Abstract
Objective
To determine shared comorbidities and to identify underrecognized or unexpected morbidities in children with leukodystrophies using an unbiased phenome-wide association study (PheWAS) analysis of a nationwide pediatric clinical and financial database.
Methods
Data were extracted from the Pediatric Health Information System database. Patients with leukodystrophy were identified with International Classification of Diseases, 10th revision, clinical modification, diagnostic codes for any of 4 specific leukodystrophies (X-linked adrenoleukodystrophy (E71.52x), Hurler disease (E76.01), Krabbe disease (E75.23), and metachromatic leukodystrophy (E75.25)) over a 3-year time period. Confirmed leukodystrophy cases (n = 553) were matched with 1659 controls. A PheWAS analysis was performed on all available ICD diagnostic codes for cases and controls. Comparisons were performed for all 4 leukodystrophies as a group and individually.
Results
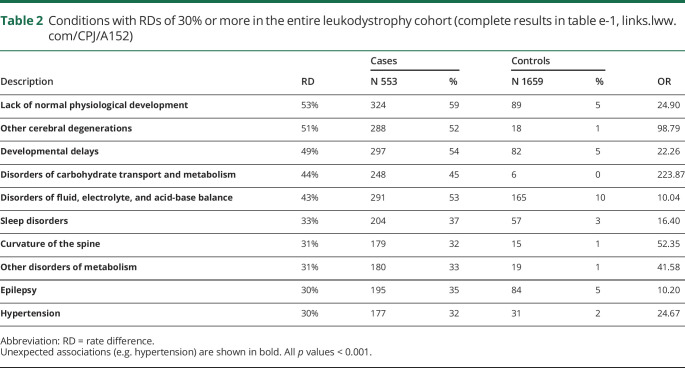
We found 174 phecodes (grouped ICD codes) associated with leukodystrophies, including 28 codes with a rate difference (RD) > 20%. Known comorbidities of leukodystrophies including developmental delay, epilepsy, and adrenal insufficiency were identified. Unexpected associations identified included hypertension (RD 30%, OR 25), hearing loss (RD 28%, OR 15), and cardiac dysrhythmias (RD 27%, OR 9). Hurler disease had a greater number of unique disease conditions.
Conclusions
PheWAS analysis from a national database demonstrates shared and unique features of leukodystrophies. Developmental delay, cardiac dysrhythmias, fluid and electrolyte disturbances, and respiratory issues were common to all 4 leukodystrophy diseases. Use of a PheWAS in leukodystrophies and other pediatric neurologic diseases offers a method for targeting improved care for patients by identification of morbidities.
Leukodystrophies are genetic diseases of the myelin that affect more than 1 in 7,500 live births with a 30% mortality by the age of 8 years.1–4 Leukodystrophies have a broad range of severe morbidities with extensive health care requirements,2,5,6 but sparse data have hampered treatment7–9 and limited clinical trial development.
The relative rarity of individual leukodystrophies, and the absence of a central data registry, has limited understanding of leukodystrophy morbidities. It is not known whether most leukodystrophies share common disease features, whether there are shared patterns of medical complications, and whether shared disease pathophysiology could be targeted for treatment.
The relative rarity of individual leukodystrophies, and absence of a central data registry, has limited understanding of leukodystrophy morbidities.
Recently, approaches using national electronic health records (EHRs) have been used to identify health care burden in leukodystrophies.6 Furthermore, with the advent of disease-specific codes for individual leukodystrophies in the International Classification of Diseases, 10th edition, clinical modification (ICD-10-CM), more granularity in diagnosis has also become feasible. A novel method for understanding disease-associated morbidities has been the development of an algorithm to determine the association of all disease “phenotypes” with a specified genotype or diagnosis.10,11 Using this method, a phenome-wide association study (PheWAS) can test for all ICD-9-CM and ICD-10-CM codes associated with a specific diagnosis (such as leukodystrophy) in the EHR. An EHR-based PheWAS has been shown to identify known associations across various disease domains and has the ability to reveal novel associations.12,13
Our objective was to determine the health conditions associated with hospitalization in patients with leukodystrophy and to identify shared and unique patterns of medical morbidities.
Methods
We conducted a retrospective case-control matched study of patients admitted to a Pediatric Health Information System (PHIS) hospital with an International Classification of Diseases, 10th Revision, clinical modification (ICD-10-CM) diagnosis of one of the following leukodystrophies: metachromatic leukodystrophy (MLD) (E75.25), X-linked adrenoleukodystrophy (ALD) (E71.52x), Krabbe disease (E75.23), and Hurler disease (E76.01). Patient identification was between October 1, 2015, through March 31, 2018.
The PHIS database contains information from 52 children's hospitals in the United States.14 Hospitals are affiliated with the Children's Hospital Association (Lenexa, KS). Although deidentified, the PHIS retains a patient-specific identifier, permitting tracking of patient medical care and use. Data collected includes patient and visit demographics such as age, sex, race, insurance type, type of visit, estimated total visit cost, length of stay, ICD coding, and detailed charge information. The hospital visits included primarily inpatient admissions with additional visit types such as emergency department, observation, ambulatory surgery, and clinic and other visit types where available.
Our objective was to determine the health conditions associated with hospitalization in leukodystrophy patients and to identify shared patterns of medical morbidities.
Patients with leukodystrophy were identified from any PHIS visit (inpatient, observation, emergency department, ambulatory surgery, and clinic visit) as having an ICD-10-CM code for one of the 4 leukodystrophies. Each patient with leukodystrophy was matched to 3 control patients. The control patients were identified from the PHIS during the same time period and did not have a leukodystrophy ICD-10-CM code. Matching was performed using patient demographics available at the first PHIS visit, using propensity score matching. The demographic variables used for matching were hospital, sex, race and ethnicity grouping, payer grouping, and patient zip code information (urban/rural, median 2010 household income). For transplanted patients with leukodystrophy, their controls consisted of patients receiving a hematopoietic stem cell transplant who did not have a leukodystrophy.
Data collected on the cases and controls included demographics at the first available PHIS visit in the collection time frame (quarter 4 of 2015 and onward) and a summary of all available PHIS visits (including visits before the patient identification time window and clinic and other visit types). ICD diagnostic codes available both before and during the patient identification window (both ICD-9-CM and ICD-10-CM) and at all PHIS visit types were also collected.
PheWAS was performed using the PheWAS R package (github.com/PheWAS/PheWAS/tree/v1.0). The ICD codes were crosswalked to phecodes using both an ICD-9-CM and ICD-10-CM crosswalk provided through the PheWAS catalog and the team at Vanderbilt University Medical Center. We truncated all available phecodes to 3–4 digits before a decimal for evaluation. We used the Fisher exact test to compare the number of patients in the case vs control groups with at least one instance of each truncated phecode. The phecodes were evaluated between cases and controls for the following 6 groups: overall patients with leukodystrophy, patients with leukodystrophy excluding any patients with a transplant, Hurler, ALD, MLD, and Krabbe. These results are represented using Manhattan plots showing the −log10(p-value) and the rate difference (RD) (control rate subtracted from case rate).
Statistical analysis
Analyses were performed with SAS 9.4 (SAS Institute, Cary, NC) using the LOGISTIC procedure for propensity score matching and in R 3.3.2 using the PheWAS package. All analyses were two sided, and a Bonferroni adjusted p value < 0.05 was considered statistically significant.
Standard protocol approvals, registrations, and patient consents
This project used deidentified data and was not considered human subjects research and was exempted by the Institutional Review Boards at the University of Utah and the Privacy Board of Intermountain Healthcare.
Data availability
All data reported in this study are published with this study.
Results
We identified 553 patients with one of 4 specific leukodystrophies in the PHIS database over the nearly 3-year study time period and 1,659 matched controls (table 1). The 4 leukodystrophies (X-linked ALD, Hurler disease, Krabbe disease, and MLD) were selected because their ICD-10-CM codes are specific to that individual leukodystrophy genetic diagnosis. By contrast, other leukodystrophies coded using different ICD-9-CM or ICD-10-CM codes such as “sphingolipidoses” (E75.3) are nonspecific6 and include a large range of genetically discrete disorders, such as vanishing white matter disease, Canavan Disease, etc.
Table 1.
Selected demographic characteristics of patients with leukodystrophy and controls in the PHIS used in PheWAS analysis
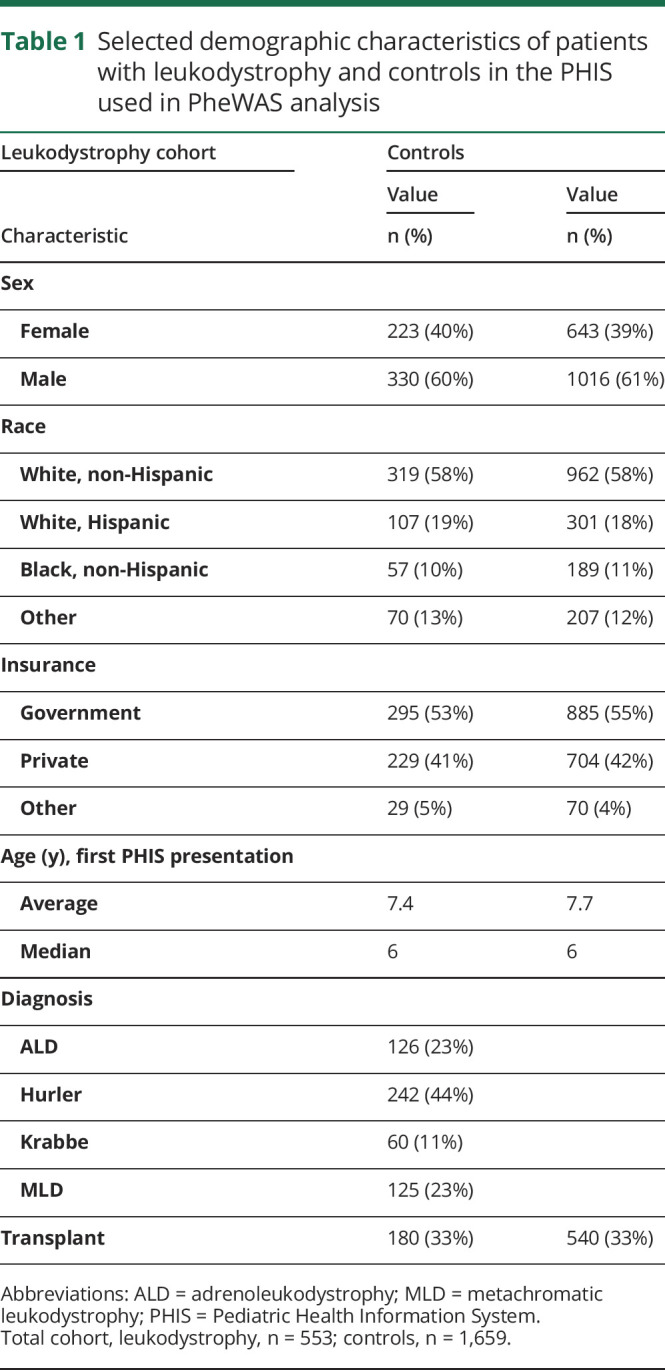
The cohort was composed of 330 men (60%), with a median age of 6 years (average age 7.4 years); the most common racial groups represented were White non-Hispanic, White Hispanic, and Black non-Hispanic (319 (58%), 107 (19%), and 57 (10%) patients, respectively,). The cohort included 126 patients with ALD (23%), 242 cases of Hurler disease (44%), 60 cases of Krabbe disease (11%), and 125 cases of MLD (23%). Hundred eighty patients (33%) had a transplant. Two hundred twenty-nine patients (41%) had private insurance.
We adopted a PheWAS algorithm11 for use of ICD-9-CM and ICD-10-CM codes and for evaluation of the PHIS EHR data. The PheWAS algorithm categorizes each ICD-10-CM code into one of 1866 PHeWAS “phecodes” (typically similar diseases or traits), which are further categorized into 17 phenotypic disease category groups, such as “dermatologic” or “neurologic.” The proportion of patients with each of the phecodes was compared between patients with leukodystrophy and controls using the Fisher exact test.
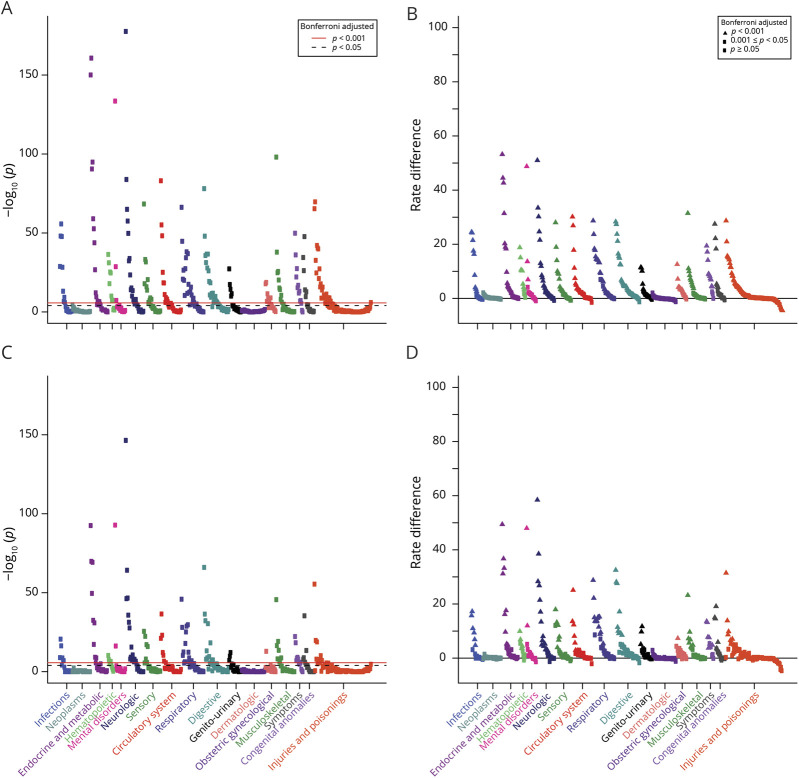
In the entire cohort of patients with leukodystrophy, 497 phecodes were identified, of which 174 were associated with leukodystrophy after Bonferroni adjustment (figure 1A; table e-1, links.lww.com/CPJ/A152). We next evaluated the RD between leukodystrophy cases and controls (figure 1B). We observed significant differences across most disease phecode groups. Disease conditions typically associated with leukodystrophies such as “developmental delay,” “epilepsy,” and infections15 were observed (table 2; table e-1, links.lww.com/CPJ/A152). However, we also noted unexpected associations including hypertension (RD 30%, OR 25), hearing loss (RD 28%, OR 15), and cardiac dysrhythmias (RD 27%, OR 9).
Figure 1. PheWAS plots.
Each panel represents 1,645 phenotypes tested for association using logistic regression assuming a model adjusted for age, sex, insurance type, and location. Phenotypes are grouped along the x-axis by categorization within the PheWAS code hierarchy. The upper red lines indicate p < 0.001 (Bonferroni false discovery rate = 0.1 for the entire PheWAS); lower black lines indicate p < 0.05. (A) Phecodes for the entire leukodystrophy cohort, displayed by p value. (B) Phecodes for the entire leukodystrophy cohort, displayed by RD. (C) Phecodes for the nontransplant leukodystrophy cohort, displayed by p value. (D) Phecodes for the nontransplant leukodystrophy cohort, displayed by rate difference. PheWAS = phenome-wide association study; RD = rate difference.
Table 2.
Conditions with RDs of 30% or more in the entire leukodystrophy cohort (complete results in table e-1, links.lww.com/CPJ/A152)
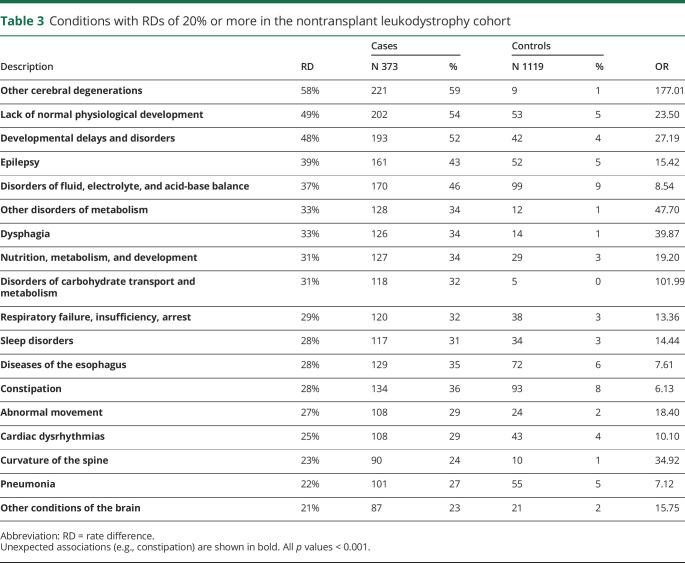
Some diseases detected in the analysis included conditions associated with transplantation, such as graft-vs-host disease. To evaluate whether the pattern and frequency of morbidities identified with PheWAS were primarily linked to transplant or to leukodystrophies, we performed a PheWAS evaluation in which we excluded patients who had received transplantation. We found a very similar pattern of disease group involvement and associations (figure 1, C and D; table e-2, links.lww.com/CPJ/A153; table 3). However, we did find a decrease in the overall number of conditions with a RD of 20% or greater.
Table 3.
Conditions with RDs of 20% or more in the nontransplant leukodystrophy cohort
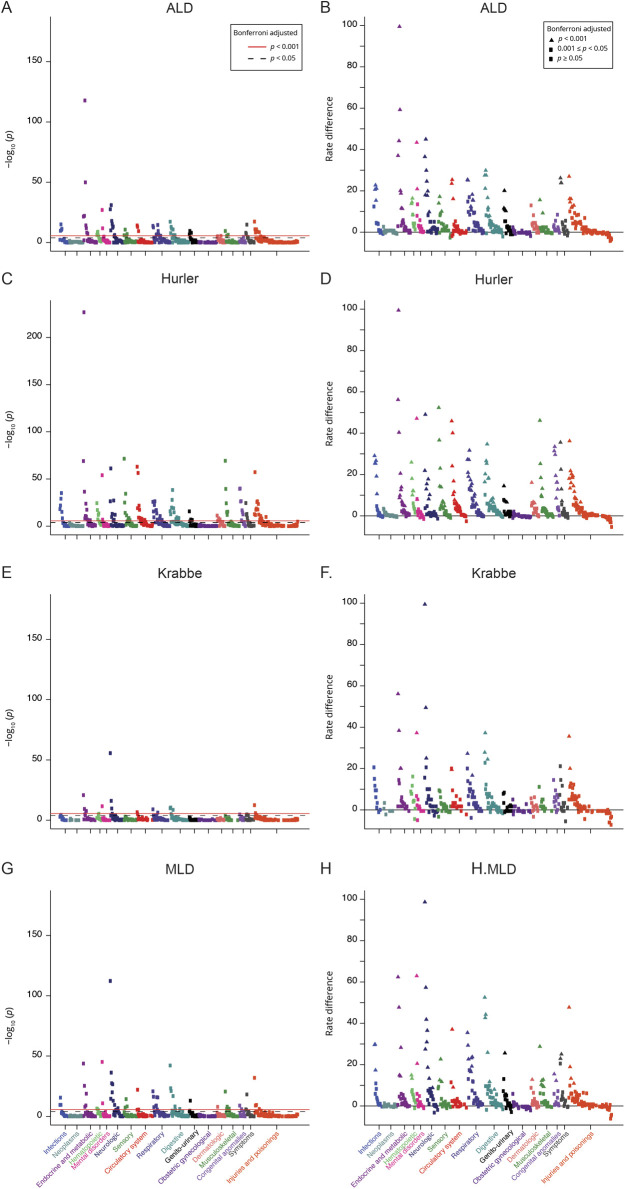
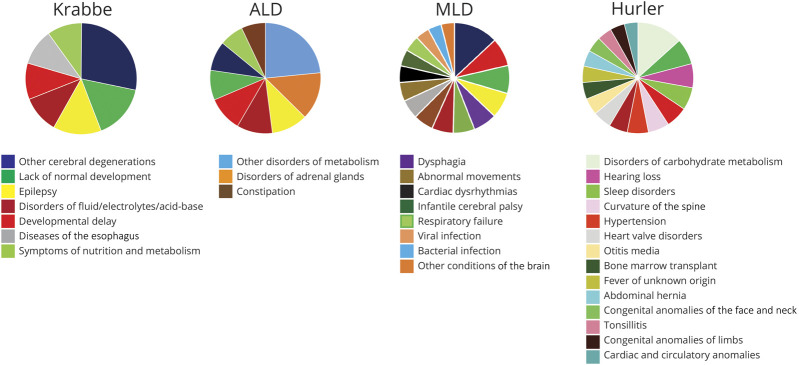
To determine whether distinct leukodystrophy diseases had unique morbidities, we performed individual PheWAS analysis of patients with ALD, Hurler, Krabbe, or MLD (figures 2 and 3; tables e3-e6, links.lww.com/CPJ/A154, links.lww.com/CPJ/A155, links.lww.com/CPJ/A156, links.lww.com/CPJ/A157). We found that all 4 of the leukodystrophies shared many common disease features, including expected features such as developmental delay and lack of normal physiological development. However, unexpected shared features included cardiac dysrhythmias, diseases of the esophagus, and nausea and vomiting. Other features which were commonly but not universally shared included epilepsy, various infection-related diagnoses, and constipation. Finally, some of the disorders had unique disease codes, such as infantile cerebral palsy for MLD or disorders of adrenal glands for ALD. Hurler disease in particular had more disease codes that were distinct from the other 3 leukodystrophies, including hearing loss, heart valve disorders, and abdominal hernia.
Figure 2. PheWAS plots for individual leukodystrophies.
Each panel represents 1,645 phenotypes tested for association, using logistic regression assuming a model adjusted for age, sex, insurance type, and location. Phenotypes are grouped along the x-axis by categorization within the PheWAS code hierarchy. The upper red lines indicate p < 0.001 (Bonferroni false discovery rate = 0.1 for the entire PheWAS); lower black lines indicate p < 0.05. Left panels, y-axis, p value; right panels, y-axis, RD. (A and B) Phecodes for ALD. (C and D) Phecodes for Hurler disease. E and F. Phecodes for Krabbe disease. G and H. Phecodes for MLD. ALD = adrenoleukodystrophy; MLD = metachromatic leukodystrophy; PheWAS = phenome-wide association study; RD = rate difference.
Figure 3. Conditions with RDs of 30% or more in the individual leukodystrophies, displayed by leukodystrophy in the order of increasing number of conditions (phecodes).
Values reported in tables e3-e6, links.lww.com/CPJ/A154, links.lww.com/CPJ/A155, links.lww.com/CPJ/A156, links.lww.com/CPJ/A157. All p values < 0.001. Phecodes are color-coded for all charts; phecode (and its color in the pie chart) is only listed the first time it is reported. RD = rate difference.
Discussion
We performed a national evaluation of medical morbidities in leukodystrophies. This PheWAS analysis of a national pediatric database demonstrates shared and unique disease features of leukodystrophies. All 4 of the leukodystrophies shared disease features, including expected features such as developmental delay and lack of normal physiological development. However, unexpected features which included cardiac dysrhythmias, diseases of the esophagus, and nausea and vomiting were also observed in all 4 leukodystrophies. These features suggest a need to evaluate cardiac rhythm features in patients with leukodystrophy and presumably that gastrointestinal issues, for example, reflux, are a more problematic feature than typically considered. Fluid and electrolyte disturbances, presumably during the course of hospitalization, were also significantly observed in all 4 leukodystrophies.
Other features were commonly but not universally shared including epilepsy, various infection-related diagnoses, and constipation. Infection-related issues are a known morbidity in leukodystrophies and can be amenable to intervention.15 Finally, some of the leukodystrophies had unique disease associations, such as infantile cerebral palsy for MLD or disorders of adrenal glands for ALD. Hurler disease in particular had more disease codes that were distinct from the other 3 leukodystrophies, including hearing loss, heart valve disorders, and abdominal hernia. This suggests that the more extensive multiorgan involvement in Hurler disease leads to a different pattern of morbidities than the predominant “CNS-only” complications of ALD, Krabbe disease, and MLD. Similarly, the greater number of significant morbidities observed in MLD might reflect that more patients received bone marrow transplantation, as compared to patients with Krabbe or ALD.
Identification of these morbidities could assist with designing patient treatment strategies and with guiding clinical trial design. Although promising treatments and cures for several leukodystrophies appear to be nearing clinical use,16,17 even in the absence of a cure, improvements in patient care can be accomplished by targeting known morbidities.18 This could help with the burden of hospitalizations in leukodystrophy: patients with leukodystrophy in children's hospitals account for $59 million per year in total costs. Individually, their treatment and care cost is 5 times the average cost for a hospitalized child.6
Specific interventions that could be considered based on the findings here could include targeted epilepsy education for families of patients with leukodystrophy, based on the observed morbidity of epilepsy; proactive treatment of constipation; and determining the issues regarding nutrition because this was common to all 4 leukodystrophies. Thus, although some of the morbidities are nonspecific to leukodystrophies, the findings offer potential for interventions that might have a meaningful impact on a patient and their family. Furthermore, treatments or care plans could be assessed for their impact on quality of life, in which symptoms such as constipation could be followed to determine whether there is improvement for patients.
Strength of this work is the use of leukodystrophy disease-specific ICD-10-CM codes. This work would not have been feasible previously because ICD-9-CM codes have low specificity to the diagnosis of an inherited leukodystrophy and complex algorithms had to be used with ICD-9-CM codes to track leukodystrophies.6 With the use of ICD-10-CM codes for ALD, MLD, Krabbe, and Hurler, we were able to track and compare specific leukodystrophies. Finally, the use of the PHIS national database provides a more comprehensive view of trends across the United States for these relatively rare diseases. Coding of PheWAS is relatively specific to the morbidity. For example, “cardiac dysrhythmias” does not indicate that an EKG was performed, but rather that a cardiac rhythm disturbance such as atrial fibrillation was present.10
Limitations include use of retrospective data and the limited temporal duration of data collection. The time limitation was necessary because the use of ICD-10-CM has only been over the past 3 years, and ICD-10-CM code use was necessary to have sufficient specificity in determining diagnoses. Furthermore, even with the use of national databases, in some instances, there were very few patients experiencing certain morbidities, thus limiting the statistical power of PheWAS. This study was not organized in a fashion to collect data on all patients with leukodystrophy with a diagnosis of ALD, MLD, Krabbe, or Hurler, and we cannot use the data to determine incidences or prevalences. However, one advantage is that it includes data on any patient with a given diagnosis during the enrollment time frame. That is, even if a patient with Krabbe was born in 2013 before the study and if they came to the hospital in 2016, they were still included in our analysis.
This work demonstrates a broad extent of shared morbidities in children with leukodystrophies. With this same methodology, many avenues of investigation are possible. For example, to investigate whether different commonly used medications in different leukodystrophies associate with the presence or absence of comorbidities, which could provide information about improved medicine choice for patients with leukodystrophy. The scope of the work was limited in that only 4 leukodystrophies could be specifically examined, and future efforts should include the adoption of more specific ICD codes for leukodystrophies. It seems countersensical that specific ICD-10-CM codes exist for “sucked into jet engine” and “injury at library,” but not to distinguish Canavan disease from vanishing white matter disease, for example. In conclusion, use of PheWAS in leukodystrophies and other pediatric neurologic diseases offers a method for characterizing disease burden and targeting improved care for patients.
Acknowledgment
We appreciate the advice and suggestions of R.J. Carroll, J.C. Denny, M. Hall, and T. Richardson.
Appendix. Authors
Footnotes
Editorial, page 375
Study funding
J.L. Bonkowsky was supported by NIH grant 3UL1TR002538 and by the Bray Presidential Chair in Child Neurology research. This investigation was supported by the University of Utah Study Design and Biostatistics Center, with funding in part from the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant 8UL1TR000105 (formerly UL1RR025764).
Disclosure
J.L. Bonkowsky was supported by NIH grant 3UL1TR002538 and by the Bray Presidential Chair in Child Neurology research; has served as a consultant to Bluebird Bio, Inc., Calico, Inc., Neurogene, Inc.; is on the Board of Directors of wFluidx, Inc.; and owns stock in Orchard Therapeutics. J. Wilkes reports no disclosures. J. Ying was supported by the University of Utah Study Design and Biostatistics Center, with funding in part from the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant 8UL1TR000105 (formerly UL1RR025764). W.-Q. Wei was supported by NHLBI R01HL133786. Full disclosure form information provided by the authors is available with the full text of this article at Neurology.org/cp.
TAKE-HOME POINTS
→ We performed a PheWAS analysis of leukodystrophy comorbidities using a national database.
→ Developmental delay, epilepsy, fluid and electrolyte disturbances, and respiratory issues were shared morbidities in leukodystrophies.
→ PheWAS identified unexpected morbidities including hypertension, hearing loss, and cardiac dysrhythmias.
→ Information from PheWAS analysis provides targets for clinical treatments and for following outcomes in patients with leukodystrophy.
References
- 1.Heim P, Claussen M, Hoffmann B, et al. Leukodystrophy incidence in Germany. Am J Med Genet 1997;71:475–478. [PubMed] [Google Scholar]
- 2.Bonkowsky JL, Nelson C, Kingston JL, Filloux FM, Mundorff MB, Srivastava R. The burden of inherited leukodystrophies in children. Neurology 2010;75:718–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hult M, Darin N, von Döbeln U, Månsson JE. Epidemiology of lysosomal storage diseases in Sweden. Acta Paediatr 2014;103:1258–1263. [DOI] [PubMed] [Google Scholar]
- 4.Ashrafi MR, Rezaei Z, Heidari M, et al. The first report of relative incidence of inherited white matter disorders in an Asian Country based on an Iranian Bioregistry System. J Child Neurol 2018;33:255–259. [DOI] [PubMed] [Google Scholar]
- 5.Nelson C, Mundorff MB, Korgenski EK, Brimley CJ, Srivastava R, Bonkowsky JL. Determinants of health care use in a population-based leukodystrophy cohort. J Pediatr 2013;162:624–e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brimley CJ, Lopez J, van Haren K, et al. National variation in costs and mortality for leukodystrophy patients in US children's hospitals. Pediatr Neurol 2013;49:156–162 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holmes L, Cornes MJ, Foldi B, Miller F, Dabney K. Clinical epidemiologic characterization of orthopaedic and neurological manifestations in children with leukodystrophies. J Pediatr Orthop 2011;31:587–593. [DOI] [PubMed] [Google Scholar]
- 8.Helman G, Van Haren K, Bonkowsky JL, et al. Disease specific therapies in leukodystrophies and leukoencephalopathies. Mol Genet Metab 2015;114:527–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adang LA, Sherbini O, Ball L, et al. Revised consensus statement on the preventive and symptomatic care of patients with leukodystrophies. Mol Genet Metab 2017;122:18–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denny JC, Ritchie MD, Basford MA, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics 2010;26:1205–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denny JC, Bastarache L, Ritchie MD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol 2013;31:1102–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carroll RJ, Bastarache L, Denny JC. RPheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics 2014;30:2375–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei WQ, Bastarache LA, Carroll RJ, et al. Evaluating phecodes, clinical classification software, and ICD-9-CM codes for phenome-wide association studies in the electronic health record. PLoS One 2017;12:e0175508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kittle K, Currier K, Dyk L, Newman K. Using a pediatric database to drive quality improvement. Semin Pediatr Surg 2002;11:60–63. [DOI] [PubMed] [Google Scholar]
- 15.Anderson HM, Wilkes J, Korgenski EK, et al. Preventable infections in children with leukodystrophy. Ann Clin Transl Neurol 2014;1:370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sessa M, Lorioli L, Fumagalli F, et al. Lentiviral haemopoietic stem-cell gene therapy in early-onset metachromatic leukodystrophy: an ad-hoc analysis of a non-randomised, open-label, phase 1/2 trial. Lancet 2016;388:476–487. [DOI] [PubMed] [Google Scholar]
- 17.Eichler F, Duncan C, Musolino PL, et al. Hematopoietic stem-cell gene therapy for cerebral adrenoleukodystrophy. N Engl J Med 2017;377:1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Haren K, Bonkowsky JL, Bernard G, et al. Consensus statement on preventive and symptomatic care of leukodystrophy patients. Mol Genet Metab 2015;114:516–526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data reported in this study are published with this study.