Abstract
Angiogenesis is a tightly regulated biological process by which new blood vessels are formed from pre-existing blood vessels. This process is also critical in diseases such as cancer. Therefore, angiogenesis has been explored as a drug target for cancer therapy. The future of effective anti-angiogenic therapy lies in the intelligent combination of multiple targeting agents with novel modes of delivery to maximize therapeutic effects. Therefore, a novel approach is proposed that utilizes dumbbell RNA (dbRNA) to target pathological angiogenesis by simultaneously targeting multiple molecules and processes that contribute to angiogenesis. In the present study, a plasmid expressing miR-34a-3p and −5p dbRNA (db34a) was constructed using the permuted intron-exon method. A simple protocol to purify dbRNA from bacterial culture with high purity was also developed by modification of the RNASwift method. To test the efficacy of db34a, pancreatic cancer cell lines PANC-1 and MIA PaCa-2 were used. Functional validation of the effect of db34a on angiogenesis was performed on human umbilical vein endothelial cells using a tube formation assay, in which cells transfected with db34a exhibited a significant reduction in tube formation compared with cells transfected with scrambled dbRNA. These results were further validated in vivo using a zebrafish angiogenesis model. In conclusion, the present study demonstrates an approach for blocking angiogenesis using db34a. The data also show that this approach may be used to targeting multiple molecules and pathways.
Keywords: circular RNA, miR-34a, dumbbell RNA, db34a, tumor-associated macrophage, angiogenesis, pancreatic cancer, inflammation
Introduction
Metastatic tumors are usually highly vascularized, and this increased vascularity aids the dissemination of tumor cells to promote metastatic events. This high vascularity is induced via the expression of pro-angiogenic cytokines, which promote the development of abnormal tumor vasculature. The abnormal vasculature is characterized by hyperpermeable vessels, increased vessel diameter and abnormally thickened basement membranes. All these factors contribute to tumor growth (1–4). Several anti-angiogenic agents, such as bevacizumab, sunitinib and vatalanib have been developed to target increased tumor vascularity (5). The goal of anti-angiogenic therapy is the obliteration of tumor-induced vasculature with the aim of decreasing vascular permeability and the perfusion of oxygen and nutrients to tumor cells. This approach has often been termed as ‘tumor starvation’. A converse approach to anti-angiogenic therapy is the use of agents to stabilize abnormal tumor vasculature by reducing blood vessel diameter and permeability, controlling vessel perfusion, reducing tumor interstitial pressure and improving tumor oxygenation with the aim of reducing tumor hypoxia and thereby controlling metastasis (6–9). Researchers have even suggested that anti-angiogenic therapy may transiently normalize tumor vasculature and its microenvironment, thus enhancing the efficacy of chemoradiotherapy (10). Unfortunately, both these approaches have failed to achieve a clinically relevant standing despite the vast amount of research being conducted. The main drawback of these approaches is that they have been directed at single targets and have not produced clinically consistent desirable outcomes. In the present study, the targeting of multiple pro-angiogenic cytokines to achieve a complete anti-angiogenic outcome is proposed. This multipronged approach aims to exploit the microRNA (miRNA, miR) machinery to attain a clinically desirable outcome. The present study may pave the way for the development of a multi-targeted strategy, as well as methodologies that could be applicable to numerous other disease conditions.
The use of RNA molecules to target angiogenesis is not new (11–14). Various approaches have been developed to target angiogenesis, including the use of RNA aptamers, miRNAs, small interfering RNAs (siRNAs) and combinations thereof. Although promising, the half-life of RNA molecules is very short. In vitro, natural RNA exhibits a half-life of a few seconds to a few minutes in various biological fluids, including human serum, whereas RNAs partially modified at the 20th position have extended half-lives of 5 sec to 15 h (15). An RNA-based anti-angiogenesis drug called Macugen has these modifications; however, although somewhat successful, it is limited in addressing the problem of angiogenesis (16). The approach used in the present study is the development of a closed circular RNA having multiple double-stranded regions coding for miRNAs, which are interrupted by loops or dumbbells. Using a circular RNA should eliminate exonuclease activity while double-stranded RNA (dsRNA), being inherently more stable than single-stranded RNA (ssRNA), should contribute to an increased half-life. To the best of our knowledge, this is the first attempt to use closed circular dumbbell RNAs (dbRNAs) to code for miRNAs/siRNAs with a therapeutic intention. The study uses miR-34a-3p and −5p in a circular form.
Materials and methods
Construction of dbRNAs
The permuted intron-exon (PIE) method was used, which is an enzyme-free RNA circularization method based on group I intron self-splicing (17). Circularization is triggered by the presence of magnesium ions and guanine nucleotides, and the production of circular RNA can be conducted using essentially any type of cell (17–19). A previously described strategy was used, in which recombinant RNA is disguised as a natural RNA and thus appropriates the host machinery to evade cellular RNases (20), but with an added RNA circularization step using the rrnC terminator (21). Sequences coding for miR-34a-3p and −5p (db34a) and its scrambled sequence (dbSCR) were synthesized with the lpp promoter and the rrnC terminator sequences (Fig. 1A and B), and inserted in pUC57-Kan plasmids (GenScript Biotech). The JM101Tr E. coli strain [Δ(lac pro), supE, thi, recA56, srl-300.:Tn10, (F′, traD36, proAB, lacIq, lacZ, ΔM15)] (20) was used for the isolation of bacterial RNA. Modeling analysis using the on line mfold tool (http://unafold.rna.albany.edu/?q=mfold/RNA-Folding-Form) was done to determine the secondary structure of db34a (22).
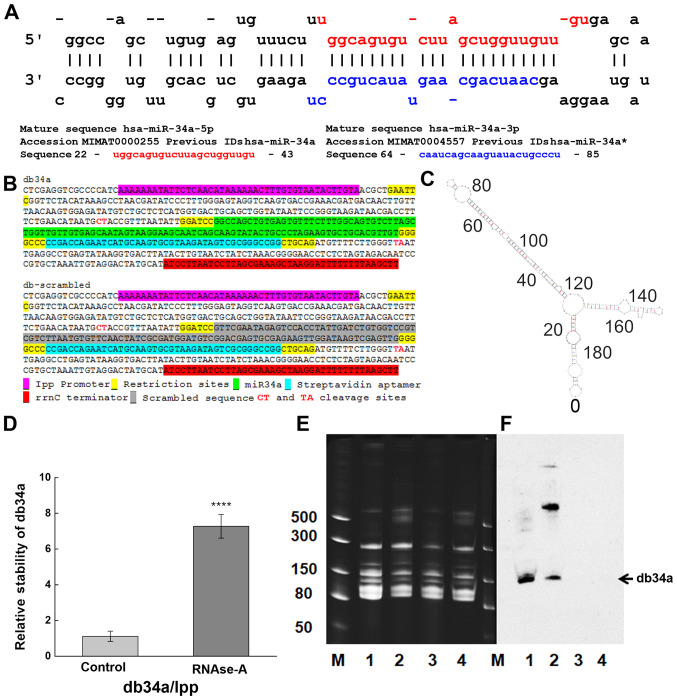
Figure 1.
Designing and purification of db34a. (A) Sequences of miR-34a-3p and miR-34a-5p used in the study. (B) Linear sequence of db34a and its scrambled version, dbSCR. (C) Modeling analysis using the mfold web server, showing the predicted structure of db34a. (D) Stability assay as performed by reverse transcription-quantitative PCR showing increased stability of the circular db34a compared with linear lpp RNA in the presence of RNase A. The significance of the difference between the control and RNase A-treated groups was evaluated by one-way ANOVA (****P=1.41627×10−4). (E) TBE-urea acrylamide gel electrophoresis of db34a in small RNA samples purified from db34a-expressing E. coli (lanes 1 and 2) and dbSCR-expressing control (lanes 3 and 4). Samples 2 and 4 were purified using RNAzol reagent, and samples 1 and 3 were purified using a modified RNASwift protocol. Lane M, dsRNA ladder (BioLabs). (F) Northern blotting analysis using a miR-34a-specific biotinylated probe showing the presence of db34a in lanes 1 and 2 but not in lanes 3 and 4. miR, microRNA; db34a, miR-34a-3p and −5p dumbbell RNA; dbSCR, scrambled dumbbell RNA; lpp, lipoprotein.
Isolation of small RNAs
Purification of small RNA was performed using a modified RNASwift protocol (23) and compared with purification performed using RNAzol reagent (Sigma-Aldrich; Merck KGaA). In the modified RNASwift method, bacteria expressing the db34a or dbSCR plasmid were grown in Luria-Bertani (LB) medium (BD Biosciences) with 50 µg/ml of kanamycin (MilliporeSigma) for 24 h at 37°C. The cultured cells (1.5×1010 cells) were spun down (3,000 × g for 20 min at 26°C), suspended in 10 ml of LB1 lysis reagent (4% sodium dodecyl sulfate, pH 7.5, 0.5 M NaCl), and lysed by incubation at 90°C for 4 min. Then, 5 ml of 5 M NaCl was added to the homogenate and the mixture was centrifuged at 16,000 × g for 25 min at 4°C. The supernatant was transferred to a new tube, and 6 ml of isopropanol was added and the mixture was centrifuged at 16,000 × g for 25 min at 4°C to precipitate out the large RNA. To the remaining supernatant, 10.5 ml of isopropanol was added and the mixture was centrifuged 20,000 × g for 25 min at 4°C to precipitate the small RNAs, which included the dbRNA product. The small RNA was washed twice with ice-cold 75% ethanol and then dissolved in sterile water. The purity of the small RNA was verified in a 1% agarose gel. The quality of RNA was determined using a NanoDrop™ spectrophotometer.
Purification of dbRNAs
The isolated small RNAs were separated on a 15% urea-polyacrylamide gel (PAGE). The gel was stained with ethidium bromide (0.5 mg/ml) for 15 min at room temperature and then destained by washing with distilled water twice. The gel portion containing the dbRNA was excised with a sharp scalpel and the RNA was purified using the ZR Small-RNA PAGE Recovery Kit (Zymo Research Corp.) according to the manufacturer's instructions. RNA quality and quantity were measured using a NanoDrop spectrophotometer.
Verification of dbRNA specificity
The isolated dbRNAs were separated on a 15% urea-PAG (TBE is the running buffer) at 20 mA followed by blotting using the iBlot™ Gel Transfer Device onto a Novex™ iBlot™ DNA Transfer Stack with a nylon membrane (both Invitrogen; Thermo Fisher Scientific, Inc.). Northern blot analysis was conducted using a miR-34a specific biotinylated probe (sequence: /5Biosg/AGGGCAGTATACTTGCTGAT) after membrane was incubated with prehybridization buffer (6X SSC buffer, 10X Denhardt's solution, 0.2% SDS; MilliporeSigma) for 1 h at 42°C on rotating shaker following 3 day incubation with hybridization buffer (6X SSC buffer, 5X Denhardt's solution, 0.2% SDS; MilliporeSigma) with probe at the same conditions. The chemiluminescent signal was detected using a Pierce™ Chemiluminescent Nucleic Acid Detection Module kit (Thermo Fisher Scientific, Inc.) according to the manufacturer's recommendations.
Determining the RNase A resistance of db34a RNA
To determine the resistance of db34a to RNase A, 1 µg total RNA from bacteria transformed with db34a-expressing plasmid was isolated and incubated with or without 0.7×10−5 U/µl RNase-A for 24 h in four independent treatments at room temperature. Since the bacterial lipoprotein (lpp) promoter (18,24) is a constitutive bacterial promoter that drives the expression of bacterial lpp, bacterial lpp RNA was used as the linear control for db34a. First, cDNA was synthesized using miScript II RT kit (Qiagen GmbH) using conditions: 37°C for 60 min and 95°C for 5 min from untreated and 24 h RNase A treated samples followed by quantitative PCR (qPCR) with the following primers: lpp1 forward, CTGTCTTCTGACGTTCAGACTC and lpp reverse, ACGAGCTGCGTCATCTTTAG. Expression of db34a was measured by qPCR with miR-34a specific primers [5′UGGCAGUGUCUUAGCUGGUUGU, cat. no. 218300; Hs_miR-34a_1 miScript Primer Assay (forward primer) and miScript Universal Primer (reverse)] using the miScript SYBR Green PCR Kit (Qiagen GmbH). The qPCR conditions used were as follows: Initial heat activation 95°C for 15 min, followed by denaturation at 94°C for 15 sec, annealing at 55°C for 30 sec and extension at 70°C for 30 sec, for 40 cycles. Four independent experiments with three replicates in each experiment were performed. ΔCq was calculating by subtracting the average Cq values of lpp from db34a for both untreated (control) and RNase A treated cells. ΔΔCq values were obtained by subtracting the average ΔCq of the control group from both the control and treatment ΔCq data followed by the calculation of 2−ΔΔCq (25). The difference between treated and control groups with reference to db34a/lpp RNA was analyzed using one-way ANOVA.
Cell culture conditions
The MIA PaCa-2 and PANC-1 pancreatic cancer cell lines were obtained from American Type Culture Collection (ATCC) and maintained under the conditions recommended by the supplier. Cells were grown in tissue culture-treated Petri plates in RPMI-1640 medium (Thermo Fisher Scientific, Inc.) with 10% FBS (VWR Corporation) and 1% penicillin-streptomycin (Thermo Fisher Scientific Inc.) in a humidified 5% CO2 atmosphere at 37°C. To evaluate immune activation, J774A.1 mouse macrophages obtained from ATCC were used. The macrophages were grown in tissue culture-treated Petri plates in DMEM (Thermo Fisher Scientific Inc.) with 10% FBS and 1% penicillin-streptomycin. Human umbilical vein endothelial cells (HUVECs) were obtained from Thermo Fisher Scientific, Inc. and cultured in Medium 200 with 1X low serum growth supplement (Thermo Fisher Scientific Inc.). HUVECs with a low passage number (3 passages) were used for the tube formation assay as described in a previous study (26).
Angiogenesis antibody array
An angiogenic antibody array analysis was performed using a RayBio® Human Angiogenesis Array C1 kit (AAH-ANG-1; RayBiotech, Inc.) according to the manufacturer's instructions. In brief, MIA PaCa-2 and PANC-1 cells were seeded (1.2×106/well, 6-well plate) in tissue culture-treated Petri plates overnight in RPMI-1640 medium (Thermo Fisher Scientific Inc.) with 10% FBS (VWR Corporation) and 1% penicillin-streptomycin (Thermo Fisher Scientific Inc.) in a humidified 5% CO2 atmosphere at 37°C. The cells were transfected with 4 µg of db34a or dbSCR using jetPRIME® transfection reagent (Polyplus-transfection SA at 37°C) or remained untransfected (control). After a 24-h incubation period, the medium was replaced with serum-free RPMI-1640 medium (Thermo Fisher Scientific Inc.) and the plates were incubated for another 24 h. Conditioned medium was then collected, and the antibody-spotted membranes from the array kit were incubated with the conditioned medium according to the manufacturer's protocol. Following incubation, the membranes were washed, 1 ml diluted biotin-conjugated antibody mix was added and the membranes were incubated at room temperature for 2 h, followed by washing and the addition of diluted HRP-conjugated streptavidin at room temperature for 2 h. Detection was performed using ECL protocols in which the membranes were exposed to X-ray films to visualize the binding of angiogenic factors. The intensities of the spots were quantified using ImageJ version 1.52a software (National Institutes of Health), and angiogenic molecules showing a significant difference in expression levels between the db34a and dbSCR conditioned media were identified. Significant differences between dbSCR and db34a conditioned media were detected using one-way ANOVA.
Inflammation array
A mouse inflammation array analysis was performed using a RayBio Mouse Inflammation Array C1 kit (cat. no. AAM-INF-1; RayBiotech, Inc.) according to the manufacturer's instructions. In brief, MIA PaCa-2 and PANC-1 cells were seeded (1.2×106/well; 6-well plate) and cultured overnight in RPMI-1640 medium (Thermo Fisher Scientific Inc.) supplemented with 10% FBS (VWR Corporation) and 1% penicillin-streptomycin (Thermo Fisher Scientific Inc.) in a humidified 5% CO2 atmosphere at 37°C. The next day, cells were transfected with 4 µg of db34a or dbSCR at 37°C using jetPRIME transfection reagent or remained untransfected (control). After 20 h, the medium was replaced with serum-free medium RPMI-1640 (Thermo Fisher Scientific Inc.) and the plates were incubated for another 16 h. Conditioned medium was then collected after centrifugation at 16,000 × g at 4°C for 10 min and transferred to freshly grown macrophages. After overnight incubation at 37°C, the macrophages were lysed with lysis buffer provided with kit and equal samples from each condition with regard to total protein quantity (100 µg per sample; total protein concentration was determined by Pierce 660 nm Protein Assay (Thermo Fisher Scientific Inc.) were applied to the antibody-spotted membranes. Following overnight incubation at 4°C, the membranes were washed and 1 ml diluted biotin-conjugated antibody mix was added to the membranes, which were kept at room temperature for 2 h. After several washes, the membranes were incubated with diluted HRP-conjugated streptavidin for 2 h. Detection was performed using ECL protocols in which the membranes were exposed to X-ray films to detect the binding of inflammatory factors. Spot intensities were quantified using ImageJ version 1.52a software. Significant differences between macrophages grown in dbSCR and db34a conditioned media were identified using one-way ANOVA.
In vitro angiogenic assay
To determine the anti-angiogenic effect of the dbRNA molecules, an endothelial cell tube formation assay was conducted using HUVECs as previously described (26). Briefly, HUVECs were stained with 2 µg/ml cell-permeant fluorescent dye (calcein-AM; Thermo Fisher Scientific, Inc.) with incubation at 37°C in the dark for ≥30 min. A Geltrex basement membrane matrix (Thermo Fisher Scientific, Inc; 40 µl) was applied to 96-well plates and allowed to solidify for 30 min at room temperature. The stained HUVECs were trypsinized, re-suspended in serum-free medium (Gibco Medium 200PR; Gibco; Thermo Fisher Scientific Inc.) and plated at a density of 20,000 cells/well in 100 µl conditioned medium from the db34a and dbSCR transfected cells. HUVECs in complete medium (Gibco Medium 200PRF supplemented with Gibco Low Serum Growth Supplement; Gibco; Thermo Fisher Scientific Inc.) were used as controls. The plates were incubated at 37°C in a humidified incubator for 3 h. The formation of angiogenic network was visualized at ×10 magnification using an inverted fluorescence microscope and quantified by ImageJ version 1.52a (n=17). The significance of difference of each HUVEC treatment group compared to control was determined using one-way ANOVA.
Zebrafish aquaculture
Transgenic VEGFR2:green-reef coral fluorescent protein (G-RCFP) zebrafish (Danio rerio) that express G-RCFP under the control of a VEGFR2/KDR promoter were obtained from ZFIN (y1Tg). The fish were maintained under standard aquaculture conditions in a circulating tank system constructed as previously described (27), and according to protocols approved by the Institutional Animal and Use Committee (IACUC) of University of Illinois College of Medicine at Peoria. Water was purified by a reverse osmosis system and supplemented with sea salts at concentration of 60 mg/l in order to provide the trace minerals that the fish required. The pH of the water was maintained between 7.6 and 8.5 by the addition of sodium bicarbonate. All experimental procedures using zebrafish were performed with approval from the IACUC of University of Illinois College of Medicine at Peoria.
In vivo zebrafish angiogenic assay
Adult male and female zebrafish were placed in spawning baskets. Newly fertilized eggs were collected soon after fertilization and rinsed at 28°C in embryo water (Milli-Q water with 60 mg/l Instant Ocean- Spectrum Brands) and immediately followed by incubation in 1% methylene blue in embryo water at 28°C for 24 h. After which the embryos were dechorionated and allowed to develop in embryo water alone for 24 h at 28°C (48 h post fertilization). Following this the embryos were incubated in embryo water alone or embryo water supplemented with 10 pmol db34a or dbSCR RNA, 5 µM sunitinib or a 1:1,000 dilution of DMSO as vehicle control at 28°C. After 24 h treatment, the 72 h old embryos were fixed in 10% buffered formaldehyde at room temperature for 10 min. The fixed embryos were mounted on glass slides and the completeness of sub-intestinal vein (SIV) vasculature was observed by the detection of G-RCFP using fluorescent inverted microscopy. The significance of differences of each treatment group compared to control was determined using one-way ANOVA.
Statistical analysis
P<0.05 was considered to indicate a statistically significant difference. The difference between treated and control groups with reference to db34a/lpp RNA was analyzed using one-way ANOVA. For the angiogenesis array, significant differences between dbSCR and db34a conditioned media were analyzed using one-way ANOVA. For the macrophage assay, significant differences between macrophages grown in dbSCR and db34a conditioned media were analyzed using one-way ANOVA. Data are presented as a mean ± standard error and calculations done with OriginPro version 8.6 software (OriginLab Corporation)
Results
db34a RNA harboring miR-34a-3p and miR-34a-5p shows increased stability compared with linear RNA
A circular RNA-expressing plasmid was developed to express miR-34a. This construct codes for miR-34a and a streptavidin-binding aptamer (Fig. 1A-C). The JM101Tr E. coli strain was used for the isolation of bacterial RNA. Upon circularization of the RNA, minimal degradation was observed, even after exposure to RNAse-A for 24 h (Fig. 1D). Stability was quantified using the 2−ΔΔCq method. A statistically significant difference between the control and RNase A-treated groups was detected (P=1.41627×10−4) with reference to linear lpp RNA vs. db34a RNA. The isolated db34a and dbSCR were characterized using urea-PAG electrophoresis. Using the modified RNASwift protocol, isolation of the circular RNAs was successfully achieved, as indicated by the low-molecular-weight bands in the urea gel (Fig. 1E). To confirm that the circular forms that were isolated were indeed db34a, northern blotting was performed using a biotinylated probe specific for miR-34a. The northern blotting results show the presence of a band for each db34a group, confirming that the methodology was specific (Fig. 1F).
db34a RNA harboring miR-34a-3p and miR-34a-5p induces the expression of C-C motif chemokine ligand 5 (CCL5) in PANC-1 and MIA PaCa-2 pancreatic cancer cells
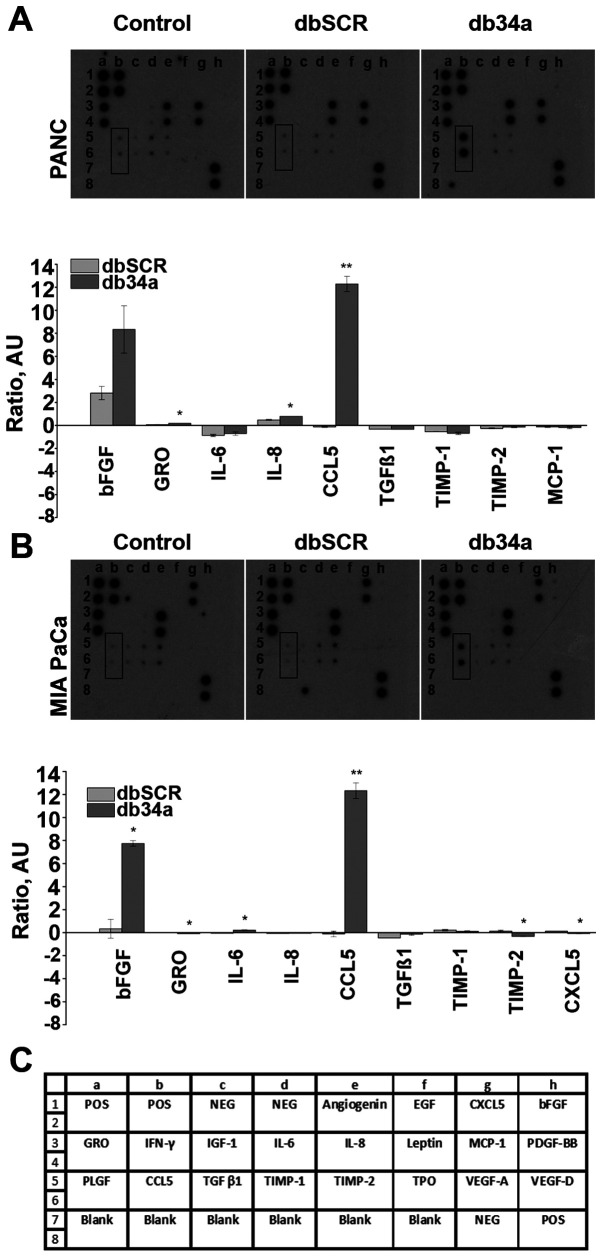
To evaluate the expression of pro-angiogenic molecules, the RayBio Human Angiogenesis Array C1 kit was used. Conditioned media collected from untransfected and db34a- or dbSCR-transfected MIA PaCa-2 and PANC-1 cells were analyzed using the array kit. From the array data, it was observed that CCL5 was significantly upregulated in PANC-1 (P=0.0028) and MIA PaCa-2 (P=0.0034) cells transfected with db34a compared with the respective dbSCR-transfected cells. Interestingly, basic fibroblast growth factor (bFGF) was also upregulated in PANC-1 and MIA PaCa-2 cells transfected with db34a, however, the upregulation was significant only in the MIA PaCa-2 cells (P=0.012; Fig. 2).
Figure 2.
Angiogenesis array results. A significant increase of CCL5 expression was detected in (A) PANC-1 (P=0.0028) and (B) MIA PaCa-2 cell lines (P=0.0034) for db34a-transfected cells vs. dbSCR-transfected control cells. *P≤0.05, **P≤0.01. (C) RayBio Human Angiogenesis Array C1 map. CCL5, C-C motif chemokine ligand 5; db34a, miR-34a-3p and −5p dumbbell RNA; dbSCR, scrambled dumbbell RNA.
db34a RNA harboring miR-34a-3p and miR-34a-5p suppresses angiogenic induction in HUVECs
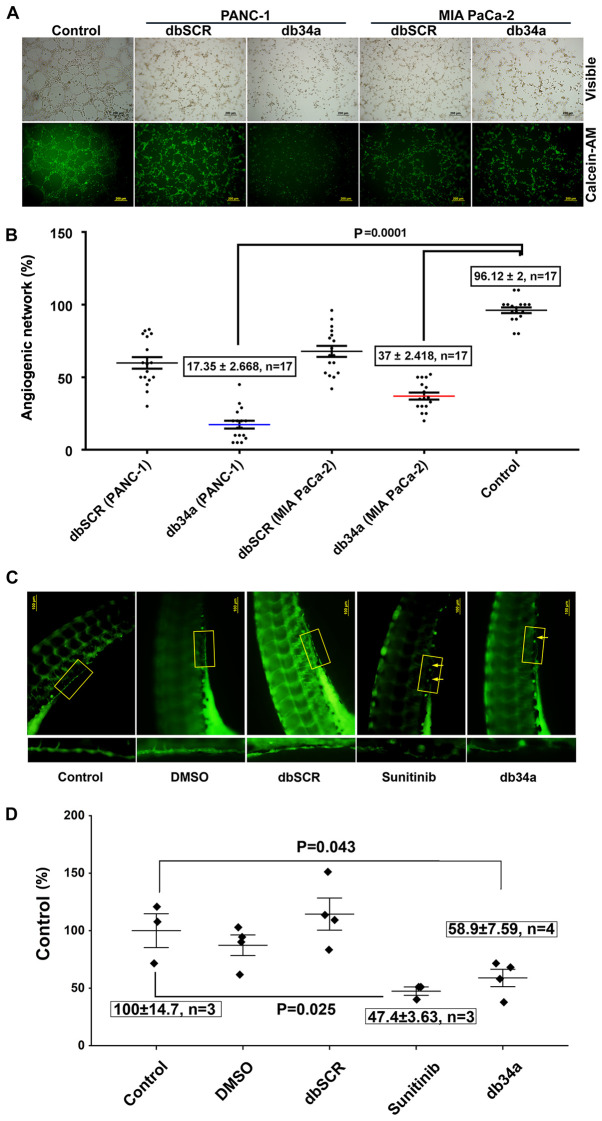
An angiogenic tube formation assay was performed using HUVECs according to a previously described protocol (26). The assay was used to determine whether db34a has the ability to alter the angiogenic potential of PANC-1 and MIA PaCa-2 cells. In this in vitro angiogenic assay, it was observed that HUVECs treated with conditioned media from db34a-transfected PANC-1 and MIA PaCa-2 cells exhibited suppressed angiogenic potential when compared with control (PANC-1, 17.35±2.66%; MIA PaCa-2, 37±2.41%; P=0.0001; Fig. 3A and B).
Figure 3.
Inhibition of angiogenesis by db34a in in vitro and in vivo models. (A) and (B) Endothelial cell tube formation assay was performed using HUVECs as a measure of angiogenesis. Phase contrast and calcein AM stained fluorescent microscope images showing reduced angiogenic tubule formation in HUVECs incubated with conditioned media from MIA PaCa-2 and PANC-1 cell lines transfected with db34a when compared with dbSCR control for 3 h. Scale bar, 200 µm. (C) In vivo angiogenesis assay was performed in zebrafish embryos with analysis using a fluorescent microscope. Dechorionated embryos were allowed to develop for 72 h then placed in embryo water alone (control), or supplemented with 10 pmol db34a or dbSCR RNA, 5 µM sunitinib or a 1:1,000 dilution of DMSO. The db34a-treated zebrafish embryos exhibited a reduction in the completeness of the SIV when compared with dbSCR and the controls. (D) Measurement of SIV completeness by the quantification of fluorescent intensity values of identical SIV areas and presented as a percentage of the control (control vs. db34a, P=0.043; control vs. sunitinib; P=0.025). HUVECs, human umbilical vein endothelial cells; db34a, miR-34a-3p and −5p dumbbell RNA; dbSCR, scrambled dumbbell RNA; SIV, sub-intestinal vein.
db34a RNA harboring miR-34a-3p and miR-34a-5p suppresses angiogenic induction in zebrafish embryos
Zebrafish strain y1tg that expresses a green fluorescent protein in the vasculature was used as a model organism. SIV vasculature was observed 24 h after treatment with 10 pmol db34a or dbSCR RNA, 5 µM sunitinib or a 1:1,000 dilution of DMSO, and images were captured using a fluorescent microscope (Fig. 3C). It was observed that the embryos treated with db34a showed reduced SIV integrity compared with the controls (58.9±7.59% of the untreated control; P=0.043), which confirmed the functionality of db34a and indicated the potential of dbRNA as a therapeutic agent. Positive control embryos treated with sunitinib also exhibited decreased SIV vasculature (47.4±3.63% of the untreated control; P=0.025).
db34a RNA harboring miR-34a-3p and miR-34a-5p induces the PANC-1 and MIA PaCa-2 pancreatic cancer cell-mediated activation of macrophage inflammatory markers
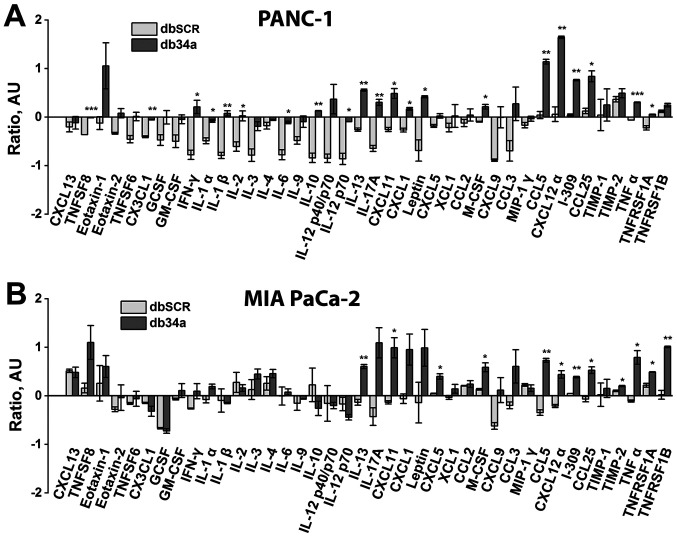
To determine whether db34a induces immune activation, conditioned media from PANC-1 and MIA PaCa-2 cells transfected with db34a were collected and the ability of the conditioned media to activate an inflammatory response in a mouse macrophage cell line was determined. It was observed that conditioned media from PANC-1 and MIA PaCa-2 cells transfected with db34a significantly induced the expression of interleukin (IL)-13, CCL5 and C-X-C motif chemokine ligand 12 α (CXCL12α) among other inflammatory response genes (Table I and Fig. 4).
Table I.
Cytokines significantly different between macrophages treated with conditioned media from PANC-1 and MIA PaCa-2 cell lines transfected with db34a compared with dbSCR.
| Cytokine | PANC-1 (fold change vs. control) | P-value | MIA PaCa-2 (fold changes vs. control) | P-value |
|---|---|---|---|---|
| CD30 ligand (TNFSF8) | 1.543449 | 4.61297×10−5 | 1.808521 | – |
| Fractalkine (CX3CL1) | 1.591752 | 0.00309 | 0.798687 | – |
| IFN-γ | 5.434441 | 0.02768 | 1.484371 | – |
| IL-1α (IL-1 F1) | 1.793997 | 0.02553 | 1.287388 | – |
| IL-1β (IL-1 F2) | 5.106111 | 0.0092 | 0.943795 | – |
| IL-2 | 2.601422 | 0.04669 | 0.909556 | – |
| IL-6 | 3.973113 | 0.02473 | 1.081643 | – |
| IL-10 | 7.051837 | 0.00978 | 0.605812 | – |
| IL-12 p70 | 6.513021 | 0.02201 | 0.673057 | – |
| IL-13 | 2.100802 | 0.00251 | 1.849357 | 0.00994 |
| IL-17A | 3.668945 | 0.00847 | 3.655409 | 0.03355 |
| I-TAC (CXCL11) | 1.996696 | 0.02114 | 2.297556 | – |
| KC (CXCL1) | 1.596054 | 0.0123 | 2.07834 | – |
| Leptin | 4.536913 | 0.0375 | 2.313414 | – |
| LIX (CXCL5) | 1.252327 | – | 1.334515 | 0.02446 |
| M-CSF | 1.339565 | 0.02349 | 1.40063 | 0.03974 |
| RANTES (CCL5) | 2.056593 | 0.00633 | 2.646111 | 0.00422 |
| SDF-1α (CXCL12α) | 2.493808 | 0.00911 | 1.820613 | 0.01686 |
| I-309 (TCA-3/CCL1) | 1.689148 | 0.00128 | 1.319467 | 0.00263 |
| TECK (CCL25) | 1.633533 | 0.02981 | 1.637408 | 0.02115 |
| TIMP-2 | 1.091021 | – | 1.089587 | 0.04152 |
| TNF-α | 1.387676 | 4.64829×10−4 | 2.003669 | 0.02406 |
| TNF RI (TNFRSF1A) | 1.355374 | 0.02447 | 1.223076 | 0.02037 |
| TNF RII (TNFRSF1B) | 1.114493 | – | 1.962633 | 0.00873 |
db34a, miR-34a-3p and −5p dumbbell RNA; dbSCR, scrambled dumbbell RNA; IL, interleukin; CCL5, C-C motif chemokine ligand 5; CXCL12α, C-X-C motif chemokine ligand 12α.
Figure 4.
Inflammatory proteins secreted by macrophages in response to conditioned medium from pancreatic cancer cells expressing db34a as detected using a mouse inflammation sp16 array. Array results for (A) PANC-1 and (B) MIA PaCa-2 cells. Expression levels were plotted as relative density ratios compared with wild-type controls. The analysis shows significant alterations in the expression of several key inflammatory cytokines and proteins in macrophages treated with db34a conditioned medium compared with dbSCR conditioned medium. *P≤0.05, **P≤0.01, ***P≤0.001 vs. dbSCR. db34a, miR-34a-3p and −5p dumbbell RNA; dbSCR, scrambled dumbbell RNA; IL, interleukin; CCL5, C-C motif chemokine ligand 5; CXCL12α, C-X-C motif chemokine ligand 12α.
Discussion
Despite decades of research, the treatment of pancreatic ductal adenocarcinoma continues to be a major challenge. According to the American Cancer Society, for all stages of pancreatic cancer combined, the 1-year survival rate is ~20% and the 5-year survival rate is 9% (28). One of the major factors that contributes to this poor prognosis is late detection, as early-stage pancreatic cancer is usually asymptomatic (29). New surgical techniques and evolving therapeutics have achieved only modest outcomes (30). A clearer understanding of targetable molecular pathways is required to design therapeutic agents and achieve a desired clinical outcome. Non-coding RNAs such as miRNAs play important roles in the regulation of gene expression. Unlike siRNAs, miRNAs target multiple genes and are usually process specific (31). Most biological processes are in some way regulated by miRNAs, which are small RNA molecules ~22 nucleotides in length that probably function as antisense regulators of other RNAs (32). The mechanisms of miRNA action have been elucidated, although not completely (33). Their capacity to regulate mRNAs and other miRNAs is being reported with increasing frequency, and they have become powerful tools for gene regulation. The use of RNA molecules to target angiogenesis continues to be explored (11–16). However, the approach described in the present study, utilizing a circular RNA molecule with two miRs is novel and, to the best of our knowledge, has not been described previously. The approach was to develop a closed circular RNA coding for miRNAs. A circular RNA is not readily degraded by exonucleases and the inherent higher stability of dsRNAs compared with ssRNAs may contribute to an increased half-life (34). Increased stability of db34a compared with linear RNA was observed in the present study. To the best of our knowledge, the present study is the first to use closed circular dbRNAs to code for both miR34-3p and −5p simultaneously with a therapeutic intention.
The present study aimed to demonstrate that circular RNA harboring miRs are functional and have significant therapeutic potential using dbRNA encoding miR-34a-3p and −5p. To construct the circular RNA expressing miR-34a-3p and −5p, the PIE method was used, which is an enzyme-free RNA circularization method based on group I intron self-splicing (17), which can be conducted using essentially any type of cell. Cells transcribe more RNA than they accumulate (35). This indicates that RNA is constantly being degraded and that a dynamic process occurs to balance RNA synthesis and degradation. The use of siRNA therapeutically is not new but has the limitations of low half-life and questionable in vivo efficacy (36,37), which is mainly due to the effects of exonuclease RNases. The present study circumvents the problem of a low half-life by designing dbRNAs that should not be degraded by the exonuclease activity of RNases. A previous study demonstrated the stability of a 35mer FITC-labeled RNA in the presence of circular dbRNA/DNA chimeric oligonucleotides to RNase H. This induced stability is most likely due to the competition of RNA/DNA chimeric with the 35mer FITC-labeled RNA (38). The stability experiments conducted in the present study demonstrate that circular RNA exposed to RNase A for 24 h was more stable than linear RNA driven by the same lpp promoter (18,24). Gene silencing by siRNA/miRNAs can be used as a therapeutic approach for the treatment of un-druggable targets. Researchers have also shown that dumbbell-shaped DNA minimal vectors can be used for small hairpin RNA expression (39,40). These studies also demonstrated the use of a dumbbell DNA vector to produce hairpin RNAs that can be used therapeutically. Another study described the generation of dumbbell-shaped nano-circular RNAs for RNA interference, using a chemical method of synthesis involving the enzyme-based circularization of small RNA molecules (41). By contrast, the present study reveals the development of a larger dbRNA and is unique in that it uses a bacterial system for large-scale dbRNA production.
Although not much is known about miR-34a-3p, miR-34a-5p is well studied and has been shown to be involved in tumor suppression, angiogenic suppression and cancer stem cell suppression (42–49); hsa-miR-34a-3p was previously known as hsa-miR-34a* and hsa-miR-34a-5p was previously known hsa-miR-34a. It was recently demonstrated that normal cells do show detectable expression levels of miR34a (50). Normal cell lines were not used in the present study and this is a limitation of the study. Since normal cells do show detectable levels of miR34a, the addition of db34a to normal cells should not, in theory, induce a notable perturbance in the existing molecular pathways of normal cells; this however requires further study. To demonstrate the possibility of using circular RNA therapeutically, in the present study circular miR-34a-3p and −5p were transfected into pancreatic cancer cells and the expression of pro-angiogenic molecules was determined. It was observed that in the two pancreatic cancer cell lines transfected with db34a, the expression levels of bFGF and CCL5 were significantly increased. miR-34a-5p expression is known to be increased by the activation of the PI3K signaling pathway (51) and, notably, mir-34a-5p has been shown to suppress the PI3K signaling pathway (52–56). These contradictory observations indicates the possibility of the existence of a tight regulatory apparatus that is not yet clearly understood. Interestingly, TNF is predicted as the target of hsa-miR-34a-3p by the miRDB online database, and TNF is a strong promoter of angiogenesis (57). This indicates that hsa-miR-34a-3p may be more therapeutically relevant in angiogenic suppression. It should be noted that both miR-34a-3p and miR-34a-5p are processed from the same pre-miR. This is consistent with the observation that angiogenesis was suppressed significantly in the in vitro and in vivo angiogenic assays of the present study. This observation appears contrary to the expected outcome; however, miR-34a-5p is a known angiogenic suppressor (43) and the quantification of other relevant angiogenesis-associated molecules will provide a clearer mechanistic profile of this angiogenesis modulation. Every biological process in an oncogenic system is tightly regulated; therefore, targeting a process rather than a single molecule would not be clinically relevant as compensatory pathways can be activated to achieve the oncogenic phenotype. The overexpression of bFGF may be one such compensatory response, which is seen in both pancreatic cell lines.
The present study also demonstrated that CCL5 is significantly upregulated in PANC-1 and MIA PaCa-2 cells transfected with db34a. CCL5, also known as regulated on activation, normal T cell expressed and secreted (RANTES), is a chemokine that is involved in T-cell activation. RANTES has been identified as a major HIV-suppressive factor. Studies have shown that recombinant human RANTES induces a dose-dependent inhibition of different strains of HIV-1, HIV-2 and simian immunodeficiency virus (SIV) (58,59). The expression of CCL5 in MIA PaCa-2 and PANC-1 cells transfected with db34a RNA indicates the involvement of a conserved pathway. Additional research is necessary to determine whether transfection with db34a can also induce the overexpression of CCL5 in other types of cells. In a recent study, it was shown that CCL5 and CCR5 expression levels are increased in pancreatic cancer tissue sections, and the overexpression of CCL5 and CCR5 increases the invasiveness and metastatic potential of pancreatic cancer cells (60). It has also been reported that progranulin (PGRN) expression levels correlate with a poor prognosis for melanoma patients, and PGRN inhibits CCL5 gene expression at the transcriptional level, showing that CCL5 is responsible for the recruitment of activated natural killer cells to the tumor microenvironment (61). Another study highlighted the importance of CCL5, demonstrating that the blockade of 1,4-α-glucan-branching enzyme in lung cancer cells promoted the secretion of CCL5, which induced the recruitment of CD8+ T lymphocytes to the tumor microenvironment (62). CCL5 expression has also been shown to restore the immune surveillance in antigen-expressing MYC;CTNNB1 hepatocellular carcinoma tumors (63). This indicates that CCL5 is necessary for tumor cells to be visible to the immune system; especially, CCL5 expression is necessary for immune recognition. However, it is also important to note that CCL5 was recently identified as a signature of poor prognosis for oral squamous cell carcinoma, indicating its multifaceted role (64). Interestingly, another study observed that the expression of CCL5 in breast cancer cells was associated with lymph node status and tumor node-metastasis stage (65). The conflicting roles of CCL5, and whether its overexpression is a predisposing factor for poor prognosis or a response to poor prognosis remain unclear. The association of CCL5 with other immune-associated molecules may provide clearer insight, and CCL5 expression alone should not be used as a measure of prognosis.
Furthermore, the present study investigated whether the transfection of pancreatic cancer cells with db34a induces the activation of immune cells. It was observed that the conditioned medium from db34a-transfected pancreatic cancer cells significantly induced the expression of IL-13, CCL5 and CXCL12α among other inflammatory response genes in mouse macrophage cell lysates. Tumor-associated macrophages (TAMs) are present in high numbers in the microenvironment of solid tumors and have been studied for their potential as targets for cancer therapy (66). TAMs suppress host immune responses and also promote tumor cell proliferation, angiogenesis, invasion and metastasis (67). The role of TAMs remains unclear; however, most evidence suggests that TAMs promote tumor progression. From the data in the present study, especially the overexpression of IL-13, it appears that the J774A.1 macrophages exposed to conditioned medium from db34a-transfected pancreatic cancer cells appeared more closely associated with the M1 phenotype than the M2 phenotype (data not shown) (68). Further investigation is required to decipher the influence of db34a on macrophages.
In conclusion, the present data suggest that the supplementation of miR-34a-3p and −5p as a circular pre-miR form is a viable method of miR replacement therapy.
Acknowledgements
Assistance for the maintenance of Zebrafish culture at the Laboratory Animal Care Facility, UICOMP, LACF-Peoria IL, by Laboratory Animal Care Supervisor Ms. Angela Daniels and, Laboratory Animal Care Technician Ms. Stephanie Sampson is acknowledged.
Funding
The Department of Internal Medicine, University of Illinois College of Medicine at Peoria; McElroy Foundation (Springfield, IL, USA) (grant no, 096726) and gift from The Theresa Tracy Strive to Survive Foundation (East Peoria, IL, USA) supported the present study.
Availability of data and materials
The plasmid constructs db34a and dbSCR are available from the corresponding author upon reasonable request in accordance with institutional MTA approvals and all applicable rules and regulations. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
MG designed and performed experiments and reviewed the manuscript. LD validated the constructs, designed the RNA stability experiments, confirmed dbRNA stability and quantified data. LG and PS were involved in designing the concept of the study. SB was involved in RNA experiments and data collection. JAN was involved in zebrafish data collection. CSG designed, and developed the concept and experiments, and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All experimental procedures using zebrafish were performed with approval from the Institutional Animal and Use Committee of University of Illinois College of Medicine at Peoria.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Boehm T, Folkman J, Browder T, O'Reilly MS. Antiangiogenic therapy of experimental cancer does not induce acquired drug resistance. Nature. 1997;390:404–407. doi: 10.1038/37126. [DOI] [PubMed] [Google Scholar]
- 2.Zetter BR. Angiogenesis and tumor metastasis. Annu Rev Med. 1998;49:407–424. doi: 10.1146/annurev.med.49.1.407. [DOI] [PubMed] [Google Scholar]
- 3.Folkman J. Antiangiogenic gene therapy. Proc Natl Acad Sci USA. 1998;95:9064–9066. doi: 10.1073/pnas.95.16.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donmez G, Sullu Y, Baris S, Yildiz L, Aydin O, Karagoz F, Kandemir B. Vascular endothelial growth factor (VEGF), matrix metalloproteinase-9 (MMP-9), and thrombospondin-1 (TSP-1) expression in urothelial carcinomas. Pathol Res Pract. 2009;205:854–7. doi: 10.1016/j.prp.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 5.Neves KB, Montezano AC, Lang NN, Touyz RM. Vascular toxicity associated with anti-angiogenic drugs. Clin Sci (Lond) 2020;134:2503–20. doi: 10.1042/CS20200308. [DOI] [PubMed] [Google Scholar]
- 6.Mattison R, Jumonville A, Flynn PJ, Moreno-Aspitia A, Erlichman C, LaPlant B, Juckett MB. A phase II study of AZD2171 (cediranib) in the treatment of patients with acute myeloid leukemia or high-risk myelodysplastic syndrome. Leuk Lymphoma. 2015;56:2061–2066. doi: 10.3109/10428194.2014.977886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong DS, Garrido-Laguna I, Ekmekcioglu S, Falchook GS, Naing A, Wheler JJ, Fu S, Moulder SL, Piha-Paul S, Tsimberidou AM, et al. Dual inhibition of the vascular endothelial growth factor pathway: A phase 1 trial evaluating bevacizumab and AZD2171 (cediranib) in patients with advanced solid tumors. Cancer. 2014;120:2164–2173. doi: 10.1002/cncr.28701. [DOI] [PubMed] [Google Scholar]
- 8.Liu JF, Tolaney SM, Birrer M, Fleming GF, Buss MK, Dahlberg SE, Lee H, Whalen C, Tyburski K, Winer E, et al. A phase 1 trial of the poly(ADP-ribose) polymerase inhibitor olaparib (AZD2281) in combination with the anti-angiogenic cediranib (AZD2171) in recurrent epithelial ovarian or triple-negative breast cancer. Eur J Cancer. 2013;49:2972–2978. doi: 10.1016/j.ejca.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Climent-Salarich M, Quintavalle M, Miragoli M, Chen J, Condorelli G, Elia L. TGFβ triggers miR-143/145 transfer from smooth muscle cells to endothelial cells, thereby modulating vessel stabilization. Circ Res. 2015;116:1753–1764. doi: 10.1161/CIRCRESAHA.116.305178. [DOI] [PubMed] [Google Scholar]
- 10.Huang G, Chen L. Tumor vasculature and microenvironment normalization: A possible mechanism of antiangiogenesis therapy. Cancer Biother Radiopharm. 2008;23:661–667. doi: 10.1089/cbr.2008.0492. [DOI] [PubMed] [Google Scholar]
- 11.Barbas AS, White RR. The development and testing of aptamers for cancer. Curr Opin Investig Drugs. 2009;10:572–578. [PubMed] [Google Scholar]
- 12.Guglielmelli T, Bringhen S, Palumbo A. Update on the use of defibrotide. Expert Opin Biol Ther. 2012;12:353–361. doi: 10.1517/14712598.2012.657623. [DOI] [PubMed] [Google Scholar]
- 13.Gatto B, Cavalli M. From proteins to nucleic acid-based drugs: The role of biotech in anti-VEGF therapy. Anticancer Agents Med Chem. 2006;6:287–301. doi: 10.2174/187152006777698178. [DOI] [PubMed] [Google Scholar]
- 14.Kanwar JR, Mahidhara G, Kanwar RK. Antiangiogenic therapy using nanotechnological-based delivery system. Drug Discov Today. 2011;16:188–202. doi: 10.1016/j.drudis.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 15.Friedman AD, Kim D, Liu R. Highly stable aptamers selected from a 20-fully modified fGmH RNA library for targeting biomaterials. Biomaterials. 2015;36:110–23. doi: 10.1016/j.biomaterials.2014.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue M, Kadonosono K, Arakawa A, Yamane S, Ishibashi T. Long-term outcome of intravitreal pegaptanib sodium as maintenance therapy in japanese patients with neovascular age-related macular degeneration. Jpn J Ophthalmol. 2015;59:173–178. doi: 10.1007/s10384-015-0374-4. [DOI] [PubMed] [Google Scholar]
- 17.Puttaraju M, Been MD. Circular ribozymes generated in Escherichia coli using group I self-splicing permuted intron-exon sequences. J Biol Chem. 1996;271:26081–26087. doi: 10.1074/jbc.271.42.26081. [DOI] [PubMed] [Google Scholar]
- 18.Umekage S, Kikuchi Y. In vivo circular RNA production using a constitutive promoter for high-level expression. J Biosci Bioeng. 2009;108:354–356. doi: 10.1016/j.jbiosc.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 19.Ford E, Ares M., Jr Synthesis of circular RNA in bacteria and yeast using RNA cyclase ribozymes derived from a group I intron of phage T4. Proc Natl Acad Sci USA. 1994;91:3117–3121. doi: 10.1073/pnas.91.8.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ponchon L, Dardel F. Recombinant RNA technology: The tRNA scaffold. Nat Methods. 2007;4:571–576. doi: 10.1038/nmeth1058. [DOI] [PubMed] [Google Scholar]
- 21.Umekage S, Kikuchi Y. In vitro and in vivo production and purification of circular RNA aptamer. J Biotechnol. 2009;139:265–272. doi: 10.1016/j.jbiotec.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 22.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nwokeoji AO, Kilby PM, Portwood DE, Dickman MJ. RNASwift: A rapid, versatile RNA extraction method free from phenol and chloroform. Anal Biochem. 2016;512:36–46. doi: 10.1016/j.ab.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 24.Chen QX, Wang WP, Zeng S, Urayama S, Yu AM. A general approach to high-yield biosynthesis of chimeric RNAs bearing various types of functional small RNAs for broad applications. Nucleic Acids Res. 2015;43:3857–3869. doi: 10.1093/nar/gkv228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Gnanamony M, Antony R, Fernandez KS, Jaime L, Lin J, Joseph PA, Gondi CS. Chronic radiation exposure of neuroblastoma cells reduces nMYC copy number. Oncol Lett. 2017;14:3363–3370. doi: 10.3892/ol.2017.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S, Carlson R, Zafreen L, Rajpurohit SK, Jagadeeswaran P. Modular, easy-to-assemble, low-cost zebrafish facility. Zebrafish. 2009;6:269–274. doi: 10.1089/zeb.2009.0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 29.Jelski W, Mroczko B. Biochemical diagnostics of pancreatic cancer-present and future. Clin Chim Acta. 2019;498:47–51. doi: 10.1016/j.cca.2019.08.013. [DOI] [PubMed] [Google Scholar]
- 30.McGuigan A, Kelly P, Turkington RC, Jones C, Coleman HG, McCain RS. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J Gastroenterol. 2018;24:4846–4861. doi: 10.3748/wjg.v24.i43.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou LY, Qin Z, Zhu YH, He ZY, Xu T. Current RNA-based therapeutics in clinical trials. Curr Gene Ther. 2019;19:172–196. doi: 10.2174/1566523219666190719100526. [DOI] [PubMed] [Google Scholar]
- 32.Ambros V. microRNAs: Tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/S0092-8674(01)00616-X. [DOI] [PubMed] [Google Scholar]
- 33.Toscano-Garibay JD, Aquino-Jarquin G. Transcriptional regulation mechanism mediated by miRNA-DNA•DNA triplex structure stabilized by Argonaute. Biochim Biophys Acta. 2014;1839:1079–1083. doi: 10.1016/j.bbagrm.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Mohammed-Elsabagh M, Paczkowski F, Li Y. Circular nucleic acids: Discovery, functions and applications. Chembiochem. 2020;21:1547–1566. doi: 10.1002/cbic.202000003. [DOI] [PubMed] [Google Scholar]
- 35.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 36.Ryther RC, Flynt AS, Phillips JA, III, Patton JG. siRNA therapeutics: Big potential from small RNAs. Gene Ther. 2005;12:5–11. doi: 10.1038/sj.gt.3302356. [DOI] [PubMed] [Google Scholar]
- 37.Bakhtiyari S, Haghani K, Basati G, Karimfar MH. siRNA therapeutics in the treatment of diseases. Ther Deliv. 2013;4:45–57. doi: 10.4155/tde.12.136. [DOI] [PubMed] [Google Scholar]
- 38.Park WS, Miyano-Kurosaki N, Abe T, Takai K, Takaku H. Properties of circular dumbbell RNA/DNA chimeric oligonucleotides containing antisense phosphodiester oligonucleotides. Nucleic Acids Symp Ser. 1999:225–226. doi: 10.1093/nass/42.1.225. [DOI] [PubMed] [Google Scholar]
- 39.Yu H, Jiang X, Tan KT, Hang L, Patzel V. Efficient production of superior dumbbell-shaped DNA minimal vectors for small hairpin RNA expression. Nucleic Acids Res. 2015;43:e120. doi: 10.1093/nar/gkv583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang X, Yu H, Teo CR, Tan GS, Goh SC, Patel P, Chua YK, Hameed NB, Bertoletti A, Patzel V. Advanced design of dumbbell-shaped genetic minimal vectors improves non-coding and coding RNA expression. Mol Ther. 2016;24:1581–1591. doi: 10.1038/mt.2016.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abe N, Abe H, Nagai C, Harada M, Hatakeyama H, Harashima H, Ohshiro T, Nishihara M, Furukawa K, Maeda M, et al. Synthesis, structure, and biological activity of dumbbell-shaped nanocircular RNAs for RNA interference. Bioconjug Chem. 2011;22:2082–2092. doi: 10.1021/bc2003154. [DOI] [PubMed] [Google Scholar]
- 42.Misso G, Di Martino MT, De Rosa G, Farooqi AA, Lombardi A, Campani V, Zarone MR, Gullà A, Tagliaferri P, Tassone P, Caraglia M. Mir-34: A new weapon against cancer? Mol Ther Nucleic Acids. 2014;3:e194. doi: 10.1038/mtna.2014.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li XJ, Ren ZJ, Tang JH. MicroRNA-34a: A potential therapeutic target in human cancer. Cell Death Dis. 2014;5:e1327. doi: 10.1038/cddis.2014.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kofman AV, Kim J, Park SY, Dupart E, Letson C, Bao Y, Ding K, Chen Q, Schiff D, Larner J, Abounader R. microRNA-34a promotes DNA damage and mitotic catastrophe. Cell Cycle. 2013;12:3500–3511. doi: 10.4161/cc.26459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar B, Yadav A, Lang J, Teknos TN, Kumar P. Dysregulation of microRNA-34a expression in head and neck squamous cell carcinoma promotes tumor growth and tumor angiogenesis. PLoS One. 2012;7:e37601. doi: 10.1371/journal.pone.0037601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tivnan A, Tracey L, Buckley PG, Alcock LC, Davidoff AM, Stallings RL. MicroRNA-34a is a potent tumor suppressor molecule in vivo in neuroblastoma. BMC Cancer. 2011;11:33. doi: 10.1186/1471-2407-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gallardo E, Navarro A, Vinolas N, Marrades RM, Diaz T, Gel B, Quera A, Bandres E, Garcia-Foncillas J, Ramirez J, Monzo M. miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis. 2009;30:1903–1909. doi: 10.1093/carcin/bgp219. [DOI] [PubMed] [Google Scholar]
- 48.Wei JS, Song YK, Durinck S, Chen QR, Cheuk AT, Tsang P, Zhang Q, Thiele CJ, Slack A, Shohet J, Khan J. The MYCN oncogene is a direct target of miR-34a. Oncogene. 2008;27:5204–5213. doi: 10.1038/onc.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 50.Bozgeyik E, Tepe NB, Bozdag Z. Identification of microRNA expression signature for the diagnosis and prognosis of cervical squamous cell carcinoma. Pathol Res Pract. 2020;216:153159. doi: 10.1016/j.prp.2020.153159. [DOI] [PubMed] [Google Scholar]
- 51.Barnes PJ, Baker J, Donnelly LE. Cellular senescence as a mechanism and target in chronic lung diseases. Am J Respir Crit Care Med. 2019;200:556–564. doi: 10.1164/rccm.201810-1975TR. [DOI] [PubMed] [Google Scholar]
- 52.Zhai L, Zhao Y, Liu Z, Wu J, Lin L. mRNA expression profile analysis reveals a C-MYC/miR-34a pathway involved in the apoptosis of diffuse large B-cell lymphoma cells induced by yiqichutan treatment. Exp Ther Med. 2020;20:2157–2165. doi: 10.3892/etm.2020.8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang C, Cheng Z, Jiang T, Xu Y, Wang B. MicroRNA-34a inhibits cell invasion and epithelial-mesenchymal transition via targeting AXL/PI3K/AKT/snail signaling in nasopharyngeal carcinoma. Genes Genomics. 2020;42:971–978. doi: 10.1007/s13258-020-00963-3. [DOI] [PubMed] [Google Scholar]
- 54.Wang S, Tang Q, Ge F, Guo Q. Typhae pollen polysaccharides protect hypoxia-induced PC12 cell injury via regulation of miR-34a/SIRT1. Int J Immunopathol Pharmacol. 2020;34:2058738420910005. doi: 10.1177/2058738420910005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Zhang YM, Wu QM, Chang LY, Liu JC. miR-34a and miR-125a-5p inhibit proliferation and metastasis but induce apoptosis in hepatocellular carcinoma cells via repressing the MACC1-mediated PI3K/AKT/mTOR pathway. Neoplasma. 2020;67:1042–1053. doi: 10.4149/neo_2020_191019N1062. [DOI] [PubMed] [Google Scholar]
- 56.Tao H, Cheng L, Yang R. Downregulation of miR-34a promotes proliferation and inhibits apoptosis of rat osteoarthritic cartilage cells by activating PI3K/akt pathway. Clin Interv Aging. 2020;15:373–385. doi: 10.2147/CIA.S241855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mahdavi Sharif P, Jabbari P, Razi S, Keshavarz-Fathi M, Rezaei N. Importance of TNF-alpha and its alterations in the development of cancers. Cytokine. 2020;130:155066. doi: 10.1016/j.cyto.2020.155066. [DOI] [PubMed] [Google Scholar]
- 58.Scarlatti G, Tresoldi E, Bjorndal A, Fredriksson R, Colognesi C, Deng HK, Malnati MS, Plebani A, Siccardi AG, Littman DR, et al. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 59.Cocchi F, DeVico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–5. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 60.Singh SK, Mishra MK, Eltoum IA, Bae S, Lillard JW, Jr, Singh R. CCR5/CCL5 axis interaction promotes migratory and invasiveness of pancreatic cancer cells. Sci Rep. 2018;8:1323. doi: 10.1038/s41598-018-19643-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Voshtani R, Song M, Wang H, Li X, Zhang W, Tavallaie MS, Yan W, Sun J, Wei F, Ma X. Progranulin promotes melanoma progression by inhibiting natural killer cell recruitment to the tumor microenvironment. Cancer Lett. 2019;465:24–35. doi: 10.1016/j.canlet.2019.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li L, Yang L, Cheng S, Fan Z, Shen Z, Xue W, Zheng Y, Li F, Wang D, Zhang K, et al. Lung adenocarcinoma-intrinsic GBE1 signaling inhibits anti-tumor immunity. Mol Cancer. 2019;18:108. doi: 10.1186/s12943-019-1027-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ruiz de Galarreta M, Bresnahan E, Molina-Sanchez P, Lindblad KE, Maier B, Sia D, Puigvehi M, Miguela V, Casanova-Acebes M, Dhainaut M, et al. β-catenin activation promotes immune escape and resistance to anti-PD-1 therapy in hepatocellular carcinoma. Cancer Discov. 2019;9:1124–1141. doi: 10.1158/2159-8290.CD-19-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang J, Wang Y, Kong F, Han R, Song W, Chen D, Bu L, Wang S, Yue J, Ma L. Identification of a six-gene prognostic signature for oral squamous cell carcinoma. J Cell Physiol. 2020;235:3056–3068. doi: 10.1002/jcp.29210. [DOI] [PubMed] [Google Scholar]
- 65.An G, Wu F, Huang S, Feng L, Bai J, Gu S, Zhao X. Effects of CCL5 on the biological behavior of breast cancer and the mechanisms of its interaction with tumorassociated macrophages. Oncol Rep. 2019;42:2499–2511. doi: 10.3892/or.2019.7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Komohara Y, Fujiwara Y, Ohnishi K, Takeya M. Tumor-associated macrophages: Potential therapeutic targets for anti-cancer therapy. Adv Drug Deliv Rev. 2016;99:180–185. doi: 10.1016/j.addr.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 67.Cui R, Yue W, Lattime EC, Stein MN, Xu Q, Tan XL. Targeting tumor-associated macrophages to combat pancreatic cancer. Oncotarget. 2016;7:50735–50754. doi: 10.18632/oncotarget.9383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bosurgi L, Cao YG, Cabeza-Cabrerizo M, Tucci A, Hughes LD, Kong Y, Weinstein JS, Licona-Limon P, Schmid ET, Pelorosso F, et al. Macrophage function in tissue repair and remodeling requires IL-4 or IL-13 with apoptotic cells. Science. 2017;356:1072–1076. doi: 10.1126/science.aai8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The plasmid constructs db34a and dbSCR are available from the corresponding author upon reasonable request in accordance with institutional MTA approvals and all applicable rules and regulations. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.