Abstract
The long noncoding RNA cancer susceptibility candidate 9 (CASC9) has been revealed to be an oncogenic gene in several types of cancer, and high CASC9 expression is related to tumorigenesis and cancer progression. However, the role of CASC9 in bladder cancer (BC), particularly during epithelial-mesenchymal transition (EMT), has not been characterized. RT-qPCR, EdU, CCK-8, wound scratch, Transwell and flow cytometric assays were performed to detect CASC9 expression, miR-758-3p expression and their functions in BC. RNA FISH was used to detect CASC9 subcellular localization. Luciferase reporter assay, RT-qPCR assay and western blotting were used to explore the relationship of CASC9, miR-758-3p and TGF-β2. In the present study, it was revealed that CASC9 regulated EMT in BC. CASC9 expression was significantly upregulated in BC cell lines and specimens compared to that in adjacent normal bladder tissues. Upregulated CASC9 was associated with increased invasion ability and poor prognosis of BC. CASC9 knockdown inhibited BC cell proliferation, migration and invasion. Furthermore, a bioinformatics study and luciferase reporter assays revealed that CASC9 functioned as a ceRNA for miR-758-3p. CASC9 inhibited microRNA (miR)-758-3p activity and resulted in the de-suppression of its target transforming growth factor (TGF)-β2. TGF-β signaling driven by TGF-β2 was crucial for CASC9 to promote EMT in BC. Collectively, these results indicated that CASC9 sponged miR-758-3p to regulate the expression of TGF-β2, which activated the TGF-β signaling pathway and promoted proliferation and EMT in BC.
Keywords: long noncoding RNA, lncRNA cancer susceptibility candidate 9, bladder cancer, microRNA-758-3p, transforming growth factor-β2, epithelial-mesenchymal transition
Introduction
Bladder cancer (BC) is a cancer arising from the urinary bladder. BC is one of the most common malignant cancers worldwide with high morbidity and mortality, representing a huge economic burden. As of 2018, BC affected approximately 1.6 million people globally with 550,000 new cases and 200,000 deaths (1). More than 50% of patients relapse within 6–12 years after initial diagnosis (2–4). The increased incidence of BC and poor outcomes underscore attempts to understand the underlying pathological mechanisms of BC progression. Emerging evidence has revealed that epithelial-mesenchymal transition (EMT) is an important process in the development of BC, and EMT-related molecules may become new targets for treatment of chemoresistance (5–7). EMT is a process in which cells lose their epithelial features and acquire mesenchymal characteristics, and where cells with mesenchymal characteristics can migrate more efficiently and invade other tissues. In cancer, EMT is associated with tumor occurrence, metastasis, tumor stemness and resistance to treatment (8,9).
EMT can be induced by several cell signaling transduction pathways. The transforming growth factor (TGF)-β pathway has been revealed to induce EMT in several cancer types (6,8,10–13). The TGF-β ligand binds to TGF-β receptors, resulting in phosphorylation of SMAD2 and SMAD3 (14). Activated SMAD2-SMAD3 forms complexes with SMAD4, and these complexes translocate to the nucleus to regulate the expression of TGF-β target genes, including a large number of genes involved in EMT, invasion, motility and proliferation (9,11,12,14). Potential cancer therapies targeting TGF-β signaling pathways have been investigated, and several promising therapies are being tested in clinical trials (6,14).
Long noncoding RNAs (lncRNAs) are noncoding RNAs longer than 200 nucleotides (nts) (15–17). lncRNAs can act as scaffolds or competing endogenous RNAs (ceRNAs) by interacting with microRNAs (miRNAs or miRs), circRNAs and proteins (18–21). In addition, lncRNAs recruit chromatin remodeling and modification complexes to guide epigenetic regulations (21,22). Several lncRNAs have been reported to serve as oncogenes in BC, including H19, MALAT1, TUG1, UCA1, and HOTAIR (23–27), highlighting the potential for lncRNAs to serve as biomarkers and therapeutic targets in BC.
Cancer susceptibility candidate 9 (CASC9) is located on human chromosome 8q21.11 (28). It was originally identified in esophageal squamous cell carcinoma (ESCC) and is predicted to be a novel putative oncogene (28). Subsequently, CASC9 expression has been reported to be aberrantly upregulated in numerous human malignancies, including esophageal cancer (28–30), pancreatic ductal adenocarcinoma (31), gastric cancer (32), nasopharyngeal carcinogenesis (33), and non-small cell lung cancer (34). The upregulation of CASC9 in human cancer indicates its potential tumorigenic properties. In ESCC, CASC9 has been revealed to facilitate cell growth by negatively regulating PDCD4 and promote metastasis through upregulating LAMC2 expression (29,30). CASC9 has been demonstrated to interact with HIF1α and enhance the stabilization of HIF1α in nasopharyngeal carcinoma (33). However, the role of CASC9 in BC has not been characterized, especially during EMT.
In this study, we aimed to elucidate the expression of CASC9 in BC tissues and cell lines, its association with the depth of bladder tumor invasion and prognosis, and to determine the role of CASC9 in the development and progression of BC.
Materials and methods
Sample collection
In total, 49 pairs of BC tissues and corresponding adjacent normal bladder tissues were collected from Peking University Shenzhen Hospital (Shenzhen, China) from January 2010 to November 2011. Tissue specimens were collected from 49 patients (aged 30 to 80 years old) with BC who underwent cystectomy; 40 male patients and 9 female patients. All patients were diagnosed as transitional cell carcinoma clinically and pathologically. The exclusion criteria included patients with other tumors, patients with a history of other cancer treatments, or patients with bladder cancer who had received chemotherapy or radiation therapy before surgery. All human tissue samples were obtained with informed consent. The Ethics Committee of Peking University Shenzhen Hospital in China approved this study (approval no. 20090017).
Cell lines and cell cultures
All cell lines were obtained from the American Type Culture Collection and maintained using standard media and conditions. Human BC cells (T24, TCCSUP, UM-UC-3, J82 and 5637), human normal bladder epithelial cells (SV-HUC-1) and 293T cells were maintained in Roswell Park Memorial Institute (RPMI)-1640 medium, Dulbecco's modified Eagle's medium (DMEM) or F-12K supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (PS) (all from Gibco; Thermo Fisher Scientific Inc.). All cells were cultured at 37°C in a 5% CO2 incubator.
Cell transfection
Cells were transfected with 100 nM small interfering (si)RNA or mimics or inhibitors using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) for 24–48 h at 37°C. Then, the transfected cells were analyzed. All siRNAs, mimics and inhibitors were synthesized by Suzhou GenePharma Co., Ltd. The sequences of siRNAs were as follows: Negative control (NC), 5′-UUCUCCGAACGUGUCACGUTT-3′; siCASC9-1, 5′-CAACUGGAUUCCAACUUUAUU-3′; siCASC9-2, 5′-CAAGAAGUUUAGUAAACCAUU-3′; siCASC9-3, 5′-GAGAUCAUUAAGCCCAGAAUU-3′; mimics NC, 5′-UUGUACUACACAAAAGUACUG-3′; inhibitor NC, 5′-CAGUACUUUUGUGUAGUACAA-3′; mimics miR-758-3p, 5′-UUUGUGACCUGGUCCACUAACC-3′; inhibitor miR-758-3p, 5′-GGUUAGUGGACCAGGUCACAAA-3′.
RNA extraction, cDNA synthesis and reverse transcription-quantitative (RT-q)PCR
Total RNA was extracted from cells or tissue specimens using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The cDNA was synthesized with random primers using PrimeScript RT reagent Kit (Takara Biotechnology, Co., Ltd.) or miScript II RT kit (Qiagen GmbH) according to the manufacturer's instructions. Quantitative RT-PCR was performed on a Roche Lightcycler 480 (Roche Diagnostics) using SYBR Premix Ex Taq kit (Takara Biotechnology, Co., Ltd.) according to the manufacturer's instructions. Quantitative RT-PCR amplification was performed according to the following thermocycling conditions: 30 sec at 95°C for initial denaturation; 40 cycles of 5 sec at 95°C for denaturation and 31 sec at 60°C for extension; and 10 min at 60°C for final extension. Human EMT RT2 Profiler PCR Array (Qiagen GmbH) was used to analyze the expression of genes involved in EMT. The relative expression levels of candidate genes were analyzed using the 2−ΔΔCq method (35). The primers for AHNAK, CTNNB1, EGFR, FN1, ITGAV, PDGFRB and SNAI3 were purchased from Qiagen, Inc. The primer sequences were as follows: AHNAK forward, 5′-CAGGCATTGGTGTTCAAGGC-3′ and reverse, 5′-TCTGCCCAGTTGGGAGTTTC-3′; CTNNB1 forward, 5′-TTGTGCGGCGCCATTTTAAG-3′ and reverse, 5′-TCCTCAGACCTTCCTCCGTC-3′; EGFR forward, 5′-AAGGCACGAGTAACAAGC-3′ and reverse, 5′-AGGGCAATGAGGACATAA-3′; FN1 forward, 5′-TGTGCCAAAGCTTTACTACTGT-3′ and reverse, 5′-TATTTCCCCCGAAGGTGTCT-3′; ITGAV forward, 5′-TCACTAAGCGGGATCTTGCC-3′ and reverse, 5′-AAGCACTGAGCAACTCCACA-3′; PDGFRB forward, 5′-GCTGTTACCCACTCTGGGAC-3′ and reverse, 5′-TGGTGTCCTTGCTGCTGATG-3′; SNAI3 forward, 5′-GCACAACTACCTCTCAGCCA-3′ and reverse, 5′-ATAGACGTGTGACATGGGGC-3′; CASC9 forward, 5′-CCAGACAGCAGCAAAGCAAT-3′ and reverse, 5′-GGAAGCAGCAAATGTGTCCAT-3′; TGF-β2 forward, 5′-CGACGAAGAGTACTACGCCA-3′ and reverse, 5′-GATGGCATTTTCGGAGGGGA-3′; GAPDH forward, 5′-CGCTCTCTGCTCCTCCTGTTC-3′ and reverse, 5′-ATCCGTTGACTCCGACCTTCAC-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-ACGCTTCACGAATTTGCGT-3′; miR-758-3p forward, 5′-ACACTCCAGCTGGGTTTGTGACCTGGTCCA-3′ and reverse, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGGTTAGTG-3′.
Cell Counting Kit 8 (CCK-8) and 5-ethynyl-2′-deoxyuridine (EdU) assays
Cell Counting Kit-8 (CCK-8; US Everbright, Inc.) and EdU assay kit (Guangzhou RiboBio Co., Ltd.) were used to assess cell proliferation. Experiments were performed as previously described (36).
Wound healing, Transwell and flow cytometric assays
Cell migration was determined using wound healing assays. Transwell assays without or with Matrigel were used to assess BC cell migration and invasion abilities, respectively. Wound healing, Transwell and flow cytometric assays were performed as previously described (36). Serum-free medium was used in the wound healing experiment.
Dual luciferase report assay
The CASC9 (or TGF-β2) fragment containing the predicted miR-758-3p binding site or a fragment with a mutated binding site were cloned into the psiCHECK-2 luciferase reporter vector (Wuhan GeneCreate Biological Engineering Co., Ltd.). Then, 1 µg/ml luciferase reporter vector psiCHECK-2-CASC9-WT (wild type) or psiCHECK-2-CASC9-MT (mutant type) or psiCHECK-2-TGF-β2-WT or psiCHECK-2-TGF-β2-MT and 100 nM miRNA mimic or mimic NC were co-transfected into 293T cells using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) for 24 h at 37°C. Renilla and firefly luciferase activities were detected using the Dual luciferase reporter assay system (Promega Corporation). miRNA mimic and mimic NC sequences were as follows: mimics NC, 5′-UUGUACUACACAAAAGUACUG-3′; and mimics miR-758-3p, 5′-UUUGUGACCUGGUCCACUAACC-3′.
Antibodies and western blotting
Anti-E-cadherin (product code ab15148), anti-N-cadherin (product code ab18203) and anti-GAPDH (product code ab9485) were purchased from Abcam. Goat anti-mouse IgG-HRP (cat. no. sc-2005) and goat anti-rabbit IgG-HRP (cat. no. sc-2004) were purchased from Santa Cruz Biotechnologies. Western blotting was performed as previously described (36).
RNA fluorescence in situ hybridization (FISH)
FISH assay was performed using the Ribo™ FISH Kit (Guangzhou RiboBio Co., Ltd.) according to the manufacturer's instructions. The lncRNA CASC9 probe was designed and synthesized by Guangzhou RiboBio Co., Ltd. and was labeled with Cy3 fluorescent dye. BC cells were seeded onto sterile coverslips until cells reached 30–60% confluence. The cells were washed with PBS, fixed with 4% paraformaldehyde for 10 min at 25°C, and then permeabilized with 0.5% Triton X-100 (PBS) for 10 min at 4°C. Next, the cells were blocked with prehybridization buffer for 30 min at 37°C and then incubated in 0.5 µM lncRNA CASC9 probe in hybridization buffer at 37°C overnight. The cells were then washed with saline sodium citrate (SSC) buffer solution and stained with DAPI for 10 min at 25°C. Finally, the cell slides were removed from the plate and fixed on a glass slide for detection by fluorescence microscopy (magnification, ×400).
Statistical analysis
The data were presented as the mean ± standard error of mean (SEM). Log-rank test, chi-square test, one-way ANOVA with Bonferroni post hoc test, paired and unpaired Student's t-test were employed for statistical analysis. Kaplan-Meier survival analysis from http://gepia.cancer-pku.cn (37) was used to reveal that the relationship between CASC9 expression and the prognosis of bladder cancer patients. Survival analysis was performed using log-rank test. Chi-square test was used to assess the association between CASC9 expression and clinicopathological characteristics of bladder cancer patients. When comparing the population means of only two groups, the Student's t-test was used, and when means of more than two groups were compared, ANOVA was selected. Paired Student's t-test was used to assess CASC9 expression in 49 pairs of BC tissues (Tumor) and matched adjacent normal tissues (Normal). The association of CASC9 expression and BC tumor invasion depth was calculated by one-way ANOVA with Bonferroni post hoc test. P<0.05 was considered to indicate a statistically significant difference. All statistical analyses were carried out with Graphpad Prism 6 (GraphPad Software, Inc.).
Data sets
Bioinformatics tools (LncBase v2 and miRDB) were used to predict potential target miRNAs of CASC9 (38,39). Computational algorithms (TargetScan 7.1 and miRDB) were used to search for potential miR-758-3p target genes (39,40).
Results
Upregulation of CASC9 in BC tissues is significantly associated with BC tumor invasion depth and poor prognosis
To investigate the role of CASC9 in BC, the expression levels of CASC9 in 49 BC tissues and adjacent normal bladder tissues from patients were first analyzed by RT-qPCR. Compared with paired adjacent normal tissues, 79.6% (39/49) of human BC tissues had upregulated CASC9 expression (Fig. 1A and B; P<0.001). In addition, the expression level of CASC9 in T3/T4 patients was higher than that in T1 patients, which may indicate that CASC9 is related to cell invasion ability (Fig. 1C; P<0.05).
Figure 1.
CASC9 is upregulated in BC tissues and cell lines. (A and B) RT-qPCR analysis of the relative expression levels of CASC9 in 49 pairs of BC tissues (Tumor) and matched adjacent normal tissues (Normal). The significance of statistical difference was calculated by paired Student's t-test. Error bars represent the SEM. (C) CASC9 upregulation was significantly associated with BC tumor invasion depth. The significance of statistical difference was calculated by one-way ANOVA with Bonferroni post hoc test. Error bars represent the SEM. (D) Survival curves of DFS. Patients were grouped into CASC9-low or CASC9-high groups based on CASC9 expression levels. Survival analysis was performed using log-rank test. *P<0.05, and ***P<0.001. CASC9, cancer susceptibility candidate 9; BC, bladder cancer; RT-qPCR, reverse transcription-quantitative PCR; SEM, standard error of mean; DFS, disease-free survival.
The correlation between CASC9 expression levels and the tumor invasion depth in BC patients was further analyzed. As revealed in Table I, CASC9 upregulation was significantly associated with BC tumor invasion depth (n=49; P<0.05) and age (n=49, P<0.05), however sex and tumor size were not associated with CASC9 expression levels. These results indicated that CASC9 may play a carcinogenic role in BC.
Table I.
Associations between CASC9 expression and clinicopathological characteristics of bladder cancer patients.
| Expression of CASC9 | ||||
|---|---|---|---|---|
| Characteristics | Total | High (n=39) | Low (n=10) | P-value |
| Sex | 0.069 | |||
| Male | 40 | 34 | 6 | |
| Female | 9 | 5 | 4 | |
| Tumor size (cm) | 0.719 | |||
| <4 | 19 | 16 | 3 | |
| ≥4 | 30 | 23 | 7 | |
| Age (years) | 0.029 | |||
| ≤60 | 22 | 14 | 8 | |
| >60 | 27 | 25 | 2 | |
| Tumor invasion depth (T) | 0.025 | |||
| Tis, Ta, T1 | 18 | 11 | 7 | |
| T2, T3 or above | 31 | 28 | 3 | |
| TNM stage | 0.247 | |||
| 0/I | 15 | 10 | 5 | |
| II/III/IV | 34 | 29 | 5 | |
TNM was according to the seventh edition of staging TNM of the Union for International Cancer Control (UICC) in 2009. Chi-square test was used to assess the association between two categorical variables. CASC9, cancer susceptibility candidate 9.
In addition, Kaplan-Meier survival analysis revealed that the disease-free survival (DFS) of patients with high CASC9 expression was significantly decreased compared with patients with low CASC9 expression (Fig. 1D; analysis from GEPIA). Collectively, the present results revealed that CASC9 was upregulated in BC, and the expression level of CASC9 could serve as a predictor of prognosis in BC patients.
CASC9 promotes BC cell proliferation
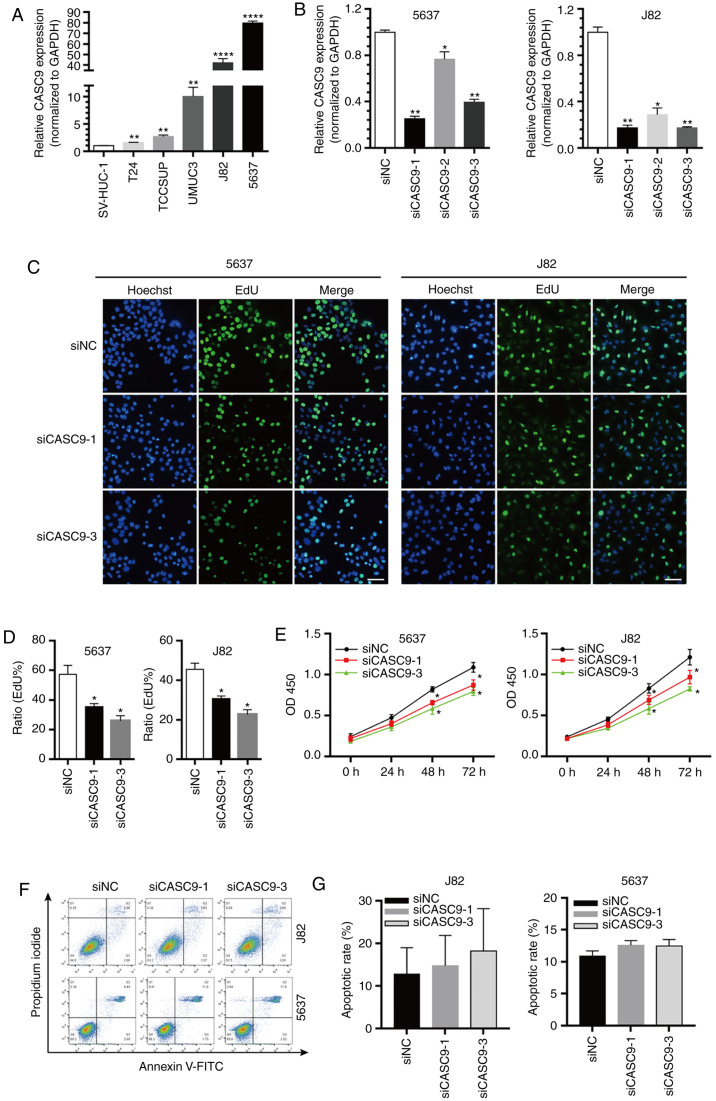
As revealed in Fig. 2A, CASC9 expression was significantly upregulated in BC cell lines (T24, TCCSUP, UM-UC-3, J82 and 5637) compared with normal human bladder epithelial cells (SV-HUC-1) (Fig. 2A). Notably, compared with SV-HUC-1 cells, the expression of CASC9 in J82 and 5637 cells was significantly increased by >40-fold (Fig. 2A). Therefore, 5637 and J82 cells were selected for further investigation.
Figure 2.
CASC9 promotes BC cell proliferation. (A) The relative expression levels of CASC9 in BC cell lines (T24, TCCSUP, UM-UC-3, J82 and 5637) compared with normal human bladder epithelial cells (SV-HUC-1). (B) The efficacy of CASC9 siRNAs. (C and D) The proliferation rate of 5637 and J82 cells transfected with 50 nM siCASC9-1, siCASC9-3 or siNC measured by EdU assay. Scale bars, 100 µm. (E) The proliferation rate of 5637 and J82 cells transfected with 50 nM siCASC9-1, siCASC9-3 or siNC measured by CKK-8 assay reported as the means ± SEM from 3 independently repeated experiments. (F and G) The apoptotic rate of siCASC9- and siNC-transfected cells (P>0.05). Unpaired Student's t-test was used and data are presented as the mean ± SEM. *P<0.05, **P<0.01 and ****P<0.0001. CASC9, cancer susceptibility candidate 9; BC, bladder cancer; siRNAs, small interfering RNAs; NC, negative control; EdU, 5-ethynyl-2′-deoxyuridine; CCK-8, Cell Counting Kit-8; SEM, standard error of mean.
The marked upregulated expression of CASC9 in BC tissues and various BC cell lines prompted us to further explore the role of CASC9 in tumorigenesis. Three siRNAs that specifically targeted CASC9 were first designed, and the knockdown efficiency of the siRNAs was quantified by RT-qPCR. siCASC9-1 and siCASC9-3 were efficient in depleting CASC9 compared with the negative control (NC treatment) or siCASC9-2 (Fig. 2B). Therefore, siCASC9-1 and siCASC9-3 were selected for further experiments. The effect of CASC9 on BC cell proliferation was further assessed using EdU and CCK-8 assays. EdU assay results revealed that CASC9 knockdown (siCASC9-1 or siCASC9-3 transfection) in 5637 and J82 cells significantly attenuated cell proliferation (Fig. 2C and D). Similar results were observed using CCK-8 assays (Fig. 2E). Collectively, the results demonstrated that CASC9 knockdown inhibited BC cell proliferation.
Flow cytometric assays were performed to assess whether CASC9 knockdown promotes BC cell apoptosis. However, no statistically significant differences in apoptotic rates were observed between 5637 cells transfected with siCASC9 and siNC (P>0.05, Fig. 2F and G). Similar results were observed in J82 cells (P>0.05; Fig. 3F and G).
Figure 3.
CASC9 promotes BC cell migration and invasion. (A) Representative images of wound healing assays of 5637 and J82 cells transfected with 50 nM siCASC9-1, siCASC9-3 or siNC. Magnification, ×200; scale bars, 100 µm. *P<0.05. (B) Representative images of Transwell assays of 5637 and J82 cells transfected with 50 nM siCASC9-1, siCASC9-3 or siNC. Magnification, ×200; scale bars, 100 µm. (C) Quantification of relative migration and invasion of 5637 and J82 cells transfected with 50 nM siCASC9-1, siCASC9-3 or siNC. Unpaired Student's t-test was used and data are presented as the mean ± SEM. *P<0.05, **P<0.01 and. CASC9, cancer susceptibility candidate 9; BC, bladder cancer; si, small interfering; NC, negative control; SEM, standard error of mean.
CASC9 promotes BC cell migration and invasion
Wound healing and Transwell assays (without Matrigel coating) were utilized to assess the effect of CASC9 on BC cell migration. In wound healing assays, the open wound area of siCASC9 (siCASC9-1 or siCASC9-3)-transfected cells was significantly increased compared with siNC-transfected cells 48 h after scratching (Fig. 3A). Transwell migration assays (without Matrigel) revealed that the number of migrated cells in siCASC9-treated groups was significantly decreased compared with siNC-treated groups (Fig. 3B and C). These results indicated that CASC9 promoted BC cell migration.
The effects of CASC9 on BC cell invasion were assessed using Transwell assays (with Matrigel). The number of invasive cells in the siCASC9 groups was reduced by ~50% in 5637 and J82 cells (Fig. 3B and C). These results indicated that CASC9 promoted BC cell invasion.
CASC9 functions as a sponge for miR-758-3p
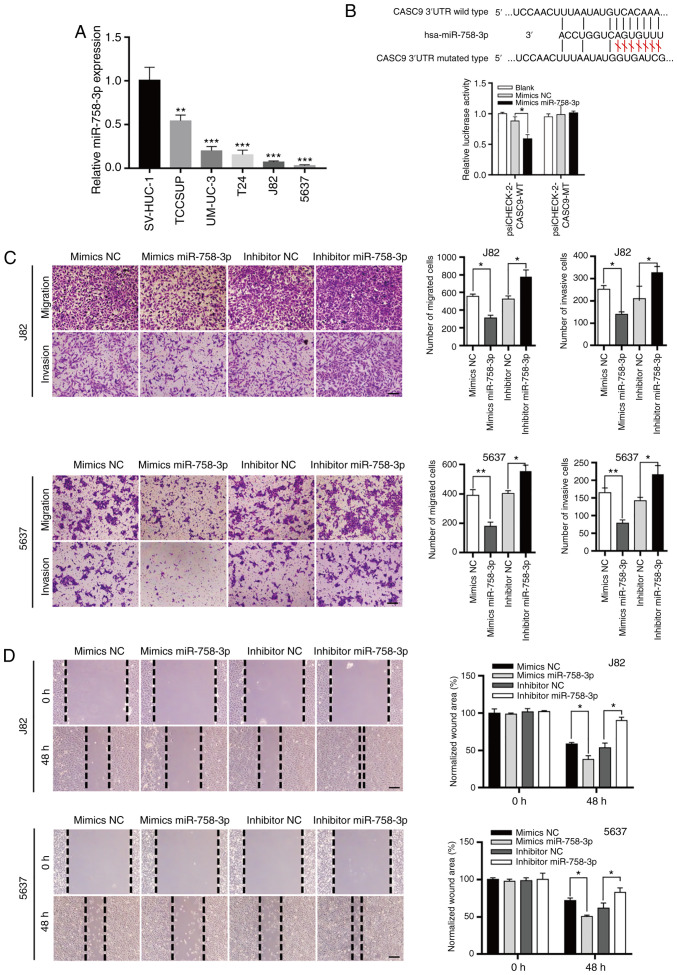
Increasing evidence indicates that lncRNAs may act as sponges for miRNAs, thereby regulating their downstream targets (19,21). Bioinformatics tools (LncBase v2 and miRDB) were used to predict potential target miRNAs of CASC9 (38,39). The predicted miRNAs were screened and the candidate to miR-758-3p was narrowed down. The results revealed that miR-758-3p expression levels were significantly decreased in BC cell lines (T24, TCCSUP, UM-UC-3, J82 and 5637) compared with normal human bladder epithelial cells (SV-HUC-1) (Fig. 4A).
Figure 4.
CASC9 functions as a sponge for miR-758-3p. (A) The relative expression levels of miR-758-3p in BC cell lines (TCCSUP, UM-UC-3, T24, J82 and 5637) compared with normal human bladder epithelial cells (SV-HUC-1). (B) CASC9 functions as a sponge for miR-758-3p. Upper panel, schematic diagrams of the mutual interactions between miR-758-3p and CASC9. Bottom panel, 293T cells were transfected with miRNA mimics in combination with luciferase reporters harboring wild-type or mutated miRNA binding sites on CASC9. The effects of miR-758-3p on luciferase activity were determined by luciferase reporter assays. The activities of firefly luciferase were normalized to Renilla luciferase. *P<0.05. (C) Transwell assay of 5637 and J82 cells transfected with mimics NC, mimics miR-758-3p, inhibitor NC or inhibitor miR-758-3p as indicated reported as the means ± SD from 3 independently repeated experiments. (D) Wound healing assays of 5637 and J82 cells transfected with NC mimics, miR-758-3p mimics, NC inhibitor or miR-758-3p inhibitor as indicated. Unpaired Student's t-test was used and data are presented as the mean ± SEM. Scale bars, 100 µm. *P<0.05, **P<0.01 and ***P<0.001. CASC9, cancer susceptibility candidate 9; miR-758-3p, microRNA-758-3p; BC, bladder cancer; NC, negative control; SEM, standard error of mean.
Subsequent luciferase reporter assays revealed that miR-758-3p overexpression reduced the luciferase activity of a luciferase reporter harboring wild-type (WT) CASC9 but not the reporter carrying mutant (MT) CASC9 (Fig. 4B). Collectively, these results indicated that CASC9 acted as a sponge for miR-758-3p.
miR-758-3p suppresses BC cell proliferation
To understand the roles of miR-758-3p in BC, wound healing and Transwell assays were used to assess the effect of miR-758-3p on BC cell migration and invasion. Consistent with the theory that CASC9 functions as an oncogene, it was revealed that miR-758-3p mimics suppressed BC cell migration and invasion, while a miR-758-3p inhibitor promoted BC cell migration and invasion (Fig. 4C and D).
The effect of miR-758-3p on cell proliferation was further evaluated using EdU assays. As revealed in Fig. 5A-C, the proliferation of BC cells transfected with miR-758-3p mimics was significantly inhibited compared to the NC group (P<0.05), while BC cell proliferation in the miR-758-3p inhibitor group was increased compared with the NC group (P<0.05). The rate of BC cell apoptosis was also quantified using flow cytometric assays. However, no statistical significance was observed between any groups of BC cells (P>0.05; Fig. 5E).
Figure 5.
miR-758-3p inhibits BC cell proliferation. (A-C) The proliferation rate of 5637 and J82 cells transfected with 50 nM mimics miR-758-3p, inhibitor miR-758-3p or NC measured by EdU assay. Scale bars, 100 µm. (D) Subcellular localization of CASC9 in 5637 and J82 detected by RNA FISH. Scale bars, 20 µm. (E) The apoptotic rate of miR-758-3p mimic-, miR-758-3p inhibitor- and NC-transfected cells (P>0.05). Unpaired Student's t-test was used and data are presented as the mean ± SEM. *P<0.05 and **P<0.01. miR-758-3p, microRNA-758-3p; BC, bladder cancer; NC, negative control; EdU, 5-ethynyl-2′-deoxyuridine; FISH, fluorescence in situ hybridization; SEM, standard error of mean.
lncRNA CASC9 sponges miR-758-3p to promote EMT of BC by upregulating TGF-β2
An RNA FISH experiment was also performed to determine the cellular localization of CASC9. CASC9 was distributed in both the cytoplasm and nucleus (Fig. 5D), indicating that CASC9 may function in the nucleus and/or cytoplasm.
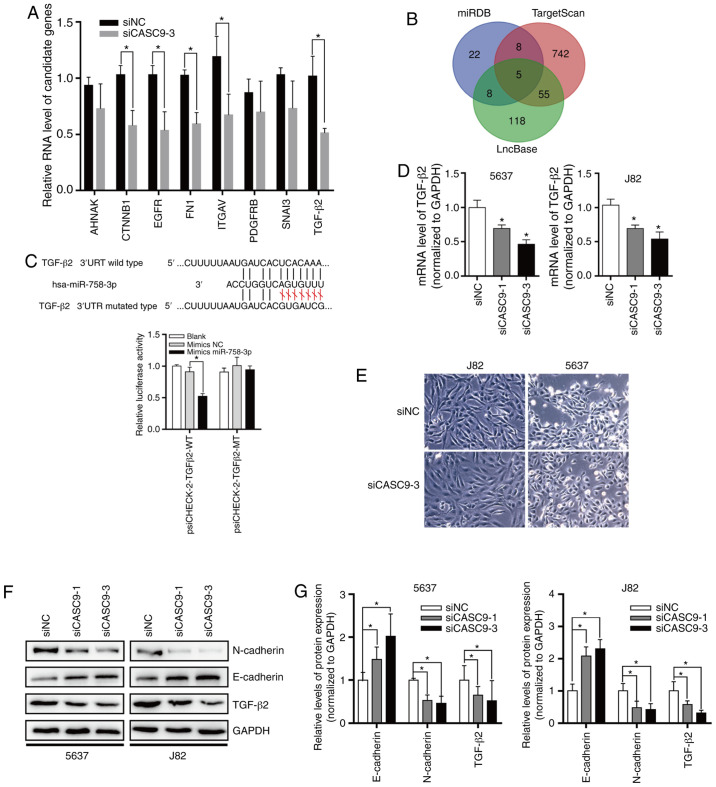
In order to explore the function of miR-758-3p, computational algorithms (TargetScan and miRDB) were used to search for potential miR-758-3p target genes (Fig. 6B). After knocking down the expression of CASC9, the expression changes of EMT-related genes were detected by the Human EMT RT2 Profiler PCR Array (Fig. 6A). A miR-758-3p-binding site was revealed in the 3′-UTR of TGF-β2 (Fig. 6C). The TGF-β pathway plays a central role in inducing EMT in BC (41). To verify whether TGF-β2 is the direct target of miR-758-3p, the TGF-β2 3′-UTR sequence was subcloned into the luciferase reporter vector pSicheck-2. It was revealed that miR-758-3p overexpression suppressed the luciferase activity of the luciferase reporter harboring wild-type (WT) TGF-β2 3′-UTR but not the reporter carrying mutant (MT) TGF-β2 (Fig. 6C).
Figure 6.
CASC9 sponges miR-758-3p to promote EMT in BC by upregulating TGF-β2. (A) After knocking down the expression of CASC9, the expression changes of EMT-related genes were detected by the Human EMT RT2 Profiler PCR Array. (B) Target genes of CASC9 predicted by miRDB, TargetScan and LncBase. (C) TGF-β2 is a direct target of miR-758-3p. Upper panel, schematic diagrams of the mutual interactions between miR-758-3p and TGF-β2 3′UTR. Lower panel, luciferase reporter assay was performed to examine the effect of CASC9 on antagonizing miR-758-3p-mediated suppression of TGF-β2 expression. (D) CASC9 was transfected into BC cells (J82 and 5637), and TGF-β2 mRNA levels were evaluated by RT-qPCR. (E) Morphological changes of 5637 and J82 after knocking down CASC9 levels. (F and G) After transfection with CASC9, the protein levels of TGF-β2 and EMT markers were evaluated by western blotting. Unpaired Student's t-test was used and data are presented as the mean ± SEM. *P<0.05. CASC9, cancer susceptibility candidate 9; miR-758-3p, microRNA-758-3p; EMT, epithelial-mesenchymal transition; BC, bladder cancer; TGF, transforming growth factor; RT-qPCR, reverse transcription-quantitative PCR; SEM, standard error of mean; si, small interfering; NC, negative control.
To assess the effect of CASC9 on TGF-β2 expression, CASC9 was knocked down in 5637 and J82 cells, and it was determined that CASC9 knockdown reduced TGF-β2 mRNA and protein levels (Fig. 6D-F). Subsequently, the protein levels of EMT markers were also determined by western blotting. The results revealed that after knockdown of CASC9, E-cadherin protein levels were significantly increased. After knocking down the expression of CASC9, 5637 and J82 cells exhibited stromal cell morphological characteristics (Fig. 6E), and N-cadherin protein expression was significantly decreased in 5637 and J82 cells (Fig. 6F and G).
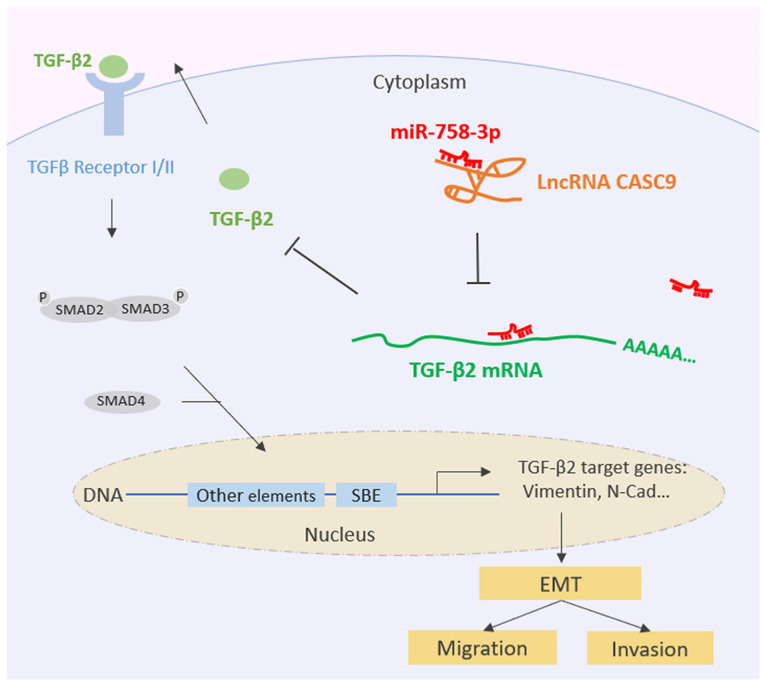
Collectively, all of the aforementioned data indicated that CASC9 sponges miR-758-3p to promote EMT in BC by upregulating TGF-β2 (Fig. 7).
Figure 7.
Schematic diagram of the lncRNA CASC9 sponging miR-758-3p to promote proliferation and EMT in BC by upregulating TGF-β2. CASC9, cancer susceptibility candidate 9; miR-758-3p, microRNA-758-3p; EMT, epithelial-mesenchymal transition; BC, bladder cancer; TGF, transforming growth factor. Table I. Associations between CASC9 expression and clinicopathological characteristics of bladder cancer patients.
Discussion
Previous research has highlighted the potential of lncRNAs as biomarkers and therapeutic targets in BC (42). The lncRNA CASC9 is located in a gene desert region that is devoid of nearby protein-coding genes (30). CASC9 is distributed in both the cytoplasm and nucleus (30), suggesting that CASC9 may play a role in the cytoplasm and nucleus and regulate gene expression in different ways. However, the relationship between CASC9 and BC remains unknown.
This study is the first, to the best of our knowledge, to explore the cellular functions of CASC9 in BC. It was revealed that CASC9 expression was increased in BC tissues, and CASC9 upregulation was significantly associated with the depth of bladder tumor invasion. CASC9 knockdown inhibited BC cell proliferation, migration and invasion. However, knockdown of CASC9 had no effect on BC cell apoptosis. Recent studies have demonstrated that upregulated CASC9 expression is a poor prognostic factor for esophageal cancer (28–30), pancreatic ductal adenocarcinoma (31), gastric cancer (32), nasopharyngeal carcinogenesis (33), and non-small cell lung cancer (34). These data support the present findings that CASC9 functions as an oncogene and plays a key role in the progression of BC.
Recently, it has been reported that lncRNAs can function as ceRNAs. Such ceRNAs regulate the distribution of miRNAs, thereby exerting an additional level of post-transcriptional regulation (43–46). In the present study, it was confirmed that CASC9 was upregulated in BC cells and CASC9 functioned as an effective miRNA (miR-758-3p) sponge. Consistent with the role of CASC9 as an oncogene, it was revealed that miR-758-3p was downregulated in BC. The downregulation of miR-758-3p has also been observed in hepatocellular carcinoma (HCC), papillary thyroid cancer (PTC) (47), gastric cancer (GC) (48) and non-small cell lung cancer (NSCLC) (49). However, the molecular mechanisms that underlie the tumor suppressive role of miR-758-3p remains unknown. In the present study, it was demonstrated that miR-758-3p downregulation activated TGF-β2 and promoted cell growth and metastasis in BC.
Increased expression of TGF-β isoforms, receptors, and signaling components is reported in high-grade invasive BC cells expressing vimentin and lacking E-cadherin (50). TGF-β activation promotes BC metastasis (12). Upregulation of phosphorylated SMAD2 has been reported in advanced invasive BC and associated with more frequent recurrence and poor survival (50). In human glioma, TGF-β2 initiated autophagy via SMAD and non-SMAD pathways, thereby promoting the invasion of glioma cells (51).
In the nucleus, CASC9 was revealed to recruit CBP and modulate H3K27ac levels of the LAMC2 promoter, thereby promoting LAMC2 transcription and stimulating ESCC cell growth (30). In the cytoplasm, CASC9 was revealed to sponge miR-758-3p to regulate the expression of TGF-β2, which activated the TGF-β signaling pathway and promoted proliferation and EMT in BC. In addition to interactions with miRNAs, lncRNAs also act as protein scaffolds to mediate protein interactions (18–21). More functions of CASC9 remain to be revealed. Notably, CASC9 appears to utilize various mechanisms to achieve its functions (28–30). Interactions with other proteins or even other nucleic acids (e.g., miRNAs, mRNAs, ncRNAs, or DNA) increase the spectrum of CASC9 functions. The function of CASC9 depends on the molecular context of the corresponding cancer cell. CASC9 plays different roles by interacting with other proteins or nucleic acids, such as miRNA, mRNA, ncRNA or DNA (28–30).
In summary, the present study revealed that CASC9 sponged miR-758-3p to regulate the expression of TGF-β2, a cytokine that activates the TGF-β signaling pathway and promoted proliferation and EMT in BC. These data allowed us to conclude that CASC9 plays an important role in the complex regulatory interaction network that controls the progression of BC. This regulatory mechanism facilitates our understanding of EMT of BC as well as other relevant human diseases.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Guangdong Basic and Applied Basic Research Fund (Guangdong Natural Science Fund, grant no. 2019A1515110766), and Longhua Science and Technology Innovation Program (grant no. 2017029).
Availability of data and materials
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Authors' contributions
JY and YL conceived the study and analyzed the findings. ZZ, FC and YL performed all of the experiments. ZZ and FC wrote the manuscript. FC, HZ, LC, TX and QD assisted in performing the experiments. QD provided partial funding support and manuscript modification. All the authors reviewed and approved the final version of the manuscript.
Ethics approval and consent to participate
The collection and use of all tissues were approved by the Ethic Committee of Peking University Shenzhen Hospital. All human tissue samples were obtained with informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Richters A, Aben KKH, Kiemeney L. The global burden of urinary bladder cancer: An update. World J Urol. 2020;38:1895–1904. doi: 10.1007/s00345-019-02984-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grayson M. Bladder cancer. Nature. 2017;551(Suppl 1):S33. doi: 10.1038/551S33a. [DOI] [PubMed] [Google Scholar]
- 3.Smith ZL, Guzzo TJ. Urinary markers for bladder cancer. F1000Prime Rep. 2013;5:21. doi: 10.12703/P5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamat AM, Hegarty PK, Gee JR, Clark PE, Svatek RS, Hegarty N, Shariat SF, Xylinas E, Schmitz-Dräger BJ, Lotan Y, et al. ICUD-EAU international consultation on bladder cancer 2012: Screening, diagnosis, and molecular markers. Eur Urol. 2013;63:4–15. doi: 10.1016/j.eururo.2012.09.057. [DOI] [PubMed] [Google Scholar]
- 5.Santamaria PG, Moreno-Bueno G, Portillo F, Cano A. EMT: Present and future in clinical oncology. Mol Oncol. 2017;11:718–738. doi: 10.1002/1878-0261.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh M, Yelle N, Venugopal C, Singh SK. EMT: Mechanisms and therapeutic implications. Pharmacol Ther. 2018;182:80–94. doi: 10.1016/j.pharmthera.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 7.Du B, Shim JS. Targeting epithelial-mesenchymal transition (EMT) to overcome drug resistance in cancer. Molecules. 2016;21:965. doi: 10.3390/molecules21070965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pastushenko I, Brisebarre A, Sifrim A, Fioramonti M, Revenco T, Boumahdi S, Van Keymeulen A, Brown D, Moers V, Lemaire S, et al. Identification of the tumour transition states occurring during EMT. Nature. 2018;556:463–468. doi: 10.1038/s41586-018-0040-3. [DOI] [PubMed] [Google Scholar]
- 9.Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20:69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 10.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F, Liu Y. TGF-β-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin Cancer Res. 2014;20:1531–1541. doi: 10.1158/1078-0432.CCR-13-1455. [DOI] [PubMed] [Google Scholar]
- 13.Calon A, Espinet E, Palomo-Ponce S, Tauriello DV, Iglesias M, Céspedes MV, Sevillano M, Nadal C, Jung P, Zhang XH, et al. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell. 2012;22:571–584. doi: 10.1016/j.ccr.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papageorgis P. TGFβ signaling in tumor initiation, epithelial-to-mesenchymal transition, and metastasis. J Oncol. 2015;2015:587193. doi: 10.1155/2015/587193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 16.Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73:2491–2509. doi: 10.1007/s00018-016-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiong T, Li J, Chen F, Zhang F. PCAT-1: A novel oncogenic Long Non-coding RNA in human cancers. Int J Biol Sci. 2019;15:847–856. doi: 10.7150/ijbs.30970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flynn RA, Chang HY. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14:752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 20.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, Zhang Y, Yang L, Shan W, He Q, et al. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell. 2015;28:529–540. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333:213–221. doi: 10.1016/j.canlet.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 24.Han Y, Liu Y, Nie L, Gui Y, Cai Z. Inducing cell proliferation inhibition, apoptosis, and motility reduction by silencing long noncoding ribonucleic acid metastasis-associated lung adenocarcinoma transcript 1 in urothelial carcinoma of the bladder. Urology. 2013;81:209.e1–e7. doi: 10.1016/j.urology.2012.08.044. [DOI] [PubMed] [Google Scholar]
- 25.Han Y, Liu Y, Gui Y, Cai Z. Long intergenic non-coding RNA TUG1 is overexpressed in urothelial carcinoma of the bladder. J Surg Oncol. 2013;107:555–559. doi: 10.1002/jso.23264. [DOI] [PubMed] [Google Scholar]
- 26.Yang C, Li X, Wang Y, Zhao L, Chen W. Long non-coding RNA UCA1 regulated cell cycle distribution via CREB through PI3-K dependent pathway in bladder carcinoma cells. Gene. 2012;496:8–16. doi: 10.1016/j.gene.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 27.Yan TH, Lu SW, Huang YQ, Que GB, Chen JH, Chen YP, Zhang HB, Liang XL, Jiang JH. Upregulation of the long noncoding RNA HOTAIR predicts recurrence in stage Ta/T1 bladder cancer. Tumour Biol. 2014;35:10249–10257. doi: 10.1007/s13277-014-2344-8. [DOI] [PubMed] [Google Scholar]
- 28.Pan Z, Mao W, Bao Y, Zhang M, Su X, Xu X. The long noncoding RNA CASC9 regulates migration and invasion in esophageal cancer. Cancer Med. 2016;5:2442–2447. doi: 10.1002/cam4.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Y, Hu L, Liang Y, Li J, Wang K, Chen X, Meng H, Guan X, Yang K, Bai Y. Up-regulation of lncRNA CASC9 promotes esophageal squamous cell carcinoma growth by negatively regulating PDCD4 expression through EZH2. Mol Cancer. 2017;16:150. doi: 10.1186/s12943-017-0715-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang Y, Chen X, Wu Y, Li J, Zhang S, Wang K, Guan X, Yang K, Bai Y. LncRNA CASC9 promotes esophageal squamous cell carcinoma metastasis through upregulating LAMC2 expression by interacting with the CREB-binding protein. Cell Death Differ. 2018;25:1980–1995. doi: 10.1038/s41418-018-0084-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu X, Lin Y, Sui W, Zou Y, Lv Z. Analysis of distinct long noncoding RNA transcriptional fingerprints in pancreatic ductal adenocarcinoma. Cancer Med. 2017;6:673–680. doi: 10.1002/cam4.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shang C, Sun L, Zhang J, Zhao B, Chen X, Xu H, Huang B. Silence of cancer susceptibility candidate 9 inhibits gastric cancer and reverses chemoresistance. Oncotarget. 2017;8:15393–15398. doi: 10.18632/oncotarget.14871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su X, Li G, Liu W. The long noncoding RNA cancer susceptibility candidate 9 promotes nasopharyngeal carcinogenesis via stabilizing HIF1α. DNA Cell Biol. 2017;36:394–400. doi: 10.1089/dna.2016.3615. [DOI] [PubMed] [Google Scholar]
- 34.Ma P, Zhang M, Nie F, Huang Z, He J, Li W, Han L. Transcriptome analysis of EGFR tyrosine kinase inhibitors resistance associated long noncoding RNA in non-small cell lung cancer. Biomed Pharmacother. 2017;87:20–26. doi: 10.1016/j.biopha.2016.12.079. [DOI] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative pcr and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Quan J, Chen F, Pan X, Zhuang C, Xiong T, Zhuang C, Li J, Huang X, Ye J, et al. MiR-31-5p acts as a tumor suppressor in renal cell carcinoma by targeting cyclin-dependent kinase 1 (CDK1) Biomed Pharmacother. 2019;111:517–526. doi: 10.1016/j.biopha.2018.12.102. [DOI] [PubMed] [Google Scholar]
- 37.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. Gepia: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paraskevopoulou MD, Vlachos IS, Karagkouni D, Georgakilas G, Kanellos I, Vergoulis T, Zagganas K, Tsanakas P, Floros E, Dalamagas T, Hatzigeorgiou AG. Diana-lncbase v2: Indexing microrna targets on non-coding transcripts. Nucleic Acids Res. 2016;44:D231–D238. doi: 10.1093/nar/gkv1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y, Wang X. MiRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48:D127–D131. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McConkey DJ, Choi W, Marquis L, Martin F, Williams MB, Shah J, Svatek R, Das A, Adam L, Kamat A, et al. Role of epithelial-to-mesenchymal transition (EMT) in drug sensitivity and metastasis in bladder cancer. Cancer Metastasis Rev. 2009;28:335–344. doi: 10.1007/s10555-009-9194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chandra Gupta S, Nandan Tripathi Y. Potential of long non-coding RNAs in cancer patients: From biomarkers to therapeutic targets. Int J Cancer. 2017;140:1955–1967. doi: 10.1002/ijc.30546. [DOI] [PubMed] [Google Scholar]
- 43.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The rosetta stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar MS, Armenteros-Monterroso E, East P, Chakravorty P, Matthews N, Winslow MM, Downward J. HMGA2 functions as a competing endogenous RNA to promote lung cancer progression. Nature. 2014;505:212–217. doi: 10.1038/nature12785. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Jeyapalan Z, Deng Z, Shatseva T, Fang L, He C, Yang BB. Expression of CD44 3′-untranslated region regulates endogenous microRNA functions in tumorigenesis and angiogenesis. Nucleic Acids Res. 2011;39:3026–3041. doi: 10.1093/nar/gkq1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen J, Xu Z, Yu C, Wu Z, Yin Z, Fang F, Chen B. MiR-758-3p regulates papillary thyroid cancer cell proliferation and migration by targeting TAB1. Pharmazie. 2019;74:235–238. doi: 10.1691/ph.2019.8933. [DOI] [PubMed] [Google Scholar]
- 48.Guo J, Zhang Z, Pan L, Zhou Y. Identification of miR-758-3p as potential modulator of CBX5 expression in gastric cancer. Technol Cancer Res Treat. 2018;17:1533033818816061. doi: 10.1177/1533033818816061. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Wang S, Jiang M. The long non-coding RNA-DANCR exerts oncogenic functions in non-small cell lung cancer via miR-758-3p. Biomed Pharmacother. 2018;103:94–100. doi: 10.1016/j.biopha.2018.03.053. [DOI] [PubMed] [Google Scholar]
- 50.Gupta S, Hau AM, Al-Ahmadie HA, Harwalkar J, Shoskes AC, Elson P, Beach JR, Hussey GS, Schiemann WP, Egelhoff TT, et al. Transforming growth factor-beta is an upstream regulator of mammalian target of rapamycin complex 2-dependent bladder cancer cell migration and invasion. Am J Pathol. 2016;186:1351–1360. doi: 10.1016/j.ajpath.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang C, Zhang X, Xu R, Huang B, Chen AJ, Li C, Wang J, Li XG. TGF-β2 initiates autophagy via Smad and non-Smad pathway to promote glioma cells' invasion. J Exp Clin Cancer Res. 2017;36:162. doi: 10.1186/s13046-017-0628-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.