Abstract
Background and aim:
Isolation of subjects with active severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is a pivotal preventive measure in the ongoing coronavirus disease 2019 (COVID-19) pandemic. A growing number of studies reported cases of recurrent SARS-CoV-2 RNA positivity following disease recovery, which were identified with a critical literature search and then meta-analyzed in this article.
Materials and Methods:
A digital search was performed in Medline and Web of Science, using the keywords “coronavirus disease 2019” OR “COVID-19” OR “severe acute respiratory disease 2” OR “SARS-CoV-2” AND “recurrence” OR “repositivization” OR “retesting”, without date or language restrictions. Recovery was defined as resolution of symptoms, with at least two consecutive negative molecular tests.
Results:
A total number of 17 studies, with 5,182 COVID-19 patients, were included. SARS-CoV-2 recurrent RNA positivity in recovered COVID-19 patients ranged between 7-23% across the studies, with follow-up testing between 1-60 days. The estimated cumulative rate of SARS-CoV-2 recurrent RNA positivity was 12% (95% confidence interval, 12-13%; I2, 74%).
Conclusions:
Repeated molecular testing on respiratory tracts specimens at 1 and 2 months after recovery from COVID-19 is strongly advisable for early identification, isolation and clinical management of subjects with SARS-CoV-2 recurrent RNA positivity. (www.actabiomedica.it)
Keywords: coronavirus, COVID-19, SARS-CoV-2, testing: recurrence
Introduction
The incessant and unstoppable spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the pathogen responsible for coronavirus disease 2019 (COVID-19) which was officially declared a pandemic infectious disease by the World Health Organization (WHO) (1), is not only jeopardizing the response capacity of many healthcare systems, but is also causing enormous societal and economic burdens all around the world (2).
The information garnered thus far on SARS-CoV-2 allows to strongly suspect that the disease originated from spill-over of a recombinant bat coronavirus, whilst SARS-CoV-2 is currently spreading from person to person through droplets emitted from the nose or mouth, particularly when an infected person breathes, coughs, sneezes, talks louds, shouts, yells, or even sings, especially in indoor spaces (3). Transmission may also potentially occur after massive inhalation of virus-containing aerosols (4), or by transporting infected material deposited on various surfaces to the eyes, nose and mouth, though droplet-conveyed transmission likely remains the prevailing modality (5).
Based on the transmission routes of SARS-CoV-2, the most effective contagion-containment strategies encompass nationwide lockdown and stay-in-place orders (6), followed by social distancing (i.e., at least 6 feet distance), widespread facemask wearing (especially in busy, indoor places), and frequent hands sanitation (7,8). Most importantly, early and accurate identification of positive cases and contact tracing is central to effective containment strategies (9,10). Isolation of infected subjects, irrespective of their symptomatic status (asymptomatic, pre-symptomatic, or mildly symptomatic, and so forth), is an essential measure for limiting viral spread, since recent data reported that more than half of total COVID-19 infections could be traced to transmission from subjects with active SARS-CoV-2 infection without clinical manifestations (11).
Recently, reports have been describing a discrete number of COVID-19 patients who have already recovered from the disease (mostly when symptoms have disappeared for at least 14 days and with two negative consecutive nucleic acid amplification tests via nasopharyngeal swabs) (12), but then turned positive again at reverse-phase polymerase chain reaction (RT-PCR) testing after a certain period of time. Some of these individuals developed symptoms, while others remained asymptomatic, thus representing another potential source of viral spread and contagion within the general population.
Therefore, we aimed to critically review the current literature and carry out a meta-analysis of published articles that have reported SARS-CoV-2 recurrent RNA positivity in patients who had previously recovered from COVID-19.
Materials and Methods
A digital search was carried out in Medline (PubMed interface) and Web of Science using the keywords “coronavirus disease 2019” OR “COVID-19” OR “severe acute respiratory disease 2” OR “SARS-CoV-2” AND “recurrence” OR “repositivization” OR “retesting” in all fields, without date or language restrictions. The title, abstract and full text of pre-identified documents were accurately examined by two independent reviewers (CM and GL). Clinical studies reporting the rate of recurrently positive RT-PCR test results in patients recovered after a diagnosis of SARS-CoV-2 infection (i.e., displaying at least two consecutive negative tests) and a total sample size >50 COVID-19 patients were selected. The reference list of all articles was also analyzed to detect other potentially eligible documents. A meta-analysis was then performed, encompassing the calculation of the pooled prevalence (95% confidence interval; 95% CI) of recurrently positive RT-PCR test results after recovering from COVID-19. Heterogeneity was evaluated with the χ2 test and I2 statistics. Publication bias was assessed using a funnel plot. The statistical analysis was carried out using MetaXL, software Version 5.3 (EpiGear International Pty Ltd., Sunrise Beach, Australia). The study was performed in accordance with the declaration of Helsinki and within the terms of the local legislation.
Results
A total of 168 documents were initially identified by our search criteria, 154 of which were excluded as they did not contain specific data on recurrently positive RT-PCR test results in patients who recovered from COVID-19 (n=109), were case reports (n=18), review articles (n=10), letters to the editor without original data (n=5) or editorial material (n=4), because re-testing was carried out in SARS-CoV-2 initially negative patients (n=4), or had an insufficient sample size (i.e., <50 total COVID-19 patients) based on our inclusion criteria (n=4). Three additional studies could be identified from the reference list of the selected articles, so that a final number of 17 studies (13-29), totaling 5182 COVID-19 patients, were included in our analysis. No inter-reviewer disagreements occurred. The characteristics of these studies are summarized in Table 1. The vast majority of the studies (14/17; 82.3%) were performed in China, and all employed respiratory and/or throat specimens for diagnosing SARS-CoV-2 re-infection, whilst fecal samples were also used in 4/17 studies (23.5%). The sample size ranged between 55 to 2,521 COVID-19 patients, the recurrence monitoring period between 1 and 60 days for the majority of COVID-19 patients, whilst the mean or median age of patients with recurrently positive tests ranged between 27 to 64 years. The prevalence of female sex among the included studies ranged between 36% to 71%.
Table 1.
Characteristics of the studies included in the meta-analysis
| Authors | Sample Size | Recurrence monitoring period1 | Setting | Samples | Age (years)2 | Females (%) |
| Abdullah MS et al. | 138 | 11 days | Brunei | Throat and respiratory specimens | 41 (24-58) | 41% |
| Cento V et al. | 2521 | 1-60 days | Italy | Respiratory specimens | NCS | NCS |
| Deng W et al. | 576 | 3-35 days | China | Respiratory and fecal specimen | 55 (43-68) | 59% |
| Hao S. et al. | 104 | NCS | China | Respiratory specimens | NCS | NCS |
| Hartman WR et al. | 75 | 19-28 days | US | Respiratory specimens | 53 (38-68) | 63% |
| Hu R et al. | 69 | 9-17 days | China | Respiratory specimens | 27 (4-58) | 36% |
| Huang J et al. | 414 | 3-12 days | China | Respiratory specimens | NCS | 59% |
| Liu T et al. | 150 | NCS | China | Throat swabs | 49 (37-62) | 45% |
| Tian M et al. | 147 | 7-47 days | China | Respiratory specimens | 37 (4-80) | 40% |
| Wang Y et al. | 94 | 1-14 days | China | Respiratory specimens | 46 (30-70) | 50% |
| Wu J et al. | 60 | 4-24 days | China | Respiratory and fecal specimen | NCS | NCS |
| Xiao AT et al. | 70 | 1-45 days | China | Throat specimens | 64 (51-73) | 40% |
| Ye G et al. | 55 | 4-17 days | China | Throat specimens | 32 (27-42) | 60% |
| Yuan B et al. | 182 | 1-14 days | China | Respiratory and fecal specimen | 40 (20-60) | 65% |
| Yuan Y et al. | 172 | 2-13 days | China | Respiratory and fecal specimen | 28 (16-42) | 68% |
| Zhu H et al. | 98 | 1-14 days | China | Respiratory specimens | 54 (44-63) | 71% |
| Zou Y et al. | 257 | 1-12 days | China | Throat specimens | 62 (29-87) | 57% |
1 For most of patients with nucleic acid amplification test (NAAT) recurrence; 2 confidence interval estimated from original data. NCS, non-clearly specified.
The single-study and pooled prevalence of recurrent RT-PCR positivity is shown in Figure 1. The rate of recurrence of positive SARS-CoV-2 viral RNA in recovered COVID-19 patients was reported at between 7% to 23% across the 17 studies. The calculated pooled rate (fixed effects) of SARS-CoV-2 recurrent RNA positivity was 12% (95% CI, 12-13%), with high heterogeneity (I2, 74%), partly attributable to the largely variable sample size among the included studies. Slightly higher rate of SARS-CoV-2 recurrent RNA positivity could be estimated with a random effects model (14%; 95% CI, 12-17%).
Figure 1.
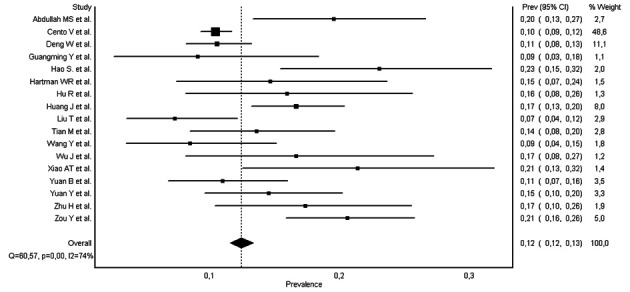
Single-study and cumulative positivity recurrence rate of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with nucleic acid amplification tests (NAATs)
After excluding the large study of Cento et al., which accounted for nearly half of the total sample weight, the cumulative prevalence modestly increased to 14% (95% CI, 13-16%), with a slightly lower heterogeneity (I2, 64%). The prevalence did not vary substantially when the calculation was carried out in studies performed outside of China, as well as in those using only respiratory tract specimens (Table 2). The funnel plot, shown in figure 2, demonstrates the presence of moderate asymmetry, suggestive of modest publication bias.
Table 2.
Cumulative prevalence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) positivity recurrence with nucleic acid amplification test (NAAT) in specific settings
| Setting | Prevalence | Heterogeneity |
| Excluding Cento et al. | 0.14 (95% CI, 0.13-0.16) | I2, 64% |
| Only respiratory tract specimens | 0.13 (95% CI 0.12-0.14%) | I2, 79% |
| Studies outside China | 0.11 (95% CI, 0.10-0.12) | I2, 80% |
Figure 2.
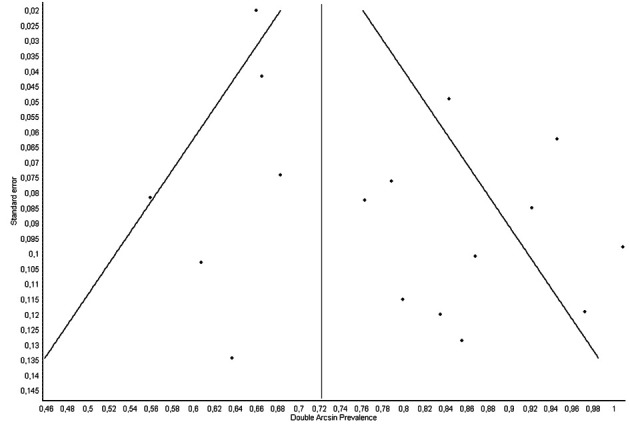
Funnel plot of the studies included in the meta-analysis
Discussion
The results of our meta-analysis attest a rate of cumulative SARS-CoV-2 recurrent RNA positivity of ~12%. No substantial differences were observed when limiting our analysis to non-Chinese studies or those in which only respiratory tract specimens were used to diagnose SARS-CoV-2 infection. This evidence underscores an important aspect in the clinical epidemiology of COVID-19, since nearly one in ten patients may be found positive again for the presence of SARS-CoV-2 RNA after recovering from the disease. Some hypotheses can be made for explaining this not so rare phenomenon (30).
The first assumption entails that some subjects might have been re-infected by SARS-CoV-2 due to (i) suboptimal therapeutic management of SARS-CoV-2 infection, (ii) because their natural immunity has declined, as recently highlighted by Long et al. (31) and Ibarrondo et al. (32), or (iii) because the virus has undergone some degree of recombination that would make the immune response somehow inefficient against this relatively new infection, thus fostering a second episode of active viral replication (33). The second hypothesis is that acute SARS-CoV-2 infection and viral shedding may have been concluded in the upper respiratory tract, while the virus may still be present in some forms in the lower respiratory tract. This hypothesis is supported by evidence that shedding of SARS-CoV-2 seems indeed prolonged in sputum compared to nasopharyngeal and oropharyngeal swabs in patients recovering from COVID-19, as reported by Wang et al (34). Systemic re-propagation or natural shedding of either the virus or infected cells from lower to the upper respiratory tract would then be associated with recurrence of nasopharyngeal and oropharyngeal swab positivity, but with lower viral load (35). It is important to state that it is still largely unclear whether this second positivity may be sustained by viable viral particles (and thereby patients may still be contagious), viral fragments or extra-viral RNA, since all these three possibilities would be associated with positivity of nucleic acid amplification tests. Finally, the impact of some technical drawbacks that may have generated false-positive results cannot be excluded, including pre-analytical and analytical mistakes, the use of different molecular assays during follow-up, characterized by variable detection limits or functional sensitivity (36), the cross-contamination with other coronaviruses (37), along with the use of molecular techniques not adequately validated and ready for prime time (38).
Whether or not SARS-CoV-2 recurrent RNA positivity after recovering from COVID-19 may represent an important source of amplified transmission and enhanced viral circulation within the general population requires further investigation with specific viral culture and cytopathic studies (39). Nevertheless, we believe that repeating molecular testing using RT-PCR analysis on respiratory tracts specimens at 1 and 2 months after recovery from COVID-19 would be advisable for several reasons. First, this policy will enable the early identification and treatment of a unique cohort of COVID-19 patients. These patients may be at particularly high risk for developing severe or critical illness, worse than in their prior infection, due to the phenomenon of antibody-dependent enhancement (ADE). With ADE, non-neutralizing antibodies generated during the active phase of the first SARS-CoV-2 infection may then contribute to amplify the internalization of immunoglobulin-bound viral particles through the Fc receptors present on the surface of many immune cells (40). This would result in the boosting of viral replication, activation of the immune cell, ultimately triggering the release of a vast array of pro-inflammatory mediators, which will contribute to elicit, propagate or aggravate the devastating “cytokine storm” that characterizes the more severe forms of COVID-19, as well as the release of new viral particles and thus increasing viral load (40).
Preventive and proactive isolation of recurrent SARS-CoV-2 positive cases is another important public health strategy in the fight against COVID-19 pandemic, since it may contribute to lower the possibility that potentially viable viral particles may be transmitted from positive subjects to their close contacts during social interactions.
In conclusion, this study found that over one in ten patients have recurrence of RT-PCR positivity within 60 days of recovery from their primary infection. While it is still unclear if this represents transmittable and infectious viral particles, given these observations, we recommend repeated testing at one and two months post-recovery. Such recurrent RT-PCR positive patients should be isolated to reduce the potential for community transmission until further data becomes available. Recurrence of RNA positivity requires urgent molecular investigations by the medical and public health community, with focused attention on specific categories of patients (i.e., asymptomatic vs. symptomatic, younger vs. older, women vs. men, ill vs. healthy).
Conflict of interest:
Each author declares that he or she has no commercial associations (e.g. consultancies, stock ownership, equity interest, patent/licensing arrangement etc.) that might pose a conflict of interest in connection with the submitted article
References
- 1.Cucinotta D, Vanelli M. WHO Declares COVID-19 a Pandemic. Acta Biomed. 2020;91:157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khullar D, Bond AM, Schpero WL. COVID-19 and the Financial Health of US Hospitals. JAMA. 2020 May 4 doi: 10.1001/jama.2020.6269. doi: 10.1001/jama.2020.6269. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 3.Borak J. Airborne Transmission of COVID-19. Occup Med (Lond) 2020;70:297–299. doi: 10.1093/occmed/kqaa080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shao S, Zhou D, He R, Li J, Zou S, Mallery K, Kumar S, Yang S, Hong J. Risk assessment of airborne transmission of COVID-19 by asymptomatic individuals under different practical settings. Version 2. ArXiv [Preprint] 2020 Jul 7 doi: 10.1016/j.jaerosci.2020.105661. arXiv:2007.03645v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lippi G, Adeli K, Ferrari M, et al. Biosafety measures for preventing infection from COVID-19 in clinical laboratories: IFCC Taskforce Recommendations. Clin Chem Lab Med. 2020;58:1053–1062. doi: 10.1515/cclm-2020-0633. [DOI] [PubMed] [Google Scholar]
- 6.Lippi G, Henry BM, Bovo C, Sanchis-Gomar F. Health risks and potential remedies during prolonged lockdowns for coronavirus disease 2019 (COVID-19) Diagnosis (Berl) 2020;7:85–90. doi: 10.1515/dx-2020-0041. [DOI] [PubMed] [Google Scholar]
- 7.López L, Rodó X. The end of social confinement and COVID-19 re-emergence risk. Nat Hum Behav. 2020;4:746–755. doi: 10.1038/s41562-020-0908-8. [DOI] [PubMed] [Google Scholar]
- 8.Chu DK, Akl EA, Duda S, Solo K, Yaacoub S, Schünemann HJ. COVID-19 Systematic Urgent Review Group Effort (SURGE) study authors. Physical distancing, face masks, and eye protection to prevent person-to-person transmission of SARS-CoV-2 and COVID-19: a systematic review and meta-analysis. Lancet. 2020;395:1973–1987. doi: 10.1016/S0140-6736(20)31142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keeling MJ, Hollingsworth TD, Read JM. Efficacy of contact tracing for the containment of the 2019 novel coronavirus (COVID-19) J Epidemiol Community Health. 2020 Jun 23 doi: 10.1136/jech-2020-214051. jech-2020-214051. doi: 10.1136/jech-2020-214051. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pradhan D, Biswasroy P, Kumar Naik P, Ghosh G, Rath G. A Review of Current Interventions for COVID-19 Prevention. Arch Med Res. 2020;51:363–374. doi: 10.1016/j.arcmed.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lippi G, Plebani M. Asymptomatic COVID-19 transmission: the importance of avoiding official miscommunication. Diagnosis (Berl) 2020 Jul 13 doi: 10.1515/dx-2020-0085. doi: 10.1515/dx-2020-0085. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 12.Foppiani A, Bertoli S, Battezzati A, Zuccotti G. Data to guide the application of the new WHO criteria for releasing COVID-19 patients from isolation. Pharmacol Res. 2020 Jul 11;160:105063. doi: 10.1016/j.phrs.2020.105063. doi: 10.1016/j.phrs.2020.105063. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abdullah MS, Chong PL, Asli R, et al. Post discharge positive re-tests in COVID-19: common but clinically non-significant. Infect Dis (Lond) 2020 Jun 24:1–3. doi: 10.1080/23744235.2020.1780309. doi: 10.1080/23744235.2020.1780309. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 14.Cento V, Colagrossi L, Nava A, et al. Persistent positivity and fluctuations of SARS-CoV-2 RNA in clinically-recovered COVID-19 patients. J Infect. 2020 Jun 20;S0163-4453(20):30405–9. doi: 10.1016/j.jinf.2020.06.024. doi: 10.1016/j.jinf.2020.06.024. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng W, Guang TW, Yang M, et al. Positive results for patients with COVID-19 discharged form hospital in Chongqing, China. Version 2. BMC Infect Dis. 2020;20:429. doi: 10.1186/s12879-020-05151-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hao S, Lian J, Lu Y, et al. Decreased B Cells on Admission Associated With Prolonged Viral RNA Shedding From the Respiratory Tract in Coronavirus Disease 2019: A Case-Control Study. J Infect Dis. 2020;222:367–371. doi: 10.1093/infdis/jiaa311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartman WR, Hess AS, Connor JP. Persistent viral RNA shedding after COVID-19 symptom resolution in older convalescent plasma donors. Transfusion. 2020 Jun 13 doi: 10.1111/trf.15927. 10.1111/trf.15927. doi: 10.1111/trf.15927. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu R, Jiang Z, Gao H, et al. Recurrent Positive Reverse Transcriptase-Polymerase Chain Reaction Results for Coronavirus Disease 2019 in Patients Discharged From a Hospital in China. JAMA Netw Open. 2020;3:e2010475. doi: 10.1001/jamanetworkopen.2020.10475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang J, Zheng L, Li Z, et al. Recurrence of SARS-CoV-2 PCR positivity in COVID-19 patients: a single center experience and potential implications. MedRxiv. 2020.05.06.20089573. Doi: 10.1101/2020.05.06.20089573. [Google Scholar]
- 20.Liu T, Wu S, Zeng G, et al. Recurrent positive SARS-CoV-2: Immune certificate may not be valid. J Med Virol. 2020 May 29 doi: 10.1002/jmv.26074. 10.1002/jmv.26074. doi: 10.1002/jmv.26074. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian M, Long Y, Hong Y, Zhang X, Zha Y. The treatment and follow-up of “recurrence” with discharged COVID-19 patients: Data from Guizhou, China. Environ Microbiol. 2020 Jul 6 doi: 10.1111/1462-2920.15156. 10.1111/1462-2920.15156. doi: 10.1111/1462-2920.15156. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Xu H, Jiang H, et al. The Clinical Features and Outcomes of Discharged Coronavirus Disease 2019 PatientsA Prospective Cohort Study. QJM. 2020 May 22 doi: 10.1093/qjmed/hcaa178. hcaa178. doi: 10.1093/qjmed/hcaa178. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu J, Liu X, Liu J, et al. Coronavirus Disease 2019 Test Results After Clinical Recovery and Hospital Discharge Among Patients in China. JAMA Netw Open. 2020;3:e209759. doi: 10.1001/jamanetworkopen.2020.9759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao AT, Tong YX, Zhang S. False negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: Rather than recurrence. J Med Virol. 2020 Apr 9 doi: 10.1002/jmv.25855. 10.1002/jmv.25855. doi: 10.1002/jmv.25855. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye G, Pan Z, Pan Y, et al. Clinical characteristics of severe acute respiratory syndrome coronavirus 2 reactivation. J Infect. 2020;80:e14–e17. doi: 10.1016/j.jinf.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan B, Liu HQ, Yang ZR, et al. Recurrence of positive SARS-CoV-2 viral RNA in recovered COVID-19 patients during medical isolation observation. Sci Rep. 2020;10:11887. doi: 10.1038/s41598-020-68782-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan J, Kou S, Liang Y, Zeng J, Pan Y, Liu L. PCR Assays Turned Positive in 25 Discharged COVID-19 Patients. Clin Infect Dis. 2020 Apr 8 doi: 10.1093/cid/ciaa398. ciaa398. doi: 10.1093/cid/ciaa398. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu H, Fu L, Jin Y, et al. Clinical features of COVID-19 convalescent patients with re-positive nucleic acid detection. J Clin Lab Anal. 2020 Jun 7:e23392. doi: 10.1002/jcla.23392. doi: 10.1002/jcla.23392. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zou Y, Wang BR, Sun L, et al. The issue of recurrently positive patients who recovered from COVID-19 according to the current discharge criteria: investigation of patients from multiple medical institutions in Wuhan, China. J Infect Dis. 2020 Jun 3 doi: 10.1093/infdis/jiaa301. jiaa301. doi: 10.1093/infdis/jiaa301. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lippi G, Henry BM, Sanchis-Gomar F, Mattiuzzi C. Updates on laboratory investigations in coronavirus disease 2019 (COVID-19) Acta BioMed. 2020 Jul 13;91(3) doi: 10.23750/abm.v91i3.10187. Epub ahead of print. Doi: 10.23750/abm.v91i3.10187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Long QX, Tang XJ, Shi QL, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020 Jun 18 doi: 10.1038/s41591-020-0965-6. doi: 10.1038/s41591-020-0965-6. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 32.Ibarrondo FJ, Fulcher JA, Goodman-Meza D, et al. Rapid Decay of Anti–SARS-CoV-2 Antibodies in Persons with Mild Covid-19. N Engl J Med. doi: 10.1056/NEJMc2025179. Published online July 21, 2020. doi:10.1056/NEJMc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tiwari M, Mishra D. Investigating the genomic landscape of novel coronavirus (2019-nCoV) to identify non-synonymous mutations for use in diagnosis and drug design. J Clin Virol. 2020;128:104441. doi: 10.1016/j.jcv.2020.104441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang K, Zhang X, Sun J, et al. Differences of SARS-CoV-2 Shedding Duration in Sputum and Nasopharyngeal Swab Specimens Among Adult Inpatients With COVID-19. Chest. 2020 Jun 20;S0012-3692(20):31718–9. doi: 10.1016/j.chest.2020.06.015. doi: 10.1016/j.chest.2020.06.015. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clementi N, Ferrarese R, Tonelli M, et al. Lower nasopharyngeal viral load during the latest phase of COVID-19 pandemic in a Northern Italy University Hospital. Clin Chem Lab Med. 2020 Jun 29 doi: 10.1515/cclm-2020-0815. doi: 10.1515/cclm-2020-0815. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 36.Muenchhoff M, Mairhofer H, Nitschko H, et al. Multicentre comparison of quantitative PCR-based assays to detect SARS-CoV-2, Germany, March 2020. Euro Surveill. 2020;25:2001057. doi: 10.2807/1560-7917.ES.2020.25.24.2001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Law SK, Leung AWN, Xu C. Is reinfection possible after recovery from COVID-19? Hong Kong Med J. 2020;26:264–265. doi: 10.12809/hkmj208601. [DOI] [PubMed] [Google Scholar]
- 38.Lippi G, Simundic AM, Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19) Clin Chem Lab Med. 2020;58:1070–1076. doi: 10.1515/cclm-2020-0285. [DOI] [PubMed] [Google Scholar]
- 39.Batisse D, Benech N, Botelho-Nevers E, et al. Clinical recurrences of COVID-19 symptoms after recovery: viral relapse, reinfection or inflammatory rebound. J Infect. 2020 Jun 30 doi: 10.1016/j.jinf.2020.06.073. S0163-4453;20:30454-0. doi: 10.1016/j.jinf.2020.06.073. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arvin AM, Fink K, Schmid MA, et al. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature. 2020 Jul 13 doi: 10.1038/s41586-020-2538-8. doi: 10.1038/s41586-020-2538-8. Epub ahead of print. [DOI] [PubMed] [Google Scholar]


