Abstract
A novel strain belonging to the family Planctomycetaceae, designated V22T, was isolated from sediment of a seawater fish tank in Braunschweig, Germany. The isolate forms pink colonies on solid medium and displays common characteristics of planctomycetal strains, such as division by budding, formation of rosettes, a condensed nucleoid and presence of crateriform structures and fimbriae. Unusual invaginations of the cytoplasmic membrane and filamentous putative cytoskeletal elements were observed in thin sections analysed by transmission electron microscopy. Strain V22T is an aerobic heterotroph showing optimal growth at 30 °C and pH 8.5. During laboratory cultivations, strain V22T reached generation times of 10 h (maximal growth rate of 0.069 h−1). Its genome has a size of 5.2 Mb and a G + C content of 54.9%. Phylogenetically, the strain represents a novel genus and species in the family Planctomycetaceae, order Planctomycetales, class Planctomycetia. We propose the name Calycomorphotria hydatis gen. nov., sp. nov. for the novel taxon, represented by the type strain V22T (DSM 29767T = LMG 29080T).
Keywords: Marine bacteria, Planctomycetes, Cell biology, Membrane invaginations, Aggregation
Introduction
Planctomycetes, an ubiquitous phylum of bacteria of mostly aquatic origin (Wiegand et al. 2018), comprises species with an uncommon physiology and morphology among bacteria (Lage et al. 2019; Wiegand et al. 2020). Phylogenetically, the phylum Planctomycetes, together with Chlamydiae, Verrucomicrobia and others, forms the PVC superphylum (van Niftrik and Devos 2017). The phylum Planctomycetes is subdivided into the classes Candidatus Brocadiae, Phycisphaerae and Planctomycetia. The taxonomy of the family Planctomycetia was recently revised, which led to further subdivision into the orders Gemmatales, Isosphaerales, Pirellulales and Planctomycetales (Dedysh et al. 2019b). Species of Candidatus Brocadiae are capable of performing anaerobic ammonium oxidation (anammox) (Strous et al. 1999) and thereby convert ammonium to dinitrogen gas (Peeters and van Niftrik 2018). Members of the class Planctomycetia have been isolated from various aquatic biotic and abiotic surfaces in the past decade (Boersma et al. 2019; Bondoso et al. 2014, 2017; Kallscheuer et al. 2020; Peeters et al. 2020; Vollmers et al. 2017). Such species can be highly abundant on marine biotic surfaces, e.g. on macroscopic phototrophs (Bengtsson and Øvreås 2010). Since oceans are typically oligotrophic, species of the class Planctomycetia probably digest complex carbon substrates derived from the biotic surfaces to which they are attached (Jeske et al. 2013; Lachnit et al. 2013). For this purpose, they may utilise a specialised machinery for the uptake and intracellular digestion of complex polysaccharides (Boedeker et al. 2017), which could be a decisive advantage during competition for nutrients in their natural habitats.
In recent years, microscopic techniques and genetic tools (Jogler et al. 2011; Jogler and Jogler 2013; Rivas-Marin et al. 2016) have enabled a detailed analysis of the cell envelope architecture of Planctomycetes. Both, Planctomycetes and Verrucomicrobia, were shown to possess peptidoglycan (Jeske et al. 2015; Rast et al. 2017; van Teeseling et al. 2015). The cell envelope architecture of Planctomycetes is therefore similar to that of Gram-negative bacteria (Boedeker et al. 2017; Devos 2014a, b). Nevertheless, Planctomycetes display uncommon cell biological features, e.g. with regard to their mode of cell division. Members of the class Planctomycetia divide by budding, whereas Phycisphaerae divide by binary fission. Some species probably even switch between both modes of division (Wiegand et al. 2020). All characterised members of the phylum lack canonical divisome proteins including the otherwise universal FtsZ (Jogler et al. 2012; Pilhofer et al. 2008). The sizes of planctomycetal genomes range between 3 and 12 Mb (Ravin et al. 2018; Wiegand et al. 2018), while typically 40–55% of the gene products are annotated as hypothetical or uncharacterised proteins. Given such values, Planctomycetes represent an attractive subject for future research.
Here, we describe a novel strain, V22T, isolated from sediment of a seawater fish tank in Braunschweig, Germany. According to the results of our analysis, the strain represents a novel species and genus in the recently revisited family Planctomycetaceae, order Planctomycetales in the class Planctomycetia (Dedysh et al. 2019b).
Material and methods
Isolation of the novel strain and cultivation
For the isolation and cultivation of strain V22T, M1H NAG ASW medium was prepared as described by Boersma et al. (2019). Sediment and water from a seawater fish tank in Braunschweig, Germany (location: 52.2689 N 10.5268 E) were mixed by shaking and the water was subsequently plated on M1H NAG ASW plates containing 8 g/L gellan gum, 1000 mg/L streptomycin, 200 mg/L ampicillin and 20 mg/L cycloheximide, which were then incubated at 20 °C for several weeks. In order to verify that obtained strains are members of the phylum Planctomycetes, the 16S rRNA gene was amplified using colony-PCR and subsequently sequenced as previously described (Rast et al. 2017).
Determination of pH and temperature optimum
Cultivations for determination of the pH optimum were performed in M1H NAG ASW medium. A concentration of 100 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) was used for cultivations at pH 7, 7.5 and 8. For cultivation at pH 5 and 6, HEPES was replaced by 100 mM 2-(N-morpholino)ethanesulfonic acid (MES), whereas 100 mM N-cyclohexyl-2-aminoethanesulfonic acid (CHES) served as a buffering agent at pH 9 and 10. Cultivations for determination of the pH optimum were performed at 28 °C. Cultivations for determination of the temperature optimum were performed in standard M1H NAG ASW medium at pH 7.5.
Microscopy protocols
Phase contrast and field emission scanning electron microscopy were performed as previously described (Boersma et al. 2019). Thin sectioning, subsequent staining and transmission electron microscopy (TEM) were performed as previously described (Jogler et al. 2011).
Genome information
The genome sequence of strain V22T is available from GenBank under accession number CP036316. The 16S rRNA gene sequence of strain V22T can be found under accession number MK554537. Sequencing of the genome of strain V22T is described in a previously published study (Wiegand et al. 2020). Numbers of carbohydrate-active enzymes were obtained from the CAZY database (Lombard et al. 2014). Gene clusters potentially involved in the production of secondary metabolites were determined using antiSMASH 4.0 (Blin et al. 2017).
Phylogenetic analysis
16S rRNA gene sequence-based phylogeny was computed for strain V22T, the type strains of all described planctomycetal species (assessed in January 2020) and all isolates recently published and described (Boersma et al. 2019; Dedysh et al. 2019a, b; Kallscheuer et al. 2019a, b, 2020; Kohn et al. 2019; Peeters et al. 2020; Rensink et al. 2020). The 16S rRNA gene sequences were aligned with SINA (Pruesse et al. 2012) and the phylogenetic inference was calculated with RAxML (Stamatakis 2014) with a maximum likelihood approach with 1000 bootstraps, nucleotide substitution model GTR, gamma distributed rate variation and estimation of proportion of invariable sites (GTRGAMMAI option). For the multi-locus sequence analysis (MLSA) the unique single-copy core genome of the analysed genomes was determined with proteinortho5 (Lechner et al. 2011) with the ‘selfblast’ option enabled. The protein sequences of the resulting orthologous groups were aligned using MUSCLE v.3.8.31 (Edgar 2004). After clipping, partially aligned C- and N-terminal regions and poorly aligned internal regions were filtered using Gblocks (Castresana 2000). The final alignment was concatenated and clustered using FastTree (Price et al. 2009). The average nucleotide identity (ANI) was calculated using OrthoANI (Lee et al. 2016). The average amino acid identity (AAI) was calculated using the aai.rb script of the enveomics collection (Rodriguez-R and Konstantinidis 2016) and the percentage of conserved proteins (POCP) was calculated as described (Qin et al. 2014). The rpoB nucleotide sequences were taken from publicly available planctomycetal genome annotations and the sequence identities were determined as described (Bondoso et al. 2013). Upon extracting only those parts of the sequence that would have been sequenced with the described primer set, the alignment and matrix calculation was performed with Clustal Omega (Sievers et al. 2011).
Results and discussion
Phylogenetic inference
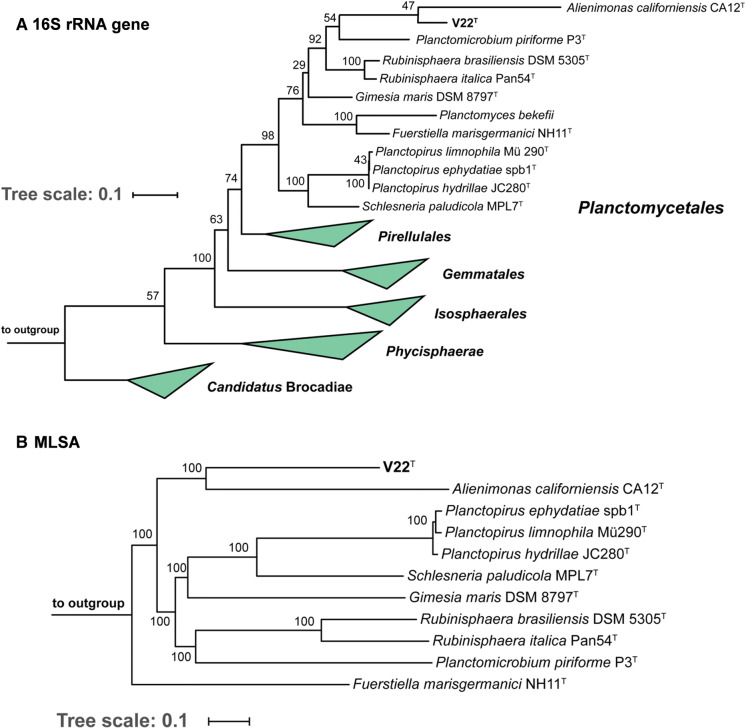
Based on 16S rRNA gene phylogeny (Fig. 1a), strain V22T belongs to the recently redefined family Planctomycetaceae, the sole family in the order Planctomycetales (Dedysh et al. 2019b). This family is currently formed by eight genera, namely Gimesia, Planctomyces, Planctopirus, Schlesneria, Planctomicrobium, Rubinisphaera, Fuerstiella and Alienimonas. Within this family, the current closest relative of strain V22T is Alienimonas californiensis CA12T (Boersma et al. 2019) as determined by both 16S rRNA gene analysis and MLSA (Fig. 1b). Strain V22T shares a 16S rRNA gene identity of 85.2% with A. californiensis, a value clearly below the genus threshold of 94.5% and even slightly below the family threshold of 86.5% (Yarza et al. 2014). This strongly supports delineation of strain V22T from the genus Alienimonas. In order to substantiate the delineation from known genera additional phylogenetic markers were taken into consideration, e.g. AAI, rpoB similarity and POCP. Similarity values obtained during comparison of strain V22T with A. californiensis and Planctomicrobium piriforme, a second close relative, are below the genus threshold values applied for rpoB (75.5–78.0%) (Kallscheuer et al. 2019b), AAI (60–80%) (Konstantinidis and Tiedje 2005), and POCP (50%) (Qin et al. 2014) (Fig. 2). ANI values of 67.2% and 67.1% confirm that strain V22T does not belong to A. californiensis or P. piriforme. Hence, all used methods support the delineation of strain V22T from members of already described genera in the family Planctomycetaceae.
Fig. 1.
Maximum likelihood phylogenetic analysis. Phylogenetic tree showing the position of strain V22T. 16S rRNA gene sequence- and MLSA-based phylogeny was computed as described in the Material and Methods section. Bootstrap values after 1,000 re-samplings (16S rRNA-based tree) are given at the nodes (in %). The outgroup consists of three 16S rRNA genes from the PVC superphylum outside of the phylum Planctomycetes (GenBank accession numbers AJ229235, NR_146840 and NR_027571). In the MLSA-based tree confidence values are given at the nodes (in %). Six species of the family Pirellulaceae served as outgroup
Fig. 2.
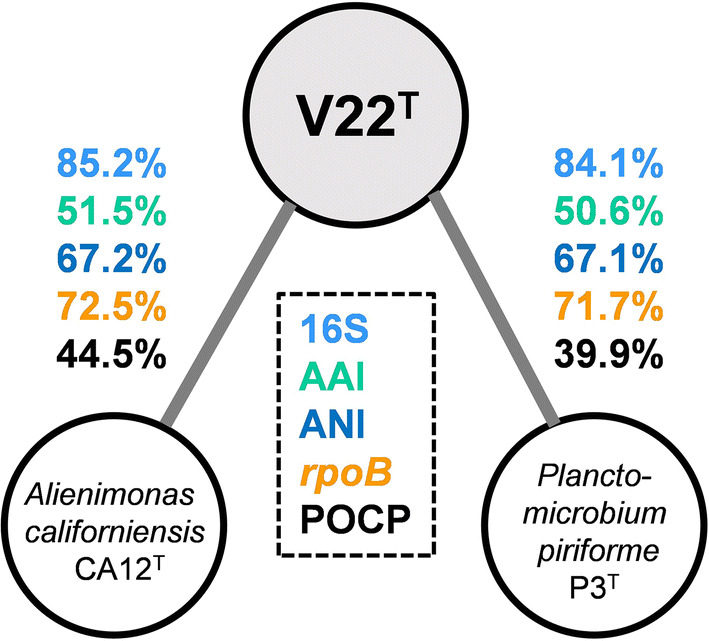
Phylogenetic marker values of strain V22T and its currently closest neighbours. The numbers give the similarity values shared between strain V22T, Alienimonas californiensis CA12T and Planctomicrobium pirifome P3T for 16S rRNA gene sequence identity (16S), average amino acid identity (AAI), average nucleotide identity (ANI), rpoB nucleotide sequence identity (rpoB) and percentage of conserved proteins (POCP)
Morphological, physiological and biochemical analyses
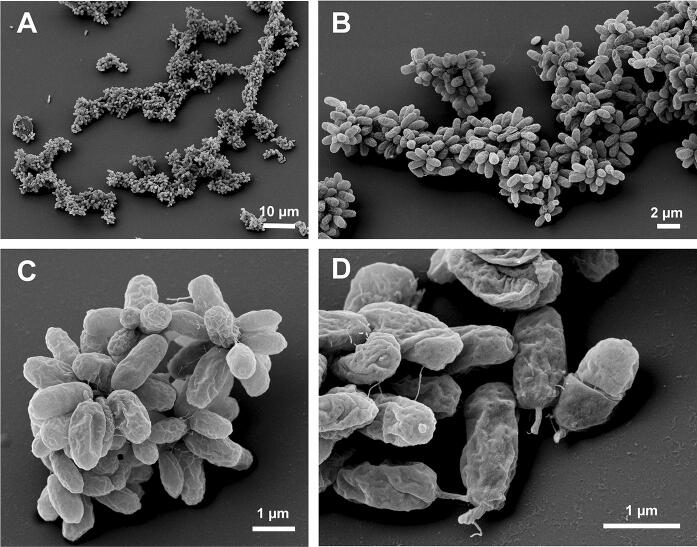
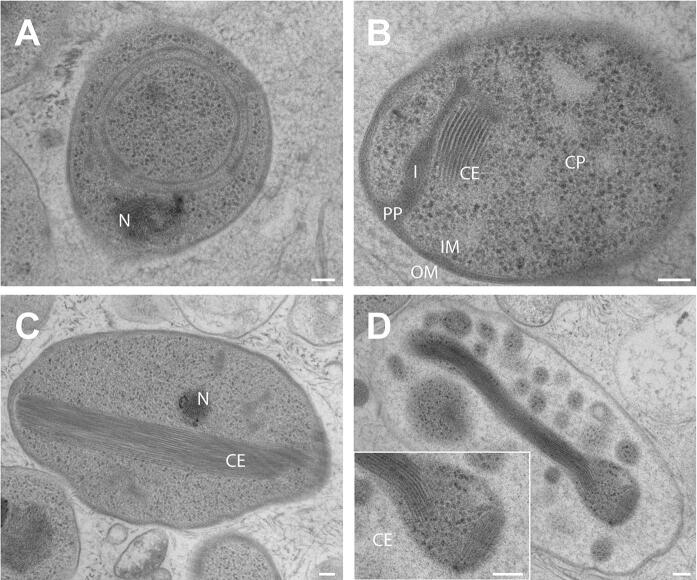
Basic features of strain V22T including cell morphology, growth and mechanism of cell division are summarised in Table 1 and compared to the currently closest neighbours A. californiensis and P. piriforme. Exponentially growing V22T cells were analysed using phase contrast, SEM and TEM analysis. Cells of strain V22T are round grain rice-shaped with an average size of 1.6 ± 0.3 µm (length) and 0.7 ± 0.2 µm (width) (Fig. 3a, c). Cells of A. californiensis CA12T and P. piriforme P3T are larger than V22T cells. Strain V22T forms rosettes of typically 6–15 cells, which assemble to characteristic linear or slightly branched chains (Fig. 4). Crateriform structures are present on the surface of all three strains, but their distribution is different (Table 1). TEM thin sections of strain V22T confirm a planctomycetal cell architecture with a condensed nucleoid and invaginations of the cytoplasmic membrane leading to an enlarged periplasmic space (Fig. 5). In the cytoplasm of strain V22T, cytoskeletal elements (CE) were observed (Fig. 5b–d). Such putative CEs form partly thick bundles (Fig. 5c), which seem to fill membrane cavities (Fig. 5d). The presence of such structures has been reported in Planctomycetes before (Lage et al. 2013).
Table 1.
Phenotypic and genotypic features of strain V22T compared to closely related strains
| Feature | V22T | A. californiensis CA12T | P. piriforme P3T |
|---|---|---|---|
| Phenotypic characteristics | |||
| Shape | Round grain rice-shaped | Spherical to ovoid | Ellipsoid to pear-shaped |
| Length [µm] | 1.6 ± 0.3 | 2.0 ± 0.2 | 1.7–2.8 |
| Width [µm] | 0.7 ± 0.2 | 1.5 ± 0.3 | 0.9–1.3 |
| Colour | Pink | Pink | White |
| Relation to oxygen | Aerobic | Aerobic | Aerobic |
| Temperature range (optimum) [°C] | 15–32 (30) | 10–36 (27) | 10–30 (20–28) |
| pH range (optimum) | 6.0–10.0 (8.5) | 6.0–9.0 (7.5) | 4.2–7.1 (6.0–6.5) |
| Division | Budding | Budding | Budding |
| Dimorphic life cycle | No | No | Yes |
| Motility | Yes | Yes | Yes |
| Crateriform structures | Overall | Overall, except for the pole at which the flagellum is located | At reproductive pole |
| Fimbriae | Few | Yes, polar | n.d. |
| Capsule | n.o. | n.o. | n.d. |
| Stalk | Short, opposite of budding pole | n.o. | Yes |
| Holdfast structure | n.o. | n.o. | n.d. |
| Genomic characteristics | |||
| Genome size [bp] | 5,163,473 | 5,475,215 | 6,317,004 |
| G + C [%] | 53.9 | 70.7 | 58.8 |
| Coding density [%] | 87.8 | 88.5 | 85.8 |
| Completeness [%] | 94.8 | 94.8 | 95.7 |
| Contamination [%] | 0 | 0 | 1.72 |
| Total genes | 4,376 | 4,382 | 5,117 |
| Protein-coding genes | 4,301 | 4,309 | 5,050 |
| Hypothetical proteins | 1,673 | 1,798 | 2,814 |
| 16S rRNA genes | 2 | 2 | 1 |
| tRNA genes | 64 | 65 | 53 |
Fig. 3.
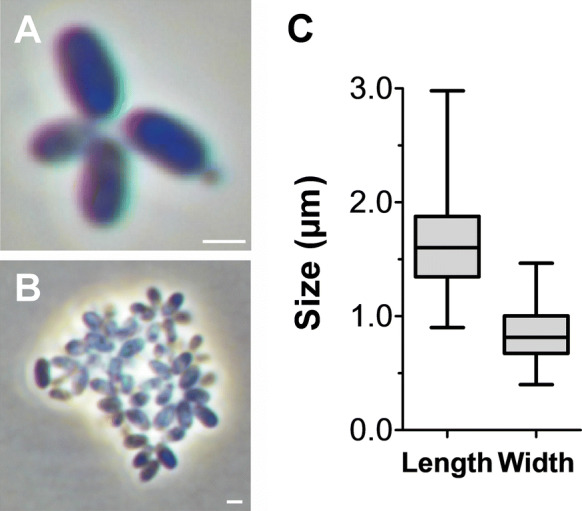
Light microscopy images and cell size plot of strain V22T. The mode of cell division (A) and a general overview of cell morphology (B) is shown in the pictures. The scale bar is 1 µm. For determination of the cell size (C) at least 100 representative cells were counted manually or by using a semi-automated object count tool
Fig. 4.
Electron microscopic images of strain V22T. Cells of strain V22T form rosettes typically comprising 6–15 cells. The rosettes assemble to linear or slightly branched chains. Separate scale bars are presented in each of the images
Fig. 5.
Transmission electron microscopy images of strain V22T. TEM images of thin sections showing the cell morphology of strain V22T. Abbreviations: CE: (potential) cytoskeletal elements, CP cytoplasm, I invagination, IM inner membrane, OM outer membrane, PP periplasmic space, N condensed nucleoid. The scale bar is 0.1 µm
The three compared strains are aerobic and motile. Strain V22T shares pink pigmentation with A. californiensis, whereas P. piriforme is non-pigmented. In M1H NAG ASW medium, strain V22T was able to growth over a temperature range of 15–32 °C and a pH range of 6.0–10.0. Strain V22T is heterotrophic with a mesophilic and slightly alkaliphilic growth profile. The new isolate originated from sediment of a seawater fish tank, which might indicate a facultatively anaerobic lifestyle. Whether growth under oxygen depletion is based on anaerobic respiration or fermentation needs further studies. The presence of menaquinone biosynthesis genes has been verified in the genome sequence, however, the production of menaquinones is not indicative of an anaerobic energy metabolism. Planctomycetales generally produce menaquinones (Sittig and Schlesner 1993). Strain V22T harbours the gene for a cobalamin-containing methionine synthase (metH, V22_33860). Since an open reading frame encoding the cobalamin-independent variant of the essential anabolic enzyme has not been identified, the organism might be a cobalamin auxotroph that salvages B12 vitamins from the environment.
Optimal growth of strain V22T was observed at 30 °C and pH 8.5 (Table 1), which led to a maximal growth rate of 0.069 h−1, corresponding to a generation time of 10 h. While the pH range allowing growth of strain V22T is similar to that of A. californiensis, P. piriforme prefers slightly acidic conditions. A. californiensis can grow up to temperatures of 36 °C, while the other two strains only grew up to 30–32 °C. Taken together, the three strains can be clearly differentiated using morphological and physiological properties.
Genomic characteristics and genome-based analysis of metabolic capabilities
The genome of strain V22T has a size of 5.2 Mb and a G + C content of 53.9%. The genome of strain V22T and A. californiensis have a comparable size, whereas the genome of P. piriforme is 1 Mb larger. The numbers of tRNA genes in the three strains range between 53–65. Planctomycetal genomes typically contain a high percentage of genes coding for hypothetical or uncharacterised proteins. The values are in a comparable range (39–42%) in case of strain V22T and A. californiensis, but lower than the 56% in P. piriforme. Hence, the latter has about 700 hypothetical proteins more than the other two strains. A. californiensis has a high G + C content of 71%, which is a distinctive criterion for delineation from strain V22T and P. piriforme, which have a moderate G + C content of 54–59%.
Based on the genome sequences of strain V22T and the two current close neighbours, we analysed the numbers of genes coding for enzymes putatively involved in the degradation of polysaccharides (carbohydrate-active enzymes) or in biosynthetic pathways for secondary metabolites (Table 2). Carbohydrate-active enzymes could not be analyzed for P. piriforme as the CAZY database only lists strains with complete genomes. Strain V22T and A. californiensis CA12T harbour 96–122 carbohydrate-active enzymes and show a similar distribution pattern with regard to the different families. Approximately 60% of the identified enzymes could be assigned to the glycosyl transferase family in both strains. Numbers in the other families vary only slightly between the two strains. Numbers of predicted clusters involved in the secondary metabolism of strain V22T are surprisingly low. The strain harbours only three terpenoid biosynthesis-related genes/gene clusters, which might be relevant for carotenoid biosynthesis in this strain. Genes coding for different types of polyketide synthases (PKSs) or non-ribosomal peptide synthetases (NRPSs) were not identified in strain V22T. The same is true for clusters putatively involved in the synthesis of ectoine, bacteriocins and resorcinol (Table 2). A. californiensis CA12T codes for an additional type III PKS compared to strain V22T, whereas a total number of 8 clusters are present in the genome of P. piriforme P3T. In conclusion, strain V22T either produces only a very small set of secondary metabolites or harbours as yet uncharacterised clusters which escaped the bioinformatic prediction.
Table 2.
Genome-based analysis of carbohydrate-active enzymes and secondary metabolite-related gene clusters
| Feature | V22T |
A. californiensis CA12T |
P. piriforme P3T |
|---|---|---|---|
| Genome size (Mb) | 5.16 | 5.48 | 6.32 |
| Carbohydrate-active enzymes | |||
| Glycoside hydrolase family | 23 | 28 | n.d. |
| Glycosyl transferase family | 53 | 71 | n.d. |
| Polysaccharide lyase family | 2 | 6 | n.d. |
| Carbohydrate esterase family | 10 | 11 | n.d. |
| Carbohydrate-binding module family | 8 | 6 | n.d. |
| Total | 96 | 122 | n.d. |
| Secondary metabolite-related gene clusters | |||
| Terpenoid | 3 | 3 | 4 |
| Type I PKS | 0 | 0 | 1 |
| Type II PKS | 0 | 0 | 0 |
| Type III PKS | 0 | 1 | 1 |
| NRPS | 0 | 0 | 0 |
| Bacteriocin | 0 | 0 | 2 |
| Ectoine | 0 | 0 | 0 |
| Resorcinol | 0 | 0 | 0 |
| Total | 3 | 4 | 8 |
Taken together, the observed differences during comparison of morphological and genomic features of strain V22T and its neighbours support the results of the phylogenetic analysis, which delineate strain V22T from members of the genera Alienimonas and Planctomicrobium. We thus conclude that strain V22T should be classified as the type strain of a novel species within a novel genus, for which we propose the name Calycomorphotria gen. nov., with the type species Calycomorphotria hydatis sp. nov.
Calycomorphotria gen. nov.
Ca.ly.co.mor.pho’tri.a. Gr. n. kalyx a bud; Gr. fem. n. morphotria a creator; N.L. fem. n. Calycomorphotria, a bacterium that generates buds.
Members of the genus have a Gram-negative cell envelope architecture, are motile aerobic heterotrophs with a mesophilic, neutrophilic or slightly alkaliphilic growth profile. Cells are round grain rice-shaped, divide by budding and contain fimbriae and crateriform structures. The genus belongs to the family Planctomycetaceae, order Planctomycetales, class Planctomycetia, phylum Planctomycetes. The type species of this genus is Calycomorphotria hydatis.
Calycomorphotria hydatis sp. nov.
hy’da.tis. Gr. n. hydor, -atos water; N.L. gen. n. hydatis of water; corresponding to the origin of the type strain from an aquatic environment.
Cells have an average size of 1.6 ± 0.3 × 0.7 ± 0.2 µm and form rosettes typically consisting of 6–15 cells. Rosettes assemble into linear or slightly branched aggregates. Cells contain crateriform structures covering the entire cell surface and a short stalk opposite of the budding pole. The cell plan features a condensed nucleoid, invaginations of the cytoplasmic membrane and cytoskeletal elements in the cytoplasm. Colonies are pink. Grows at ranges of 15–32 °C (optimum 30 °C) and at pH 6.0–10.0 (optimum 8.5). The G + C content of the type strain is 53.9%.
The type strain, V22T (DSM 29767T = LMG 29080T), was isolated from water and sediment of a seawater fish tank in June 2013. The type strain genome (acc. no. CP036316) and 16S rRNA gene sequence (acc. no. MK554537) are available from GenBank. The genome size of the type strain is 5,163,473 bp.
Acknowledgements
Open Access funding provided by Projekt DEAL. Part of this research was funded by the Deutsche Forschungsgemeinschaft grants KA 4967/1–1 and JO 893/4–1, grant ALWOP.308 of the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO), SIAM (Soehngen Institute for Anaerobic Microbiology) grant no. 024002002 and the Radboud Excellence fellowship. We thank Ina Schleicher for skilful technical assistance. Brian Tindall and Regine Fähnrich from the DSMZ as well as the BCCM/LMG Bacteria collection we thank for support during strain deposition. We are grateful to Elizabeth Benecchi from Harvard Medical School, EM facility, for supporting thin section and TEM observations.
Author contributions
T.S. and N.K. wrote the manuscript and analysed genome-encoded features, S.W. performed the phylogenetic analysis, A.H. and M.J. isolated the strains and performed the initial cultivation and strain deposition, S.H.P. and C.B. performed the light microscopic analysis and prepared the pictures, C.B. produced thin sections and analysed them by TEM. M.S.M.J. contributed to text preparation and revised the manuscript, M.R. performed the electron microscopic analysis and prepared the SEM pictures, C.J. supervised A.H. and C.B. and planned and managed the study. All authors read and approved the final version of the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
This article does not contain any studies with animals performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Bengtsson MM, Øvreås L. Planctomycetes dominate biofilms on surfaces of the kelp Laminaria hyperborea. BMC Microbiol. 2010;10:261. doi: 10.1186/1471-2180-10-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K, Wolf T, Chevrette MG, Lu X, Schwalen CJ, Kautsar SA, Suarez Duran HG, de Los Santos EL, Kim HU, Nave M. antiSMASH 4.0—improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017;45:W36–W41. doi: 10.1093/nar/gkx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boedeker C, Schuler M, Reintjes G, Jeske O, van Teeseling MC, Jogler M, Rast P, Borchert D, Devos DP, Kucklick M, Schaffer M, Kolter R, van Niftrik L, Engelmann S, Amann R, Rohde M, Engelhardt H, Jogler C. Determining the bacterial cell biology of Planctomycetes. Nat Commun. 2017;8:14853. doi: 10.1038/ncomms14853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boersma AS, Kallscheuer N, Wiegand S, Rast P, Peeters SH, Mesman RJ, Heuer A, Boedeker C, Jetten MS, Rohde M, Jogler M. Alienimonas californiensis gen. nov. sp. nov., a novel Planctomycete isolated from the kelp forest in Monterey Bay. Antonie van Leeuwenhoek. Jogler C. 2019 doi: 10.1007/s10482-019-01367-4. [DOI] [PubMed] [Google Scholar]
- Bondoso J, Balague V, Gasol JM, Lage OM. Community composition of the Planctomycetes associated with different macroalgae. FEMS Microbiol Ecol. 2014;88:445–456. doi: 10.1111/1574-6941.12258. [DOI] [PubMed] [Google Scholar]
- Bondoso J, Godoy-Vitorino F, Balague V, Gasol JM, Harder J, Lage OM. Epiphytic Planctomycetes communities associated with three main groups of macroalgae. FEMS Microbiol Ecol. 2017;93:fiw255. doi: 10.1093/femsec/fiw255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondoso J, Harder J, Lage OM. rpoB gene as a novel molecular marker to infer phylogeny in Planctomycetales. Antonie van Leeuwenhoek. 2013;104:477–488. doi: 10.1007/s10482-013-9980-7. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Dedysh SN, Henke P, Ivanova AA, Kulichevskaya IS, Philippov DA, Meier-Kolthoff JP, Göker M, Huang S, Overmann J. 100-year-old enigma solved: identification, genomic characterization and biogeography of the yet uncultured Planctomyces bekefii. Environ Microbiol. 2019;22:198–211. doi: 10.1111/1462-2920.14838. [DOI] [PubMed] [Google Scholar]
- Dedysh, SN, Kulichevskaya IS, Beletsky AV, Ivanova AA, Rijpstra WIC, Damsté JSS, Mardanov AV, Ravin NV (2019b) Lacipirellula parvula gen. nov., sp. nov., representing a lineage of Planctomycetes widespread in low-oxygen habitats, description of the family Lacipirellulaceae fam. nov. and proposal of the orders Pirellulales ord. nov., Gemmatales ord. nov. and Isosphaerales ord. nov. Syst Appl Microbiol 43: 126050. [DOI] [PMC free article] [PubMed]
- Devos DP. PVC bacteria: variation of, but not exception to, the gram-negative cell plan. Trends Microbiol. 2014;22:14–20. doi: 10.1016/j.tim.2013.10.008. [DOI] [PubMed] [Google Scholar]
- Devos DP. Re-interpretation of the evidence for the PVC cell plan supports a gram-negative origin. Antonie Van Leeuwenhoek. 2014;105:271–274. doi: 10.1007/s10482-013-0087-y. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeske O, Jogler M, Petersen J, Sikorski J, Jogler C. From genome mining to phenotypic microarrays: Planctomycetes as source for novel bioactive molecules. Antonie Van Leeuwenhoek. 2013;104:551–567. doi: 10.1007/s10482-013-0007-1. [DOI] [PubMed] [Google Scholar]
- Jeske O, Schüler M, Schumann P, Schneider A, Boedeker C, Jogler M, Bollschweiler D, Rohde M, Mayer C, Engelhardt H, Spring S, Jogler C. Planctomycetes do possess a peptidoglycan cell wall. Nat Commun. 2015;6:7116. doi: 10.1038/ncomms8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jogler C, Glockner FO, Kolter R. Characterization of Planctomyces limnophilus and development of genetic tools for its manipulation establish it as a model species for the phylum Planctomycetes. Appl Environ Microbiol. 2011;77:5826–5829. doi: 10.1128/AEM.05132-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jogler C, Waldmann J, Huang X, Jogler M, Glöckner FO, Mascher T, Kolter R. Identification of proteins likely to be involved in morphogenesis, cell division, and signal transduction in Planctomycetes by comparative genomics. J Bacteriol. 2012;194:6419–6430. doi: 10.1128/JB.01325-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jogler M, Jogler C. Towards the development of genetic tools for Planctomycetes. In: Fuerst JA, editor. Planctomycetes: cell structure. Origins and Biology: Springer; 2013. pp. 141–164. [Google Scholar]
- Kallscheuer N, Jogler M, Wiegand S, Peeters SH, Heuer A, Boedeker C, Jetten MS, Rohde M, Jogler C. Three novel Rubripirellula species isolated from plastic particles submerged in the Baltic Sea and the estuary of the river Warnow in northern Germany. Antonie Van Leeuwenhoek. 2019 doi: 10.1007/s10482-019-01368-3. [DOI] [PubMed] [Google Scholar]
- Kallscheuer, N, Wiegand S, Heuer A, Rensink S, Boersma AS, Jogler M, Boedeker C, Peeters SH, Rast P, Jetten MS, Rohde M, Jogler C (2020) Blastopirellula retiformator sp. nov. isolated from the shallow-sea hydrothermal vent system close to Panarea Island. Antonie van Leeuwenhoek, 10.1007/s10482-019-01377-2 [DOI] [PubMed]
- Kallscheuer, N, Wiegand S, Peeters SH, Jogler M, Boedeker C, Heuer A, Rast P, Jetten MS, Rohde M, Jogler C (2019b) Description of three bacterial strains belonging to the new genus Novipirellula gen. nov., reclassificiation of Rhodopirellula rosea and Rhodopirellula caenicola and readjustment of the genus threshold of the phylogenetic marker rpoB for Planctomycetaceae. Antonie van Leeuwenhoek, 10.1007/s10482-019-01374-5 [DOI] [PubMed]
- Kohn T, Wiegand S, Boedeker C, Rast P, Heuer A, Schüler M, Rohde C, Müller R-W, Brümmer F, Rohde M, Engelhardt H, Jogler M, Jogler C. Planctopirus ephydatiae, a novel planctomycetal species isolated from a freshwater sponge. Syst Appl Microbiol. 2019;43:126022. doi: 10.1016/j.syapm.2019.126022. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. Towards a genome-based taxonomy for prokaryotes. J Bacteriol. 2005;187:6258–6264. doi: 10.1128/JB.187.18.6258-6264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachnit T, Fischer M, Kunzel S, Baines JF, Harder T. Compounds associated with algal surfaces mediate epiphytic colonization of the marine macroalga Fucus vesiculosus. FEMS Microbiol Ecol. 2013;84:411–420. doi: 10.1111/1574-6941.12071. [DOI] [PubMed] [Google Scholar]
- Lage OM, Bondoso J, Lobo-da-Cunha A. Insights into the ultrastructural morphology of novel Planctomycetes. Antonie Van Leeuwenhoek. 2013;104:467–476. doi: 10.1007/s10482-013-9969-2. [DOI] [PubMed] [Google Scholar]
- Lage OM, van Niftrik L, Jogler C, Devos DP (2019) Planctomycetes. In: Thomas M. Schmidt (ed) Reference module in life sciences, Elsevier, pp.614–626. 4th edn 10.1016/B978-0-12-809633-8.90689-7
- Lechner M, Findeiss S, Steiner L, Marz M, Stadler PF, Prohaska SJ. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinform. 2011;12:124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Ouk Kim Y, Park SC, Chun J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66:1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters SH, van Niftrik L. Trending topics and open questions in anaerobic ammonium oxidation. Curr Opin Chem Biol. 2018;49:45–52. doi: 10.1016/j.cbpa.2018.09.022. [DOI] [PubMed] [Google Scholar]
- Peeters SH, Wiegand S, Kallscheuer N, Jogler M, Heuer A, Jetten MS, Rast P, Boedeker C, Rohde M, Jogler C. Three marine strains constitute the novel genus and species Crateriforma conspicua in the phylum Planctomycetes. Antonie Van Leeuwenhoek. 2020 doi: 10.1007/s10482-019-01375-4. [DOI] [PubMed] [Google Scholar]
- Pilhofer M, Rappl K, Eckl C, Bauer AP, Ludwig W, Schleifer KH, Petroni G. Characterization and evolution of cell division and cell wall synthesis genes in the bacterial phyla Verrucomicrobia, Lentisphaerae, Chlamydiae, and Planctomycetes and phylogenetic comparison with rRNA genes. J Bacteriol. 2008;190:3192–3202. doi: 10.1128/JB.01797-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Peplies J, Glöckner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Q-L, Xie B-B, Zhang X-Y, Chen X-L, Zhou B-C, Zhou J, Oren A, Zhang Y-Z. A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol. 2014;196:2210–2215. doi: 10.1128/JB.01688-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rast, P, Glockner I, Boedeker C, Jeske O, Wiegand S, Reinhardt R, Schumann P, Rohde M, Spring S, Glockner FO, Jogler C, Jogler M (2017) Three Novel Species with Peptidoglycan Cell Walls form the New Genus Lacunisphaera gen. nov. in the Family Opitutaceae of the Verrucomicrobial Subdivision 4. Front Microbiol 8: 202 [DOI] [PMC free article] [PubMed]
- Ravin NV, Rakitin AL, Ivanova AA, Beletsky AV, Kulichevskaya IS, Mardanov AV, Dedysh SN. Genome analysis of Fimbriiglobus ruber SP5T, a planctomycete with confirmed chitinolytic capability. Appl Environ Microbiol. 2018;84:e02645–e2717. doi: 10.1128/AEM.02645-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensink, S, Wiegand S, Kallscheuer N, Rast P, Peeters SH, Heuer A, Boedeker C, Jetten MS, Rohde M, Jogler M, Jogler C (2020) Description of the novel planctomycetal genus Bremerella, containing Bremerella volcania sp. nov., isolated from an active volcanic site, and reclassification of Blastopirellula cremea as Bremerella cremea comb. nov. Antonie van Leeuwenhoek. 10.1007/s10482-019-01378-1 [DOI] [PubMed]
- Rivas-Marin E, Canosa I, Santero E, Devos DP. Development of genetic tools for the manipulation of the planctomycetes. Front Microbiol. 2016;7:914. doi: 10.3389/fmicb.2016.00914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-R LM, Konstantinidis KT (2016) The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Preprints 4:e1900v1
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittig M, Schlesner H. Chemotaxonomic investigation of various prosthecate and/or budding bacteria. Syst Appl Microbiol. 1993;16:92–103. doi: 10.1016/S0723-2020(11)80253-5. [DOI] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strous M, Fuerst JA, Kramer EH, Logemann S, Muyzer G, van de Pas-Schoonen KT, Webb R, Kuenen JG, Jetten MS. Missing lithotroph identified as new planctomycete. Nature. 1999;400:446–449. doi: 10.1038/22749. [DOI] [PubMed] [Google Scholar]
- van Niftrik L, Devos DP. Planctomycetes-Verrucomicrobia-Chlamydiae bacterial superphylum: new model organisms for evolutionary cell biology. Front Microbiol. 2017;8:1458. doi: 10.3389/fmicb.2017.01458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Teeseling MC, Mesman RJ, Kuru E, Espaillat A, Cava F, Brun YV, VanNieuwenhze MS, Kartal B, van Niftrik L. Anammox Planctomycetes have a peptidoglycan cell wall. Nat Commun. 2015;6:6878. doi: 10.1038/ncomms7878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmers J, Frentrup M, Rast P, Jogler C, Kaster AK. Untangling genomes of novel planctomycetal and verrucomicrobial species from monterey bay kelp forest metagenomes by refined binning. Front Microbiol. 2017;8:472. doi: 10.3389/fmicb.2017.00472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand S, Jogler M, Boedeker C, Pinto D, Vollmers J, Rivas-Marín E, Kohn T, Peeters SH, Heuer A, Rast P, Oberbeckmann S, Bunk B, Jeske O, Meyerdierks A, Storesund JE, Kallscheuer N, Lücker S, Lage OM, Pohl T, Merkel BJ, Hornburger P, Müller R-W, Brümmer F, Labrenz M, Spormann AM, Op den Camp HJM, Overmann J, Amann R, Jetten MSM, Mascher T, Medema MH, Devos DP, Kaster A-K, Øvreås L, Rohde M, Galperin MY, Jogler C. Cultivation and functional characterization of 79 planctomycetes uncovers their unique biology. Nat Microbiol. 2020;5:126–140. doi: 10.1038/s41564-019-0588-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand S, Jogler M, Jogler C. On the maverick Planctomycetes. FEMS Microbiol Rev. 2018;42:739–760. doi: 10.1093/femsre/fuy029. [DOI] [PubMed] [Google Scholar]
- Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, Schleifer KH, Whitman WB, Euzeby J, Amann R, Rossello-Mora R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol. 2014;12:635–645. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]