- 49.Liang QC J, Chen Y, Huang Y, Higashimori A, Fang J, Brim H, Ashktorab H, Chien Ng S, Ng SSM, Zheng S, Chan FKL, Sung JJY, Yu J. Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clin Cancer Res. 2017;23(8):2061–2070. doi: 10.1158/1078-0432.CCR-16-1599. [DOI] [PubMed] [Google Scholar]
- 50.Mira-Pascual L, Cabrera-Rubio R, Ocon S, et al. Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J Gastroenterol. 2015;50(2):167–179. doi: 10.1007/s00535-014-0963-x. [DOI] [PubMed] [Google Scholar]
- 51.Repass JRPC, Biology, Iorns E, Denis A, Williams SR, Perfito N, Errington TM. Replication study: Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. eLife. 2018;7:13. doi: 10.7554/eLife.25801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Russo EB G, Chiellini C, Fagorzi C, Niccolai E, Taddei A, Ricci F, Ringressi MN, Borrelli R, Melli F, Miloeva M, Bechi P, Mengoni A, Fani R, Amedei A. Preliminary comparison of oral and intestinal human microbiota in patients with colorectal cancer: a pilot study. Front Microbiol. 2017;8(2699):2699. doi: 10.3389/fmicb.2017.02699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vogtmann EH X, Zeller G, Sunagawa S, Voigt AY, Hercog R, Goedert JJ, Shi J, Bork P, Sinha R. Colorectal cancer and the human gut microbiome: reproducibility with whole-genome shotgun sequencing. PLoS One. 2016;11(5):e0155362. doi: 10.1371/journal.pone.0155362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang HFL LF, Guo SH, Zeng QY, Ning F, Liu WL, Zhang G. Evaluation of antibody level against Fusobacterium nucleatum in the serological diagnosis of colorectal cancer. Sci Rep. 2016;6:10. doi: 10.1038/s41598-016-0003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang TC, G.; Qiu, Y.; Fei, N.; Zhang, M.; Pang, X.; Jia, W.; Cai, S.; Zhao, L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. Isme J. 2012;6(2):320–329. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wong SHK TNY, Chow TC, Luk AKC, Dai RZW, Nakatsu G, Lam TYT, Zhang L, Wu JCY, Chan FKL, Ng SSM, Wong MCS, Ng SC, Wu WKK, Yu J, Sung JJY. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut. 2017;66(8):1441–1448. doi: 10.1136/gutjnl-2016-312766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu NY X, Zhang R, Li J, Xiao X, Hu Y, Chen Y, Yang F, Lu N, Wang Z, Luan C, Liu Y, Wang B, Xiang C, Wang Y, Zhao F, Gao GF, Wang S, Li L, Zhang H, Zhu B. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol. 2013;66(2):462–470. doi: 10.1007/s00248-013-0245-9. [DOI] [PubMed] [Google Scholar]
- 58.Xie YHGQY, Cai GX, Sun XM, Zou TH, Chen HM, Yu SY, Qiu YW, Gu WQ, Chen XY, Cui Y, Sun D, Liu ZJ, Cai SJ, Xu J, Chen YX, Fang JY. Fecal Clostridium symbiosum for noninvasive detection of early and advanced colorectal cancer: test and validation studies. EBioMed. 2017;25:32–40. doi: 10.1016/j.ebiom.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu JC, Y.; Fu, X.; Zhou, X.; Peng, Y.; Shi, L.; Chen, T.; Wu, Y. Invasive Fusobacterium nucleatum may play a role in the carcinogenesis of proximal colon cancer through the serrated neoplasia pathway. Int J Cancer. 2016;139(6):1318–1326. doi: 10.1002/ijc.30168. [DOI] [PubMed] [Google Scholar]
- 60.Yu JF Q, Wong SH, Zhang D, Yi Liang Q, Qin Y, Tang L, Zhao H, Stenvang J, Li Y, Wang X, Chen N, Wu WKK, Al-Aama J, Nielsen HJ, Kiilerich P, Jensen BAH, Yau TO, Lan Z, Jia H, Li J, Xiao L, Lam TYT, Ng SC, Cheng ASL, Wong VWS, Chan FKL, Xu X, Yang H, Madsen L, Datz C, Tilg H, Brunner N, Kristiansen K, Arumugam M, Sung JJY, Wang J. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2017;66(1):70–78. doi: 10.1136/gutjnl-2015-309800. [DOI] [PubMed] [Google Scholar]
- 61.Tunsjo HS, Gundersen G, Rangnes F, et al. Detection of Fusobacterium nucleatum in stool and colonic tissues from Norwegian colorectal cancer patients (vol 38, pg 1367, 2019) Eur J Clin Microbiol Infect Dis. 2019;39(1):213. doi: 10.1007/s10096-019-03712-x. [DOI] [PubMed] [Google Scholar]
- 62.Liu X, Cheng Y, Shao L, Ling Z. Alterations of the predominant fecal microbiota and disruption of the gut mucosal barrier in patients with early-stage colorectal cancer. Biomed Res Int. 2020;2020:2948282. doi: 10.1155/2020/2948282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kashani N, Bezmin Abadi AT, Rahimi F, Forootan M. FadA-positive Fusobacterium nucleatum is prevalent in biopsy specimens of Iranian patients with colorectal cancer. N Microbes N Infect. 2020;34:100651. doi: 10.1016/j.nmni.2020.100651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Strauss J, Kaplan GG, Beck PL, et al. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis. 2011;17(9):1971–1978. doi: 10.1002/ibd.21606. [DOI] [PubMed] [Google Scholar]
- 65.Rezasoltani S, Asadzadeh Aghdaei H, Dabiri H, et al. The association between fecal microbiota and different types of colorectal polyp as precursors of colorectal cancer. Microb Pathog. 2018;124:244–249. doi: 10.1016/j.micpath.2018.08.035. [DOI] [PubMed] [Google Scholar]
- 66.McCoy AN, Araujo-Perez F, Azcarate-Peril A, et al. Fusobacterium is associated with colorectal adenomas. Plos One. 2013;8(1):8. doi: 10.1371/journal.pone.0053653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ye XC, Wang R, Bhattacharya R, et al. Fusobacterium nucleatum subspecies animalis influences proinflammatory cytokine expression and monocyte activation in human colorectal tumors. Cancer Prev Res. 2017;10(7):398–409. doi: 10.1158/1940-6207.CAPR-16-0178. [DOI] [PubMed] [Google Scholar]
- 68.Huang Q, Peng Y, Xie F. Fecal fusobacterium nucleatum for detecting colorectal cancer: a systematic review and meta-analysis. Int J Biol Markers. 2018;1724600818781301. 10.1177/1724600818781301. Epub ahead of print. PMID: 29968498. [DOI] [PubMed]
- 69.Nishijima S, Suda W, Oshima K, et al. The gut microbiome of healthy Japanese and its microbial and functional uniqueness. DNA Res. 2016;23(2):125–133. doi: 10.1093/dnares/dsw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jalanka J, Salonen A, Salojarvi J, et al. Effects of bowel cleansing on the intestinal microbiota. Gut. 2015;64(10):1562–1568. doi: 10.1136/gutjnl-2014-307240. [DOI] [PubMed] [Google Scholar]
- 71.Nie S, Tian B, Wang X, et al. Fusobacterium nucleatum subspecies identification by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2015;53(4):1399–1402. doi: 10.1128/JCM.00239-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ye X, Wang R, Bhattacharya R, et al. Fusobacterium nucleatum subspecies animalis influences proinflammatory cytokine expression and monocyte activation in human colorectal tumors. Cancer Prev Res (Phila) 2017;10(7):398–409. doi: 10.1158/1940-6207.CAPR-16-0178. [DOI] [PubMed] [Google Scholar]
- 73.Komiya Y, Shimomura Y, Higurashi T, et al. Patients with colorectal cancer have identical strains of Fusobacterium nucleatum in their colorectal cancer and oral cavity. Gut. 2019;68(7):1335–1337. doi: 10.1136/gutjnl-2018-316661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu D. Molecular detection of human bacterial pathogens: CRC Press. 2011. [Google Scholar]
- 75.Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10(8):575–582. doi: 10.1038/nrmicro2819. [DOI] [PubMed] [Google Scholar]
- 76.Gao Z, Guo B, Gao R, Zhu Q, Qin H. Microbiota disbiosis is associated with colorectal cancer. Front Microbiol. 2015;6:20. doi: 10.3389/fmicb.2015.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Flynn KJ, Baxter NT, Schloss PD. Metabolic and community synergy of oral bacteria in colorectal cancer. Msphere. 2016;1(3):e00102–e00116. doi: 10.1128/mSphere.00102-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. PRISMA 2009 checklist.
Additional file 2. Supplementary material.