SUMMARY
Multiple factors influence translation termination efficiency, including nonsense codon identity and immediate context. To determine whether the relative position of a nonsense codon within an open reading frame (ORF) influences termination efficiency, we quantitate the production of prematurely terminated and/or readthrough polypeptides from 26 nonsense alleles of 3 genes expressed in yeast. The accumulation of premature termination products and the extent of readthrough for the respective premature termination codons (PTCs) manifest a marked dependence on PTC proximity to the mRNA 3′ end. Premature termination products increase in relative abundance, whereas readthrough efficiencies decrease progressively across different ORFs, and readthrough efficiencies for a PTC increase in response to 3′ UTR lengthening. These effects are eliminated and overall translation termination efficiency decreases considerably in cells harboring pab1 mutations. Our results support a critical role for poly(A)-binding protein in the regulation of translation termination and also suggest that inefficient termination is a trigger for nonsense-mediated mRNA decay (NMD).
Graphical Abstract
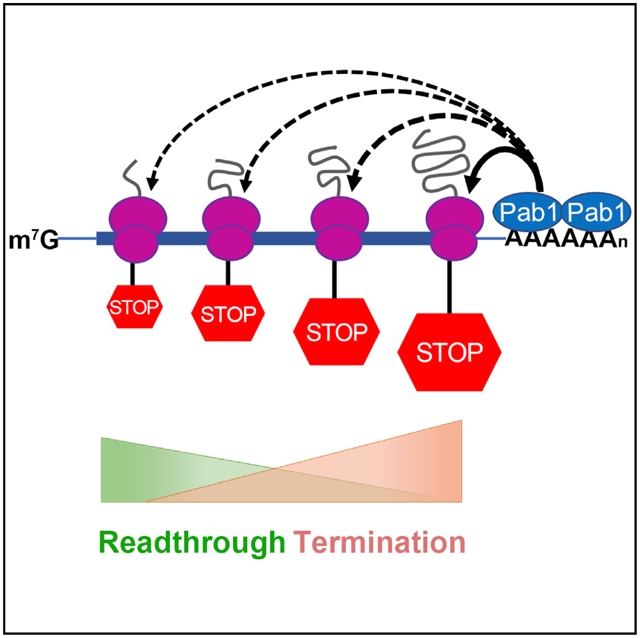
In Brief
Premature termination codons (PTCs) trigger translational termination and can promote mRNA decay. Wu et al. insert PTCs at multiple locations in yeast genes and find that their termination efficiencies increase with proximity to ORF 3′ ends and that this positional effect on termination is modulated through mRNA-associated poly(A)-binding protein.
INTRODUCTION
Translation termination in eukaryotes is orchestrated by the release factors eRF1 and eRF3 when any of the three stop codons (UAA, UAG, or UGA) in an mRNA occupies the A site of a ribosome (Alkalaeva et al., 2006; Stansfield et al., 1995; Zhouravleva et al., 1995). eRF1 recognizes A-site-localized stop codons and hydrolyzes peptidyl-tRNA, whereas eRF3 interacts with eRF1 and stimulates termination by its GTPase activity (Alkalaeva et al., 2006; Salas-Marco and Bedwell, 2004). eRF1 and near-cognate tRNAs (nc-tRNAs), i.e., tRNAs capable of base pairing with stop codons at two of the three standard codon positions, compete for binding to the ribosomal A site (Brown et al., 2015). Although this competition is inefficient for nc-tRNA, when successful, it leads to the insertion of an amino acid and continuation of translational elongation until the next in-frame stop codon is encountered (Roy et al., 2015, 2016). Such a bypass of translation termination is designated nonsense suppression or readthrough (Brenner et al., 1965; Keeling et al., 2014; Peltz et al., 2013).
Multiple factors appear to influence the competition between nc-tRNAs and eRF1 and, hence, the extent of nonsense suppression, including (1) the identity of the stop codon, with UGA promoting higher levels of readthrough than UAG or UAA (Howard et al., 2000; Loughran et al., 2014; Manuvakhova et al., 2000); (2) the immediate context of a stop codon, with the highest read-through levels typically occurring when adenine precedes and cytosine follows the stop codon (Bonetti et al., 1995; McCaughan et al., 1995; Mottagui-Tabar et al., 1998; Tork et al., 2004); (3) specific sequences or structures 3′ to the stop codon (Anzalone et al., 2019; Cridge et al., 2018; Harrell et al., 2002; Namy et al., 2001; Skuzeski et al., 1991); and (4) the integrity of the release factors and the ribosome (Carnes et al., 2003; Chernoff et al., 1994; Liu and Liebman, 1996; Loenarz et al., 2014; Serio and Lindquist, 1999; Singleton et al., 2014; Velichutina et al., 2000). Another important determinant of readthrough efficiency may be the relative position of the stop codon within the open reading frame (ORF). Experiments in yeast suggested that termination at premature termination codons (PTCs) is slower than that at normal termination codons (NTCs) (Amrani et al., 2004). This apparent difference predicts that readthrough should occur more readily at PTCs than at NTCs, an expectation borne out by results obtained when patients, animals, or cultured cells are treated with the read-through-promoting drug ataluren (Hirawat et al., 2007; Welch et al., 2007), as well as by recent studies of non-canonical genetic codes in ciliates (Heaphy et al., 2016; Swart et al., 2016; Záhonová et al., 2016).
These functional differences between PTCs and NTCs are consistent with the notion that the position of a PTC within an ORF may influence its termination efficiency and suggest that PTCs closest to the 3′ end of an ORF, i.e., those most likely to have NTC-like contexts, may be the most likely to have high efficiencies of termination, whereas those more 5′ proximal are likely to have reduced termination efficiencies and higher extents of readthrough. Here, we have investigated the role of PTC position within an ORF in the regulation of termination and read-through in yeast. We constructed PTCs at multiple positions of the TPI1, LUC, and PGK1 ORFs; expressed these alleles in yeast cells inactive for the nonsense-mediated mRNA decay (NMD) pathway; and assessed termination efficiencies and/or read-through at each ORF position by quantitating the premature termination products and/or full-length protein expressed from each PTC allele. Our results revealed a position-dependent effect for termination and readthrough efficiencies and suggested that nonsense codon proximity to the mRNA 3′ end was an important determinant of termination efficiency. Consistent with results of earlier studies implicating a role for poly(A)-binding protein in the regulation of translation termination (Ivanov et al., 2016), we find that deletion of the yeast PAB1 gene markedly reduces premature termination efficiency, thus yielding substantial enhancement of PTC readthrough.
RESULTS
Efficiency of Premature Termination Increases across the TPI1 ORF
We constructed a set of reporters in which PTCs were positioned from codons 9 to 233 of the TPI1 ORF. Each allele has a stop codon at its designated position, a 3×-hemagglutinin (HA) tag at the ORF N terminus, and a StrepII-FLAG (SF) tag at the ORF C terminus. All TPI1 cassettes were cloned into the pRS315 yeast centromere vector, flanked by the promoter, 5′ UTR, and 3′ UTR of the TPI1 gene. We used a weak terminator and weak context (UGA CAA) (Bonetti et al., 1995) for each PTC reporter while including a strong terminator (UAA) at the NTC.
Upf1 is a key regulator of NMD, and deletion of its gene in yeast not only stabilizes nonsense-containing mRNAs but also enhances nonsense codon readthrough and inhibits the degradation of prematurely terminated polypeptides (He and Jacobson, 2015; Johansson and Jacobson, 2010; Kuroha et al., 2009; Leeds et al., 1991). Hence, to stabilize premature termination products, augment detection of readthrough, and minimize variability in mRNA levels, we performed all experiments in upf1Δ cells. Each TPI1-PTC reporter and an empty vector was transformed into upf1Δ cells, and the resulting strains were grown, harvested, and analyzed in parallel. Premature termination products, identified and quantitated by western blotting (WB) with anti-HA antibodies, could not be detected when a PTC was proximal to the TPI1 ORF N terminus (i.e., PTC9 to PTC72), but their levels increased progressively as PTC positions approached the C terminus of the TPI1 ORF (see PTC90 to PTC233, anti-HA, Figure 1B; Table S3, sheet 1).
Figure 1. PTC Termination Efficiency Increases across the TPI1 ORF.
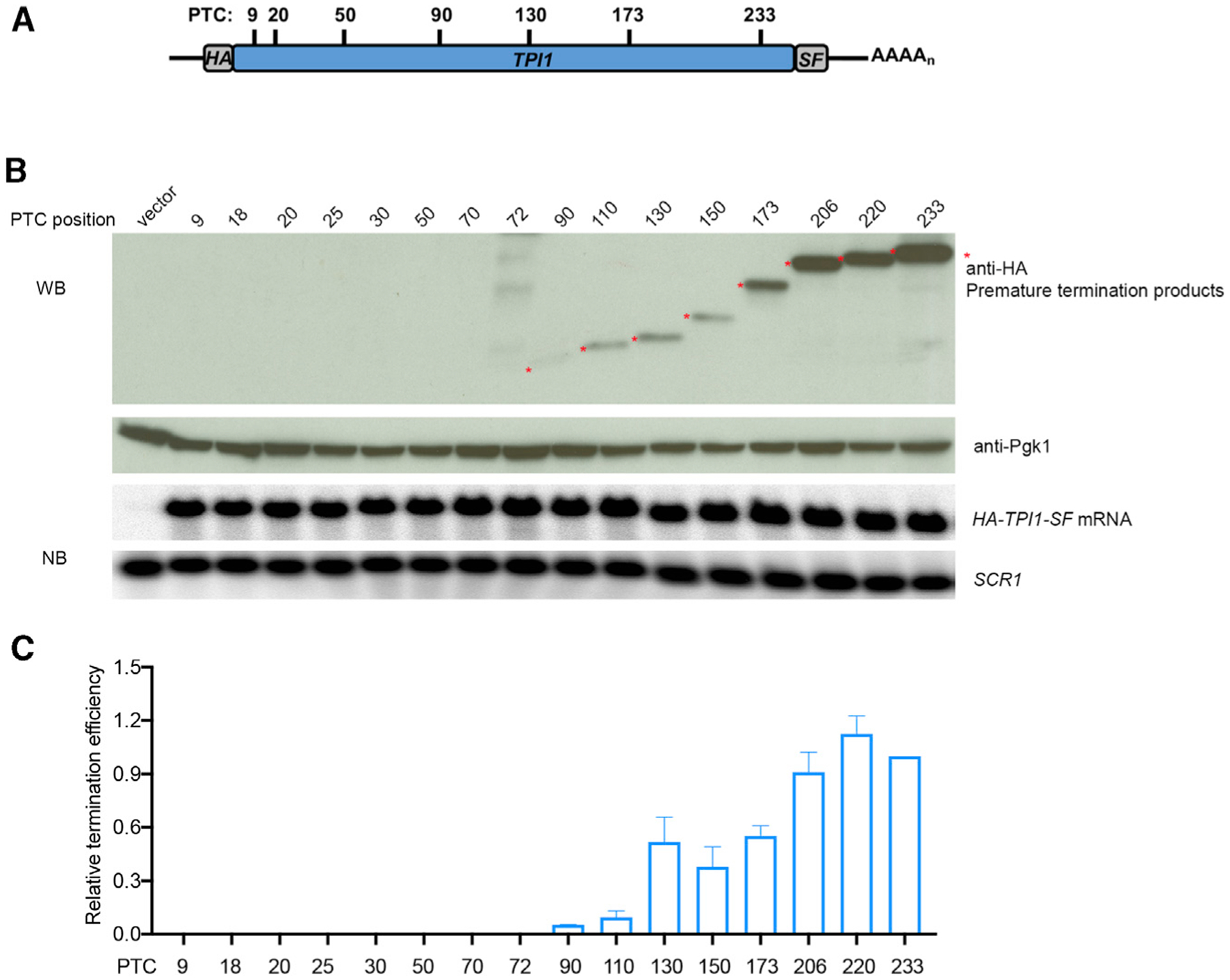
(A) TPI1-PTC readthrough reporters. Schematic of a subset of the HA-TPI1-SF PTC constructs used to analyze translation termination efficiency. HA, S, and F represent HA, StrepII, and FLAG epitope tags, respectively.
(B) Western and northern analyses of prematurely terminated polypeptides and mRNAs expressed from TPI1-PTC alleles. Yeast upf1Δ cells expressing each of the TPI1-PTC alleles were harvested, lysed, and analyzed by western blotting (WB). Anti-HA antibodies detected premature termination products (asterisks), and anti-Pgk1 antibodies detected the Pgk1 control protein. Northern blotting (NB) and phosphorimaging were used to measure the levels of TPI1-PTC mRNA and SCR1 RNA in each sample.
(C) Relative termination efficiency at each TPI1 PTC. Densitometry was used to quantitate the western blots and the northern blots in (B). Relative termination efficiency was calculated as the relative truncated Tpi1 protein level (anti-HA divided by Pgk1 control) normalized to the relative TPI1-PTC mRNA level (mRNA divided by SCR1 RNA). The efficiency of TPI1-PTC233 was set as 1. The results shown are the average of three independent experiments ± SEM.
Relative termination efficiencies at TPI1 PTCs (Figure 1C) were determined by normalizing anti-HA WB results to the level of an internal control protein (Pgk1) in the same samples (Figure 1B, bottom WB panel) and to the level of the corresponding TPI1-PTC mRNA in each sample (determined by northern blotting [NB] and normalized to a control RNA, SCR1; Figure 1B, NB panels; Table S3, sheet 1). The relative termination efficiencies of TPI1 PTCs showed a position effect in which termination efficiency was too low to measure within early parts of the TPI1 ORF but increased progressively from PTC90 to the end of the TPI1 ORF (Figure 1C).
PTC Readthrough Efficiency Decreases across the LUC ORF
Our observation that the efficiency of premature termination appears to improve at least 15-fold as the position of a PTC approaches the 3′ end of the TPI1 ORF (Figure 1) suggested that the readthrough efficiency of PTCs should manifest the inverse, i.e., a systematic decrease across an ORF. Accordingly, we constructed a set of reporters with PTCs positioned at six locations within the firefly luciferase (LUC) ORF (Figure 2A). These LUC-PTC reporters have the same regulatory elements, epitope tags, and termination codon usage as the TPI1-PTC reporters. LUC-PTC reporters, an NTC reporter (lacking any PTC in the coding region), and a vector control were transformed into upf1Δ cells; and the resulting strains were analyzed as in Figure 1. Readthrough efficacy was assessed by WB with anti-FLAG antibodies to monitor the accumulation of full-length luciferase protein (Figure 2B, top panel). Relative readthrough efficiencies (Figure 2D; Table S3, sheet 2) were determined by normalizing the anti-FLAG WB results to the level of a control protein (Pgk1) (Figure 2B, bottom panel) and to the level of LUC mRNA in each sample (determined by NB and also normalized to a control RNA, SCR1; Figure 2C). This experiment showed that the extent of readthrough was highest when a PTC was proximal to the N terminus of the LUC ORF (PTC20 and PTC41 showed ~2% of Luc-wt levels) and diminished progressively as the PTC positions approached the C terminus of the ORF (Figure 2B, top panel). After quantitation, the relative readthrough efficiencies of PTCs showed a clear position effect in which readthrough efficiency decreased across the LUC ORF (Figure 2D), i.e., readthrough efficiency at each PTC was inversely correlated with its distance to the ORF 3′ end. These variations in readthrough efficiency were unlikely to be due to differences in the stability of the respective full-length readthrough proteins (Kuroha et al., 2009) because treating cells with the proteasome inhibitor MG132 did not alter the relative recovery of readthrough proteins (Figures S2A and S2B).
Figure 2. PTC Readthrough Efficiency Decreases across the LUC ORF.
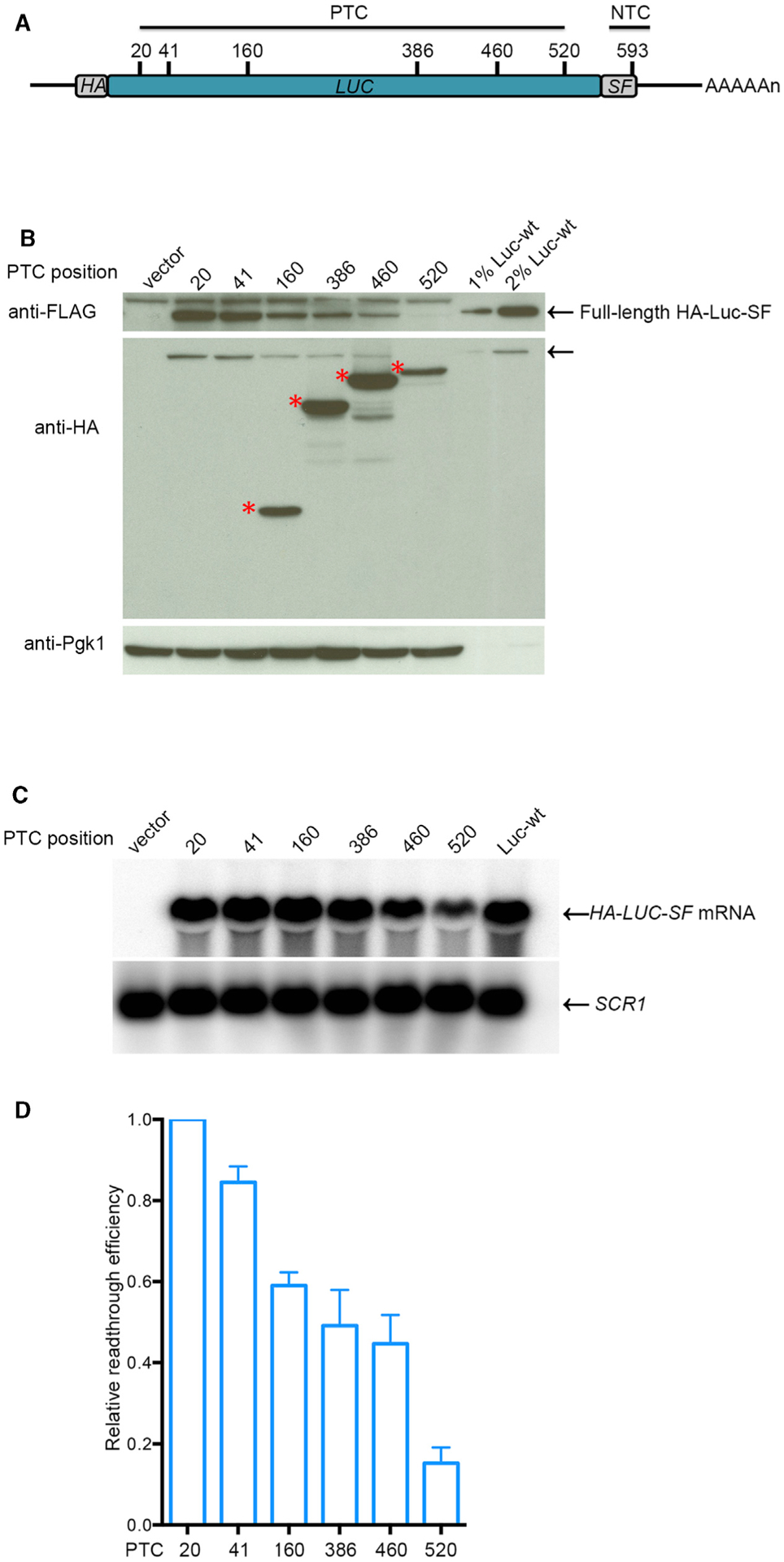
(A) LUC-PTC readthrough reporters. Schematic of the positions of PTCs and the NTC in HA-LUC-SF constructs. HA, S, and F represent HA, StrepII, and FLAG epitope tags, respectively.
(B) Western analysis of full-length readthrough proteins and prematurely terminated polypeptides expressed from LUC-PTC alleles. Yeast upf1Δ cells expressing a LUC-PTC allele, a vector control, or a WT LUC allele were harvested, lysed, and analyzed by WB by using anti-FLAG antibodies to detect full-length readthrough products, anti-HA antibodies to detect premature termination products, and anti-Pgk1 antibodies to detect the Pgk1 control protein. The arrows (top and middle panels) indicate full-length readthrough protein, and the asterisks (middle panel) indicate premature termination products. Figure S1A shows the complete blot used for the top panel.
(C) Northern analyses of LUC mRNA levels in cells expressing PTC alleles. NB and phosphorimaging were used to measure the level of LUC mRNA and SCR1 RNA in each sample.
(D) Relative readthrough efficiency at each of the six LUC PTCs. Densitometry was used to quantitate the blots of (B) and (C). Relative read-through efficiency of each LUC-PTC allele was calculated as the ratio of full-length Luc protein level (anti-FLAG) normalized to Pgk1 level divided by LUC mRNA level normalized to the level of SCR1 RNA. The efficiency of LUC-PTC20 was set as 1. The results shown are the average of three independent experiments ± SEM. 1% and 2% Luc-wt are aliquots of extracts from cells expressing a wild-type LUC allele that were 1/100th and 1/50th the volume of samples from cells expressing PTC alleles.
In parallel with our assays for full-length readthrough proteins, we also used WB with anti-HA antibodies to detect prematurely terminated LUC polypeptides. As was seen for early TPI1 PTC alleles (Figure 1), premature termination products could not be detected from LUC PTC20 and PTC41 but were readily detected from PTC160, PTC386, PTC460, and PTC520 (Figure 2B, middle panel, and Figure S2A, middle panel; see red asterisks). Corrected for the Pgk1 control protein and LUC mRNA levels, the recovery of prematurely terminated polypeptides showed a general upward trend from PTC160 to PTC460, a result supported by our observation of decreased readthrough across the same region of the LUC ORF (Figures 2, S2A, and S2B) and with similar measurements of premature termination efficiencies in TPI1-PTC reporters (Figure 1).
Our inability to detect any premature termination products from early PTCs in the TPI1 and LUC mRNAs was not likely to be due to their susceptibility to proteasomal degradation (Figures S2A and S2B), their failure to be recovered by WB, or to some previously unknown mechanism that degrades short nascent polypeptides while they are still associated with tRNA. The latter two conclusions follow from the WB detection of (1) a synthetic polypeptide containing the 3×-HA epitope and the first 19 amino acids of luciferase (i.e., the polypeptide that would be generated by premature termination at PTC20) (Figure S2C); and (2) peptidyl-tRNAs from LUC-PTC20 and LUC-PTC41 (Figures S2D and S2E).
The Ratios of Full-Length Readthrough Product to the Corresponding Prematurely Terminated Product at Each PTC Vary as a Function of ORF Position
As an independent approach for assessing changes in read-through efficiency across an ORF, we measured changes in the ratio of prematurely terminated polypeptide relative to the accumulation of full-length readthrough protein, and both were simultaneously detected with the same antibody. The anti-HA western blot of Figure 2B used to identify premature termination products also detected full-length Luc protein derived from PTC readthrough (middle panel, top band, see arrow). As shown in Figure S1B, the ratio of recoverable premature termination products to the full-length readthrough protein increased substantially from PTC160 to PTC520, a result consistent with a progressive improvement in termination efficiency as the ribosome approached the NTC. Variable recoveries of the full-length readthrough product precluded a similar analysis for TPI1, but we were able to determine the ratio of prematurely terminated products to full-length read-through protein with the PGK1 gene. We constructed a set of PGK1 reporters flanked by the promoter, 5′ UTR, and 3′ UTR of the PGK1 gene and with PTCs positioned at codons 37, 84, 219, and 309 (Figure 3A). Each PGK1-PTC reporter had the same weak terminator, epitope tags, and plasmid backbone as the TPI1-PTC reporters. PGK1-PTC reporters and the control vector were transformed into upf1Δ cells that were analyzed by the same procedures used above for the TPI1-PTC and LUC-PTC reporters. This experiment showed that the extent of readthrough was highest when a PTC was proximal to the N terminus of the PGK1 ORF and diminished progressively as the PTC positions approached the C terminus of the ORF (Figure 3B, top bands in anti-HA blot), whereas the extent of termination showed the opposite trend (Figure 3B, bottom bands in anti-HA blot). After being normalized to PGK1-PTC mRNA levels (Figure 3C), the relative readthrough and termination efficiencies of PTCs showed clear position effects in which readthrough efficiency decreased and termination efficiency increased across the PGK1 ORF (Figures 3D and 3E), consistent with the LUC-PTC and TPI1-PTC results. Importantly, the ratio of premature termination product to readthrough product of each allele also showed an upward trend across the PGK1 ORF, indicating a progressive improvement of termination efficiency as PTCs approached the ORF C terminus (Figure 3F).
Figure 3. PTC Position Effect on Translation Termination and Readthrough Efficiencies across the PGK1 ORF.
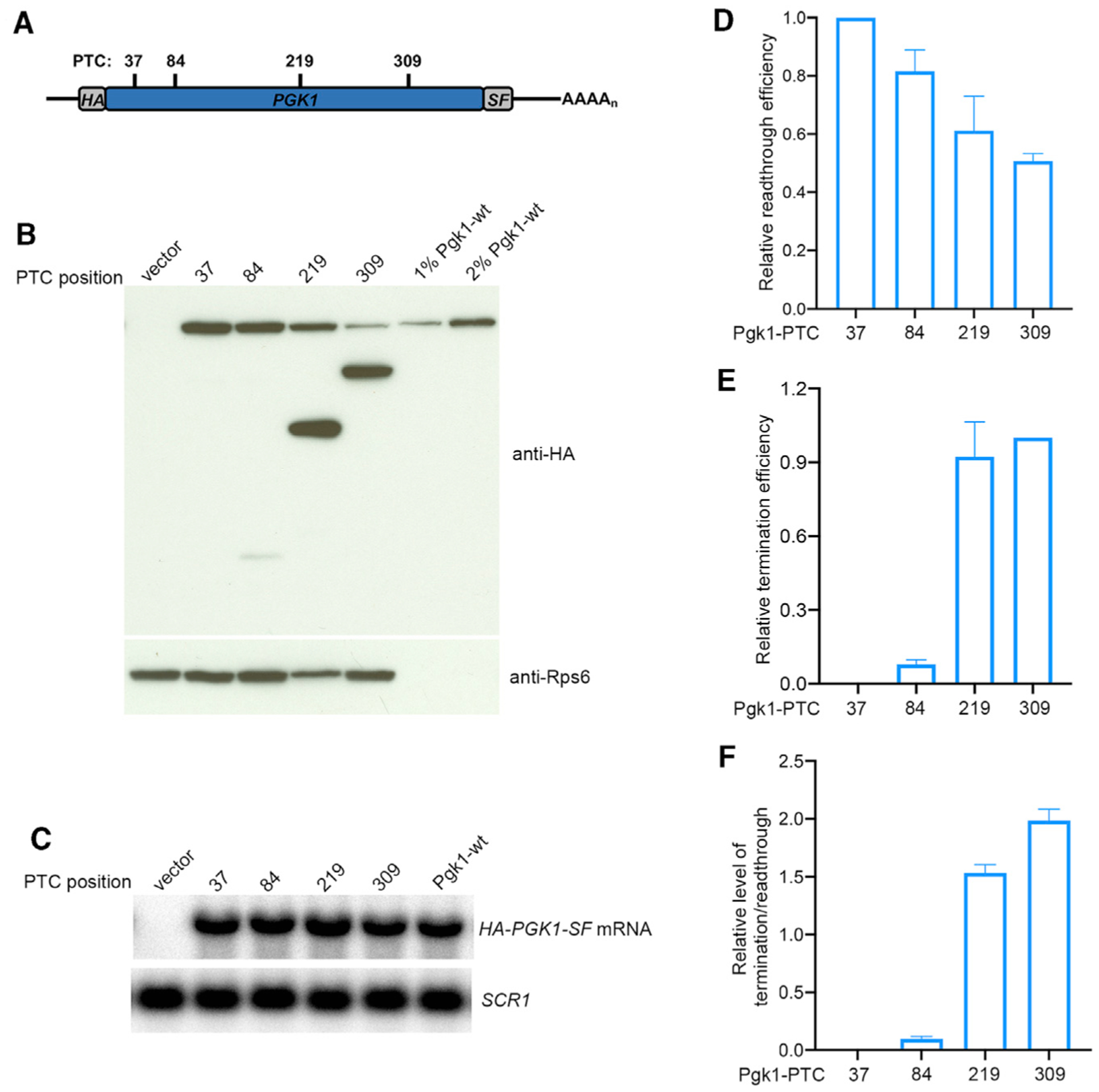
(A) PGK1-PTC readthrough reporters. Schematic of PTCs within individual HA-PGK1-SF constructs. HA, S, and F represent HA, StrepII, and FLAG epitope tags, respectively.
(B) Western analysis of full-length readthrough proteins and prematurely terminated polypeptides expressed from PGK1-PTC alleles. Yeast upf1Δ cells expressing a PGK1-PTC allele, a vector control, or a wild-type PGK1 allele were harvested, lysed, and analyzed by WB by using anti-HA antibodies to detect both readthrough and premature termination products and by using anti-Rps6 antibodies to detect the Rps6 control protein.
(C) NB analyses of PGK1-PTC mRNA levels in cells expressing different PTC alleles. NB and phosphorimaging were used to measure the level of PGK1-PTC mRNA and SCR1 RNA in each sample.
(D) Relative readthrough efficiency at each of the four PGK1 PTCs. Densitometry was used to quantitate the blots of (B) and (C). Relative readthrough efficiency of each PGK1-PTC allele was calculated as the ratio of full-length Pgk1 protein level (anti-HA, top band) normalized to Rps6 level divided by PGK1-PTC mRNA level normalized to the level of SCR1 RNA. The efficiency of PGK1-PTC37 was set as 1. The results shown are the average of three independent experiments ± SEM. 1% and 2% Pgk1-wt are aliquots of extracts from cells expressing a wild-type PGK1 allele that were 1/100th and 1/50th the volume of samples from cells expressing PTC alleles.
(E) Relative termination efficiency at each of the four PGK1 PTCs. Densitometry was used to quantitate the blots of (B) and (C). Relative termination efficiency of each PGK1-PTC allele was calculated as the ratio of truncated Pgk1 protein level (anti-HA, bottom band) normalized to Rps6 level divided by PGK1-PTC mRNA level normalized to the level of SCR1 RNA. The efficiency of PGK1-PTC309 was set as 1. The results shown are the average of three independent determinations ± SEM.
(F) Ratio of termination to readthrough efficiency at each of the PGK1 PTCs. Relative termination efficiency at each PTC was calculated as the ratio of termination efficiency in (E) divided by readthrough efficiency in (D).
3′ UTR Length Regulates the Efficiency of PTC Readthrough
Earlier studies suggested that interactions between mRNA-associated poly(A)-binding protein (Pab1 in yeast) and eRF3 enhance translation termination efficiency (Amrani et al., 2004; Heaphy et al., 2016; Ivanov et al., 2016; Roque et al., 2015; Swart et al., 2016; Záhonová et al., 2016). To test whether the progressive changes in termination and readthrough efficiencies we observed with TPI1, LUC, and PGK1 PTC mutations reflected changes in PTC proximity to Pab1 (or any other 3′ UTR-localized regulatory factor), we began with the LUC reporter manifesting the least efficient readthrough (LUC-PTC520) and constructed a series of LUC-PTC520 alleles having 3′ UTR lengths ranging from 60 to 600 nucleotides (nt) (Figure 4A). Quantitation of the expression of these alleles in upf1Δ cells showed that the relative readthrough efficiencies increased progressively with 3′ UTR length (Figures 4B and 4C), i.e., the results indicate that the length of the 3′ UTR associated with the LUC mRNA plays an important cis-acting role in regulating translational readthrough of PTCs in yeast.
Figure 4. The Efficiency of Readthrough of LUC-PTC520 mRNA Varies Directly with the Length of Its Associated 3′ UTR.
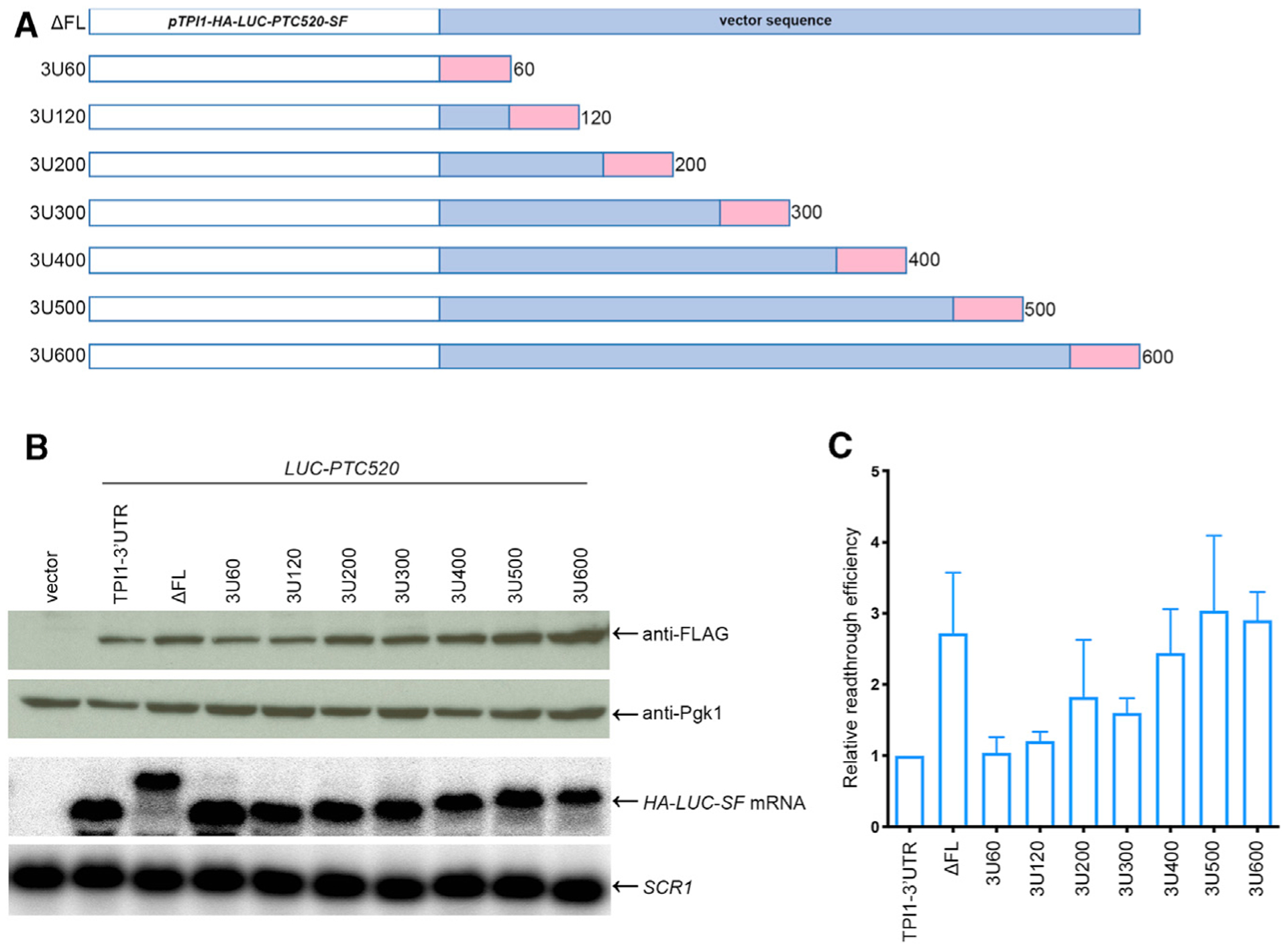
(A) Schematic representation of LUC-PTC520 alleles with defined 3′ UTR lengths. The GAL7 cleavage/polyadenylation signal (pink rectangles) was inserted at different locations downstream of the LUC-PTC520 ORF (DFL allele, full-length TPI1 3′ UTR was deleted) to generate a set of LUC-PTC 520 alleles that were transformed into upf1Δ cells for an assessment of LUC mRNA and protein expression. mRNAs from this set of alleles have 3′ UTR lengths ranging from 60 to 600 nt. pTPI1, TPI1 promoter; SF, StrepII-FLAG tag; FL, full-length TPI1 3′ region.
(B) Analyses of the mRNAs and readthrough products generated by LUC-PTC520 alleles with defined 3′ UTR lengths. LUC-PTC520 alleles harboring 3′ UTRs of 60–600 bp were expressed in upf1Δ cells and lysates of the respective cells were analyzed by WB and NB as in Figure 2.
(C) Relative readthrough efficiencies of LUC-PTC520 alleles with 3′ UTRs of defined lengths. Relative readthrough efficiencies were calculated as in Figure 2D. The results shown are the average of four independent experiments ± SEM. The efficiency of TPI1–3′ UTR was set as 1.
Poly(A)-Binding Protein Restricts PTC Readthrough and Controls the PTC Position Effect
In light of the correlation between 3′ UTR length and PTC read-through seen in Figure 4 and earlier studies addressing Pab1:eRF3 interactions (Roque et al., 2015), we sought to directly test whether the observed readthrough phenotypes reflected PTC proximity to the mRNA-associated Pab1 protein. PAB1 is an essential gene in yeast (Sachs et al., 1987), but its absence can be studied in several suppressor mutants. Pbp1 was identified as a Pab1-interacting protein, and pbp1Δ cells suppress a pab1Δ allele without having significant effects on mRNA translation or decay (Kato et al., 2019; Mangus et al., 1998; Yang et al., 2019). Accordingly, we generated pab1Δ pbp1Δ upf1Δ yeast cells and transformed them with the pRS316-LUC-PTC reporters and control plasmids equivalent to those analyzed in the experiments of Figure 2. As additional controls, each reporter was also transformed into pbp1Δ upf1Δ cells. Readthrough efficiencies were determined in upf1Δ, pbp1Δ upf1Δ, and pab1Δ pbp1Δ upf1Δ cells; and the results of these analyses are shown in Figure 5. Consistent with previous conclusions about the modest impact of the pbp1Δ allele on mRNA translation and decay, we found the relative readthrough efficiencies of LUC-PTC reporters in pbp1Δ upf1Δ cells to be quite similar to those seen in upf1Δ cells (Figure 5C). However, in pab1Δ pbp1Δ upf1Δ cells, the relative readthrough efficiencies of all LUC-PTC reporters were much higher than those in upf1Δ or pbp1Δ upf1Δ cells (Figure 5C), with readthrough efficiencies of LUC-PTCs in pab1Δ pbp1Δ upf1Δ cells approximately 2- to 8-fold higher than those observed in the two other strains. Readthrough efficiencies across the LUC ORF in upf1Δ cells continuously decreased as the ribosome progressed from 5′ to 3′, whereas in pab1Δ pbp1Δ upf1Δ cells, this gradient was absent and was replaced by a “bell-shaped” distribution of readthrough activity across the ORF. Furthermore, in contrast to the increased levels of LUC readthrough proteins (Figure 5A, top panel), the premature termination products were much lower in pab1Δ pbp1Δ upf1Δ cells than in the other two strains (Figure 5A, middle panel), with several premature termination products beyond detection when western blots were exposed for similar times. The presence of prematurely terminated products in pab1Δ pbp1Δ upf1Δ cells could be detected if blots were exposed for longer times (Figure S3). Although the termination efficiencies of LUC-PTCs in pab1Δ pbp1Δ upf1Δ cells could not be compared properly in parallel with those in upf1Δ or pbp1Δ upf1Δ cells, termination efficiencies in pbp1Δ upf1Δ cells could be compared to those in upf1Δ cells, and they were found to be 1.5- to 2-fold lower (Figure S4A).
Figure 5. Deletion of PAB1 Enhances Translational Readthrough and Disrupts PTC Position Effects.
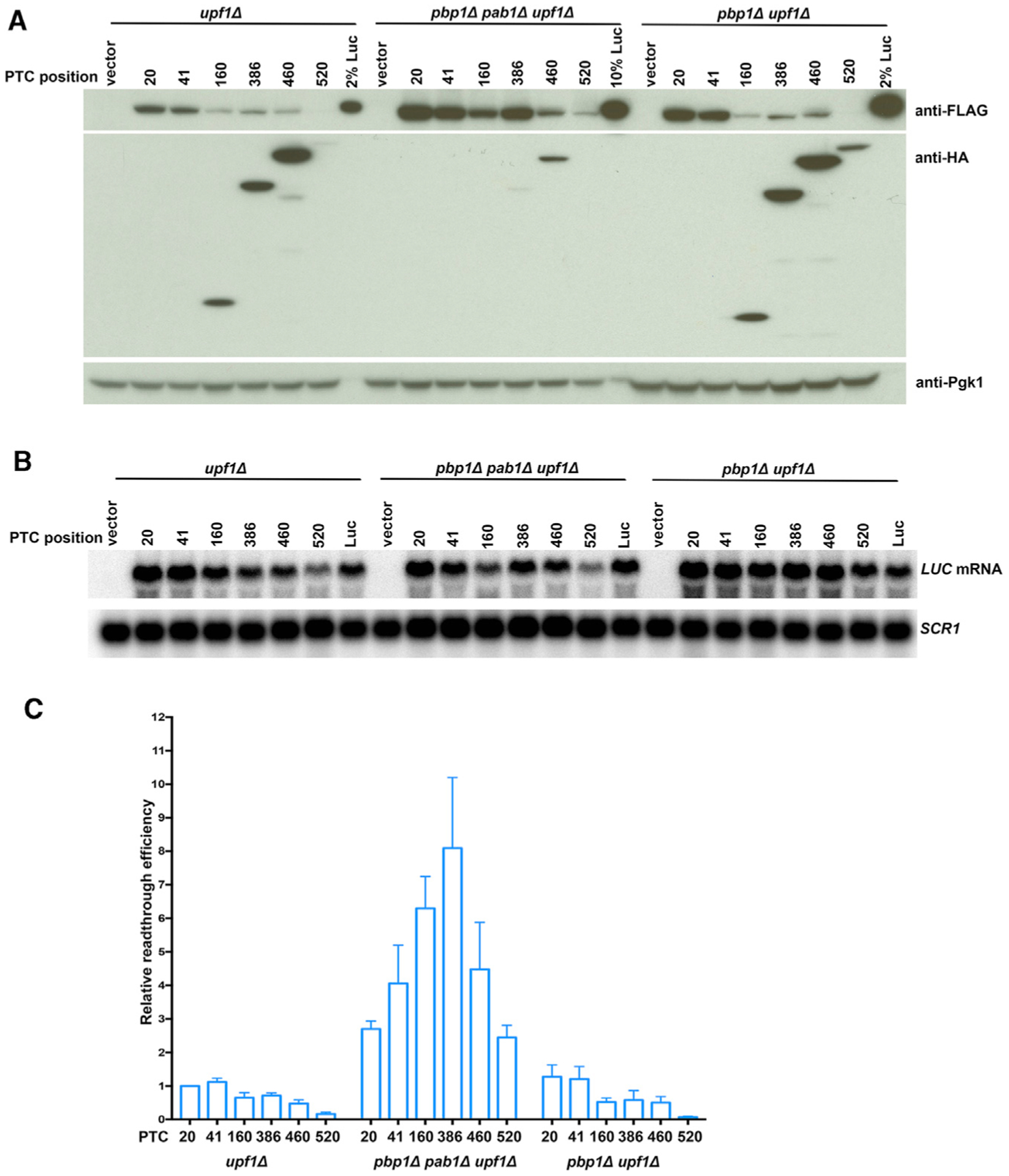
(A) Western analyses of LUC-PTC reporters expressed in upf1Δ, pab1Δ pbp1Δ upf1Δ, and pbp1Δ upf1Δ cells. Yeast upf1Δ, pab1Δ pbp1Δ upf1Δ, or pbp1Δ upf1Δ cells expressing a LUC PTC allele, a vector control, or wild-type LUC were harvested, lysed, and analyzed by WB as in Figure 2. 10% Luc is an aliquot of extract from cells expressing a wild-type LUC allele that was 1/10th the total protein of samples from cells expressing PTC alleles.
(B) Northern analyses of LUC-PTC reporter mRNA expression in upf1Δ, pab1Δ pbp1Δ upf1Δ, and pbp1Δ upf1Δ cells. Yeast cells shown in (A) were analyzed for LUC mRNA levels as in Figure 2.
(C) Relative readthrough efficiencies of different LUC-PTC reporters in cells with and without Pab1. Relative readthrough efficiencies of the LUC-PTC alleles in upf1Δ, pab1Δ pbp1Δ upf1Δ, and pbp1Δ upf1Δ cells were calculated as in Figure 2D. The results shown are the average of three independent experiments ± SEM.
The results of Figure 5 demonstrated that Pab1 enhanced termination and restricted PTC readthrough in vivo. In order to assign that function to specific Pab1 domains, we examined the consequences of deleting the C-terminal domain of the protein, the region thought to interact with eRF3 (Roque et al., 2015). This deletion does not impair yeast cell viability (Sachs et al., 1987). We constructed pab1ΔC upf1Δ cells, transformed them with LUC-PTC and control reporters, and evaluated PTC read-through efficiency in these cells. Figure S5A (top panel) shows that the yields of full-length proteins from all six LUC-PTCs in pab1ΔC upf1Δ cells were higher than in upf1Δ cells. After correction for protein and mRNA controls (Figures S5A, bottom panel, and S5B), the relative readthrough efficiencies of PTCs in pab1ΔC upf1Δ cells were 2- to 4-fold higher than those of the same alleles in upf1Δ cells (Figure S5C). Readthrough efficiencies in pab1ΔC upf1Δ cells were flat across the first two-thirds of the LUC ORF but decreased for the last two alleles. Although premature termination products were detectable in pab1ΔC upf1Δ cells, their termination efficiencies were lower than those in upf1Δ cells (Figure S4B).
DISCUSSION
PTC Position Effects in the TPI1, LUC, and PGK1 ORFs Are Associated with Variations in the Efficiency of Translation Termination
The mechanisms regulating termination efficiency and nonsense codon readthrough remain poorly understood. Here, by using three independent reporter assays as criteria for the extent of termination at a given PTC, we demonstrate that the position of a PTC within an ORF dictates its termination and/or read-through efficiency. In support of this conclusion, we find that (1) PTCs proximal to the 5′ end of an ORF have higher read-through efficiencies than PTCs proximal to the 3′ end of the same ORF (Figures 2B, 2D, 3B, 3D, 5A, 5C, S1A, S2A, S2B, S5A, and S5C); (2) premature termination efficiencies largely appear to increase across the same regions (Figures 1B, 1C, 2B, 3B, 3E, 5A, S2A, S3, S4A, S4B, and S5A); and (3) consistent with the respective descending and ascending efficiencies of readthrough and termination efficiency across an ORF, the ratio of termination products to readthrough products increases progressively toward the mRNA NTC (Figures S1B and 3F). Note that values for all the readthrough and termination efficiencies are relative, not absolute percentages.
Premature termination products could not be detected from TPI1, LUC, and PGK1 mRNAs with “early” PTCs, a shortcoming that we could not account for by deficiencies in protein stability, gel transfer, or accumulation of the respective peptidyl-tRNAs (Figure S2). The consistency of the phenomenon with three different sets of premature termination products suggests that the detection of short prematurely terminated polypeptides may be limited by the overall expression levels of the respective genes or that they are targeted for degradation by a specific proteolytic pathway that does not use the proteasome. In light of the complementary and confirmatory results we obtained from read-through assays and from the ratio of premature termination products to readthrough products, we do not consider the selective loss of a subset of premature termination products to have weakened the argument for an ORF-wide gradient of termination efficiency.
The position dependence of nonsense codon function observed in our experiments is reminiscent of the non-canonical genetic code use in some species of protozoa, in which all three nonsense codons function as sense codons unless they are located near ORF 3′ ends, where they still serve their usual termination function (Heaphy et al., 2016; Swart et al., 2016; Záhonová et al., 2016). The decoding of UAA, UAG, and UGA in these atypical genetic codes appears to be mediated in part by mutations that lessen the specificity of eRF1 and make it an ineffective competitor with tRNAs capable of nonsense codon recognition (Swart et al., 2016). The enhanced specificity of the same eRF1 molecules when ribosomes are translating near the end of ORFs is postulated to arise from proximity to 3′-end-associated poly(A)-binding protein and its stimulatory effects on eRF3, which is the release factor that normally augments eRF1 function (Swart et al., 2016). As discussed below, the poly(A)-binding protein also appears to be a critical regulator of the PTC position effects in yeast.
Yeast Pab1 Is a Key Determinant of the PTC Position Effect
The multiple experimental approaches described here implicate a role for Pab1 in the regulation of termination efficiency and PTC readthrough. First, we observed a trend toward higher efficiencies of premature translation termination as PTCs approached the 3′ ends of three different ORFs (Figures 1B, 1C, 3B, 3E, 3F, 5A, S1B, S2A, S3, S4A, S4B, and S5A). Second, we observed an inverse correlation between the extent of read-through at a given PTC and the distance of that PTC from the respective mRNA 3′ end, i.e., the position of the poly(A) tail (Figures 2B, 2D, 3B, 3D, 4B, 4C, 5A, 5C, S1A, S2A, S2B, S5A, and S5C). This was true for PTCs at different ORF positions (e.g., Figures 2 and 3), as well as for LUC PTC520 when it was associated with 3′ UTRs of different lengths (Figure 4). Although these results provide only indirect evidence for a Pab1 role in termination regulation, an additional set of experiments provided direct evidence for that role. Figure 5 shows that deletion of the PAB1 gene leads to large increases in readthrough efficiency at all PTC positions, comparably large reductions in termination efficiency, and loss of the progressive PTC position effects across the LUC ORF. Furthermore, the experiments of Figure 5 demonstrate that the pbp1Δ mutation required for the maintenance of viability in pab1Δ cells does not contribute to these effects.
A role for Pab1 in restricting readthrough is consistent with results of several earlier studies in multiple systems, including those reporting (1) position dependence of the non-canonical genetic codes in ciliates (see above) (Heaphy et al., 2016; Swart et al., 2016; Záhonová et al., 2016); (2) interactions between eRF3 and PABP from mammalian, Xenopus, and yeast cells, and identification of specific interacting domains in the respective proteins (Cosson et al., 2002a, 2002b; Hoshino et al., 1999; Hosoda et al., 2003; Jerbi et al., 2016; Kononenko et al., 2010; Roque et al., 2015; Uchida et al., 2002); and (3) direct stimulation of peptidyl-tRNA hydrolysis dependent on a eRF3:PABP interaction in a reconstituted cell-free system (Ivanov et al., 2016). However, our results differ from those of Roque et al. (2015) who found that readthrough efficiency in a dual-luciferase assay in yeast decreased when the Pab1:eRF3 interaction was interrupted. A significant difference between our study and that of Roque et al. (2015) that may account for this difference was their inclusion of suppressor tRNA in the strains being tested, i.e., their assays focused on the strong but artificial readthrough signals generated by a cognate suppressor tRNA, whereas ours addressed the competition between eRF1 and nc-tRNAs that is typical of conventional PTC readthrough (Roy et al., 2015, 2016).
The Inefficiency of Premature Termination May Define Susceptibility to NMD
Our results have important implications for NMD. In addition to providing in vivo substantiation of our earlier in vitro studies that indicated fundamental mechanistic differences between normal and premature translation termination (Amrani et al., 2004), our results suggest further insights. Earlier work on NMD in yeast showed that PTCs within the first half to two-thirds of an ORF triggered NMD, whereas those in the latter part of an ORF had little to no mRNA destabilizing effects (Hagan et al., 1995; Hennigan and Jacobson, 1996, 1997; Peltz et al., 1993, 1994; Peltz and Jacobson, 1993). We observed a similar phenomenon when the relative abundance of LUC mRNAs harboring the six PTCs studied here was analyzed in wild-type (WT) cells (Figure S1C). It has been shown that such PTC position effects on NMD could be mimicked and manipulated by shortening or lengthening mRNA 3′ UTRs (Amrani et al., 2004; Kebaara and Atkin, 2009; Muhlrad and Parker, 1999) and that NMD can be antagonized by tethering Pab1 or eRF3 proximal to a PTC (Amrani et al., 2004). These results, and toeprinting experiments indicating that termination at PTCs had a much longer dwell time than that at NTCs (Amrani et al., 2004), led to the formulation of the faux-UTR model for NMD that postulated that the inefficient termination occurring at PTCs distant from 3′-end-localized Pab1 allowed for mRNA binding of the Upf factors that trigger NMD (Amrani et al., 2004, 2006a, 2006b; Amrani and Jacobson, 2006). Although the experiments of this study were carried out in NMD-deficient cells, the results reported here and the earlier data supporting the faux-UTR model strongly suggest that a threshold level of termination inefficiency is a trigger for NMD, possibly because the ribosome is transiently in a state that allows association of the Upf factors (He and Jacobson, 2015).
STAR⋆METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Allan Jacobson (allan.jacobson@umassmed.edu).
Materials Availability
All plasmids and yeast strains generated in this study are available upon request to the lead contact.
Data and Code Availability
This study did not generate any datasets or code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Yeast strains used in this study have the W303 background and are listed in the Key Resources Table. Cells were grown in synthetic complete medium lacking the amino acids required to maintain specific plasmids at 30°C.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-FLAG | Sigma-Aldrich | Cat#F7425; RRID:AB_439687 |
| Mouse monoclonal anti-OctA-Probe (F-tag-01) | Santa Cruz | Cat#sc-51590; RRID:AB_677316 |
| Mouse monoclonal anti-Pgk1 (22C5D8) | Thermo Fisher | Cat#459250; RRID:AB_2532235 |
| Donkey polyclonal anti-Mouse Secondary | Thermo Fisher | Cat#A24506; RRID:AB_2535975 |
| Donkey anti-Rabbit Secondary | Sigma-Aldrich | Cat#GENA934; RRID:AB_2722659 |
| Mouse monoclonal anti-HA | Sigma-Aldrich | Cat#H3663; RRID:AB_262051 |
| Phospho-S6 Ribosomal Protein (Ser235/236) | Cell Signaling | Cat#4858; RRID:AB_916156 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Protease inhibitor tablets, EDTA-free | Thermo Fisher | Cat#A32965 |
| dCTP, [α−32P]-6000Ci/mmol | Perkin Elmer | Cat#BLU513Z |
| MG-132 | Sigma-Aldrich | Cat#474790 |
| RNasecure | Thermo Fisher | Cat#7006 |
| Synthetic HA-Luc19: MGYPYDVPDYAGYPYDVPDYAGSYPYDVPDYAMEDAKNIKKGPAPFYPLED | GenScript | This paper |
| Critical Commercial Assays | ||
| Random Primed DNA Labeling Kit | Sigma-Aldrich | Cat#11004760001 |
| Experimental Models: Organisms/Strains | ||
| S. cerevisiae: upf1Δ (HFY871): MATa ade2–1 his3–11,15 leu2–3,112 trp1–1 ura3–1 can1–100 upf1::HIS3 | Jacobson lab | He et al., 1997 |
| S. cerevisiae: pab1Δ pbp1Δ upf1Δ: MATa ade2–1 his3–11,15 leu2–3,112 trp1–1 ura3–1 can1–100 pab1::HIS3 pbp1::LEU2 upf1::KanMX6 | Jacobson lab | This paper |
| S. cerevisiae: pab1ΔC upf1Δ: MATa ade2–1 his3–11,15 Ieu2–3,112 trp1–1 ura3–1 can1–100 pab1ΔAC::HIS3 upf1::KanMX6 | Jacobson lab | This paper |
| S. cerevisiae: pbp1Δ upf1Δ: MATa ade2–1 his3–11,15 leu2–3,112trp1–1 ura3–1 can1–100 pbp1::LEU2 upf1::KanMX6 | Jacobson lab | This paper |
| Software and Algorithms | ||
| MultiGauge software | Fujifilm | Science lab 2005 |
METHOD DETAILS
Yeast strains and plasmid constructs
The upf1Δ strain was described previously (He et al., 1997). Strains combining additional deletions with upf1Δ, i.e., pbp1Δ upf1Δ, pab1Δ pbp1Δ upf1Δ, and pab1ΔC upf1Δ were upf1::KanMX6 derivatives of yDM146, yDM206, and YAS2239 (Kessler and Sachs, 1998; Mangus et al., 1998) and were constructed by PCR-mediated strategies with pFA6a-KanMX6 as the template (Longtine et al., 1998). Fragments amplified by the primer pair upf1-KanMX6-F and upf1-KanMX6-R were transformed into the respective yeast strains by the high-efficiency method (Schiestl and Gietz, 1989). Each genomic DNA deletion was confirmed by PCR analysis with the oligonucleotide pair upf1-SF and upf1-SR.
All of the plasmids and oligonucleotides used in this study are listed in Tables S1 and S2. The pRS315-TPI1 plasmid was generated by direct PCR cloning of the TPI1 gene amplified by primer pair TPI1-F(NcoI)/TPI1-R(XhoI). The PCR fragments were digested by NcoI/XhoI and cloned into pRS315-LUC digested by NcoI/XhoI. pRS315-TPI1-PTC plasmids were generated by overlap PCRs using a common outside primer pair, TPI1-promoter-F (PstI)/TPI13U-R (NotI), and an internal primer pair that introduced PTCs at specific codons: codon 9 (TPI1-PTC9-F/R), codon 18 (TPI1-PTC18-F/R), codon 20 (TPI1-PTC20-F/R), codon 25 (TPI1-PTC25-F/R), codon 30 (TPI1-PTC30-F/R), codon 50 (TPI1-PTC50-F/R), codon 70 (TPI1-PTC70-F/R), codon 72 (TPI1-PTC72-F/R), codon 90 (TPI1-PTC90-F/R), codon 110 (TPI1-PTC110-F/R), codon 130 (TPI1-PTC130-F/R), codon 150 (TPI1-PTC150-F/R), codon 173 (TPI1-PTC173-F/R), codon 206 (TPI1-PTC206-F/R), codon 220 (TPI1-PTC220-F/R), and codon 233 (TPI1-PTC233-F/R). In each case, the final PCR products were digested by PstI/NotI and cloned into pRS315 digested by PstI/NotI.
The plasmid pRS315-LUC was generated through subcloning from plasmid YEplac181-HA-LUC-SF (Roy et al., 2015) as follows: First, a PstI-XbaI fragment was isolated from YEplac181-HA-LUC-SF and cloned into the pRS315 vector digested by PstI/XbaI. Then, a TPI1 3′-UTR fragment was amplified from YEplac181-HA-LUC-SF by the primer pair TPI13U-F(XbaI)/TPI13U-R(NotI), digested by XbaI/NotI, and ligated into pRS315-TPI1 promoter-3X-HA-LUC-StrepII-FLAG digested by XbaI/NotI.
The pRS315-LUC-PTC plasmids were generated by overlap PCRs using a common outside primer pair, TPI1-promoter-F (PstI)/TPI13U-R (NotI), and an internal primer pair that introduced PTCs at specific codons: codon 20 (LUC-PTC20-F/R), codon 41 (LUC-PTC41-F/R), codon 160 (LUC-PTC160-F/R), codon 386 (LUC-PTC386-F/R), codon 460 (LUC-PTC460-F/R), and codon 520 (LUC-PTC520-F/R). In each case, the final PCR products were digested by PstI/NotI and cloned into pRS315 digested by PstI/NotI.
The pRS315-PGK1 plasmid was constructed in four steps. First, a pRIP1 vector containing the PGK1 promoter and 3′-UTR region (pRIP1-PGK1 promoter-PGK1 3′-UTR) was generated from pRIP1PGK1 (Peltz et al., 1993) by overlap PCRs using a common outside primer pair, M13-F/R, and a specific internal primer pair, PGK1–3U-F(BclI-NotI)/PGK1-pro-R(NotI-BclI). The final PCR products were digested by BamH1/HindIII and cloned into pRIP1PGK1 digested by BamH1/HindIII. Second, the PGK1 coding region was amplified from pRIP1PGK1 by the primer pair PGK1-F-PciI/PGK1-R-XhoI, digested by PciI/XhoI, and ligated into pRS315-LUC previously digested by NcoI/XhoI, resulting in pRS315-TPI1 promoter-3X-HA-PGK1-StrepII-FLAG-TPI1 3′-UTR. Third, we generated pRIP1-PGK1 promoter-HA-PGK1-SF-PGK1 3′UTR by direct PCR. The PCR fragment was amplified from pRS315-TPI1 promoter-3X-HA-PGK1-StrepII-FLAG-TPI1 3′-UTR by the primer pair HA-F-BclI/SF-R-NotI, digested by BclI/NotI, ligated into pRIP1-PGK1 promoter-PGK1 3′-UTR digested by BclI/NotI. Fourth, a SmaI-HindIII fragment was isolated from pRIP1-PGK1 promoter-HA-PGK1-SF-PGK1 3′UTR and cloned into pRS315 digested by SmaI/HindIII.
The pRS315-PGK1-PTCs were generated by overlap PCR using a common outside primer pair, M13-F/M13-R, and an internal primer pair that introduced PTCs at specific codons: codon 37 (PGK1-PTC37-F/R), codon 84 (PGK1-PTC84-F/R), codon 219 (PGK1-PTC219-F/R), codon 309 (PGK1-PTC309-F/R). In each case, the final PCR products were digested by SmaI/HindIII and cloned into pRS315 digested by SmaI/HindIII.
pRS315 plasmids containing LUC-PTC520–3U120, −3U200, −3U300, −3U400, −3U500, and −3U600 were all constructed by overlap PCRs in two steps. First, using a common outside primer pair (RS315-F-SalI/RS315-R-KasI) and a specific internal primer pair (3U120-F/3U120-R, 3U200-F/3U200-R, 3U300-F/3U300-R, 3U400-F/3U400-R, 3U500-F/3U500-R, and 3U600-F/3U600-R), we generated a set of pRS315 vectors with a GAL7 polyadenylation signal (Bucheli et al., 2007) inserted at different locations of the 1 kb SalI-KasI fragment. Second, a PstI-XbaI fragment was isolated from pRS315-LUC-PTC520 and ligated into this set of modified pRS315 vectors digested by PstI/XbaI. This set of LUC-PTC520-xxUTR alleles from pRS315 plasmids generates transcripts with 3′-UTR lengths ranging from 120 to 600 nt.
pRS316 plasmids containing LUC or LUC-PTC alleles were constructed in two steps. First, a HindIII-XbaI DNA fragment was isolated from the respective pRS315-LUC or LUC-PTC plasmids and cloned into pRS316. Then a XbaI-NotI fragment containing the TPI1 3′ region was cloned downstream of the LUC or LUC-PTC ORF.
pRS315 derived plasmids were transformed into upf1Δ cells, and pRS316 derived plasmids were transformed into pab1Δ pbp1Δ upf1Δ or pbp1Δ upf1Δ cells, both by the high-efficiency method (Schiestl and Gietz, 1989).
Cell growth and western analysis
Cells were grown at 30°C in 25 mL of synthetic complete media lacking leucine or uracil to an optical density at 600nm (OD600) of 0.7. For each culture expressing a specific TPI1-PTC, LUC-PTC, PGK1-PTC, or control allele, 10 OD600 units were harvested for western analysis and 4 OD600 units from the same culture were collected for RNA extraction. Cell pellets for western analyses were resuspended in 110 μl RIPA buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% NP-40, 1 mM EDTA, 5% glycerol) with 1mM PMSF and 1X protease inhibitor mixture (Thermo Scientific, #A32965). The cell suspensions were lysed by vortexing for 30 s with 0.1g pre-washed glass beads (Sigma-Aldrich, #8772), followed by 30 s on ice, for 8 cycles. The lysates were centrifuged at 12,000rpm for 15 min in a Sorvall Legend X1R centrifuge, at 4°C. Aliquots (90 μl) of the supernatants were mixed with Laemmli’s SDS-sample buffer (Boston BioProducts, #BP-110R) followed by boiling for 5 min. The samples were then resolved by 10% SDS-PAGE, transferred to Immobilon-P membrane (Millipore, #IPVH0010), and incubated with anti-HA antibodies (Sigma-Aldrich, #H3663, 1:2,000), anti-FLAG antibodies (Sigma-Aldrich, #F7425, 1:1,000 or SANTA CRUZ, #sc-51590, 1:100), anti-Rps6 (Cell Signaling, #4858, 1:2,000), or anti-Pgk1 antibodies (Thermo Fisher, #459250, 1:4,000) overnight at 4°C. The membrane was washed with PBST for 7 min, 3 times, then incubated with anti-Mouse secondary antibodies (Thermo Fisher, #A24506, 1:5,000–1:10,000) or anti-Rabbit secondary antibodies (Sigma-Aldrich, #GENA934, 1:5,000) for 45 min at RT. Then the membrane was washed with PBST for 7 min, 3 times. Proteins were detected using ECL reagents and Hyperfilm ECL (GE, #28906835). The signal was analyzed with MultiGauge software.
Detection of peptidyl-tRNA
Peptidyl-tRNA was detected with LUC-PTC20 and LUC-PTC41 whole cell lysates. 35 OD600 units were harvested and resuspended in 250 μl RIPA buffer pretreated with RNasecure (Thermo Fisher, #AM7006) and supplemented with 1mM PMSF and 1X protease inhibitor. Cell lysates were then prepared as described above. To prepare cell lysates treated with cycloheximide, cells were incubated in the presence of cycloheximide (0.1mg/ml) for 5 min before harvest, and lysed in buffer supplemented with cycloheximide. To preserve peptidyl-tRNA ester linkage, cell lysates were denatured with Laemmli’s sample buffer which was pretreated with RNasecure. To release the peptides, cell lysates were treated with 0.1 M NaOH for 5 min at 65°C, then neutralized with equimolar HCl. Samples were run without prior heat treatment on NuPAGE Bis-Tris gels with MES-SDS running buffer. To detect 35S-fmet-tRNAfmet, 35S-fmet-tRNAfmet molecules (a gift from Dr. A. Korostelev) were mixed with RNasecure treated Laemmli’s sample buffer, run on NuPAGE Bis-Tris gels with MES-SDS running buffer, and detected by phosphorimaging using a Fujifilm bio-imaging analyzer (BAS-2500).
RNA preparation and northern analysis
Total RNA from cell pellets was isolated as described previously (Herrick et al., 1990). Briefly, pellets were resuspended in 500 μl buffer A (50 mM NaOAc pH5.2, 10 mM EDTA, 1% SDS, 1% DEPC), extracted with 500 μl RNA-phenol (phenol saturated with 50 mM NaOAc pH5.2, 10 mM EDTA) by vortexing for 10 s, followed by 50 s incubation in a 65°C water bath, for 6 cycles. Tubes were centrifuged at 12,000rpm in an Eppendorf 5417C centrifuge for 10 min at RT. The aqueous layers were recovered and followed by another phenol extraction. Phenol/chloroform (500 μl) was added to the recovered aqueous layer, and the mixture was vortexed for 2 min at RT followed by 10 min centrifugation at 12,000rpm. The aqueous layers were recovered and subjected to another phenol/chloroform extraction. The RNAs were precipitated by adding 40 μl NaOAc (3 M, pH 5.2) and 1 mL ethanol and incubated for 2 hours at −80°C. Tubes were centrifuged at 12,000rpm for 15 min at 4°C. Pellets were washed with 70% ethanol and re-dissolved in 50 μl RNase free distilled water. Aliquots (15 μg) of each RNA sample were loaded onto a 1% agarose/formaldehyde/MOPS gel and electrophoresed, blotted, and hybridized as described previously (He and Jacobson, 1995). Random-primed DNA probes made from NcoI-XbaI LUC fragments were used to detect HA-LUC-SF mRNAs, probes made from StrepII-FLAG fragments were used to detect HA-TPI1-PTC-SF and HA-PGK1-PTC-SF mRNAs, and full-length SCR1 probes were used to detect the SCR1 RNA (loading control). [α−32P]-dCTP (Perkin Elmer, Blu513Z) and a random primed DNA labeling kit (Roche, # 11-004-760-001) were used to generate probes according to the manufacturer’s protocol. Signals from Northern blots were detected and analyzed by phosphorimaging using a Fujifilm bio-imaging analyzer (BAS-2500) and MultiGauge software.
QUANTIFICATION AND STATISTICAL ANALYSIS
The following formulas were used to determine the relative efficiencies of PTC readthrough and termination:
In these formulas FLAG, Pgk1, Rps6, and HA respectively represent the amounts of full-length FLAG-tagged protein, Pgk1 protein, Rps6, and prematurely terminated HA-tagged protein determined by western blotting, and mRNA and SCR1 RNA respectively designate the levels of these two transcripts determined by Northern blotting. The results shown in the figures are the average of three independent experiments ± SEM unless otherwise indicated.
Supplementary Material
Highlights.
Premature translational termination manifests position effects in yeast mRNAs
Termination increases and readthrough decreases as stop codons approach the ORF 3′ end
Stop codon proximity to 3′ poly(A)-binding protein regulates termination efficiency
Positional variation and termination dependence on Pab1 support an NMD faux-UTR model
ACKNOWLEDGMENTS
This work was supported by grants to A.J. (5R01 GM27757-37 and 1R35GM122468-04) from the U.S. National Institutes of Health. We thank Robin Ganesan, Kotchaphorn Mangkalaphiban, Andrei Korostelev, Christine Carbone, and Denis Susorov for helpful discussions.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108399.
DECLARATION OF INTERESTS
A.J. is co-founder, director, and SAB chair of PTC Therapeutics Inc. B.R. is an employee of New England Biolabs.
REFERENCES
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, and Pestova TV (2006). In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell 125, 1125–1136. [DOI] [PubMed] [Google Scholar]
- Amrani N, and Jacobson A (2006). All termination events are not equal: premature termination is aberrant and triggers NMD In Nonsense-mediated mRNA decay, Maquat LE, ed. (Landes Bioscience; ), pp. 15–25. [Google Scholar]
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, and Jacobson A (2004). A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432, 112–118. [DOI] [PubMed] [Google Scholar]
- Amrani N, Dong S, He F, Ganesan R, Ghosh S, Kervestin S, Li C, Mangus DA, Spatrick P, and Jacobson A (2006a). Aberrant termination triggers nonsense-mediated mRNA decay. Biochem. Soc. Trans 34, 39–42. [DOI] [PubMed] [Google Scholar]
- Amrani N, Sachs MS, and Jacobson A (2006b). Early nonsense: mRNA decay solves a translational problem. Nat. Rev. Mol. Cell Biol 7, 415–425. [DOI] [PubMed] [Google Scholar]
- Anzalone AV, Zairis S, Lin AJ, Rabadan R, and Cornish VW (2019). Interrogation of Eukaryotic Stop Codon Readthrough Signals by in Vitro RNA Selection. Biochemistry 58, 1167–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetti B, Fu L, Moon J, and Bedwell DM (1995). The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J. Mol. Biol 251, 334–345. [DOI] [PubMed] [Google Scholar]
- Brenner S, Stretton AO, and Kaplan S (1965). Genetic code: the ‘nonsense’ triplets for chain termination and their suppression. Nature 206, 994–998. [DOI] [PubMed] [Google Scholar]
- Brown A, Shao S, Murray J, Hegde RS, and Ramakrishnan V (2015). Structural basis for stop codon recognition in eukaryotes. Nature 524, 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucheli ME, He X, Kaplan CD, Moore CL, and Buratowski S (2007). Polyadenylation site choice in yeast is affected by competition between Npl3 and polyadenylation factor CFI. RNA 13, 1756–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes J, Jacobson M, Leinwand L, and Yarus M (2003). Stop codon suppression via inhibition of eRF1 expression. RNA 9, 648–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Vincent A, and Liebman SW (1994). Mutations in eukaryotic 18S ribosomal RNA affect translational fidelity and resistance to aminoglycoside antibiotics. EMBO J 13, 906–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson B, Berkova N, Couturier A, Chabelskaya S, Philippe M, and Zhouravleva G (2002a). Poly(A)-binding protein and eRF3 are associated in vivo in human and Xenopus cells. Biol. Cell 94, 205–216. [DOI] [PubMed] [Google Scholar]
- Cosson B, Couturier A, Chabelskaya S, Kiktev D, Inge-Vechtomov S, Philippe M, and Zhouravleva G (2002b). Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI(+)] propagation. Mol. Cell. Biol 22, 3301–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cridge AG, Crowe-McAuliffe C, Mathew SF, and Tate WP (2018). Eukaryotic translational termination efficiency is influenced by the 3′ nucleotides within the ribosomal mRNA channel. Nucleic Acids Res 46, 1927–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan KW, Ruiz-Echevarria MJ, Quan Y, and Peltz SW (1995). Characterization of cis-acting sequences and decay intermediates involved in nonsense-mediated mRNA turnover. Mol. Cell. Biol 15, 809–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrell L, Melcher U, and Atkins JF (2002). Predominance of six different hexanucleotide recoding signals 3′ of read-through stop codons. Nucleic Acids Res 30, 2011–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, and Jacobson A (1995). Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev 9, 437–454. [DOI] [PubMed] [Google Scholar]
- He F, and Jacobson A (2015). Nonsense-mediated mRNA decay: Degradation of defective transcripts is only part of the story. Annu. Rev. Genet 49, 339–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Brown AH, and Jacobson A (1997). Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol. Cell. Biol 17, 1580–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaphy SM, Mariotti M, Gladyshev VN, Atkins JF, and Baranov PV (2016). Novel Ciliate Genetic Code Variants Including the Reassignment of All Three Stop Codons to Sense Codons in Condylostoma magnum. Mol. Biol. Evol 33, 2885–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennigan AN, and Jacobson A (1996). Functional mapping of the translation-dependent instability element of yeast MATalpha1 mRNA. Mol. Cell. Biol 16, 3833–3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennigan AN, and Jacobson A (1997). A genetic approach to mapping coding region determinants of mRNA stability in yeast In mRNA Formation and Function, Richter JD, ed. (Academic Press; ), pp. 149–161. [Google Scholar]
- Herrick D, Parker R, and Jacobson A (1990). Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol 10, 2269–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirawat S, Welch EM, Elfring GL, Northcutt VJ, Paushkin S, Hwang S, Leonard EM, Almstead NG, Ju W, Peltz SW, and Miller LL (2007). Safety, tolerability, and pharmacokinetics of PTC124, a nonaminoglycoside nonsense mutation suppressor, following single- and multiple-dose administration to healthy male and female adult volunteers. J. Clin. Pharmacol 47, 430–444. [DOI] [PubMed] [Google Scholar]
- Hoshino S, Hosoda N, Araki Y, Kobayashi T, Uchida N, Funakoshi Y, and Katada T (1999). Novel function of the eukaryotic polypeptide-chain releasing factor 3 (eRF3/GSPT) in the mRNA degradation pathway. Biochemistry (Mosc.) 64, 1367–1372. [PubMed] [Google Scholar]
- Hosoda N, Kobayashi T, Uchida N, Funakoshi Y, Kikuchi Y, Hoshino S, and Katada T (2003). Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J. Biol. Chem 278, 38287–38291. [DOI] [PubMed] [Google Scholar]
- Howard MT, Shirts BH, Petros LM, Flanigan KM, Gesteland RF, and Atkins JF (2000). Sequence specificity of aminoglycoside-induced stop condon readthrough: potential implications for treatment of Duchenne muscular dystrophy. Ann. Neurol 48, 164–169. [PubMed] [Google Scholar]
- Ivanov A, Mikhailova T, Eliseev B, Yeramala L, Sokolova E, Susorov D, Shuvalov A, Schaffitzel C, and Alkalaeva E (2016). PABP enhances release factor recruitment and stop codon recognition during translation termination. Nucleic Acids Res 44, 7766–7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerbi S, Jolles B, Bouceba T, and Jean-Jean O (2016). Studies on human eRF3-PABP interaction reveal the influence of eRF3a N-terminal glycin repeat on eRF3-PABP binding affinity and the lower affinity of eRF3a 12-GGC allele involved in cancer susceptibility. RNA Biol 13, 306–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MJ, and Jacobson A (2010). Nonsense-mediated mRNA decay maintains translational fidelity by limiting magnesium uptake. Genes Dev 24, 1491–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Yang YS, Sutter BM, Wang Y, McKnight SL, and Tu BP (2019). Redox State Controls Phase Separation of the Yeast Ataxin-2 Protein via Reversible Oxidation of Its Methionine-Rich Low-Complexity Domain. Cell 177, 711–721.e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebaara BW, and Atkin AL (2009). Long 3′-UTRs target wild-type mRNAs for nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Nucleic Acids Res 37, 2771–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling KM, Xue X, Gunn G, and Bedwell DM (2014). Therapeutics Based on Stop Codon Readthrough. Annu. Rev. Genomics Hum. Genet 15, 371–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler SH, and Sachs AB (1998). RNA recognition motif 2 of yeast Pab1p is required for its functional interaction with eukaryotic translation initiation factor 4G. Mol. Cell. Biol 18, 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kononenko AV, Mitkevich VA, Atkinson GC, Tenson T, Dubovaya VI, Frolova LY, Makarov AA, and Hauryliuk V (2010). GTP-dependent structural rearrangement of the eRF1:eRF3 complex and eRF3 sequence motifs essential for PABP binding. Nucleic Acids Res 38, 548–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroha K, Tatematsu T, and Inada T (2009). Upf1 stimulates degradation of the product derived from aberrant messenger RNA containing a specific nonsense mutation by the proteasome. EMBO Rep 10, 1265–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds P, Peltz SW, Jacobson A, and Culbertson MR (1991). The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev 5, 2303–2314. [DOI] [PubMed] [Google Scholar]
- Liu R, and Liebman SW (1996). A translational fidelity mutation in the universally conserved sarcin/ricin domain of 25S yeast ribosomal RNA. RNA 2, 254–263. [PMC free article] [PubMed] [Google Scholar]
- Loenarz C, Sekirnik R, Thalhammer A, Ge W, Spivakovsky E, Mackeen MM, McDonough MA, Cockman ME, Kessler BM, Ratcliffe PJ, et al. (2014). Hydroxylation of the eukaryotic ribosomal decoding center affects translational accuracy. Proc. Natl. Acad. Sci. USA 111, 4019–4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, and Pringle JR (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Loughran G, Chou MY, Ivanov IP, Jungreis I, Kellis M, Kiran AM, Baranov PV, and Atkins JF (2014). Evidence of efficient stop codon read-through in four mammalian genes. Nucleic Acids Res 42, 8928–8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus DA, Amrani N, and Jacobson A (1998). Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol. Cell. Biol 18, 7383–7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuvakhova M, Keeling K, and Bedwell DM (2000). Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. RNA 6, 1044–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaughan KK, Brown CM, Dalphin ME, Berry MJ, and Tate WP (1995). Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc. Natl. Acad. Sci. USA 92, 5431–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottagui-Tabar S, Tuite MF, and Isaksson LA (1998). The influence of 5′ codon context on translation termination in Saccharomyces cerevisiae. Eur. J. Biochem 257, 249–254. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, and Parker R (1999). Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA 5, 1299–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O, Hatin I, and Rousset J-P (2001). Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep 2, 787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltz SW, and Jacobson A (1993). mRNA Turnover in Saccharomyces cerevisiae (Academic Press; ). [Google Scholar]
- Peltz SW, Brown AH, and Jacobson A (1993). mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev 7, 1737–1754. [DOI] [PubMed] [Google Scholar]
- Peltz SW, He F, Welch E, and Jacobson A (1994). Nonsense-mediated mRNA decay in yeast. Prog. Nucleic Acid Res. Mol. Biol 47, 271–298. [DOI] [PubMed] [Google Scholar]
- Peltz SW, Morsy M, Welch EM, and Jacobson A (2013). Ataluren as an agent for therapeutic nonsense suppression. Annu. Rev. Med 64, 407–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roque S, Cerciat M, Gaugué I, Mora L, Floch AG, de Zamaroczy M, Heurgué-Hamard V, and Kervestin S (2015). Interaction between the poly(A)-binding protein Pab1 and the eukaryotic release factor eRF3 regulates translation termination but not mRNA decay in Saccharomyces cerevisiae. RNA 21, 124–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, Leszyk JD, Mangus DA, and Jacobson A (2015). Nonsense suppression by near-cognate tRNAs employs alternative base pairing at codon positions 1 and 3. Proc. Natl. Acad. Sci. USA 112, 3038–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, Friesen WJ, Tomizawa Y, Leszyk JD, Zhuo J, Johnson B, Dakka J, Trotta CR, Xue X, Mutyam V, et al. (2016). Ataluren stimulates ribosomal selection of near-cognate tRNAs to promote nonsense suppression. Proc. Natl. Acad. Sci. USA 113, 12508–12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs AB, Davis RW, and Kornberg RD (1987). A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol. Cell. Biol 7, 3268–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas-Marco J, and Bedwell DM (2004). GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol. Cell. Biol 24, 7769–7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl RH, and Gietz RD (1989). High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet 16, 339–346. [DOI] [PubMed] [Google Scholar]
- Serio TR, and Lindquist SL (1999). [PSI+]: an epigenetic modulator of translation termination efficiency. Annu. Rev. Cell Dev. Biol 15, 661–703. [DOI] [PubMed] [Google Scholar]
- Singleton R, Liu-Yi P, Formenti F, Ge W, Sekirnik R, Fischer R, Adam J, Pollard P, Wolf A, Thalhammer A, et al. (2014). OGFOD1 catalyzes prolyl hydroxylation of RPS23 and is involved in translation control and stress granule formation. Proc. Natl. Acad. Sci. USA 111, 4031–4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuzeski JM, Nichols LM, Gesteland RF, and Atkins JF (1991). The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J. Mol. Biol 218, 365–373. [DOI] [PubMed] [Google Scholar]
- Stansfield I, Jones KM, Kushnirov VV, Dagkesamanskaya AR, Poznyakovski AI, Paushkin SV, Nierras CR, Cox BS, Ter-Avanesyan MD, and Tuite MF (1995). The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J 14, 4365–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart EC, Serra V, Petroni G, and Nowacki M (2016). Genetic Codes with No Dedicated Stop Codon: Context-Dependent Translation Termination. Cell 166, 691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tork S, Hatin I, Rousset JP, and Fabret C (2004). The major 5′ determinant in stop codon read-through involves two adjacent adenines. Nucleic Acids Res 32, 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N, Hoshino S, Imataka H, Sonenberg N, and Katada T (2002). A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in Cap/Poly(A)-dependent translation. J. Biol. Chem 277, 50286–50292. [DOI] [PubMed] [Google Scholar]
- Velichutina IV, Dresios J, Hong JY, Li C, Mankin A, Synetos D, and Liebman SW (2000). Mutations in helix 27 of the yeast Saccharomyces cerevisiae 18S rRNA affect the function of the decoding center of the ribosome. RNA 6, 1174–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch EM, Barton ER, Zhuo J, Tomizawa Y, Friesen WJ, Trifillis P, Paushkin S, Patel M, Trotta CR, Hwang S, et al. (2007). PTC124 targets genetic disorders caused by nonsense mutations. Nature 447, 87–91. [DOI] [PubMed] [Google Scholar]
- Yang YS, Kato M, Wu X, Litsios A, Sutter BM, Wang Y, Hsu CH, Wood NE, Lemoff A, Mirzaei H, et al. (2019). Yeast Ataxin-2 Forms an Intracellular Condensate Required for the Inhibition of TORC1 Signaling during Respiratory Growth. Cell 177, 697–710.e617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Záhonová K, Kostygov AY, Ševčíková T, Yurchenko V, and Eliáš M (2016). An Unprecedented Non-canonical Nuclear Genetic Code with All Three Termination Codons Reassigned as Sense Codons. Curr. Biol 26, 2364–2369. [DOI] [PubMed] [Google Scholar]
- Zhouravleva G, Frolova L, Le Goff X, Le Guellec R, Inge-Vechtomov S, Kisselev L, and Philippe M (1995). Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J 14, 4065–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any datasets or code.


