Abstract
Pan44T, a novel strain belonging to the phylum Planctomycetes, was isolated from a red biofilm in a hydrothermal area close to the island Panarea in the Tyrrhenian Sea north of Sicily, Italy. The strain forms white colonies on solid medium and displays the following characteristics: cell division by budding, formation of rosettes, presence of matrix or fimbriae and long stalks. The cell surface has an interesting and characteristic texture made up of triangles and rectangles, which leads to a pine cone-like morphology of the strain. Strain Pan44T is mesophilic (temperature optimum 26 °C), slightly alkaliphilic (pH optimum 8.0), aerobic and heterotrophic. The strain has a genome size of 6.76 Mb with a G + C content of 63.2%. Phylogenetically, the strain is a member of the family Planctomycetaceae, order Planctomycetales, class Planctomycetia. Our analysis supports delineation of strain Pan44T from all known genera in this family, hence, we propose to assign it to a novel species within a novel genus, for which we propose the name Caulifigura coniformis gen. nov., sp. nov., represented by Pan44T (DSM 29405T = LMG 29788T) as the type strain.
Keywords: Marine bacteria, Mediterranean Sea, Biotic surfaces, Planctomycetes, Panarea
Introduction
Planctomycetes is a bacterial phylum displaying exceptional cell biological features (Rivas-Marin and Devos 2018; Wiegand et al. 2018, 2020). Together with Chlamydiae, Verrucomicrobia and other sister phyla, the phylum Planctomycetes forms the PVC superphylum and several of its members have environmental, medical or biotechnological relevance (Spring et al. 2016; Wagner and Horn 2006). The phylum itself is subdivided into the classes Phycisphaerae, Planctomycetia and Candidatus Brocadiae, which display differences in their cell biology, e.g. mode of cell division and metabolism (Wiegand et al. 2020). One example are species of Cand. Brocadiae, which perform unique reactions during anaerobic ammonium oxidation (anammox) (Strous et al. 1999). These reactions are e.g. exploited for converting ammonium to dinitrogen gas during N-elimination in wastewater treatment plants (Peeters and van Niftrik 2018). The class Phycisphaerae comprises strains that form spherical cells and divide by binary fission (Fukunaga et al. 2009). This is a decisive difference compared to budding as the observed mode of cell division in the other two classes. Similar to Phycisphaerae, species belonging to genera within the class Planctomycetia have been often isolated from aquatic biotic and abiotic surfaces (Bondoso et al. 2014, 2017; Kohn et al. 2016; Vollmers et al. 2017), on which they can be highly abundant (Bengtsson and Øvreås 2010). Such species likely use complex polysaccharides derived from biotic surfaces, in particular macroscopic phototrophs, as source of carbon and energy (Jeske et al. 2013; Lachnit et al. 2013). However, the dominance of planctomycetal species on such surfaces is remarkable given their rather slow growth compared to natural competitors in this ecological niche, e.g. members of the ‘Roseobacter group’ (Frank et al. 2014). Underlying mechanisms allowing Planctomycetes to compensate for lower growth rates may include the capability to produce bioactive small molecules (Graça et al. 2016; Jeske et al. 2016; Kallscheuer et al. 2019b), resistance against several antibiotics (Cayrou et al. 2010; Godinho et al. 2019) and a specialised machinery for the uptake and intracellular digestion of complex polysaccharides. The latter is suspected to be facilitated by unique pili-forming crateriform structures and an extremely enlarged periplasmic space (Boedeker et al. 2017).
The application of novel microscopic techniques and genetic tools for Planctomycetes (Jogler et al. 2011; Jogler and Jogler 2013; Rivas-Marin et al. 2016b) has given more detailed insights into their cell envelope architecture and the mode of cell division (Rivas-Marin et al. 2020a). Planctomycetes were shown to possess peptidoglycan (Jeske et al. 2015; van Teeseling et al. 2015), which led to the conclusion that the cell envelope architecture of Planctomycetes resembles that of Gram-negative bacteria (Boedeker et al. 2017; Devos 2014). Nevertheless, Planctomycetes are still exceptional. Members of the orders Gemmatales, Isosphaerales, Planctomycetales and Pirellulales, as well as of the class Cand. Brocadiae, divide by budding while binary fission is the observed mode of division in the class Phycisphaerae (Wiegand et al. 2020). All known Planctomycetes lack many of the canonical divisome proteins including the otherwise universal FtsZ (Jogler et al. 2012; Pilhofer et al. 2008, Rivas-Marin et al. 2016a). Given their fascinating cell biology, several novel strains have been described in the recent year (Boersma et al. 2019; Dedysh et al. 2019b; Kallscheuer et al. 2019a, 2019c, 2020a, 2020b, Kohn et al. 2020; Kovaleva et al. 2019; Kulichevskaya et al. 2019; Peeters et al. 2020; Rensink et al. 2020), which led to an updated taxonomy and more precise definition of threshold values of phylogenetic markers in the class Planctomycetia (Dedysh et al. 2019b; Kallscheuer et al. 2019c).
As an additional contribution, here we describe the novel strain Pan44T isolated from a red biofilm sampled in the shallow-water hydrothermal vent system close to Panarea Island, Italy.
Materials and methods
Isolation of the novel strain and cultivation
M1 medium with 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) as buffering agent and additionally supplemented with N-acetyl glucosamine (NAG) and artificial seawater (ASW) was used for the isolation and cultivation of strain Pan44T. The medium, designated M1H NAG ASW, was prepared as described (Boersma et al. 2019). Isolation of strain Pan44T from a red biofilm sampled in a hydrothermal area close to Panarea island (exact location 38.5568 N 15.1097 E) was previously described (Wiegand et al. 2020). The biofilm was isolated on the 9th of September 2013. Briefly, a piece of the natural biofilm was scraped off into sterile natural seawater using single-use scalpels. 20 µL of the biofilm suspension was streaked on M1H NAG ASW agar plates containing 500 mg/L streptomycin, 200 mg/L ampicillin and 20 mg/L cycloheximide, which were incubated at 20 °C for at least four weeks. The 16S rRNA gene of colonies obtained was amplified by PCR and sequenced following an established protocol (Rast et al. 2017). This step was performed in order to ensure that isolated strains selected for further characterisation indeed represent members of the phylum Planctomycetes.
Determination of pH and temperature optimum
Cultivations for determination of the pH optimum were performed in M1H NAG ASW medium with 100 mM HEPES for cultivations at pH 7.0, 7.5 and 8.0. For cultivation at pH 5.0 and 6.0 HEPES was replaced by 100 mM 2-(N-morpholino)ethanesulfonic acid (MES), whereas 100 mM N-cyclohexyl-2-aminoethanesulfonic acid (CHES) served as a buffering agent at pH 9.0 and 10.0. Cultivations for determination of the pH optimum were performed at 28 °C. Cultivations for determination of the temperature optimum were performed in standard M1H NAG ASW medium at pH 8.0. Cell densities were measured as optical density at 600 nm (OD600).
Microscopy protocols
Phase contrast and field emission scanning electron microscopy (SEM) were performed as previously described (Boersma et al. 2019). Transmission electron microscopy was performed according to a previously published protocol (Kohn et al. 2016).
Genome information
The genome and 16S rRNA gene sequence of strain Pan44T are available from GenBank under accession numbers CP036271 and MK554532, respectively. Sequencing of the genome is described in a previous study (Wiegand et al. 2020).
Phylogenetic analysis
16S rRNA gene sequence-based phylogeny was computed for strain Pan44T, the type strains of all described planctomycetal species (assessed in January 2020) including all isolates recently published and described (Boersma et al. 2019; Dedysh et al. 2019a, 2019b; Kallscheuer et al. 2019a, 2019c, 2020a, 2020b; Kohn et al. 2020; Peeters et al. 2020; Rensink et al. 2020). The 16S rRNA gene sequences were aligned with SINA (Pruesse et al. 2012) and the phylogenetic inference was calculated with RAxML (Stamatakis 2014) with a maximum likelihood approach with 1000 bootstraps, nucleotide substitution model GTR, gamma distributed rate variation and estimation of proportion of invariable sites (GTRGAMMAI option). Three 16S rRNA genes of bacterial strains from the PVC superphylum, outside of the phylum Planctomycetes (Opitutus terrae, acc. no. AJ229235; Kiritimatiella glycovorans, acc. no. NR_146840 and Lentisphaera araneosa, acc. no. NR_027571), were used as outgroup. For the multi-locus sequence analysis (MLSA), the unique single-copy core genome of the analysed genomes was determined with proteinortho5 (Lechner et al. 2011) with the ‘selfblast’ option enabled. The protein sequences of the resulting orthologous groups were aligned using MUSCLE v.3.8.31 (Edgar 2004). After clipping, partially aligned C- and N-terminal regions and poorly aligned internal regions were filtered using Gblocks (Castresana 2000). The final alignment was concatenated and clustered using the maximum likelihood method implemented by RaxML (Stamatakis 2014) with the ‘rapid bootstrap’ method and 500 bootstrap replicates. Five planctomycetal genomes from the order Pirellulales served as outgroup. The average nucleotide identity (ANI) was calculated using OrthoANI (Lee et al. 2016). The average amino acid identity (AAI) was calculated using the aai.rb script of the enveomics collection (Rodriguez-R and Konstantinidis 2016) and the percentage of conserved proteins (POCP) was calculated as described (Qin et al. 2014). The rpoB nucleotide sequences were taken from publicly available planctomycetal genome annotations and the sequence identities were determined as described (Bondoso et al. 2013). Upon extracting only those parts of the sequence that would have been sequenced with the described primer set, the alignment and matrix calculation was done with Clustal Omega (Sievers et al. 2011).
Results and discussion
Phylogenetic inference
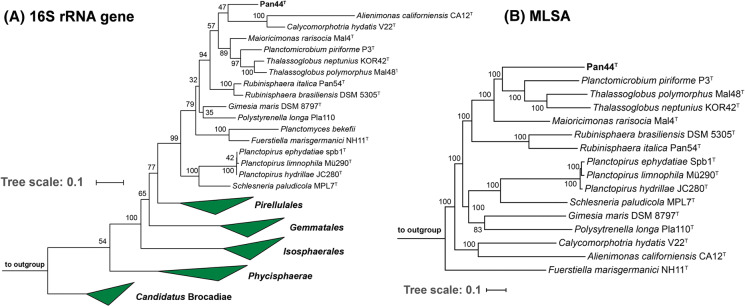
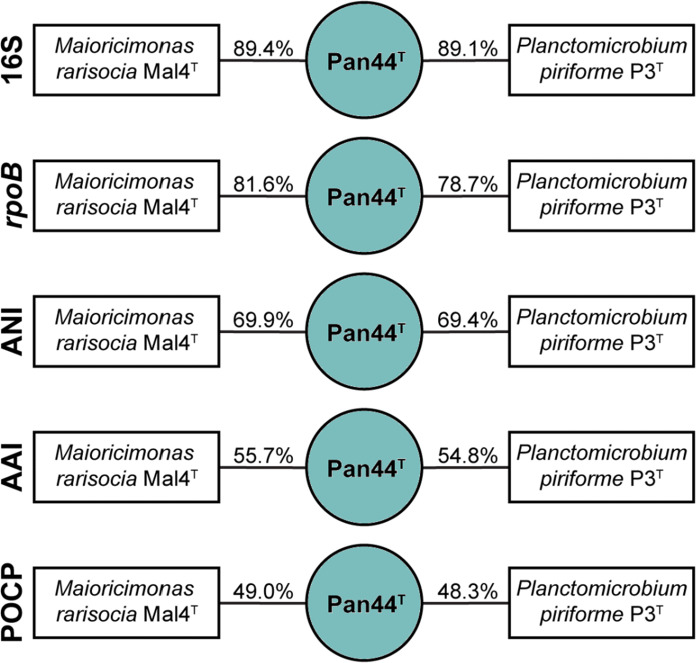
In the phylogenetic trees obtained from 16S rRNA gene sequence analysis and MLSA (Fig. 1), strain Pan44T was observed to cluster within the family Planctomycetaceae, which is currently the sole family within the order Planctomycetales. All investigated phylogenetic markers (16S rRNA gene identity, rpoB identity, AAI, ANI and POCP) suggest Maioricimonas rarisocia Mal4T and Planctomicrobium piriforme P3T to be the current closest neighbours (Kulichevskaya et al. 2015; Rivas-Marin et al. 2020b). ANI values of 69.9% and 69.4%, respectively, indicate that strain Pan44T is not a member of the species M. rarisocia or P. piriforme. The 16S rRNA gene sequence identity of strain Pan44T compared to both strains is < 90% and thus falls below the proposed genus threshold of 94.5% (Yarza et al. 2014) (Fig. 2), thereby suggesting clear delineation of strain Pan44T from members of the two genera. This conclusion is further supported by analysis of additional phylogenetic markers. Comparison of strain Pan44T with M. rarisocia Mal4T and P. piriforme P3T yielded AAI and POCP values below the respective genus thresholds of 60% and 50%, respectively (Konstantinidis and Tiedje 2005; Qin et al. 2014) (Fig. 2). During analysis of a partial sequence of the rpoB gene (Fig. 2), we obtained identity values slightly above the proposed genus threshold of 75.5–78% (Kallscheuer et al. 2019c). This, however, should not overrule the overall conclusion based on the other phylogenetic markers, which are in line with the delineation of strain Pan44T from known genera in the family Planctomycetaceae.
Fig. 1.
Maximum likelihood phylogenetic analysis. Phylogenetic trees showing the position of strain Pan44T. 16S rRNA gene- and MLSA-based phylogeny was computed as described in the Materials and methods section. Bootstrap values after 1000 re-samplings (16S rRNA gene)/500 re-samplings (MLSA) are given at the nodes (in %). The outgroup for the 16S rRNA-based tree consists of three 16S rRNA genes from the PVC superphylum. For the MLSA tree Rubripirellula obstinata, Rhodopirellula baltica, Roseimaritima ulvae, Bythopirellula goksoyri and Thermogutta terrifontis were used as outgroup
Fig. 2.
Analysis of phylogenetic markers. The figure shows the comparison of strain Pan44T to its current closest relatives. Analysed markers include 16S rRNA gene identity (16S), rpoB gene identity, average nucleotide identity (ANI), average amino acid identity (AAI) and percentage of conserved proteins (POCP)
Morphological and physiological analyses
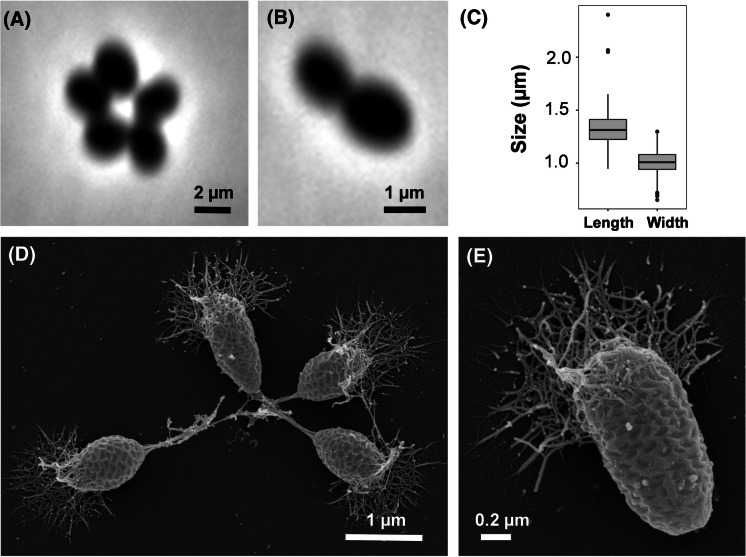
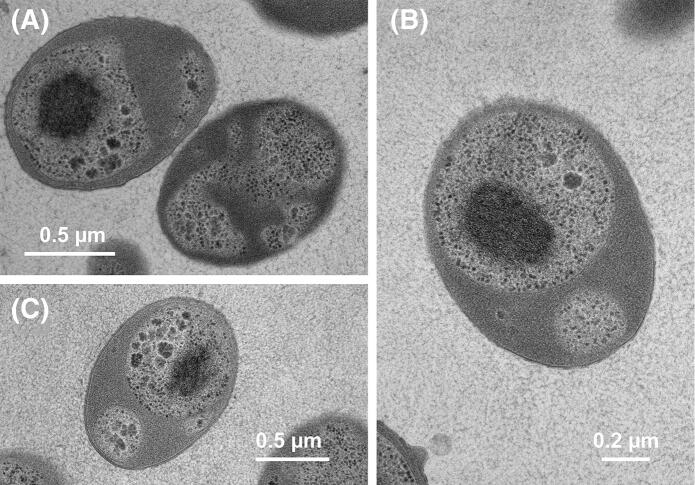
Basic features of strain Pan44T regarding its physiology and morphology are summarised in Table 1 and compared to the close relatives M. rarisocia Mal4T and P. piriforme P3T. For the analysis of morphological features, Pan44T cells were harvested in the exponential growth phase and were analysed using phase contrast light microscopy and SEM (Fig. 3). Strain Pan44T forms pear-shaped cells with an average size of 1.3 ± 0.2 × 1.0 ± 0.1 µm (Fig. 3a, c), which either appear as single cells or form rosettes or larger aggregates (Fig. 3d, e). Similar to P. piriforme, strain Pan44T forms long stalks on one of the cell poles, which can reach a length of up to 0.8 µm (Fig. 3d). Stalks are particularly visible in smaller aggregates with less than ten connected cells. In contrast, stalks of M. rarisocia Mal4T are much shorter. On the opposite pole of Pan44T cells, fimbriae or matrix are usually formed. The cell surface has a characteristic texture comprised of triangles or rectangles, which resembles a pine cone (Fig. 3d, e). Such depressions can be artefacts of critical point drying during SEM specimen preparation. However, we optimised our preparation protocol for Planctomycetes and have never observed such an unusual pine cone texture in any other planctomycetal species described thus far (Boersma et al. 2019; Kallscheuer et al. 2019a, 2019c, 2020a, 2020b; Kohn et al. 2020; Peeters et al. 2020; Rensink et al. 2020). If SEM artefacts occur, planctomycetal cells tend to appear crescent-shaped, indicating that the pine cone texture of strain Pan44T might be real rather than artefactual. Transmission electron micrographs of thin sections show typical planctomycetal features of Pan44T cells, such as a condensed nucleoid and invaginations of the cytoplasmic membrane (Fig. 4).
Table 1.
Phenotypic and genotypic features of strain Pan44T compared to the closely related strains Maioricimonas rarisocia Mal4T and Planctomicrobium piriforme P3T
| Feature | Pan44T | Maioricimonas rarisocia Mal4T | Planctomicrobium piriforme P3T |
|---|---|---|---|
| Phenotypic features | |||
| Shape | Pear-shaped | Pear-shaped | Ellipsoid to pear-shaped |
| Length (µm) | 1.3 ± 0.2 | 2.0 | 1.7–2.8 |
| Width (µm) | 1.0 ± 0.1 | 1.4 | 0.9–1.3 |
| Colour | White | Orange | White |
| Temperature range (optimum) (°C) | 15–30 (26) | 10–39 (31) | 10–30 (20–28) |
| pH range (optimum) | 5.0–10.0 (8.0) | 6.5–9.0 (7.5) | 4.2–7.1 (6.0–6.5) |
| Aggregates | Yes, rosettes | Yes, rarely | Yes, rosettes |
| Division | Budding | Budding | Budding |
| Dimorphic life cycle | n.o. | n.o. | Yes |
| Flagella | n.o. | n.o. | Yes |
| Crateriform structures | n.o. | Yes, overall | At reproductive pole |
| Fimbriae | Yes | Yes, overall matrix or fibre | Yes |
| Stalk | Yes | Yes | Yes |
| Holdfast structure | n.o. | n.o. | n.o. |
| Genomic features | |||
| Genome size (bp) | 6,761,146 | 7,744,989 | 6,317,004 |
| Plasmids | No | No | n.o. |
| G + C content (%) | 63.2 | 63.4 | 58.8 ± 1.7 |
| Completeness (%) | 96.55 | 98.28 | 95.69 |
| Contamination (%) | 1.72 | 0 | 1.72 |
| Total genes | 5587 | 5915 | 5117 |
| Genes/Mb | 826 | 764 | 810 |
| Giant genes | 0 | 1 | 1 |
| Protein-coding genes | 5524 | 5829 | 5050 |
| Proteins-coding genes/Mb | 817 | 753 | 799 |
| Hypothetical proteins | 2357 | 2257 | 2814 |
| Coding density (%) | 86.9 | 85.9 | 85.8 |
| tRNAs | 51 | 55 | 53 |
| 16S rRNA genes | 2 | 2 | 1 |
Fig. 3.
Microscopy images and cell size plot of strain Pan44T. The mode of cell division (b) and a general overview of cell morphology (a, d, e) is shown in the pictures, respectively. For determination of the cell size (c) at least 100 representative cells were counted manually or by using a semi-automated object count tool
Fig. 4.
Thin sections of Pan44T cells. Transmission electron micrographs show a condensed nuceloid and invaginations of the cytoplasmic membrane of Pan44T cells. Separate scales bars are provided for each of the photographs
Strain Pan44T divides by budding with the daughter cell having the same shape as the mother cell (Fig. 3b). Rosettes formed by strain Pan44T look similar to those formed by P. piriforme, while M. rarisocia Mal4T mostly occurs in the form of single cells and only in rare cases forms aggregates. Cell width for the three compared strains is similar, however, cells of strain Pan44T are slightly shorter. Strain Pan44T and P. piriforme P3T lack pigmentation, whereas M. rarisocia Mal4T is one of the rare examples of an orange-pigmented Planctomycete.
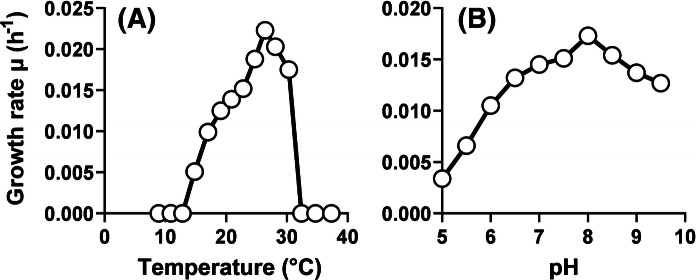
In M1H NAG ASW medium, strain Pan44T was found to grow over a range of 15–30 °C, with optimal growth at 26 °C (Fig. 5a). The temperature profile for growth is comparable to that of P. piriforme (range: 10–30 °C, optimum: 20–28 °C) (Table 1). In contrast, large differences were observed for the pH range. P. piriforme is slightly acidiphilic with a pH optimum of 6.0–6.5, whereas strain Pan44T showed optimal growth under slightly alkaline conditions (pH 8.0). The strain is able to grow over a range of pH 5–10, while maintaining more than 60% of the maximal growth rate at pH 6.0 and 10.0 (Fig. 5b). The notably broad pH range of strain Pan44T might be an indication of fluctuating pH values in its natural environment. The highest observed growth rate of strain Pan44T in M1H NAG ASW medium was established to be 0.022 h−1, corresponding to a doubling time of 32 h.
Fig. 5.
Temperature and pH optimum of strain Pan44T. The graphs show the average growth rates obtained from cultivation in M1H NAG ASW medium in biological triplicates. Cultivations at different pH values were conducted at 28 °C and cultivations at different temperatures were performed at pH 8.0
Genomic characteristics
The genome of strain Pan44T has a size of 6.76 Mb with a G + C content of 63%. Its genome is 7% larger than the P. piriforme P3T genome, which has a slightly lower G + C content (59%). The genome is 1 Mb smaller compared to M. rarisocia Mal4T, but the G + C content is nearly identical. Comparable numbers were observed for protein-coding genes per Mb and coding densities (Table 1). 43% of the putative protein-encoding genes found in strain Pan44T are of unknown function, which is in the range of 40–55% calculated for most of the planctomycetal genomes sequenced so far (Bordin et al. 2018). With 2814 hypothetical proteins out of a total number 5050 protein-encoding genes (56%) this number is notably higher in P. piriforme. The number of tRNAs in the three strains is comparable. Strain Pan44T and M. rarisocia Mal4T harbour two copies of the 16S rRNA gene, while a single 16S rRNA gene was found in P. piriforme P3T.
Although displaying similarities in cell morphology and genome properties, significant differences between the three compared strains were observed, e.g. with regard to colony colour, pH range and optimum, number of hypothetical proteins, the unusual pine cone texture of the cell surface and length of the stalk. Together with the results of the phylogenetic analysis, the data justifies delineation of strain Pan44T from the genera Maioricimonas and Planctomicrobium. Hence, we conclude that the novel isolate Pan44T (= DSM 29405T = LMG 29788T) represents a novel species belonging to a novel genus, for which we propose the name Caulifigura coniformis gen. nov., sp. nov.
Caulifigura gen. nov.
Caulifigura (Cau.li.fi.gu’ra. L. masc. n. caulis a stalk, stem; L. fem. n. figura a form, a figure; N.L. fem. n. Caulifigura a bacterium shaped like a stalk).
Members of the genus have a Gram-negative cell envelope architecture, are aerobic, mesophilic, neutrophilic to alkaliphilic and heterotrophic. Cells lack pigmentation, divide by budding and produce matrix or fimbriae originating from one of the cell poles. The genus belongs to the family Planctomycetaceae, order Planctomycetales, class Planctomycetia, phylum Planctomycetes. The type species of the genus is Caulifigura coniformis.
Caulifigura coniformis sp. nov.
Caulifigura coniformis (co.ni.for’mis. L. masc. n. conus a pine cone; L. masc. adj. suff. -formis –like, in the shape of; N.L. fem. adj. coniformis shaped like a pine cone, describing the morphology of the cells).
In addition to the genus characteristics, cells are pear-shaped (average size of 1.3 ± 0.2 × 1.0 ± 0.1 µm), occur as single cells, rosettes or larger aggregates and have a characteristic textured cell surface resembling a pine cone. Cells form long stalks. Cells of the type strain grow over ranges of 10–30 °C (optimum 26 °C) and pH 5.0–10.0 (optimum 8.0). Colonies are white. The genome size of the type strain is 6.76 Mb with a G + C content of 63.2%.
The type strain is Pan44T (= DSM 29405T = LMG 29788T), isolated from a red biofilm in a hydrothermal are close to the island Panarea, Italy in September 2013. The type strain genome (acc. no. CP036271) and 16S rRNA gene sequence (acc. no. MK554532) are available from GenBank.
Acknowledgements
Open Access funding provided by Projekt DEAL. Part of this research was funded by the Deutsche Forschungsgemeinschaft (DFG) Grants KA 4967/1-1 and JO 893/4-1, Grant ALWOP.308 of the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO), SIAM (Soehngen Institute for Anaerobic Microbiology) Grant No. 024002002 and the Radboud Excellence fellowship. We thank Ina Schleicher for skillful technical assistance. We thank Brian Tindall and Regine Fähnrich from the DSMZ as well as the staff from the BCCM/LMG Bacteria collection for support during strain deposition. We also thank the Scientific Diving Center of the Bergakademie Freiberg, Germany, Thomas Pohl, Peter Hornburger and all participants of the 2013 Panarea Expedition for sampling support.
Author contributions
NK wrote the manuscript and analysed the cultivation data, SW performed the genomic and phylogenetic analysis, AH and MJ isolated the strain and performed the initial cultivation and strain deposition, SHP and CB performed the light microscopic analysis and prepared the LM pictures, MSMJ contributed to text preparation and revised the manuscript, MR performed the electron microscopic analysis and prepared the SEM pictures, CJ took the samples, supervised AH and the study. All authors read and approved the final version of the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Bengtsson MM, Øvreås L. Planctomycetes dominate biofilms on surfaces of the kelp Laminaria hyperborea. BMC Microbiol. 2010;10:261. doi: 10.1186/1471-2180-10-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boedeker C, Schuler M, Reintjes G, Jeske O, van Teeseling MC, Jogler M, Rast P, Borchert D, Devos DP, Kucklick M, Schaffer M, Kolter R, van Niftrik L, Engelmann S, Amann R, Rohde M, Engelhardt H, Jogler C. Determining the bacterial cell biology of Planctomycetes. Nat Commun. 2017;8:14853. doi: 10.1038/ncomms14853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boersma AS, Kallscheuer N, Wiegand S, Rast P, Peeters SH, Mesman RJ, Heuer A, Boedeker C, Jetten MS, Rohde M, Jogler M, Jogler C. Alienimonas californiensis gen nov. sp. nov., a novel Planctomycete isolated from the kelp forest in Monterey Bay. Antonie van Leeuwenhoek. 2019 doi: 10.1007/s10482-019-01367-4. [DOI] [PubMed] [Google Scholar]
- Bondoso J, Harder J, Lage OM. rpoB gene as a novel molecular marker to infer phylogeny in Planctomycetales. Antonie Van Leeuwenhoek. 2013;104:477–488. doi: 10.1007/s10482-013-9980-7. [DOI] [PubMed] [Google Scholar]
- Bondoso J, Balague V, Gasol JM, Lage OM. Community composition of the Planctomycetes associated with different macroalgae. FEMS Microbiol Ecol. 2014;88:445–456. doi: 10.1111/1574-6941.12258. [DOI] [PubMed] [Google Scholar]
- Bondoso J, Godoy-Vitorino F, Balague V, Gasol JM, Harder J, Lage OM. Epiphytic Planctomycetes communities associated with three main groups of macroalgae. FEMS Microbiol Ecol. 2017;93:fiw255. doi: 10.1093/femsec/fiw255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordin N, González-Sánchez JC, Devos DP. PVCbase: an integrated web resource for the PVC bacterial proteomes. Database. 2018;2018:bay042. doi: 10.1093/database/bay042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Cayrou C, Raoult D, Drancourt M. Broad-spectrum antibiotic resistance of Planctomycetes organisms determined by Etest. J Antimicrob Chemother. 2010;65:2119–2122. doi: 10.1093/jac/dkq290. [DOI] [PubMed] [Google Scholar]
- Dedysh SN, Henke P, Ivanova AA, Kulichevskaya IS, Philippov DA, Meier-Kolthoff JP, Göker M, Huang S, Overmann J. 100-year-old enigma solved: identification, genomic characterization and biogeography of the yet uncultured Planctomyces bekefii. Environ Microbiol. 2019;22:198–211. doi: 10.1111/1462-2920.14838. [DOI] [PubMed] [Google Scholar]
- Dedysh SN, Kulichevskaya IS, Beletsky AV, Ivanova AA, Rijpstra WIC, Damsté JSS, Mardanov AV, Ravin NV. Lacipirellula parvula gen. nov., sp. nov., representing a lineage of planctomycetes widespread in low-oxygen habitats, description of the family Lacipirellulaceae fam. nov. and proposal of the orders Pirellulales ord. nov., Gemmatales ord. nov. and Isosphaerales ord. nov. Syst Appl Microbiol. 2019;43:126050. doi: 10.1016/j.syapm.2019.126050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos DP. PVC bacteria: variation of, but not exception to, the Gram-negative cell plan. Trends Microbiol. 2014;22:14–20. doi: 10.1016/j.tim.2013.10.008. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank O, Michael V, Pauker O, Boedeker C, Jogler C, Rohde M, Petersen J. Plasmid curing and the loss of grip: The 65-kb replicon of Phaeobacter inhibens DSM 17395 is required for biofilm formation, motility and the colonization of marine algae. Syst Appl Microbiol. 2014;38:120–127. doi: 10.1016/j.syapm.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Fukunaga Y, Kurahashi M, Sakiyama Y, Ohuchi M, Yokota A, Harayama S. Phycisphaera mikurensis gen. nov., sp. nov., isolated from a marine alga, and proposal of Phycisphaeraceae fam. nov., Phycisphaerales ord. nov. and Phycisphaerae classis nov. in the phylum Planctomycetes. J Gen Appl Microbiol. 2009;55:267–275. doi: 10.2323/jgam.55.267. [DOI] [PubMed] [Google Scholar]
- Godinho O, Calisto R, Ovreas L, Quinteira S, Lage OM. Antibiotic susceptibility of marine Planctomycetes. Antonie Van Leeuwenhoek. 2019;112:1273–1280. doi: 10.1007/s10482-019-01259-7. [DOI] [PubMed] [Google Scholar]
- Graça AP, Calisto R, Lage OM. Planctomycetes as Novel Source of Bioactive Molecules. Front Microbiol. 2016;7:1241. doi: 10.3389/fmicb.2016.01241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeske O, Jogler M, Petersen J, Sikorski J, Jogler C. From genome mining to phenotypic microarrays: planctomycetes as source for novel bioactive molecules. Antonie Van Leeuwenhoek. 2013;104:551–567. doi: 10.1007/s10482-013-0007-1. [DOI] [PubMed] [Google Scholar]
- Jeske O, Schüler M, Schumann P, Schneider A, Boedeker C, Jogler M, Bollschweiler D, Rohde M, Mayer C, Engelhardt H, Spring S, Jogler C. Planctomycetes do possess a peptidoglycan cell wall. Nat Commun. 2015;6:7116. doi: 10.1038/ncomms8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeske O, Surup F, Ketteniß M, Rast P, Förster B, Jogler M, Wink J, Jogler C. Developing techniques for the utilization of Planctomycetes as producers of bioactive molecules. Front Microbiol. 2016;7:1242. doi: 10.3389/fmicb.2016.01242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jogler M, Jogler C. Towards the development of genetic tools for Planctomycetes. In: Fuerst JA, editor. Planctomycetes: cell structure, origins and biology. Berlin: Springer; 2013. pp. 141–164. [Google Scholar]
- Jogler C, Glöckner FO, Kolter R. Characterization of Planctomyces limnophilus and development of genetic tools for its manipulation establish it as a model species for the phylum Planctomycetes. Appl Environ Microbiol. 2011;77:5826–5829. doi: 10.1128/AEM.05132-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jogler C, Waldmann J, Huang X, Jogler M, Glöckner FO, Mascher T, Kolter R. Identification of proteins likely to be involved in morphogenesis, cell division, and signal transduction in Planctomycetes by comparative genomics. J Bacteriol. 2012;194:6419–6430. doi: 10.1128/JB.01325-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallscheuer N, Jogler M, Wiegand S, Peeters SH, Heuer A, Boedeker C, Jetten MS, Rohde M, Jogler C. Three novel Rubripirellula species isolated from plastic particles submerged in the Baltic Sea and the estuary of the river Warnow in northern Germany. Antonie Van Leeuwenhoek. 2019 doi: 10.1007/s10482-019-01368-3. [DOI] [PubMed] [Google Scholar]
- Kallscheuer N, Moreira C, Airs R, Llewellyn CA, Wiegand S, Jogler C, Lage OM. Pink-and orange-pigmented Planctomycetes produce saproxanthin-type carotenoids including a rare C45 carotenoid. Environ Microbiol Rep. 2019;11:741–748. doi: 10.1111/1758-2229.12796. [DOI] [PubMed] [Google Scholar]
- Kallscheuer N, Wiegand S, Peeters SH, Jogler M, Boedeker C, Heuer A, Rast P, Jetten MS, Rohde M, Jogler C. Description of three bacterial strains belonging to the new genus Novipirellula gen. nov., reclassificiation of Rhodopirellula rosea and Rhodopirellula caenicola and readjustment of the genus threshold of the phylogenetic marker rpoB for Planctomycetaceae. Antonie van Leeuwenhoek. 2019 doi: 10.1007/s10482-019-01374-5. [DOI] [PubMed] [Google Scholar]
- Kallscheuer N, Wiegand S, Boedeker C, Peeters SH, Jogler M, Rast P, Heuer A, Jetten MS, Rohde M, Jogler C. Aureliella helgolandensis gen. nov., sp. nov., a novel Planctomycete isolated from a jellyfish at the shore of the island Helgoland. Antonie van Leeuwenhoek. 2020 doi: 10.1007/s10482-020-01403-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallscheuer N, Wiegand S, Heuer A, Rensink S, Boersma AS, Jogler M, Boedeker C, Peeters SH, Rast P, Jetten MS, Rohde M, Jogler C. Blastopirellula retiformator sp. nov. isolated from the shallow-sea hydrothermal vent system close to Panarea Island. Antonie van Leeuwenhoek. 2020 doi: 10.1007/s10482-019-01377-2. [DOI] [PubMed] [Google Scholar]
- Kohn T, Heuer A, Jogler M, Vollmers J, Boedeker C, Bunk B, Rast P, Borchert D, Glöckner I, Freese HM, Klenk H-P, Overmann J, Kaster A-K, Rohde M, Wiegand S, Jogler C. Fuerstia marisgermanicae gen. nov., sp. nov., an unusual member of the phylum Planctomycetes from the German Wadden Sea. Front Microbiol. 2016;7:2079. doi: 10.3389/fmicb.2016.02079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn T, Wiegand S, Boedeker C, Rast P, Heuer A, Schüler M, Rohde C, Müller R-W, Brümmer F, Rohde M, Engelhardt H, Jogler M, Jogler C. Planctopirus ephydatiae, a novel planctomycete isolated from a freshwater sponge. Syst Appl Microbiol. 2020;43:126066. doi: 10.1016/j.syapm.2019.126022. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. Towards a genome-based taxonomy for prokaryotes. J Bacteriol. 2005;187:6258–6264. doi: 10.1128/JB.187.18.6258-6264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovaleva OL, Elcheninov AG, Toshchakov SV, Novikov AA, Bonch-Osmolovskaya EA, Kublanov IV. Tautonia sociabilis gen. nov., sp. nov., a novel thermotolerant planctomycete, isolated from a 4000 m deep subterranean habitat. Int J Syst Evol Microbiol. 2019;69:2299–2304. doi: 10.1099/ijsem.0.003467. [DOI] [PubMed] [Google Scholar]
- Kulichevskaya IS, Ivanova AA, Detkova EN, Rijpstra WIC, Damste JSS, Dedysh SN. Planctomicrobium piriforme gen. nov., sp. nov., a stalked planctomycete from a littoral wetland of a boreal lake. Int J Syst Evol Microbiol. 2015;65:1659–1665. doi: 10.1099/ijs.0.000154. [DOI] [PubMed] [Google Scholar]
- Kulichevskaya IS, Naumoff DG, Miroshnikov KK, Ivanova AA, Philippov DA, Hakobyan A, Rijpstra WIC, Damsté JSS, Liesack W, Dedysh SN. Limnoglobus roseus gen. nov., sp. nov., a novel freshwater planctomycete with a giant genome from the family Gemmataceae. Int J Syst Evol Microbiol. 2019;70:1240–1249. doi: 10.1099/ijsem.0.003904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachnit T, Fischer M, Kunzel S, Baines JF, Harder T. Compounds associated with algal surfaces mediate epiphytic colonization of the marine macroalga Fucus vesiculosus. FEMS Microbiol Ecol. 2013;84:411–420. doi: 10.1111/1574-6941.12071. [DOI] [PubMed] [Google Scholar]
- Lechner M, Findeiss S, Steiner L, Marz M, Stadler PF, Prohaska SJ. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinform. 2011;12:124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Ouk Kim Y, Park SC, Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66:1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- Peeters SH, van Niftrik L. Trending topics and open questions in anaerobic ammonium oxidation. Curr Opin Chem Biol. 2018;49:45–52. doi: 10.1016/j.cbpa.2018.09.022. [DOI] [PubMed] [Google Scholar]
- Peeters SH, Wiegand S, Kallscheuer N, Jogler M, Heuer A, Jetten MS, Rast P, Boedeker C, Rohde M, Jogler C. Three marine strains constitute the novel genus and species Crateriforma conspicua in the phylum Planctomycetes. Antonie Van Leeuwenhoek. 2020 doi: 10.1007/s10482-019-01375-4. [DOI] [PubMed] [Google Scholar]
- Pilhofer M, Rappl K, Eckl C, Bauer AP, Ludwig W, Schleifer KH, Petroni G. Characterization and evolution of cell division and cell wall synthesis genes in the bacterial phyla Verrucomicrobia, Lentisphaerae, Chlamydiae, and Planctomycetes and phylogenetic comparison with rRNA genes. J Bacteriol. 2008;190:3192–3202. doi: 10.1128/JB.01797-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Peplies J, Glöckner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Q-L, Xie B-B, Zhang X-Y, Chen X-L, Zhou B-C, Zhou J, Oren A, Zhang Y-Z. A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol. 2014;196:2210–2215. doi: 10.1128/JB.01688-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rast P, Glockner I, Boedeker C, Jeske O, Wiegand S, Reinhardt R, Schumann P, Rohde M, Spring S, Glockner FO, Jogler C, Jogler M. Three novel species with peptidoglycan cell walls form the new genus Lacunisphaera gen. nov. in the family Opitutaceae of the verrucomicrobial subdivision 4. Front Microbiol. 2017;8:202. doi: 10.3389/fmicb.2017.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensink S, Wiegand S, Kallscheuer N, Rast P, Peeters SH, Heuer A, Boedeker C, Jetten MS, Rohde M, Jogler M, Jogler C. Description of the novel planctomycetal genus Bremerella, containing Bremerella volcania sp. nov., isolated from an active volcanic site, and reclassification of Blastopirellula cremea as Bremerella cremea comb. nov. Antonie van Leeuwenhoek. 2020 doi: 10.1007/s10482-019-01378-1. [DOI] [PubMed] [Google Scholar]
- Rivas-Marin E, Devos DP. The paradigms they are a-changin’: past, present and future of PVC bacteria research. Antonie Van Leeuwenhoek. 2018;111:785–799. doi: 10.1007/s10482-017-0962-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas-Marin E, Canosa I, Devos DP. Evolutionary cell biology of division mode in the bacterial Planctomycetes-Verrucomicrobia-Chlamydiae superphylum. Front Microbiol. 2016;7:1964. doi: 10.3389/fmicb.2016.01964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas-Marin E, Canosa I, Santero E, Devos DP. Development of genetic tools for the manipulation of the planctomycetes. Front Microbiol. 2016;7:914. doi: 10.3389/fmicb.2016.00914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas-Marin E, Peeters SH, Fernández LC, Jogler C, van Niftrik L, Wiegand S, Devos DP. Non-essentiality of canonical cell division genes in the planctomycete Planctopirus limnophila. Sci Rep. 2020;10:1–8. doi: 10.1038/s41598-019-56978-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas-Marin E, Wiegand S, Kallscheuer N, Jogler M, Peeters S, Heuer A, Jetten MS, Boedeker C, Rohde M, Devos DP, Jogler C (2020b) Maioricimonas rarisocia gen. nov., sp. nov., a novel planctomycete isolated from marine sediments close to Mallorca Island. Antonie van Leeuwenhoek. 10.1007/s10482-020-01436-z [DOI] [PMC free article] [PubMed]
- Rodriguez-R LM, Konstantinidis KT (2016) The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes, PeerJ Preprints: e1900v1
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring S, Bunk B, Spröer C, Schumann P, Rohde M, Tindall BJ, Klenk H-P. Characterization of the first cultured representative of Verrucomicrobia subdivision 5 indicates the proposal of a novel phylum. ISME J. 2016;10:2801. doi: 10.1038/ismej.2016.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strous M, Fuerst JA, Kramer EH, Logemann S, Muyzer G, van de Pas-Schoonen KT, Webb R, Kuenen JG, Jetten MS. Missing lithotroph identified as new planctomycete. Nature. 1999;400:446–449. doi: 10.1038/22749. [DOI] [PubMed] [Google Scholar]
- van Teeseling MC, Mesman RJ, Kuru E, Espaillat A, Cava F, Brun YV, VanNieuwenhze MS, Kartal B, van Niftrik L. Anammox Planctomycetes have a peptidoglycan cell wall. Nat Commun. 2015;6:6878. doi: 10.1038/ncomms7878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmers J, Frentrup M, Rast P, Jogler C, Kaster AK. Untangling genomes of novel planctomycetal and verrucomicrobial species from monterey bay kelp forest metagenomes by refined binning. Front Microbiol. 2017;8:472. doi: 10.3389/fmicb.2017.00472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M, Horn M. The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr Opin Biotechnol. 2006;17:241–249. doi: 10.1016/j.copbio.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Wiegand S, Jogler M, Jogler C. On the maverick Planctomycetes. FEMS Microbiol Rev. 2018;42:739–760. doi: 10.1093/femsre/fuy029. [DOI] [PubMed] [Google Scholar]
- Wiegand S, Jogler M, Boedeker C, Pinto D, Vollmers J, Rivas-Marín E, Kohn T, Peeters SH, Heuer A, Rast P, Oberbeckmann S, Bunk B, Jeske O, Meyerdierks A, Storesund JE, Kallscheuer N, Lücker S, Lage OM, Pohl T, Merkel BJ, Hornburger P, Müller R-W, Brümmer F, Labrenz M, Spormann AM, Op den Camp HJM, Overmann J, Amann R, Jetten MSM, Mascher T, Medema MH, Devos DP, Kaster A-K, Øvreås L, Rohde M, Galperin MY, Jogler C. Cultivation and functional characterization of 79 Planctomycetes uncovers their unique biology. Nat Microbiol. 2020;5:126–140. doi: 10.1038/s41564-019-0588-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, Schleifer KH, Whitman WB, Euzeby J, Amann R, Rossello-Mora R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol. 2014;12:635–645. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]