Abstract
Esophageal squamous cell carcinoma (ESCC) is a highly malignant and deadly tumor. Radiation therapy is one of the primary treatments for locally advanced ESCC. However, the biomarkers for prognosis of definitive radiation remain undefined. Peripheral blood circulating tumor (ct)DNA provides information of tumor genetic alterations and has been confirmed as a potential non-invasive biomarker for several types of cancer. The present study investigated the clinical implications of ctDNA detection in patients with ESCC and receiving definitive radiation therapy. Patients with locally advanced ESCC were retrospectively recruited. Plasma samples were collected before, during and following radiation therapy. Next-generation sequencing was performed to identify somatic mutations in 180 genes. A total of 69 baseline and post-radiation plasma samples were collected from 25 patients. A total of 59 non-silent single nucleotide variants were present in 33 genes. All pre-radiation and 58.3% (14/24) of post-radiation samples had at least one mutation. Patients with lymph node metastases (LNM) exhibited a higher number of pre-radiation mutations compared with those without LNM. The variables, progression-free survival (PFS) and overall survival (OS) of the patients with one baseline mutation were not significantly different compared with that in patients with more than one baseline mutation. Patients with initial ctDNA-positive post-radiation samples exhibited significantly reduced PFS (P=0.047) and OS (P=0.005) compared with that in patients with ctDNA-negative samples. The post-radiation plasma ctDNA status was an independent prognostic factor from univariate and multivariate analyses. Dynamic monitoring of ctDNA during follow-up was examined. The results indicated that ctDNA was a predictive and prognostic marker in patients with ESCC and receiving definitive radiation therapy, which may guide subsequent treatment.
Keywords: biomarker, circulating tumor DNA, esophageal squamous cell carcinoma, prognosis, radiation therapy
Introduction
Esophageal cancer is one of the most prevalent malignancies in the world and accounted for >500,000 deaths in 2018 (1). Approximately half of all esophageal cancer cases occur in China (2). Squamous cell carcinoma represents ~90% of these cases (3). Currently, there are limited treatment options for esophageal squamous cell carcinoma (ESCC), resulting in a five-year survival rate of 17.9% according to the Surveillance, Epidemiology, and End Results database (4). Definitive radiation therapy or chemoradiotherapy (CRT) are treatment options for patients that are not able to undergo surgery (5). However, patients with locally advanced ESCC demonstrate poor outcomes following radiation therapy, with a median survival of 16–20 months worldwide (6–8). It is unclear whether additional treatment results in reduced or lack of recurrence and metastases following radiation. Therefore, identification of clinically viable biomarkers that predict survival following radiation therapy is urgently required. Previous studies have focused on clinical and pathological factors, such as sex, age, type, TNM stage, treatment, molecular markers and the general condition of the patient (9–11). However, to the best of our knowledge, widely accepted markers that predict outcomes in patients with ESCC following radiation remain unidentified.
Circulating tumor (ct)DNA is a tumor-derived DNA found in the plasma of patients with cancer, that could be used as a potential biomarker (12). This non-invasive approach (13) could be used to monitor tumor burden during treatment, regardless of tumor heterogeneity (14–17). Using a cancer-personalized profiling strategy, ctDNA was detected in 100% of patients with stages II–IV and 50% of patients with stage I non-small cell lung cancer (18). In a prospective study of 230 patients with early-stage colorectal cancer, ctDNA was assessed at the first follow-up visit following surgical resection. The results indicated that recurrence-free survival at 3 years was 0% for the ctDNA-positive group and 90% for the ctDNA-negative group (19). Previous studies have reported an interval of 7.9–11.0 months between ctDNA detection and clinical relapse (14,16,20). In addition, ctDNA was detected in the plasma of patients with ESCC and was used for dynamic monitoring (longitudinally monitored during treatment at different time point) (21).
In the present study, a unique gene panel was used to evaluate consecutively enrolled patients with ESCC. Plasma samples were sequenced and analyzed prior to and following radiation therapy using next-generation sequencing (NGS) techniques. In addition, ctDNA was monitored using a non-invasive approach to predict disease outcome following radiation therapy and identify specific mutations.
Materials and methods
Patient selection and sample collection
Eligible patients with ESCC were retrospectively recruited at the Department of Gastrointestinal Oncology, The Fifth Medical Centre, Chinese People's Liberation Army General Hospital (Beijing, China). The main inclusion criteria were pathologically diagnosed ESCC and Eastern Cooperative Oncology Group (ECOG) performance status ≤2. Only patients who were found to have unresectable ESCC, as evaluated by a thoracic surgeon and those who were candidates for definitive radiation therapy or chemoradiation therapy were included. Patients had received enhanced computed tomography (CT) scan of the chest and abdomen, and gastroscopy and endoscopic ultrasonography when suitable, for the evaluation of primary tumor. The patients were evaluated using the American Joint Committee on Cancer 7th staging system (22). The patients with ESCC who presented with other malignant tumors and/or other severe or uncontrolled diseases were excluded. A total of 29 patients were initially recruited, and 69 plasma samples were collected from 25 patients with ESCC, who were enrolled between July 2013 and May 2016 (Table SI). All patients provided signed informed consent and the study protocol was approved by PLA 307 Hospital Institutional Review Board. Clinical features were obtained from the patients' medical records. Peripheral blood samples were collected prior to (pre-radiation or baseline), during, and following (post-radiation) radiation therapy, as well as during follow-up visits every three months for ctDNA, CT scan and tumor marker (CEA, CA19-9, and CA72-4) analyses. CEA, CA19-9 and CA72-4 were routinely tested by chemiluminescence immunoassay in our hospital and the upper limits of normal were 4.3 ng/ml, 27 and 6.9 U/ml, respectively. Patients were divided according to baseline ctDNA status (1 vs. >1), abnormal and normal biomarker levels and according to the positive and negative ctDNA status post-radiation. Plasma was separated by centrifugation at 1,600 × g for 10 min, transferred to new microcentrifuge tubes, and centrifuged at 16,000 × g for 10 min to remove cell debris at room temperature. Peripheral blood lymphocytes (PBLs) from the first centrifugation were used for the extraction of germline genomic DNA.
DNA extraction and quality control
Cell-free (cf)DNA was isolated from 0.6–1.8 ml plasma using a QIAamp Circulating Nucleic Acid kit (Qiagen, Inc.). Baseline PBL DNA was extracted using the QIAamp DNA Blood Mini kit (Qiagen, Inc.) (23). cfDNA and PBL DNA concentration levels were measured using a Qubit 3.0 fluorometer and a Qubit dsDNA High Sensitivity Assay kit (both from Thermo Fisher Scientific Inc.). The fragments of cfDNA were analyzed on an Agilent 2100 BioAnalyzer using the Agilent High Sensitivity DNA kit (both from Agilent Technologies, Inc.) and only samples with a fragment peak at ~170 bp passed quality control. Qualified PBL DNA was identified using agarose gel electrophoresis.
Target capture and NGS
A panel, including 180 genes was designed. Briefly, the panel consisted of top 100 recurrent potential driver genes in ESCC identified from the COSMIC database (24) (http://cancer.sanger.ac.uk/cosmic), top 100 oncogenes and tumor suppressor genes (TSG), associated with tumorigenesis and metastasis in ESCC (25–27), and top 100 genes associated with other types of cancer were identified from The Cancer Genome Atlas database (https://cancergenome.nih.gov) (28).
Sequencing libraries of both plasma cfDNA and baseline PBL DNA were constructed using the KAPA LTP DNA Library Preparation kit#KK8230 (Kapa Biosystems), as previously described (23). For each sample, at least 30 ng cfDNA and 1.0 µg PBL DNA were used for library input. The library length was measured using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc). The libraries were analyzed by using an Applied Biosystems 7500 Real-Time PCR system (Thermo Fisher Scientific Inc.) and 2 n moles of the loading concentration of the final library was used. Additional 15 pre-capture PCR cycles were performed on samples close to the minimum input requirement to generate sufficient PCR product for hybridization. The libraries were hybridized to custom-designed biotinylated oligonucleotide probes (Kapa Biosystems) covering ~0.5 Mbp. DNA sequencing was performed on the HiSeq3000 Sequencing System (Illumina, Inc.) with 2×100-bp paired-end reads using TruSeq PE Cluster Generation Kit v3 and the TruSeq SBS Kit v3 (Illumina, Inc.).
Sequencing data analysis
Terminal adaptor sequences and low-quality reads were removed from the raw sequencing data. The reads were then aligned with the human genome build GRCh37 using a Burrows-Wheeler aligner (29). Picard tools (http://broadinstitute.github.io/picard/) were used to mark PCR duplicates. Single nucleotide variants (SNVs), and insertions and deletions were identified using MuTect (version 1.1.4) (30) and Genome Analysis Toolkit (version 3.4-46-gbc02625) softwares (31), respectively. Sequencing results of cfDNA samples were used to identify somatic mutations, while PBL DNA was used to filter the germline mutations. All candidate somatic mutations identified using the bioinformatics pipeline were manually reviewed in the Integrative Genomics Viewer interactive tool (32) by assessing the quality of bases the mapping quality of reads and the overall read depth at each mutation site. A mutation was identified as somatic if: i) a variant allele fraction (VAF) ≥0.1%; and ii) at least 5 high-quality reads (Phred score, ≥30; mapping quality, ≥30, without paired-end reads bias) were found. For a specific variant in the plasma cfDNA, VAF was calculated using the following formula: VAF=sequencing read count of alternate alleles/(sequencing read count of reference alleles + sequencing read count of alternate alleles) ×100%. The mutations were annotated to genes using the ANNOVAR software (version 77) (33) to identify the mutated protein-coding position, and filter intronic and silent changes.
Statistical analysis
Overall survival (OS) was defined as the survival time from the initiation of radiation therapy to the date of death from any cause. Progression-free survival (PFS) was defined as the survival time from the initiation of radiation therapy to the date of disease progression or death from any cause. Cases without progression or death events were censored at the date of last follow-up. Survival curves were determined using the Kaplan-Meier method and univariate analysis of these calculations were performed using the log-rank test. Multivariate analysis was performed with manual backward stepwise Cox regression modeling. The number of mutations was compared using the Mann-Whitney U test. The Fisher's exact test was used to compare proportions between the two groups. All reported P-values were two-sided and P<0.05 was considered to indicate a statistically significant difference. All statistical analyses were performed using the SPSS version 21.0 (IBM Corp.) and PFS and OS were shown by the GraphPad Prism version 5.0 (GraphPad Software Inc.) softwares.
Results
Patient characteristics
All patients received definitive esophagus radiation therapy or CRT. The plasma samples were obtained prior to radiation for all patients and following radiation for 24 patients. The clinical characteristics of the patients are shown in Table I. The median age at diagnosis was 60 years (range, 40–82 years), and males accounted for 88% of the patients. A total of 52% of the primary tumors were located in the upper third of the thoracic esophagus, while 72% of patients were diagnosed with clinical stage T3 or T4 tumors and with regional LN involvement. A total of 10 patients (40%) received CRT. The mean duration of follow-up was 18.0±2.4 months, with a median of 14.7 months. During this period, 14 patients were documented with disease progression, including local recurrence and/or distant metastases.
Table I.
Clinical characteristics of 25 patients with ESCC before radiation.
| Clinical characteristics | Value | Percentage |
|---|---|---|
| Sex | ||
| Male | 22 | 88.0 |
| Female | 3 | 12.0 |
| Median age, years (range) | 60 (40–82) | |
| ECOG performance status | ||
| 0 | 3 | 12.0 |
| 1 | 19 | 76.0 |
| 2 | 3 | 12.0 |
| Location | ||
| Upper thoracic | 13 | 52.0 |
| Mid-thoracic | 10 | 40.0 |
| Lower thoracic | 2 | 8.0 |
| Differentiation | ||
| Well | 1 | 4.0 |
| Moderate | 15 | 60.0 |
| Poor | 8 | 32.0 |
| Unknown | 1 | 4.0 |
| T stagea | ||
| T1 | 1 | 4.0 |
| T3 | 17 | 68.0 |
| T4 | 1 | 4.0 |
| Unknown | 6 | 24.0 |
| N stage | ||
| Positive | 18 | 72.0 |
| Negative | 7 | 28.0 |
| Chemotherapy before radiation | ||
| Yes | 14 | 56.0 |
| No | 11 | 44.0 |
| Chemoradiotherapy | ||
| Yes | 10 | 40.0 |
| No | 15 | 60.0 |
| Tumor biomarkersb | ||
| Normal | 14 | 56.0 |
| Abnormal | 11 | 44.0 |
T staging was based on endoscopic ultrasonography.
Tumor markers included CEA, CA19-9 and CA72-4. A total of 14 patients exhibited disease progression during follow-up with local recurrence or distant metastases. ESCC, esophageal squamous cell carcinoma; ECOG, Eastern Cooperative Oncology Group.
Identification of mutations in the patients with ESCC
NGS was performed on baseline and post-radiation plasma samples from patients with ESCC. Following quality control and removal of duplicates, a mean coverage depth of ×1,337 (range, 379–2,222) was achieved. Somatic mutations were recorded, with 59 non-silent SNVs identified in 33 genes (Table SII).
The baseline mutation spectrum was analyzed. At least one somatic mutation was detected per sample. The average mutation burden at baseline was 1.9 (range, 1–6). A total of 13 patients had one mutation, while 12 patients harbored more than one mutation. The most frequent mutation was noted in TP53, which was mutated in 9 (36%) pre-treatment cancer cases. All of the TP53 variations were either missense or non-sense and were annotated in the COSMIC (Table SII) database. Other frequently mutated genes included NOTCH1 (n=4), CCND1 (n=3), CNKN2A (n=3) and NOTCH2 (n=3). Notably, the majority of the mutated genes with a high prevalence rate were TSGs.
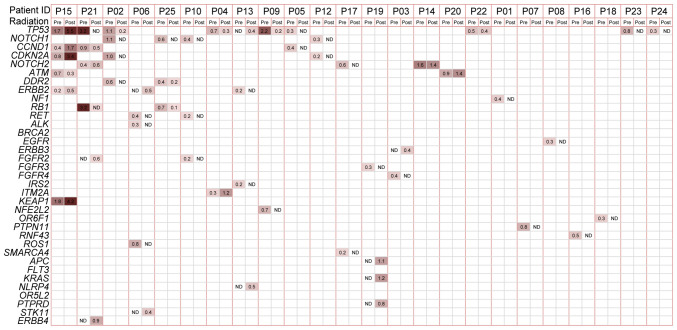
The mutation spectrum in the initial post-radiation plasma samples was compared with that in the baseline samples (Fig. 1). Plasma mutations were altered by radiation therapy. Baseline and matched post-radiation samples were available for 24 patients, with the exception of one patient (P11), for whom a post-treatment plasma sample was not obtained. The mutation burden was lower in the first post-radiation plasma sample, with an average of 1.1 (range, 0–6) compared with that in the baseline sample. A total of 15 patients (62.5%) exhibited a reduced mutation burden following therapy. A total of 10 (41.7%) patients did not exhibit mutations following radiation (ctDNA-negative). As the most frequently mutated gene, TP53 was tracked following radiation in 5 patients and ATM, CCND1, ERBB2 and NOTCH2 were also repeatedly found. The most apparent difference was noted in the NOTCH1 gene, in which no mutations occurred in the initial post-radiation samples of all 4 patients with baseline mutations. In addition, several mutations were detected in the first post-radiation samples, whereas these were not present in the baseline samples. These mutations were the following: STK11 1062C>G (F354L) in P06 and KRAS c.38G>A (p.G13D) missense and APC c.4285C>T (p.Q1429*) truncation in P19.
Figure 1.
Mutations in plasma samples from patients with esophageal squamous cell carcinoma identified using next-generation sequencing panel testing. The mutation status of baseline and post-treatment plasma samples from 24 patients were used to create the heat map. Patient ID is shown at the top of the figure. The gene names are labeled on the left and the most recurrent baseline genes are ranked upwards. The VAF values are recorded in squares and the darker color was associated with the larger VAF. VAF, variant allele fraction; Pre, baseline plasma; post, first post-treatment plasma; ND, not detected.
Association between ctDNA and clinical features
The association between baseline ctDNA and the clinical features of the patients was examined. No association was noted between sex, ECOG performance status, location, or differentiation and baseline mutation burden (data not shown). A total of 12 out of 18 patients (67%) with lymph node metastasis (LNM) (LN-positive) exhibited >1 mutation at baseline compared with that in patients who were LN-negative and only had one mutation (Fig. S1A). In addition, 46.2% (6/13) of patients with one mutation and 100% of patients with more than one mutation were LN-positive (Fisher's exact test, patients with one mutation vs. those with more than one mutation; P=0.005).
Furthermore, the status of ctDNA in the first post-radiation plasma samples was further examined. These samples were collected within 2 months of radiation for 19 patients, within 2–6 months following radiation for 3 patients and within 6 months following radiation for 2 patients. The patients received no further treatment between the end of radiation therapy and the first plasma collection following radiation. A total of 12 out of 17 (71%) LN-positive patients and 29% (2/7) of LN-negative patients harbored mutations in the first post-radiation plasma sample, and more mutations were detected in patients with LNM (Fig. S1B). In the 14 ctDNA-positive (at least one mutation detected) patients, 90.9% (10/11) had documented disease recurrence, while three patients exhibited no recurrence status and did not survive following 10 months of disease progression. In the 10 ctDNA-negative patients, 50% (4/8) had documented disease recurrence while two patients did not exhibit recurrence status (data not shown).
Predictive and prognostic values
To determine the prognostic value of ctDNA, the association of the baseline mutations with patient outcome was examined. No significant differences were noted for PFS (P=0.221) or OS (P=0.579) times (Fig. S2A and B) between patients harboring one and more than one mutation. The specific baseline mutation of P53 was also evaluated and it was found not to be a prognostic marker for PFS (P=0.055) and OS (P=0.147; data not shown).
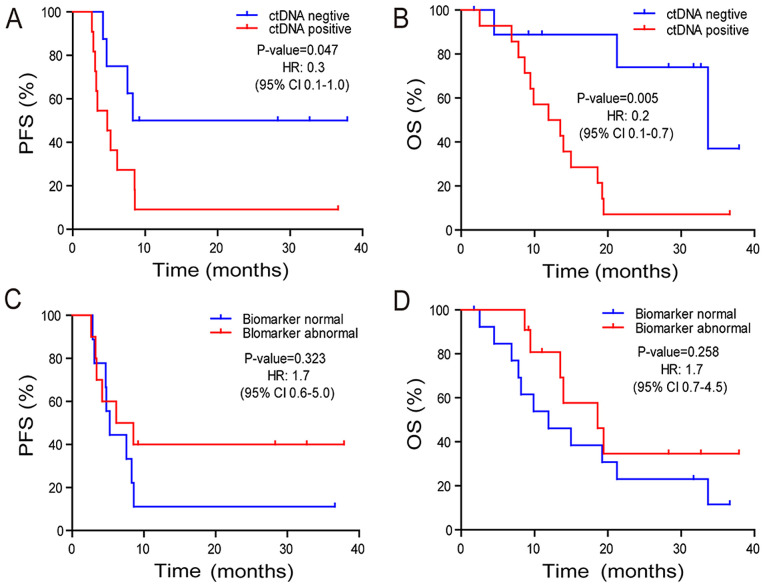
Subsequently, the association between post-radiation ctDNA status and PFS and OS times was investigated. Patients who were ctDNA-positive exhibited a marginally significant reduction in PFS (P=0.047) time and a significantly decreased OS (P=0.005) time compared with those in patients who were ctDNA-negative (Fig. 2A and B). The median OS time was 33.7 and 11.9 months for the ctDNA-negative and -positive groups, respectively. The two-year survival rate was 74.1 and 7.1% in the ctDNA-negative and -positive groups (odds ratio, 24.8; 95% CI, 1.7–1,625.0; P=0.0055) (Fig. 2B). Abnormal tumor biomarkers CEA, CA19-9, and CA72-4 did not exhibit a significant association with PFS (P=0.323) or OS (P=0.258) times (Fig. 2C and D) compared with normal tumor biomarkers.
Figure 2.
PFS and OS times of patients stratified by post-radiation ctDNA or tumor biomarker status. Kaplan-Meier plots of (A) PFS and (B) OS times in patients with ESCC and treated with radiation therapy stratified by ctDNA status in the initial post-radiation plasma samples. Kaplan-Meier plots of (C) PFS and (D) OS times in patients with ESCC and treated with radiation therapy stratified by clinical biomarker status in the initial post-radiation plasma samples. PFS, progression free survival; OS, overall survival; ct, circulating tumor; ESCC, esophageal squamous cell carcinoma; HR, hazard ratio.
In addition to the aforementioned data, the histopathological variables were integrated with the ctDNA status in the initial post-radiation plasma samples and a multivariate analysis was conducted. According to the univariate analysis, only ctDNA status was significantly associated with OS time (Table II). Multivariate Cox proportional hazard regression analysis revealed that the presence of ctDNA in the initial post-radiation plasma sample was an independent prognostic factor for patients with ESCC (P=0.011; Table III).
Table II.
Univariate analysis of clinical characteristics affecting OS time.
| Clinical characteristics | Number (n=25) | OS (95% CI) | P-value |
|---|---|---|---|
| Sex | 0.373 | ||
| Male | 22 | 14.97 (7.75–22.19) | |
| Female | 3 | 9.87 | |
| ECOG performance status | 0.174 | ||
| 0/1 | 22 | 18.62 (10.86–26.38) | |
| 2 | 3 | 9.87 (9.18–10.56) | |
| Tumor location | 0.482 | ||
| Upper 1/3 | 13 | 14.97 (3.73–26.21) | |
| Middle 1/3 | 10 | 13.52 (1.34–25.70) | |
| Lower 1/3 | 2 | 9.44 | |
| Differentiation | 0.221 | ||
| Well | |||
| Moderate | 15 | 11.91 (5.22–18.60) | |
| Poor | 8 | 21.11 (6.01–35.17) | |
| Unknown | 1 | ||
| N stage | 0.131 | ||
| Positive | 18 | 12.93 (7.63–18.22) | |
| Negative | 7 | 23.07 (7.96–36.20) | |
| Tumor biomarkers | 0.258 | ||
| Normal | 14 | 11.91 (3.91–19.91) | |
| Abnormal | 11 | 18.62 (8.50–28.74) | |
| Chemotherapy before radiation | 0.350 | ||
| Yes | 14 | 14.97 (6.27–23.67) | |
| No | 11 | 13.95 (0.94–26.96) | |
| Chemoradiotherapy | 0.552 | ||
| Yes | 10 | 18.62 (10.80–26.44) | |
| No | 15 | 13.52 (4.82–22.22) | |
| ctDNA status in first post-treatment plasma | 0.005 | ||
| Negative | 10 | 33.68 (15.66–51.70) | |
| Positive | 14 | 11.91 (5.22–18.60) |
OS, overall survival; ECOG, Eastern Cooperative Oncology Group; ct, circulating tumor.
Table III.
Multivariate analysis of clinical characteristics affecting OS.
| Clinical characteristics | HR (95% CI) | P-value |
|---|---|---|
| ECOG (0/1 vs. 2) | 0.691 (0.176–2.710) | 0.596 |
| N stage (negative vs. positive) | 0.466 (0.110–1.983) | 0.302 |
| ctDNA status in first post-radiation plasma (negative vs. positive) | 0.183 (0.049–0.678) | 0.011 |
OS, overall survival; HR, hazard ratio; ECOG, Eastern Cooperative Oncology Group; ct, circulating tumor.
ctDNA dynamic monitoring
ctDNA may also be used as a longitudinal monitoring biomarker during and following treatment to evaluate disease progression (Fig. S3). Mutations in both P1 and P10 were undetectable following radiation therapy, which was consistent with the clinical findings, demonstrating that both patients were disease-free during follow-up. The frequency of the NOTCH2 mutation in P14 was decreased following radiation; however, it was still detectable and was increased afterwards, which was consistent with multiple LNMs detected by CT scan, at that time point. Elevated VAF values were detected in several driver genes (TP53, KEAP1 and CDKN2A) in P15 following radiation; however, disease progression was detected from imaging analysis 4 months following radiation.
Discussion
Optimal treatment for the upper esophagus or locally advanced disease remains problematic for patients with ESCC (34). Esophagectomy can cause serious postoperative complications, such as anastomotic leakage and pneumonia (35,36). Clinical trial results, that compared chemoradiation followed by surgery with chemoradiation alone, indicated similar median survival rates and 2-year survival rates, of no more than 40% for both groups (7). These data suggested that the majority of the patients who received surgery remained vulnerable to local recurrence or distant metastasis. Thus, how to select the right patient for surgery is essential. The establishment of a multi-disciplinary and more effective treatment modality for patients with ESCC would improve disease outcomes. The development of a viable marker that could predict outcome following radiation therapy would facilitate more informed treatment decisions.
ctDNA is a highly sensitive biomarker, that has been used in multiple types of cancer, including melanoma (37), breast cancer (38) and colorectal cancer (39). ctDNA reflects tumor burden and dynamic alterations, that can be monitored non-invasively during treatment (40). The presence of ctDNA following curative therapy indicated minimal residual disease and predicted recurrence (19,20). In ESCC, Luo et al (21) designed a panel of 90 genes, which accounted for at least one mutation in 94% of patients and provided promising results for ctDNA detection. However, the prognostic value of ctDNA was not evaluated in ESCC. Therefore, to the best of our knowledge, the present study was the first to report ctDNA detection in a cohort of patients with locally advanced ESCC. The results demonstrated that the mutations found in the plasma ctDNA, following radiation therapy, were the most efficacious independent prognostic factors for patients with ESCC. Both LNM and baseline mutation number were not prognostic factors in the present study. Therefore, the outcome from radiation was affected by the ctDNA biomarker following radiation and was not associated with the pre-radiation disease.
A marginally significant difference in PFS was observed for patients who were ctDNA-positive and -negative, following radiation. Furthermore, the high recurrence rate of patients who were ctDNA-positive indicated a positive predictive value of ctDNA, suggesting that they should be monitored carefully. In contrast to the patients who were ctDNA-positive, 50% of the patients who were ctDNA-negative exhibited disease recurrence, which indicated a considerably lower negative predictive value (NPV) of ctDNA. However, 3 out of 4 patients with ctDNA-negative status exhibited a relatively long OS time (>20 months). Therefore, ctDNA negativity was not a strong predictor of prolonged PFS time; however, it may be an indicator of longer survival time. Based on a higher recurrence rate and worse OS time, patients who are ctDNA-positive may require additional treatments following radiation, including chemotherapy or surgery. In addition, it could be hypothesized that immunotherapy performed in non-small cell lung cancer (41,42) could also be applied in ESCC (43) in the future. Furthermore, due to the poor NPV, the ctDNA assays require further optimization by addition of other genes, notably those associated with radiation resistance, such as OBSCN and DGKK (44).
Patients with ESCC, in the present study, indicated heterogeneous mutant gene profiles. The most prevalent mutated genes encoded tumor suppressor proteins. The absence of hot-spot mutations increases the suitability of using NGS and a large gene panel for the identification of molecular markers. A notable difference was noted in the number of mutations in the post-radiation samples was the disappearance of NOTCH1 mutations, which were only detected in baseline samples. This result suggested that cancer cell populations harboring NOTCH1 mutations may be more sensitive to radiation therapy, as previously described (44). Future large-scale studies should aim to investigate the efficacy of radiation in patients harboring NOTCH1 mutations and notably clonal NOTCH1. The presence of new mutations after radiation, such as STK11, which have been previously associated with radioresistance (45), warrants future evaluation, with respect to their association with disease progression.
The VAF values of the mutations, in this study, were low, reflecting the low plasma concentration of ctDNA. This may be due to the relatively early stage of tumors, that were used in the present study, which included locally advanced cancers lacking metastases at baseline. Plasma ctDNA concentration has been previously associated with tumor size (15) and stage (12). The analysis of tumor tissue volume, measured using CT, and plasma ctDNA VAF indicated that tumor size was associated with mean plasma VAF of clonal SNVs (15). In a previous study, the quantification of tumor mutations in each patient, using digital droplet PCR revealed that patients with stage I disease rarely exhibited >10 copies per 5 ml plasma. By contrast, patients with advanced prostate, ovarian, or colorectal cancer exhibited a median concentration of 100–1,000 copies per 5 ml plasma (12). In addition, the amount of ctDNA released from different tissue-derived tumors into the blood was distinct. For example, a higher number of ctDNA copies was detected per 5 ml plasma in patients with advanced colorectal cancer compared with that in patients with advanced gastroesophageal cancer (12). We found that the general VAF may be relatively low in ESCC compared with that in colorectal and ovarian cancers (46). Nevertheless, the use of integrated digital error suppression technology, based on a molecular barcode, ensures that ctDNA mutations can be detected at VAFs as low as 0.1%, such as in lung cancer (47).
High heterogeneity among patients with ESCC has been reported in several large-scale genomic studies (25,26,48,49). Several hundreds of somatic mutations exist; however, no single mutation has been found to be widespread among all patients. The limitations of the present study can be summarized as follows: Firstly, the small sample size and the small number of mutations detected prevented analysis of the significance of individual genes (such as TP53), including prognostic value, presence of local recurrence or metastases and predictive value of chemotherapy or radiation therapy. In addition, the small number of baseline mutations and LNM led to poor statistical power with respect to the detection of significant differences in disease prognosis. Secondly, since the present study was retrospective, several inconsistencies may lead to potentially substantial confounding recurrence outcomes, such as treatment heterogeneity (including the concurrent therapy with radiation and different chemotherapy regimens) and the heterogeneity of time points at which the ctDNA was acquired following therapy. These deviations could be corrected in a future prospective study including a larger cohort of patients.
In patients with locally advanced ESCC, ctDNA was detectable prior to and following radiation therapy. The ctDNA status in the first post-radiation plasma samples could be used as an independent prognostic factor for patients with locally advanced ESCC.
Supplementary Material
Acknowledgements
The abstract of the study has been presented in the 2018 54th American Society of Clinical Oncology annual meeting in Chicago, United States of America, June 1st to June 5th and published as an abstract no. e16053 in Journal of Clinical Oncology 2018 36:15_suppl, e16053.
Funding
The present study was supported by the Application of Clinical Features of Capital City (grant no. Z141107002514105).
Availability of data and materials
The details of the mutations in each patient are presented in Table SII. The other datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Author's contributions
JMX, RJ and CHZ conceived the project and designed experiments. CHZ, RRL, YZ and HEC contributed to the acquisition of the data. RJ, CHZ and PSL contributed to the raw data collection and organization. PSL, LPC, YHG, YFG and XY performed the experiments and analyzed the data. RJ, PSL and JMX wrote the manuscript with assistance and feedback of all the other co-authors. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the PLA 307 Hospital Institutional Review Board. All patients provided signed informed consent.
Patient consent for publication
All patients whose data were included in the current study provided informed consent for publication.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Feng RM, Zong YN, Cao SM, Xu RH. Current cancer situation in China: Good or bad news from the 2018 global cancer statistics? Cancer Commun (Lond) 2019;39:22. doi: 10.1186/s40880-019-0368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Worni M, Martin J, Gloor B, Pietrobon R, D'Amico TA, Akushevich I, Berry MF. Does surgery improve outcomes for esophageal squamous cell carcinoma? An analysis using the surveillance epidemiology and end results registry from 1998 to 2008. J Am Coll Surg. 2012;215:643–651. doi: 10.1016/j.jamcollsurg.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y, Ye J, Zhu Z, Zhao W, Zhou J, Wu C, Tang H, Fan M, Li L, Lin Q, et al. Comparing paclitaxel plus fluorouracil versus cisplatin plus fluorouracil in chemoradiotherapy for locally advanced esophageal squamous cell cancer: A randomized, multicenter, phase III clinical trial. J Clin Oncol. 2019;37:1695–1703. doi: 10.1016/S0959-8049(01)00182-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rawat S, Kumar G, Kakria A, Sharma MK, Chauhan D. Chemoradiotherapy in the management of locally advanced squamous cell carcinoma esophagus: Is surgical resection required? J Gastrointest Cancer. 2013;44:277–284. doi: 10.1007/s12029-013-9477-7. [DOI] [PubMed] [Google Scholar]
- 7.Bedenne L, Michel P, Bouche O, Milan C, Mariette C, Conroy T, Pezet D, Roullet B, Seitz JF, Herr JP, et al. Chemoradiation followed by surgery compared with chemoradiation alone in squamous cancer of the esophagus: FFCD 9102. J Clin Oncol. 2007;25:1160–1168. doi: 10.1200/JCO.2005.04.7118. [DOI] [PubMed] [Google Scholar]
- 8.Conroy T, Galais MP, Raoul JL, Bouché O, Gourgou-Bourgade S, Douillard JY, Etienne PL, Boige V, Martel-Lafay I, Michel P, et al. Definitive chemoradiotherapy with FOLFOX versus fluorouracil and cisplatin in patients with oesophageal cancer (PRODIGE5/ACCORD17): Final results of a randomised, phase 2/3 trial. Lancet Oncol. 2014;15:305–314. doi: 10.1016/S1470-2045(14)70028-2. [DOI] [PubMed] [Google Scholar]
- 9.Chen MF, Yang YH, Lai CH, Chen PC, Chen WC. Outcome of patients with esophageal cancer: A nationwide analysis. Ann Surg Oncol. 2013;20:3023–3030. doi: 10.1245/s10434-013-2935-4. [DOI] [PubMed] [Google Scholar]
- 10.Chen MF, Chen PT, Lu MS, Lee CP, Chen WC. Survival benefit of surgery to patients with esophageal squamous cell carcinoma. Sci Rep. 2017;7:46139. doi: 10.1038/srep46139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stahl M, Lehmann N, Walz MK, Stuschke M, Wilke H. Prediction of prognosis after trimodal therapy in patients with locally advanced squamous cell carcinoma of the oesophagus. Eur J Cancer. 2012;48:2977–2982. doi: 10.1016/j.ejca.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 12.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murtaza M, Dawson SJ, Pogrebniak K, Rueda OM, Provenzano E, Grant J, Chin SF, Tsui DWY, Marass F, Gale D, et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun. 2015;6:8760. doi: 10.1038/ncomms9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olsson E, Winter C, George A, Chen Y, Howlin J, Tang MH, Dahlgren M, Schulz R, Grabau D, van Westen D, et al. Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease. EMBO Mol Med. 2015;7:1034–1047. doi: 10.15252/emmm.201404913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, Le Quesne J, Moore DA, Veeriah S, Rosenthal R, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017;545:446–451. doi: 10.1038/nature22364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reinert T, Scholer LV, Thomsen R, Tobiasen H, Vang S, Nordentoft I, Lamy P, Kannerup AS, Mortensen FV, Stribolt K, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut. 2016;65:625–634. doi: 10.1136/gutjnl-2014-308859. [DOI] [PubMed] [Google Scholar]
- 17.Pectasides E, Stachler MD, Derks S, Liu Y, Maron S, Islam M, Alpert L, Kwak H, Kindler H, Polite B, et al. Genomic heterogeneity as a barrier to precision medicine in gastroesophageal adenocarcinoma. Cancer Discov. 2018;8:37–48. doi: 10.1158/2159-8290.CD-17-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, Liu CL, Neal JW, Wakelee HA, Merritt RE, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, Silliman N, Tacey M, Wong HL, Christie M, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8:346ra92. doi: 10.1126/scitranslmed.aaf6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, Cheang M, Osin P, Nerurkar A, Kozarewa I, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. 2015;7:302ra133. doi: 10.1126/scitranslmed.aab0021. [DOI] [PubMed] [Google Scholar]
- 21.Luo H, Li H, Hu Z, Wu H, Liu C, Li Y, Zhang X, Lin P, Hou Q, Ding G, et al. Noninvasive diagnosis and monitoring of mutations by deep sequencing of circulating tumor DNA in esophageal squamous cell carcinoma. Biochem Biophys Res Commun. 2016;471:596–602. doi: 10.1016/j.bbrc.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 22.Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A, editors. (7th edition) Springer; New York, NY: 2010. AJCC Cancer Staging Manual; pp. 103–115. [Google Scholar]
- 23.Yang X, Chu Y, Zhang R, Han Y, Zhang L, Fu Y, Li D, Peng R, Li D, Ding J, et al. Technical validation of a next-generation sequencing assay for detecting clinically relevant levels of breast cancer-related single-nucleotide variants and copy number variants using simulated Cell-Free DNA. J Mol Diagn. 2017;19:525–536. doi: 10.1016/j.jmoldx.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 24.Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, et al. COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song Y, Li L, Ou Y, Gao Z, Li E, Li X, Zhang W, Wang J, Xu L, Zhou Y, et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509:91–95. doi: 10.1038/nature13176. [DOI] [PubMed] [Google Scholar]
- 26.Gao YB, Chen ZL, Li JG, Hu XD, Shi XJ, Sun ZM, Zhang F, Zhao ZR, Li ZT, Liu ZY, et al. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet. 2014;46:1097–1102. doi: 10.1038/ng.3076. [DOI] [PubMed] [Google Scholar]
- 27.Qin HD, Liao XY, Chen YB, Huang SY, Xue WQ, Li FF, Ge XS, Liu DQ, Cai Q, Long J, et al. Genomic characterization of esophageal squamous cell carcinoma reveals critical genes underlying tumorigenesis and poor prognosis. Am J Hum Genet. 2016;98:709–727. doi: 10.1016/j.ajhg.2016.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cancer Genome Atlas Research Network. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh D, Kim JH. The current evidence on neoadjuvant therapy for locally advanced esophageal squamous cell carcinoma. Korean J Thorac Cardiovasc Surg. 2020;53:160–167. doi: 10.5090/kjtcs.2020.53.4.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu PC, Posner MC. The role of surgery in the management of oesophageal cancer. Lancet Oncol. 2003;4:481–488. doi: 10.1016/S1470-2045(03)01167-7. [DOI] [PubMed] [Google Scholar]
- 36.Stahl M, Stuschke M, Lehmann N, Meyer HJ, Walz MK, Seeber S, Klump B, Budach W, Teichmann R, Schmitt M, et al. Chemoradiation with and without surgery in patients with locally advanced squamous cell carcinoma of the esophagus. J Clin Oncol. 2005;23:2310–2317. doi: 10.1200/JCO.2005.00.034. [DOI] [PubMed] [Google Scholar]
- 37.Tsao SC, Weiss J, Hudson C, Christophi C, Cebon J, Behren A, Dobrovic A. Monitoring response to therapy in melanoma by quantifying circulating tumour DNA with droplet digital PCR for BRAF and NRAS mutations. Sci Rep. 2015;5:11198. doi: 10.1038/srep11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 39.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Zhao C, Chang L, Jia R, Liu R, Zhang Y, Gao X, Li J, Chen R, Xia X, et al. Circulating tumor DNA analyses predict progressive disease and indicate trastuzumab-resistant mechanism in advanced gastric cancer. EBioMedicine. 2019;43:261–269. doi: 10.1016/j.ebiom.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Antonia SJ, Villegas A, Daniel D, Vicente D, Murakami S, Hui R, Kurata T, Chiappori A, Lee KH, de Wit M, et al. Overall survival with durvalumab after chemoradiotherapy in stage III NSCLC. N Engl J Med. 2018;379:2342–2350. doi: 10.1056/NEJMoa1809697. [DOI] [PubMed] [Google Scholar]
- 42.Antonia SJ, Villegas A, Daniel D, Vicente D, Murakami S, Hui R, Yokoi T, Chiappori A, Lee KH, de Wit M, et al. Durvalumab after chemoradiotherapy in stage III non-small-cell lung cancer. N Engl J Med. 2017;377:1919–1929. doi: 10.1056/NEJMoa1709937. [DOI] [PubMed] [Google Scholar]
- 43.Kelly RJ, Lockhart AC, Jonker DJ, Melichar B, Andre T, Chau I, Clarke SJ, Cleary JM, Doki Y, Franke FA, et al. CheckMate 577: A randomized, double-blind, phase 3 study of nivolumab (Nivo) or placebo in patients (Pts) with resected lower esophageal (E) or gastroesophageal junction (GEJ) cancer. J Clin Oncol. 2017;35(4 Suppl):TPS212. doi: 10.1200/JCO.2017.35.15_suppl.TPS4131. [DOI] [Google Scholar]
- 44.Yang L, Zhang X, MacKay M, Foox J, Hou Q, Zheng X, Zhou R, Huang M, Jing Z, Mason CE, Wu S. Identification of radioresponsive genes in esophageal cancer from longitudinal and single cell exome sequencing. Int J Radiat Oncol Biol Phys. 2020;108:1103–1114. doi: 10.1016/j.ijrobp.2020.06.015. [DOI] [PubMed] [Google Scholar]
- 45.He Q, Li J, Dong F, Cai C, Zou X. LKB1 promotes radioresistance in esophageal cancer cells exposed to radiation, by suppression of apoptosis and activation of autophagy via the AMPK pathway. Mol Med Rep. 2017;16:2205–2210. doi: 10.3892/mmr.2017.6852. [DOI] [PubMed] [Google Scholar]
- 46.Phallen J, Sausen M, Adleff V, Leal A, Hruban C, White J, Anagnostou V, Fiksel J, Cristiano S, Papp E, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. 2017;9:eaan2415. doi: 10.1126/scitranslmed.aan2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, Stehr H, Liu CL, Bratman SV, Say C, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol. 2016;34:547–555. doi: 10.1038/nbt.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin DC, Hao JJ, Nagata Y, Xu L, Shang L, Meng X, Sato Y, Okuno Y, Varela AM, Ding LW, et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat Genet. 2014;46:467–473. doi: 10.1038/ng.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hao JJ, Lin DC, Dinh HQ, Mayakonda A, Jiang YY, Chang C, Jiang Y, Lu CC, Shi ZZ, Xu X, et al. Spatial intratumoral heterogeneity and temporal clonal evolution in esophageal squamous cell carcinoma. Nat Genet. 2016;48:1500–1507. doi: 10.1038/ng.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The details of the mutations in each patient are presented in Table SII. The other datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.