Abstract
In colorectal cancer (CRC), high expression of trefoil factor 3 (TFF3) is associated with tumor progression and reduced patient survival; however, bioinformatics analyses of public ‘omics’ databases show low TFF3 expression in CRCs as compared to normal tissues. Thus, we examined TFF3 expression in CRCs and matching normal tissues to evaluate its role in CRC progression. TFF3 gene expression was characterized using the bioinformatics portal UALCAN (http://ualcan.path.uab.edu). Tissue microarrays (TMAs) of archival CRC specimens (n=96) were immunostained with anti-human TFF3 antibodies. Immunohistochemical (IHC) staining intensity was semi-quantitatively scored. For this cohort, the median follow-up was 5.4 years. Associations between clinical and pathological variables were determined using Chi-square or Fisher's exact tests. Univariate disease-free survival was estimated by the Kaplan-Meier method. Omics data analyses by UALCAN showed downregulation of TFF3 expression in CRC relative to normal tissue at protein (χ2, P<0.0001) levels. There was a similar decreasing trend of TFF3 expression in the pathologic stages of the CRCs (RNA, χ2, P=0.88 and protein, χ2 P<0.0001). UALCAN data analysis showed that TFF3 exhibited 27% lower mRNA expression in tumors with mutant TP53 (P=0.007). Confirming the findings of omics analyses, IHC analysis of TMAs exhibited lower TFF3 expression in 95.6% (65 of 68) of the available normal-tumor matching pairs (χ2, P<0.0001). There was no statistically significant association of tumor TFF3 expression with patient sex, race/ethnicity, tumor location within the colorectum, Tumor, Node, Metastasis (TNM) stage, lymph node metastasis, or surgical margins. However, low TFF3 IHC staining in tumor tissue was associated with histological grade (P=0.026). Kaplan-Meier survival analysis showed no prognostic value of low TFF3 expression relative to those with high expression (log-rank, P=0.605). Our findings demonstrate low expression of TFF3 in CRCs. Association between low TFF3 and histopathological features suggests involvement of this molecule in progression of CRC.
Keywords: TFF3, trefoil factor 3, colorectal cancer, RNA expression, immunohistochemical expression
Introduction
In the United States, colorectal cancer (CRC) is a leading cause of cancer-related deaths (1). The diagnosis of CRC is based on the tumor-node-metastasis (TNM) stage, a system that has limitations due to varying genetic and epigenetic backgrounds. Biological markers can improve early detection and guide clinicians in subsequent therapies.
Trefoil factor (TFF) proteins include family members TFF1, TFF2, and TFF3, which are characterized by the presence of at least one 40-amino acid protein domain defining a three clover-leaf structure termed the trefoil motif (2). TFF3 is a small, 9-kDa, stable protein expressed in epithelia of the trachea, lungs, salivary glands, and in the small intestine (3). In the gastrointestinal (GI) tract, TFF3 is expressed in goblet cells of the small and large intestines (4) and is secreted, together with mucus, by mucus-secreting cells (3,5). TFF3 promotes normal cell migration and maintains colonic epithelial homeostasis (6,7).
TFF3 is involved in tumorigenesis and the progression of various solid tumors (8–16). The function of this protein in CRCs, however, remains unclear. It has been suggested that patients with high TFF3 tumor levels have a lower survival rate than those with lower expression (17). In addition, higher tumor expression of TFF3 correlates with lymph node metastasis (LNM), i.e., a more advanced stage (17). Furthermore, high TFF3 levels in serum are associated with poor tumor differentiation and a more advanced clinical TNM stage (18). By contrast, expression of TFF3 mRNA, analyzed by real-time quantitative PCR (RT-PCR), does not differ for paired CRCs and normal colonic mucosa (19). However, TFF3 transcripts are elevated in CRCs with LNM compared with non-LNM tumors (19). As assessed by immunohistochemistry (IHC), TFF3 expression in CRCs with LNM is elevated relative to that in non-LNM CRCs (19). Nevertheless, IHC of TFF3 in normal colonic mucosa and CRCs shows no correlation of TFF3 expression with patient sex, cancer differentiation status, or cancer stage (19).
Due to the conflicting findings related to TFF3, further clarification is needed to elucidate the clinical value of TFF3 as a marker of aggressive disease. Therefore, in the present study, we analyzed TFF3 expression levels at the RNA and protein levels in publicly available databases and validated its protein expression in our collection of CRCs and their matching normal tissues. Additionally, correlations between TFF3 expression, molecular (expression of TP53), clinical and pathological characteristics of patients (including patient survival) were assessed.
Materials and methods
Bioinformatics analysis
The bioinformatics portal UALCAN (http://ualcan.path.uab.edu) was used to access TFF3 RNA and protein levels in normal colon and rectum tissue and in CRCs of patients. This resource for expression analysis uses data from The Cancer Gene Atlas (TCGA) (transcriptome by RNA sequencing) (20) and the Clinical Proteomic Tumor Analysis Consortium (CPTAC) Confirmatory/Discovery datasets (proteomics by mass-spectrometry) (21). We checked for under-expressed genes in TP53 mutant CRCs by use of heat-maps, which demonstrated that TFF3 was one of the 25 genes having low expression in TP53 mutant cancers as compared to TP53-wild-type tumors. RNA data are expressed as transcripts per million and protein data as Z-values, representative of standard deviations from the median across samples for the given cancer type. Log2 spectral count ratio values from CPTAC were first normalized within each sample profile, then normalized across samples.
Patients and tissue samples
The population of this study was derived from the University of Mississippi Medical Center (UMMC), Department of Pathology research database. All tissue specimens (collected between 2006 and 2016) were obtained at the time of surgery and had complete clinicopathological data. The tissue samples were de-identified and assigned a study number. Clinical and pathological characteristics of the study subjects are provided in Table I. These data include sex, race, TNM stage, histological grade, evidence of LNM, surgical margins, survival times, and status. Tumor and adjacent normal colonic tissues from patients routinely obtained during surgery were included in this study. A total of 96 cases were assessed by board-certified pathologists (CS and VS). At the time of surgery, patients were staged according to the guidelines of the American Joint Committee on Cancer. The median follow-up for the 96 patients was 5.4 years (range 0.1–10.3 years). All procedures of this study involving human materials were approved by the UMMC Institutional Review Board approval and performed according to the ethical standards with the Declaration of Helsinki.
Table I.
Clinicopathological characteristics of the CRC patients.
| Characteristic | Data |
|---|---|
| Age, years, mean (range) | 59.2 (23–87) |
| Sex, n (%) | |
| Male | 50 (52.1) |
| Female | 46 (47.9) |
| Race/ethnicity, n (%) | |
| African-Americans | 56 (58.3) |
| Non-Hispanic Whites | 40 (41.7) |
| Site, n (%) | |
| Colon | 62 (64.6) |
| Rectum | 34 (35.4) |
| TNM stage, n (%) | |
| I | 11 (11.4) |
| II | 30 (31.3) |
| III | 35 (36.5) |
| IV | 20 (20.8 |
| Histological grade, n (%) | |
| Well-differentiated | 6 (6.3) |
| Moderately differentiated | 78 (81.3) |
| Poorly differentiated | 7 (7.3) |
| Unknown | 5 (5.1%) |
| Lymph node metastasis, n (%) | |
| Negative | 36 (37.5) |
| Positive | 50 (52.1) |
| Unknown | 10 (10.4) |
| Surgical margins, n (%) | |
| Negative | 74 (77.1) |
| Positive | 18 (18.8) |
| Unknown | 4 (4.1) |
| Follow-up time (years), median (range) | 5.4 (0.1–10.3) |
CRC, colorectal cancer.
Construction of tissue microarrays
Tissue microarrays (TMA) were created using tumor stage-matched CRC tissues. For each patient, representative formalin-fixed paraffin-embedded (FFPE) tissue blocks, consisting of a normal block and a tumor block were selected, giving a total of 192 samples for TMA construction. Selection of the FFPE blocks was accomplished on the basis of verified histological features, which then were topographically correlated with the corresponding paraffin blocks by pathologists. Cylindrical cores (2-mm) from selected sites of the primary FFPE block were transferred to composite paraffin blocks to construct the TMA using a Beecher MTA1 Manual Tissue Arrayer (Beecher Instruments). The resulting TMA composite blocks were then sectioned at 5-µm thickness for IHC staining and analysis.
Immunohistochemistry
IHC was performed on the FFPE tissue samples using ABC Kits (Vector Laboratories Inc.) following the manufacturer's instructions. Slides with the TMA sections were placed in a 60°C oven for 1 h to allow the tissues to adhere. The sections were deparaffinized in xylene, rehydrated with graded ethanol, and washed with PBS, followed by antigen retrieval (Vector Laboratories Inc., #H-3300, citrate-based antigen unmasking solution) with citrate buffer (pH 6.0) for 20 min. Then, slides were incubated with 3% hydrogen peroxide for 10 min to inactivate the endogenous peroxidases, rinsed with PBS for 10 min, and incubated for 12 min with protein blocking solution (Dako #X0590). Using mouse Vectastain Elite ABC kits (Vector Laboratories, Inc.), blocking serum was prepared using normal serum, and blocking of the tissue sections was performed for 1 h at room temperature. Next, the slides were incubated with blocking serum and primary antibody anti human-TFF3 (mouse monoclonal diluted 1/25; cat # MAB4407, R&D Systems) overnight at 4°C. Following extensive washing, antigen-antibody complexes were detected using Vectastain Elite ABC kits (Vector Laboratories, Inc.) according to the manufacturer's protocol. By use of the ABC kits, anti-mouse IgG biotinylated secondary antibody was prepared with blocking serum and incubated for 30 min, followed by incubation with ABC Reagents A and B for another 30 min. For color development, sections were processed with 3,3-diaminobenzidine peroxidase substrate kits (Vector Laboratories, Inc.). Sections were then counterstained in Gill's hematoxylin and dehydrated in ascending grades of ethanol before clearing in xylene and being mounted under coverslips using Cytoseal™ XYL (Thermo Fisher Scientific, Inc.; #83124). Subcellular localizations of TFF3 were examined by a pathologist, defined as cytoplasmic/membranous or globular staining, and scored.
Immunostaining evaluation
Evaluation of the IHC TMA was accomplished by two independent evaluators blinded for specific diagnosis or prognosis for each individual case. Tumor cells showing cytoplasm stained in brown were considered positive. To provide a semi-quantitative assessment of the expression of TFF3 in each core, colonic epithelial cells were individually scored from 0–3 for the intensity of immunostaining and 1–3 for the area of immunostaining. The product of intensity and area was used as the combined intensity score, which ranged from 0–9, with 0 defining no expression and 9 defining high expression. Values are expressed as median (interquartile range). To assess the association between TFF3 expression and clinical features in the CRC cases, patients were divided into two groups, with low and high TFF3 tumor expression, based on the optimal cutoff point calculated as the value with the most significant log-rank test split (4.5 for combined intensity score).
Statistical analysis
Data were analyzed using SPSS software, version 13.0 (SPSS Inc.) and SAS 9.4 (SAS Inc.). The difference in TFF3 gene or protein expression between normal tissue and tumor tissue or for any other pairwise comparison obtained using bioinformatics analyzes was evaluated by Student's t-test (for comparisons between two groups), or one-way ANOVA followed by Dunnett's multiple comparisons (when three or more groups were compared). Pairwise comparisons were always relative to normal tissue. Pearson's correlation coefficient (r) was used to evaluate association between variables. For IHC data, differences were compared by Mann-Whitney U test for non-matched data or Wilcoxon matched-pairs signed rank test. Two-sided P-values were determined via Chi-square or Fisher's exact tests for categorical variables. Overall survival was analyzed by the Kaplan-Meier method with the use of the log-rank test to compare survival of groups. For all analyses, the level of significance was set at P<0.05.
Results
Bioinformatics analyses of RNA expression of TFF3 in the CRC tissues
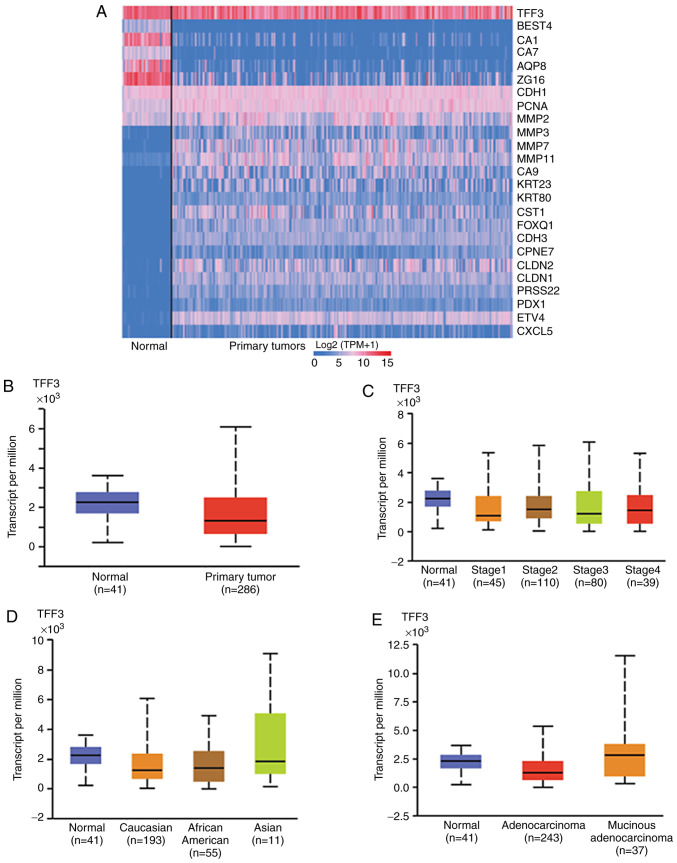
Using UALCAN we generated an interactive heat-map of differentially expressed genes in normal tissue or CRCs and included TFF3 as a query gene. The heat-map demonstrated differential expression of various genes, including TFF3, in normal and CRC tissues (Fig. 1A). Further, as determined with the UALCAN database, mRNA data for CRCs suggested TFF3 overexpression [NS (not significant), P=0.72] in normal (n=41) as compared to CRC tissues (n=286) (Fig. 1B). Data mining for different pathologic stages [stage 1 (n=45), stage 2 (n=110), stage 3 (n=80), and stage 4 (n=39)] suggested lower TFF3 expression in CRCs as compared to normal colon (n=41), irrespective of the stage (NS, P=0.88) (Fig. 1C). Moreover, UALCAN-acquired TCGA data showed lower expression of TFF3 in tumors relative to normal colonic tissues irrespective of patient race [Caucasian (n=193), African-American (n=55), and Asian (n=11)] (Fig. 1D); however, race-related values were not statistically different (P=0.56). A similar trend was observed for tumor histologic types [adenocarcinoma (n=243) and mucinous adenocarcinoma (n=37)] (Fig. 1E, P=0.053), nodal status [N0 (n=166), N1 (n=70), and N2 (n=47)] (Fig. S1A, P=0.77), patient sex [male (n=156) vs. female (n=127)] (Fig. S1B, P=0.51), and age [21–40 years (n=12), 41–60 years (n=90), 61–80 years (n=149), and 81–100 years (n=32)] (Fig. S1C, P=0.53).
Figure 1.
TFF3 RNA expression in CRC tissues. (A) Heat-map plot of 25 differentially expressed genes in CRCs and normal colonic tissue from TCGA dataset generated by web-portal UALCAN. (B) Box plots showing relative expression of TFF3 mRNA in CRC and normal tissues (Student's t-test). The mRNA expression levels were normalized as transcripts per million reads. (C) The expression of TFF3 for various stages of CRC and normal colon. (D) TFF3 expression in CRCs on the basis of patient race and (E) tumor histologic types (C-E, one-way ANOVA with Dunnett's multiple comparisons test). Pairwise comparisons relative to normal tissue. CRC, colorectal cancer; TFF3, trefoil factor 3; TCGA, The Cancer Gene Atlas.
Bioinformatics CPTAC analyses of protein expression of TFF3 in normal colonic and tumor tissues and correlation with tumor and patient characteristics
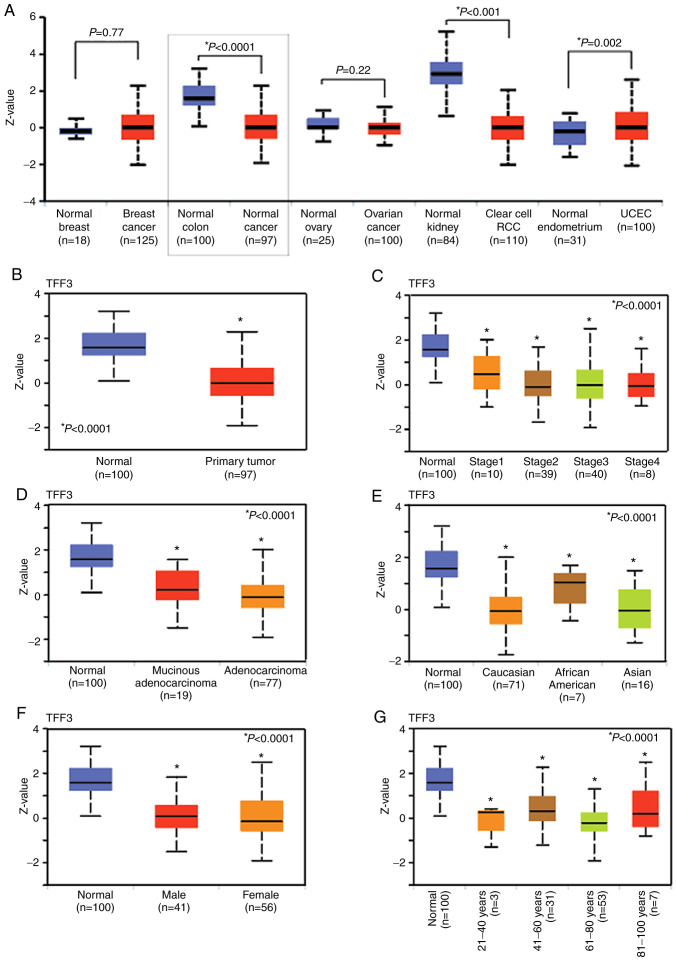
UALCAN bioinformatics analysis was used to analyze protein levels of TFF3 in colonic tissues. The protein expression data was obtained from the mass-spectrometry proteomic profiles, generated by the Clinical Proteomic Tumor Analysis Consortium (CPTAC). The results showed that the expression of TFF3 was high in normal colonic tissue (n=100) and low in CRCs (n=97) (Fig. 2A and B) (P<0.0001). In addition, there was high expression in uterine corpus endometrial carcinoma (UCEC) (P=0.002), low expression in clear cell renal cell carcinoma (P<0.001), but no difference in breast cancers (P=0.77) and ovarian cancer (P=0.22) (Fig. 2A). TFF3 expression was significantly lower (P<0.0001) in late-stage CRCs [stage 3 (n=40); stage 4 (n=8)] compared to early stages [stage 1 (n=10); stage 2 (n=39)], suggesting a role for TFF3 in tumor suppression (Fig. 2C). Expression was low, particularly for adenocarcinoma (n=77), but also for mucinous adenocarcinoma (n=19), as compared to normal tissue, P<0.0001 (Fig. 2D). Furthermore, TFF3 expression was low when tumors of Caucasian (n=71), African-American (n=7), and Asian (n=16) CRC patients (P<0.0001) (Fig. 2E) were compared to normal tissue. Next, we found downregulation of TFF3 in CRCs of patients for both sexes [male (n=41) vs. female (n=56), P<0.0001] (Fig. 2F) and for age groups [21–40 years (n=3), 41–60 years (n=31), 61–80 years (n=53), and 81–100 years (n=7), P<0.0001] (Fig. 2G). These data showed downregulation of TFF3 protein expression in CRCs as compared to normal colon irrespective of the tumor stage; histologic type; and patient race, sex, and age.
Figure 2.
Protein expression of TFF3 in CRC tissues. (A) The graphs show the pan-cancer subtype for various cancers. Z-values represent standard deviations from the median across samples for the given cancer type. Log2 spectral count ratio values from CPTAC were first normalized within each sample profile, and then normalized across samples (Student's t-test). (B) Box-whisker plots showing the protein levels of TFF3 in normal tissue and tumors (Student's t-test). (C) Plots showing correlations between TFF3 protein expression with pathologic tumor stages, (D) tumor histologic types, (E) patient race, (F) patient sex, and (G) age (C-G, one-way ANOVA with Dunnett's multiple comparisons test). Pairwise comparisons relative to normal tissue. CRC, colorectal cancer; TFF3, trefoil factor 3; CPTAC, Clinical Proteomic Tumor Analysis Consortium. UALCAN website that used two-sided Students t-test provided P-values, indicated by asterisk (*). P-value <0.05 was defined as statistical significant.
Correlation of TFF3 RNA expression with TP53-mutational status using bioinformatics
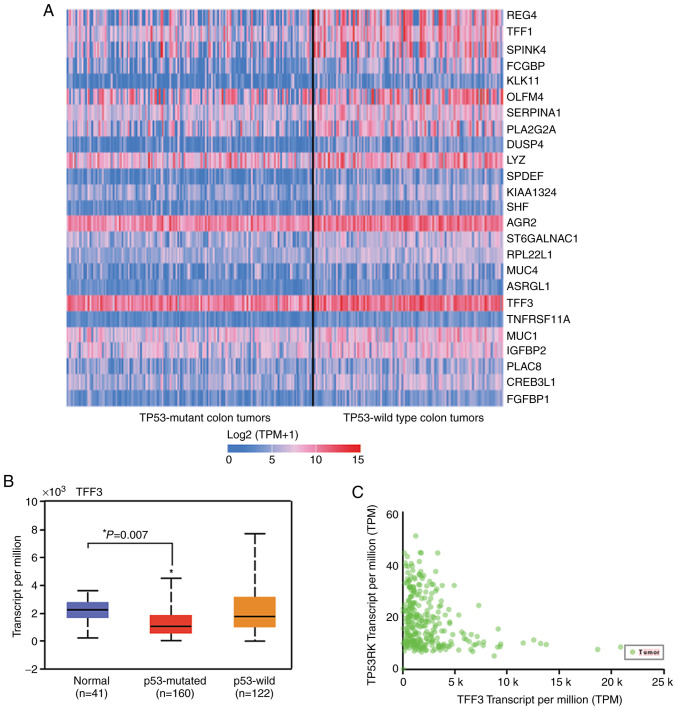
Since P53 mutations occur in 50–60% of CRCs, we explored the correlation between TFF3 and p53 RNA expression and generated an interactive heat-map using the UALCAN database (Fig. 3A). The graph shows 27% lower TFF3 RNA expression in TP53-mutant tumors (n=162) (P=0.007) as compared to normal tissue (n=41). No significant changes in transcripts of TFF3 were noted in TP53 wild-type tumors (n=122), as compared to those observed in normal epithelium (n=41) (Fig. 3B). UALCAN correlation analysis showed an inverse correlation of TFF3 expression with TP53 regulating kinase (TP53RK) (Pearson correlation coefficient, r=−0.32) (Fig. 3C).
Figure 3.
TFF3 RNA expression is inversely correlated with TP53 mutation. (A) Heat-map showing differential expression of genes in TP53 mutant and wild-type CRC. Each column in the heat-map represents a biological sample of TP53 mutant or wild-type, and each row represents a gene. The depth of the color indicates the expression levels of genes. (B) Box plot displaying the expression levels of TFF3 in TP53-mutant CRCs, TP53 wild-type CRCs, and normal samples. Pairwise comparisons (ANOVA with Dunnett's multiple comparisons test), relative to normal tissue. P-value was provided by UALCAN website that used two-sided Students t-test and are indicated by asterisk (*). P-value <0.05 was defined as statistical significant. (C) Correlation analysis between TFF3 and TP53RK (Pearson correlation coefficient, r=−0.32). CRC, colorectal cancer; TFF3, trefoil factor 3; TP53RK, TP53 regulating kinase.
Validation of bioinformatics findings of TFF3 protein expression by immunohistochemical (IHC) profiling of normal colonic and tumor tissue
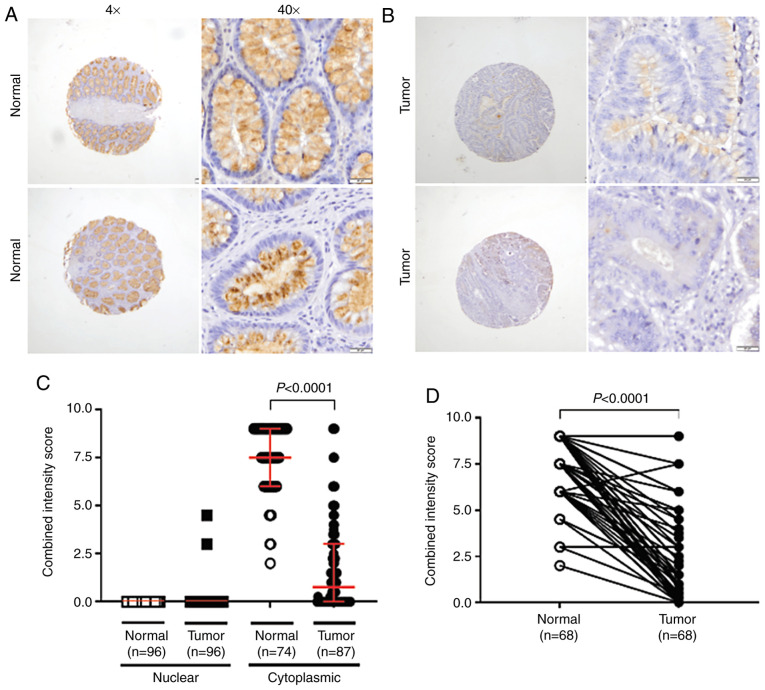
We evaluated TFF3 expression by IHC analysis of normal tissues and CRCs. Positive staining was evident for 74 normal cores. In both the basal and luminal portions of colonic crypts, cytoplasmic staining was present mainly in the supra- and perinuclear cytoplasm of epithelial cells (Fig. 4A). In CRCs, although globular staining was present in 2.1% (2 of 96) of specimens, TFF3 staining was predominantly cytoplasmic, as noted in 71.9% (60 of 87) of the positively stained cores. Mainly supra-nuclear (Fig. 4B), cytoplasmic TFF3 immunoreactivity in CRCs [0.75 (3.0)] (Fig. 4C) was lower by 90% (P<0.0001) relative to staining in normal tissues, when independent samples were compared. Analysis of the cytoplasmic immunostaining in normal samples revealed the absence of globular TFF3 expression; the normal combined intensity score, however, was 7.5 (3.0) (Fig. 4C). Lower TFF3 expression was noted in 95.6% (65 of 68) of available normal-tumor matching pairs (P<0.0001) (Fig. 4D).
Figure 4.
IHC staining of TFF3. The slides were viewed by light microscopy (×40 magnification). (A) For normal colonic mucosa, TFF3 cytoplasmic staining was mainly associated with the supra- and perinuclear cytoplasm of epithelial cells in both the basal and luminal portions of the colonic crypts. Low to null staining was evident at the nuclear/globular level. (B) TFF3 was lower in cancer cells, and expressed mainly in the supra-nuclear cytoplasm. (C) Scatter plot showing that TFF3 IHC staining is mostly cytoplasmic and lower in tumors, relative to normal mucosa (P<0.0001, Mann-Whitney U test for non-matched data). (D) TFF3 expression was lower in 95.6% of available normal-tumor matching pairs (P<0.0001, Wilcoxon matched-pairs signed rank test). TFF3, trefoil factor 3; IHC, immunohistochemistry.
Patients grouped by IHC expression of TFF3 and association with clinicopathologic features
There was no significant association of TFF3 expression with patient sex (P=0.656), race/ethnicity (P=0.383), tumor site (P=0.819), TNM stage (P=0.120), LNM (P=0.081), or surgical margins (P=0.682) (Table II). However, low TFF3 tumor immunoreactivity was associated with moderately differentiated histological grade (P=0.026).
Table II.
Correlation of the clinicopathologic findings with TFF3 expression in the CRC cases.
| Characteristic | TFF3 low (score ≤4.75) | TFF3 high (score >4.75) | χ2 or Fisher's exact test P-value |
|---|---|---|---|
| Sex | |||
| Male | 40 | 5 | 0.656 |
| Female | 36 | 6 | |
| Race/ethnicity | |||
| African-Americans | 46 | 5 | 0.383 |
| Non-Hispanic Caucasians | 30 | 6 | |
| Site | |||
| Colon | 31 | 29 | 0.819 |
| Rectum | 13 | 14 | |
| TNM stage | |||
| I | 9 | 2 | 0.120 |
| II | 21 | 6 | |
| III | 30 | 1 | |
| IV | 16 | 2 | |
| Histological grade | |||
| Well-differentiated | 2 | 2 | 0.026a |
| Moderately differentiated | 64 | 7 | |
| Poorly differentiated | 5 | 2 | |
| Lymph node metastasis | |||
| Negative | 25 | 7 | 0.081 |
| Positive | 44 | 3 | |
| Surgical margins | |||
| Negative | 57 | 10 | 0.682 |
| Positive | 15 | 1 |
Significant difference, P<0.05. TFF3, trefoil factor 3; CRC, colorectal cancer.
Survival analysis
There was no significant difference for high and low IHC expression of TFF3 and overall survival (Fig. 5).
Figure 5.
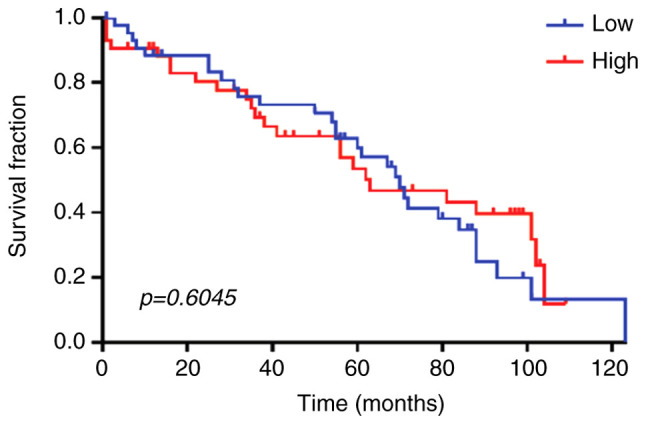
Overall survival curves for patients with CRC according to the IHC expression of TFF3. Plots, censored at 123 months, were generated using the Kaplan-Meier method, and survival distributions were compared using log-rank tests for patients with low vs. high tumor expression of TFF3. Patients with high or low TFF3 expression had similar prognosis (P=0.6045). CRC, colorectal cancer; TFF3, trefoil factor 3; IHC, immunohistochemistry.
Discussion
Initiation and progression of CRC involves several molecular alterations. Thus, finding new molecular targets remains an essential milestone toward understanding the pathobiology and progression of human cancers. Meta-analysis using available web-tools to find molecular targets have helped cancer researchers in achieving these milestones. By using UALCAN, a user-friendly web-tool for analyzing cancer omics data from TCGA (RNA expression by sequencing) and CPTAC (protein expression), the current study established that TFF3 expression was significantly downregulated in CRCs. Our study also validated, by IHC assays, low phenotypic expression levels of TFF3 in CRCs as compared to normal tissue; the expression was independent of pathologic tumor stage and histologic type and patient race, age, or sex. In addition, the present study showed, by bioinformatics analysis, that low levels of TFF3 RNA expression were strongly associated with tumors exhibiting p53 mutations. Collectively, our data showed that TFF3 is downregulated in CRCs and indicate that loss of TFF3 could be involved in the pathogenesis and progression of the disease.
TFF proteins maintain GI mucosal homeostasis in response to mechanical and/or chemical mucosal injury (22–26). Through anti-apoptotic effects, TFFs protect epithelia from death and promote cell migration (6,9,27,28). TFFs are involved in the development and progression of various types of cancer (8–10,12–14,16), including CRC (17–19). The role of TFF3 in the pathogenesis of CRC, however, remains unclear. Previously studied in different malignancies, a discrepancy was observed regarding protein and RNA levels of TFF3. Interestingly, similar to our findings, two earlier studies in CRC reported reduced TFF3 expression at RNA and protein levels in tumors relative to normal epithelium (11,19). Specifically, Huang et al (19) found higher mRNA of TFF3 in metastatic lesions of CRCs than those CRCs confined to the colorectum, which is in agreement with our data that mRNA levels of TFF3 were increased in advanced CRC stage relative to low stage tumors. Additionally, Huang et al demonstrated decreased mRNA levels of TFF3 organ-confined CRCs relative to normal tissues. However, this association was not statistically significant (19). Moreover, our data obtained from publicly available RNA (TCGA data) and protein expression (proteomics data) of TFF3 (obtained by using UALCAN) agrees with our findings. Our data using UALCAN as a tool to analyze RNA (TCGA data) and protein expression (proteomics), agrees with the reports showing organ-specific tumor expression of TFF3. Moreover, our analysis reproduces the expression patterns for TFF3 and CRC revealed by previous studies. Although different from some previous findings, we are confident about the relevance of our results since our validation strategy using the UALCAN expression platform, queried for the CPTAC Confirmatory/Discovery dataset, revealed lower TFF3 expression in tumors relative to normal epithelium, similar to what we found in our tissue sample cohort. Collectively, our data suggest that protein expression of TFF3, a modulator of cell differentiation and glandular structure, is low in CRCs relative to normal mucosa.
In CRCs, p53 signaling is frequently dysregulated (29,30). Moreover, tumors with p53 mutations are resistant to various therapies. The current bioinformatics analysis showed that CRC tumors exhibiting p53 mutations had low expression of TFF3 but that p53 wild-type tumors did not, indicating an inverse relationship of p53-mutated CRCs with low TFF3 expression. Additionally, UALCAN correlation analysis showed an inverse correlation of TFF3 expression with TP53 regulating kinase (TP53RK), suggesting a link between TFF3 and TP53. Indeed, a recent study of retinoblastoma cells showed that TFF3 overexpression activates TP53 and that TP53 is a downstream target of TFF3 signaling (31). Moreover, the heat map data showed reduced transcripts of TFF3 in TP53-mutated CRCs, suggest a functional interactome linking the signaling pathways which relate the expression of these two molecules. Thus, future research is warranted focusing on establishing the molecular basis of the functional interaction between TFF3 and TP53 in CRC and would provide tools to molecularly categorize histologic types (e.g., mucinous tumors), known to have high expression of TFF3. Such multi-molecular classifiers could add prognostic value to that offered by traditional single-molecule classifiers.
Concerning the association between TFF3 expression and clinicopathological variables, we did not find a correlation between tumor expression of TFF3 and survival, even upon stratification by clinicopathological variables. Since we did not have a separate validation cohort for our IHC study, we performed survival analyses based on mRNA of TFF3. We utilized two data mining portals: UALCAN (TCGA dataset) and prognostics database ProGeneV2 (32). For ProGenV2 analysis we used datasets: GSE28814, GSE17536, GSE17537, GSE12945, GSE16125, GSE24551, GSE28722, GSE30378, GSE41258, GSE29621, GSE38832, and GSE39582. In none of the studied datasets, statistically significant association between expression of TFF3 and survival was found (data not shown). These results demonstrate concordance between the UALCAN and ProGeneV2 platforms, and suggest lack of prognostic value of TFF3 transcripts to anticipate survival in CRC. Future study in a large CRC tissue cohort, with extended follow-up, may provide the prognostic value of TFF3 in CRC. Our findings add to the complexity related to the prognostic value of TFF3 in CRC. For example, Yusup et al found poor overall survival of CRC patients with high TFF3 expression relative to those with low TFF3 expression (17). Similarly, there was lower survival of patients with CRCs positive for TFF3 in comparison to those with CRCs negative for TFF3 (33). In addition, in the cited studies, higher TFF3 expression was found in cancer tissue relative to corresponding normal mucosa. We, on the contrary, found lower expression of TFF3 in CRCs relative to normal mucosa. Despite the differences and the need of further investigation, we and others have support to propose that, in tumors with low or lack of expression of TFF3, epithelial restitution and wound healing effects mediated by this protein, as seen in normal mucosa, would not be prevalent. Under these conditions, low TFF3 in the tumor may not help to maintain epithelial integrity, as occurs in non-tumor tissue (34). Appropriate exploration of the molecules related with CRC tumorigenesis or survival will help to test this hypothesis. Although our findings differ with most of those in the literature (17,33) and suggest that tumor expression of TFF3 is not a prognostic factor for poor survival of patients with CRC, this matter is an issue for debate.
The limitations of our study include its retrospective nature as well as its small sample size. Furthermore, the results require validation in larger, independent cohorts. It would be valuable to determine the prognostic value of serum TFF3, previously determined to be associated with poor tumor differentiation and clinical TNM stage (18).
In summary, our findings provide evidence that TFF3, a factor involved in cell migration and integrity of the GI mucosa, is downregulated in CRCs relative to normal epithelium and that it may be involved in disease progression. We believe that relevance of our findings is high due to similar patterns of low TFF3 expression in tumors identified in large, publicly available omics databases. However, the prognostic value of TFF3 in CRCs is an issue needing further research.
Supplementary Material
Acknowledgements
We are grateful to Elizabeth Tarsi, Tara Craft, Eldrin Bhanat, Jaswinder Kaur, and Joy King for establishing and maintaining the databases, and to Jesus Monico and Lisa Sullivan for establishing preparatory TMAs. We also thank Richard Summers, Srinivasan Vijayakumar, and Roy Duhe (UMMC) for their continued support. Anne Dautenhahn Martin and Jennifer Reneker at UMMC, and Donald Hill at the University of Alabama at Birmingham edited the manuscript.
Funding
This study was supported in part by the Office of Research and Sponsored Programs, University of Mississippi Medical Center (CRG and IE); the University of Mississippi School of Medicine Medical Student Research Program (LF); Coordination for the Improvement of the Higher Education Personnel (CAPES) foundation, scholarship #13603-13-2 (MJS), and the impact funds from School of Medicine and the Department of Pathology, University of Alabama at Birmingham (UM).
Availability of data and materials
The data supporting the findings reported in this study are available from the corresponding author upon reasonable request.
Authors' contributions
IE, SA, AR, VS, CS, MS, LF, TP, XZ, WSO, SB, and SAD contributed to the conceptualization of the research concept, performing the experiments, formal analyses, and writing the original draft. CL, UM, and CRG contributed to the conceptualization of the research concept, study design, supervision, reviewing and editing the original draft, and funding acquisition. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent for publication
The UMMC Institutional Review Board approved the study and all authors have given consent for publication. Written informed consent was obtained from all of the patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 2.Thim L. A new family of growth factor-like peptides. ‘Trefoil’ disulphide loop structures as a common feature in breast cancer associated peptide (pS2), pancreatic spasmolytic polypeptide (PSP), and frog skin peptides (spasmolysins) FEBS Lett. 1989;250:85–90. doi: 10.1016/0014-5793(89)80690-8. [DOI] [PubMed] [Google Scholar]
- 3.Madsen J, Nielsen O, Tornøe I, Thim L, Holmskov U. Tissue localization of human trefoil factors 1, 2, and 3. J Histochem Cytochem. 2007;55:505–513. doi: 10.1369/jhc.6A7100.2007. [DOI] [PubMed] [Google Scholar]
- 4.Podolsky DK, Lynch-Devaney K, Stow JL, Oates P, Murgue B, DeBeaumont M, Sands BE, Mahida YR. Identification of human intestinal trefoil factor. Goblet cell-specific expression of a peptide targeted for apical secretion. J Biol Chem. 1993;268:6694–6702. [PubMed] [Google Scholar]
- 5.Xiao P, Ling H, Lan G, Liu J, Hu H, Yang R. Trefoil factors: Gastrointestinal-Specific proteins associated with gastric cancer. Clin Chim Acta. 2015;450:127–134. doi: 10.1016/j.cca.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 6.Taupin D, Podolsky DK. Trefoil factors: Initiators of mucosal healing. Nat Rev Mol Cell Biol. 2003;4:721–732. doi: 10.1038/nrm1249. [DOI] [PubMed] [Google Scholar]
- 7.Hernandez C, Santamatilde E, McCreath KJ, Cervera AM, Diez I, Ortiz-Masia D, Martínez N, Calatayud S, Esplugues JV, Barrachina MD. Induction of trefoil factor (TFF)1, TFF2 and TFF3 by hypoxia is mediated by hypoxia inducible factor-1: Implications for gastric mucosal healing. Br J Pharmacol. 2009;156:262–272. doi: 10.1111/j.1476-5381.2008.00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samson MH. Quantitative measurements of trefoil factor family peptides: Possibilities and pitfalls. Scand J Clin Lab Invest. 2013;73:193–202. doi: 10.3109/00365513.2013.765962. [DOI] [PubMed] [Google Scholar]
- 9.Aihara E, Engevik KA, Montrose MH. Trefoil factor peptides and gastrointestinal function. Annu Rev Physiol. 2017;79:357–380. doi: 10.1146/annurev-physiol-021115-105447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong WM, Poulsom R, Wright NA. Trefoil peptides. Gut. 1999;44:890–895. doi: 10.1136/gut.44.6.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Babyatsky M, Lin J, Yio X, Chen A, Zhang JY, Zheng Y, Twyman C, Bao X, Schwartz M, Thung S, et al. Trefoil factor-3 expression in human colon cancer liver metastasis. Clin Exp Metastasis. 2009;26:143–151. doi: 10.1007/s10585-008-9224-9. [DOI] [PubMed] [Google Scholar]
- 12.Garraway IP, Seligson D, Said J, Horvath S, Reiter RE. Trefoil factor 3 is overexpressed in human prostate cancer. Prostate. 2004;61:209–214. doi: 10.1002/pros.20096. [DOI] [PubMed] [Google Scholar]
- 13.Kannan N, Kang J, Kong X, Tang J, Perry JK, Mohankumar KM, Miller LD, Liu ET, Mertani HC, Zhu T, et al. Trefoil factor 3 is oncogenic and mediates anti-estrogen resistance in human mammary carcinoma. Neoplasia. 2010;12:1041–1053. doi: 10.1593/neo.10916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirikoshi H, Katoh M. Expression of TFF1, TFF2 and TFF3 in gastric cancer. Int J Oncol. 2002;21:655–659. [PubMed] [Google Scholar]
- 15.Kim WG, Kim JY, Park DY. Simple classifiers for molecular subtypes of colorectal cancer. Arab J Gastroenterol. 2017;18:191–200. doi: 10.1016/j.ajg.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 16.Jahan R, Shah A, Kisling SG, Macha MA, Thayer S, Batra SK, Kaur S. Odyssey of trefoil factors in cancer: Diagnostic and therapeutic implications. Biochim Biophys Acta Rev Cancer. 2020;1873:188362. doi: 10.1016/j.bbcan.2020.188362. [DOI] [PubMed] [Google Scholar]
- 17.Yusup A, Huji B, Fang C, Wang F, Dadihan T, Wang HJ, Upur H. Expression of trefoil factors and TWIST1 in colorectal cancer and their correlation with metastatic potential and prognosis. World J Gastroenterol. 2017;23:110–120. doi: 10.3748/wjg.v23.i1.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Q, Wang K, Su C, Fang J. Serum trefoil factor 3 as a protein biomarker for the diagnosis of colorectal cancer. Technol Cancer Res Treat. 2017;16:440–445. doi: 10.1177/1533034616674323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang YG, Li YF, Wang LP, Zhang Y. Aberrant expression of trefoil factor 3 is associated with colorectal carcinoma metastasis. J Cancer Res Ther. 2013;9:376–380. doi: 10.4103/0973-1482.119308. [DOI] [PubMed] [Google Scholar]
- 20.Chandrashekar DS, Bashel B, Balasubramanya SA, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, Varambally S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen F, Chandrashekar DS, Varambally S, Creighton CJ. Pan-Cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers. Nat Commun. 2019;10:5679. doi: 10.1038/s41467-019-13528-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mashimo H, Wu DC, Podolsky DK, Fishman MC. Impaired defense of intestinal mucosa in mice lacking intestinal trefoil factor. Science. 1996;274:262–265. doi: 10.1126/science.274.5285.262. [DOI] [PubMed] [Google Scholar]
- 23.Farrell JJ, Taupin D, Koh TJ, Chen D, Zhao CM, Podolsky DK, Wang TC. TFF2/SP-deficient mice show decreased gastric proliferation, increased acid secretion, and increased susceptibility to NSAID injury. J Clin Invest. 2002;1091:193–204. doi: 10.1172/JCI0212529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carrasco R, Pera M, May FE, Westley BR, Martinez A, Morales L. Trefoil factor family peptide 3 prevents the development and promotes healing of ischemia-reperfusion injury in weanling rats. J Pediatr Surg. 2004;39:1693–1700. doi: 10.1016/j.jpedsurg.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 25.Serwin NM, Wisniewska M, Jesionowska A, Skwirczyńska E, Marcinowska Z, Dołęgowska B. Serum levels of 12 renal function and injury markers in patients with glomerulonephritis. Pol Arch Med Wewn. 2016;126:483–493. doi: 10.20452/pamw.3477. [DOI] [PubMed] [Google Scholar]
- 26.Schulze U, Hampel U, Sel S, Goecke TW, Thäle V, Garreis F, Paulsen F. Fresh and cryopreserved amniotic membrane secrete the trefoil factor family peptide 3 that is well known to promote wound healing. Histochem Cell Biol. 2012;138:243–250. doi: 10.1007/s00418-012-0943-2. [DOI] [PubMed] [Google Scholar]
- 27.Taupin DR, Kinoshita K, Podolsky DK. Intestinal trefoil factor confers colonic epithelial resistance to apoptosis. Proc Natl Acad Sci USA. 2000;97:799–804. doi: 10.1073/pnas.97.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinoshita K, Taupin DR, Itoh H, Podolsky DK. Distinct pathways of cell migration and antiapoptotic response to epithelial injury: Structure-function analysis of human intestinal trefoil factor. Mol Cell Biol. 2000;20:4680–4690. doi: 10.1128/MCB.20.13.4680-4690.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li XL, Zhou J, Chen ZR, Chng WJ. P53 mutations in colorectal cancer-molecular pathogenesis and pharmacological reactivation. World J Gastroenterol. 2015;21:84–93. doi: 10.3748/wjg.v21.i1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manne U, Myers RB, Moron C, Poczatek RB, Dillard S, Weiss H, Brown D, Srivastava S, Grizzle WE. Prognostic significance of bcl-2 expression and p53 nuclear accumulation in colorectal adenocarcinoma. Int J Cancer. 1997;74:346–358. doi: 10.1002/(SICI)1097-0215(19970620)74:3<346::AID-IJC19>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 31.Busch M, Klein S, Große-Kreul J, Scheiner O, Metz K, Stephan H, Dünker N. P53, miR-34a and EMP1-newly identified targets of TFF3 signaling in Y79 retinoblastoma cells. Int J Mol Sci. 2019;20:4129. doi: 10.3390/ijms20174129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goswami CP, Nakshatri H. PROGgeneV2: Enhancements on the existing database. BMC Cancer. 2014;14:970. doi: 10.1186/1471-2407-14-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morito K, Nakamura J, Kitajima Y, Kai K, Tanaka T, Kubo H, Miyake S, Noshiro H. The value of trefoil factor 3 expression in predicting the longterm outcome and early recurrence of colorectal cancer. Int J Oncol. 2015;46:563–568. doi: 10.3892/ijo.2014.2755. [DOI] [PubMed] [Google Scholar]
- 34.Scholven J, Taras D, Sharbati S, Schön J, Gabler C, Huber O, zum Büschenfelde DM, Blin N, Einspanier R. Intestinal expression of TFF and related genes during postnatal development in a piglet probiotic trial. Cell Physiol Biochem. 2009;23:143–156. doi: 10.1159/000204103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings reported in this study are available from the corresponding author upon reasonable request.