Abstract
The COVID-19 pandemic has resulted in a tremendous need for access to the latest scientific information, leading to both corpora for COVID-19 literature and search engines to query such data. While most search engine research is performed in academia with rigorous evaluation, major commercial companies dominate the web search market. Thus, it is expected that commercial pandemic-specific search engines will gain much higher traction than academic alternatives, leading to questions about the empirical performance of these tools. This paper seeks to empirically evaluate two commercial search engines for COVID-19 (Google and Amazon) in comparison with academic prototypes evaluated in the TREC-COVID task. We performed several steps to reduce bias in the manual judgments to ensure a fair comparison of all systems. We find the commercial search engines sizably underperformed those evaluated under TREC-COVID. This has implications for trust in popular health search engines and developing biomedical search engines for future health crises.
Keywords: information retrieval, COVID-19, coronavirus, TREC-COVID
BACKGROUND AND SIGNIFICANCE
The COVID-19 pandemic has resulted in a surge of scientific study. A systematic effort to consolidate the flood of such information content (mostly scientific articles), along with past studies on related coronaviruses, is being carried out in the form of CORD-191.1 Meanwhile, the TREC-COVID challenge was introduced to evaluate the capabilities of search engines for meeting the information needs of biomedical researchers using CORD-19.2,3 The challenge involved an information retrieval (IR) task to retrieve relevant articles for a given query. Similar to TREC-COVID, major technology companies Amazon and Google also developed their own systems for exploring CORD-19.
Both Amazon and Google have made recent forays into biomedical natural language processing (NLP). Amazon launched Amazon Comprehend Medical (ACM) for processing unstructured medical text.4–7 This same technology is incorporated into their search engine for CORD-19. Similarly, BERT from Google8 is enormously popular. BERT is a powerful language model that is trained on large raw text datasets to learn the nuances of natural language in an efficient manner. BERT’s training methodology helps transfer knowledge from vast raw data sources to other domains, such as biomedicine. Several works have explored the efficacy of BERT models in the biomedical domain for tasks such as information extraction9 and question answering.10 Many biomedical and scientific variants of the model have also been built, such as BioBERT,11 Clinical BERT,12 and SciBERT.13 Google has incorporated BERT into their web search engine (Google.com)14 as well as their CORD-19 search engine.
However, despite the popularity of these companies’ products, no formal evaluation of these systems is made available by the companies. Also, neither participated in TREC-COVID. In this paper, we aim to evaluate these two IR systems and compare them against the runs submitted to TREC-COVID to gauge the efficacy of what are likely highly utilized search engines.
METHODS
Information retrieval systems
We evaluate two commercial IR systems targeted toward CORD-19, from Amazon (CORD-19 Search2) and Google (COVID-19 Research Explorer3). We hereafter refer to these systems by their corporation names. Both systems take a natural language query as input and return a ranked list of CORD-19 links.
Amazon’s system uses an enriched version of CORD-19, constructed by passing it through a language processing service called Amazon Comprehend Medical (ACM).15 ACM is a machine learning-based NLP pipeline that extracts clinical concepts from unstructured text.4 The data is further mapped to clinical topics related to COVID-19, such as immunology, clinical trials, and virology, using multilabel classification and inference models. After enrichment, the data is indexed using Amazon Kendra, which also uses machine learning to provide natural language querying capabilities.
Google’s system is based on a semantic search mechanism powered by BERT.16 Semantic search, unlike lexical term-based search, which performs phrasal matching, focuses on understanding the meaning of user queries. However, deep learning models like BERT require substantial amounts of annotated data to be tuned to a specific task/domain. Biomedical articles have very different linguistic features than the general domain, upon which BERT is built. Thus, it needs to be tuned for the target domain using annotated data. For this they use the BioASQ data4. Due to the smaller size of these datasets, they use a synthetic query generation technique for data augmentation.17 Finally, these expanded datasets are used to fine-tune the neural model. They further enhance their system by combining term- and neural-based retrieval models by balancing memorization and generalization dynamics.18
Evaluation
We use the topics from Round 1 of the TREC-COVID challenge for our evaluation.2,3 These topics are information need statements for important COVID-19 topic areas. Each topic consists of three fields with increasing granularity: a (keyword-based) query; a (natural language) question; and a (longer descriptive) narrative. Four example topics are presented in Table 1 . Participants return a “run” consisting of a ranked list of documents for each topic. Round 1 used 30 topics and evaluated against the April 10, 2020 release of CORD-19.
Table 1.
Four example topics from Round 1 of the TREC-COVID challenge. A category is assigned (for this paper, not TREC-COVID) to each topic based on both the topic’s research field and function, which allows us to classify the performance of the systems on certain kinds of topics.
| Topic 10 |
Query: coronavirus social distancing impact Question: has social distancing had an impact on slowing the spread of COVID-19? Narrative: seeking specific information on studies that have measured COVID-19’s transmission in one or more social distancing (or non-social distancing) approaches. Categories:Research Field - Public Health Function - Prevention |
| Topic 13 |
Query: how does coronavirus spread Question: what are the transmission routes of coronavirus? Narrative: looking for information on all possible ways to contract coronavirus from people, animals and objects. Categories:Research Field - Biological Function - Transmission |
| Topic 22 |
Query: coronavirus heart impacts Question: are cardiac complications likely in patients with COVID-19? Narrative: seeking information on the types, frequency and mechanisms of cardiac complications caused by coronavirus. Categories:Research Field - Clinical Function - Effect |
| Topic 30 |
Query: coronavirus remdesivir Question: is remdesivir an effective treatment for COVID-19? Narrative: seeking specific information on clinical outcomes in COVID-19 patients treated with remdesivir. Categories:Research Field - Clinical Function - Treatment |
We use the question and narrative fields to query the systems following the recommendations of the companies to use fully formed queries with questions and context. We use two variants for querying the systems: question only and question+narrative.
As we accessed these systems in the first week of May 2020, the systems could be using the latest version of CORD-19 at that time (May 1 release). Thus, we filter the result list, only including those from the April 10 release. We compare the performance of the Amazon and Google systems with the five top submissions to TREC-COVID Round 1 (on the basis of bpref [binary preference] scores). It is valid to compare Amazon and Google systems with the submissions from Round 1 because all these systems are similarly built without using any relevance judgments from TREC-COVID.
Relevance judgments (or assessments) for TREC-COVID are carried out by individuals with biomedical expertise. Pooling is used to select documents for assessment, consisting of the top-ranked results from different submissions. A document is judged as relevant, partially relevant, or not relevant. Since the two evaluated systems did not participate in the pooling, the official TREC-COVID judgments do not include many of their top documents. It has recently been shown that pooling effects can negatively impact post hoc evaluation of systems that did not participate in the pooling.19 Therefore, to create a level ground for comparison, we performed additional relevance assessments for the evaluated systems such that the top 10 documents from all the commercial runs are judged (following the pooling strategy of TREC-COVID for the submitted runs with priority 1). In total, 141 documents were assessed by two individuals involved in performing the relevance judgments for TREC-COVID.
TREC-COVID runs can contain up to 1000 documents per topic. Due to the restrictions imposed by the commercial systems, we could only fetch up to 100 documents per query. This number further decreases when we remove the documents that are not part of the April 10 CORD-19 release. Thus, to ensure a fair comparison, we calculate the minimum number of documents per topic (we call it “topic-minimum”) across the different variations of querying the evaluated systems (i.e., question or question+narrative). We then use this topic-minimum as a threshold for the maximum number of documents per topic for all evaluated systems (both commercial and TREC-COVID). This ensures each system returns the same number of documents for each topic.
We use the standard evaluation measures employed for TREC-COVID: bpref (binary preference), NDCG@10 (normalized discounted cumulative gain with top 10 documents), and P@5 (precision at 5 documents). Here, bpref only uses judged documents in calculation while the other two measures assume the nonjudged documents to be not relevant. Additionally, we calculate MAP (mean average precision), NDCG, and P@10. Note that we can precisely calculate measures that cut the number of documents to 10 since we have ensured that all the evaluated commercial systems have their top 10 documents manually judged.
To better understand system differences, we created different topic and error categories. We use these categories to compare the performance of the four commercial variants and the best run from TREC-COVID. Given the wide variety of TREC-COVID topics, we created two topic categorizations based on research field and function (Table 1). We use P@10 and NDCG@10 for topic-level comparisons to ensure the top 10 documents for all systems are annotated. For error analysis, we pooled all partially relevant and not relevant documents from all five systems’ top 10 documents. This resulted in 660 documents that were additionally annotated with the following error categories: NA to COVID-19 (not applicable); tangential (not relevant at all); partially tangential (not relevant but there is a common link with the topic, eg, quarantine); partially relevant (answers only a part of the topic); and relevant (provides an answer to the topic). We keep partially relevant because the documents previously judged partially relevant may or may not fall into the other error categories. The category relevant is more of an error in the available relevance annotations. More detailed descriptions and examples of these topic and error categories are included in the supplementary material.
All manual annotations created for this evaluation (141 additional relevance annotations and 660 error classifications) are provided in the supplementary material.
RESULTS
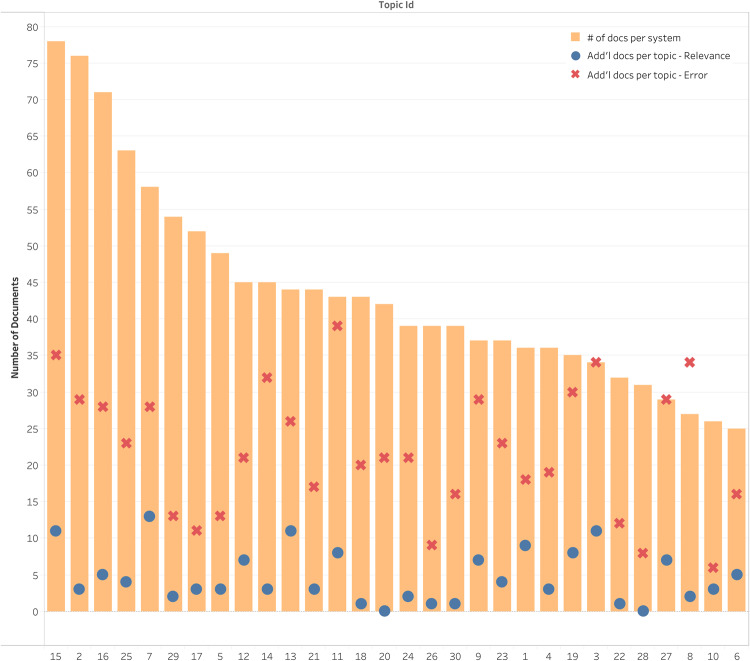
The numbers of documents used for each topic (topic-minimums) are shown in Figure 1. Approximately, an average of 43 documents are evaluated per topic with a median of 40.5. This is another reason for using a topic-wise minimum rather than cutting off all the systems to the same level as the lowest return count (which would be 25 documents). Having a topic-wise cut-off allowed us to evaluate runs with the maximum possible depth while keeping the evaluation fair. The topic-wise count of newly annotated documents for relevance and error analysis is also included in Figure 1.
Figure 1.
A bar chart with the number of documents for each topic as used in our evaluations (after filtering the documents based on the April 10 release of the CORD-19 dataset and setting a threshold at the minimum number of documents for any given topic). The total numbers of documents annotated additionally for relevance and error analysis are shown as circle and cross marks on the bars corresponding to each topic. Note that these additional documents are at topic level and thus can be more than the number of documents per system shown in the figure using bars.
The evaluation results of our study are presented in Table 2. Among the commercial systems that we evaluated, the Amazon question+narrative variant consistently performed better than any other variant in all the measures other than bpref. For bpref, the Google question-only variant performed best. Note that the best run from TREC-COVID (a run from the sabir team), after cutting using topic-minimums, still performed better than the other four TREC-COVID runs included in our evaluation. Interestingly, this best run also performed substantially better than all the variants of both commercial systems on all calculated metrics. We discuss more about this system below.
Table 2.
Evaluation results after setting a threshold at the number of documents per topic using a minimum number of documents present for each individual topic. The relevance judgments used are a combination of Rounds 1 and 2 of TREC-COVID and our additional relevance assessments. The highest scores for the evaluated and TREC-COVID systems are underlined.
| System | P@5 | P@10 | NDCG@10 | MAP | NDCG | bpref | |
|---|---|---|---|---|---|---|---|
| Amazon | question | 0.6733 | 0.6333 | 0.5390 | 0.0722 | 0.1838 | 0.1049 |
| question + narrative | 0.7200 | 0.6400 | 0.5583 | 0.0766 | 0.1862 | 0.1063 | |
| question | 0.5733 | 0.5700 | 0.4972 | 0.0693 | 0.1831 | 0.1069 | |
| question + narrative | 0.6067 | 0.5600 | 0.5112 | 0.0687 | 0.1821 | 0.1054 | |
| TREC-COVID | 1. sab20.1.meta.docs | 0.7800 | 0.7133 | 0.6109 | 0.0999 | 0.2266 | 0.1352 |
| 2. sab20.1.merged | 0.6733 | 0.6433 | 0.5555 | 0.0787 | 0.1971 | 0.1154 | |
| 3. UIowaS_Run3 | 0.6467 | 0.6367 | 0.5466 | 0.0952 | 0.2091 | 0.1279 | |
| 4. smith.rm3 | 0.6467 | 0.6133 | 0.5225 | 0.0914 | 0.2095 | 0.1303 | |
| 5. udel_fang_run3 | 0.6333 | 0.6133 | 0.5398 | 0.0857 | 0.1977 | 0.1187 | |
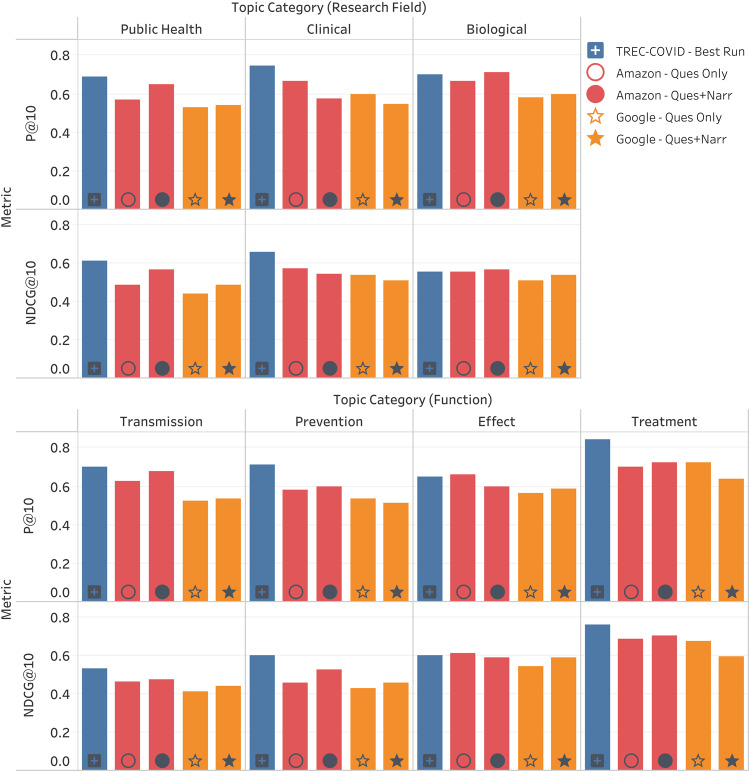
The system performances (of all the commercial runs and the best run from TREC-COVID, referred to here as “sabir”) using some of the standard evaluation metrics as classified by the topic categories are shown in Figure 2. The Amazon system performed better than the Google system on almost all topic categories. In the functional category, all systems performed the best on “Treatment”, whereas among the research field-based categories the best results were different for TREC-COVID and the commercial runs (sabir performed best on the “Clinical” category while most of the commercial variants performed best on “Biological”). Sabir consistently outperformed the commercial system variants on all categories except “Biological” (among the research field categories) and “Effect” (among the function categories), in both of which a commercial system had an edge.
Figure 2.
Analysis of system performances on the basis of different categories of topics. Research Field – categories based on the field of study in biomedical informatics. Function – based on the functional aspect of COVID-19 as expressed in the topic’s information need.
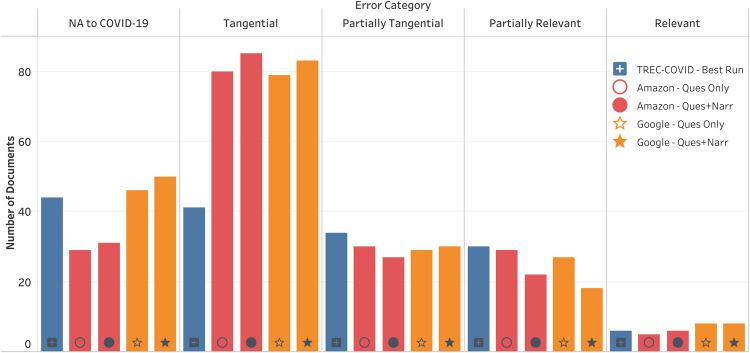
The results from our error analysis are shown in Figure 3. The commercial systems made about twice as many tangential errors as sabir. The commercial variants with the narrative part made slightly more errors in the first three categories than the corresponding variants with only the question. Note that the number of documents annotated as relevant during the error analysis is roughly the same for all the systems (thus not creating an unfair situation for any particular system).
Figure 3.
Total number of documents retrieved by the systems (among the top 10 documents per topic) based on different categories of errors. NA to COVID-19 – document not applicable to COVID-19. Tangential – not relevant at all. Partially Tangential – not relevant but there is a common link with the topic (e.g., quarantine). Partially Relevant – answers only a part of the topic. Relevant – provides an answer to the topic.
DISCUSSION
We evaluated two commercial IR systems targeted toward CORD-19. For comparison, we also included the five best runs from TREC-COVID. We annotated an additional 141 documents from the commercial system runs to ensure a fair comparison with the TREC-COVID runs. We found the best system from TREC-COVID in terms of bpref outperformed all commercial system variants on all evaluated measures. We illustrated the system performances in light of different categories of topics and further annotated a set of 660 documents to conduct an error analysis.
The commercial systems often employ cutting-edge technologies, such as ACM and BERT, as part of their systems. Also, the availability of computational resources such as CPUs and GPUs may be better in industry than in academic settings. This follows a common concern in academia, namely that the resource requirements for advanced machine learning methods (eg, GPT-320) are well beyond the capabilities available to the vast majority of researchers. Instead, these results demonstrate the potential pitfalls of deploying a deep learning-based system without proper tuning. The sabir (sab20.*) system does not use machine learning at all: it is based on the very old SMART system21 and does not utilize any biomedical resources. It is instead manually tuned based on an analysis of the data fields available in CORD-19. Subsequent rounds of TREC-COVID have since overtaken sabir (based indeed on machine learning with relevant training data). The lesson, then, for future emerging health events is that deploying “state-of-the-art” methods without event-specific data may be dangerous, and in the face of uncertainty simple may still be best. On the other hand, the strengths of the commercial systems must be acknowledged: they are capable of serving large numbers of users and can be rapidly disseminated, and while their performance suffers compared with simpler systems, their performance is good enough that they are still likely “useful,” though this term is much debated in IR research.
As evident from Figure 1, many documents retrieved by the commercial systems were not part of the April 10 CORD-19 release. We queried these systems after another version of the CORD-19 dataset was released. This may have led to the retrieval of more articles from the new release of CORD-19. However, the relevance judgments used here are from the initial rounds of TREC-COVID and thus would not include all documents from the latest version of CORD-19. Thus, for a fair comparison, we pruned the document list and performed additional relevance judgments. We have included the evaluation results that would have resulted without our modifications in the supplementary material, which makes the commercial systems look far more inferior. Yet, as addressed, this would not have been a “fair” comparison and thus the corrective measures described above were necessary to ensure a scientifically valid comparison.
CONCLUSION
We evaluated two commercial IR systems against the TREC-COVID data. To facilitate fair comparison, we cut all runs at different thresholds and performed more relevance judgments beyond those provided by TREC-COVID. We found the top performing system from TREC-COVID remained the best-performing system, outperforming the commercial systems on all metrics. Interestingly, this best-performing run comes from a simple system that does not apply machine learning. Thus, blindly applying machine learning without specific labeled data (a condition that may be necessary in a rapidly emerging health crisis) may be detrimental to system performance.
FUNDING
This work was supported in part by the National Science Foundation (NSF) under award OIA-1937136.
AUTHOR CONTRIBUTIONS
KR conceived of the overall project. SS and KR decided upon the experimental design. SS carried out the experiment and drafted the initial manuscript. SS and KR approved the final manuscript.
CONFLICT OF INTEREST STATEMENT
None.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
7The authors thank Meghana Gudala and Jordan Godfrey-Stovall for conducting the additional retrieval assessments.
Footnotes
https://www.semanticscholar.org/cord19
https://cord19.aws
https://covid19-research-explorer.appspot.com
http://bioasq.org
REFERENCES
- 1. Wang LL, Lo K, Chandrasekhar Y, et al. CORD-19: The Covid-19 Open Research Dataset. arXiv: 2004.10706v2 [Published online 2020]. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7251955/ Accessed June 18, 2020
- 2. Roberts K, Alam T, Bedrick S, et al. TREC-COVID: rationale and structure of an information retrieval shared task for COVID-19. J Am Med Inform Assoc 2020; 27: 1431–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Voorhees E, Alam T, Bedrick S, et al. TREC-COVID: constructing a pandemic information retrieval test collection. ACM SIGIR Forum 2020; 54: 1–12. [Google Scholar]
- 4. Kass-Hout TA, Wood M.. Introducing medical language processing with Amazon Comprehend Medical. AWS Mach Learn Blog 2018. https://aws.amazon.com/blogs/machine-learning/introducing-medical-language-processing-with-amazon-comprehend-medical/ Accessed June 25, 2020 [Google Scholar]
- 5. Bhatia P, Celikkaya B, Khalilia M, et al. Comprehend Medical: a named entity recognition and relationship extraction web service. In: 2019 18th IEEE International Conference on Machine Learning and Applications (ICMLA). 2019: 1844–51; Boca Raton, FL. doi: 10.1109/ICMLA.2019.00297
- 6. Guzman B, Metzger I, Aphinyanaphongs Y, et al. Assessment of Amazon Comprehend Medical: medication information extraction. [Published online 2 February 2020]. https://arxiv.org/abs/2002.00481v1 Accessed July 15, 2020
- 7. Heider PM, Obeid JS, Meystre SM.. A comparative analysis of speed and accuracy for three off-the-shelf de-identification tools. AMIA Jt Summits Transl Sci Proc 2020; 2020: 241–50. [PMC free article] [PubMed] [Google Scholar]
- 8. Devlin J, Chang M-W, Lee K, et al. BERT: pre-training of deep bidirectional transformers for language understanding In: proceedings of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. 2019: 4171–86; Minneapolis, MN. doi: 10.18653/v1/N19-1423. [Google Scholar]
- 9. Wu S, Roberts K, Datta S, et al. Deep learning in clinical natural language processing: a methodical review. J Am Med Inform Assoc 2020; 27 (3): 457–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Soni S, Roberts K. Evaluation of dataset selection for pre-training and fine-tuning transformer language models for clinical question answering. In: proceedings of the 12th International Conference on Language Resources and Evaluation. Marseille, France: European Language Resources Association, 2020: 5534–40.
- 11. Lee J, Yoon W, Kim S, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2019; 36 (4): 1–7. doi: 10.1093/bioinformatics/btz682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Alsentzer E, Murphy J, Boag W, et al. Publicly available clinical BERT embeddings. In: proceedings of the 2nd Clinical Natural Language Processing Workshop. Minneapolis, MN: Association for Computational Linguistics; 2019: 72–8.
- 13. Beltagy I, Lo K, Cohan A. SciBERT: a pretrained language model for scientific text. In: proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP). Hong Kong, China: Association for Computational Linguistics; 2019: 3615–3620.
- 14. Nayak P. Understanding searches better than ever before. Google Blog - Search. 2019. https://blog.google/products/search/search-language-understanding-bert/ Accessed July 23, 2020
- 15. Kass-Hout TA, Snively B. AWS launches machine learning enabled search capabilities for COVID-19 dataset. AWS Public Sect Blog. 2020. https://aws.amazon.com/blogs/publicsector/aws-launches-machine-learning-enabled-search-capabilities-covid-19-dataset/ Accessed June 18, 2020
- 16. Hall K. An NLU-powered tool to explore COVID-19 scientific literature. Google AI Blog. 2020. http://ai.googleblog.com/2020/05/an-nlu-powered-tool-to-explore-covid-19.html Accessed June 18, 2020
- 17. Ma J, Korotkov I, Yang Y, et al. Zero-shot neural retrieval via domain-targeted synthetic query generation. ArXiv2004.14503 Cs [Published online April 29, 2020]. http://arxiv.org/abs/2004.14503 Accessed June 25, 2020
- 18. Jiang Z, Zhang C, Talwar K, et al. Characterizing structural regularities of labeled data in overparameterized models. ArXiv2002.03206 Cs Stat [Published online June 10, 2020]. http://arxiv.org/abs/2002.03206 Accessed June 18, 2020
- 19. Yilmaz E, Craswell N, Mitra B, et al. On the reliability of test collections for evaluating systems of different types. In: proceedings of the 43rd International ACM SIGIR Conference on Research and Development in Information Retrieval. 2020.
- 20. Brown TB, Mann B, Ryder N, et al. Language models are few-shot learners. ArXiv2005.14165 Cs [Published online June 4, 2020]. http://arxiv.org/abs/2005.14165 Accessed July 2, 2020
- 21. Buckley C. Implementation of the SMART information retrieval system. Department of Computer Science, Cornell University; 1985. https://ecommons.cornell.edu/bitstream/handle/1813/6526/85-686.pdf
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.