Abstract
The aim of this study is to assess the association among species of bacteria and to identify the presence of clusters of patients in sub intensive care unit with different profiles of infection, and to study the relationship between such profiles and patient demographics (gender, age), kind of investigations and material used to detect the infection. The findings need to analyse a bigger amount of data in the same setting to make evident that it is constant the infection only with Escherichia coli and Staphylocossus epidemidis and a third case in which more bacteria are inlvolved. (www.actabiomedica.it)
Keywords: Covid 19 Hospital Acquired Infection Subintensive Unit
In Italy, Corona Virus 19 (COVID-19) had its first patient in Lombardy Region and then it circulated all over the Country and at the moment (27.07.2020) there are 244.708 confirmed cases, 34.126 deaths with 14% as case fatality rate (1,2). As it was recorded from of this beginning of this epidemic up to now almost 10% of patients with COVID-19 experimented a hospital admission and 9% of them needed to stay in intensive care units but the numbers were very different at the beginning of the epidemic when the patients with critical conditions were much more (1).
In a research and teaching hospital located in the centre of Milan with active 716 beds, 84 of them were turned up in sub intensive care to admit patients who needed less intensive care (3). In total from March 9th to June 6th 2020, 246 patients were admitted to these 84 beds for sub intensive care and 80 of whom perished, all these patients were admitted with a very high appropriateness level (data from the hospital administrative records) (4,5).
During their stay in hospital these patients, as all the others, faced also a considerable threat for their safety caused by healthcare associated infections (HAIs) possibly related to individual habits which might have determined adverse clinical outcomes and aggressive antimicrobial therapies with further resistance selection (6-13).
As reported in another study all these 246 patients were routinely followed with the usual local infection control surveillance program to detect colonization by multidrug-resistant bacteria, namely MRSA (Methicillin-resistant Staphylococcus aureus), multidrug-resistant Gram-negative bacteria and VRE (Vancomycin-resistant enterococci) in addiction received all the microbiological investigation in case of infectious symptoms (3).
The aim of this study is to assess the association among species of bacteria and to identify the presence of clusters of patients in sub intensive care unit with different profiles of infection, and to study the relationship between such profiles and patient demographics (gender, age), kind of investigations and material used to detect the infection.
Statistical Methods
To the aim of evaluating the association among different species of bacteria, a dataset of 74 rows (patients) and 27 columns (species of bacteria) was considered; for each bacteria the data was classified as absence or presence of the species of bacteria for each individual. All the species of bacteria presented in at most one subject were excluded from the analysis, because it was not reasonable to use such data to detect the co-presence with other species of bacteria. The association was described by Multiple Correspondence Analysis (MCA) methods (14,15). MCA yields graphical representations of subjects and variables in bidimensional scatterplots which preserve the greatest possible amount of global variability within the data. The axes of the plot represent the first factorial dimensions. The amount of variability explained by the figure is evaluated by indices called “proportions of explained inertia”. Such indices vary within the range 0%-100%, and the higher the explained inertia, the higher will be the overall level of association among the types of bacteria considered. Within the plot, the individuals are represented by points; presence and absence of each bacteria are represented also by points, and labeled with species id. In order to interpret the patterns of association within the plot, the points that are closest one another indicate which subjects have similar profiles (i.e. species of bacteria present) and which categories are mostly associated.
To identify patients profiles a cluster analysis was performed to identify groups of sampling points with homogeneous bacterial patterns. For determining the clusters, we used only the coordinates of points that represent the patients on the first factorial axis, since it explains the most relevant explained proportion of variability. The clustering algorithm was the hierarchical clustering with Euclidean distance metric and Ward linkage.
The results of cluster analysis were used to study the relationships between patient profiles and the reminder variables: patient demographics (gender, age), kind of investigations and the material used to detect the infection. Furthermore, as an aid to interpreting the such relationship, the projections of the remainder variables categories (considered as supplementary variables) were also represented within the MCA plot.
Results
74 patients were considered for the study with 186 exams, 58 (31%) in females and 128 (69%) in males, median and mean age were 66 years with 39-89 range and 58-79 1st and 3rd quartiles, respectively. Table 1 reports the specific material for the examinations and corresponding frequencies. The kind of investigations is reported in Table 2. The detected microorganisms are reported in Table 3.
Table 1.
Material for the investigation
| Material | N | Perccentage |
| Bronchial aspirate | 2 | 1.08% |
| Sputum | 7 | 3.76% |
| Arterial blood from catheter | 7 | 3.76% |
| Venous blood from catheter | 56 | 30.11% |
| Blood from vein | 54 | 29.03% |
| Arterial blood from catheter in situ (art) | 2 | 1.08% |
| Venuos blood from catheter in situ | 5 | 2.69% |
| Nasal swab | 2 | 1.08% |
| Urine form catheter | 39 | 20.97% |
| Middle jet urine | 9 | 4.84% |
| Urine from Stoma | 3 | 1.61% |
Table 2.
Kind of investigations
| Investigation | N | Perccentage |
| Blood culture AER 1 SET (Bact/ALERT) | 63 | 33.87% |
| Blood culture AER 2 SET (Bact/ALERT) | 10 | 5.38% |
| Blood culture ANA 1 SET (Bact/ALERT) | 43 | 23.12% |
| Blood culture ANA 2 SET (Bact/ALERT) | 8 | 4.30% |
| Lower respiratory tract sputum culture Colturale | 9 | 4.84% |
| Culture of nasopharyngeal swabs | 2 | 1.08% |
| Urine culture | 51 | 27.42% |
Table 3.
Detected microorganisms
| Microorganisms | N Investigations | Perccentage | N of Patients | Code |
| Aerococcus viridans | 1 | 0.54% | 1 | av |
| Bacillus clausii | 3 | 1.61% | 3 | bc |
| Citrobacter koseri | 1 | 0.54% | 1 | ck |
| Corynebacterium amycolatum | 1 | 0.54% | 1 | ca |
| Corynebacterium striatum | 1 | 0.54% | 1 | cs |
| Corynebacterium urealyticum | 1 | 0.54% | 1 | cu |
| Enterobacter aerogenes | 1 | 0.54% | 1 | ea |
| Enterococcus faecalis | 29 | 15.59% | 20 | ef |
| Enterococcus faecium | 13 | 6.99% | 9 | efaec |
| Escherichia coli | 22 | 11.83% | 17 | ec |
| K. pneumoniae resistente ai carbapenemi | 1 | 0.54% | 1 | kpr |
| Klebsiella pneumoniae | 8 | 4.30% | 7 | kp |
| Morganella morganii | 2 | 1.08% | 2 | mm |
| Proteus mirabilis | 8 | 4.30% | 5 | pm |
| Providencia stuartii | 2 | 1.08% | 2 | ps |
| Pseudomonas aeruginosa | 10 | 5.38% | 4 | pa |
| Serratia marcescens | 3 | 1.61% | 1 | smar |
| Stafilococco aureo Meticillino Resistente | 5 | 2.69% | 1 | samr |
| Stafilococco aureo Meticillino Sensibile | 1 | 0.54% | 1 | sams |
| Staphylococcus aureus | 15 | 8.06% | 9 | sa |
| Staphylococcus epidermidis | 29 | 15.59% | 18 | se |
| Staphylococcus haemolyticus | 11 | 5.91% | 10 | sha |
| Staphylococcus hominis | 14 | 7.53% | 10 | sho |
| Staphylococcus pettenkoferi | 1 | 0.54% | 1 | sp |
| Stenotrophomonas maltophilia | 1 | 0.54% | 1 | smal |
| Str. beta emol. Gr.F | 1 | 0.54% | 1 | sbegf |
| cocco-bacilli Gram positivi | 1 | 0.54% | 1 | cbgp |
Species which were found on only a single patient were: Aerococcus viridans, Citrobacter koseri, Corynebacterium amycolatum, Corynebacterium striatum, Corynebacterium urealyticum, Enterobacter aerogenes, K. pneumoniae resistente ai carbapenemi, Serratia marcescens, Stafilococco aureo Meticillino Resistente, Stafilococco aureo Meticillino Sensibile, Staphylococcus pettenkoferi, Stenotrophomonas maltophilia, Str. beta emol. Gr.F, cocco-bacilli Gram positive. They were excluded from further analyses. Concerning occurrences, 42 patients had a single species, and 17, 9, 5, 1 had 2,3,4,6 co-occurrent species respectively.
MCA was conducted to assess associations related to the concurrent infections on 13 bacterial species (strains): Bacillus clausii, Enterococcus faecalis, Enterococcus faecium, Escherichia coli, Klebsiella pneumonia, Morganella morganii, Proteus mirabilis, Providencia stuartii, Pseudomonas aeruginosa, Staphylococcus aureus, Staphylococcus epidermidis, Staphylococcus haemolyticus, Staphylococcus hominis. Furthermore the additional variables age, gender, ward, material and kind of investigations, were considered as supplementary variables.
Figure 1 shows the MCA plot for the first two axes. The variability explained by the first two axes was 24.8%, and 14.4% for the first and the second one respectively. The total explained variability was of 39.2%: this result suggests indicated that the overall association between the presence and absence among the thirteen bacteria was limited. For describing the profiles of infection we used only the first axis, because it explains the greatest proportions of variability and provides a better quality of representation of categories than the second axis.
Figure 1.

MCA Plot for describing the association among species of bacteria. The positions of the categories (0 for absence or 1 for presence) are indicated by triangles; the labels used for species of bacteria are reported in table 3 (column: Code). Patients are indicated by black points. The arrows represent the original position in the graph of the species of bacteria which are near to the origin.
Considering the categories well represented in the plot (as indicated by square cosine statistics at least 0.5 on the firs axis), the species of bacteria that were better represented were: Escherichia coli, Staphylococcus epidermidis, Staphylococcus hominis and Staphylococcus haemolyticus.
The position of the presence and the absence of the species above in the MCA plot suggested that the first axis diversified subjects who had the Escherichia coli and not the other three species from subjects who had not the Escherichia coli and had the other three species. In fact, 17 patients, out of 18 infected by Staphylococcus epidermidis, were not infected by Escherichia coli, 10 patients, out of 10 with Staphylococcus hominis, did not present Escherichia coli, and 8 patients, out of 9 with Staphylococcus haemolyticus, did not present Escherichia coli. Concerning the co-occurences between species: 6 times out of 10 Staphylococcus hominis was present when Staphylococcus haemolyticus was absent, 12 times out of 18 Staphylococcus epidermidis was present when Staphylococcus haemolyticus was absent and 14 times out of 18 Staphylococcus epidermidis was present when Staphylococcus hominis was absent). Therefore, we could support the pattern that in patients infected by Escherichia coli, Staphylococcus epidermidis, Staphylococcus hominis and Staphylococcus haemolyticus were generally not present, and vice-versa. On the contrary, we could not support that patients infected by one among Staphylococcus epidermidis, Staphylococcus hominis and Staphylococcus haemolyticus, are also infected by the remainder ones.
In cluster analysis four major infection profiles were identified; they are represented in Fig. 2.
Figure 2.
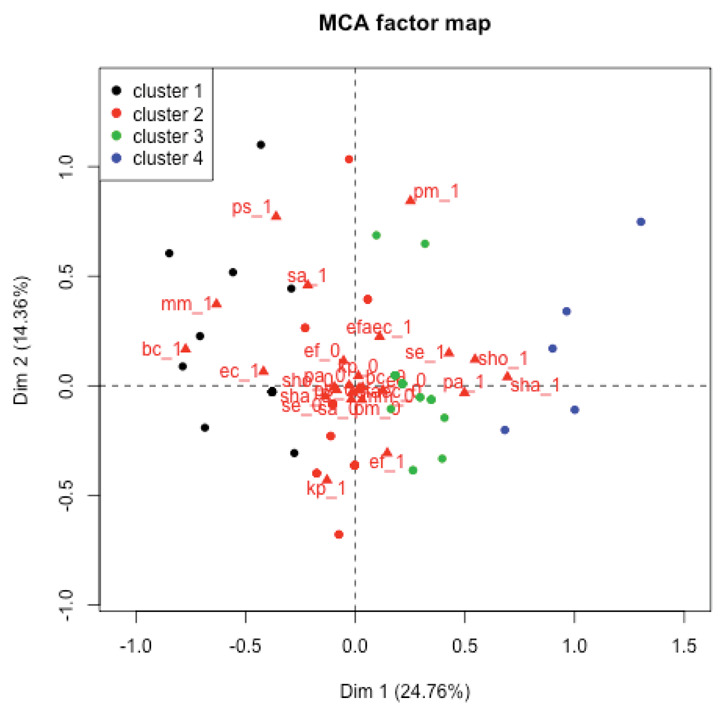
Cluster analysis on the MCA scores. Patients within clusters are projected on the MCA plane and points of patients within the same cluster are represented with the same color.
Cluster 1 had 18 subjects mainly characterized by the presence of Escherichia coli (16/18), cluster 2 included 28 subjects and it was mainly characterized by absence of every of the four species considered (only Staphylococcus epidermidis was found in 3 out of 28 subjects), cluster 3 had 23 subjects infected by either Staphylococcus epidermidis, Staphylococcus hominis or Staphylococcus haemolyticus and not infected by Escherichia coli; (1 subjects had Escherichia coli, 4 subjects had Staphylococcus haemolyticus, 6 subjects had Staphylococcus hominis and 10 had Staphylococcus epidermidis) cluster 4 had 5 subjects characterized by strong co-occurrence of Staphylococcus haemolyticus (5 subjects out of 5), Staphylococcus hominis (4 subjects out of 5) and Staphylococcus epidermidis (5 subjects out of 5).
Concerning supplementary variables, age suggested a very low general association with the first dimension of MCA. Considering the relationship between age and the four clusters it can be shown that age tended to decrease from cluster 1 to cluster 4: median age was equal to 72.00 for cluster 1, was 64.50 for cluster 2, was 63.00 for cluster 3 and was 59.0 for cluster 4 (see Figure 3).
Figure 3.
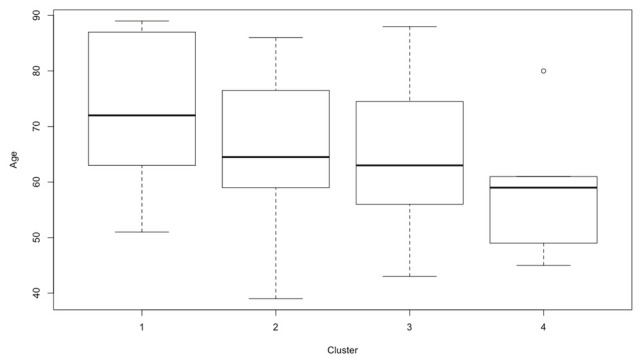
Age distribution according to the clusters
Sex was not well represented in the first dimension of MCA (had a squared cosine equal to 0.157). Anyway, cluster 1 had 10/18 males (55.6%), cluster 2 had 20/28 (71.4%) males, cluster 3 had 16/23 (69.6%) males and cluster 4 had 4/5 male (80.0%).
Materials that were well represented in the first dimension of the MCA were: arterial catheter blood (squared cosine equal to 0.568), venous catheter blood (cosine squared equal to 0.542) and in situ catheter blood (venous) (squared cosine equal to 0.581). However arterial catheter blood and in situ catheter blood (venous) were picked up in very few subjects (4 and 3 respectively). arterial catheter blood was not picked up not used for subjects in cluster 1, while in situ catheter blood (venous) was picked up only in subjects in cluster 3 and 4.
Cluster 1 and 2 were characterized by absence of the venous catheter blood (in 16 subjects out of 18 for the first and in 22 subjects out of 28 for the second this material had not been picked up); in contrast with clusters 3 and 4 which are characterized by large use of this material (11/23 and 5/5 picked up in cluster 3 and 4 respectively).
Concerning kind of investigation, only ANA 1 SET (Bact / ALERT) was well represented in the first dimension (squared cosine: 0.625),
In the first cluster 16/18 subjects (88.9%) had not the Blood culture ANA 1 SET (Bact / ALERT), in cluster 2, 22/28 subjects (78.6%); in contrast with clusters 3 and 4 with 10/23 and 5/5 subject with Blood culture ANA 1 SET (Bact / ALERT).
Discussion
All the patients admitted in hospital are routinally followed with the usual local infection control surveillance program to detect colonization by multidrug-resistant bacteria, namely MRSA (Methicillin-resistant Staphylococcus aureus), multidrug-resistant Gram-negative bacteria and VRE (Vancomycin-resistant enterococci) in addiction received all the microbiological investigation in case of infectious symptoms. This program were ensured also for all the COVID-19 positive patients. We studied the results of this program in 246 patients admitted to the sub intensive care unit in a research and teaching hospital located in the centre of Milan. 74 patients were considered in ous study because they presented infection symptoms after the performance of the surveillance program. Their mean age was 66 years. The most frequent positive material sent in microbiological laboratory was blood from venous catheter (56 – 30,11%) and consequently the most frequent investigation was blood culture (121 – 66,67%). The most frequent bacterium isolated was Staphylococcus epidermidis (29 – 15,59%).
MCA was conducted to assess the associations related to the concurrent infections on 13 bacterial species (strains) The overall association was low. Only 4 of the 13 species were well represented by the MCA plot, so the patterns of association were based only on these species. More specifically, it is possible to support the pattern of almost mutual exclusivity between Escherichia coli on one and Staphylococcus epidermidis, Staphylococcus hominis or Staphylococcus haemolyticus on the other one.
The evaluation of age and sex suggests a a very low general association in particular age is increasing from cluster 1 to cluster 4: median age was equal to 72.00 for cluster 1, was 64.50 for cluster 2,was 63.00 for cluster 3 and was 59.0 for cluster 4 (see Figure 3).
These considerations need to be study on a bigger amount of data in the same setting to make evident that it is constant the infection only with Escherichia coli and Staphylocossus epidemidis and a third case in which more bacteria are inlvolved.
Conflict of interest:
Each author declares that he or she has no commercial associations (e.g. consultancies, stock ownership, equity interest, patent/licensing arrangement etc.) that might pose a conflict of interest in connection with the submitted article
References
- 1. https://www.epicentro.iss.it/coronavirus/bollettino/Bollettino-sorveglianza-integrata-COVID-19_21-luglio-2020.pdf. (last view 27.7.2020) [Google Scholar]
- 2.Rivieccio BA, Luconi E, Boracchi P, Pariani E, Romanò L, Salini S, Castaldi S, Biganzoli E, Galli M. Heterogeneity of COVID-19 outbreak in Italy. Acta Biomed. 2020;91(2):31–34. doi: 10.23750/abm.v91i2.9579. DOI: 10.23750/abm.v91i2.9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auxilia F, Maraschini A, Bono P, Ungaro R, Luconi E, Biganzoli E, Castaldi S. COVID-19: new scenario old problems. Acta Biomed. 2020;91(Suplement 9):90–91. doi: 10.23750/abm.v91i9-S.10119. DOI: 10.23750/abm.v91i2-S.10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brownlee S, Chalkidou K, Doust J, et al. Evidence for overuse of medical services around the world. Lancet. 2017;390:156–68. doi: 10.1016/S0140-6736(16)32585-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castaldi S, Bevilacqua L, Arcari G, Cantù AP, Visconti U, Auxilia F. How appropriate is the use of rehabilitation facilities? Assessment by an evaluation tool based on the AEP protocol. Journal of Preventive Medicine and Hygiene. 2010;51(3):116–120. [PubMed] [Google Scholar]
- 6.Brusaferro S, Arnoldo L, Finzi G, et al. Board; Group. Hospital Hygiene and Infection Prevention and Control in Italy: state of the art and perspectives. Ann Ig. 2018 Sep-Oct;30(5 Supple 2):1–6. doi: 10.7416/ai.2018.2245. doi: 10.7416/ai.2018.2245 37. [DOI] [PubMed] [Google Scholar]
- 7.Montagna MT, Mascipinto S, Pousis C, et al. Knowledge, experiences, and attitudes toward Mantoux test among medical and health professional students in Italy: a crosssectional study. Ann Ig. 2018 Sep-Oct;30(5 Supple 2):86–98. doi: 10.7416/ai.2018.2253. Doi: 10.7416/ai.2018.2253. [DOI] [PubMed] [Google Scholar]
- 8.Mellace L, Consonni D, Jacchetti G, Del Medico M, Colombo R, Velati M, et al. Epidemiology of Clostridium difficile-associated disease in internal medicine wards in northern Italy. Intern Emerg Med. 2013;8(8):717–723. doi: 10.1007/s11739-012-0752-6. [DOI] [PubMed] [Google Scholar]
- 9.Ardoino I, Zangirolami F, Iemmi D, Lanzoni M, Cargnelutti M, Biganzoli E, et al. Risk factors and epidemiology of Acinetobacter baumannii infections in a university hospital in Northern Italy: A case-control study. Am J Infect Control. 2016;44(12):1600–1605. doi: 10.1016/j.ajic.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 10.Capobussi M, Sabatino G, Donadini A, Tersalvi CA, Castaldi S. Control of scabies outbreaks in an Italian hospital: An information-centered management strategy. Am J Infect Control. 2014;42(3):316–320. doi: 10.1016/j.ajic.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 11.Prigitano A, Romanò L, Auxilia F, Castaldi S, Tortorano AM. Antibiotic resistance: Italian awareness survey 2016. J Infect Public Health. 2018;11(1):30–34. doi: 10.1016/j.jiph.2017.02.010. IF 2,118 Q2. [DOI] [PubMed] [Google Scholar]
- 12.Burriel MS, Keys M, Campillo-Artero C, Agodi A, Barchitta M, Gikas A, Palos C, Lopez-Casasnovas G. Impact of multi-drug resistant bacteria on economic and clinical outcomes of healthcare-associated infections in adults: Systematic review and meta-analysis. PLoS One. 2020;15(1):e0227139. doi: 10.1371/journal.pone.0227139. doi: 10.1371/journal.pone.0227139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pasquarella C, Veronesi L, Napoli C, Castaldi S, Pasquarella M.L, Saccani E, Colucci M.E, Auxilia F, Galle F, Di Onofrio V, Tafuri S, Signorelli C, Liguori G. What about behaviours in swimming pools? Results of an Italian multicentre study. Microchem. J. 2014;112:190–195. doi: 10.1016/j.puhe.2013.01.014. https://doi.org/10.1016/j.microc.2013. 09.024 . [DOI] [PubMed] [Google Scholar]
- 14.Lebart L, Morineau A, Piron M. Vol. 3. Paris: Dunod; 1995. Statistique exploratoire multidimensionnelle. [Google Scholar]
- 15.Husson F, Lê S, Pagès J. Exploratory multivariate analysis by example using R. CRC press. 2017 [Google Scholar]


