Abstract
The coronavirus disease 2019 (COVID-19) pandemic is still spreading worldwide, affecting several million people. Unlike the previous two coronavirus outbreaks, COVID-19 has caused several thousand deaths for respiratory and multiple organ failure. As specifically concerns this latest infectious pathology, laboratory medicine can provide a substantial contribution to diagnosing an acute severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection through molecular testing, establishing the presence and extent of an immune response against the virus, mostly through serological testing. However, it can also help to predict the risk of unfavorable disease progression by measuring some conventional laboratory tests and, last but not least, can provide reliable therapeutic guidance. This article is hence aimed at offering recent updates on the important role and value of laboratory investigations in COVID-19, also providing information on some hot topics such as virus RNA detection in different biological samples, causes of recurrent positivity of reverse-transcription polymerase chain reaction (RT-PCR), potential strategies for enhancing the throughput of molecular testing (i.e., pre-test probability assessment, sample pooling, use of rapid tests), as well as pragmatic indications for enhancing the quality and value of serological testing and laboratory-based monitoring. (www.actabiomedica.it)
Keywords: coronavirus, COVID-19, laboratory medicine, laboratory test
Introduction
The coronavirus disease 2019 (COVID-19) pandemic continues to spread across the globe, afflicting millions of people (1). This latest outbreak, which follows the severe acute respiratory syndrome (SARS) in 2002-2003 and the Middle-East respiratory syndrome in 2012, is caused by another member of the coronaviridae family, called the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) to distinguish it from the former homologous coronavirus which caused the SARS epidemic (i.e., SARS-CoV-1) (2). Unlike the two previous outbreaks, COVID-19 has enormously and rapidly spread worldwide, thus contributing to cause over a half-million deaths via both respiratory and multiorgan failure, especially among genetically or clinically predisposed individuals (3, 4). As previously highlighted, laboratory medicine is at the very core of SARS-CoV-2 infection diagnostics (5), and this article is hence aimed at providing recent updates on the important role and value of laboratory investigations in COVID-19, with particular focus on some hot topics that will be specifically addressed in the following parts of this manuscript.
Laboratory investigations in COVID-19
A modern and pragmatic definition of laboratory medicine is that recently proposed by Lippi and Plebani, according to which in vitro diagnostics tests are used for screening, diagnosing, staging, prognosticating and monitoring the vast majority of human disease (6), most certainly including COVID-19 (7). Laboratory medicine can provide a substantial contribution to the diagnosis of acute SARS-CoV-2 infection via molecular testing, establishing the presence and extent of immune response against the virus, mostly using serological testing, as well as helping to predict the risk of unfavorable disease progression by measuring a number of conventional laboratory tests, and, last but not least, can provide a reliable guidance on many therapeutic options (8). On a research level, laboratory medicine is helping to advance our understanding of the mechanisms by which SARS-CoV-2 cause disease, propelling us towards development of targeted therapies.
Molecular testing
As currently endorsed by the World Health Organization (WHO), the identification of viral RNA through nucleic acid amplification tests (NAATs), such as reverse-transcription polymerase chain reaction (RT-PCR) in a patient’s biological materials, remains the gold standard for identifying acute SARS-CoV-2 infection. A final diagnosis of COVID-19 could hence be made with a positive result of NAATs targeting at least two different genes, one of which is SARS-CoV-2 specific (e.g., one among N, E, S and RdRP genes) (9). The preferred approach for detecting the virus is based on the collection of upper or lower respiratory tract specimens, thus encompassing (i) two oropharyngeal and nasopharyngeal swabs in ambulatory patients and/or (ii) sputum bronchoalveolar lavage or endotracheal aspirate in patients with severe respiratory illness, (9). The value of providing quantitative results of nucleic acid testing is an important aspect that needs to be emphasized with respect to clinical management of COVID-19. For example, in an interesting study carried out by Magleby and colleagues (10), a higher viral load expressed as cycle threshold (Ct) values from a RT-PCR assay on nasopharyngeal swabs samples collected upon patient admission was found to be associated with an over 6-fold higher risk of death (odds ratio [OR], 6.05; 95% CI, 2.92-12.52) and with a nearly 3-fold higher risk of intubation (OR, 2.73; 95% CI, 1.68-4.44). Further evidence in support of providing quantitative results and viral load monitoring both intra- and inter-individually has been provided by Clementi et al. (11), who demonstrated that the significant reduction of hospitalizations and admissions to the intensive care unit (ICU) recorded in two different periods in Milan, the Italian epicenter of the COVID-19 crisis, can also be explained by a remarkable reduction in the viral load among subjects infected with SARS-CoV-2. No sufficient evidence exists to suggest that rapid immunoassays for SARS-CoV-2 antigen detection would have any role in the COVID-19 diagnostics, as the diagnostic performance of many commercially available tests remains regrettably poor, with a sensitivity frequently around 30% (12-14).
Although molecular testing remains the most used approach worldwide for diagnosing SARS-CoV-2 infection, the diagnostic accuracy of NAAT is still suboptimal (15) as it can be plagued by several biological, pre-analytical, analytical and post-analytical complications. Besides the variable detectability of SARS-CoV-2 in different sample matrices, a variable that will be more specifically addressed below, additional sources of bias in diagnosing COVID-19 with molecular biology techniques encompass anatomic problems (e.g., nasal septal deformities or obstructions), inadequate procedures for collecting, handling, transport, storing, and preparing the samples, the collection of inappropriate material for amount or quality, the presence of various substances that may interfere with nucleic acids amplification, clerical and instrumental errors, sample contamination, analysis carried out outside the diagnostic window, genetic rearrangement recombination of the virus (leading to impairment of probes annealing), use of non-analytically and/or non-clinically validated tests, poor harmonization of reagents (e.g., probes and primers), as well as misinterpretation of test results (e.g., for adoption of unsuitable diagnostic thresholds and/or reference ranges) (16).
It should be clearly noted that a negative RT-PCT test on oropharyngeal and nasopharyngeal swabs does not rule out the possibility of being infected by SARS-CoV-2, especially among patients with high clinical suspicion. In such cases, clinics and imaging testing should guide the clinical decision making, and molecular testing should be repeated at least twice, whereby repeated testing has been clearly shown to considerably increase the likelihood of a positive result, reaching 99% sensitivity at the fourth consecutive test (17).
SARS-CoV-2 RNA detection in biological samples
The heterogeneous detection of viral RNA in various biological materials is perhaps most important of limitation of molecular testing, as summarized in table 1. Although SARS-CoV-2 RNA can be detected in bronchoalveolar lavage fluid in around 90% of COVID-19 patients, nucleic acid detectability in other biological materials is lower. In particular, the positivity rate of nasopharyngeal and oropharyngeal swabs is between 40-70% and 30-50%, respectively, yielding to a combined diagnostic sensitivity of around 70-80% (18). This implies that between 20-30% of patients with acute SARS-CoV-2 infection may go undetected using this diagnostic strategy, thus persuading the WHO to affirm that one or more negative NAAT results will not exclude the potential of SARS-CoV-2 infection (9). With respect to other biological materials, the presence of SARS-CoV-2 in blood has been comprehensively assessed in a recent meta-analysis published by Andersson and colleagues (19). According to the preliminary literature review, inclusive of 22 different studies, 10% (95% CI, 5-18%) positivity of RT-PCR for SARS-CoV-2 was observed in the blood of COVID-19 patients. Viremia could be more frequently observed in association with severe illness, especially in patients needing intensive care or with deterioration of clinical status. In a subsequent investigation published in the same article and based on data of 212 consecutive COVID-19 patients, SARS-CoV-2 RNA could be detected by RT-PCR in 12.7% serum samples, a percentage decreasing to 0% after 28 days. Interestingly, among the PCR-positive samples, the Ct value was generally low, suggesting low viral RNA copy numbers. This study also found that inoculation of PCR-positive samples in cell culture failed to generate cytopathic effects, thus suggesting low infectivity. Although the rate of fecal specimens positivity for SARS-CoV-2 in patients with COVID-19 is a matter of debate, a recent meta-analysis has concluded that it may be observed in 34-44%, with a trend towards a higher positivity rate in patients with severe as opposed to mild illness (20). Controversial evidence has also been published on the rate of urine positivity for SARS-CoV-2 in patients with COVID-19. In a meta-analysis published by Chan et al. (21), the percentage of positive urine samples has been calculated at ~6%, with a confidence of interval comprised between 3-9%. Overall, the risk of detecting SARS-CoV-2 in urine seems more likely in patients with renal colonization and concurrent kidney injury. Importantly, it must always be clearly acknowledged that the identification of viral RNA in a particular biological sample may not reflect the concomitant presence of viable viral particles (22).
Table 1.
Rate of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) reverse-transcription polymerase chain reaction (RT-PCR) positivity in different biological sources collected from patients with coronavirus disease 2019 (COVID-19)
| Biological source | Detection rate |
| Bronchoalveolar lavage fluid | >90% |
| Saliva | 80-100% |
| Sputum | 70-80% |
| Nasopharyngeal AND oropharyngeal swabs | 70-80% |
| Nasal swabs | 40-70% |
| Pharyngeal swabs | 30-50% |
| Stool | 30-45% |
| Throat wash | ~30% |
| Blood | 5-20% |
| Urine | 3-9% |
| Breastmilk | <1% |
| Peritoneal fluid | <1% |
| Semen | <1% |
Recurrence of RT-PCR positivity
Recurrence of SARS-CoV-2 positivity is an important issue, which refers to the possibility that a patient who tested negative with one or more nasopharyngeal and oropharyngeal swabs, may then turn positive again, later. Some studies have been published on this matter, with recurrence rates of SARS-CoV-2 positivity comprised between 9.1% and 21.4% (23-27). On average, it can hence be estimated that the mean risk of retesting positive for SARS-CoV-2 after a negative test would be ~12%. In most patients, this later recurrence of RT-PCR positivity may occur within one month from the last negative swab. Several potential causes can be identified for justifying the recurrence of SARS-CoV-2 positivity, which can be basically attributed to SARS-CoV-2 reinfection, delayed viral shedding in the lower respiratory tract, or technical issues, as summarized in Table 2.
Table 2.
Leading causes of recurrence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) reverse-transcription polymerase chain reaction (RT-PCR) positivity in nasopharyngeal and oropharyngeal swabs
|
Enhancing the throughput of molecular testing
The need to enhance the throughput of molecular testing in certain areas and under some critical situations is an essential aspect in diagnosing SARS-CoV-2 infections. Some interesting strategies have been proposed, such as assessing pre-test probability, using kits without RNA extraction and purification, and/or sample pooling.
The first interesting approach to improve the throughput of molecular testing entails the integration of some predictive demographic, clinical and diagnostic, with calculation of a final score summarizing the likelihood that one subject would test positive with SARS-CoV-2 RT-PCR. One of the first and perhaps most widely used of these models, called “Corona Score,” incorporates age, sex, presence of infiltrate at chest X-ray, along with the laboratory values of C reactive protein (CRP), lactate dehydrogenase (LDH), ferritin, neutrophil and lymphocyte counts. In a real-life scenario, this model was found to have 91% accuracy, 0.96 sensitivity and 0.95 specificity for discriminating patients testing positive or negative for SARS-CoV-2 by RT-PCR at emergency department admission (28). In another study, Qin et al developed the Predicted Score for COVID-19 (PSC-19) (29), which is based on history of exposure, leukocyte count, peripheral lesions and crazy-paving pattern on chest computed tomography (CT). This model displayed 91% accuracy, 0.88 sensitivity and 0.92 specificity for identifying COVID-19-positive cases. In another model, developed by Jehi et al. (30), the authors also integrated a number of environmental, behavioral, demographic, clinical and laboratory variables, to generate a final score which would also reflect the probability of being infected by SARS-CoV-2. This model exhibited 84% accuracy, 0.77 sensitivity and 0.96 specificity. Further validation will be necessary to determine whether these resources are reliable and can be clinically useful. However, in the meantime, it should be acknowledged that the use of such testing algorithms appears a promising approach.
Since fast and accurate identification of SARS-CoV-2 infection is pivotal for case isolation and/or clinical management, some innovative NAAT kits have been developed, which halve the turnaround time by eliminating some pre-analytical steps, such as RNA extraction and purification. Preliminary data shows that some of these assays display sufficient accuracy for being used as screening tests in clinical practice (e.g., concordance of over 90% and correlation of around 0.84 with the reference RT-PCR technique, especially in nasopharyngeal swabs and saliva) (31).
Sample pooling is another interesting opportunity, recently proposed for purpose of enhancing the throughput of SARS-CoV-2 molecular testing (32). Briefly, this strategy encompasses the process of pooling and then screening a variable number of clinical specimens, whilst the individual samples that have been used for the pool are only assayed when their cumulative result is positive. Some published reports have recently highlighted that, under some specific conditions, this strategy may be feasible for counteracting the local shortage of reagents or limited test availability in intensive testing areas to enhance the throughput and reduce the turnaround time, well as for supporting cost-saving policies. Cherif and colleagues calculated that an optimal strategy would encompass the creation of pools with an optimal size of 13 patients in a real-life scenario, where the prevalence of COVID-19 is around 1%, and the RT-PCR on oropharyngeal and nasopharyngeal swabs has a detection limit of at least 1100 RNA copies/mL, displaying 70% diagnostic sensitivity (33). Merging this evidence with that of some other studies (32, 34, 35), including the recent technical note endorsed by the European Center for Disease Prevention and Control (ECDC) (36), some practical considerations can be delineated for sample pooling, as summarized in more details in Figure 1 and Table 3. Among the various aspects that need to be considered in practicing this process, the foremost issues that need to be clearly highlighted for safeguarding the quality of testing is that (i) this strategy should be used for population screening, but not for testing people with high suspicion of SARS-CoV-2 infection (i.e., symptomatic, reporting strict contacts with COVID-19 patients), (ii) the assay should have been accurately validated for this purpose (e.g., characterized by adequate analytical sensibility), and (iii) traceability to the original sample (with availability of a second aliquot, that can be tested when the pool is positive) must be guaranteed. It is then evident that local protocols must be specifically designed according to clinical purpose, disease prevalence, testing capacity, resource availability, laboratory organization and assay characteristics (37).
Figure 1.
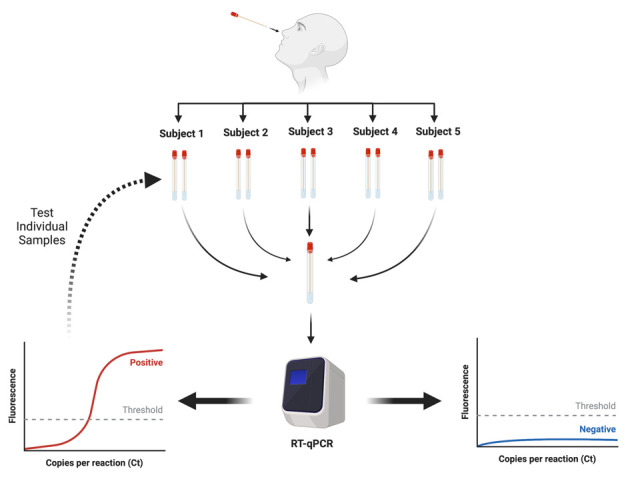
Sample pooling procedure for screening severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with reverse-transcription polymerase chain reaction (RT-PCR)
Table 3.
Pragmatic indications for pooling samples for molecular diagnosis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)
|
Serological testing
Serological testing can be theoretically outlined as a type of diagnostic testing aimed at detecting the presence, nature and even the extent of a humoral immune response against a particular pathogen (5). In keeping with this congenial definition, serological testing in patients with SARS-CoV-2 infection cannot be used as a surrogate of viral RNA identification for specific etiological diagnosis of COVID-19, but instead for determining whether or not a person has been infected by SARS-CoV-2 and has then developed an immune response against the virus itself (8). Some critical biological and technical issues need to be clarified before serological testing can be diffusely used.
The first important aspect is the different rates of positivity and the kinetics of the different classes of antibodies. Although it seems now virtually certain that anti-SARS-CoV-2 immunoglobulins (Ig) G will develop in virtually all patients (i.e., >99%) after 2-3 weeks from the start of viral infection, the rate of positivity and kinetics of IgM is instead more heterogeneous. Several studies showed that the percentage of IgM-positive COVID-19 patients is comprised between 50% and 80%, and these immunoglobulins begin to be measurable between three and ten days after symptom onset. Interestingly, IgG also tends to become measurable within the first week after the onset of symptoms. Therefore, whether or not assessing the IgM response may help in COVID-19 diagnostics and serological surveillance remains a matter of debate. This consideration is supported by the findings of an interesting study published by Xu and colleagues (38), who clearly described an early and sustained onset of IgG titer in patients with SARS-CoV-2 infection, greater than that of IgM. The rate of positivity after seven days was found to be only slightly higher than 50% for IgG and 40% of IgM, increasing to approximately 90% for IgG and 75% for IgM after two weeks, thus highlighting that the immune response generated in patients with SARS-CoV-2 infection may not be comparable with that of other viral infections. Compelling evidence has also emerged, that the low IgM positivity is perhaps attributable to attenuated disease severity since over 80% of severe COVID-19 cases can be found positive for this immunoglobulin class, while IgM positivity is nearly half that (e.g., slightly higher than 40%) in patients with mild illness (39).
The exponential growth of laboratory-based immunoassays is a second import aspect in SARS-CoV-2 serology. Theoretically, these assays can be substantially divided into laboratory-based and rapid tests. A comprehensive analysis of the diagnostic performance of many different anti-SARS-CoV-2 immunoassays is available in the Cochrane Database of Systematic Reviews (40). Within 7 days from the onset of symptoms, the diagnostic sensitivity of IgG, IgM, IgA and total antibodies was found to be 29.7%, 23.2%, 28.4% and 24.5%, respectively, thus largely insufficient because the measurement of any of these immunoglobulins could be not considered a valuable diagnostic tool in this phase of the infection. The diagnostic sensitivity of these antibodies then increased to 66.5% for IgG, 58.4% for IgM, 78.1% for IgA and 84.0% for total antibodies between 1 and 2 weeks since the onset of symptoms, then to 88.2% for IgG, 75.4% for IgM, 98.7% for IgA and 98.1% for total antibodies between 2 and 3 weeks, and then further to 86.7% for IgG, 53.9% for IgM, 100% for IgA and 79.0% for total antibodies after 1 months since the onset of symptoms. The cumulative diagnostic sensitivity against the current gold standard, which is the diagnosis of SARS-CoV-2 based on viral RNA detection through RT-PCR, was 87.9% for IgG, 70.8% for IgM and 90.6% for total antibodies, respectively. The diagnostic sensitivity stratified according to the assay technique revealed much better performance of chemiluminescent and ELISA techniques for both IgG and IgM compared to lateral flow immunoassays, whose sensitivity remained around 76% for IgG and around 50% for IgM. The cumulative diagnostic specificity of the currently available diagnostic immunoassays seems instead optimal for all the different immunoglobulin classes, consistently higher than 98% and, more specifically, 99.1% for IgG, 98.7% for IgM and 98.5% for IgA. These findings have been recently confirmed in another meta-analysis published by Lisboa Bastos et al. (41), who also calculated a pooled diagnostic sensitivity of commercial lateral flow immunoassays (LFIAs) as low as 0.66 (95% CI, 0.49-0.79). Conversely, the pooled diagnostic sensitivity of chemiluminescent and ELISA tests was 0.98 (95% CI, 0.46-1.00) and 0.84 (95% CI, 0.76-0.91). The use of COVID-19 serology tests with the LFIA technique seems unsupported mainly by current evidence. Similar data on the so-called “rapid tests” have recently been published by the Norwegian Organization for Quality Improvement of Laboratory Examinations (42), which evaluated many of these devices for measuring anti-SARS-CoV-2 IgM and IgG. They used three different cohorts of patients, the first based on hospitalized patients with PCR-confirmed COVID-19, the second encompassing non-hospitalized patients with PCR-confirmed COVID-19 and the third consisting of random participants being tested at primary care visits. Consistent with what has been previously highlighted for the diagnostic performance of IgM, a huge variability of clinical performance for IgM rapid tests was observed, with diagnostic sensitivity ranging between 15-80% in the population of hospitalized patients, thus reinforcing the concept that measuring this class of immunoglobulins is of questionable clinical significance for diagnosing COVID-19. On the contrary, IgG’s diagnostic performance appeared more satisfactory, with most kits displaying values around 80% or higher in the most representative population of hospitalized patients with positive RT-PCR. Nonetheless, some of these tests still displayed insufficient sensitivity, with values as low as 60%, thus further emphasizing the real need that serological testing kits must be validated before being used in clinical practice.
Some interim conclusions on SARS-CoV-2 serology can hence be proposed (Table 4). What can be said at this point is that the current approach for diagnosing SARS-CoV-2 infection relies on identification of viral RNA with molecular biology in upper or lower respiratory tracts specimens. Moreover, serological testing will not replace molecular testing; however it may perhaps support viral RNA detection for diagnosing acute SARS-CoV-2 infection, especially in certain dubious cases. Some paradigmatic examples of the subsidiary role of SARS-CoV-2 serology in diagnosing COVID-19 have been published. For example, Li et al. (43) described the case of a patient with suggestive clinical and radiological features of SARS-CoV-2 infection, who tested negative for three consecutive times with RT-PCR on nasopharyngeal and oropharyngeal swabs, but who then displayed a clear IgG and IgM positivity against the virus, which enabled to make a final diagnosis of SARS-CoV-2 infection. Benoit et al. (44) used serology screening to help identify suspected cases of COVID-19 among RT-PCR negative sick controls in a clinical study.
Table 4.
Pragmatic indications for serological testing of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
|
Disease prediction and monitoring
Early and accurate identification of patients at higher risk of requiring intensive care support or death is essential in the COVID-19 pandemic, in order to establish strict monitoring, maintaining contingency care or more aggressive therapeutic management, as well as optimizing the use of technical and human resource and preventing the collapse of ICUs in a time of limited resources, insistently plagued by cuts of healthcare budgets worldwide (45, 46). Notably, a recent meta-analysis based on 24 observational studies and including over ten thousand patients from around the world, calculated cumulative ICU mortality as high as 42% (95% CI 34-50%) (47), thus reinforcing the concept that predicting the risk of developing severe or critical illness is a mainstay in patients with SARS-CoV-2 infections.
Since it is now rather clear that many demographic and clinical factors contribute to influencing the outcome of COVID-19 (48, 49), the use of accurate and reliable prediction models may provide an excellent value for stratifying the risk of unfavorable outcomes. In an interesting study, Jang et al. compared the accuracy of three conventional algorithms for predicting the clinical progression of patients with COVID-19 (50). They found that both the SIRS (Systemic Inflammatory Response Syndrome) and qSOFA (Quick Sepsis-related Organ Failure Assessment) score, two widespread models for estimating the outcome of patients with sepsis, had a cumulative diagnostic accuracy lower than 80%. Conversely, the National Early Warning Score (NEWS) displayed a better performance, with an accuracy of 90% for predicting many critical outcomes. The sensitivity and specificity of NEWS for predicting critical outcomes were also as high as 0.87 and 0.91, respectively. This alternative model encompasses seven different parameters, such as temperature, pulse and respiration rate, systolic blood pressure, pulse oximetry, oxygen and central nervous system status. A score between 0-3 points is assigned to each parameter, which cumulative yield to three risk classes, entailing a low risk between 0 and 4, a medium risk between 5 and 6, and high risk when the final score is 7 or higher.
Several reports are now available on the role of many laboratory tests in predicting progression towards severe or critical COVID-19 illness (51). A recent meta-analysis showed that progression towards severe disease might be associated with higher values of white blood cells and neutrophils, a higher concentration of tissue injury biomarkers such as aminotransferases, total bilirubin, urea and creatinine, creatine kinase (CK), LDH, myoglobin cardiac troponin I and CK isoenzyme MB, with increased values of hemostasis tests such as prothrombin time and D-dimer, but also with higher values of many inflammatory biomarkers such as the erythrocyte sedimentation rate, CRP, ferritin, interleukins and procalcitonin (52). Similarly, unfavorable disease progression has also found to be associated with lymphopenia, eosinopenia, thrombocytopenia and anemia, as well as with lower values of albumin. Important evidence has also been recently obtained by measuring presepsin in COVID-19 patients. In particular, Zaninotto and colleagues found that the value of this biomarker during hospitalization was significantly higher in patients who died than in those who survived, as well as in those who needed intensive care unit admission compared to those who did not (53). Presepsin exhibited 72% accuracy for predicting mortality, while in-hospital stay was found to be nearly double in COVID-19 patients with biomarker values >250 ng/L.
Very simple algorithms have also been proposed for risk stratification of SARS-CoV-2 infection. For example, Yan et al. (54) used three elementary laboratory tests such as lactate dehydrogenase, high-sensitivity CRP and lymphocyte count for predicting the risk of developing severe COVID-19 illness. The accuracy of the algorithm incorporating these three parameters was as high as 95% in the validation set, thus confirming that some easy, fast, widespread and almost inexpensive laboratory tests may have fundamental impact on managed care of COVID-19. Another similar algorithm has been developed by Dong et al. (55), based on lymphocyte count, D-dimer and erythrocyte sedimentation rate. The combination of these three parameters has enabled to predict COVID-19 severity with up to 84% accuracy.
Altogether, this evidence allows for the defining of specific test profiles, as that suggested by Lippi and Favaloro (56), that can be easily constructed within the hospital information system, and which may enable rapid ordering of useful laboratory tests for in-hospital monitoring of COVID-19 patients.
Conclusions
With the COVID-19 pandemic still unremittingly progressing, accurate, efficient, rapid and cost-effective diagnostic tools are needed for timely identification, isolation and/or management of patients with SARS-CoV-2 infection. Regardless of the virtually unpredictable evolution of this outbreak, laboratory medicine remains central to the diagnostic reasoning and clinical decision making in COVID-19. Nonetheless, it must be acknowledged that the current gold standard for diagnosing SARS-CoV-2 infection, entailing the identification of viral RNA with RT-PCR in specimens collected from either the upper respiratory tract is not foolproof and may be associated with a rather high number of false negative test results, up to 30%. On the other hand, neither the clinics nor diagnostic imaging can either replace diagnostic testing for identifying SARS-CoV-2 infections. Besides some peculiar features, such as smell and taste abnormalities, the clinical signs and symptoms of COVID-19 infection do not differ that much from those of other respiratory diseases, such that their interpretation remains challenging, especially during other outbreaks of other milder infectious respiratory pathologies such as common cold and influenza (57). With respect to diagnostic imaging, chest CT helps in identifying initial pulmonary involvement and monitoring disease severity and recovery. Nevertheless, this technique is characterized by poor specificity and may also display suboptimal sensitivity in the early stages of COVID-19 or in patients presenting without respiratory symptoms (i.e. gastrointestinal symptoms, etc.) (58). A potentially feasible strategy would hence encompass a reasonable integration of all these different branches of medicine, as outlined, for example, in the flow-chart summarized in Figure 2. Further studies will be necessary to assess whether this or other tentative algorithms may be clinically valid and cost-effective in a real-life scenario.
Figure 2.
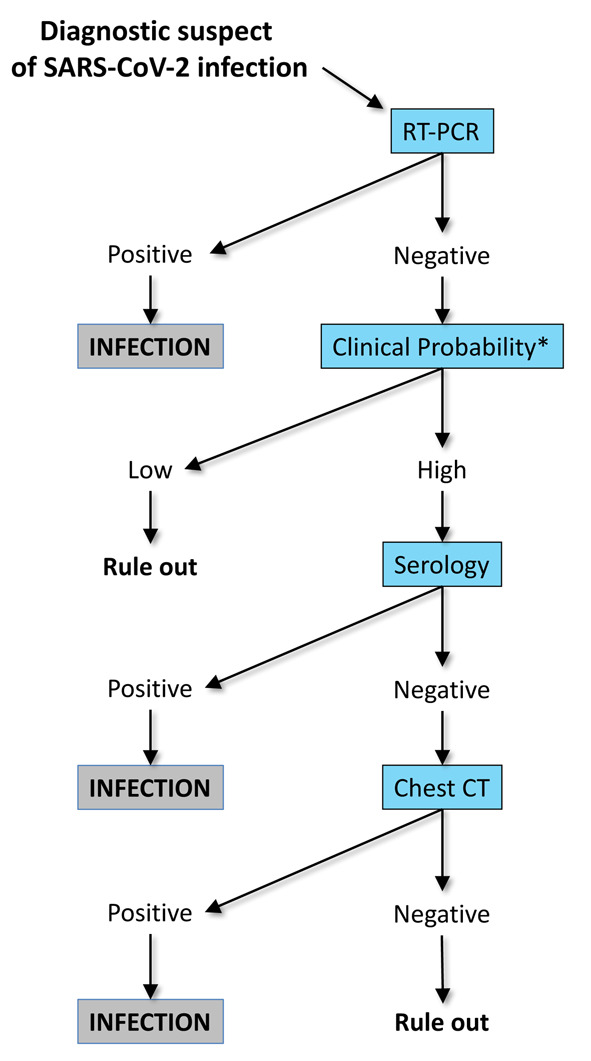
Tentative algorithm with integration of molecular testing (reverse-transcription polymerase chain reaction; RT-PCR), clinics, serology and diagnostic imaging (chest computed tomography; CT) for diagnosing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection.
* Clinical probability can be assessed with the Corona Score
Conflict of interest:
Each author declares that he or she has no commercial associations (e.g. consultancies, stock ownership, equity interest, patent/licensing arrangement etc.) that might pose a conflict of interest in connection with the submitted article
References
- 1.Cucinotta D, Vanelli M. WHO Declares COVID-19 a Pandemic. Acta Biomed. 2020;91(1):157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coronaviridae Study Group of the International Committee on Taxonomy of V. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lippi G, Sanchis-Gomar F, Henry BM. Coronavirus disease 2019 (COVID-19): the portrait of a perfect storm. Ann Transl Med. 2020;8(7):497. doi: 10.21037/atm.2020.03.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lippi G, Lavie CJ, Henry BM, Sanchis-Gomar F. Do genetic polymorphisms in angiotensin converting enzyme 2 (ACE2) gene play a role in coronavirus disease 2019 (COVID-19) Clin Chem Lab Med. 2020 doi: 10.1515/cclm-2020-0727. doi: 10.1515/cclm-2020-0727. [DOI] [PubMed] [Google Scholar]
- 5.Lippi G, Mattiuzzi C, Bovo C, Plebani M. Current laboratory diagnostics of coronavirus disease 2019 (COVID-19) Acta Biomed. 2020;91(2):137–145. doi: 10.23750/abm.v91i2.9548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lippi G, Plebani M. A modern and pragmatic definition of Laboratory Medicine. Clin Chem Lab Med. 2020;58(8):1171. doi: 10.1515/cclm-2020-0114. [DOI] [PubMed] [Google Scholar]
- 7.Lippi G, Plebani M. The critical role of laboratory medicine during coronavirus disease 2019 (COVID-19) and other viral outbreaks. Clin Chem Lab Med. 2020;58(7):1063–1069. doi: 10.1515/cclm-2020-0240. [DOI] [PubMed] [Google Scholar]
- 8.Bohn MK, Lippi G, Horvath A, Sethi S, Koch D, Ferrari M, et al. Molecular, serological, and biochemical diagnosis and monitoring of COVID-19: IFCC taskforce evaluation of the latest evidence. Clin Chem Lab Med. 2020;58:1037–1052. doi: 10.1515/cclm-2020-0722. [DOI] [PubMed] [Google Scholar]
- 9.World Health Organization. Geneva: 2020. Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases: interim guidance, 2 March 2020. [Google Scholar]
- 10.Magleby R, Westblade LF, Trzebucki A, Simon MS, Rajan M, Park J, et al. Impact of SARS-CoV-2 Viral Load on Risk of Intubation and Mortality Among Hospitalized Patients with Coronavirus Disease 2019. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa851. doi: 10.1093/cid/ciaa851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clementi N, Ferrarese R, Tonelli M, Amato V, Racca S, Locatelli M, et al. Lower nasopharyngeal viral load during the latest phase of COVID-19 pandemic in a Northern Italy University Hospital. Clin Chem Lab Med. 2020 doi: 10.1515/cclm-2020-0815. doi: 10.1515/cclm-2020-0815. [DOI] [PubMed] [Google Scholar]
- 12.Scohy A, Anantharajah A, Bodeus M, Kabamba-Mukadi B, Verroken A, Rodriguez-Villalobos H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J Clin Virol. 2020;129:104455. doi: 10.1016/j.jcv.2020.104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blairon L, Wilmet A, Beukinga I, Tre-Hardy M. Implementation of rapid SARS-CoV-2 antigenic testing in a laboratory without access to molecular methods: Experiences of a general hospital. J Clin Virol. 2020;129:104472. doi: 10.1016/j.jcv.2020.104472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mak GC, Cheng PK, Lau SS, Wong KK, Lau CS, Lam ET, et al. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J Clin Virol. 2020;129:104500. doi: 10.1016/j.jcv.2020.104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plebani M, Lippi G. Molecular diagnostics at the times of SARS-CoV-2 outbreak. Diagnosis (Berl) 2020 doi: 10.1515/dx-2020-0050. doi: 10.1515/dx-2020-0050. [DOI] [PubMed] [Google Scholar]
- 16.Lippi G, Simundic AM, Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19) Clin Chem Lab Med. 2020;58(7):1070–1076. doi: 10.1515/cclm-2020-0285. [DOI] [PubMed] [Google Scholar]
- 17.Zhang JJ, Cao YY, Dong X, Wang BC, Liao MY, Lin J, et al. Distinct characteristics of COVID-19 patients with initial rRT-PCR-positive and rRT-PCR-negative results for SARS-CoV-2. Allergy. 2020 doi: 10.1111/all.14316. doi. 10.1111/all.14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohammadi A, Esmaeilzadeh E, Li Y, Bosch RJ, Li J. SARS-CoV-2 Detection in Different Respiratory Sites: A Systematic Review and Meta-Analysis. medRxiv. 2020 doi: 10.1016/j.ebiom.2020.102903. 2020.05.14.20102038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersson M, Arancibia Carcamo CV, Auckland K, Baillie JK, Barnes E, Beneke T, et al. SARS-CoV-2 RNA detected in blood samples from patients with COVID-19 is not associated with infectious virus. medRxiv. 2020 doi: 10.12688/wellcomeopenres.16002.1. 2020.05.21.20105486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong MC, Huang J, Lai C, Ng R, Chan FKL, Chan PKS. Detection of SARS-CoV-2 RNA in fecal specimens of patients with confirmed COVID-19: A meta-analysis. J Infect. 2020 doi: 10.1016/j.jinf.2020.06.012. doi: 10.1016/j.jinf.2020.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan VW, Chiu PK, Yee CH, Yuan Y, Ng CF, Teoh JY. A systematic review on COVID-19: urological manifestations, viral RNA detection and special considerations in urological conditions. World J Urol. 2020 doi: 10.1007/s00345-020-03246-4. doi: 10.1007/s00345-020-03246-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walsh KA, Jordan K, Clyne B, Rohde D, Drummond L, Byrne P, et al. SARS-CoV-2 Detection, Viral Load and Infectivity over the Course of an Infection: SARS-CoV-2 Detection, Viral Load and Infectivity. J Infect. 2020 doi: 10.1016/j.jinf.2020.06.067. doi: 10.1016/j.jinf.2020.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye G, Pan Z, Pan Y, Deng Q, Chen L, Li J, et al. Clinical characteristics of severe acute respiratory syndrome coronavirus 2 reactivation. J Infect. 2020;80(5):e14–e17. doi: 10.1016/j.jinf.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cento V, Colagrossi L, Nava A, Lamberti A, Senatore S, Travi G, et al. Persistent positivity and fluctuations of SARS-CoV-2 RNA in clinically-recovered COVID-19 patients. J Infect. 2020 doi: 10.1016/j.jinf.2020.06.024. doi: 10.1016/j.jinf.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao AT, Tong YX, Zhang S. False-negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: Rather than recurrence. J Med Virol. 2020 doi: 10.1002/jmv.25855. doi: 10.1002/jmv.25855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan J, Kou S, Liang Y, Zeng J, Pan Y, Liu L. PCR Assays Turned Positive in 25 Discharged COVID-19 Patients. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa398. doi: 10.1093/cid/ciaa398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang J, Zheng L, Li Z, Hao S, Ye F, Chen J, et al. Recurrence of SARS-CoV-2 PCR positivity in COVID-19 patients: a single center experience and potential implications. medRxiv. 2020 2020.05.06.20089573. [Google Scholar]
- 28.Kurstjens S, van der Horst A, Herpers R, Geerits MWL, Kluiters-de Hingh YCM, Gottgens EL, et al. Rapid identification of SARS-CoV-2-infected patients at the emergency department using routine testing. Clin Chem Lab Med. 2020 doi: 10.1515/cclm-2020-0593. doi: 10.1515/cclm-2020-0593. [DOI] [PubMed] [Google Scholar]
- 29.Qin L, Yang Y, Cao Q, Cheng Z, Wang X, Sun Q, et al. A predictive model and scoring system combining clinical and CT characteristics for the diagnosis of COVID-19. Eur Radiol. 2020 doi: 10.1007/s00330-020-07022-1. doi: 10.1007/s00330-020-07022-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jehi L, Ji X, Milinovich A, Erzurum S, Rubin B, Gordon S, et al. Individualizing Risk Prediction for Positive COVID-19 Testing: Results from 11,672 Patients. Chest. 2020 doi: 10.1016/j.chest.2020.05.580. doi: 10.1016/j.chest.2020.05.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fukumoto T, Iwasaki S, Fujisawa S, Hayasaka K, Sato K, Oguri S, et al. Efficacy of a novel SARS-CoV-2 detection kit without RNA extraction and purification. Int J Infect Dis. 2020 doi: 10.1016/j.ijid.2020.06.074. doi: 10.1016/j.ijid.2020.06.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pilcher CD, Westreich D, Hudgens MG. Group Testing for Sars-Cov-2 to Enable Rapid Scale-Up of Testing and Real-Time Surveillance of Incidence. J Infect Dis. 2020 doi: 10.1093/infdis/jiaa378. doi: 10.1093/infdis/jiaa378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cherif A, Grobe N, Wang X, Kotanko P. Simulation of Pool Testing to Identify Patients With Coronavirus Disease 2019 Under Conditions of Limited Test Availability. JAMA Netw Open. 2020;3(6):e2013075. doi: 10.1001/jamanetworkopen.2020.13075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ben-Ami R, Klochendler A, Seidel M, Sido T, Gurel-Gurevich O, Yassour M, et al. Large-scale implementation of pooled RNA extraction and RT-PCR for SARS-CoV-2 detection. Clin Microbiol Infect. 2020 doi: 10.1016/j.cmi.2020.06.009. doi: 10.1016/j.cmi.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yelin I, Aharony N, Shaer Tamar E, Argoetti A, Messer E, Berenbaum D, et al. Evaluation of COVID-19 RT-qPCR test in multi-sample pools. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa531. doi: 10.1093/cid/ciaa531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stockholm: ECDC; 2020. European Centre for Disease Prevention and Control. Methodology for estimating point prevalence of SARSCoV-2 infection by pooled RT-PCR testing. [Google Scholar]
- 37.Nalbantoglu OU. Group testing performance evaluation for SARS-CoV-2 massive scale screening and testing. BMC Med Res Methodol. 2020;20(1):176. doi: 10.1186/s12874-020-01048-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu X, Sun J, Nie S, Li H, Kong Y, Liang M, et al. Seroprevalence of immunoglobulin M and G antibodies against SARS-CoV-2 in China. Nat Med. 2020 doi: 10.1038/s41591-020-0949-6. doi: 10.1038/s41591-020-0949-6. [DOI] [PubMed] [Google Scholar]
- 39.Liu ZL, Liu Y, Wan LG, Xiang TX, Le AP, Liu P, et al. Antibody profiles in mild and severe cases of COVID-19. Clin Chem. 2020 doi: 10.1093/clinchem/hvaa137. doi: 10.1093/clinchem/hvaa137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Spijker R, Taylor-Phillips S, et al. Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database Syst Rev. 2020;6:CD013652. doi: 10.1002/14651858.CD013652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lisboa Bastos M, Tavaziva G, Abidi SK, Campbell JR, Haraoui LP, Johnston JC, et al. Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. BMJ. 2020;370:m2516. doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tollanes MC, Bakken Kran AM, Abildsnes E, Jenum PA, Breivik AC, Sandberg S. Evaluation of eleven rapid tests for detection of antibodies against SARS-CoV-2. Clin Chem Lab Med. 2020 doi: 10.1515/cclm-2020-0628. doi: 10.1515/cclm-2020-0628. [DOI] [PubMed] [Google Scholar]
- 43.Li H, Pan J, Su Y, Wang B, Ge J. SARS-CoV-2 lgM/lgG antibody detection confirms the infection after three negative nucleic acid detection. J Cell Mol Med. 2020 doi: 10.1111/jcmm.15275. doi: 10.1111/jcmm.15275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benoit J, Benoit S, Lippi G, Henry BM. False negative RT-PCR or false positive serological testing in SARS-CoV-2 diagnostics? Navigating between Scylla and Charybdis to prevent misclassification bias in COVID-19 clinical investigations. Diagnosis (Berl) 2020 doi: 10.1515/dx-2020-0091. [DOI] [PubMed] [Google Scholar]
- 45.Inoue K, Ohira Y, Takeshita H. The risk of the collapse of public health centres under the current system to prevent the spread of COVID-19. Int Marit Health. 2020;71(2):149. doi: 10.5603/IMH.2020.0026. [DOI] [PubMed] [Google Scholar]
- 46.Wurmb T, Scholtes K, Kolibay F, Schorscher N, Ertl G, Ernestus RI, et al. Hospital preparedness for mass critical care during SARS-CoV-2 pandemic. Crit Care. 2020;24(1):386. doi: 10.1186/s13054-020-03104-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Armstrong RA, Kane AD, Cook TM. Outcomes from intensive care in patients with COVID-19: a systematic review and meta-analysis of observational studies. Anaesthesia. 2020 doi: 10.1111/anae.15201. doi: 10.1111/anae.15201. [DOI] [PubMed] [Google Scholar]
- 48.Lippi G, Sanchis-Gomar F, Henry BM. COVID-19: unravelling the clinical progression of nature’s virtually perfect biological weapon. Ann Transl Med. 2020;8(11):693. doi: 10.21037/atm-20-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lauretani F, Ravazzoni G, Roberti MF, Longobucco Y, Adorni E, Grossi M, et al. Assessment and treatment of older individuals with COVID 19 multi-system disease: Clinical and ethical implications. Acta Biomed. 2020;91(2):150–168. doi: 10.23750/abm.v91i2.9629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jang JG, Hur J, Hong KS, Lee W, Ahn JH. Prognostic Accuracy of the SIRS, qSOFA, and NEWS for Early Detection of Clinical Deterioration in SARS-CoV-2 Infected Patients. J Korean Med Sci. 2020;35(25):e234. doi: 10.3346/jkms.2020.35.e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lippi G, Plebani M. Laboratory abnormalities in patients with COVID-2019 infection. Clin Chem Lab Med. 2020;58(7):1131–1134. doi: 10.1515/cclm-2020-0198. [DOI] [PubMed] [Google Scholar]
- 52.Henry BM, de Oliveira MHS, Benoit S, Plebani M, Lippi G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin Chem Lab Med. 2020;58:1021–1028. doi: 10.1515/cclm-2020-0369. [DOI] [PubMed] [Google Scholar]
- 53.Zaninotto M, Mion MM, Cosma C, Rinaldi D, Plebani M. Presepsin in risk stratification of SARS-CoV-2 patients. Clin Chim Acta. 2020;507:161–163. doi: 10.1016/j.cca.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan L, Zhang H-T, Goncalves J, Xiao Y, Wang M, Guo Y, et al. An interpretable mortality prediction model for COVID-19 patients. Nat Mach Intell. 2020;2(5):283–288. [Google Scholar]
- 55.Dong Y, Zhou H, Li M, Zhang Z, Guo W, Yu T, et al. A novel simple scoring model for predicting severity of patients with SARS-CoV-2 infection. Transbound Emerg Dis. 2020 doi: 10.1111/tbed.13651. doi: 10.1111/tbed.13651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Favaloro EJ, Lippi G. Recommendations for Minimal Laboratory Testing Panels in Patients with COVID-19: Potential for Prognostic Monitoring. Semin Thromb Hemost. 2020;46(3):379–382. doi: 10.1055/s-0040-1709498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lippi G, Mattiuzzi C. Diagnostic and clinical significance of “atypical” symptoms in coronavirus disease 2019. Pol Arch Intern Med. 2020;130(6):478–480. doi: 10.20452/pamw.15448. [DOI] [PubMed] [Google Scholar]
- 58.Harahwa TA, Lai Yau TH, Lim-Cooke MS, Al-Haddi S, Zeinah M, Harky A. The optimal diagnostic methods for COVID-19. Diagnosis (Berl) 2020 doi: 10.1515/dx-2020-0058. doi: 10.1515/dx-2020-0058. [DOI] [PubMed] [Google Scholar]


