Abstract
Targeting protein for Xenopus kinesin-like protein 2 (TPX2) has been identified as an oncogene in multiple cancers. However, the associations among TPX2 expression, prognosis, and tumor immunity in hepatic cell cancer (HCC) have not been explored. We analyzed TPX2 expression by multiple gene expression databases, including Oncomine, TIMER, and UALCAN. The prognosis effect of TPX2 was analyzed by Kaplan--Meier plotter. The coexpressed genes with TPX2 were analyzed using Linked Omics. The association among TPX2 and immune infiltrates and immune checkpoints was determined by TIMER. It was found that TPX2 expression was notably upregulated in multiple HCC tissues. Overexpression of TPX2 has associations with race, age, weight, clinical stage and tumor grade, as well as poor prognosis in overall survival (OS), progression-free survival (PFS), disease-free survival (DFS), and disease-specific survival (DSS). In addition, TPX2 expression has a positive association with the infiltration of immune cells and the expression of immune checkpoint molecules. Coexpressed genes and functional network analysis suggested several potential mechanisms of TPX2 affecting HCC progression. The findings reveal that TPX2 has associations with prognosis and infiltration of immune cells in HCC patients, which has laid a basis for in-depth study of TPX2 role in HCC.
Keywords: bioinformatics, hepatic cell cancer, prognosis, TPX2
1. Introduction
Hepatic cell cancer (HCC) is one of the most frequently occurring digestive malignant tumors in the world. As the modern medical technology is developing quickly, the methods for diagnosing and treating HCC have been gradually improved, but the 5-year recurrence rate is still nearly 80% after surgical operation.[1] A study of American Cancer Society made a prediction that about 42,810 new HCC cases and more than 30,000 deaths will occur in the United States in 2020.[2] Whatever, the lack of effective diagnostic strategies and therapeutic targets are the main reason for the poor prognosis of HCC patients. Therefore, it is still crucial to further investigate more efficient potential biomarkers to predict and monitor the prognosis of HCC patients.
Targeting protein for Xenopus kinesin-like protein 2 (TPX2) is taken as a microtubule-related protein, which can activate protein kinases, thus regulating cell proliferation, apoptotic processes, cell division, etc.[3] TPX2 has been identified as oncogenic factor in multiple cancers.[4–6] For example, upregulated expression of TPX2 enhances metastases of breast cancer by mediating MMP2 and MMP9 expressions.[7] In addition, TPX2 can inhibit cellular proliferation and enhance cellular apoptosis through blocking of the PI3k/AKT/p21 pathway and activation of p53 pathway in breast cancer.[8] Although increasing numbers of studies turn attention to the molecular functions and clinical associations of TPX2 in multiple human cancers, its exact prognostic value of TPX2 and association with tumor immunity in HCC have not been defined.
In the past decades, many user-friendly and interactive online TCGA-based platforms and other large-scale sequencing databases obviously enhance the efficiency of bioinformatics assay, and a growing number of cancer-related indicators have been determined based on the strengths of the above websites.[9–11] In this study, these interactive platforms were used to determine the expression and prognosis effect of TPX2 in HCC. Consequently, our study systematically summarized the expression of TPX2 and discusses the potential prognostic value and association with immune infiltration of TPX2 in HCC.
2. Materials and methods
2.1. Oncomine database analysis
Oncomine (https://www.oncomine.org/) provides an effective analysis methodology for determining gene expression signatures.[12] In this study, Oncomine was adopted to assess mRNA expression level of TPX2 in cancers and normal tissues. Regarding the differential expression analysis of TPX2 between HCC and normal tissue, the thresholds were established as analysis type: cancer vs normal tissues; cancer type: hepatic cancer; gene rank: the top 10%; P value: .05; fold-change: all; data type: mRNA.
2.2. HCCDB database analysis
HCCDB (http://lifeome.net/database/hccdb/home.html) is a web database of HCC expression atlas including 15 public HCC gene expression datasets and involving a total of 3917 samples.[13] HCCDB visualizes the results of multiple database analyses such as differential expression, prognosis, and tissue and cancer-specific expression analyses. We employed HCCDB for in-depth study of TPX2 expression in HCC.
2.3. Human protein atlas database analysis
Human Protein Atlas (HPA, http://www.proteinatlas.org/) is established for mapping all human proteins within cells, tissues, and organs using integrated multiple omics technologies such as antibody-based imaging, mass spectrometry-based proteomics, transcriptomics, and systems biology.[14,15] This database provides open access to all data for both academic and industrial scientists to explore the human proteome. In this study, the definition of protein expression level was obtained from the HPA database, and the TPX2 protein expression level was compared between normal hepatic tissue and HCC tissue using the data from HPA.
2.4. Kaplan--Meier plotter database analysis
Kaplan--Meier Plotter (KM Plotter, http://kmplot.com/analysis/) is a web database capable of storing and retrieving large amounts of data on the effect of 54k genes on survival in 21 cancer types.[16] The prognosis effect of TPX2 in HCC was analyzed using KM Plotter. The patients were divided into different groups according to the median value of mRNA expression level of TPX2. Comparison among all groups were performed using KM survival plots. Hazard ratio (HR), 95% confidence interval (95% CI), and log-rank P value were computed and displayed using this web database.
2.5. Cox regression analysis in the TCGA-LIHC cohort
The level 3 data of TCGA-LIHC and clinical data were obtained from the cBioPortal (https://www.cbioportal.org/). A total of 348 cases containing both TPX2 mRNA expression RSEM and clinical information were reserved to further analysis. On the basis of the expression of TPX2, total cases were ranked from low expression to high expression. The bottom 50% of patients were divided into the low expression group and the top 50% belonged to the high expression group. Next, the independent prognostic value of TPX2 was analyzed by univariate and multivariate analyses.
2.6. TIMER database analysis
TIMER is a web database for comprehensive assessments of the clinical effect of different immune cells in diversified cancer types,[17] in which, a deconvolution algorithm is used to predict the type and proportion of tumor-infiltrating immune cells (TIICs) according to gene expression profiles.[18] We analyzed TPX2 expression in multiple cancers and the association between TPX2 expression and abundance of immune infiltrates, involving B, CD4+ T, and CD8+ T cells, neutrophils, macrophages, and dendritic cells (DCs), as well as cancer purity in HCC. In addition, the association between TPX2 and immune checkpoints expression was also analyzed by TIMER.
2.7. Linked omics database analysis
Linked Omics (http://www.linkedomics.org/login.php) is a web database comprising multi-omics datasets for analysis of 32 TCGA cancer types.[19] Pearson correlation coefficient was used to statistically analyze the genes coexpressed with TPX2, and the results were presented in volcano plot, heat map, or scatter plot, respectively. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed by gene set enrichment analysis (GSEA) based on function modules of Linked Omics. The false discovery rate (FDR) was less than 0.05 and 500 simulations were performed.
2.8. Statistical analysis
Statistical analyses were mainly conducted using the above-mentioned web databases. The expressions of TPX2 in different HCC tissues were compared using t test or one-way analysis of variance (ANOVA). Kaplan--Meier survival plots were constructed to visualize the comparison of survival curves using log-rank test. Univariate and multivariate analyses were used to evaluate the independent prognostic role of TPX2 in HCC. It would be concluded that there was a statistically obvious difference when P value was lower than .05.
3. Results
3.1. Pan-cancer analysis of TPX2 expression levels
Expression levels of TPX2 in cancerous and normal tissues of multiple cancer patients were analyzed using Oncomine. Figure 1A indicated that TPX2 was highly expressed in most solid tumors, while TPX2 exhibited contradictory expression in leukemia. The TCGA RNA-seq data from TIMER was applied to confirm the expression levels of TPX2 in cancers. TPX2 expression was also significantly up-regulated in most types of solid cancers (Fig. 1B). These data exhibited that TPX2 was significantly upregulated most solid cancers, suggesting that TPX2 exerted oncogenic functions in various tumors. Moreover, we analyzed the prognostic values of TPX2 in multiple cancers. As shown in Table 1, TPX2 was a potential prognostic indicator for both OS and RFS in renal papillary cell carcinoma, HCC, pancreatic ductal adenocarcinoma, and uterine corpus endometrial carcinoma. Given the high morbidity and mortality of HCC, we next analyzed the feasibility of TPX2 as a therapeutic target and prognostic biomarker in HCC.
Figure 1.
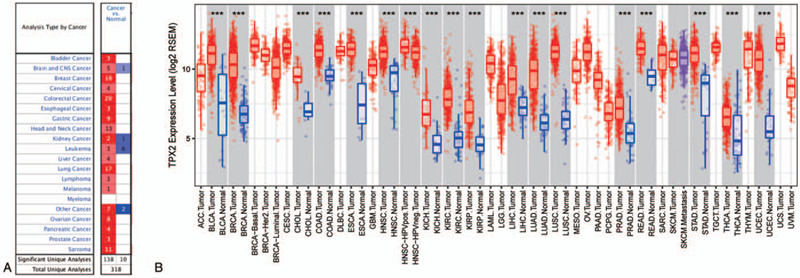
Expression of TPX2 in different types of human cancers. (A) Dysregulated expressions of TPX2 in datasets of different cancers compared with normal tissues are examined by Oncomine. (B) The TPX2 expression levels in different tumor types from the TCGA database in TIMER. ∗P < .05, †P < .01, ‡P < .001. “Total Unique Analyses” is defined as total analyses included in the Oncomine database. “Total Unique Analyses” is defined as analyses that meet our stated threshold values.
Table 1.
Pan-cancer analysis of prognostic values of TPX2 for OS and RFS generated by the Kaplan--Meier Plotter database.
| OS | RFS | |||||
| Cancer types | HR | 95% CI | P | HR | 95% CI | P |
| Bladder carcinoma | 1.10 | 0.82–1.48 | .509 | 0.88 | 0.43–1.78 | .717 |
| Breast cancer | 1.20 | 0.87–1.65 | .265 | 1.49 | 0.96–2.30 | .070 |
| Cervical squamous cell carcinoma | 0.79 | 0.50–1.26 | .323 | 0.97 | 0.45–2.09 | .932 |
| Esophageal adenocarcinoma | 2.60 | 1.33–5.09 | .004 | 1.24 | 0.17–9.09 | .831 |
| Esophageal squamous cell carcinoma | 0.32 | 0.13–0.79 | .010 | 0.48 | 0.18–1.30 | .139 |
| Head-neck squamous cell carcinoma | 1.06 | 0.82–1.39 | .645 | 0.72 | 0.34–1.52 | .385 |
| Kidney renal clear cell carcinoma | 1.97 | 1.44–2.69 | <.001 | 1.42 | 0.48–4.24 | .525 |
| Kidney renal papillary cell carcinoma | 3.72 | 1.92–7.24 | <.001 | 2.80 | 1.26–6.23 | .009 |
| Liver hepatocellular carcinoma | 2.03 | 1.43–2.90 | <.001 | 1.62 | 1.16–2.25 | .004 |
| Lung adenocarcinoma | 1.59 | 1.18–2.13 | .002 | 1.13 | 0.74–1.71 | .569 |
| Lung squamous cell carcinoma | 0.90 | 0.69–1.18 | .461 | 0.98 | 0.59–1.62 | .936 |
| Ovarian cancer | 0.87 | 0.67–1.13 | .307 | 0.87 | 0.61–1.24 | .445 |
| Pancreatic ductal adenocarcinoma | 1.76 | 1.16–2.67 | .007 | 3.02 | 1.23–7.40 | .011 |
| Pheochromocytoma and paraganglioma | 4.05 | 0.46–35.39 | .172 | 3.06 | 0.32–29.48 | .308 |
| Rectum adenocarcinoma | 0.77 | 0.35–1.69 | .517 | 1.66 | 0.30–9.09 | .557 |
| Sarcoma | 1.38 | 0.92–2.06 | .114 | 1.61 | 0.98–2.64 | .057 |
| Stomach adenocarcinoma | 1.02 | 0.73–1.41 | .915 | 0.84 | 0.44–1.62 | .608 |
| Testicular germ cell tumor | 1.79 | 0.16–19.79 | .629 | 1.78 | 0.81–3.90 | .143 |
| Thymoma | 0.18 | 0.04–0.89 | .020 | / | / | / |
| Thyroid carcinoma | 2.12 | 0.74–6.11 | .154 | 3.43 | 1.38–8.54 | .005 |
| Uterine corpus endometrial carcinoma | 1.91 | 1.24–2.94 | .003 | 2.04 | 1.17–3.57 | .011 |
95% CI = 95% confidence interval, HR = hazard ratio, OS = overall survival, RFS = relapse-free survival, TPX2 = targeting protein for Xenopus kinesin-like protein 2.
Bold values indicated statistical significance.
3.2. Elevated expression of TPX2 in HCC
We next evaluated TPX2 transcription levels in multiple HCC studies from HCCDB database. The results indicated that mRNA expression of TPX2 was obviously enhanced in HCC tissues in comparison with that in normal tissues (Fig. 2A). We further used HPA database to examine TPX2 protein expression. We found that TPX2 protein expression in the HCC tissue was higher than that in the normal hepatic tissue, in which, TPX2 was identified by antibody HPA006487 (Fig. 2B, C). Taken together, we confirmed that both mRNA and protein expression levels of TPX2 were obviously upregulated in HCC tissue.
Figure 2.
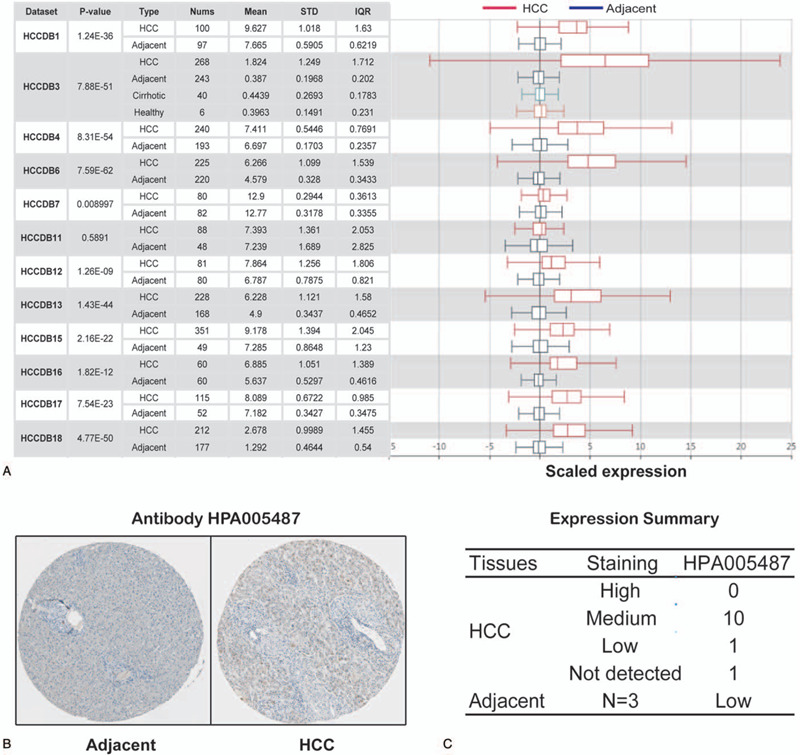
TPX2 expression level in HCC. (A) Chart and box plot showing the expression of TPX2 in cancerous tissues and the adjacent normal tissues, according to t test in HCCDB. (B) The protein expression of TPX2 in HCC from HPA. (C) Distribution of TPX2 protein expression level in HCC in HPA.
3.3. Association of TPX2 expression with clinical features in HCC patients
After high expression of TPX2 was confirmed in HCC, it was speculated that overexpression of TPX2 may be associated with clinical features of HCC patients. Therefore, the association of TPX2 expression with clinical features of HCC patients was analyzed, including gender, race, age, weight, clinical stage, and tumor grades. As exhibited in Figure 3, TPX2 expression was significantly associated with race, age, and weight, but had no relationship with gender in general characteristics (Fig. 3A--D). More importantly, the HCC patients at advanced clinical stages with higher tumor grades tended to express higher TPX2 mRNA. The highest expression of TPX2 was found in stage 3 and grade 4 cancer patients, respectively (Fig. 3E, F). The reason why the mRNA expression of TPX2 in stage 3 HCC patients seemed to be higher than that in stage 4 HCC patients might be caused by the fact that there was a limited number of stage 4 HCC patients (merely 6 HCC patients at stage 4). Overall, the above results suggested that mRNA expression level of TPX2 was obviously associated with the clinical features of HCC patients and might act as a potential prognosis predictor for diagnosis of HCC at advanced stages with a poorer differentiation.
Figure 3.
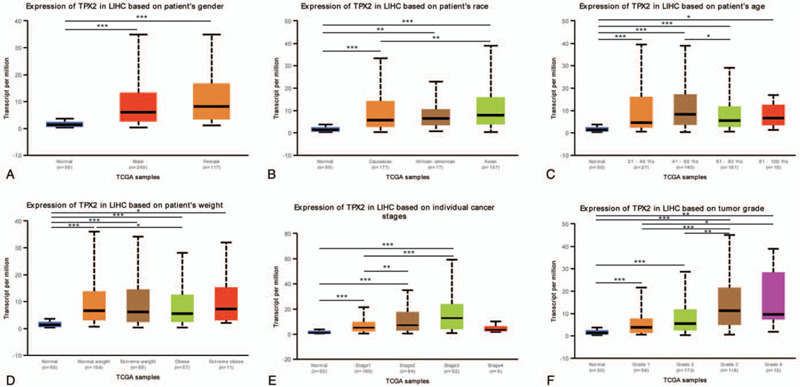
TPX2 transcriptional level in subgroups of patients with HCC. (A) Boxplot showing expression level of TPX2 in normal individuals of either gender and male or female HCC patients, respectively. (B) Boxplot showing expression level of TPX2 in normal, African American, Caucasian, and Asian HCC patients. (C) Boxplot showing expression level of TPX2 in normal individuals of any age or in HCC patients aged 21-40, 41-60, 61-80, or 81–100 yr. (D) Boxplot showing expression level of TPX2 in normal individuals of any weight or in HCC patients with normal weight, extreme weight, obese or extreme obese. (E) Boxplot showing expression level of TPX2 in normal individuals or in HCC patients at stages 1, 2, 3, or 4. (F) Boxplot showing expression level of TPX2 in normal individuals or HCC patients with grade 1, 2, 3, or 4 tumors. ∗P < .05, †P < .01, ‡P < .001.
3.4. Prognosis effects of TPX2 expression in HCC
Moreover, we also evaluated the prognosis effect of TPX2. As shown in Figure 4, high mRNA expression of TPX2 had obvious associations with poorer overall survival (OS), progression-free survival (PFS), disease-free survival (DFS), and disease-specific survival (DSS) in HCC patients (Fig. 4A–D). Besides, high mRNA expression of TPX2 also had obvious associations with a poorer OS in HCCDB18 group [Supplemental Digital Content (figure S1)]. These findings uncovered that TPX2 was an unfavorable prognostic indicator in HCC patients.
Figure 4.
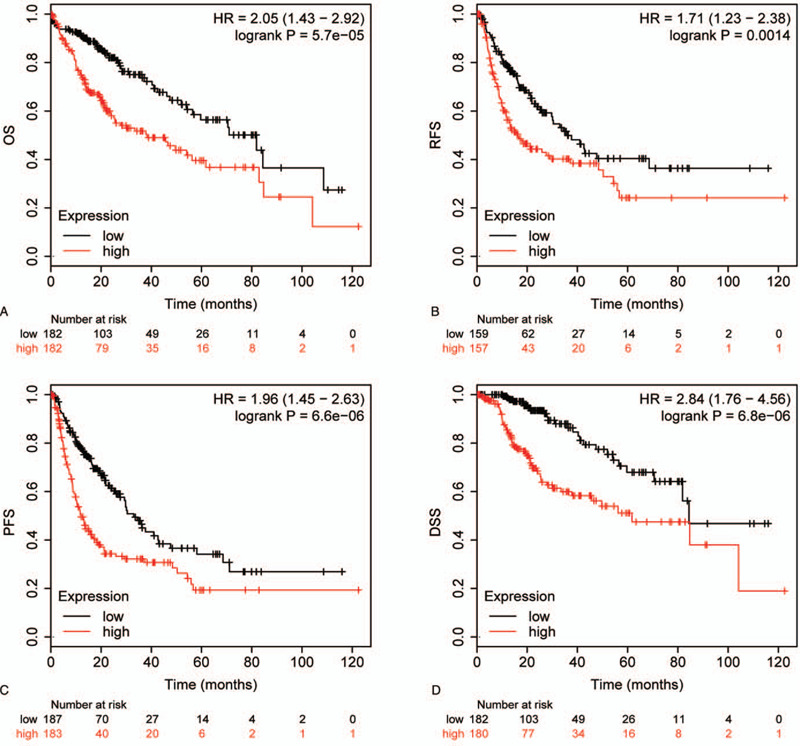
The prognosis effect of TPX2 expression in HCC. (A) OS curve showing the prognostic difference between patients with high expression of TPX2 and patients with low expression of TPX2. (B) PFS curve showing the prognostic difference between patients with high expression of TPX2 and patients with low expression of TPX2. (C) PFS curve showing the prognostic difference between patients with high expression of TPX2 and patients with low expression of TPX2. (D) DSS curve showing the prognostic difference between patients with high expression of TPX2 and patients with low expression of TPX2.
To further evaluate whether TPX2 expression was an independent prognostic factor for patients with HCC, the Cox regression model was applied for univariate and multivariate survival analyses. Univariate analysis exhibited that TPX2 expression status as well as T stage and clinical stage were associated with OS (HR = 1.86, 95% CI: 1.29–2.69, P = .001), PFS (HR = 1.86, 95% CI: 1.36–2.54, P < .001), DFS (HR = 1.72, 95% CI: 1.22–2.43, P = .002), and DSS (HR = 2.73, 95% CI: 1.66–4.49, P < .001) (Table 2). In multivariate analysis, we confirmed that TPX2 expression was an independent prognostic indicator for OS (HR = 1.66, 95% CI: 1.13–2.45, P = .011), PFS (HR = 1.66, 95% CI: 1.20–2.29, P = .002), DFS (HR = 1.56, 95% CI: 1.09–2.23, P = .016), and DSS (HR = 2.51, 95% CI: 1.45–4.33, P = .001) in HCC patients (Table 2).
Table 2.
Univariate and multivariate analysis of survival in patients with HCC.
| Univariate analysis | Multivariate analysis | |||||
| Clinicopathological factors | HR | 95% CI | P | HR | 95% CI | P |
| OS | ||||||
| Gender | 0.78 | 0.54–1.13 | .186 | |||
| Age | 1.01 | 1.00–1.03 | .075 | |||
| T stage | 1.66 | 1.37–2.00 | <.001 | 1.23 | 0.65–2.32 | .520 |
| Clinical stage | 1.64 | 1.34–2.01 | <.001 | 1.28 | 0.67–2.46 | .454 |
| Histologic grade | 1.13 | 0.89–1.45 | .324 | |||
| TPX2 expression | 1.86 | 1.29–2.69 | .001 | 1.66 | 1.13–2.45 | .011 |
| PFS | ||||||
| Gender | 1.00 | 0.72–1.38 | .977 | |||
| Age | 1.00 | 0.98–1.01 | .478 | |||
| T stage | 1.60 | 1.37–1.88 | <.001 | 0.86 | 0.49–1.51 | .603 |
| Clinical stage | 1.65 | 1.38–1.92 | <.001 | 1.84 | 1.04–3.25 | .038 |
| Histologic grade | 1.13 | 0.92–1.39 | .244 | |||
| TPX2 expression | 1.86 | 1.36–2.54 | <.001 | 1.66 | 1.20–2.29 | .002 |
| DFS | ||||||
| Gender | 1.18 | 0.82–1.71 | .370 | |||
| Age | 1.00 | 0.98–1.01 | .610 | |||
| T stage | 1.65 | 1.36–2.00 | <.001 | 0.96 | 0.46–2.02 | .958 |
| Clinical stage | 1.68 | 1.36–2.06 | <.001 | 1.69 | 0.79–3.61 | .176 |
| Histologic grade | 1.21 | 0.97–1.53 | .099 | |||
| TPX2 expression | 1.72 | 1.22–2.43 | .002 | 1.56 | 1.09–2.23 | .016 |
| DSS | ||||||
| Gender | 0.85 | 0.53–1.37 | .510 | |||
| Age | 1.00 | 0.99–1.02 | .748 | |||
| T stage | 2.01 | 1.58–2.55 | <.001 | 1.15 | 0.57–2.32 | .691 |
| Clinical stage | 2.02 | 1.54–2.65 | <.001 | 1.65 | 0.81–3.37 | .170 |
| Histologic grade | 1.13 | 0.82–1.54 | .464 | |||
| TPX2 expression | 2.73 | 1.66–4.49 | <.001 | 2.51 | 1.45–4.33 | .001 |
95% CI = 95% confidence interval, DFS = disease-free survival, DSS = disease-specific survival, HR = hazard ratio, OS = overall survival, PFS = progression-free survival, TPX2 = targeting protein for Xenopus kinesin-like protein 2.
Bold values indicated statistical significance.
3.5. Association of TPX2 expression with infiltration of immune cells in HCC
The survival times of patients in several cancers is determined by the quantity and activity status of tumor-infiltrating lymphocytes.[20–22] Therefore, we explored the association among TPX2 expression and the infiltrating immune cells in HCC using TIMER database, and the finding exhibited obvious associations of TPX2 expression with cancer purity and dominant infiltration of immune cells such as B, CD4+T and CD8+T cells, neutrophils, macrophages, and DCs (Fig. 5).
Figure 5.
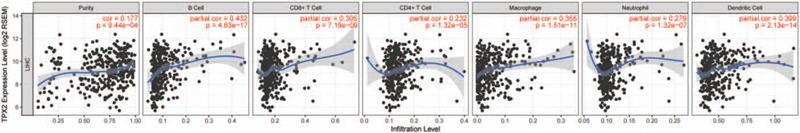
Association analysis of TPX2 expression and infiltration levels of immune cells in HCC. TPX2 expression in HCC tissues was positively associated with tumor purity and infiltration levels of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and DCs.
3.6. Association of TPX2 with immune checkpoints expressions in HCC
Immunotherapy has been recognized as a crucial strategy to limit the progression of malignant tumors. Among these immunotherapies, immune checkpoint inhibitors are the most promising immunotherapy strategies.[23] Given the fact that TPX2 was associated with immune cells infiltration in HCC, we determined the association of TPX2 expression with the abundance of several immune checkpoint molecules published by an authoritative review.[24] As cancer purity (percentage of malignant cells in a tumor tissue) of clinical samples affected the analysis of immune cell infiltration, the correlation analysis was adjusted for purity. The results showed that TPX2 expression was obviously associated with the expression of immune checkpoints in cancer cells, including PDL1, CD80, CD86, FGL1, LGALS3, and CEACAM1 (Table 3). In addition, TPX2 expression was also associated with the expression of immune checkpoints expressed on immune cells, such as PD1, CTLA4, CD28, LAG3, TIGIT, CD96, BTLA, and TIM3 (Table 3). Overall, these findings implied that TPX2 expression might be a novel indicator to measure the abundance of several immune checkpoint molecules.
Table 3.
Correlation analysis between TPX2 and immune checkpoints expression.
| None | Purity | None | Purity | ||||||
| Expressed on tumor cells | Core | P | Core | P value | Expressed on immune cells | Core | P | Core | P |
| PDL1 | 0.060 | .248 | 0.145 | .006 | PD1 | 0.230 | <.001 | 0.338 | <.001 |
| PDL2 | -0.070 | .181 | 0.037 | .497 | CTLA4 | 0.256 | <.001 | 0.376 | <.001 |
| CD80 | 0.245 | <.001 | 0.380 | <.001 | CD28 | 0.106 | .041 | 0.204 | <.001 |
| CD86 | 0.132 | .011 | 0.291 | <.001 | LAG3 | 0.217 | <.001 | 0.266 | <.001 |
| FGL1 | -0.239 | <.001 | -0.193 | <.001 | CD226 | -0.039 | .457 | 0.061 | .262 |
| LGALS3 | 0.141 | .006 | 0.188 | <.001 | TIGIT | 0.202 | <.001 | 0.334 | <.001 |
| CD112 | 0.037 | .472 | 0.033 | .545 | CD96 | 0.050 | .341 | 0.178 | <.001 |
| CD115 | -0.040 | .440 | 0.085 | .113 | BTLA | 0.048 | .359 | 0.182 | <.001 |
| HVEM | 0.034 | .520 | 0.061 | .261 | VISTA | -0.048 | .376 | 0.020 | .706 |
| CEACAM1 | -0.215 | <.001 | -0.196 | <.001 | TIM3 | 0.152 | .003 | 0.315 | <.001 |
Tumor purity is defined as percentage of malignant cells in a tumor tissue.
Bold values indicated statistical significance.
3.7. Coexpressed networks of TPX2 in HCC
To obtain an insight into the biological effect of TPX2 on HCC, the function modules of Linked Omics were used to examine the genes coexpressed with TPX2 in TCGA-LIHC cohort. Genes significantly associated with TPX2 were exhibited in Figure 6A. The top 50 significant genes positively and negatively associated with TPX2 were shown in the heat map (Fig. 6B, C). Thereafter, we carried out GO analysis, including biological process (BP), cellular component (CC), and molecular function (MF) analyses, as well as KEGG analysis on TPX2 and its coexpressed genes by GSEA. BP term annotation showed that TPX2 coexpressed genes participate primarily in chromosome segregation, mitotic cell cycle phase transition, and negative regulation of cell cycle process, while protein activation cascade, fatty acid metabolic process, and peroxisomal transport were inhibited (Fig. 6D). CC term annotation showed that TPX2 coexpressed genes were mainly located in chromosomal region, heterochromatin, and microtubule (Fig. 6E). MF term annotation showed that TPX2 coexpressed genes were mainly associated with positive regulation of motor activity, helicase activity, and DNA secondary structure binding, and negative regulation of electron transfer activity, vitamin binding, and antioxidant activity (Fig. 6F). KEGG term annotation showed that genes coexpressed with TPX2 mostly participate in positively regulating cell cycle and negatively regulating complement and coagulation cascades (Fig. 6G). These results suggested several potential mechanisms of TPX2 affecting HCC progression.
Figure 6.
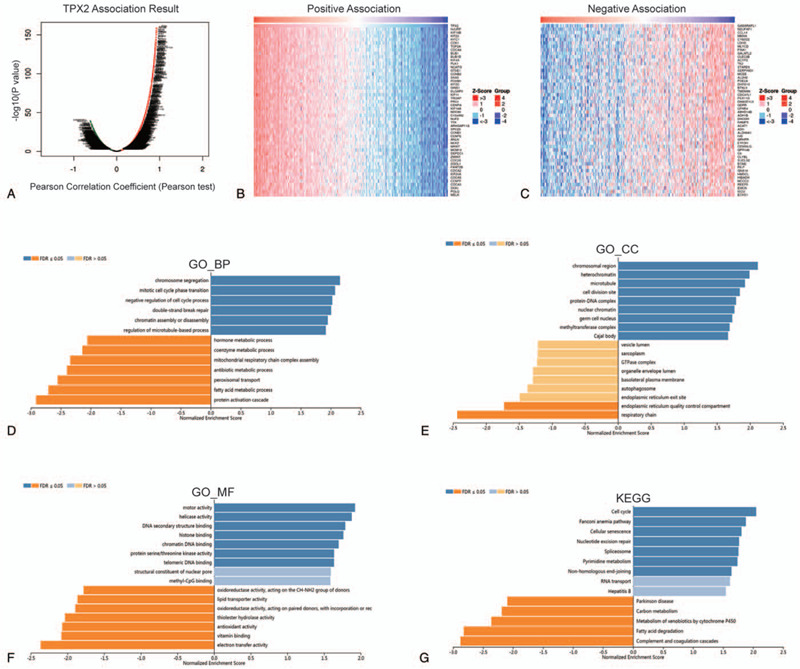
The genes coexpressed with TPX2 in HCC. (A) The global TPX2 highly associated genes identified by Pearson test in HCC cohort. (B) Heat maps showing top 50 genes positively associated with TPX2 in HCC. (C) Heat maps showing top 50 genes negatively associated with TPX2 in HCC. (D) Significantly enriched GO_BP annotations of TPX2 in HCC group. (E) Significantly enriched GO_CC annotations of TPX2 in HCC group. (F) Significantly enriched GO_MF annotations of TPX2 in HCC group. (G) Significantly enriched KEGG pathways of TPX2 in HCC group.
4. Discussion
It can be observed that TPX2 is abnormally expressed in certain nonphysiological conditions, especially in cancerous diseases. It was reported that TPX2 expression was notably upregulated in gastric cancer, esophageal cancer, prostate cancer, etc.[25–27] In addition, Cai et al[28] identified TPX2 as a novel prognosis predictor to predict the risk of metastasis in breast cancer. A study indicated that TPX2 expression is significantly greater in HCC tissues in comparison with that in normal tissues and enhanced TPX2 expression is evidently associated with poorer 5-year OS and DFS in HCC patients.[29] Moreover, CDK5-mediated stabilization of TPX2 can promote HCC tumorigenesis, and inhibiting TPX2 can exert tumor suppressive effects by suppressing the PI3K/AKT signaling pathway in HCC.[30,31] Obviously, these studies revealed that TPX2 is an unfavorable marker and promising therapeutic target in HCC.
Although increasing numbers of studies concentrated on the clinical associations and functions of TPX2 in human cancers, its exact prognosis effect on HCC has not been defined. To obtain more comprehensive insights into the potential prognostic ability of TPX2 in HCC, we conducted a series of systematic bioinformatics analyses of public data. Our current study uncovered that TPX2 was significantly upregulated in HCC tissues and the upregulation of TPX2 was associated with a poorer prognosis in patients, and was also associated with clinical characteristics such as advanced stage and poor differentiation. Taken together, our studies confirmed the oncogenic role of TPX2 in HCC by multilevel analyses, and implied the significant value of TPX2 acting as a diagnostic and prognostic biomarker in HCC.
This study also demonstrated that TPX2 expression was associated with the infiltration status of immune cells in HCC. Our results exhibited that TPX2 expression had significantly positive correlations with cancer purity and the dominant infiltration of immune cells, including B, CD4+T and CD8+T cells, neutrophils, macrophages, and DCs. TPX2 was a microtubule-related protein and participates in regulating cell cycle. Limited researches observed the association between microtubule-related protein and the infiltration and activation of immune cells. Bernasconi et al[32] implied that KIF4 has an association with activated T cells and may participate in cytoskeletal modifications associated with in situ T-cell activation. To our knowledge, we found that TPX2 was a novel indicator to predict infiltration of immune cells HCC for the first time. In addition, we also reported that TPX2 expression was correlated with the expressions of several immune checkpoints. Encouragingly, detection of TPX2 expression may reflect the abundance of several immune checkpoint molecules in HCC, suggesting that TPX2 expression might be a novel indicator to measure the immune status. However, further studies based on patients’ cohorts should be conducted to confirm the critical role of TPX2 expression and assess the association between its expression and response to immunotherapy.
Whatever, the current study also has several unavoidable limitations. First of all, the study is based on bioinformatics analysis, and there are no recruited cohorts for verification of diagnostic and prognostic value of TPX2 in HCC; second, TPX2 is upregulated in almost every cancer type, and its value as a prognostic or diagnostic biomarker for HCC dramatically decreases. Finally, gene markers used to estimate abundance of TIICs always show positive correlation with each other, thus the correlations between kinds of TIICs are also positive [Supplemental Digital Content (figure S2)]. We speculate that the general effect of TPX2 that TPX2 has a widespread association with the tumor-infiltrating immune cells in HCC may be misleading to some extent.
5. Conclusion
To sum up, TPX2 expression has a positive association with a poorer prognosis and can act as a promising diagnostic and prognostic indicator for HCC. In addition, TPX2 expression also has an association with the infiltration of immune cells and immune checkpoint expression. Our results suggest a potential novel immune regulatory role of TPX2 in cancer immunity and lay a foundation for subsequent studies to validate the important role of TPX2 in HCC.
Author contributions
YL and JZ designed the research. HZ, JL, JF, QZ, and TB acquired the data. HZ, JL, JF, QZ, XL, and HS analyzed and interpreted the data. HZ drafted the manuscript. YL and JZ critically revised the manuscript for intellectual content.
Conceptualization: Hongjun Zhu.
Data curation: Hongjun Zhu, Jian Liu, Jia Feng.
Formal analysis: Hongjun Zhu, Jian Liu, Jia Feng, Hui Sun.
Funding acquisition: Jianguo Zhang, Yifei Liu.
Methodology: Hongjun Zhu, Jia Feng.
Project administration: Jianguo Zhang, Yifei Liu.
Validation: Hongjun Zhu, Qing Zhang, Tingting Bian, Xiaoli Li, Jianguo Zhang, Yifei Liu.
Visualization: Qing Zhang, Xiaoli Li.
Writing – original draft: Hongjun Zhu, Jian Liu, Tingting Bian.
Writing – review & editing: Jianguo Zhang, Yifei Liu.
Supplementary Material
Supplementary Material
Footnotes
Abbreviations: 95%CI = 95% confidence interval, BP = biological process, CC = cellular component, DCs = dendritic cells, DFS = disease-free survival, DSS = disease-specific survival, FDR = the false discovery rate, GO = Gene Ontology, GSEA = gene set enrichment analysis, HCC = hepatic cell cancer, HPA = Human Protein Atlas, HR = hazard ratio, KEGG = Kyoto Encyclopedia of Genes and Genomes, KM = Plotter Kaplan--Meier Plotter, MF = molecular function, OS = overall survival, PFS = progression-free survival, TIICs = tumor-infiltrating immune cells, TPX2 = targeting protein for Xenopus kinesin-like protein 2.
How to cite this article: Zhu H, Liu J, Feng J, Zhang Q, Bian T, Li X, Sun H, Zhang J, Liu Y. Overexpression of TPX2 predicts poor clinical outcome and is associated with immune infiltration in hepatic cell cancer. Medicine. 2020;99:49(e23554).
This study was funded by grants from Key Scientific and Technological Projects in Nantong of Jiangsu (MS22018001, MS22019015) and Jiangsu Post-doctoral Foundation Research Project (2019Z142).
The data used to support the findings of this study are available from the relative bioinformatics database.
The main results of this research are based on bioinformatics analysis, thus the ethics approval is not applicable.
The authors declare that they have no conflicts of interest relevant to this study.
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
Supplemental digital content is available for this article.
References
- [1].Kudo M. Adjuvant therapy after curative treatment for hepatocellular carcinoma. Oncology 2011;81: Suppl 1: 50–5. [DOI] [PubMed] [Google Scholar]
- [2].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7–30. [DOI] [PubMed] [Google Scholar]
- [3].Wieczorek M, Bechstedt S, Chaaban S, et al. Microtubule-associated proteins control the kinetics of microtubule nucleation. Nat Cell Biol 2015;17:907–16. [DOI] [PubMed] [Google Scholar]
- [4].Neumayer G, Belzil C, Gruss OJ, et al. TPX2: of spindle assembly, DNA damage response, and cancer. Cell Mol Life Sci 2014;71:3027–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Yan L, Li Q, Yang J, et al. TPX2-p53-GLIPR1 regulatory circuitry in cell proliferation, invasion, and tumor growth of bladder cancer. J Cell Biochem 2018;119:1791–803. [DOI] [PubMed] [Google Scholar]
- [6].Jiang T, Sui D, You D, et al. MiR-29a-5p inhibits proliferation and invasion and induces apoptosis in endometrial carcinoma via targeting TPX2. Cell Cycle 2018;17:1268–78. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [7].Yang Y, Li DP, Shen N, et al. TPX2 promotes migration and invasion of human breast cancer cells. Asian Pac J Trop Med 2015;8:1064–70. [DOI] [PubMed] [Google Scholar]
- [8].Chen M, Zhang H, Zhang G, et al. Targeting TPX2 suppresses proliferation and promotes apoptosis via repression of the PI3k/AKT/P21 signaling pathway and activation of p53 pathway in breast cancer. Biochem Biophys Res Commun 2018;507:74–82. [DOI] [PubMed] [Google Scholar]
- [9].Mei J, Hao L, Liu X, et al. Comprehensive analysis of peroxiredoxins expression profiles and prognostic values in breast cancer. Biomark Res 2019;7:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sun CC, Li SJ, Hu W, et al. Comprehensive analysis of the expression and prognosis for E2Fs in human breast cancer. Mol Ther 2019;27:1153–65. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [11].Xu R, Pan J, Mei J, et al. Systematic characterization of prognostic values of peroxiredoxin family in gastric cancer. BioMed Res Int 2020;2020:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004;6:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lian Q, Wang S, Zhang G, et al. HCCDB: a database of hepatocellular carcinoma expression atlas. Genomics Proteomics Bioinformatics 2018;16:269–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Uhlen M, Oksvold P, Fagerberg L, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 2010;28:1248–50. [DOI] [PubMed] [Google Scholar]
- [15].Uhlen M, Fagerberg L, Hallstrom BM, et al. Proteomics. Tissue-based map of the human proteome. Science 2015;347:1260419. [DOI] [PubMed] [Google Scholar]
- [16].Lanczky A, Nagy A, Bottai G, et al. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat 2016;160:439–46. [DOI] [PubMed] [Google Scholar]
- [17].Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res 2017;77:e108–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li B, Severson E, Pignon JC, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol 2016;17:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 2018;46:D956–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Denkert C, von Minckwitz G, Darb-Esfahani S, et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol 2018;19:40–50. [DOI] [PubMed] [Google Scholar]
- [21].Almangush A, Ruuskanen M, Hagstrom J, et al. Tumor-infiltrating lymphocytes associate with outcome in nonendemic nasopharyngeal carcinoma: a multicenter study. Hum Pathol 2018;81:211–9. [DOI] [PubMed] [Google Scholar]
- [22].Chen TH, Zhang YC, Tan YT, et al. Tumor-infiltrating lymphocytes predict prognosis of breast cancer patients treated with anti-Her-2 therapy. Oncotarget 2017;8:5219–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ma W, Gilligan BM, Yuan J, et al. Current status and perspectives in translational biomarker research for PD-1/PD-L1 immune checkpoint blockade therapy. J Hematol Oncol 2016;9:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Qin S, Xu L, Yi M, et al. Novel immune checkpoint targets: moving beyond PD-1 and CTLA-4. Mol Cancer 2019;18:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Tomii C, Inokuchi M, Takagi Y, et al. TPX2 expression is associated with poor survival in gastric cancer. World J Surg Oncol 2017;15:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sui C, Song Z, Yu H, et al. Prognostic significance of TPX2 and NIBP in esophageal cancer. Oncol Lett 2019;18:4221–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zou J, Huang RY, Jiang FN, et al. Overexpression of TPX2 is associated with progression and prognosis of prostate cancer. Oncol Lett 2018;16:2823–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Cai Y, Mei J, Xiao Z, et al. Identification of five hub genes as monitoring biomarkers for breast cancer metastasis in silico. Hereditas 2019;156:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Liu Q, Tu K, Zhang H, et al. TPX2 as a novel prognostic biomarker for hepatocellular carcinoma. Hepatol Res 2015;45:906–18. [DOI] [PubMed] [Google Scholar]
- [30].Huang DH, Jian J, Li S, et al. TPX2 silencing exerts antitumor effects on hepatocellular carcinoma by regulating the PI3K/AKT signaling pathway. Int J Mol Med 2019;44:2113–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wang F, Zhao W, Gao Y, et al. CDK5-mediated phosphorylation and stabilization of TPX2 promotes hepatocellular tumorigenesis. J Exp Clin Cancer Res 2019;38:286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bernasconi P, Cappelletti C, Navone F, et al. The kinesin superfamily motor protein KIF4 is associated with immune cell activation in idiopathic inflammatory myopathies. J Neuropathol Exp Neurol 2008;67:624–32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


