Abstract
Genomic imprinting and X chromosome inactivation (XCI) are classic epigenetic phenomena that involve transcriptional silencing of one parental allele. Germline-derived differential DNA methylation is the best-studied epigenetic mark that initiates imprinting, but evidence indicates that other mechanisms exist. Recent studies have revealed that maternal trimethylation of H3 on lysine 27 (H3K27me3) mediates autosomal maternal allele-specific gene silencing and has an important role in imprinted XCI through repression of maternal Xist. Furthermore, loss of H3K27me3-mediated imprinting contributes to the developmental defects observed in cloned embryos. This novel maternal H3K27me3-mediated noncanonical imprinting mechanism further emphasizes the important role of parental chromatin in development, and could provide the basis for improving the efficiency of embryo cloning.
ToC blurb
The role of DNA methylation in genomic imprinting and X chromosome inactivation (XCI) is well-documented, but other imprinting mechanisms exist. Here, the authors review the role of oocyte-derived histone H3 lysine 27 trimethylation in establishing autosomal imprinting and imprinted XCI.
Introduction
Most autosomal genes in diploid cells are transcribed at similar levels from both alleles. However, for a small subset of genes, one parental allele is transcriptionally silenced by genomic imprinting, and expression depends on whether the allele is inherited from the oocyte or the sperm 1. In addition to autosomal imprinting, the paternal X chromosome is preferentially silenced in female mouse preimplantation embryos and placental lineages by a process known as imprinted X-chromosome inactivation (XCI) (a process distinct from random XCI, which occurs in post-implantation embryonic lineages in mouse and other mammals, BOX1)2. As these imprints can persist from gametes to the next generation, genomic imprinting and imprinted XCI represent two examples of intergenerational epigenetic inheritance. Together, these two processes are critical for controlling gene dosage during embryonic development, and their dysregulation can cause developmental defects and diseases. For example, loss of imprinting contributes to childhood disorders such as the Prader-Willi/Angelman and Beckwith-Wiedemann/Silver-Russell syndromes 2,3.
BOX1. Random and imprinted X inactivation.
X-chromosome inactivation (XCI) is a mechanism of dosage compensation by which one of the X chromosomes of XX females is transcriptionally silenced so that expression levels of X-linked genes are equalized between XX female and XY male cells 146. In somatic cells, XCI is random, with either the maternal (Xm) or paternal (Xp) X being silenced 146. However, in mouse pre-implantation embryos, XCI is imprinted so that Xp is preferentially repressed 85. After implantation, Xp remains inactive in the extraembryonic lineages that contribute to the placenta whereas it gets reactivated in the epiblast, which gives rise to the embryo proper and in which random XCI subsequently takes place 89,90,147. Once random XCI is complete, the inactive X remains stably silenced during cell propagation.
The long non-coding RNA (lncRNA) X-inactive specific transcript (Xist) is only expressed from the future inactive X and is required to initiate both imprinted and random XCI in cis 16,148,149. Although the details of Xist-induced silencing are not fully understood, it is well established that Xist associates with a number of partners to inactivate the entire X chromosome. For example, a recent study revealed that a region of the Xist RNA, the repeat A element, recruits the RNA-binding protein SPEN at the onset of XCI to elicit gene silencing. Protein interactome analyses of the SPEN effector domain suggest that SPEN mediates gene silencing by recruiting transcriptional co-repressors to the X chromosome 150. In addition, the Xist RNA repeat B element associates with another RNA-binding protein, hnRNPK, which recruits variant Polycomb Repressive Complexes 1.3 and 1.5 (vPRC1.3/1.5) to deposit the transcriptional repressive chromatin mark H2AK119Ub 151–153. Together with other mechanisms, these processes lead to the formation of facultative heterochromatin and stable XCI through depletion of active histone marks (such as H3K4me3, H3K27ac, and H3K9ac) and establishment of the repressive histone marks (such as H2AK119Ub, H3K27me3, and H3K9me2) 2,154. For more detailed information on how Xist induces XCI, readers can refer to recent reviews 155,156.
The unequal contributions of parental genomes during development was first demonstrated by elegant pronuclear transfer experiments in the 1980s 4,5; bi-maternal and bi-paternal mouse embryos generated in these studies were found to be non-viable, indicating that both maternal and paternal genomes are required for normal development. The first imprinted genes were identified in the early 1990s 6–9, and shortly afterwards parental-allele-specific DNA methylation was found to be critical for imprinted gene expression 10. Parental-allele-specific DNA methylation originates from differential DNA methylation between oocytes and sperm and is maintained throughout development 11. These germline differentially methylated regions (DMRs) are the primary signals for establishing secondary allele-specific epigenetic features such as histone modifications and somatic DMRs that help to achieve imprinted expression 12,13. Germline DNA methylation-dependent allele-specific expression is the classic form of genomic imprinting, and is therefore referred to here as canonical imprinting.
However, several paternally-expressed imprinted genes in mouse placenta do not harbor germline DMRs and their imprinted expression is independent of oocyte DNA methylation 14,15. Furthermore, germline DNA methylation does not regulate the paternal-allele-specific expression of the long noncoding RNA (lncRNA) X-inactive specific transcript (Xist) in mouse extraembryonic cells, which causes the paternal-allele-specific silencing of most X-linked genes in this lineage 2,16,17. Taken together, these observations indicate the existence of a germline DNA methylation-independent imprinting mechanism.
Recently, low-input epigenomic profiling techniques 18 have been used to demonstrate that Polycomb Repressive Complex 2 (PRC2)-mediated trimethylation of H3 on lysine 27 (H3K27me3) in mouse oocytes is the cause of maternal-allele-specific silencing of both the autosomal imprinted genes and the imprinted Xist 19,20 in the placenta (BOX 2). Because this new imprinting mechanism uses oocyte-inherited H3K27me3, rather than DNA methylation, to distinguish parental alleles in pre-implantation embryos, it is mechanistically different from classic imprinting and is therefore termed noncanonical imprinting. It should be noted that the noncanonical imprinting referred to here is different from the phenomenon of modest parental expression bias observed in specific brain regions, which has also been referred to as noncanonical imprinting in some contexts 21.
BOX2. Polycomb Repressive Complexes 1 and 2.
Polycomb repressive complexes 1 and 2 (PRC1 and PRC2) are multi-subunit protein complexes that mediate transcriptional repression, mainly by altering chromatin activity 137. The catalytic core of PRC1 contains one of two E3 ubiquitin ligases, RING1A or RING1B, and one of six PCGF proteins (PCGF1–6) 157. PRC1 deposits mono-ubiquitin to lysine 119 on histone H2A (H2AK119Ub) 158,159. PRC1 can be sub-divided into canonical PRC1 (cPRC1) and variant PRC1 (vPRC1) based on their distinct accessory subunits. The cPRC1 is composed of either PCGF2 or 4 and one of the CBX subunits that can recognize H3K27me3 157,160,161. By contrast, vPRC1 can utilize any of the six PCGF proteins but incorporates either YAF2 or RYBP instead of the CBX subunit 157,162,163. Therefore, unlike cPRC1, vPRC1 cannot recognize H3K27me3.
The core subunits of PRC2 include one of two histone methyltransferases, EZH1 or EZH2, and the regulatory subunits EED, SUZ12, and either RBAP46 or RBAP48. PRC2 is responsible for mono-, di-, and tri-methylation at lysine 27 on histone H3 (H3K27me1/2/3) 161,164–166. PRC2 can be further divided into PRC2.1 and PRC2.2. PRC2.1 associates with the PCL1, PCL2 or PCL3 subunits that are known to bind CpG islands 167, whereas PRC2.2 contains AEBP2 and JARID2; JARID2 recognizes H2AK119Ub deposited by PRC1 168.
How PRC1 and PRC2 are recruited to their specific targets and how they exert transcriptional silencing is not fully understood. In the extensively studied mouse embryonic stem cell (mESC) model, vPRC1, but not cPRC1 or PRC2, mediates transcriptional silencing of the majority of Polycomb-group protein (PcG) targets 169. However, it should be noted that PRC function is context-dependent as PRC2 and cPRC1 are critical in other processes 170–172. In mESCs, a compelling model for PRC recruitment is that vPRC1 is first recruited to PcG targets where it deposits H2AK119Ub, which then serves as a docking site for JARID2-mediated recruitment of PRC2.2 168,173. Subsequently, the chromo domain of CBX may bind to PRC2-deposited H3K27me3 to recruit cPRC1 161. Whether this ‘vPRC1-PRC2-cPRC1’ model applies to other systems remains to be determined. For more detailed information on PRC1 and PRC2 regulation and function, readers may refer to recent reviews 137,174.
In this Review, we will first briefly summarize the mechanisms involved in canonical imprinting. We will then describe our current understanding of noncanonical imprinting and compare and contrast it to canonical imprinting in terms of its establishment and maintenance. In addition, the role of noncanonical imprinting in imprinted XCI, placental development, and animal cloning will be discussed. Lastly, we will discuss how noncanonical imprinting might be conserved during evolution. Unless otherwise specified, both imprinting mechanisms will be discussed in the context of the mouse.
Mechanisms of canonical imprinting
Canonically imprinted genes typically are found in clusters of more than three genes and span genomic regions ranging in size from several Kb to a few Mb 11. The allele-specific expression of the transcripts within each cluster is regulated by a cis-regulatory element known as the imprinting control region (ICR)11. ICRs exhibit germline-derived differential DNA methylation between parental alleles and genetic manipulation of the ICRs in either in vitro cell culture or in vivo mouse studies can cause loss of imprinting of all genes in an imprinted cluster 11. Given its essential role, the establishment, maintenance and erasure of allelic DNA methylation at ICRs is controlled by multiple regulators. In addition, ICRs use diverse cis-regulatory mechanisms to control imprinted gene expression. However, most mechanisms are not fully understood and, even for well-studied mechanisms such as the insulator model and lncRNA model described below, it is not clear how applicable they might be to other imprinted loci. For a more comprehensive discussion of canonical imprinting, readers are directed to excellent reviews of the topic 11,22,3,23.
Establishment of canonical imprinting during gametogenesis.
Primary imprinting marks need to be established during gametogenesis, a developmental window when the parental genomes are in separate compartments and are subject to different epigenetic modifications (Figure 1A). At this stage, both global de novo DNA methylation and methylation at individual germline DMRs are deposited by the DNA methyltransferase DNMT3A and its essential non-catalytic co-factor DNMT3L 24,25. Loss of DNMT3A or DNMT3L in oocytes causes maternal imprinting defects and embryonic lethality, and lack of either protein in the male germline leads to spermatogenesis defects and de novo methylation failure at two of the three paternally methylated DMRs (that is, the H19/Igf2 and Gtl2/Dlk1 ICRs) 24,25. The other paternally methylated DMR, Rasgrf1, depends on the piwi-interacting RNA pathway and the recently identified rodent-specific DNMT3C 26–28.
Figure 1. Germline inherited DNA methylation governs canonical imprinting.
A) During oogenesis, transcription across imprinting control regions (ICRs) recruits the histone methylase SETD2 to deposit H3K36me3, which then guides the de novo DNA methyltransferases DNMT3A and DNMT3L to establish DNA methylation. Removal of H3K4me2/3 by the demethylases KDM1A/1B is also required for de novo DNA methylation. During spermatogenesis, DNA methylation is dependent on either DNMT3A and DNMT3L (for example, at the H19/Igf2 and Gtl2/Dlk1 ICRs) or DNMT3C and the piRNA pathway (for example, at the Rasgrf1 ICR). Differential DNA methylation at ICRs is protected from global DNA demethylation during pre-implantation development by ZFP57, ZFP445 and TRIM28, which bind to the methylated ICRs and recruit the maintenance methyltransferase DNMT1 and its cofactor UHRF1. N-alpha-acetyltransferase 10 protein (NAA10P) is also required to facilitate binding of DNMT1 to the methylated allele for imprinting maintenance. During implantation, the presence of active histone marks such as H3K4me3 (and possibly others) may prevent unmethylated ICRs from gaining DNA methylation in the wave of global de novo methylation. Secondary allelic DNA methylation (that is, somatic differentially methylated regions (DMRs)) is established at some imprinted genes during this period. To reset imprints for the next generation, allelic DNA methylation at ICRs is erased in primordial germ cells (PGCs). This demethylation process is mainly mediated by passive dilution and TET1-mediated oxidation of 5-methylcytosine at ICRs.
B) The insulator model of imprinted gene regulation is illustrated by the H19/Igf2 cluster. At this locus, the ICR for the long noncoding RNA (lncRNA) H19 is paternally DNA methylated. In the conceptus, DNA methylation extends to the H19 promoter to silence its transcription on the paternal allele. DNA methylation at the H19 ICR also prevents binding of CTCF to the ICR, which results in formation of a topologically associated domain (TAD, blue triangle) that permits transcriptional activation of Igf2 by the downstream enhancers (long double-headed arrow). On the maternal allele, CTCF-binding to the unmethylated H19 ICR forms two sub-TADs (pink triangles) that prevent the interaction between Igf2 and the enhancers and Igf2 remains transcriptionally repressed. CTCF-binding also facilitates the initiation of H19 transcription by preventing gain of DNA methylation on the maternal allele.
C) The long non-coding RNA (lncRNA) model of imprinted gene regulation is illustrated by the Kcnq1 cluster. Here, the ICR (also known as KvDMR1) serves as the promoter for the lncRNA Kcnq1ot1. On the paternal allele, the unmethylated KvDMR1 allows Kcnq1ot1 transcription, which recruits Polycomb repressive complexes 1 and 2 (PRC1/2) to deposit H3K27me3 and H2AK119Ub, respectively, to silence flanking protein coding genes. On the maternal allele, Kcnq1ot1 is repressed by the methylated ICR, allowing expression of the flanking genes. At this cluster, while Kcnq1ot1 and Cdkn1c exhibit ubiquitous imprinting, Slc22a18 and Tssc4 are only imprinted in placental lineages. The size and signal of H3K27me3 domains, and the allelic gene expression are drawn based on publicly available datasets 20,67,70. Not all genes in this cluster are shown for simplicity.
Whereas paternally methylated DMRs acquire DNA methylation prenatally, maternal DMRs are methylated postnatally during oocyte growth 11. Despite extensive studies, some aspects of de novo DNA methylation during oogenesis remain elusive. The current working model is that transcription elongation causes an enrichment of dimethylation and trimethylation of histone H3 at lysine 36 (H3K36me2/3) at the transcribed regions, and these histone modifications recruit the DNMT3A/3L complex to establish DNA methylation in oocytes 29–31. In support of this model, premature termination of transcription at germline DMRs in oocytes leads to reduced H3K36me3 levels and a failure of de novo DNA methylation at these loci 29,31,32. Furthermore, depletion of the H3K36 methyltransferase SETD2 in oocytes causes genome-wide loss of H3K36me3 and DNA hypomethylation, including at germline DMRs 33. In addition, removal of histone modifications that antagonize DNA methylation is also important for imprinting establishment as loss of the H3K4 demethylases KDM1A or KDM1B causes a substantial increase of H3K4me2 in oocytes and results in defective establishment of DNA methylation at maternally methylated DMRs 30,34.
Maintenance of canonical imprinting during development.
In canonical imprinting, parental allele-specific DNA methylation at ICRs needs to survive two waves of DNA methylation reprogramming; global demethylation during pre-implantation development, and the subsequent re-methylation at implantation 35 (Figure 1A). Genome-wide DNA methylation profiling has revealed that half of the sperm and oocyte genomes are differentially methylated; however, most of these DMRs become hypomethylated on both parental alleles before implantation 36. Germline DMRs that overlap ICRs are protected from this global DNA demethylation by the Krüppel-associated box (KRAB)-containing zinc finger proteins (KZFP) ZFP57 and ZFP445 37,38, as mouse embryos that lack ZFP57 and ZFP445 fail to maintain DNA methylation at most ICRs 37,38. Mechanistic studies in mouse embryonic stem cells (mESCs) indicate that ZFP57 and ZFP445 bind to the methylated allele at ICRs and recruits the co-factor KAP1 (also known as TRIM28) 38,39. ZFP57/KAP1 complex also associates with other epigenetic modifiers including the DNA methylation maintenance machinery DNMT1 and UHRF1 and the H3K9 methyltransferase SETDB1 to protect allele-specific DNA methylation at ICRs 39,40. Recently, the N-alpha-acetyltransferase 10 protein (NAA10P) has been shown to facilitate DNMT1 binding to the methylated alleles and loss of NAAP10P causes DNA hypomethylation at ICRs in both mouse embryos and mESCs 41. How NAA10P recruits DNMT1 and interacts with other imprinting maintenance factors such as ZFP57 and KAP1 remains to be determined.
In addition to the methylated allele at ICRs escaping global DNA demethylation in pre-implantation development, it is equally important for the unmethylated allele to survive genome-wide re-methylation during implantation 42. Although the underlying mechanism remains unclear, it is believed that the unmethylated allele at ICRs is protected from de novo DNA methylation by the presence of histone marks that can antagonize DNA methylation machinery, such as H3K4me3 and/or other modifications 42,43.
Erasure of canonical imprinting in primordial germ cells.
In order to re-establish DNA methylation in the germline according to the sex of the embryo, the allelic DNA methylation at ICRs must first be erased. The erasure of DNA methylation at ICRs occurs as part of the global DNA demethylation process in the primordial germ cells PGCs), which involves passive demethylation of the bulk of the genome by DNA replication followed by active demethylation mainly of imprinted loci and germline-specific genes by the Ten-Eleven Translocation (TET) family enzymes 44,45,46 (Figure 1A). TET enzymes can convert 5-methylcytosine (5mC) into 5-hydroxymethylcytosine (5hmC) and its derivatives, which are then removed by replication-dependent dilution or by the DNA base excision repair pathway 44. Genetic studies in mouse indicate that TET1 deficiency causes aberrant DNA hyper-methylation at only a subset of ICRs in germ cells and somatic tissues and results in dysregulated imprinted gene expression 46,47. For example, DNA demethylation at the Snrpn ICR is unaffected even in TET1/TET2 double mutants 47,48, suggesting that demethylation at Snrpn ICR occurs through passive dilution but not active demethylation. The mechanism underlying ICR-specific dependency on TET proteins remains unknown, but it has been suggested that the sequence composition of the Snrpn ICRs could explain why, it does not undergo active demethylation like other imprinted loci 47.
The insulator model of imprinted gene regulation.
The insulator model of imprinted gene regulation is best exemplified by the H19/Igf2 locus, which has been the subject of a series of elegant mouse genetic studies. H19 is a long non-coding RNA and is maternally expressed whereas the insulin-like growth factor Igf2 is paternally expressed 6,8,9. The H19/Igf2 ICR is located between the H19 and Igf2 genes and is methylated on the paternal allele but unmethylated on the maternal allele 49,50(Figure 1B). Deletion of the paternally inherited H19/Igf2 ICR causes de-repression of paternal H19 and reduced levels of Igf2, whereas deletion of the maternally transmitted ICR leads to activation of maternal Igf2 and repression of H19 expression 51. The key to imprinting regulation by the H19/Igf2 ICR is the DNA methylation-sensitive CCCTC-binding factor (CTCF), which binds only to the unmethylated maternal ICR 52,53. On the maternal allele, CTCF acts as an insulator and blocks interactions between the Igf2 promoter and the downstream shared enhancers, preventing Igf2 expression 52,53. In addition, CTCF binding facilitates initiation of H19 expression and prevents ectopic DNA methylation on the unmethylated maternal ICR 54. Maternal inheritance of an H19/Igf2 ICR that contains mutated CTCF binding sites abolishes maternal CTCF binding and causes DNA hypermethylation on the maternal allele, preventing H19 expression 54. On the paternal allele, however, DNA methylation at the ICR prevents CTCF binding, which allows the enhancers to interact with the Igf2 promoter to activate Igf2 expression 52,53. In addition, DNA methylation at the ICR spreads into the H19 promoter to silence paternal H19 expression 51.
The differential access of H19 and Igf2 to the shared downstream enhancers indicates distinct three-dimensional conformations between parental alleles at this locus. Using 4C-seq and DNA-fluorescence in situ hybridization (FISH), a recent study indicated that, in addition to allelic CTCF binding at the ICR, bi-allelic CTCF binding to sites that flank the H19/Igf2 locus are also involved in modulating allelic chromatin looping in mESCs 55. Specifically, the bi-allelic CTCF binding correlates with a topologically associated domain (TAD) that is common to both alleles. However, on the maternal allele, the additional CTCF binding at the H19/Igf2 ICR contributes to a sub-TAD, which can override the higher-level TAD and restrict the interaction between Igf2 and the enhancers (Figure 1B). It remains to be shown if this allelic TAD model is universal for other imprinted clusters.
The lncRNA model of imprinted gene regulation.
One of the best-characterized imprinted clusters that illustrates the lncRNA model of gene regulation is the Kcnq1 imprinted cluster. The Kcnq1 ICR, known as KvDMR1, is unmethylated on the paternal allele and methylated on the maternal allele (Figure 1C) 56,57. KvDMR1 contains a promoter for the paternally expressed lncRNA Kcnq1ot1, which recruits repressive histone modifications H3K27me3 and H3K9me2 to silence ten flanking maternally expressed protein-coding genes including Cdkn1c, Slc22a18, and Tssc458–62. On the maternal allele, however, DNA methylation of KvDMR1 prevents Kcnq1ot1 expression, thereby allowing the transcription of flanking genes. Deletion of the Kcnq1ot1 promoter or premature termination of the lncRNA on the paternal allele causes de-repression of the neighboring protein coding genes in mouse embryos 63,64. By contrast, maternal transmission of the KvDMR1 deletion has no effect on imprinted regulation at this cluster 63,64.
Although the role of Kcnq1ot1 in regulating imprinted gene expression is well established, how it recruits epigenetic modifiers and induces chromatin changes remain unknown. It is also unclear how Kcnq1ot1 mediates gene silencing in a tissue- and stage-specific manner. For example, although Kcnq1ot1 is ubiquitously imprinted, Slc22a18 and Tssc4 are imprinted only in the placenta and not the embryo. Evidence from mouse trophoblast stem cells (mTSCs) indicates that the stability and abundance of lncRNAs seem to be critical for determining the level of H3K27me3 enrichment at imprinted loci 65 as overexpression or knockdown of another imprinted lncRNA, Airn, in mTSCs causes enhanced or reduced H3K27me3 levels at the imprinted cluster, respectively 65. In addition, genomic structures (such as DNA loops and TADs) and DNA sequences (such as CpG islands) also seem to be involved in shaping the H3K27me3 domains 65. Therefore, the capacity of lncRNAs to induce gene silencing can be influenced by complex factors and their variable activities in different cell lineages may explain how lncRNAs can mediated tissue- and stage-specific imprinting.
Mechanisms of noncanonical imprinting
Oocyte H3K27me3 and noncanonical imprinting.
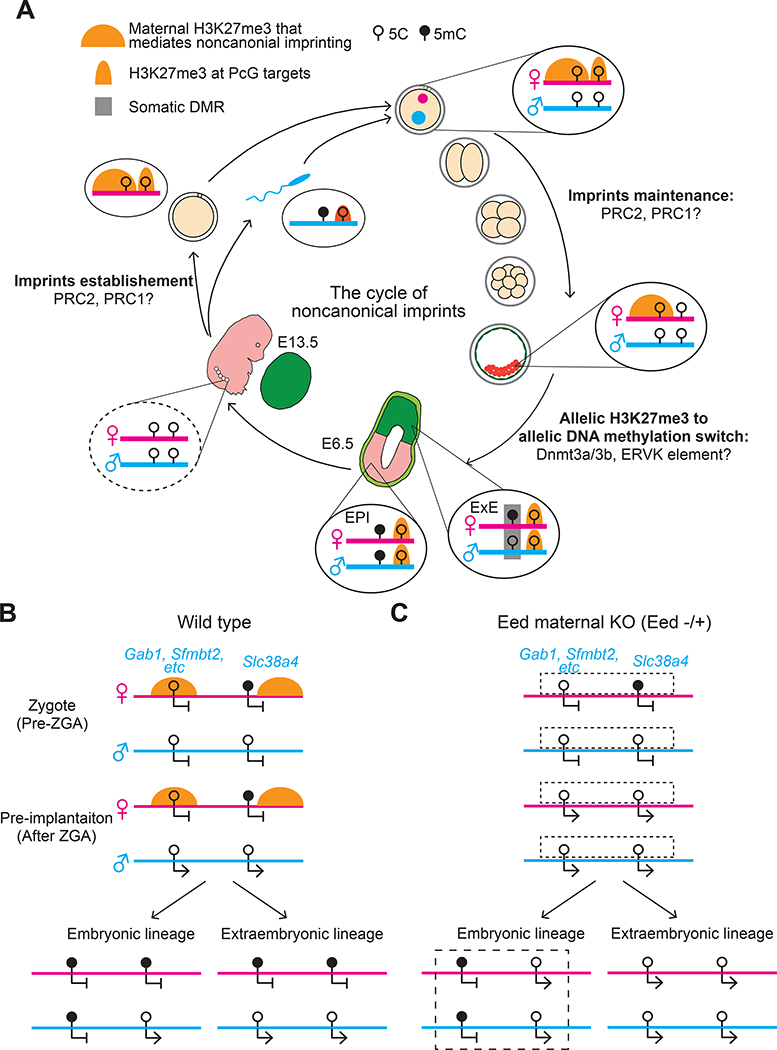
Although the very different epigenetic landscapes of the sperm and egg become largely equalized during preimplantation development, allelic analyses of DNase I hypersensitivity sites (DHSs) in preimplantation mouse embryos has revealed that known ICRs exhibit differential chromatin accessibility, with the hypomethylated allele showing a higher DHS signal 66. In addition to these known ICRs, a substantial number of paternal allele-specific DHSs (Ps-DHSs) were detected that are hypomethylated on both alleles, indicating that mechanisms other than DNA methylation determine the allele-specificity of these Ps-DHSs in early embryos 20. Notably, some of these Ps-DHSs are associated with paternally expressed genes known to be independent of oocyte-derived DNA methylation, such as Gab1, Sfmbt2, and Slc38a4 14,15. Further analyses indicated that the Ps-DHSs harbor maternal allele-specific H3K27me3 that is inherited from oocytes 67, suggesting that maternal H3K27me3 may reduce chromatin accessibility of the corresponding regions on the maternal allele 20. Acute depletion of H3K27me3 in mouse pre-implantation embryos by overexpressing the demethylase KDM6B causes bi-allelic DHSs and gene expression at these loci, demonstrating that maternally inherited H3K27me3 contributes to the Ps-DHSs and paternal allele-specific gene expression 20. Taken together, these observations suggest that oocyte H3K27me3 can serve as a primary epigenetic mark for imprinted gene expression (Figure 2).
Figure 2. Oocyte inherited H3K27me3 initiates noncanonical imprinting.
A) The figure depicts the dynamics of H3K27me3 at noncanonical imprinting loci and at genomic targets of Polycomb group (PcG) proteins. Polycomb repressive complex 2 (PRC2) mediates H3K27me3 deposition during oogenesis; whether PRC1-mediated H2AK119ub is involved in PRC2 function in oogenesis remains unknown. After fertilization, H3K27me3 at PcG targets is largely reprogrammed, but maternally inherited H3K27me3 at noncanonically imprinted loci is maintained during pre-implantation development and is responsible for silencing the maternal allele of these genes. After implantation, H3K27me3 is re-established at PcG targets whereas the maternal H3K27me3 that initiates noncanonical imprinting disappears from both the epiblast (EPI) and extraembryonic ectoderm (EXE). The maintenance of some noncanonical imprinting in EXE depends on the acquisition of somatic differentially methylated regions (DMRs) during implantation via the DNA methyltransferases DNMT3A and DNMT3B. Active endogenous virus-K (ERVK) long terminal repeats (LTR) in the somatic DMRs may have a role in the maintenance of noncanonical imprinting in the placental lineage. In the epiblast, both alleles at noncanonically imprinted loci are repressed by DNA methylation. In primordial germ cells (PGCs), DNA methylation at noncanonical imprinting loci is expected to be erased (dashed line) during the wave of global DNA demethylation mediated by TET1 and DNA replication.
B) The typical dynamics of H3K27me3 and DNA methylation at noncanonically imprinted loci (represented by Gab1) in wild type mice (Ba) and Eed maternal knockout (KO) mice (Bb) are shown. Oocyte-specific depletion of EED, an essential subunit of PRC2, causes loss of H3K27me3 in mature oocytes. Embryos that develop from Eed-null oocytes (that is, Eed maternal KO embryos) lack maternally-provided H3K27me3 and lose noncanonical imprinting in both pre-implantation embryos and extraembryonic cells. Furthermore, somatic DMRs are unmethylated on both alleles in EXE of these embryos. The Slc38a4 locus differs from other noncanonically imprinted loci because its DMR is established during oogenesis. However, it becomes hypomethylated in Eed maternal KO EXE, suggesting that maternal H3K27me3 is essential to maintain differential DNA methylation at this locus. DNA methylation status of the loci in embryonic lineages of Eed maternal KO has not been analyzed, but predicted patterns are included and indicated by dashed boxes. ZGA: zygotic genome activation.
A few differences should be noted between canonical and noncanonical imprinting mechanisms. For canonical imprinting, the imprints (that is, DNA methylation) that govern allele-specific gene expression can be inherited from either oocytes or sperm cells 11 (Figure 1A). However, the H3K27me3 that mediates noncanonical imprinting is only inherited from oocytes because most sperm DNA is packaged by protamines rather than histones and the minor amount of paternal H3K27me3 is completely reprogrammed at fertilization 67. The oocyte-inherited H3K27me3 also differs from the allelic H3K27me3 implicated in canonical imprinting, which is secondary to the germline DMRs 68. Lastly, while canonical imprinting is maintained in both embryonic and extraembryonic lineages, most noncanonical imprinting is transient, with only some genes important for placental development maintaining their imprinted expression in extraembryonic cells 20. As noncanonical imprinting is not maintained in the epiblast lineage that gives rise to germ cells, noncanonical imprints do not need to be erased in PGCs (Figure 2A); by contrast, canonical imprints must be erased in PGCs to reset imprinting for the next generation (Figure 1A).
Establishment of noncanonical imprinting during oocyte growth.
Analogous to DNA methylation, most H3K27me3 is deposited by PRC2 during oocyte growth 67 (Figure 2A, Box 2). However, H3K27me3 is generally anticorrelated with DNA methylation and H3K36me3 in oocytes 33. In addition, unlike somatic cells, H3K27me3 in oocytes is present not only at the classic genomic targets of Polycomb group (PcG) proteins, such as developmental gene promoters, but also at non-transcribed regions that can be several Mb in size 67,69. This acquisition of H3K27me3 during oogenesis is independent of DNA methylation as H3K27me3 domains are largely unaffected in Dnmt3l knockout mice 33. Furthermore, embryos derived from DNA methylation-deficient oocytes maintain intact maternal H3K27me3 domains, explaining why these embryos show normal noncanonical imprinting but abnormal canonical imprinting 70,71. Conversely, DNA methylation acquisition in oocytes is independent of H3K27me3 as embryos from H3K27me3-deficient oocytes (that is, oocytes from conditional PRC2 null mice) exhibit proper canonical, but not noncanonical, imprinting 72. Therefore, canonical and noncanonical imprints are independently established during oogenesis.
One intriguing question is how genomic regions are selected for DNA methylation or H3K27me3 during oogenesis, which will determine whether a gene, if imprinted, will be regulated by canonical or noncanonical imprinting. It is likely that H3K27me3 promiscuously marks transcriptionally inactive regions during oogenesis and is antagonized by H3K36me3 at the actively transcribed regions. In support of this notion, H3K27me3 can ectopically occupy regions that are normally marked by H3K36me3 in Setd2-null oocytes 33. However, H3K36me3 cannot be the sole mechanism that defines H3K27me3 boundaries because not all non-transcribed regions in oocytes are marked by H3K27me3 33. Given that disruption of Polycomb repressive complex 1 (PRC1, a repressive complex that ubiquitinylates lysine 119 of histone H2A (H2AK119ub), BOX2) causes more severe defects in oogenesis than disruption of PRC2 72–75, it is likely that PRC1 recruitment may be upstream of H3K27me3 acquisition during oocyte development. Indeed, Kdm2b (also known as Fbxl10), an H3K36 demethylase 76,77, binds to unmethylated CpG islands and recruits the PRC1 member RING1B to mediate H2AK119ub deposition in mESCs 78–80. In addition, Kdm2b is responsible for protecting genes bound by PRC1 and PRC 2 from ectopic de novo DNA methylation in mESCs 81. However, a role for Kdm2b in recruiting PRC1 and antagonizing DNA methylation during oogenesis remains to be demonstrated.
Maintenance of noncanonical imprinting during development.
In contrast to DNA methylation at ICRs, which is generally maintained throughout development, the maternally inherited H3K27me3 domains that mediate noncanonical imprinting are only temporarily maintained in pre-implantation embryos (Figure 2A) 70,71. This maintenance depends on genomic context. For example, H3K27me3 profiling in mouse early embryos indicates that H3K27me3 at typical PcG targets is erased by the late 1-cell stage and then re-established at implantation 67. Notably, RNA-sequencing based analyses revealed that PcG targets remain inactive even in the absence of H3K27me3 67, suggesting that either transcription factors required for gene activation are not present or additional repressive epigenetic mechanisms compensate for the loss of H3K27me3 to silence PcG targets in early embryos. Nonetheless, maternally inherited H3K27me3 is essential at this developmental stage to preserve the parental allele specificity at noncanonically imprinted loci, as acute depletion of H3K27me3 by overexpressing the demethylase KDM6B in mouse pre-implantation embryos results in loss of imprinted expression of these genes 20.
Maternally inherited H3K27me3 diminishes during pre-implantation development and is largely absent after implantation (Figure 2)70,71, possibly explaining why most noncanonical imprinting is transient and not maintained beyond implantation 20. However, the fact that some noncanonically imprinted genes do maintain their imprinted expression in the placental lineage suggests that an additional epigenetic modification takes over from H3K27me3 to repress maternal allele transcription at these loci. Analyses of allelic DNA methylome and H3K27me3 in pre-implantation embryos and post-implantation placental lineages revealed that although these genes lose their maternally inherited H3K27me3, they acquire DNA methylation (that is, somatic DMRs) specifically on the maternal allele to maintain imprinted expression in extraembryonic cells (Figure 2B) 70,71. Furthermore, the somatic DMR acquisition depends on the zygotic de novo DNA methyltransferases DNMT3A/3B as DNMT3A/3B double mutant embryos fail to acquire the somatic DMRs and show de-repression of the maternal allele at the noncanonical imprinting loci 70. It should be noted that this switch from a dependence on maternal H3K27me3 in pre-implantation embryos to allelic DNA methylation after implantation is the opposite of the placenta-specific canonical imprinting that occurs at the Kcnq1 cluster, in which imprinted expression initially depends on allelic DNA methylation but then switches to H3K27me3 to maintain imprinting (Figure 1C) 59,61,62.
It remains unclear how these few genes are selected to acquire somatic DMRs and maintain imprinted expression in the placenta. It has been observed that the noncanonically imprinted loci that preferentially acquire somatic DMRs overlap active endogenous retrovirus-K (ERVK) long terminal repeats (LTR) 71. These ERVK LTRs seem to have a role in maintaining noncanonical imprinting as disruption of the ERVK promoter at the Gab1 locus caused weakened paternal gene expression bias, although DNA methylation at the promoter was not determined 71. Furthermore, the somatic DMRs become hypermethylated on both alleles in the epiblast, which explains why noncanonical imprinting is not maintained in this lineage 71. However, it remains unclear how insertions of ERVK LTRs, but not other repeat types, can maintain imprinting and what placenta-specific transcription factors protect the paternal allele from global de novo DNA methylation at implantation. In addition, it is not known why Sfmbt2 retains allelic H3K27me3 in early post-implantation development and acquires somatic DMRs later than other noncanonically imprinted loci 70. Nonetheless, the switch from allelic H3K27me3 to allelic DNA methylation indicates that transient allelic histone modifications in early embryos can have long-term consequences in mouse embryonic development.
Mechanisms of imprinted X inactivation
Xist imprinting by oocyte H3K27me3.
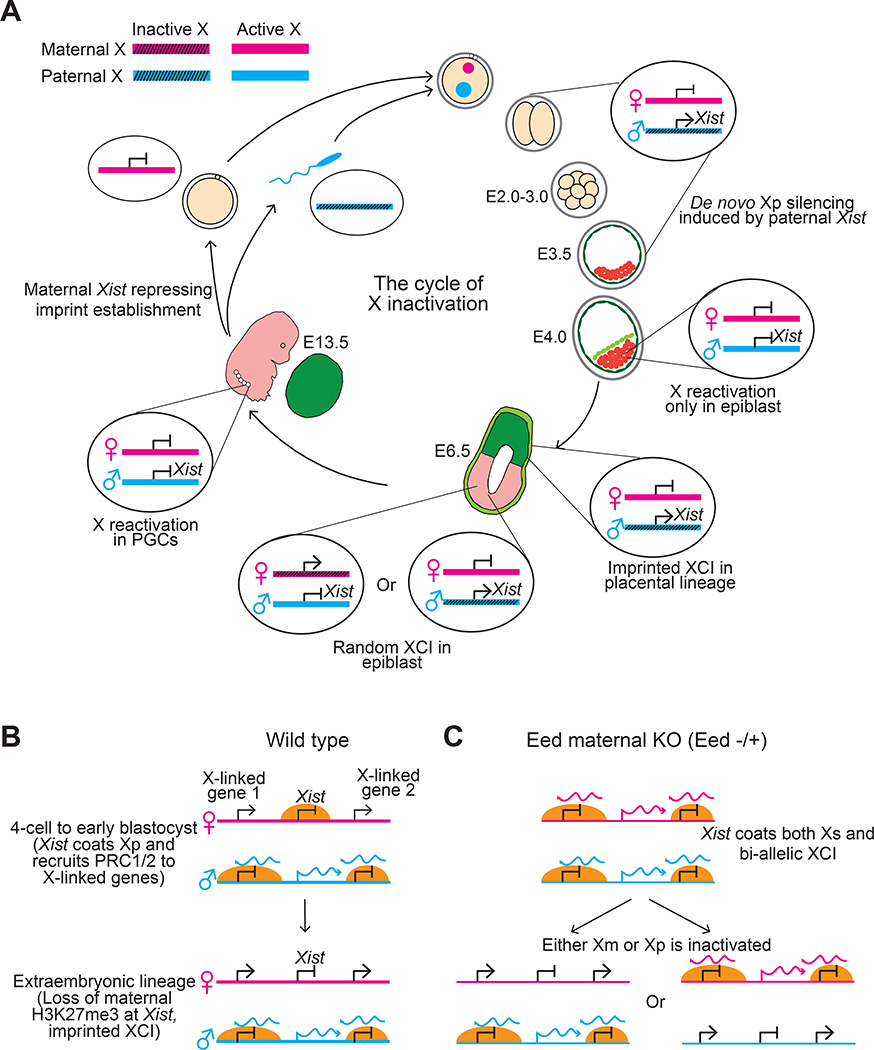
What controls imprinted X inactivation in mouse pre-implantation embryos has been a long-standing question. In mouse embryos generated by nuclear transfer using either non-growing oocytes or fully-grown oocytes, the X-chromosome derived from the non-growing oocyte, which resembles a normal Xp, is preferentially silenced 82. This observation suggests that a maternal imprint is established during oocyte growth to prevent Xm from being silenced in early embryos. Consistent with this hypothesis, Xist initially remains silenced until the morula stage in diploid bi-maternal mouse embryos generated by parthenogenetic activation 83. By contrast, it has also bene proposed that Xp could inherit a pre-inactive state from the male germ line, in which meiotic sex chromosome inactivation (MSCI) occurs 84. Although these two possibilities are not mutually exclusive, results from further studies argue against the pre-inactivation of Xp prior to imprinted XCI. An Xist transgene on autosomes (which do not undergo MSCI in the male germline) can still cause imprinted in cis inactivation when paternally inherited 85, indicating that MSCI is not required for imprinted XCI. In addition, single-cell RNA sequencing of mouse pre-implantation embryos reveals that Xp silencing begins at the 4-cell stage instead of being pre-activated 16. Furthermore, mouse embryos in which the paternal allele of Xist has been deleted cannot initiate Xp inactivation 16. These results suggest that Xp inactivation occurs de novo after zygotic genome activation (ZGA) and is fully dependent on expression of Xist from the paternal allele 16. By contrast, the maternal allele of Xist remains repressed in early embryos to keep Xm active (Figure 3A).
Figure 3. Maternal H3K27me3 controls imprinted X inactivation by repressing maternal Xist.
A) Depicted are the life cycle of X-chromosome inactivation (XCI) and the allelic expression dynamics of Xist. After fertilization, Xist is paternally expressed in female embryos and induces paternal X chromosome inactivation (Xp) during pre-implantation development. At the late blastocyst stage, the silenced Xp becomes reactivated in the epiblast and then both X chromosomes undergo random X inactivation in the embryonic lineage. However, XCI remains imprinted in the extraembryonic lineages. X reactivation also takes place in primordial germ cells (PGCs). During oogenesis, H3K27me3 is established at the Xist locus, which then represses maternal Xist in pre-implantation embryos. During spermatogenesis, X and Y chromosomes are condensed into the sex body and become inaccessible to transcriptional machinery, which is referred to as meiotic sex chromosome inactivation (MSCI) and is independent of Xist 145.
B) The figure shows how ectopic maternal XCI occurs in Eed (an essential component of Polycomb repressive complex 2, PRC2) maternal knock out (KO) embryos. In wild type female pre-implantation embryos (Ba), Xist on the maternal X-chromosome (Xm) is repressed by oocyte-inherited H3K27me3. Xist on the paternal X-chromosome (Xp) is expressed, which recruits PRC1 and PRC2 to deposit repressive H2AK119Ub and H3K27me3 respectively to silence X-linked genes on Xp. Although maternally inherited H3K27me3 is no longer present at Xist in extraembryonic lineages, Xist is still only expressed from the paternal allele and XCI remains imprinted in this lineage. In Eed maternal KO embryos (Bb), Xp undegoes XCI normally. However, the lack of maternally-provided H3K27me3 at Xist leads to ectopic Xist expression from Xm, leading to its inactivation. However, the ectopically expressed Xist is silenced at the blastocyst stage and random XCI takes place in the extraembryonic lineage.
With this in mind, what is the epigenetic imprint that represses maternal Xist? Recent studies in early mouse embryos indicate that oocyte-inherited H3K27me3 silences maternal Xist expression, whereas paternally-expressed Xist silences Xp in cis (Figure 3B)19,72,86. This conclusion is supported by several pieces of evidence. Firstly, H3K27me3, but not DNA methylation, is gradually established at the Xist locus during oocyte growth and maternally inherited H3K27me3 is maintained until the blastocyst stage 19. Secondly, acute depletion of H3K27me3 by overexpressing the histone demethylase KDM6B causes loss of maternal H3K27me3 at the Xist locus, ectopic maternal Xist expression, and aberrant XmCI in both male and female mouse embryos 19. Lastly, depletion of EED, a core PRC2 subunit, in oocytes causes loss of maternal H3K27me3, ectopic expression of maternal Xist, and aberrant XmCI in embryos of both sexes 72,86. Therefore, after fertilization, the oocyte-inherited H3K27me3 silences maternal Xist and protects Xm from being inactivated. By contrast, Xist on the paternal allele is transcriptionally accessible and is expressed to induce Xp silencing in cis 16(Figure 3B).
In addition to H3K27me3, maternal H3K9me3 has been proposed to prevent activation of maternal Xist in early embryos because acute depletion of H3K9me3 by overexpressing the H3K9me3 demethylase KDM4B caused a partial de-repression of Xist in diploid parthogenetic 4-cell embryos 87. However, this result is not reproducible in bi-parental embryos generated by in vitro fertilization 19. Importantly, neither the Xist promoter nor the gene body is enriched for H3K9me3 in fully grown oocytes 88. Therefore, oocyte H3K9me3 may not be the imprint that suppresses maternal Xist in early embryos.
Reactivation of Xp in the inner cell mass.
Xist-induced paternal XCI is complete by around the 32-cell stage (~E3.0), and Xp then initiates reactivation in the inner cell mass (ICM) of early blastocysts (~E3.5) 89,90. Single-cell RNA sequencing-based analyses of early and mid ICM revealed that X-linked genes undergo reactivation at different kinetics, with some genes reactivating early at E3.5 and others only fully reactivating at E4.0–4.5 when the epiblast has formed 91. Notably, the early-reactivated genes in the ICM undergo re-silencing in ICM-derived primitive endoderm (PrE), thus maintaining imprinted XCI in PrE that will develop into the yolk sac 91. Meanwhile, in the ICM-derived epiblast, Xp is fully reactivated and random XCI occurs shortly after.
What controls Xp reactivation remains largely unknown. Initiation of Xp reactivation has been linked to Xist repression by pluripotency factors expressed in ICM 92,93. In naive female mESCs, in which both Xs are active, pluripotent factors NANOG, OCT4, and SOX2 bind to Xist intron 1 and repress Xist transcription 94. In addition, loss of PRDM14, a guardian of naïve pluripotency 95, causes defective Xp reactivation in mouse blastocysts 92. Mechanistic studies in mESCs suggest that PRDM14 represses Xist by directly binding to Xist intron 1 and indirectly silencing the Xist activator RNF12 92. Therefore, the relationships between the pluripotent factors and Xist seem to be complex. To what extent the insights obtained in mESCs is applicable to Xp reactivation in embryos remains to be determined. It is also intriguing that some X-linked genes initiate reactivation before the loss of Xist coating and repressive H3K27me3 in ICM, two cytological hallmarks of Xp reactivation 91,96. It is unclear how the early reactivation is initiated, although transcription factors such as MYC have been proposed to play a part in driving transcriptional activation of these genes in early blastocysts 91. On the other hand, erasure of H3K27me3 by histone demethylase KDM6A (also known as UTX) contributes to the transcription of late-reactivated genes 91. How removal of additional repressive chromatin marks associated with paternal XCI, such as H3K9me2 and H2AK119ub, contributes to Xp reactivation remains to be studied.
Maintenance of imprinted Xist in placenta.
Following Xp reactivation, random XCI occurs in the embryo proper with Xist expressed from either the maternal or paternal X. By contrast, Xist imprinting is maintained in extraembryonic lineages by the maternally expressed lncRNA Tsix, which is transcribed in an antisense direction from the Xist locus and represses Xist transcription in cis 97–99. When a Tsix knock-out allele is maternally inherited, maternal Xist is ectopically expressed in extraembryonic lineages, leading to aberrant maternal XCI and embryonic lethality 98,99. Given the essential role of Tsix in Xist imprinting and the absence of H3K27me3 from the Xist region after implantation, it is likely that oocyte H3K27me3-mediated maternal repression of Xist is replaced by Tsix-mediated repression in early post-implantation development. It should be noted that Tsix does not initiate maternal Xist silencing because it is not expressed until the morula stage 99,100.
As occurs at autosomal noncanonically imprinted loci, the Xist promoter becomes differentially methylated in extraembryonic lineages after implantation 101. However, disruption of DNMT1 does not affect the imprinted expression of an X-linked reporter gene in extraembryonic lineages, indicating that this DNA methyltransferase is not responsible for maintaining Xist imprinting in this lineage 102. Furthermore, simultaneous disruption of both DNMT3A and DNMT3B, de novo DNA methyltransferases that potentially compensate for DNMT1 in Dnmt1 mutants 102, does not affect Xist coating and only one X is inactivated 103. These observations imply that, unlike autosomal noncanonical imprinting, Xist maintains monoallelic expression in the absence of de novo DNA methylation in extraembryonic lineages.
Another notable difference between autosomal noncanonical imprinting and Xist imprinting is the developmental consequences of loss-of-imprinting in Eed maternal knock-out embryos. For noncanonical imprinting on autosomes, loss of maternal EED causes ectopic expression of the maternal allele in both pre-implantation embryos and placental lineages (Figure 2B) 72. By contrast, in Eed maternal knock-out embryos both X-chromosomes in females and the sole X-chromosome in males are silenced at the morula stage owing to ectopic expression of maternal Xist, but aberrant Xist imprinting and XCI is resolved at the blastocyst stage 72,86. Interestingly, Xist and XCI are no longer imprinted but either Xm or Xp is inactivated in placental lineages of Eed maternal knock-out female embryos (Figure 3B) 72. Analogously, androgenetic XpXp embryos, which do not have oocyte H3K27me3 to repress either Xist allele, also show bi-allelic XCI in early embryos but only one X-chromosome is randomly inactivated in later development 104. The correction of abnormal Xist imprinting in Eed maternal knock-out and XpXp embryos indicates that an X-chromosome counting mechanism exists in early embryos to ensure that a single X-chromosome is active regardless of its parental origin. However, aberrant XmCI in pre-implantation embryos already causes down-regulation of X-linked genes 72 and may contribute to the developmental defects observed in these mouse models.
Noncanonical imprinting in placenta
As the majority of canonically imprinted genes are expressed prenatally, their functions have been best characterized in fetal development and placental biology 3. Recently, roles for genomic imprinting in neuronal processes and adult behaviors have been reported 23, which is consistent with the brain, along with the placenta, being one of the organs with the highest number of imprinted genes 105. The physiological functions of canonical imprinting has been thoroughly reviewed elsewhere 2,3,23,106, so here we focus on the role of noncanonical imprinting in development.
Most noncanonical imprinting is transient in pre-implantation embryos, with several genes maintaining imprinted expression in the placenta 20. The function of the transient noncanonical imprinting is unclear and whether it has any long-term effects on development remains to be shown. However, evidence from canonical imprinting indicates that transient imprinting in early embryos can regulate somatic DMR acquisition, which affects later physiological processes 107. Specifically, a transient maternal germline DMR (the Gpr1/Zdbf2 DMR) causes paternal allele-specific expression of a lncRNA Liz in early mouse embryos 108. Liz is required to promote a paternally methylated intergenic somatic DMR (~10kb upstream of Zdbf2), which can antagonize H3K27me3-mediated repression of Zdbf2 107. Mouse embryos that lack early transient Liz expression fail to acquire the somatic DMR and to activate Zdbf2 in the postnatal brain. These animals also show ~20% body weight reduction through adult life 107. Therefore, although some transient noncanonical imprinting may be a by-product of transient asymmetric parental H3K27me3 in early embryos, a functional role with life-long consequences remains possible.
Of the noncanonical imprinted genes whose imprinting state is maintained in the placenta (Table 1), Slc38a4, Sfmbt2 and Gab1 are the best characterized. Knock-out mouse models for each of these genes develop placenta hypoplasia and show lethality or sub-lethality 109–111. For Slc38a4 and Sfmbt2, placental development is defective only when the knock-out allele is paternally inherited, consistent with imprinting and silencing of the maternal allele 109,110. SLC38A4 is an amino acid transporter that is likely involved in transporting amino acids at the maternal-fetal interface 109, which is consistent with the placental hypoplasia, and subsequent small-body phenotype, observed in mutant mice110. SFMBT2 is a mammalian homologue of the Drosophila Polycomb group protein Sfmbt but its molecular function remains poorly characterized 112. Paternal inheritance of a Sfmbt2 knock-out allele results in embryonic lethality at mid-gestation due to severe placenta defects 110. Intron 10 of Sfmbt2 harbors one of the largest miRNA clusters in the mouse, which is imprinted like Sfmbt2 113. Deletion of this miRNA cluster on the paternal allele severely impairs placental function and approximately one third of the paternal knock-out pups die around mid-gestation 113. Therefore, Sfmtb2 regulates placental development through both the SFMBT2 protein and the associated miRNA cluster 113. Lastly, GAB1 functions as an adaptor protein downstream of tyrosine kinase signaling and GAB1 homozygous mutant embryos die at late gestation and display developmental defects in placenta and other organs such as heart and skin 111,114.
Table 1.
List of noncanonically imprinted genes in extraembryonic cells.
| Gene | Molecular function | Mouse knock-out phenotype | Antisense imprinted lncRNA? | Germline DMR? | Somatic DMR? | DMR overlapped repeat element |
|---|---|---|---|---|---|---|
| Gab1 | A docking protein involved in cell signaling | Embryonic lethality; placenta, heart, and skin defects111,114 | Yes, paternal | No | Yes, E6.5 | ERVK:RLTR15 |
| Sfmbt2 | A Polycomb group protein | Embryonic lethality due to severe placenta defects110 | Yes, paternal | No | Yes, E7.5 | ERVK:RLTR11B |
| Slc38a4 | An amino acid transporter | Placenta hypoplasia, reduced fetal weight, 20% survival rate109 | No | Yes | NA* | ERVK:MLTR31F |
| Phf17 | A co-factor involved in histone acetylation | NA | No | No | Yes, E6.5 | ERVK:RLTR20C and RLTR31B |
| Smoc1 | A matricellular protein involved in cell signaling | Perinatal lethality144 | Yes, paternal | No | Yes, E6.5 | ERVK:RLTR11B |
| Platr20 | A lncRNA with unknown function | NA | Yes, paternal | No | Yes, E6.5 | ERVK:RLTR15 |
| Gm32885 | A lncRNA with unknown function | NA | No | No | Yes, E6.5 | ERVK:RLTR31A |
Slc38a4 germline DMR maintenance requires maternal H3K27me3 and zygotic DNMT3A/3B.
NA: Not applicable
Although Gab1, Sfmbt2, and Slc38a4 are required for normal mouse placental development, it has not yet been demonstrated whether these genes need to be imprinted. The maternal alleles at all noncanonically imprinted loci are derepressed in Eed maternal knock-out embryos and this mouse model has a variety of developmental defects, including embryonic sub-lethality, growth retardation at gastrulation, and postnatal overgrowth 72,74. However, these defects could be a combined effect of aberrant imprinted XCI, loss of noncanonical imprinting on autosomes, and imprinting-independent functions related to maternal H3K27me3 depletion. Given that Gab1, Sfmbt2, and Slc38a4 mutant mice show placental hypoplasia, it is possible that bi-allelic expression of these genes may cause enlarged placenta. Indeed, mouse embryos derived from somatic cell nuclear transfer (SCNT) always express Gab1, Sfmbt2, and Scl38a4 biallelically and show placenta hyperplasia 115. However, mouse models with either bi-allelic expression or paternal-duplication of the individual loci are needed to further clarify the role of autosomal noncanonical imprinting in placental development.
Aberrant imprinting and XCI in SCNT
SCNT is a technique by which a differentiated somatic cell nucleus is reprogramed by an enucleated oocyte to acquire totipotency (Figure 4A). SCNT makes possible not only reproductive cloning but also derivation of embryonic stem cells from cloned blastocysts 116. Therefore, SCNT holds great potential for regenerative medicine and the agricultural industry. However, the efficiency of the process has remained low in the 20 years since it was first used to successfully clone the first mammal 117. Aberrant genomic imprinting and imprinted XCI are two of the major known barriers impeding post-implantation development of cloned animals 116.
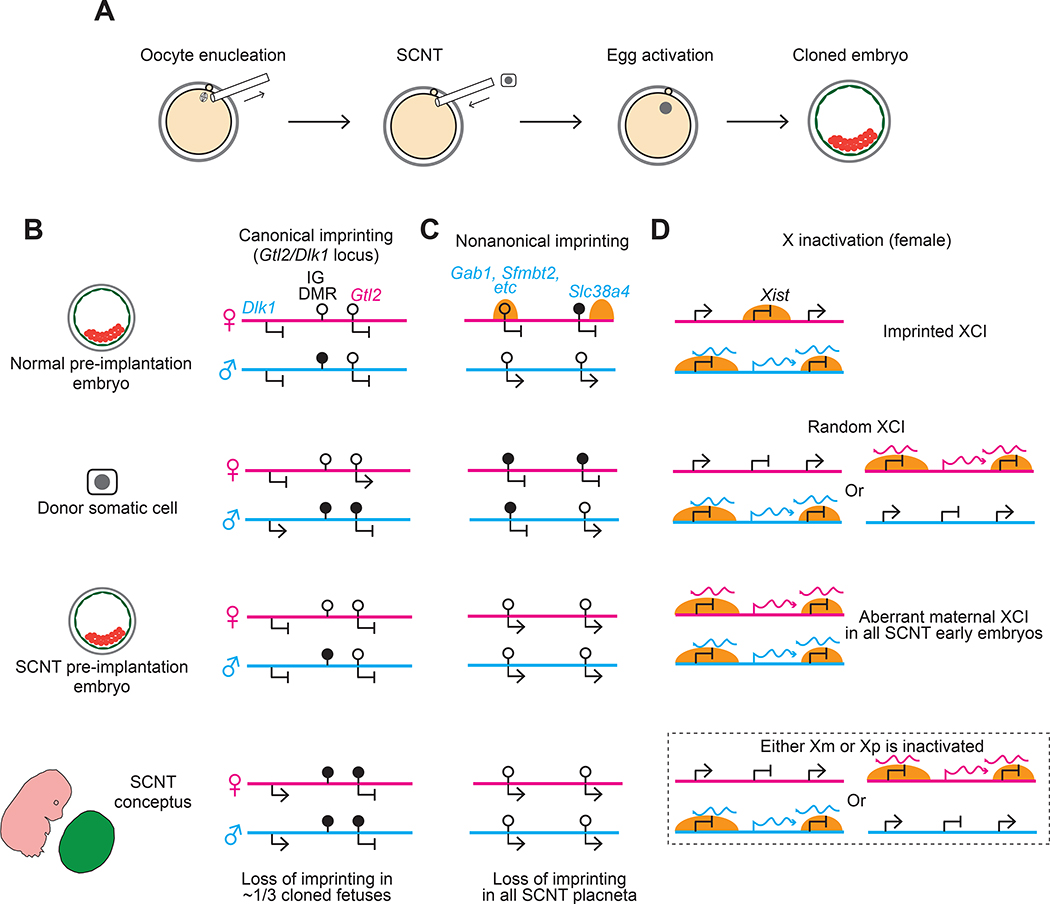
Figure 4. Defects in genomic imprinting and imprinted X inactivation occur in somatic cell nuclear transfer reprogramming.
A) Depicted are the general processes involved in somatic cell nuclear transfer (SCNT). A metaphase II oocyte is first enucleated and a donor cell nucleus from a differentiated somatic cell is transferred to the enucleated oocyte. The oocyte is then artificial activated by applying a chemical or electrical stimulus to initiate the developmental program to form cloned embryos.
B) Stochastic loss of canonical imprinting occurs at the Gtl2/Dlk1 locus in SCNT embryos. At this locus, the ICR is known as IG DMR and paternal-specific DNA methylation of the donor somatic cell is maintained in pre-implantation embryos derived by SCNT. However, after implantation, around one third of SCNT embryos exhibit gain of DNA methylation on the normally unmethylated maternal allele. The abnormal gain of DNA methylation is associated with biallelic expression of Dlk1 and biallelic repression of Gtl2, and with fetal lethality.
C) Imprinting is consistently lost at noncanonically imprinted loci in SCNT embryos. In donor cells such as cumulus and sertoli cells, the typical noncanonical imprinted loci (represented by Gab1) are marked by neither maternal H3K27me3 nor somatic differentially methylated regions (DMRs). Therefore, all placenta derived by SNCT show loss of noncanonical imprinting; they are also enlarged, which may be caused by disrupted noncanonical imprinting. Although the DMR is maintained at the noncanonically imprinted Slc38a4 locus in the donor cell, Slc38a4 becomes bi-allelically expressed in all cloned embryos, suggesting that the Slc38a4 DMR cannot be maintained without maternal H3K27me3 in early embryos.
D) Aberrant X inactivation occurs in SCNT embryos. In donor cells, one X-chromosome is randomly inactivated and Xist is not marked by maternal H3K27me3. Therefore, cloned embryos always express ectopic maternal Xist in addition to paternal Xist, and both X-chromosomes undergo XCI. Analogous to Eed maternal knockout embryos or XpXp androgenetic embryos, the bi-allelic inactivation of XCI is likely to be resolved at the late blastocyst stage (dashed boxes). However, insufficient expression of X-linked genes in pre-implantation development may still contribute to the post-implantation defects of cloned embryos, and correction of Xist expression has been shown to increase cloning efficiency by about 10-fold 122
Loss of canonical imprinting in SCNT embryos.
The initial assessment of canonical imprinting in cloned mouse embryos revealed that SCNT only alters transcript abundance but not allelic expression of imprinted genes 118. However, this study was based on the analyses of only a few imprinted genes. Later, a comprehensive RNA sequencing-based study indicated that canonical imprinting is stochastically disrupted in the brain and the placenta of cloned mice and the aberrant imprinting involves both loss of monoallelic gene expression and alterations of transcriptional abundance 15. It should be noted that some of the imprinting errors in cloned embryos may not be solely caused by SCNT reprogramming as the assisted reproductive techniques (ARTs) used in SCNT, such as superovulation and embryo culture, are also known to induce epimutations 119.
Nonetheless, at least some imprinting errors, such as those seen at the Gtl2/Dlk1 locus are likely caused by SCNT as they are rarely observed in embryos generated with the use of ART 120. It has been previously have shown that one third of SCNT embryos lose imprinting at the Gtl2/Dlk1 locus. In these embryos, Dlk1 becomes biallelically expressed and Gtl2 becomes biallelically repressed (Figure 4B)15 and their loss of allele-specific expression is associated with the gain of DNA methylation on the normally unmethylated maternal ICR 15. It remains unknown how such epimutations are caused by SCNT reprogramming. It is likely that ectopic gain of maternal DNA methylation occurs during the wave of global de novo DNA methylation at implantation as the Gtl2/Dlk1 maternal ICR is still hypomethylated at the blastocyst stage of SCNT embryos 115. Loss of imprinting at the Gtl2/Dlk1 locus is strongly correlated with lethality of SCNT embryos 15, so understanding the mechanisms underlying this epimutation could uncover the means to mitigate its effects and thereby improve cloning efficiency
Loss of noncanonical imprinting in SCNT embryos.
Noncanonically imprinted genes always show bi-allelic expression in mouse SCNT pre-implantation embryos, placenta and the derived TSCs because somatic cells do not retain the primary imprint, the oocyte-derived H3K27me3 (Figure 4C) 15,115,121. Indeed, maternal-biased H3K27me3 domains that normally exist during pre-implantation development have been shown to be absent in SCNT morula stage embryos 115. Intriguingly, the germline DMR on the maternal allele of Slc38a4 is maintained in SCNT donor cells, but all resulting embryos exhibit bi-allelic expression and loss of maternal-specific DNA methylation of this locus by the blastocyst stage (Figure 4C) 15. This observation suggests that the Slc38a4 germline DMR cannot mediate imprinting in pre-implantation embryos in the absence of maternally-inherited H3K27me3. As noted above, Gab1, Slc38a4 and Sfmbt2 knock-out mice show placenta hypoplasia, and bi-allelic expression of these genes may contribute to the enlarged placenta observed in all cloned mouse embryos 115. Whether using donor cells that are heterozygous for knock-out alleles of all three of these genes can reverse the enlarged placenta phenotype of SCNT embryos awaits to be shown.
Similar to noncanonical imprinting on autosomes, maternal H3K27me3 domains at the Xist locus also do not persist after implantation. Therefore, all SCNT-derived early embryos ectopically express Xist from the maternal allele, resulting in XmCI (Figure 4D) 122. Remarkably, cloning efficiency (in terms of live pup rate) can be increased by around 10-fold by correcting Xist expression in SCNT embryos, either by using Xist knock-out donor cells or by knocking down Xist expression via siRNA injection at the 1-cell stage 122,123. Correcting Xist expression in SCNT embryos both reverses the downregulation of X-linked genes owing to aberrant XmCI and reduces the number of differentially expressed genes on autosomes 122. These observations suggest that abnormal XCI in SCNT embryos disturbs the expression of both autosomal and X-linked genes. Similarly, aberrant XmCI may also contribute to the embryonic sub-lethality observed in the Eed maternal knock-out mouse model 72,74,86.
Conservation of noncanonical imprinting
Noncanonical imprinting is not conserved in humans.
In general, genomic imprinting in mice and humans is less conserved in the placenta than in the fetus 124,125. The oocyte H3K27me3-controlled mouse imprinted genes that have human orthologs such as Gab1 and Sfmbt2 are also not imprinted in the human placenta 14. Recently, comprehensive profiling of histone modifications during human early embryonic development revealed that H3K27me3 is globally depleted on both parental alleles at the 8-cell stage 126. These results indicate that oocyte-derived H3K27me3 in humans is unable to preserve allele-specificity throughout development and is therefore unlikely to serve as an imprinting mark 126. Although oocyte H3K27me3 does not mediate imprinted gene expression in humans as it does in mice, paternal-specific expression of genes not associated with germline DMRs can occur in human morula embryos, suggesting that a DNA methylation-independent imprinting mechanism may exist 127. Recently, data from a comprehensive survey of allele-specific gene expression that compared transcriptomes between bi-maternal and bi-paternal human early embryos suggested that around half of maternally- or paternally-biased gene expression cannot be explained by differential DNA methylation between parental alleles 128. Whether other epigenetic mechanisms modulate this allele-specific gene expression independently of DNA methylation remains to be investigated.
XCI dynamics in human early development is also distinct from mouse, although the detailed mechanisms remain elusive, partly owing to conflicting data. It was first proposed that one X chromosome is inactive in female human pre-implantation embryos as RNA-FISH detected XIST coating and some X-linked gene foci on only one of the two X chromosomes 129. However, another RNA-FISH based study reported that XIST coats both X-chromosomes in female and the sole X in male human early embryos 130. The discrepancies could be due to the different FISH conditions, which may compromise the detection of FISH signal on both alleles 130. Intriguingly, the XIST-coated X-chromosome lacks H3K27me3 and a few examined X-linked genes do not undergo silencing at these stages 130. Recently, data from single-cell RNA sequencing analyses indicated that dosage compensation of X-linked genes is achieved by reducing gene expression levels on both X chromosomes in female embryos 131, although this model was later challenged when the dataset was reanalyzed using different computational criteria 132. Despite the conflicting results and analyses, these studies highlight that important differences exist between XCI in human and mouse, and support the view that imprinted XCI is not conserved in human pre-implantation embryos. Interested readers are directed to a detailed review of human XCI dynamics 133.
Germline histone-mediated imprinting occurs in flowering plants.
Maternal H3K27me3 has been implicated as a primary imprint in the endosperm of flowering plants 134. Analogous to the mammalian placenta, the endosperm does not contribute to the next generation but is required for nourishment of the embryos. Similar to noncanonical imprinting in mice, H3K27me3-controlled imprinting in angiosperms is asymmetrically established in gametes and can persist in the endosperm 134. In addition, maternal H3K27me3 can recruit additional repressive epigenetic marks, including CHG methylation and H3K9me2, which may enforce gene silencing 135,136. In support of this notion, co-enrichment of H3K27me3, H3K9me2 and CHG methylation was observed on the maternal allele at paternally expressed genes in Arabidopsis endosperm 136. Furthermore, lack of PRC2 causes reduced CHG methylation suggesting that maternal CHG methylation depends on PRC2 activity 136. Given that the primary organs for germline H3K27me3-mediated imprinting in both plants and mice are involved in nutrient transfer, it is possible that this imprinting mechanism has evolved to respond to a similar selective pressure.
Conclusions and future perspectives
Recent advances in low-input epigenomic profiling have greatly enhanced our understanding of chromatin dynamics during the mammalian parental-to-zygotic transition. Accumulating evidence indicates that histone modifications can be transmitted from gametes to fertilized embryos to exert transcriptional regulation in the next generation. In particular, oocyte-inherited H3K27me3 can govern imprinted XCI and some placenta-specific imprinted genes in mice. These findings expand the known mechanisms by which intergenerational epigenetic inheritance occurs and provide an opportunity to fully understand epigenetic reprogramming and totipotency acquisition in early development.
Although much has been discovered in recent years about the mechanisms underlying noncanonical imprinting, including how it is established and maintained, many details remain to be clarified. Firstly, it remains unknown whether PRC1-mediated H2AK119Ub plays a part in regulating noncanonical imprinting. PRC1-catalyzed H2AK119Ub usually overlaps with PRC2-mediated H3K27me3 in mESCs and plays a predominant role in silencing PcG targets and maintaining pluripotency 137. By contrast, removal of H3K27me3 alone can cause loss of noncanonical imprinting 20,70,72, suggesting a distinct interplay between PRC1 and PRC2 in early embryos, at least at the oocyte H3K27me3-controlled imprinted genes. Secondly, it is not clear why noncanonical imprinting cannot be maintained in the embryonic lineage after implantation. Thirdly, imprinted antisense lncRNAs have been identified upstream of the promoters of Gab1, Sfmbt2, and Smoc1 and whether these lncRNAs are involved in imprinting regulation remains to be determined 138. At least for Sfmbt2, the transcription and/or splicing of its antisense RNA contributes to Sfmbt2 activation, potentially by modulating the chromatin state at the Sfmbt2 promoter 139. Fourthly, it remains a point of debate whether expression of the Slc38a4 gene, which has a germline DMR that maintains paternal allele-expression in the epiblast, is controlled by canonical or noncanonical imprinting. The observation that Slc38a4 imprinting is compromised in Eed, but not Dnmt3l or Dnmt3a/3b maternal knock-out embryos indicates that this gene is regulated by the noncanonical mechanism 15,20,70,72. However, it was reported recently that local oocyte DNA hypomethylation at the Slc83a4 DMR can cause bi-allelic expression of Slc38a4 in the placenta 140. Whether this discrepancy is caused by alternative promoter usage or lineage-specific imprinting regulation remains to be determined 71,140. Lastly, it remains challenging to correct canonical or noncanonical imprinting errors to rescue post-implantation defects in cloned embryos. It is unclear whether the modified epigenome of donor cells can persist to the next generation during the dynamic SCNT reprogramming in early embryos. In addition, although targeted DNA methylation or demethylation in oocytes and early embryos has been achieved 141,142, fixing the imprinting errors in SCNT embryos in an allele-specific manner is still challenging.
Beyond the role of maternal histones in genomic imprinting, the precise mechanisms and the extent to which parental chromatin affects the next generation remains unclear. For example, although oocyte-provided PRC2 in Drosophila melanogaster prevents precocious activation of some developmental regulators at ZGA by restricting enhancer function 143, it remains unknown whether a similar transcriptional repressive mechanism exists in mammals because there is no evidence to support that maternal H3K27me3 in mouse performs an analogous role. Notably, despite considerable achievements in mapping the chromatin landscape in mammalian early development, the dynamic control of this process remains unclear 18. Thus, the function of, and regulatory mechanisms underlying, parental chromatin dynamics in gametogenesis and early development will remain important areas of research for years to come.
Key points:
Germline differential DNA methylation governs canonical imprinting.
Oocyte H3K27me3 initiates noncanonical imprinting independent of germline DNA methylation.
Oocyte-derived H3K27me3 controls imprinted X inactivation by repressing maternal Xist.
Noncanoical impmrinting on autosomes are implicated in placental development.
Loss of both canonical and noncanonical imprinting impede mouse cloning efficiency.
Acknowledgements
The authors thank Amanda Liefeld for critical reading of the manuscript. This work was supported by HHMI and NIH (R01HD092465). Y.Z. is an Investigator of the Howard Hughes Medical Institute.
Glossary
- pronuclear transfer
A technique that involves moving one or both pronuclei (which are formed from the sperm and oocyte genomes shortly after fertilization) from a fertilized 1-cell embryo to a different recipient embryo.
- DNA methylation
An epigenetic modification in which a methyl group is added to the 5th carbon of a cytosine in a DNA molecule. DNA methylation at gene promoters is generally associated with transcriptional silencing.
- somatic DMRs
Also known as secondary DMRs, somatic DMRs are regions of the genome containing allele-specific DNA methylation that is established after fertilization.
- primordial germ cells (PGCs)
precursors of the gametes that are specified from the somatic lineage during gastrulation.
- 4C-seq
A sequencing-based method that allows unbiased detection of all genomic regions that interact with a genomic region of interest.
- topologically associated domain (TAD)
A major form of chromatin organization that represents genomic regions with high frequencies of self-interacting events.
- CpG islands
Genomic regions with a high density of CpG dinucleotides. In mammalian genomes, CpG islands usually extend from 200bp to a few Kbp.
- DNase I hypersensitivity sites
Chromatin regions that are less condensed and more sensitive to DNase I enzyme-mediated cleavage than other regions.
- protamines
Basic proteins that replace histones in mature sperm cells and are involved in sperm DNA condensation.
- epiblast
One of the two lineages that are derived from the inner cell mass (ICM) of the blastocyst. The epiblast contributes to all three primary germ layers. Primitive endoderm (PrE), the other lineage derived from ICM, contributes to the yolk sac.
- parthenogenetic activation
A procedure that mimics sperm stimuli to trigger egg activation to initiate embryo development without the contribution of the paternal genome.
- meiotic sex chromosome inactivation (MSCI)
The process of silencing X and Y chromosomes during the meiotic phase of spermatogenesis.
- CHG methylation
DNA methylation typically occurs in a CpG context. In CHG methylation, H correspond to A, T or C, but not G.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Bartolomei MS & Ferguson-Smith AC Mammalian genomic imprinting. Cold Spring Harb Perspect Biol 3, doi: 10.1101/cshperspect.a002592 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee JT & Bartolomei MS X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell 152, 1308–1323, doi: 10.1016/j.cell.2013.02.016 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Monk D, Mackay DJG, Eggermann T, Maher ER & Riccio A Genomic imprinting disorders: lessons on how genome, epigenome and environment interact. Nature reviews. Genetics 20, 235–248, doi: 10.1038/s41576-018-0092-0 (2019). [DOI] [PubMed] [Google Scholar]
- 4.McGrath J & Solter D Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 37, 179–183 (1984). [DOI] [PubMed] [Google Scholar]
- 5.Surani MA, Barton SC & Norris ML Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature 308, 548–550, doi: 10.1038/308548a0 (1984). [DOI] [PubMed] [Google Scholar]
- 6.Bartolomei MS, Zemel S & Tilghman SM Parental imprinting of the mouse H19 gene. Nature 351, 153–155, doi: 10.1038/351153a0 (1991). [DOI] [PubMed] [Google Scholar]
- 7.Barlow DP, Stoger R, Herrmann BG, Saito K & Schweifer N The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature 349, 84–87, doi: 10.1038/349084a0 (1991). [DOI] [PubMed] [Google Scholar]
- 8.Ferguson-Smith AC, Cattanach BM, Barton SC, Beechey CV & Surani MA Embryological and molecular investigations of parental imprinting on mouse chromosome 7. Nature 351, 667–670, doi: 10.1038/351667a0 (1991). [DOI] [PubMed] [Google Scholar]
- 9.DeChiara TM, Robertson EJ & Efstratiadis A Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64, 849–859 (1991). [DOI] [PubMed] [Google Scholar]
- 10.Li E, Beard C & Jaenisch R Role for DNA methylation in genomic imprinting. Nature 366, 362–365, doi: 10.1038/366362a0 (1993). [DOI] [PubMed] [Google Scholar]
- 11.Barlow DP & Bartolomei MS Genomic imprinting in mammals. Cold Spring Harb Perspect Biol 6, doi: 10.1101/cshperspect.a018382 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanli I & Feil R Chromatin mechanisms in the developmental control of imprinted gene expression. Int J Biochem Cell Biol 67, 139–147, doi: 10.1016/j.biocel.2015.04.004 (2015). [DOI] [PubMed] [Google Scholar]
- 13.John RM & Lefebvre L Developmental regulation of somatic imprints. Differentiation 81, 270–280, doi: 10.1016/j.diff.2011.01.007 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Okae H et al. Re-investigation and RNA sequencing-based identification of genes with placenta-specific imprinted expression. Human molecular genetics 21, 548–558, doi: 10.1093/hmg/ddr488 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Okae H et al. RNA sequencing-based identification of aberrant imprinting in cloned mice. Human molecular genetics 23, 992–1001, doi: 10.1093/hmg/ddt495 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Borensztein M et al. Xist-dependent imprinted X inactivation and the early developmental consequences of its failure. Nat Struct Mol Biol 24, 226–233, doi: 10.1038/nsmb.3365 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiba H et al. De novo DNA methylation independent establishment of maternal imprint on X chromosome in mouse oocytes. Genesis 46, spc one, doi: 10.1002/dvg.20496 (2008).This study and the 2012 study by Okae et al demonstrated the presence of germline DNA methylation-independent autosomal and X-chromosomal imprinting.
- 18.Eckersley-Maslin MA, Alda-Catalinas C & Reik W Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nature reviews. Molecular cell biology 19, 436–450, doi: 10.1038/s41580-018-0008-z (2018). [DOI] [PubMed] [Google Scholar]
- 19.Inoue A, Jiang L, Lu F & Zhang Y Genomic imprinting of Xist by maternal H3K27me3. Genes & development 31, 1927–1932, doi: 10.1101/gad.304113.117 (2017).This study demonstrated that maternal Xist is repressed by oocyte H3K27me3, thus causing imprinted X inactivation.
- 20.Inoue A, Jiang L, Lu F, Suzuki T & Zhang Y Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 547, 419–424, doi: 10.1038/nature23262 (2017).This study demonstrated that maternal H3K27me3 can serve as a primary imprinting mark.
- 21.Bonthuis PJ et al. Noncanonical Genomic Imprinting Effects in Offspring. Cell reports 12, 979–991, doi: 10.1016/j.celrep.2015.07.017 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Ferguson-Smith AC Genomic imprinting: the emergence of an epigenetic paradigm. Nature reviews. Genetics 12, 565–575, doi: 10.1038/nrg3032 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Tucci V, Isles AR, Kelsey G, Ferguson-Smith AC & Erice Imprinting G Genomic Imprinting and Physiological Processes in Mammals. Cell 176, 952–965, doi: 10.1016/j.cell.2019.01.043 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Bourc’his D, Xu GL, Lin CS, Bollman B & Bestor TH Dnmt3L and the establishment of maternal genomic imprints. Science 294, 2536–2539, doi: 10.1126/science.1065848 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Kaneda M et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429, 900–903, doi: 10.1038/nature02633 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Barau J et al. The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science 354, 909–912, doi: 10.1126/science.aah5143 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Jain D et al. rahu is a mutant allele of Dnmt3c, encoding a DNA methyltransferase homolog required for meiosis and transposon repression in the mouse male germline. PLoS genetics 13, e1006964, doi: 10.1371/journal.pgen.1006964 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watanabe T et al. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science 332, 848–852, doi: 10.1126/science.1203919 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chotalia M et al. Transcription is required for establishment of germline methylation marks at imprinted genes. Genes & development 23, 105–117, doi: 10.1101/gad.495809 (2009).This study demonstrated that transcription through a maternally methylated DMR is required for de novo DNA methylation during oogenesis.
- 30.Stewart KR et al. Dynamic changes in histone modifications precede de novo DNA methylation in oocytes. Genes & development 29, 2449–2462, doi: 10.1101/gad.271353.115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Veselovska L et al. Deep sequencing and de novo assembly of the mouse oocyte transcriptome define the contribution of transcription to the DNA methylation landscape. Genome biology 16, 209, doi: 10.1186/s13059-015-0769-z (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith EY, Futtner CR, Chamberlain SJ, Johnstone KA & Resnick JL Transcription is required to establish maternal imprinting at the Prader-Willi syndrome and Angelman syndrome locus. PLoS genetics 7, e1002422, doi: 10.1371/journal.pgen.1002422 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu Q et al. SETD2 regulates the maternal epigenome, genomic imprinting and embryonic development. Nature genetics 51, 844–856, doi: 10.1038/s41588-019-0398-7 (2019).This study demonstrated the critial role of histone methyltransferase SETD2 in regulating the oocyte epigenome, including the establishment of maternal imprints.
- 34.Ciccone DN et al. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 461, 415–418, doi: 10.1038/nature08315 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Chen Z & Zhang Y Role of Mammalian DNA Methyltransferases in Development. Annual review of biochemistry, doi: 10.1146/annurev-biochem-103019-102815 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Smith ZD et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484, 339–344, doi: 10.1038/nature10960 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X et al. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Developmental cell 15, 547–557, doi: 10.1016/j.devcel.2008.08.014 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi N et al. ZNF445 is a primary regulator of genomic imprinting. Genes & development 33, 49–54, doi: 10.1101/gad.320069.118 (2019).This study and Li et al (2008) demonstrated that the KRAB-containing zinc finger proteins ZFP57 and ZFP445 maintain allelic DNA methylation specifically at imprinting control regions during the global wave of DNA demethylation in early embryos.
- 39.Quenneville S et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Molecular cell 44, 361–372, doi: 10.1016/j.molcel.2011.08.032 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Messerschmidt DM et al. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science 335, 1499–1502, doi: 10.1126/science.1216154 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Lee CC et al. The Role of N-alpha-acetyltransferase 10 Protein in DNA Methylation and Genomic Imprinting. Molecular cell 68, 89–103 e107, doi: 10.1016/j.molcel.2017.08.025 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Proudhon C et al. Protection against de novo methylation is instrumental in maintaining parent-of-origin methylation inherited from the gametes. Molecular cell 47, 909–920, doi: 10.1016/j.molcel.2012.07.010 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanna CW & Kelsey G The specification of imprints in mammals. Heredity (Edinb) 113, 176–183, doi: 10.1038/hdy.2014.54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu X & Zhang Y TET-mediated active DNA demethylation: mechanism, function and beyond. Nature reviews. Genetics 18, 517–534, doi: 10.1038/nrg.2017.33 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Yamaguchi S et al. Tet1 controls meiosis by regulating meiotic gene expression. Nature 492, 443–447, doi: 10.1038/nature11709 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamaguchi S, Shen L, Liu Y, Sendler D & Zhang Y Role of Tet1 in erasure of genomic imprinting. Nature 504, 460–464, doi: 10.1038/nature12805 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.SanMiguel JM, Abramowitz LK & Bartolomei MS Imprinted gene dysregulation in a Tet1 null mouse model is stochastic and variable in the germline and offspring. Development 145, doi: 10.1242/dev.160622 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dawlaty MM et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Developmental cell 24, 310–323, doi: 10.1016/j.devcel.2012.12.015 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bartolomei MS, Webber AL, Brunkow ME & Tilghman SM Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes & development 7, 1663–1673, doi: 10.1101/gad.7.9.1663 (1993). [DOI] [PubMed] [Google Scholar]
- 50.Ferguson-Smith AC, Sasaki H, Cattanach BM & Surani MA Parental-origin-specific epigenetic modification of the mouse H19 gene. Nature 362, 751–755, doi: 10.1038/362751a0 (1993). [DOI] [PubMed] [Google Scholar]
- 51.Thorvaldsen JL, Duran KL & Bartolomei MS Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes & development 12, 3693–3702 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bell AC & Felsenfeld G Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405, 482–485, doi: 10.1038/35013100 (2000). [DOI] [PubMed] [Google Scholar]
- 53.Hark AT et al. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405, 486–489, doi: 10.1038/35013106 (2000). [DOI] [PubMed] [Google Scholar]
- 54.Engel N, Thorvaldsen JL & Bartolomei MS CTCF binding sites promote transcription initiation and prevent DNA methylation on the maternal allele at the imprinted H19/Igf2 locus. Human molecular genetics 15, 2945–2954, doi: 10.1093/hmg/ddl237 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Lleres D et al. CTCF modulates allele-specific sub-TAD organization and imprinted gene activity at the mouse Dlk1-Dio3 and Igf2-H19 domains. Genome biology 20, 272, doi: 10.1186/s13059-019-1896-8 (2019).This study investigated how allele-specific TAD formation is involved in imprinted gene activity.
- 56.Lee MP et al. Loss of imprinting of a paternally expressed transcript, with antisense orientation to KVLQT1, occurs frequently in Beckwith-Wiedemann syndrome and is independent of insulin-like growth factor II imprinting. Proceedings of the National Academy of Sciences of the United States of America 96, 5203–5208, doi: 10.1073/pnas.96.9.5203 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smilinich NJ et al. A maternally methylated CpG island in KvLQT1 is associated with an antisense paternal transcript and loss of imprinting in Beckwith-Wiedemann syndrome. Proceedings of the National Academy of Sciences of the United States of America 96, 8064–8069, doi: 10.1073/pnas.96.14.8064 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Terranova R et al. Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Developmental cell 15, 668–679, doi: 10.1016/j.devcel.2008.08.015 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Pandey RR et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Molecular cell 32, 232–246, doi: 10.1016/j.molcel.2008.08.022 (2008). [DOI] [PubMed] [Google Scholar]
- 60.Wagschal A et al. G9a histone methyltransferase contributes to imprinting in the mouse placenta. Molecular and cellular biology 28, 1104–1113, doi: 10.1128/MCB.01111-07 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Umlauf D et al. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nature genetics 36, 1296–1300, doi: 10.1038/ng1467 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Lewis A et al. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nature genetics 36, 1291–1295, doi: 10.1038/ng1468 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Fitzpatrick GV, Soloway PD & Higgins MJ Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nature genetics 32, 426–431, doi: 10.1038/ng988 (2002). [DOI] [PubMed] [Google Scholar]
- 64.Mancini-Dinardo D, Steele SJ, Levorse JM, Ingram RS & Tilghman SM Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes & development 20, 1268–1282, doi: 10.1101/gad.1416906 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schertzer MD et al. lncRNA-Induced Spread of Polycomb Controlled by Genome Architecture, RNA Abundance, and CpG Island DNA. Molecular cell 75, 523–537 e510, doi: 10.1016/j.molcel.2019.05.028 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu F et al. Establishing Chromatin Regulatory Landscape during Mouse Preimplantation Development. Cell 165, 1375–1388, doi: 10.1016/j.cell.2016.05.050 (2016).This study described a low-input DNase I sequencing method and demonstrated that imprinting control regions exhibit allelic chromatin accessibility bias before onset of allelic expression in mouse early embryos.
- 67.Zheng H et al. Resetting Epigenetic Memory by Reprogramming of Histone Modifications in Mammals. Molecular cell 63, 1066–1079, doi: 10.1016/j.molcel.2016.08.032 (2016). [DOI] [PubMed] [Google Scholar]
- 68.Mager J, Montgomery ND, de Villena FP & Magnuson T Genome imprinting regulated by the mouse Polycomb group protein Eed. Nature genetics 33, 502–507, doi: 10.1038/ng1125 (2003). [DOI] [PubMed] [Google Scholar]
- 69.Liu X et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 537, 558–562, doi: 10.1038/nature19362 (2016).This study and Zheng et al profiled H3K27me3 in mouse development and revealed noncanonical H3K27me3 domains that are unique for oocyte and early embryos.
- 70.Chen Z, Yin Q, Inoue A, Zhang C & Zhang Y Allelic H3K27me3 to allelic DNA methylation switch maintains noncanonical imprinting in extraembryonic cells. Science Advances 5, doi: 10.1126/sciadv.aay7246 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hanna CW et al. Endogenous retroviral insertions drive non-canonical imprinting in extraembryonic tissues. Genome biology 20, 225, doi: 10.1186/s13059-019-1833-x (2019).This study and Chen et al demonstrated that maintainance of noncanonical imprinting in extraembryonic cells involves a switch from allelic H3K27me3 to allelic DNA methylation.
- 72.Inoue A, Chen Z, Yin Q & Zhang Y Maternal Eed knockout causes loss of H3K27me3 imprinting and random X inactivation in the extraembryonic cells. Genes & development, doi: 10.1101/gad.318675.118 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Posfai E et al. Polycomb function during oogenesis is required for mouse embryonic development. Genes & development 26, 920–932, doi: 10.1101/gad.188094.112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Prokopuk L et al. Loss of maternal EED results in postnatal overgrowth. Clinical epigenetics 10, 95, doi: 10.1186/s13148-018-0526-8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Du Z et al. Polycomb Group Proteins Regulate Chromatin Architecture in Mouse Oocytes and Early Embryos. Molecular cell, doi: 10.1016/j.molcel.2019.11.011 (2019). [DOI] [PubMed] [Google Scholar]
- 76.He J, Kallin EM, Tsukada Y & Zhang Y The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b). Nat Struct Mol Biol 15, 1169–1175, doi:nsmb.1499 [pii] 10.1038/nsmb.1499 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.He J, Nguyen AT & Zhang Y KDM2b/JHDM1b, an H3K36me2-specific demethylase, is required for initiation and maintenance of acute myeloid leukemia. Blood 117, 3869–3880, doi:blood-2010–10-312736 [pii] 10.1182/blood-2010-10-312736 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu X, Johansen JV & Helin K Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Molecular cell 49, 1134–1146, doi: 10.1016/j.molcel.2013.01.016 (2013). [DOI] [PubMed] [Google Scholar]
- 79.Farcas AM et al. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. Elife 1, e00205, doi: 10.7554/eLife.00205 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.He J et al. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nature cell biology 15, 373–384, doi: 10.1038/ncb2702 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boulard M, Edwards JR & Bestor TH FBXL10 protects Polycomb-bound genes from hypermethylation. Nature genetics 47, 479–485, doi: 10.1038/ng.3272 (2015). [DOI] [PubMed] [Google Scholar]
- 82.Tada T et al. Imprint switching for non-random X-chromosome inactivation during mouse oocyte growth. Development 127, 3101–3105 (2000). [DOI] [PubMed] [Google Scholar]
- 83.Nesterova TB, Barton SC, Surani MA & Brockdorff N Loss of Xist imprinting in diploid parthenogenetic preimplantation embryos. Dev Biol 235, 343–350, doi: 10.1006/dbio.2001.0295 (2001). [DOI] [PubMed] [Google Scholar]
- 84.Huynh KD & Lee JT Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature 426, 857–862, doi: 10.1038/nature02222 (2003). [DOI] [PubMed] [Google Scholar]
- 85.Okamoto I et al. Evidence for de novo imprinted X-chromosome inactivation independent of meiotic inactivation in mice. Nature 438, 369–373, doi: 10.1038/nature04155 (2005). [DOI] [PubMed] [Google Scholar]
- 86.Harris C et al. Conversion of random X-inactivation to imprinted X-inactivation by maternal PRC2. Elife 8, doi: 10.7554/eLife.44258 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fukuda A et al. The role of maternal-specific H3K9me3 modification in establishing imprinted X-chromosome inactivation and embryogenesis in mice. Nature communications 5, 5464, doi: 10.1038/ncomms6464 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang C et al. Reprogramming of H3K9me3-dependent heterochromatin during mammalian embryo development. Nature cell biology 20, 620–631, doi: 10.1038/s41556-018-0093-4 (2018). [DOI] [PubMed] [Google Scholar]
- 89.Mak W et al. Reactivation of the paternal X chromosome in early mouse embryos. Science 303, 666–669, doi: 10.1126/science.1092674 (2004). [DOI] [PubMed] [Google Scholar]
- 90.Okamoto I, Otte AP, Allis CD, Reinberg D & Heard E Epigenetic dynamics of imprinted X inactivation during early mouse development. Science 303, 644–649, doi: 10.1126/science.1092727 (2004). [DOI] [PubMed] [Google Scholar]
- 91.Borensztein M et al. Contribution of epigenetic landscapes and transcription factors to X-chromosome reactivation in the inner cell mass. Nature communications 8, 1297, doi: 10.1038/s41467-017-01415-5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Payer B et al. Tsix RNA and the germline factor, PRDM14, link X reactivation and stem cell reprogramming. Molecular cell 52, 805–818, doi: 10.1016/j.molcel.2013.10.023 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Navarro P et al. Molecular coupling of Tsix regulation and pluripotency. Nature 468, 457–460, doi: 10.1038/nature09496 (2010). [DOI] [PubMed] [Google Scholar]
- 94.Navarro P et al. Molecular coupling of Xist regulation and pluripotency. Science 321, 1693–1695, doi: 10.1126/science.1160952 (2008). [DOI] [PubMed] [Google Scholar]
- 95.Yamaji M et al. PRDM14 ensures naive pluripotency through dual regulation of signaling and epigenetic pathways in mouse embryonic stem cells. Cell stem cell 12, 368–382, doi: 10.1016/j.stem.2012.12.012 (2013). [DOI] [PubMed] [Google Scholar]
- 96.Williams LH, Kalantry S, Starmer J & Magnuson T Transcription precedes loss of Xist coating and depletion of H3K27me3 during X-chromosome reprogramming in the mouse inner cell mass. Development 138, 2049–2057, doi: 10.1242/dev.061176 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee JT, Davidow LS & Warshawsky D Tsix, a gene antisense to Xist at the X-inactivation centre. Nature genetics 21, 400–404, doi: 10.1038/7734 (1999). [DOI] [PubMed] [Google Scholar]
- 98.Lee JT Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell 103, 17–27, doi: 10.1016/s0092-8674(00)00101-x (2000). [DOI] [PubMed] [Google Scholar]
- 99.Sado T, Wang Z, Sasaki H & Li E Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development 128, 1275–1286 (2001). [DOI] [PubMed] [Google Scholar]
- 100.Debrand E, Chureau C, Arnaud D, Avner P & Heard E Functional analysis of the DXPas34 locus, a 3’ regulator of Xist expression. Molecular and cellular biology 19, 8513–8525, doi: 10.1128/mcb.19.12.8513 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ohhata T, Senner CE, Hemberger M & Wutz A Lineage-specific function of the noncoding Tsix RNA for Xist repression and Xi reactivation in mice. Genes & development 25, 1702–1715, doi: 10.1101/gad.16997911 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sado T et al. X inactivation in the mouse embryo deficient for Dnmt1: distinct effect of hypomethylation on imprinted and random X inactivation. Dev Biol 225, 294–303, doi: 10.1006/dbio.2000.9823 (2000). [DOI] [PubMed] [Google Scholar]
- 103.Sado T, Okano M, Li E & Sasaki H De novo DNA methylation is dispensable for the initiation and propagation of X chromosome inactivation. Development 131, 975–982, doi: 10.1242/dev.00995 (2004). [DOI] [PubMed] [Google Scholar]
- 104.Okamoto I, Tan S & Takagi N X-chromosome inactivation in XX androgenetic mouse embryos surviving implantation. Development 127, 4137–4145 (2000). [DOI] [PubMed] [Google Scholar]
- 105.Andergassen D et al. Mapping the mouse Allelome reveals tissue-specific regulation of allelic expression. Elife 6, doi: 10.7554/eLife.25125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Peters J The role of genomic imprinting in biology and disease: an expanding view. Nature reviews. Genetics 15, 517–530, doi: 10.1038/nrg3766 (2014). [DOI] [PubMed] [Google Scholar]
- 107.Greenberg MV et al. Transient transcription in the early embryo sets an epigenetic state that programs postnatal growth. Nature genetics 49, 110–118, doi: 10.1038/ng.3718 (2017).This study demonstrated the important biological functions of a transient germline DMR.
- 108.Duffie R et al. The Gpr1/Zdbf2 locus provides new paradigms for transient and dynamic genomic imprinting in mammals. Genes & development 28, 463–478, doi: 10.1101/gad.232058.113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Matoba S et al. Paternal knockout of Slc38a4/SNAT4 causes placental hypoplasia associated with intrauterine growth restriction in mice. Proceedings of the National Academy of Sciences of the United States of America 116, 21047–21053, doi: 10.1073/pnas.1907884116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Miri K et al. The imprinted polycomb group gene Sfmbt2 is required for trophoblast maintenance and placenta development. Development 140, 4480–4489, doi: 10.1242/dev.096511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sachs M et al. Essential role of Gab1 for signaling by the c-Met receptor in vivo. The Journal of cell biology 150, 1375–1384 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Klymenko T et al. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes & development 20, 1110–1122, doi: 10.1101/gad.377406 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Inoue K et al. The Rodent-Specific MicroRNA Cluster within the Sfmbt2 Gene Is Imprinted and Essential for Placental Development. Cell reports 19, 949–956, doi: 10.1016/j.celrep.2017.04.018 (2017). [DOI] [PubMed] [Google Scholar]
- 114.Itoh M et al. Role of Gab1 in heart, placenta, and skin development and growth factor- and cytokine-induced extracellular signal-regulated kinase mitogen-activated protein kinase activation. Molecular and cellular biology 20, 3695–3704, doi: 10.1128/mcb.20.10.3695-3704.2000 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Matoba S et al. Loss of H3K27me3 Imprinting in Somatic Cell Nuclear Transfer Embryos Disrupts Post-Implantation Development. Cell stem cell 23, 343–354 e345, doi: 10.1016/j.stem.2018.06.008 (2018).This study demonstrated that loss of noncanonical imprinting contributes to placenta defects observed in cloned embryos.
- 116.Matoba S & Zhang Y Somatic Cell Nuclear Transfer Reprogramming: Mechanisms and Applications. Cell stem cell 23, 471–485, doi: 10.1016/j.stem.2018.06.018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wilmut I, Schnieke AE, McWhir J, Kind AJ & Campbell KH Viable offspring derived from fetal and adult mammalian cells. Nature 385, 810–813, doi: 10.1038/385810a0 (1997). [DOI] [PubMed] [Google Scholar]
- 118.Inoue K et al. Faithful expression of imprinted genes in cloned mice. Science 295, 297, doi: 10.1126/science.295.5553.297 (2002). [DOI] [PubMed] [Google Scholar]
- 119.Rhon-Calderon EA, Vrooman LA, Riesche L & Bartolomei MS The effects of Assisted Reproductive Technologies on genomic imprinting in the placenta. Placenta 84, 37–43, doi: 10.1016/j.placenta.2019.02.013 (2019). [DOI] [PubMed] [Google Scholar]
- 120.de Waal E et al. In vitro culture increases the frequency of stochastic epigenetic errors at imprinted genes in placental tissues from mouse concepti produced through assisted reproductive technologies. Biology of reproduction 90, 22, doi: 10.1095/biolreprod.113.114785 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hirose M et al. Aberrant imprinting in mouse trophoblast stem cells established from somatic cell nuclear transfer-derived embryos. Epigenetics : official journal of the DNA Methylation Society 13, 693–703, doi: 10.1080/15592294.2018.1507199 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Inoue K et al. Impeding Xist expression from the active X chromosome improves mouse somatic cell nuclear transfer. Science 330, 496–499, doi: 10.1126/science.1194174 (2010).This study demonstrated that aberrant imprinted X inactivation is a major barrier in somatic cell nuclear transfer and that impeding Xist expression can greatly improve cloning efficiency.
- 123.Matoba S et al. RNAi-mediated knockdown of Xist can rescue the impaired postimplantation development of cloned mouse embryos. Proceedings of the National Academy of Sciences of the United States of America 108, 20621–20626, doi: 10.1073/pnas.1112664108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Monk D et al. Limited evolutionary conservation of imprinting in the human placenta. Proceedings of the National Academy of Sciences of the United States of America 103, 6623–6628, doi: 10.1073/pnas.0511031103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Frost JM & Moore GE The importance of imprinting in the human placenta. PLoS genetics 6, e1001015, doi: 10.1371/journal.pgen.1001015 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xia W et al. Resetting histone modifications during human parental-to-zygotic transition. Science, doi: 10.1126/science.aaw5118 (2019). [DOI] [PubMed] [Google Scholar]
- 127.Zhang W et al. Maternal-biased H3K27me3 correlates with paternal-specific gene expression in the human morula. Genes & development 33, 382–387, doi: 10.1101/gad.323105.118 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Leng L et al. Single-Cell Transcriptome Analysis of Uniparental Embryos Reveals Parent-of-Origin Effects on Human Preimplantation Development. Cell stem cell 25, 697–712 e696, doi: 10.1016/j.stem.2019.09.004 (2019). [DOI] [PubMed] [Google Scholar]
- 129.van den Berg IM et al. X chromosome inactivation is initiated in human preimplantation embryos. Am J Hum Genet 84, 771–779, doi: 10.1016/j.ajhg.2009.05.003 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Okamoto I et al. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature 472, 370–374, doi: 10.1038/nature09872 (2011). [DOI] [PubMed] [Google Scholar]
- 131.Petropoulos S et al. Single-Cell RNA-Seq Reveals Lineage and X Chromosome Dynamics in Human Preimplantation Embryos. Cell 167, 285, doi: 10.1016/j.cell.2016.08.009 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Moreira de Mello JC, Fernandes GR, Vibranovski MD & Pereira LV Early X chromosome inactivation during human preimplantation development revealed by single-cell RNA-sequencing. Sci Rep 7, 10794, doi: 10.1038/s41598-017-11044-z (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Patrat C, Ouimette JF & Rougeulle C X chromosome inactivation in human development. Development 147, doi: 10.1242/dev.183095 (2020). [DOI] [PubMed] [Google Scholar]
- 134.Batista RA & Kohler C Genomic imprinting in plants-revisiting existing models. Genes & development 34, 24–36, doi: 10.1101/gad.332924.119 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Klosinska M, Picard CL & Gehring M Conserved imprinting associated with unique epigenetic signatures in the Arabidopsis genus. Nat Plants 2, 16145, doi: 10.1038/nplants.2016.145 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Moreno-Romero J, Del Toro-De Leon G, Yadav VK, Santos-Gonzalez J & Kohler C Epigenetic signatures associated with imprinted paternally expressed genes in the Arabidopsis endosperm. Genome biology 20, 41, doi: 10.1186/s13059-019-1652-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Schuettengruber B, Bourbon HM, Di Croce L & Cavalli G Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 171, 34–57, doi: 10.1016/j.cell.2017.08.002 (2017). [DOI] [PubMed] [Google Scholar]
- 138.Calabrese JM, Starmer J, Schertzer MD, Yee D & Magnuson T A survey of imprinted gene expression in mouse trophoblast stem cells. G3 (Bethesda) 5, 751–759, doi: 10.1534/g3.114.016238 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Engreitz JM et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452–455, doi: 10.1038/nature20149 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bogutz AB et al. Evolution of imprinting via lineage-specific insertion of retroviral promoters. Nature communications 10, 5674, doi: 10.1038/s41467-019-13662-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Horii T et al. Successful generation of epigenetic disease model mice by targeted demethylation of the epigenome. Genome biology 21, 77, doi: 10.1186/s13059-020-01991-8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wei Y et al. DNA methylation analysis and editing in single mammalian oocytes. Proceedings of the National Academy of Sciences of the United States of America 116, 9883–9892, doi: 10.1073/pnas.1817703116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zenk F et al. Germ line-inherited H3K27me3 restricts enhancer function during maternal-to-zygotic transition. Science 357, 212–216, doi: 10.1126/science.aam5339 (2017). [DOI] [PubMed] [Google Scholar]
- 144.Rainger J et al. Loss of the BMP antagonist, SMOC-1, causes Ophthalmo-acromelic (Waardenburg Anophthalmia) syndrome in humans and mice. PLoS genetics 7, e1002114, doi: 10.1371/journal.pgen.1002114 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.McCarrey JR et al. X-chromosome inactivation during spermatogenesis is regulated by an Xist/Tsix-independent mechanism in the mouse. Genesis 34, 257–266, doi: 10.1002/gene.10163 (2002). [DOI] [PubMed] [Google Scholar]
- 146.Lyon MF Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190, 372–373 (1961). [DOI] [PubMed] [Google Scholar]
- 147.Takagi N & Sasaki M Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature 256, 640–642, doi: 10.1038/256640a0 (1975). [DOI] [PubMed] [Google Scholar]
- 148.Brown CJ et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349, 38–44, doi: 10.1038/349038a0 (1991). [DOI] [PubMed] [Google Scholar]
- 149.Penny GD, Kay GF, Sheardown SA, Rastan S & Brockdorff N Requirement for Xist in X chromosome inactivation. Nature 379, 131–137, doi: 10.1038/379131a0 (1996). [DOI] [PubMed] [Google Scholar]
- 150.Dossin F et al. SPEN integrates transcriptional and epigenetic control of X-inactivation. Nature, doi: 10.1038/s41586-020-1974-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Almeida M et al. PCGF3/5-PRC1 initiates Polycomb recruitment in X chromosome inactivation. Science 356, 1081–1084, doi: 10.1126/science.aal2512 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pintacuda G et al. hnRNPK Recruits PCGF3/5-PRC1 to the Xist RNA B-Repeat to Establish Polycomb-Mediated Chromosomal Silencing. Molecular cell 68, 955–969.e910, doi: 10.1016/j.molcel.2017.11.013 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Colognori D, Sunwoo H, Kriz AJ, Wang CY & Lee JT Xist Deletional Analysis Reveals an Interdependency between Xist RNA and Polycomb Complexes for Spreading along the Inactive X. Molecular cell 74, 101–117 e110, doi: 10.1016/j.molcel.2019.01.015 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zylicz JJ et al. The Implication of Early Chromatin Changes in X Chromosome Inactivation. Cell 176, 182–197 e123, doi: 10.1016/j.cell.2018.11.041 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Galupa R & Heard E X-Chromosome Inactivation: A Crossroads Between Chromosome Architecture and Gene Regulation. Annu Rev Genet 52, 535–566, doi: 10.1146/annurev-genet-120116-024611 (2018). [DOI] [PubMed] [Google Scholar]
- 156.Jegu T, Aeby E & Lee JT The X chromosome in space. Nature reviews. Genetics 18, 377–389, doi: 10.1038/nrg.2017.17 (2017). [DOI] [PubMed] [Google Scholar]
- 157.Gao Z et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Molecular cell 45, 344–356, doi: 10.1016/j.molcel.2012.01.002 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.de Napoles M et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Developmental cell 7, 663–676, doi: 10.1016/j.devcel.2004.10.005 (2004). [DOI] [PubMed] [Google Scholar]
- 159.Wang H et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878, doi: 10.1038/nature02985 (2004). [DOI] [PubMed] [Google Scholar]
- 160.Wang L et al. Hierarchical recruitment of polycomb group silencing complexes. Molecular cell 14, 637–646, doi: 10.1016/j.molcel.2004.05.009 (2004). [DOI] [PubMed] [Google Scholar]
- 161.Cao R et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298, 1039–1043, doi: 10.1126/science.1076997 (2002). [DOI] [PubMed] [Google Scholar]
- 162.Tavares L et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 148, 664–678, doi: 10.1016/j.cell.2011.12.029 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Morey L, Aloia L, Cozzuto L, Benitah SA & Di Croce L RYBP and Cbx7 define specific biological functions of polycomb complexes in mouse embryonic stem cells. Cell reports 3, 60–69, doi: 10.1016/j.celrep.2012.11.026 (2013). [DOI] [PubMed] [Google Scholar]
- 164.Czermin B et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111, 185–196, doi: 10.1016/s0092-8674(02)00975-3 (2002). [DOI] [PubMed] [Google Scholar]
- 165.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P & Reinberg D Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes & development 16, 2893–2905, doi: 10.1101/gad.1035902 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Muller J et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111, 197–208, doi: 10.1016/s0092-8674(02)00976-5 (2002). [DOI] [PubMed] [Google Scholar]
- 167.Li H et al. Polycomb-like proteins link the PRC2 complex to CpG islands. Nature 549, 287–291, doi: 10.1038/nature23881 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Cooper S et al. Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nature communications 7, 13661, doi: 10.1038/ncomms13661 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Fursova NA et al. Synergy between Variant PRC1 Complexes Defines Polycomb-Mediated Gene Repression. Molecular cell 74, 1020–1036 e1028, doi: 10.1016/j.molcel.2019.03.024 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.O’Carroll D et al. The polycomb-group gene Ezh2 is required for early mouse development. Molecular and cellular biology 21, 4330–4336, doi: 10.1128/MCB.21.13.4330-4336.2001 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Akasaka T et al. Mice doubly deficient for the Polycomb Group genes Mel18 and Bmi1 reveal synergy and requirement for maintenance but not initiation of Hox gene expression. Development 128, 1587–1597 (2001). [DOI] [PubMed] [Google Scholar]
- 172.Moussa HF et al. Canonical PRC1 controls sequence-independent propagation of Polycomb-mediated gene silencing. Nature communications 10, 1931, doi: 10.1038/s41467-019-09628-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Blackledge NP, Rose NR & Klose RJ Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nature reviews. Molecular cell biology 16, 643–649, doi: 10.1038/nrm4067 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kuroda MI, Kang H, De S & Kassis JA Dynamic Competition of Polycomb and Trithorax in Transcriptional Programming. Annual review of biochemistry, doi: 10.1146/annurev-biochem-120219-103641 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]