Abstract
Objectives
Patients with colorectal cancer (CRC) may be susceptible to the coronavirus disease-2019 (COVID-19). However, anti-CRC/COVID-19 treatment options are currently unavailable. Since niacin is a vitamin with cytoprotective and anti-inflammatory functions, this study aimed to evaluate the possible functional roles and underlying mechanisms of action of niacin as an anti-COVID-19 and -CRC therapy.
Interventions
We used a series of network pharmacology-based and computational analyses to understand and characterize the binding capacity, biological functions, pharmacological targets and therapeutic mechanisms of niacin in CRC/COVID-19.
Measurements and main results
We revealed the clinical characteristics of CRC patients and COVID-19 patients, including predisposing genes, survival rate and prognosis. Moreover, the results of molecular docking analysis indicated that niacin exerted effective binding capacity in COVID-19. Further, we disclosed the targets, biological functions and signaling pathways of niacin in CRC/COVID-19. The analysis indicated that niacin could help in treating CRC/COVID-19 through cytoprotection, enhancement of immunologic functions, inhibition of inflammatory reactions and regulation of cellular microenvironment. Furthermore, five core pharmacological targets of niacin in CRC/COVID-19 were also identified, including BCL2L1, PTGS2, IL1B, IFNG and SERPINE1.
Conclusions
This study, for the first time, revealed the niacin-associated molecular functions and pharmacological targets for treating CRC/COVID-19, as COVID-19 remains a serious pandemic. But the findings were not validated in actual CRC patients infected with COVID-19, so further investigation is needed to confirm the potential use of niacin for treating CRC/COVID-19.
Keywords: colorectal neoplasms, COVID-19, niacin, network pharmacology, computational biology, prognosis
BACKGROUND
Coronavirus disease-19 (COVID-19), a fatal pandemic, has shown a gigantic increase in incidence and mortality rates worldwide [1]. As severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is newly-found, effective anti-COVID-19 medicines are limited, especially during the early outbreak in China [2]. Hydroxychloroquine is being administered as a potential treatment for COVID-19, and some of its curative effects can be found in clinical practice [3]; however, other pieces of evidence show limited therapeutic effectiveness [4]. Therefore, potential bioactive agents against COVID-19 need to be urgently screened and validated. Additionally, patients with cancer, such as colorectal cancer (CRC), lung cancer and breast cancer, have shown a high incidence of COVID-19 than those with noncancer diseases in China [5]. CRC is a severe, life-threatening neoplasm that can induce malignant invasiveness or metastasis [6]. In China, the incidence of CRC has increased in recent years due to the urbanized lifestyle and modern diet [7]. As per the 2020 cancer statistics, incidence rates for CRC rank among the top three cancer cases worldwide [8]. Further, during the early outbreak of COVID-19, hospitals were the main place for infection [9]. Thus, in-hospital patients with CRC are at an increased risk of COVID-19 infection. If CRC patients get infected with COVID-19, treatment for these patients will be difficult due to poor immunity and lack of medicines against COVID-19 [10]. Therefore, effective treatments should be screened in such patients.
Niacin, a B-type vitamin, is essential for normal functions in the gastrointestinal tract and nervous system [11]. It is a functional precursor of nicotinamide adenine dinucleotide, a bioactive coenzyme responsible for maintaining cell respiration and metabolism [12]. Moreover, niacin helps regulate energy metabolism, methylation, DNA rehabilitation and immune function [13]. Reports suggest that deficiency of niacin can increase the risk of neurodegeneration, immunological disorder and inflammation stress [14]. Additionally, niacin exerts its anti-inflammatory effect by suppressing the NF-κB pathway [15]. A high concentration of niacin has antiproliferative activity against colon cancer stem cells [16]. Furthermore, it can effectively inhibit inflammatory reaction in the colon tissue and tumorigenesis by activating G-Protein Coupled Receptor 109A (GPR109A) [17]. Interestingly, niacin is a potent antipneumonia agent that modulates specific genes, such as niaR, niaX and pnuC [18]. However, the pharmacological targets and mechanism by which niacin can combat COVID-19 in CRC patients remain to be studied in detail.
Network pharmacology is a useful bioinformatics tool to discover all candidate targets, functions and mechanisms of bioactive components to treat disease [19, 20]. Our previous bioinformatics analyses identified core targets, biological functions and signaling pathways downstream to few bioactive compounds, such as plumbagin and Vitamin-C in clinical disorders, including liver cancer, pneumonia and sepsis [21–23]. In the present study, using the network pharmacology approach, we aimed to elucidate the detailed anti-COVID-19/CRC mechanisms of niacin for network visualization. Further, a visible graphical abstract for the current flowchart is provided to demonstrate the anti-COVID-19/CRC functions and mechanisms exerted by niacin (Figure 1).
Figure 1.
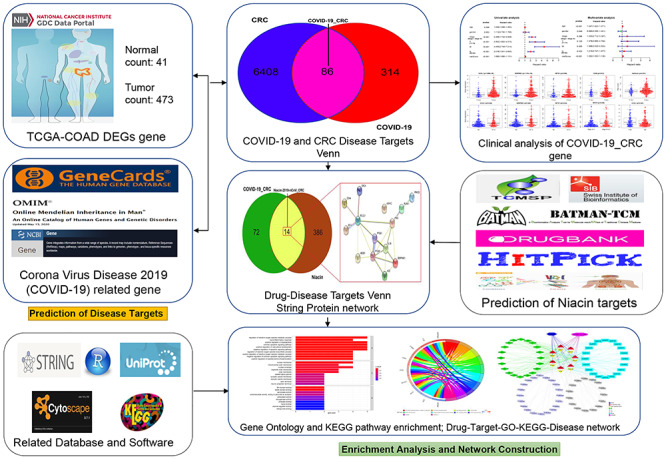
Workflow of the study. The figure indicates the antiviral action and mechanism of niacin against CRC/COVID-19 using the network pharmacology and computational bioinformatics analysis approach.
Materials and methods
Identification of CRC/COVID-19–associated genes
To identify the CRC/COVID-19–associated genes, transcriptome profiles of CRC patients were downloaded from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/) on 05 May 2020. The differential genes in CRC were screened and obtained using the ‘limma’ package of R-language Bioconductor with false discovery rate < 0.05 and | logfold change (FC) | > 1. Furthermore, genes related to COVID-19 were selected from the Genecard database, OMIM database and NCBI gene function module. Finally, these genes were compared to obtain the overlapping targets in CRC and COVID-19 [28, 29].
Clinicopathological analysis of CRC and COVID-19–associated genes
The ‘survival’ package in R package was used to analyze the correlation of CRC/COVID-19-associated genes and survival rates in patients with CRC/COVID-19. Prognostic analyses were performed using univariate Cox proportional hazards regression. Moreover, different characteristics of diseased genes and patients with CRC/COVID-19 were analyzed using a multivariate Cox proportional hazards regression model. Finally, based on the average risk score, patients were categorized into low- and high-risk groups [30].
Acquisition of niacin-pharmacological target in CRC/COVID-19
All pharmacological targets of niacin were screened and collected from accessible online tools, such as Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP), Swiss Target Prediction, TargetNet, Batman, Drugbank and HitPick [31]. The overlapping candidate genes were identified after data correction using reviewed (Swiss-Prot) and Human settings in the Uniprot database [32].
Enrichment analyses and network visualization
R-language packages, including ‘ClusterProfiler,’ ‘ReactomePA,’ ‘org.Hs.eg.Db’ and ‘GOplot,’ were used for enrichment analysis and visualization of gene ontology (GO) biological process (BP) and KEGG pathways of the overlapping gene targets of niacin in CRC/COVID-19. GO information was obtained using "org.Hs.eg.Db." P-value cutoff = 0.05 and q-value cutoff = 0.05 were set for enrichment, and output was used for plotting the bubble chart, bar chart and Circos plot [33, 34]. Cytoscape software (3.7.1 version) was used to construct a graphical representation of drug-target-GO function-pathway-disease for creating GO BP and pathway in response to targeting CRC and COVID-19 intersecting genes [35].
Identifying core targets of niacin against CRC/COVID-19
The overlapping targets of niacin treatment and CRC/COVID-19 were queried in the STRING database (version 11.0) to obtain the interaction network of the target–target function-related proteins, target interaction in protein–protein interactions (PPI) network map, and .tsv data. The Network-Analyzer setting from Cytoscape software was applied for analyzing the topological parameters. The median degree of freedom of the target network was 3.231, and the maximum freedom of degree was 7. Thus, the core target screening condition range was set to 4–7. Further, core targets were collected according to the Degree-value of the algorithm [36].
Molecular docking
Using molecular docking analysis, the binding situation and interaction force of protein and small molecules can be predicted and obtained. The molecular structure of niacin was obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) [24]. The protein structure of COVID-19 associated proteins was obtained from the PDB database (https://www.rcsb.org/) [25]. MM2 force field optimization was performed using the three-dimensional structure of the compound downloaded by the ChemBio3D Draw module in the ChemBioOffice software (version 2010). The Protein Data Bank, Partial Charge (Q), & Atom Type (T) (PDBQT) structure file necessary for virtual screening were created using the Raccoon software [26]. The supporting tool MGLTools 1.5.6 of Autodock Vina software was used to process the corresponding protein, hydrogenation and Gasteiger charge for merging nonpolar hydrogen atoms [27]. The original pdb file format was converted to the pdbqt file format recognized by the Autodock Vina program, providing a ligand basis for molecular docking. Further, the docking active center (including all residues around the original ligand) was set through the grid box function in the software, based on the size of the root mean square deviation (RMSD) of the docked ligand molecule and original ligand molecule with a reasonable setting of docking parameters. RMSD ≤ 4 was considered the threshold for conformation of the ligand after docking to match the conformation of the original ligand.
Results
Identification of COVID-19/CRC targets
First, network pharmacology collected 400 genes associated with COVID-19 from the Genecard, OMIM and NCBI databases (Figure 2A). Whereas, 6494 CRC-associated differentially expressed genes were identified using the TCGA database (Figure 2A). A comparison of these two gene clusters identified 86 intersection genes in COVID-19 and CRC (Figure 2A). Further, we checked the differential gene expression (DGE) of the intersected genes using R-language, in which 39 genes were upregulated and 47 were down-regulated in CRC (Figure 2B).
Figure 2.
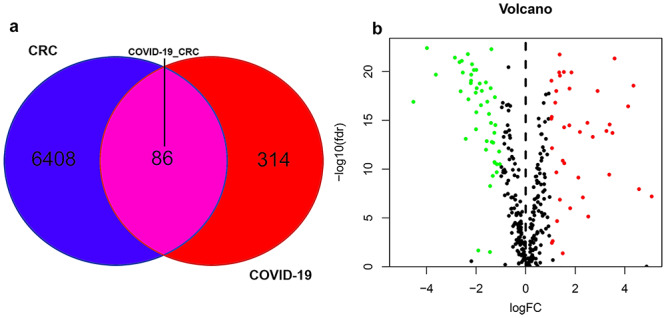
Analysis of intersecting genes in CRC/COVID-19. (A) Venn diagram depicting intersecting genes in CRC/COVID-19. (B) Volcano-plot representation of differential gene expression (DGE).
Clinicopathological analysis of CRC/ COVID-19-associated genes
To understand the correlation of CRC/COVID-19-associated genes and pathology of CRC/COVID-19, the 86 differential genes were subjected to univariate and multivariate Cox analyses. First, the univariate Cox analysis identified 13 genes, including IL16, SERPINE1, CR2, FCER2, NPTX1, TLR10, UCHL1, NCF1, CD79A, CD36, FGF2, CD40LG and ADA to be significantly (P < 0.05) associated with CRC/COVID-19 (Figure 3A, Table 1). Whereas, multivariate Cox analysis identified seven targeting genes out of 13, including IL16, SERPINE1, FCER2, NPTX1, UCHL1, CD36 and ADA (Table 2). Moreover, based on the coef values of Multivariate Cox proportional hazards regression analysis representing patient risk (Table 2), we categorized the patients into high- and low-risk groups. Next, in the overall survival analysis, no significant differences were observed between the high- and low-risk groups (Figure 3B). Further, each CRC patient's risk score, survival status and expression distribution of the seven genes mentioned above were analyzed. The results showed that greater risk value in patients correlated with higher risk score (Figure 3C) and increased expression levels of the three genes—UCHL1, SERPINE1 and ADA (Figure 3E). Furthermore, we conducted an independent single factor and multifactor prognostic analysis with the seven genes. Moreover, the clinical prognostic analysis of the seven genes showed that the expression of four genes—CD36, NPTX1, SERPINE1 and UCHL1—was closely associated with advanced stages of CRC. The expression of NPTX1, SERPINE1 and UCHL1 was also related to the number and scope of lymph node metastasis and spread in the lymph node area, while the expression of NPTX1 in CRC stage I and II disease was higher than that in stage III and IV. Additionally, the expression of UCHL1 was higher in older than that in younger patients (Figure 4, Table 3).
Figure 3.
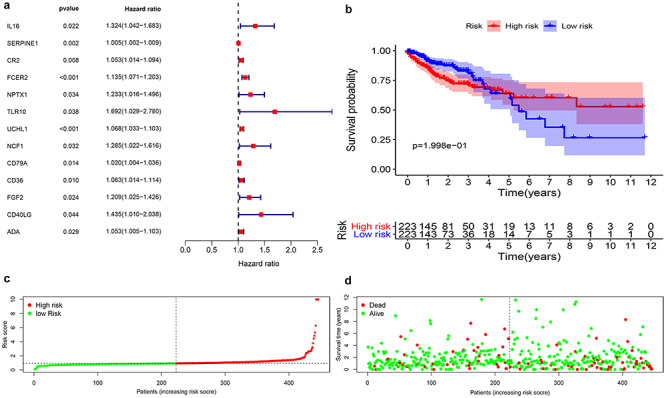
Prognostic value of CRC/COVID-19 associated genes. (A) Univariate Cox analysis identified 13 genes, including IL16, SERPINE1, CR2, FCER2, NPTX1, TLR10, UCHL1, NCF1, CD79A, CD36, FGF2, CD40LG and ADA. (B) Survival analysis indicated no difference in overall survival between high- and low-risk groups. (C) Analysis of patients’ risk score using Cox proportional hazards regression. (D) Expression levels of genes in the high- and low-risk groups.
Table 1.
Univariate Cox proportional hazards regression analysis of COVID-19 combined with CRC gene
| Symbol | HR | HR.95 L | HR.95H | P-value |
|---|---|---|---|---|
| IL16 | 1.324457 | 1.042209 | 1.683142 | 0.021558 |
| SERPINE1 | 1.005443 | 1.002007 | 1.00889 | 0.001883 |
| CR2 | 1.053227 | 1.013614 | 1.094388 | 0.008018 |
| FCER2 | 1.134922 | 1.070732 | 1.20296 | 2.04E-05 |
| NPTX1 | 1.232645 | 1.015844 | 1.495716 | 0.034071 |
| TLR10 | 1.691648 | 1.029342 | 2.780101 | 0.038074 |
| UCHL1 | 1.067599 | 1.033462 | 1.102865 | 7.98E-05 |
| NCF1 | 1.28472 | 1.021642 | 1.615543 | 0.032104 |
| CD79A | 1.019596 | 1.003902 | 1.035537 | 0.014209 |
| CD36 | 1.063087 | 1.014473 | 1.11403 | 0.010417 |
| FGF2 | 1.208832 | 1.024961 | 1.425689 | 0.02427 |
| CD40LG | 1.435032 | 1.010346 | 2.038229 | 0.043648 |
| ADA | 1.052907 | 1.005433 | 1.102622 | 0.028514 |
Table 2.
Multivariate Cox proportional hazards regression analysis
| Symbol | coef | HR | HR.95 L | HR.95H | P-value |
|---|---|---|---|---|---|
| IL16 | −0.42431 | 0.654221 | 0.392304 | 1.091003 | 0.103915 |
| SERPINE1 | 0.005727 | 1.005743 | 1.00178 | 1.009722 | 0.004473 |
| FCER2 | 0.355835 | 1.427372 | 1.093811 | 1.862653 | 0.008787 |
| NPTX1 | −0.49767 | 0.607946 | 0.342335 | 1.079639 | 0.089422 |
| UCHL1 | 0.045768 | 1.046832 | 0.998118 | 1.097923 | 0.059771 |
| CD36 | 0.058166 | 1.059891 | 0.989731 | 1.135024 | 0.095998 |
| ADA | 0.051072 | 1.052398 | 1.003059 | 1.104165 | 0.037102 |
Figure 4.
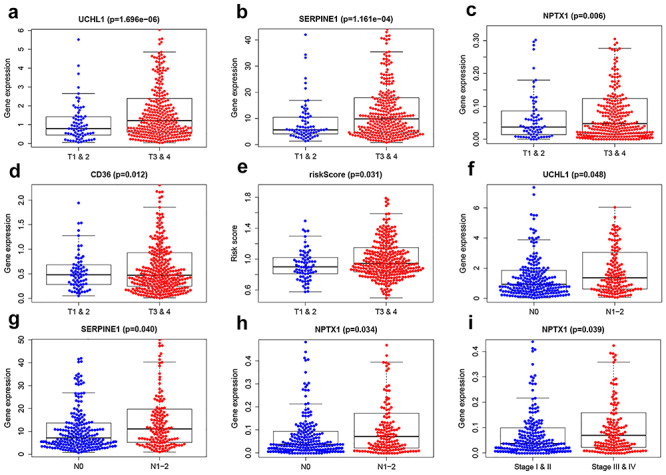
Clinical prognostic analysis of the 7 genes. (A–D) Association of gene expression of UCHL1, SERPINE1, NPTX1 and CD36 with tumor stage and metastasis in colorectal cancer (CRC) patients. (E) Association of risk score with tumor stage of CRC. (F–H) Association of expression of SERPINE1, UCHL1 and NPTX1 with the number of lymph node metastasis. (I) Association of expression of NPTX1 with staging in CRC.
Table 3.
Clinical correlation analysis
| Symbol | Age (≤65 versus >65) | Gender (male versus female) | Stage (stage I and II versus stage III & IV) | T (T1 & 2 versus T3 & 4) | M (M0 versus M1) | N (N0 versus N1–2) |
|---|---|---|---|---|---|---|
| IL16 | 0.981(0.327) | −0.546(0.586) | 0.517(0.605) | −0.16(0.873) | 0.489(0.626) | 0.156(0.876) |
| SERPINE1 | −0.048(0.962) | −0.18(0.857) | −1.922(0.056) | −3.894(1.161e-04) | −1.265(0.211) | −2.071(0.040) |
| FCER2 | 1.217(0.225) | −1.346(0.180) | −0.908(0.365) | −1.342(0.181) | −0.85(0.399) | −0.969(0.334) |
| NPTX1 | 1.038(0.300) | −0.665(0.506) | −2.084(0.039) | −2.787(0.006) | −0.975(0.333) | −2.14(0.034) |
| UCHL1 | 2.048(0.042) | 0.05(0.960) | −1.713(0.088) | −4.862(1.696e-06) | −1.416(0.162) | −1.989(0.048) |
| CD36 | 1.725(0.086) | 1.07(0.285) | −1.437(0.152) | −2.515(0.012) | −1.128(0.263) | −1.638(0.102) |
| ADA | −0.586(0.558) | −0.132(0.895) | 0.024(0.981) | 0.025(0.980) | −0.416(0.679) | −0.16(0.873) |
| Risk score | 0.884(0.378) | −1.425(0.156) | −1.48(0.141) | −2.172(0.031) | −1.507(0.137) | −1.507(0.134) |
Identifying niacin targets and intersection with COVID-19 and CRC
TCMSP databases and Swiss Target Prediction software were used to determine the pharmacological targets of niacin. After biological correction and depletion of repetitive genes using the UniProt database, 400 niacin-associated targets were obtained. An overlap of COVID-19/CRC genes with niacin-associated targets identified 14 intersection genes of niacin against COVID-19/CRC (Figure 5A; Supplementary Table 1, Supplemental Digital Content 1). The 14 intersection genes were submitted for GO and KEGG enrichment analyses, which indicated niacin to affect a series of BPs, including regulation of reactive oxygen species metabolic process, neuroinflammatory response, positive regulation of angiogenesis, extrinsic apoptotic signaling pathway, positive regulation of vasculature development, negative regulation of apoptotic signaling pathway, regulation of reactive oxygen species biosynthetic process, positive regulation of reactive oxygen species metabolic process, negative regulation of extrinsic apoptotic signaling pathway, positive regulation of peptidyl-serine phosphorylation, prostaglandin biosynthetic process, prostanoid biosynthetic process, positive regulation of alcohol biosynthetic process, reactive oxygen species metabolic process, reactive oxygen species biosynthetic process, peptidyl-serine phosphorylation, negative regulation of signal transduction in the absence of ligand, negative regulation of extrinsic apoptotic signaling pathway in the absence of ligand, peptidyl-serine modification, and regulation of peptidyl-serine phosphorylation (Figure 5B; Supplementary Table 2, Supplemental Digital Content 2). Additionally, in the KEGG pathway analysis, 48 KEGG pathways related to all core targets were found (P-adjust < 0.05). These included the AGE–RAGE signaling pathway in diabetic complications, NF-kappa B signaling pathway, p53 signaling pathway, Leishmaniasis, small cell lung cancer, Chagas disease (American trypanosomiasis), HIF-1 signaling pathway, African trypanosomiasis, measles, influenza A, chronic myeloid leukemia, EGFR tyrosine kinase inhibitor resistance, human cytomegalovirus infection, IL-17 signaling pathway, Amoebiasis, Th17 cell differentiation, toxoplasmosis, serotonergic synapse, apoptosis-multiple species, and MicroRNAs in cancer (Figure 5C; Supplementary Table 3, Supplemental Digital Content 3).
Figure 5.
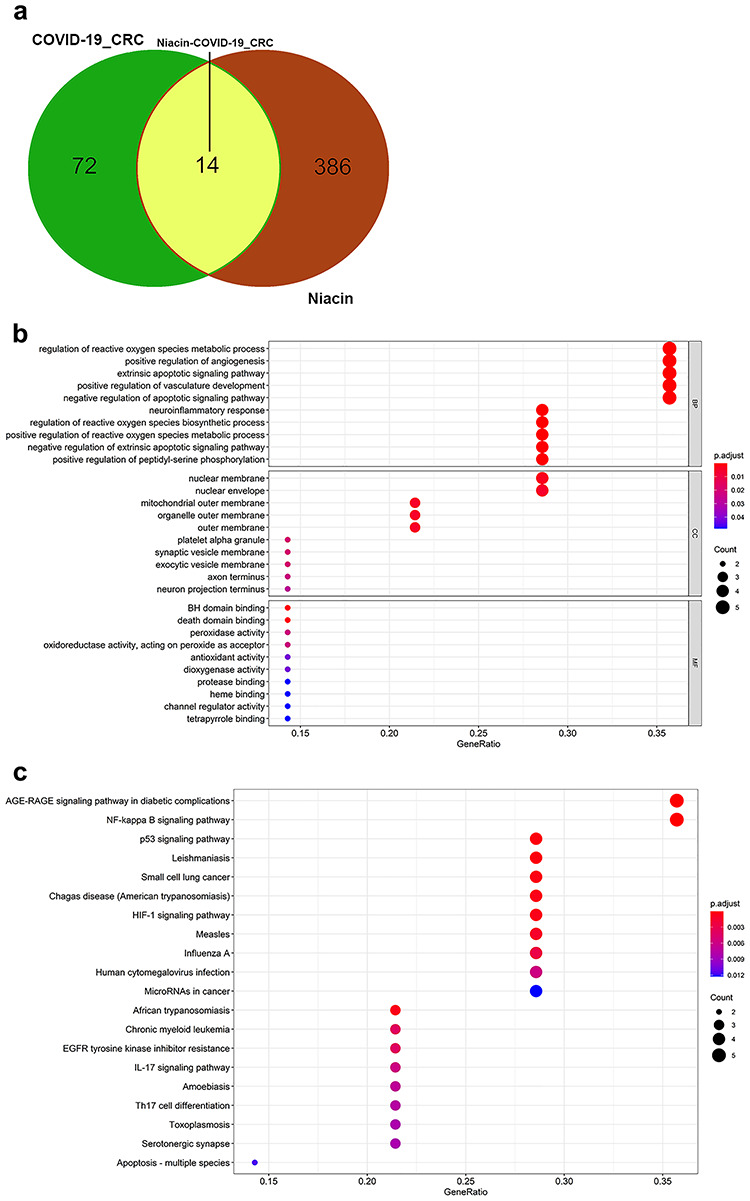
Functional characterization of niacin against CRC/COVID-19 intersecting genes. (A) Venn diagram depicting intersecting genes of niacin and CRC/COVID-19. (B) Gene ontology analysis of intersecting genes of niacin and CRC/COVID-19. (C) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway of intersecting genes of niacin and CRC/COVID-19.
Identification of core targets of niacin against COVID-19 and CRC
Next, STRING analysis was used to determine the PPI-networking mediated by 14 intersection targets of niacin against COVID-19/CRC (Figure 6A). All mapped intersecting proteins were input to the Cytoscape software to calculate the topological parameters of PPI-network related to niacin against COVID-19/CRC. The analysis identified five core gene targets, including BCL2L1, PTGS2, IL1B, IFNG and SERPINE1 (Figure 6B; Supplementary Table 4, Supplemental Digital Content 4).
Figure 6.
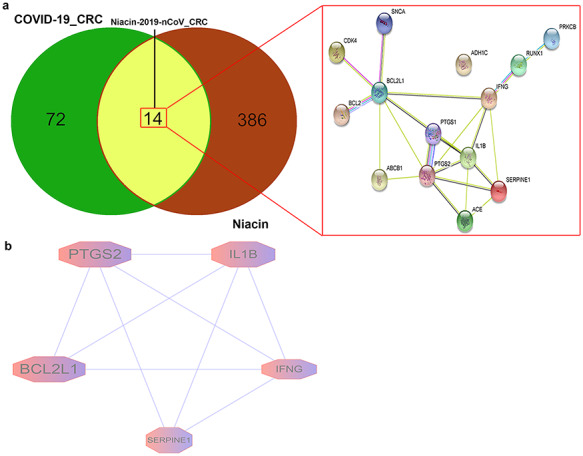
Gene network analysis of niacin against CRC/COVID-19. (A) STRING analysis indicating protein–protein interaction networking mediated by 14 intersecting targets of niacin against COVID-19/CRC. (B) Cytoscape analysis representing the protein interaction network related to the action of niacin against COVID-19/CRC. Five core targets—BCL2L1, PTGS2, IL1B, IFNG and SERPINE1—are highlighted.
Binding of niacin to COVID-19 intersecting potential target genes
In order to determine the possible binding of niacin with the COVID-19 and the identified core targets, molecular docking analysis was performed. The crystal structure of the COVID-19 main protease was collected from the PDB database (http://www.rcsb.org/structure/5R84) with PDB ID 5R84, for docking analysis with niacin (Figure 7). The active cavity box parameter setting center_x, y, z was 10.162, 0.119 and 20.548, respectively, and size_x, y, z was 11.25, whereas RMSD of the original ligand was 2.304 Å. Thus, the hydrogen bond between the proto-ligand 2-cyclohexyl-~{N}-pyridin-3-yl-ethanamide (GWS) of 5R84 and 5R84 protein acted on two amino acid residues, viz. HIS-163 (1.7 Å) and GLU-166 (2.1 Å) (Figure 7A). Our result showed that niacin forms a hydrogen bond with the amino acid residue HIS-163 (2.0 Å) of 5R84 (CONVID-19) (Figure 7B), the bond action suggested that niacin had a good binding activity with the 5R84 protein (COVID-19). Further, we analyzed the possible binding between niacin and the five COVID-19/CRC core targets (BCL2L1, PTGS2, IL1B, IFNG and SERPINE1) identified from STRING analysis. Our result showed that niacin only bind to IL1B. We collected the crystal structure of IL1B from the PDB database (http://www.rcsb.org/structure/5R85), the active cavity box parameter setting center_x, y, z was 41.653, 4.192 and 71.476, respectively, and size_x, y, z was 11.25, whereas RMSD of the original ligand was 2.328 Å. The analysis suggested that hydrogen bonding between pro-ligand 3,3,3-tris(fluoranyl)-1-piperazin-1-yl-propan-1-one (S7A) of 5R85 and the 5R85 protein acted on the amino acid residue LEU-26 (2.3 Å) (Figure 7C). Moreover, the binding of niacin was consistent with pro-ligand S7A’s hydrogen-bonding amino acid residue Leu-26 (2.3 Å) of IL1B (Figure 7D), indicating its high-affinity association between niacin and IL1B.
Figure 7.
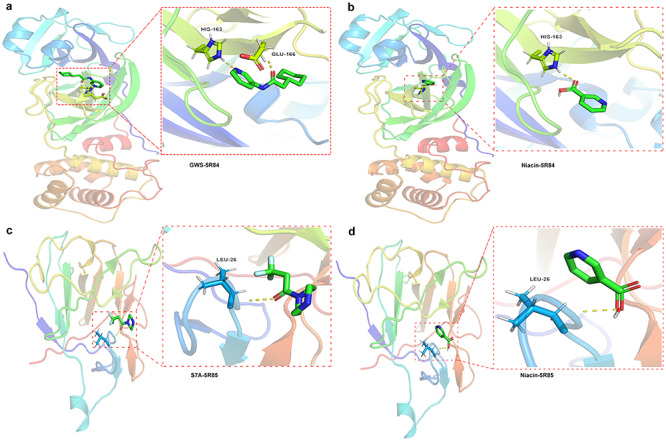
Binding of niacin to COVID-19 and the core target IL1B using molecular docking analysis. (A) Sites of niacin with SARS-CoV-2 main protease (PDB ID 5R84). (B) Hydorgen bond formed between niacin and and 5R84 protein of COVID-19 on HIS-163. (C) Sites of niacin with Human Interleukin-1 Beta (PDB ID: 5R85). (D) Hydorgen bond formed between niacin and and 5R85 protein of IL1B on LEU-26.
DISCUSSION
COVID-19 is an emerging, rapidly evolving pandemic with an increasing infection and death rate worldwide [37]. Moreover, specific therapy against COVID-19 remains undeveloped to overcome this life-threatening epidemic disease. Additionally, COVID-19 in humans is increasing every day [38]. As per statistics, the COVID-19-infected population is huge in several countries globally [39], including China, India and Brazil. In developing countries, the incidence of malignant diseases, such as cancers, has rapidly increased in recent decades [40]. As the immune system of cancer patients is weakened due to immunosuppression and immunological dysfunction [41], any external infection can deteriorate their health. However, given the current COVID-19 prevalence, hospital in-patients with cancer may have an increased risk of being infected with the novel coronavirus (SARS-CoV-2), as hospitals are a high-risk place for exposure to coronavirus [42]. As per cancer statistics in 2020, CRC is the second leading cause of death by cancer in the USA [43]. Additionally, the COVID-19 infected cases in the USA are increasing [44]. Moreover, CRC patients may be susceptible to infection with COVID-19 under current ineffective management. Thus, the incidence of COVID-19 in CRC patients may worsen the treatment outcome and decrease the survival rate in such patients.
In a previous reference analysis, niacin showed beneficial action against CRC cells [16]. Thus, based on the viral inhibitory effects of niacin [45], we hypothesize that niacin may exert potent pharmacological activity in patients with CRC combined with COVID-19. In the current computational analysis, we first screened out and identified hub genes and 86 intersecting genes of COVID-19 combined with CRC. The DGE analysis showed 39 upregulated and 47 downregulated genes in patients with CRC and/or COVID-19. The DGE-based analysis may be used for determining clinical characteristics in CRC patients with COVID-19. As per the independent prognostic and survival analyses, few of the important DGE, including IL16, SERPINE1, CR2, FCER2, NPTX1, TLR10, UCHL1, NCF1, CD79A, CD36, FGF2, CD40LG and ADA, may function as potent biomarkers for screening and characterizing different stages of CRC patients with COVID-19. Taken together, these 86 intersection genes in CRC and COVID-19 may be used as potential treatment targets. Further, using the network pharmacology approach, we identified 14 intersection genes with niacin treatment against CRC and COVID-19 before DGE analysis. The DGE-analysis identified significant differences in the expression of SERPINE1 and BCL2L1. Moreover, CRC and COVID-19 showed increased expression of SERPINE1 and BCL2L1, along with a lower survival rate. SERPINE1 is a serpin peptidase inhibitor that was found to be induced in recurrent CRC tumors compared to nonrecurrent tumors [46]. It has been reported that SERPINE1 interacted with different growth factors such as AKT1, VEGFA and TGFB1 in CRC [47]. Further, increased expression of SERPINE1 was associated with tumor cell migration and invasion through the activation of the PI3K-Akt pathway [48]. BCL2L1 is an oncogenic driver and antiapoptotic factor in CRC [49–50]. It was reported that the induction of BCL2L1 led to a reduction of apoptosis in CRC cell lines [51]. These findings indicated that these 14 intersection genes might be potent pharmacological targets of niacin against CRC and COVID-19. Based on the GO and KEGG enrichment analysis, the findings suggested that anti-CRC and anti-COVID-19 effects exerted by niacin were directed via cytoprotection, antiviral and anti-inflammatory actions, immunoregulation, as well as regulation of related signaling pathways, including the NF-kappa B, p53, HIF-1 and IL-17, Th17 cell differentiation, and MicroRNAs in cancer. Moreover, the anti-CRC/COVID-19 action of niacin could be modulated by core molecules/genes, including BCL2L1, PTGS2, IL1B, IFNG and SERPINE1. Using molecular docking analysis, we identified best binding activities of niacin with GWS_5R84 structures in COVID-19 and S7A_5R85 in the core target IL1B, indicating that niacin can effectively bind to specific proteins in the novel coronavirus. These bindings suggested that niacin may be able to bring the IL1B to target the COVID-19. Additionally, we believe that adjuvant supplementation of niacin may enhance the therapeutic efficacy of current clinical antiviral agents and immunotherapy to treat the fatal COVID-19, or CRC combined with COVID-19.
Conclusions
Taken together, the bioinformatics and computational findings highlight antivirus, anti-inflammation and immunity-modulation as key targets/pathways of niacin treatment in CRC/COVID-19 (Figure 8). Further, niacin may be used clinically to treat CRC/COVID-19, based on the identified BPs (pharmacological functions) and signaling pathways (therapeutic mechanisms). Moreover, the potent pharmacological targets of niacin against CRC/COVID-19 were identified, paving the way for further clinical trials.
Figure 8.
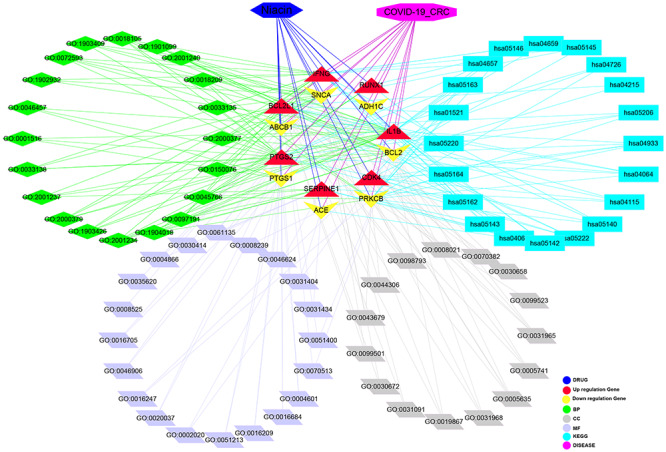
Interaction network showing core biotargets, pharmacological functions, and signaling pathways of niacin against CRC/COVID-19.
Authors' contributions
R.L, K.P.L. and M.S. conceived and designed the study. X.L. and L.M. performed data analysis and data interpretation. X.L., L.Y. and L.M. conducted the bioinformatics and statistical analyses. R.L and K.P.L prepared the manuscript.
Key Points
The clinical characteristics of colorectal cancer (CRC) patients and COVID-19 patients, including predisposing genes, survival rate and prognosis, were uncovered.
Molecular docking analysis indicated that niacin exerted effective binding capacity in COVID-19.
The targets, biological functions and signaling pathways of niacin in CRC/COVID-19 were revealed.
Five core pharmacological targets of niacin in CRC/COVID-19, including BCL2L1, PTGS2, IL1B, IFNG and SERPINE1, were identified.
Supplementary Material
Acknowledgments
Keng Po Lai was supported by Hong Kong SAR, Macao SAR, and Taiwan Province Talent Young Scientist Program of Guangxi.
Rong Li is a researcher at Guilin Medical University. He plotted the figures based on network pharmacology, using online databases.
Yu Li is a student of Guilin Medical University. He conducted data analysis and performed literature searches.
Xiao Liang and Lu Yang are master’s students of Guilin Medical University who contributed to data collection and figure plotting.
Professor Min Su is a professor at Guilin Medical University who worked on analyzing data and literature searches.
Professor Keng Po Lai is a researcher at Guilin Medical University. His research is focused on revealing mechanism networks underlying complex diseases through deep sequencing of transcriptomes, small RNA transcriptomes, and methylomes, which is then integrated with data from COVID-19.
Contributor Information
Rong Li, Gyuilin Medical University.
Yu Li, Gyuilin Medical University.
Xiao Liang, Gyuilin Medical University.
Lu Yang, Gyuilin Medical University.
Min Su, Gyuilin Medical University.
Keng Po Lai, Gyuilin Medical University.
Funding
National Natural Science Foundation of China (No. 81560134, 81660091) and the National Natural Science Foundation of Guangxi (No. 2019GXNSFBA185015, 2018GXNSFAA281242). Keng Po Lai was supported by the Hong Kong SAR, Macao SAR, and Taiwan Province Talented Young Scientist Program of Guangxi.
Abbreviations
CRC colorectal cancer
COVID-19 coronavirus disease-2019
SARS-CoV-2 severe acute respiratory syndrome coronavirus 2
GPR109A G-Protein Coupled Receptor 109A
TCGA The Cancer Genome Atlas
GO gene ontology
BP biological process
KEGG Kyoto Encyclopedia of Genes and Genomes
PDB Protein Data Bank
PDBQT Protein Data Bank, Partial Charge (Q), & Atom Type (T)
RMSD root mean square deviation
TCMSP Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform.
References
- 1. Velavan TP, Meyer CG. The COVID-19 epidemic. Trop Med Int Health 2020;25:278–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang W, Zhao Y, Zhang F, et al. The use of anti-inflammatory drugs in the treatment of people with severe coronavirus disease 2019 (COVID-19): the perspectives of clinical immunologists from China. Clin Immunol 2020;214:108393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Okour M, Al-Kofahi M, Austin D. Hydroxychloroquine and azithromycin as potential treatments for COVID-19; clinical status impacts the outcome. J Pharmacokinet Pharmacodyn 2020;47:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tang W, Cao Z, Han M, et al. Hydroxychloroquine in patients with mainly mild to moderate coronavirus disease 2019: open label, randomised controlled trial. BMJ 2020;369:m1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liang W, Guan W, Chen R, et al. Cancer patients in SARS-CoV-2 infection: a nationwide analysis in China. Lancet Oncol 2020;21:335–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dekker E, Tanis PJ, Vleugels JLA, et al. Colorectal cancer. Lancet 2019;394:1467–80. [DOI] [PubMed] [Google Scholar]
- 7. Zhu J, Tan Z, Hollis-Hansen K, et al. Epidemiological trends in colorectal cancer in China: an ecological study. Dig Dis Sci 2017;62:235–43. [DOI] [PubMed] [Google Scholar]
- 8. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7–34. [DOI] [PubMed] [Google Scholar]
- 9. Wang X, Fang J, Zhu Y, et al. Clinical characteristics of non-critically ill patients with novel coronavirus infection (COVID-19) in a Fangcang hospital. Clin Microbiol Infect 2020;26:30177–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kong Q, Xiang ZG, Wu Y, et al. Analysis of the susceptibility of lung cancer patients to SARS-CoV-2 infection. Mol Cancer 2020;19:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Meyer-Ficca M, Kirkland JB. Niacin. Adv Nutr 2016;7:556–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. MacKay D, Hathcock J, Guarneri E. Niacin: chemical forms, bioavailability, and health effects. Nutr Rev 2012;70:357–66. [DOI] [PubMed] [Google Scholar]
- 13. Gasperi V, Sibilano M, Savini I, et al. Niacin in the central nervous system: an update of biological aspects and clinical applications. Int J Mol Sci 2019;20:974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ikenouchi-Sugita A, Sugita K. Niacin deficiency and cutaneous immunity. Nihon Rinsho Meneki Gakkai Kaishi 2015;38:37–44. [DOI] [PubMed] [Google Scholar]
- 15. Si Y, Zhang Y, Zhao J, et al. Niacin inhibits vascular inflammation via downregulating nuclear transcription factor-κB Signaling pathway. Mediators Inflamm 2014;2014:263786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sen U, Shenoy PS, Bose B. Opposing effects of low versus high concentrations of water soluble vitamins/dietary ingredients vitamin C and niacin on colon cancer stem cells (CSCs). Cell Biol Int 2017;41:1127–45. [DOI] [PubMed] [Google Scholar]
- 17. Singh N, Gurav A, Sivaprakasam S, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014;40:128–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Afzal M, Kuipers OP, Shafeeq S. Niacin-mediated gene expression and role of NiaR as a transcriptional repressor of niaX, nadC, and pnuC in Streptococcus pneumoniae. Front Cell Infect Microbiol 2017;7:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ge B, Guo C, Liang YJ, et al. Network analysis, and human and animal studies disclose the Anticystitis Glandularis effects of vitamin C. Biofactors 2019;45:912–9. [DOI] [PubMed] [Google Scholar]
- 20. Li R, Ma XY, Song YQ, et al. Anti-colorectal cancer targets of resveratrol and biological molecular mechanism: analyses of network pharmacology, human and experimental data. J Cell Biochem 2019;120:11265–73. [DOI] [PubMed] [Google Scholar]
- 21. Zhou R, Wu K, Su M, et al. Bioinformatic and experimental data decipher the pharmacological targets and mechanisms of Plumbagin against hepatocellular carcinoma. Environ Toxicol Pharmacol 2019;70:103200. [DOI] [PubMed] [Google Scholar]
- 22. Li R, Guo C, Li Y, et al. Therapeutic target and molecular mechanism of vitamin C-treated pneumonia: a systematic study of network pharmacology. Food Funct 2020;11:4765–72. [DOI] [PubMed] [Google Scholar]
- 23. Li R, Guo C, Li Y, et al. Therapeutic targets and signaling mechanisms of vitamin C activity against sepsis: a bioinformatics study. Brief Bioinform 2020;bbaa079. doi: 10.1093/bib/bbaa079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang Y, Bryant SH, Cheng T, et al. PubChem BioAssay: 2017 update. Nucleic Acids Res 2017;45:955–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Burley SK, Berman HM, Kleywegt GJ, et al. Protein data Bank (PDB): the single global macromolecular structure archive. Methods Mol Biol 2017;1607:627–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Forli S, Huey R, Pique ME, et al. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc 2016;11:905–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010;31:455–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu K, Wei P, Liu M, et al. To reveal pharmacological targets and molecular mechanisms of curcumol against interstitial cystitis. J Adv Res 2019;20:43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li R, Song Y, Ji Z, et al. Pharmacological biotargets and the molecular mechanisms of oxyresveratrol treating colorectalcancer: network and experimental analyses. Biofactors 2020;46:158–67. [DOI] [PubMed] [Google Scholar]
- 30. Fisher LD, Lin DY. Time-dependent covariates in the cox proportional-hazards regression model. Annu Rev Public Health 1999;20:145–57. [DOI] [PubMed] [Google Scholar]
- 31. Su M, Guo C, Liu M, et al. Therapeutic targets of vitamin C on liver injury and associated biological mechanisms: a study of network pharmacology. Int Immunopharmacol 2019;66:383–7. [DOI] [PubMed] [Google Scholar]
- 32. Li R, Ma X, Song Y, et al. Anti-colorectal cancer targets of resveratrol and biological molecular mechanism: analyses of network pharmacology, human and experimental data. J Cell Biochem 2019;120:11265–73. [DOI] [PubMed] [Google Scholar]
- 33. Li J, Guo C, Lu X, et al. Anti-colorectal cancer biotargets and biological mechanisms of puerarin: study of molecular networks. Eur J Pharmacol 2019;858:172483. [DOI] [PubMed] [Google Scholar]
- 34. Liang Y, Zhou R, Liang X, et al. Pharmacological targets and molecular mechanisms of Plumbagin to treat colorectal cancer: a systematic pharmacology study. Eur J Pharmacol 2020;881:173227. [DOI] [PubMed] [Google Scholar]
- 35. Pan Q, Zhou R, Su M, et al. The effects of Plumbagin on pancreatic cancer: a mechanistic network pharmacology approach. Med Sci Monit 2019;25:4648–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xiao H, Qin X, Wan J, et al. Pharmacological targets and the biological mechanisms of Formononetin for Alzheimer's disease: a network analysis. Med Sci Monit 2019;25:4273–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. CDC COVID-19 Response Team Severe outcomes among patients with coronavirus disease 2019 (COVID-19) - United States, February 12-march 16, 2020. MMWR Morb Mortal Wkly Rep 2020;69:343–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhai P, Ding Y, Wu X, et al. The epidemiology, diagnosis and treatment of COVID-19. Int J Antimicrob Agents 2020;55:105955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sohrabi C, Alsafi Z, O'Neill N, et al. World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19). Int J Surg 2020;76:71–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pereira I, Katz MS, Simcock R. Developing cancer care institutions for the developing world. Lancet Oncol 2018;19:1436. [DOI] [PubMed] [Google Scholar]
- 41. Gonzalez H, Hagerling C, Werb Z. Roles of the immune system in cancer: from tumor initiation to metastatic progression. Genes Dev 2018;32:1267–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Al-Quteimat OM, Amer AM. The impact of the COVID-19 pandemic on cancer patients. Am J Clin Oncol 2020;43:452–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin 2020 May;70(3):145–64. [DOI] [PubMed] [Google Scholar]
- 44. CDC COVID-19 Response Team Geographic differences in COVID-19 cases, deaths, and incidence-United States, February 12-April 7, 2020. MMWR Morb Mortal Wkly Rep 2020;69:465–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Murray MF. Niacin as a potential AIDS preventive factor. Med Hypotheses 1999;53:375–9. [DOI] [PubMed] [Google Scholar]
- 46. Cheng X, Hu M, Chen C, et al. Computational analysis of mRNA expression profiles identifies a novel triple-biomarker model as prognostic predictor of stage II and III colorectal adenocarcinoma patients. Cancer Manag Res 2018;10:2945–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liang Y, Zhang C, Ma MH, et al. Identification and prediction of novel non-coding and coding RNA-associated competing endogenous RNA networks in colorectal cancer. World J Gastroenterol 2018;24:5259–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Balsara RD, Castellino FJ, Ploplis VA. A novel function of plasminogen activator inhibitor-1 in modulation of the AKT pathway in wild-type and plasminogen activator inhibitor-1-deficient endothelial cells. J Biol Chem 2006;281:22527–36. [DOI] [PubMed] [Google Scholar]
- 49. Zhang H, Zong Y, Qiu G, et al. Silencing Lin28 promotes apoptosis in colorectal cancer cells by upregulating let-7c targeting of antiapoptotic BCL2L1. Mol Med Rep 2018;17:5143–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Scherr AL, Gdynia G, Salou M, et al. Bcl-xL is an oncogenic driver in colorectal cancer. Cell Death Dis 2016;7:e2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lizárraga-Verdugo E, Ruiz-García E, López-Camarillo C, et al. Cell survival is regulated via SOX9/BCL2L1 Axis in HCT-116 colorectal cancer cell line. J Oncol 2020;2020:5701527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


