ABSTRACT
Introduction: Antibiotic resistance is widely found even among bacterial populations not having been exposed to selective pressure by antibiotics, such as tetracycline. In this study we analyzed the tetracycline-resistant subgingival microbiota of healthy subjects and of patients with periodontitis, comparing the prevalence of tet genes and their multidrug resistance profiles.
Methods: Samples from 259 volunteers were analyzed, obtaining 813 tetracycline-resistant isolates. The prevalence of 12 antibiotic resistance genes was assessed, and multidrug profiles were built. Each isolate was identified by 16S rRNA sequencing. Differences in qualitative data and quantitative data were evaluated using the chi-square test and the Mann-Whitney-U test, respectively.
Results: tet(M) was the most frequently detected tet gene (52.03%). We observed significant differences between the prevalence of tet(M), tet(W), tet(O), tet(32) and tet(L) in both populations studied. Multidrug resistance was largely observed, with resistance to kanamycin being the most detected (83.64%). There were significant differences between the populations in the prevalence of kanamycin, chloramphenicol, and cefotaxime resistance. Resistant isolates showed significantly different prevalence between the two studied groups.
Conclusion: The high prevalence of multidrug resistance and tetracycline resistance genes found in the subgingival microbiota, highlights the importance of performing wider and more in-depth analysis of antibiotic resistance in the oral microbiota.
KEYWORDS: Antibiotic resistance, tetracycline, periodontitis, subgingival microbiota, multidrug resistance
Introduction
Antibiotics have been used in clinical practice since their discovery, saving countless lives. Tetracycline was one of the first antibiotics to be discovered, in the 1940s, and its broad-spectrum activity and relatively few side effects made it a widely used antibiotic [1]. Its appealing properties led to extensive use, exerting a great deal of selective pressure on bacteria, which did not take long to become resistant [2]. Despite the rise of resistance, tetracyclines were and still are used in the treatment of some human infections [3], and especially in the cattle industry as growth promoters due to their anti-inflammatory effects on the gastrointestinal tract of the animals [4]. It has been reported that the use of antimicrobials in the cattle industry has an impact on the microbiota that lives in soils or sediments where all the wastes of such industry are left, increasing the prevalence of antimicrobial resistance genes in bacterial populations that had previously had little or no contact with antibiotics [5]. This abuse and misuse of antimicrobials may have repercussions on human health, increasing people’s chances of carrying antimicrobial-resistant bacteria without having been exposed to the antimicrobials in question. Such is the case of the oral environment, in which bacteria carrying tetracycline resistance genes have been previously detected [6,7]. Antibiotics have been used as adjuvants in certain dental treatments such as the treatment of periodontitis, which has been reported to benefit from the action of antimicrobials [8,9]. Currently, the main antibiotics used in periodontal treatment are amoxicillin and metronidazole [10], however, tetracyclines have been widely used in the past and there are reports of tetracycline resistance in the oral microbiota [7,11].
Resistance to tetracycline can be expressed through efflux pumps, ribosomal protection proteins (RPPs) and inactivation enzymes. Tetracycline resistance genes code for these mechanisms and are widespread among oral bacteria, where tet(M) is the most common [1]. Other genes coding for RPPs have been described in the oral environment, such as tet(Q), tet(O), tet(S) and tet(W), and genes that code for efflux pumps such as tet(B), tet(32), tet(K) and tet(L) can also be found within the oral microbiota [1]. Several genes, including tet(37) and tet(X), have been described as genes coding for inactivating enzymes, but so far only tet(37) has been found in the oral environment [12,13].
Furthermore, the strong presence of Tn916/1545 family transposons in the oral microbiota, which can carry tetracycline, macrolide and/or aminoglycoside resistance genes, among others [14], means that those tetracycline-resistant microorganisms may also be resistant to other antimicrobials. Multidrug resistance is an increasingly troubling issue for the health authorities [15], and although multiple efforts are being made to provide more information, little is known regarding multidrug resistance in the oral environment.
It has been widely described that the subgingival microbiota of patients with periodontitis differs from that of healthy subjects [16] and therefore, the prevalence of antimicrobial resistance genes and the prevalence of multidrug resistance among these bacterial populations could be different.
The use of antibiotics from the past to treat infections caused by antibiotic-resistant bacteria has been proposed, on the grounds that due to the lack of selective pressure, the characteristics that once conferred resistance might have disappeared [17]. This might be the case with tetracyclines, whose use has declined over the years. There is limited knowledge regarding the distribution of tetracycline resistance in the subgingival microbiota, and the few papers that address this topic are outdated or focus on either particular genes or specific microorganisms. Therefore, the objective of this study was to analyze the prevalence and distribution of 11 tet genes and the multidrug resistance profiles of the tetracycline-resistant subgingival microbiota isolated from healthy volunteers and patients with periodontitis.
Materials and methods
Sample collection and culture
The samples included in this work were part of two previous studies [18,19], whose research protocols were approved by the Ethics Committee of the Universitat Internacional de Catalunya (UIC), (Barcelona, Spain) with study numbers: PER-ECL-2011-06-NF and ODO-2014-01. Both research protocols complied with the principles of the Declaration of Helsinki. Subgingival samples were taken from 259 volunteers including 129 periodontally healthy subjects and 130 subjects with periodontitis. Samples were obtained at the Department of Periodontology of the UIC. All volunteers signed an Institutional review board-approved informed consent form. Patients with periodontitis were diagnosed with generalized severe chronic periodontitis [20] or stage III or IV generalized grade B or C periodontitis [21]. To be included in the study, periodontally healthy subjects had to have at least six teeth per quadrant, probing depths ≤ 3 mm and absence of moderate or severe gingivitis. Smokers of more than five cigarettes per day, wearers of orthodontic appliances and pregnant or breastfeeding women were not included in the study. None of the volunteers took antibiotics or nonsteroidal anti-inflammatory drugs at least three months prior to the sampling, presented any systemic disease or took any chronic medication.
Subgingival samples were taken by placing two sterile paper points in the deepest site of each quadrant for 20 seconds and stored in 2 ml of reduced transport fluid without ethylenediaminetetraacetic acid (EDTA) [22]. Vials with the paper points were sent to the laboratory at 4°C to be processed within the same day. Subgingival samples were dispersed by vortex for 60 seconds. Serial dilutions of each sample were plated on blood agar plates (Blood agar base No. 2; Oxoid Ltd, Basingstoke, UK) containing 5% horse blood, hemin (5 mg/L) and menadione (1 mg/L) and on blood agar plates with and without 8 μg/ml tetracycline. All plates were incubated under anaerobic conditions (10% H2, 10% CO2 and 80% N2) at 37°C for 72 h. All of the morphologically different colonies were isolated and re-plated to obtain pure cultures that were preserved at −80°C in a 30% glycerol solution.
In vitro antibiotic resistance testing
Resistance to six other antibiotics was tested using blood agar plates containing 1 μg/ml of erythromycin (ERY), 64 μg/ml of kanamycin (KAN), 8 μg/ml of chloramphenicol (CHL), 128 μg/ml of streptomycin (STR), 2 μg/ml of cefotaxime (CTX), and 8 μg/ml of amoxicillin (AMX) (all antimicrobials were obtained as pure powder from Sigma Aldrich, St. Louis, MO, USA). Antimicrobial breakpoint concentrations were set according to the recommendations of the Clinical and Laboratory Standards Institute (CLSI) [23] and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) [24]. However, most of the species present in the oral environment are not covered by any of these organizations. Therefore, antibiotic concentrations were chosen based on taxonomic relatedness to oral bacteria, using the higher concentration of antibiotics when in doubt (Table S1). Incubation was performed at 37°C under anaerobic conditions for 72 h.
DNA extraction
DNA extraction was performed on each isolate using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions with some modifications. Cells were suspended in 180 μl of a 20 mg/ml lysozyme solution (20 mM Tris-Hcl, pH 8.0, 2 mM EDTA, 1.2% Triton X-100) and incubated for 30 min at 36°C. Then, 200 μl of Buffer AL (provided in the kit), 10 μl of RNase A (20 mg/ml) and 10 μl of proteinase K (20 mg/ml) were added and incubated for 30 min at 56°C. Further steps were conducted according to the manufacturer’s protocol. Lastly, DNA was re-suspended in 100 μl of buffer AE (provided in the kit), visualized in a 0.5% agarose gel and quantified using a Nanodrop 2000 C UV-vis spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA).
Sequencing of the 16S rRNA gene
The 16S rRNA gene was amplified by the polymerase chain reaction (PCR) using universal primers 27 F and 1544 R (Table 1). PCR amplification was carried out applying a T3000 Thermocycler (Biometra, Goettingen, Germany) under the following conditions: 5 min at 95°C, followed by 35 cycles at 95°C for 60 sec, 57°C for 60 sec and 72°C for 60 sec; followed by another 10 min at 72°C. Sequencing of the gene 16S rRNA was performed in Macrogen, Inc. (Amsterdam, The Netherlands). The sequences obtained were aligned to form a single contig and were identified by comparison with those available at the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov) using BLAST software (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Only sequences with > 99% of similarity were accepted to identify isolates at species level and the 16S rRNA sequences were deposited in GenBank (accession numbers MT807114-MT807900).
Table 1.
List of primers used in this study
| Primers | Sequence 5ʹ-3’ | Size (bp) | Tm | Tm Multiplex | Multiplex group | Ref. |
|---|---|---|---|---|---|---|
| tetMF tetMR |
GCG TAC AAG CAC AGA CTC GT AGC CAT AGC GTA TCC CCT CC |
1142 | 61 | 64 | 1 | [6] |
| tetWF tetWR |
GAG AGC CTG CTA TAT GCC AGC GGG CGT ATC CAC AAT GTT AAC |
168 | 64 | [57] | ||
| intF intR |
GGC TAC AGA CCG AGT ACC AGC GGA ACT TGA CGT TCG CCA CT |
684 | 61 | [6] | ||
| ermBF ermBR |
GGT AAA GGG CAT TTA ACG AC CGA TAT TCT CGA TTG ACC CA |
494 | 55 | 60 | 2 | [58] |
| tetQF tetQR |
AGA ATC TGC TGT TTG CCA GTG CGG AGT GTC AAT GAT ATT GCA |
167 | 50 | [57] | ||
| tet32F tet32R |
GAA CCA GAT GCT GCT CTT CAT AGC CAC GCC CAC ATG AT |
620 | 57 | [59] | ||
| tetLF tetLR |
TCG TTA GCG TGC TGT CAT TC GTA TCC CAC CAA TGT AGC CG |
267 | 55 | [60] | ||
| tetOF tetOR |
AAC TTA GGC ATT CTG GCT CAC TCC CAC TGT TCC ATA TCG TCA |
515 | 55 | 58 | 3 | [61] |
| tetSF tetSR |
GAA AGC TTA CTA TAC AGT AGC AGG AGT ATC TAC AAT ATT TAC |
168 | 50 | [57] | ||
| tet31F tet31R |
CAA TCA CGC CCA AAA GAA TGT GCC ATC CCA GTT TGT |
564 | 53 | [62] | ||
| tetBF tetBR |
AAT AGC CAC TAA ATG GGG CG ATA ACA CCG GTT GCA TTG GT |
243 | 58 | 56.5 | 4 | [6] |
| tetKF tetKR |
TCG ATA GGA ACA GCA GTA CAG CAG ATC CTA CTC CTT |
169 | 52.8 | [60] | ||
| tet37F tet37R |
ATG GTT CGC TAT TAC TCT AAC ATC AGT CTC ATA TTT CGA CA |
170 | 50 | [6] | ||
| 27 F 1544 R |
GAG TTT GAT CCT GGC TCA G AGA AAG GAG GTG ATC CAG CC |
approx. 1500 | 57 | - | - | [63] |
Detection of antibiotic resistance genes
For the detection of antibiotic resistance genes, four multiplex-PCR reactions were performed using four sets of primers (Table 1) as previously described [6]. The amplified products were evaluated by electrophoresis using a 2% agarose gel. Some of these amplified products were sequenced (Macrogen, Inc., Amsterdam, The Netherlands), and used as positive controls. The GenBank accession numbers for the positive controls are available in the supplementary material.
Statistical analyses
Qualitative data were obtained when screening for genes and when testing the ability of the isolates to grow in media containing a breakpoint antimicrobial concentration. Isolates were considered resistant to the antimicrobial if they were able to grow in the medium containing antibiotic and susceptible if they were not. To compare the prevalence of antibiotic resistance and of antibiotic resistance genes in both healthy subjects and subjects with periodontitis, data were analyzed using the chi-square test. To compare the bacterial loads resistant to tetracycline, the non-parametric Mann–Whitney U test was used. A nominal significance level of 5% (p < 0.05) was applied for both tests.
Results
One hundred and twenty-nine subgingival samples from periodontally healthy subjects (SPHS) and 130 subgingival samples from subjects with periodontitis (SSP) were collected. SPHS were from patients between the ages of 19–24 (mean of 21.5 ± 3.3) years and SSP were from patients 24 to 82 (mean of 51.25 ± 11.97) years of age.
SSP grown on blood agar showed a mean bacterial load of 6.54 log10 colony-forming units per milliliter (cfu/ml) (± 0.91), while SPHS showed a mean bacterial load of 5.64 log10 cfu/ml (± 0.8). When grown on media with tetracycline, SSP showed a mean of 4.87 log10 cfu/ml (± 2.09), 2.14% of the total bacterial load, versus the 3.66 log10 cfu/ml (±1.63) for SPHS, 1.05% of the total bacterial load, which did not represent a significant difference (p = 0.76). Eighty-six-point eighty-two percent (86.82%) (n = 112) of SPHS and 86.15% (n = 112) of SSP harbored tetracycline-resistant bacteria, from which we obtained a total of 813 isolates, belonging 448 to SPHS and 365 to SSP.
Streptococcus sp. were the most frequently isolated species, representing 75.62% in SSP and 75.89% in SPHS. In both groups, Streptococcus oralis was the most frequently isolated species (n = 83), followed by other streptococcal species such as Streptococcus mitis (n = 81), Streptococcus intermedius (n = 79) and Streptococcus constellatus (n = 58). The most frequently isolated non-streptococcal species was Prevotella intermedia (n = 31) followed by Prevotella nigrescens (n = 25).
When comparing the prevalence of certain species between the two groups of subjects, significant differences were observed: Gemella haemolysans (5-fold, p = 0.016), S. oralis (3-fold, p < 0.01), Streptococcus pneumoniae (4-fold, p < 0.01) and Streptococcus sanguinis (4-fold, p < 0.01) were more prevalent in SPHS, while Streptococcus anginosus (2-fold, p = 0.036), S. constellatus (6-fold, p < 0.01), Streptococcus gordonii (2-fold, p = 0.022), Streptococcus parasanguinis (3-fold, p = 0.017) and Streptococcus tigurinus (7-fold, p = 0.049) were more prevalent in SSP (Table S2).
A list of the identified species of the tetracycline-resistant microorganisms isolated in this study, the prevalence of the antibiotic resistance genes screened and the multidrug resistance profile for the six antibiotics tested is displayed in Table 2. The genes tet(M) (52.03%), tet(32) (8.24%) and tet(O) (7.75%) were the most frequently detected tet genes. Significant differences were also observed in the prevalence of tet(M) (p < 0.01), tet(W) (p < 0.01), and tet(O) (p < 0.01), which were higher in SPHS, and tet(32) (p < 0.01) and tet(L) (p < 0.01), which were higher in SSP. The intTn gene, which codes for an integrase located in transposons of the family Tn916/1545 was widely detected (79.58%), being more prevalent in SPHS (81.92%) than in SSP (76.71%).
Table 2.
Species identified from SPHS and SSP and the number of isolates carrying each antimicrobial resistance gene and their phenotypical resistance. The asterisk indicates significant differences between SPHS and SSP isolates. AMX: amoxicillin, CTX: cefotaxime, CHL: chloramphenicol, STR: streptomycin, ERY: erythromycin, KAN: kanamycin
| Antimicrobial resistance genes | Antimicrobial resistance | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | N | intTn | tet(W) | tet(O) | tet(M) | tet [31] | tet [32] | tet(B) | tet(L) | tet(S) | tet(Q) | tet [37] | tet(K) | erm(B) | AMX | CTX | CHL | STR | ERY | KAN | |
| SSP | Actinomyces sp. | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 |
| Bacteroides sp. | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | 2 | 2 | |
| Gemella sp. * | 2 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 2 | 2 | |
| Granulicatella sp. | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Lachnoanaerobaculum sp. | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 2 | 0 | 2 | 2 | 2 | |
| Mogibacterium sp. | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | |
| Neisseria sp. | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | |
| Peptoniphilus sp. | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Peptostreptococcus sp. | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Prevotella sp. * | 48 | 16 | 0 | 0 | 3 | 0 | 18 | 7 | 0 | 0 | 27 | 0 | 2 | 3 | 28 | 36 | 2 | 15 | 24 | 42 | |
| Proteus sp. | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Pseudomonas sp. | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 2 | 2 | 2 | 2 | |
| Rothia sp. | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 2 | |
| Slackia sp. | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | |
| Staphylococcus sp. | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | |
| Streptococcus sp. | 276 | 248 | 0 | 18 | 116 | 0 | 31 | 0 | 12 | 0 | 3 | 0 | 0 | 128 | 20 | 56 | 29 | 48 | 188 | 271 | |
| Veillonella sp. | 6 | 6 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 4 | 0 | 4 | 5 | 2 | |
| Not identified | 10 | 2 | 0 | 0 | 2 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 2 | 1 | 1 | 3 | 4 | |
| SPHS | Abiotrophia sp. | 5 | 5 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 1 | 0 | 5 | 2 |
| Bacilus sp. | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | |
| Butyrivibrio sp. | 4 | 1 | 0 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 2 | 1 | 4 | |
| Eubacterium sp. | 3 | 2 | 1 | 0 | 3 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 3 | 3 | |
| Gemella sp. * | 13 | 13 | 1 | 0 | 8 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 4 | 0 | 5 | 4 | |
| Granulicatella sp. | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | |
| Haemophilus sp. | 4 | 2 | 1 | 0 | 2 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 4 | 1 | |
| Neisseria sp. | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 2 | 0 | |
| Prevotella sp. * | 25 | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 0 | 0 | 16 | 0 | 1 | 0 | 16 | 15 | 4 | 13 | 18 | 22 | |
| Rothia sp. | 4 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | |
| Staphylococcus sp. | 5 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 4 | 3 | 3 | 2 | 3 | 4 | 0 | |
| Streptococcus sp. | 340 | 305 | 44 | 42 | 250 | 0 | 12 | 2 | 0 | 1 | 5 | 0 | 0 | 218 | 39 | 12 | 68 | 59 | 227 | 283 | |
| Veillonella sp. | 12 | 11 | 5 | 1 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 3 | 5 | 2 | 4 | 12 | 6 | |
| Not identified | 27 | 24 | 3 | 0 | 17 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 20 | 7 | 0 | 3 | 7 | 27 | 20 | |
When assessing resistance to six different antimicrobials, a significant difference was found between the groups, where the SSP group showed three times more isolates. Significant differences were also observed between the isolates for susceptibility to all the antimicrobials, where SPHS isolates showed two times higher susceptibility. Moreover, we found that 91% of all the isolates were resistant to other antibiotics besides tetracycline (Table 3). Resistance to KAN was the most frequently observed (83.64%), followed by ERY (67.16%), STR (20.66%), CTX (18.08%), AMX (15.50%), and CHL (14.64%). Prevalence and significant differences of the studied genes and resistances between the two groups are shown in Figure 1.
Table 3.
Number of isolates resistant to other antimicrobials. The asterisk indicates significant differences between samples from periodontally healthy subjects (SPHS) and the samples from subjects with periodontitis (SSP) isolates. In parentheses, the percentage of isolates over the total number of isolates from each population. Define meaning of asterisk
| Number of other antimicrobials | SPHS isolates (%) | SSP isolates (%) | |||
|---|---|---|---|---|---|
| 0* | 48 | (10.71) | 21 | (5.75) | |
| 1 | 82 | (18.30) | 68 | (18.63) | |
| 2 | 179 | (39.96) | 140 | (38.36) | |
| 3 | 93 | (20.76) | 77 | (21.10) | |
| 4 | 31 | (6.92) | 37 | (10.14) | |
| 5 | 12 | (2.68) | 13 | (3.56) | |
| 6* | 3 | (0.67) | 9 | (2.47) | |
Figure 1.
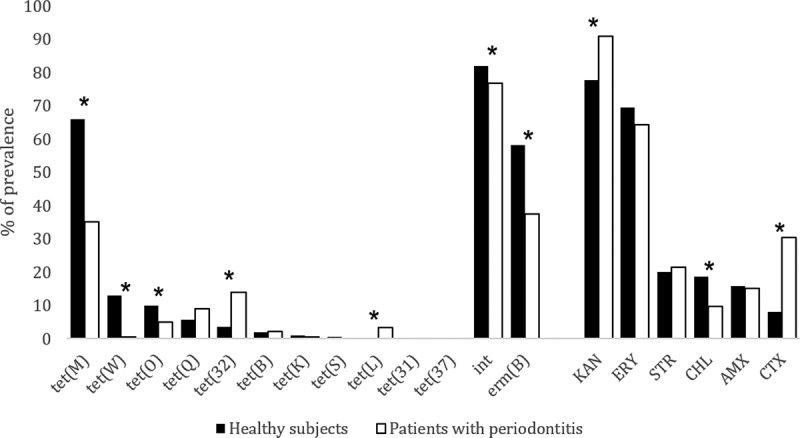
Prevalence (%) of tet, int and erm(B) genes and the prevalence (%) of subgingival isolates resistant to different antimicrobials (KAN: kanamycin, ERY: erythromycin, STR: streptomycin, CHL: Chloramphenicol, AMX: Amoxicillin, CTX: cefotaxime. The asterisk indicates significant differences between the isolates obtained from samples from periodontally healthy subjects and from samples from subjects with periodontitis
All the identified species and their prevalence of the antibiotic resistance genes screened and the resistance to six antibiotics are presented in Table S2.
Discussion
The present study investigated the tetracycline-resistant bacteria in subgingival samples of 129 periodontally healthy subjects and 130 patients with periodontitis, finding high loads of tetracycline-resistant bacteria, and a high prevalence of tet genes and multidrug resistance in both populations.
Bacterial counts, both total and that of resistant bacteria, were 10 times higher in SSP than in SPHS. This agrees with previous studies [25,26], in which the bacterial load in the periodontal pocket of patients with periodontitis was higher than that observed from periodontally healthy subjects. A higher percentage of tetracycline-resistant bacteria was also observed in SSP, probably due to the higher bacterial load of those samples.
Although the percentages of total tetracycline-resistant streptococci were similar in both groups of subjects, differences were observed at species level (Table S2). S. sanguinis, S. oralis and S. pneumoniae, which in our study were significantly more prevalent in SPHS, have been previously associated to commensal biofilms [27,28], while S. anginosus, S. constellatus, S. gordonii, S. tigurinus and S. parasanguinis, which in our study were significantly more prevalent in SSP, have been related to a periodontitis-associated microbiota [29–33]. However, the role of these species in periodontitis is not yet clear [34,35].
The prevalence of G. haemolysans was significantly higher in SPHS. This species has been described by metagenomic studies as an early colonizer of the oral biofilm [36,37] and therefore as part of the commensal microbiota. However, it has also been linked to opportunistic infections [38].
Isolates of the genus Prevotella were significantly more prevalent in SSP (2-fold). The increased richness of Prevotella species in SSP was responsible for this, increasing the total number of Prevotella isolates in SSP. However, P. intermedia and P. nigrescens, two Prevotella species that were present in both populations, did not show significantly different prevalences. The genus Prevotella has been linked to the progression of periodontitis, and therefore, a higher prevalence of this genus was expected in SSP. However, the increased richness of tetracycline-resistant Prevotella species observed in SSP is worth mentioning, since most reports have focused their attention on P. intermedia and P. nigrescens [39,40], and little is known about the prevalence of tetracycline resistance in other oral Prevotella species.
The prevalence of Gram-negative anaerobes was low, and species usually isolated from patients with periodontitis, such as Porphyromonas gingivalis or Fusobacterium nucleatum, were not detected in this study. This might be due to the high susceptibility that these organisms have to tetracycline, in most cases showing minimum inhibition concentrations lower than the 8 μg/ml tetracycline breakpoint concentration used in this study [41,42], which makes them unlikely to be isolated in such conditions, as seen in previous studies [7].
In this study, we found the genes tet(W), tet(O), tet(32), tet(B), tet(Q) and tet(K), distributed among the genera Eubacterium, Gemella, Haemophilus, Veillonella, Butyrivibrio and Prevotella which, according to the tetracycline resistance genes database available at http://faculty.washington.edu/marilynr/, had not been previously reported (Table 2). However, further studies are needed to confirm these results. For instance, the detection of the gene tet(B) in streptococci, as recently described [43], or new tet genes being described in the oral environment [44] show that there is still much to be unveiled regarding tetracycline resistance genes. The prevalence of the gene tet(M) among the SPHS in our study was similar (65.85%) to what has been previously described [7,45]. Nevertheless, the prevalence of this gene in SSP was much lower (35.07%) than what other authors found in Greek and US subjects with periodontitis [45,46], but similar to the percentages found in Dominican patients [6]. These differences might be explained by the geographical constraints of the populations involved in the studies or due to their methodological approaches, such as pooling the samples, the DNA or different selection criteria of the isolates. As previously discussed, the streptococcal species were significantly different between SPHS and SSP, which might be the cause for the differences in prevalence of tet(M) between the two groups studied. While 73.53% of the streptococci isolated from SPHS showed tet(M), only 42.02% of the streptococci isolated from SSP did, indicating that those streptococci associated to a healthy biofilm might be more susceptible to carry tet(M). In our study, the prevalence of tet [32] was higher in SSP, as previously described [6], which might be due to the increased prevalence of this gene in periodontitis-associated species such as S. constellatus and P. intermedia¸ although other authors have also detected this gene in some commensal bacteria [47]. The gene tet(W) can be frequently detected in the DNA from pooled saliva and plaque samples [7,48,49], but its distribution within the subgingival biofilm has only been studied by Collins et al. [6]. In that study, tet(W) was, on average, twice as prevalent in SSP than in SPHS. In our study, tet(W) was more ubiquitous in SPHS, since S. intermedius and S. oralis, which were more prevalent in SPHS, were some of the species that carried tet(W) most often.
Transposons of the Tn916/1545 family are frequently found in the oral microbiota [50]. These transposons usually carry tetracycline resistance genes, and in some cases, genes that confer resistance to macrolides and/or aminoglycosides [51,52]. In order to estimate the presence of these transposons among the isolates, we used PCR to screen the intTn gene, which codes for an integrase located at the 3ʹ- ends of these transposons [14], the tet(M) gene, and the erm(B) gene, which confers resistance to macrolides and is often found in these conjugative elements [53]. The results showed a high prevalence of both genes in both groups, although they were significantly more prevalent in SPHS. The previously discussed differing prevalence of streptococcal species when comparing SPHS and SSP might be the reason for these differences, given that the streptococci of this study showed different profiles of antibiotic resistance genes. These results suggest a high prevalence of transposons of the Tn916/1545 family in the tetracycline-resistant oral microbiota of healthy subjects and patients with periodontitis.
Most of the tetracycline-resistant isolates obtained in this study showed resistance to other antibiotics. Resistance to two antimicrobials besides tetracycline was the most common pattern, and a large part of this multidrug resistance was to KAN and ERY, which could be linked to the presence of the previously mentioned transposons [51]. Levels of multidrug resistance were high and similar in both populations, showing that the tetracycline-resistant subgingival microbiota is an important reservoir of antimicrobial resistance, which might be specially striking in subjects who have received little or no antibiotic therapy. The high prevalence of isolates resistant to antibiotics not commonly used, and therefore not exposed to selective pressure, might be due to the low fitness costs associated with the acquisition and maintenance of some mobile genetic elements that carry antibiotic resistance genes [54,55].
Significant differences were found when testing KAN and CHL resistance between both groups. The former was observed to be more prevalent in SSP isolates than in the SPHS isolates, where the contrary was observed for the latter. Streptococcus spp. and Prevotella spp. isolates might be accountable for these differences, since the Prevotella genus, which is known to show resistance to β-lactams and KAN [56], was twice as prevalent in SSP compared to SPHS. On the other hand, the differences in CHL resistance might be due to the unequal distribution of CHL resistance among streptococcal species, resulting in different percentages of this resistance between both groups (Table S2). The highest ratio of P. intermedia and P. nigrescens and the high prevalence of S. constellatus CTX-resistant in SSP were determinant for the higher prevalence of CTX resistance in SSP. Although the genus Prevotella is already known for its β-lactam resistance, high prevalence of CTX resistance among oral streptococci has not been previously reported; thus, it might pose a serious health issue, even more when considering their multidrug resistance capabilities.
While the differences found in this study were based on the periodontal diagnoses of the subjects, more certainty about the causes of the differences observed would most likely be achieved by including either the age or the sex of the subjects as variables of interest. Nonetheless, this study has shown high loads of bacteria exhibiting multidrug resistance and a variety of tetracycline resistance genes to be present in Spanish SSPs and SPHSs. However, significant differences were detected between the two groups in terms of i) bacterial species resistant to tetracycline; ii) prevalence of the screened genes and iii) multidrug resistance profiles. Moreover, the presence of some tet genes was detected in certain bacterial genera, which had not been previously described. Overall, we found that in Spanish subjects, although the use of tetracycline has been declining for many years, resistance to this antibiotic is still present in subgingival bacteria, which are a reservoir of tetracycline resistance genes and multidrug resistance, which, coupled with the high prevalence of conjugative transposons in the oral environment, might foster further increased spread of antimicrobial resistance.
Supplementary Material
Acknowledgments
We would like to express our gratitude to all the volunteers that participated in the study and would also like to thank Ann Bangle for editing the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Chopra I, Roberts M.. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;65(2):232–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Chopra I, Hawkey PM, Hinton M. Tetracyclines, molecular and clinical aspects. J Antimicrob Chemother. 1992;29(3):245–277. [DOI] [PubMed] [Google Scholar]
- [3].Grossman TH. Tetracycline Antibiotics and Resistance. Cold Spring Harb Perspect Med. 2016;6:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Broom LJ. The sub-inhibitory theory for antibiotic growth promoters. Poult Sci. 2017;96(9):3104–3108. [DOI] [PubMed] [Google Scholar]
- [5].Lu L, Liu J, Li Z, et al. Occurrence and distribution of tetracycline antibiotics and resistance genes in longshore sediments of the three gorges reservoir, China. Front Microbiol. 2018;9:1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Collins JR, Arredondo A, Roa A, et al. Periodontal pathogens and tetracycline resistance genes in subgingival biofilm of periodontally healthy and diseased Dominican adults. Clin Oral Investig. 2016;20(2):349–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Villedieu A, Diaz-Torres ML, Hunt N, et al. Prevalence of tetracycline resistance genes in oral bacteria. Antimicrob Agents Chemother. 2003;47(3):878–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Herrera D, Alonso B, León R, et al. Antimicrobial therapy in periodontitis: the use of systemic antimicrobials against the subgingival biofilm. J Clin Periodontol. 2008;35:45–66. [DOI] [PubMed] [Google Scholar]
- [9].Goodson JM, Haffajee AD, Socransky SS, et al. Control of periodontal infections: A randomized controlled trial I. The primary outcome attachment gain and pocket depth reduction at treated sites. J Clin Periodontol. 2012;39(6):526–536. [DOI] [PubMed] [Google Scholar]
- [10].Herrera D, Sanz M, Jepsen S, et al. A systematic review on the effect of systemic antimicrobials as an adjunct to scaling and root planing in periodontitis patients. J Clin Periodontol. 2002;29(S3):136–159. [DOI] [PubMed] [Google Scholar]
- [11].Olsvik B, Tenover FC. Tetracycline resistance in periodontal pathogens. Clin Infect Dis. 1993;16(S4):310–313. [DOI] [PubMed] [Google Scholar]
- [12].Diaz-Torres ML, McNab R, Spratt DA, et al. Novel tetracycline resistance determinant from the oral metagenome. Antimicrob Agents Chemother. 2003;47(4):1430–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Thaker M, Spanogiannopoulos P, Wright GD. The tetracycline resistome. Cell Mol Life Sci. 2010;67(3):419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Roberts AP, Mullany P. A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol. 2009;17(6):251–258. [DOI] [PubMed] [Google Scholar]
- [15].Long KS, Vester B. Resistance to linezolid caused by modifications at its binding site on the ribosome. Antimicrob Agents Chemother. 2012;56(2):603–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Diaz PI, Hoare A, Hong B-Y. Subgingival microbiome shifts and community dynamics in periodontal diseases. J Calif Dent Assoc. 2016;44(7):421–35. [PubMed] [Google Scholar]
- [17].Cassir N, Rolain J-M, Brouqui P. A new strategy to fight antimicrobial resistance: the revival of old antibiotics. Front Microbiol. 2014;5:551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Arredondo A, Blanc V, Mor C, et al. Resistance to β-lactams and distribution of β-lactam resistance genes in subgingival microbiota from Spanish patients with periodontitis. Clin Oral Investig. 2020. DOI: 10.1007/s00784-020-03333-1 [DOI] [PubMed] [Google Scholar]
- [19].Mor C, Pascual A, Nart J, et al. Inhibition of de novo plaque growth by a new 0.03 % clorhexidine mouth rinse formulation applying a non-brushing model: a randomized, double blind clinical trial. Clin Oral Investig. 2015;20:1459–1467. [DOI] [PubMed] [Google Scholar]
- [20].Armitage GC. Development of a classification system for periodontal diseases and conditions. Ann Periodontol. 1999;4(1):1–6. [DOI] [PubMed] [Google Scholar]
- [21].Caton JG, Armitage G, Berglundh T, et al. A new classification scheme for periodontal and peri-implant diseases and conditions - Introduction and key changes from the 1999 classification. J Clin Periodontol. 2018;45(S20):S1–S8. [DOI] [PubMed] [Google Scholar]
- [22].Syed SA, Loesche WJ. Survival of human dental plaque flora in various transport media. Appl Microbiol. 1972;24(4):638–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Clinical & Laboratory Standards Institute (CLSI) . Performance standards for antimicrobial susceptibility testing (27th ed). Wayne, PA: Clinical and Laboratory Standards Institute; 2017. [Google Scholar]
- [24].The European Committee on Antimicrobial Susceptibility Testing (EUCAST) . (2018). Breakpoint tables for interpretation of MICs and zone diameters, version 8.0. https://eucast.org/clinical_breakpoints/
- [25].Abusleme L, Dupuy AK, Dutzan N, et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. Isme J. 2013;7(5):1016–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Checchi L, Gatto MR, Checchi V, et al. Bacteria prevalence in a large Italian population sample: a clinical and microbiological study. J Biol Regul Homeost Agents. 2016;30(S2):199–208. [PubMed] [Google Scholar]
- [27].Kolenbrander PE, Palmer RJ, Rickard AH, et al. Bacterial interactions and successions during plaque development. Periodontol. 2006;42:47–79. [DOI] [PubMed] [Google Scholar]
- [28].Vieira Colombo AP, Magalhães CB, Hartenbach FA, et al. Periodontal-disease-associated biofilm: A reservoir for pathogens of medical importance. Microb Pathog. 2016;94:27–34. [DOI] [PubMed] [Google Scholar]
- [29].Chávez de Paz L, Svensäter G, Dahlén G, et al. Streptococci from root canals in teeth with apical periodontitis receiving endodontic treatment. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2005;100(2):232–241. [DOI] [PubMed] [Google Scholar]
- [30].Rams TE, Feik D, Mortensen JE, et al. Antibiotic Susceptibility of Periodontal Streptococcus constellatus and Streptococcus intermedius Clinical Isolates. J Periodontol. 2014;85(12):1792–1798. [DOI] [PubMed] [Google Scholar]
- [31].Abiko Y, Sato T, Mayanagi G, et al. Profiling of subgingival plaque biofilm microflora from periodontally healthy subjects and from subjects with periodontitis using quantitative real-time PCR. J Periodontal Res. 2010;45(3):389–395. [DOI] [PubMed] [Google Scholar]
- [32].Dhotre SV, Mehetre GT, Dharne MS, et al. Isolation of Streptococcus tigurinus - a novel member of Streptococcus mitis group from a case of periodontitis. FEMS Microbiol Lett. 2014;357(2):131–135. [DOI] [PubMed] [Google Scholar]
- [33].Fine DH, Markowitz K, Fairlie K, et al. A consortium of Aggregatibacter actinomycetemcomitans, Streptococcus parasanguinis, and Filifactor alocis Is present in sites prior to bone loss in a longitudinal study of localized aggressive periodontitis. J Clin Microbiol. 2013;51(9):2850–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Aas JA, Paster BJ, Stokes LN, et al. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43(11):5721–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zbinden A, Bostanci N, Belibasakis GN. The novel species Streptococcus tigurinus and its association with oral infection. Virulence. 2015;6(3):177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Heller D, Helmerhorst EJ, Gower AC, et al. Molecular characterization of the microbial diversity in the early, In Vivo-Formed, Dental Biofilm. Appl Environ Microbiol. 2016;82(6):1881–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kirst ME, Li EC, Alfant B, et al. Dysbiosis and alterations in predicted functions of the subgingival microbiome in chronic periodontitis. Appl Environ Microbiol. 2015;81(2):783–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].García López E, Martín-Galiano AJ. The versatility of opportunistic infections caused by gemella isolates is supported by the carriage of virulence factors from multiple origins. Front Microbiol. 2020;11:524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Larsen JM. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology. 2017;151(4):363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].de Aquino SG, Abdollahi-Roodsaz S, Koenders MI, et al. Periodontal pathogens directly promote autoimmune experimental arthritis by inducing a TLR2- and IL-1-Driven Th17 response. J Immunol. 2014;192(2):4103–4111. [DOI] [PubMed] [Google Scholar]
- [41].Kulik EM, Lenkeit K, Chenaux S, et al. Antimicrobial susceptibility of periodontopathogenic bacteria. J Antimicrob Chemother. 2008;61(5):1087–1091. [DOI] [PubMed] [Google Scholar]
- [42].Veloo ACM, Seme K, Raangs E, et al. Antibiotic susceptibility profiles of oral pathogens. Int J Antimicrob Agents. 2012;40(5):450–454. [DOI] [PubMed] [Google Scholar]
- [43].Arredondo A, Àlvarez G, Nart J, et al. Detection and expression analysis of tet(B) in Streptococcus oralis. J Oral Microbiol. 2019;11:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Warburton PJ, Ciric L, Lerner A, et al. TetAB(46), a predicted heterodimeric ABC transporter conferring tetracycline resistance in Streptococcus australis isolated from the oral cavity. J Antimicrob Chemother. 2013;68(1):17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ioannidis I, Sakellari D, Spala A, et al. Prevalence of tetM, tetQ, nim and blaTEM genes in the oral cavities of Greek subjects: a pilot study. J Clin Periodontol. 2009;36(7):569–574. [DOI] [PubMed] [Google Scholar]
- [46].Kim S-M, Kim HC, Lee S-WS. Characterization of antibiotic resistance determinants in oral biofilms. J Microbiol. 2011;49(4):595–602. [DOI] [PubMed] [Google Scholar]
- [47].Lancaster H, Bedi R, Wilson M, et al. The maintenance in the oral cavity of children of tetracycline-resistant bacteria and the genes encoding such resistance. J Antimicrob Chemother. 2005;56(3):524–531. [DOI] [PubMed] [Google Scholar]
- [48].Seville LA, Patterson AJ, Scott KP, et al. Distribution of tetracycline and erythromycin resistance genes among human oral and fecal metagenomic DNA. Microb Drug Resist. 2009;15(3):159–166. [DOI] [PubMed] [Google Scholar]
- [49].Diaz-Torres ML, Villedieu A, Hunt N, et al. Determining the antibiotic resistance potential of the indigenous oral microbiota of humans using a metagenomic approach. FEMS Microbiol Lett. 2006;258(2):257–262. [DOI] [PubMed] [Google Scholar]
- [50].Roberts AP, Cheah G, Ready D, et al. Transfer of Tn916-like elements in microcosm dental plaques. Antimicrob Agents Chemother. 2001;45(10):2943–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Cochetti I, Tili E, Mingoia M, et al. erm(B)-Carrying elements in tetracycline-resistant pneumococci and correspondence between Tn1545 and Tn6003. Antimicrob Agents Chemother. 2008;52(4):1285–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Roberts AP, Mullany P. Tn916-like genetic elements: a diverse group of modular mobile elements conferring antibiotic resistance. FEMS Microbiol Rev. 2011;35(5):856–871. [DOI] [PubMed] [Google Scholar]
- [53].Santoro F, Vianna ME, Roberts AP. Variation on a theme; an overview of the Tn916/Tn1545 family of mobile genetic elements in the oral and nasopharyngeal streptococci. Front Microbiol. 2014;5:535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Pallecchi L, Bartoloni A, Paradisi F, et al. Antibiotic resistance in the absence of antimicrobial use: mechanisms and implications. Expert Rev Anti Infect Ther. 2008;6:725–732. [DOI] [PubMed] [Google Scholar]
- [55].Roberts AP, Mullany P. Oral biofilms: a reservoir of transferable, bacterial, antimicrobial resistance. Expert Rev Anti Infect Ther. 2010;8(12):1441–1450. [DOI] [PubMed] [Google Scholar]
- [56].Holbrook WP, Ogston SA, Ross PW. A method for the isolation of Bacteroides melaninogenicus from the human mouth. J Med Microbiol. 1978;11(2):203–207. [DOI] [PubMed] [Google Scholar]
- [57].Aminov RI, Garrigues-Jeanjean N, Mackie RI. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl Environ Microbiol. 2001;67(1):22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Warburton PJ, Palmer RM, Munson MA, et al. Demonstration of in vivo transfer of doxycycline resistance mediated by a novel transposon. J Antimicrob Chemother. 2007;60(5):973–980. [DOI] [PubMed] [Google Scholar]
- [59].Melville CM, Scott KP, Mercer DK, et al. Novel tetracycline resistance gene, tet (32), in the Clostridium-related human colonic Anaerobe K10 and Its Transmission In Vitro to the Rumen Anaerobe Butyrivibrio fibrisolvens. Antimicrob Agents Chemother. 2001;45(11):3246–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Ng L-K, Martin I, Alfa M, et al. Multiplex PCR for the detection of tetracycline resistant genes. Mol Cell Probes. 2001;15:4,209–215. [DOI] [PubMed] [Google Scholar]
- [61].Manch-Citron JN, Lopez GH, Dey A, et al. PCR monitoring for tetracycline resistance genes in subgingival plaque following site-specific periodontal therapy. A preliminary report. J Clin Periodontol. 2000;27(6):437–446. [DOI] [PubMed] [Google Scholar]
- [62].Bojesen AM, Bager RJ, Ifrah D, et al. The rarely reported tet(31) tetracycline resistance determinant is common in Gallibacterium anatis. Vet Microbiol. 2011;149(3–4):497–499. [DOI] [PubMed] [Google Scholar]
- [63].Shigematsu T, Hayashi M, Kikuchi I, et al. A culture-dependent bacterial community structure analysis based on liquid cultivation and its application to a marine environment. FEMS Microbiol Lett. 2009;293(2):240–247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


