ABSTRACT
Sporotrichosis is a subcutaneous infection caused by fungi from the genus Sporothrix. It is transmitted by inoculation of infective particles found in plant-contaminated material or diseased animals, characterizing the classic sapronotic and emerging zoonotic transmission, respectively. Since 1998, southeastern Brazil has experienced a zoonotic sporotrichosis epidemic caused by S. brasiliensis, centred in the state of Rio de Janeiro. Our observation of feline sporotrichosis cases in Brasília (Midwestern Brazil), around 900 km away from Rio de Janeiro, led us to question whether the epidemic caused by S. brasiliensis has spread from the epicentre in Rio de Janeiro, emerged independently in the two locations, or if the disease has been present and unrecognized in Midwestern Brazil. A retrospective analysis of 91 human and 4 animal cases from Brasília, ranging from 1993 to 2018, suggests the occurrence of both sapronotic and zoonotic transmission. Molecular typing of the calmodulin locus identified S. schenckii as the agent in two animals and all seven human patients from which we were able to recover clinical isolates. In two other animals, the disease was caused by S. brasiliensis. Whole-genome sequence typing of seven Sporothrix spp. strains from Brasília and Rio de Janeiro suggests that S. brasiliensis isolates from Brasília are genetically distinct from those obtained at the epicentre of the outbreak in Rio de Janeiro, both in phylogenomic and population genomic analyses. The two S. brasiliensis populations seem to have separated between 2.2 and 3.1 million years ago, indicating independent outbreaks or that the zoonotic S. brasiliensis outbreak might have started earlier and be more widespread in South America than previously recognized.
KEYWORDS: Sporothrix, sporotrichosis, Sporothrix brasiliensis, Sporothrix schenckii, zoonotic transmission, Brasília
Introduction
The emergence of fungal pathogens worldwide is an important and frequently neglected public health issue [1]. One of the most frequent genera of pathogenic fungi is Sporothrix (Sordariomycetes, Ascomycota), which causes the subcutaneous granulomatous disease called sporotrichosis [2,3]. The most common etiologic agent of the disease, S. schenckii sensu lato, was recently shown to harbour three cryptic species: S. schenckii sensu stricto, S. globosa, and the emerging species S. brasiliensis [4,5,6]. The three species are saprophytes and produce sympodial and sessile conidia that switch their morphology to the pathogenic yeast phase after a transcutaneous injury [7]. The three species differ in their virulence. S. brasiliensis is more virulent in animal models than S. schenckii and S. globosa, and causes localized to disseminated sporotrichosis that can be fatal to both humans and cats [8]. Lower antifungal susceptibility and refractory sporotrichosis occurs in some Brazilian cases related to the zoonotic epidemics [9,10]. Previous population genetics surveys reveal that S. brasiliensis is largely clonal with strong biogeographic patterns of distribution [6]. On the other hand, its sister taxon S. schenckii shows higher rates of recombination and little to no population structure among Brazilian strains [6].
Currently, sporotrichosis is especially frequent in Latin America, South Africa, Madagascar, China, India, and Japan [3], but it can also be observed elsewhere [11]. In addition to endemic disease, multiple sporotrichosis outbreaks have been reported in the last century [2,12,13]. Before the 1990s, sporotrichosis was considered a disease of low-to-moderate endemicity in Brazil. Importantly, over the past two decades, it has become an urgent health problem [7,14]. In 1997, three individuals from the same family got infected by Sporothrix spp. after feline zoonotic transmission events in Rio de Janeiro, Brazil [15]. Since 1998, over 5,100 cats have been diagnosed with sporotrichosis in the state of Rio de Janeiro; >5,000 human cases were reported in 2015, suggesting high incidence of the disease in the state [16]. Rio de Janeiro is currently considered one of the three (along with China and South Africa) human sporotrichosis hyperendemic regions [15,17,18]. Rio de Janeiro also shows a high incidence of feline sporotrichosis and zoonotic transmission from cats to humans [6,19].
Reports of sporotrichosis have increased across South America. Recently, sporotrichosis caused by S. brasiliensis was described in Northeast Brazil, and primarily via zoonotic transmission [20,21,22,23]. Additionally, infections with S. brasiliensis were identified in Argentina and Paraguay, the first reports of this species outside Brazil [24,25]. These recent reports of S. brasiliensis-associated sporotrichosis in places with few or no cases reported prior to the early 2000s raises the question of whether the outbreak has recently spread from Rio de Janeiro, or if zoonotic sporotrichosis cases have been unrecognized. This question is crucial to establish public health policy to deal with emergent sporotrichosis. In this work, we address this question prompted by our observation in 2015 of a case of sporotrichosis in Brasília, in a cat whose owner also had a lesion that was suggestive of the disease.
In this study, additional animal cases and retrospective study of a series of sporotrichosis cases in the University Hospital of Brasília were investigated for evidence of zoonotic human infections. The genomes of six S. brasiliensis strains, including those from Brasília and from the epicentre Rio de Janeiro, were sequenced and compared to the reference S. brasiliensis 5110 genome [26] and other Sporothrix spp. We report for the first time the occurrence of S. brasiliensis infections in animals at the capital of Brazil, Brasília, located in the Midwestern portion of the country. Finally, genetic diversity in the S. brasiliensis strains in Brasília was observed, which suggests that independent outbreaks caused by S. brasiliensis occur and might have started earlier and be more widespread than previously recognized.
Material and Methods
Human cases
A cross-sectional study of a series of sporotrichosis cases in the University Hospital of Brasília, from 1993 to 2018 was completed. This study was previously approved by the Ethics Committee of the Faculty of Medicine, University of Brasília (protocol CAAE: 873718.0.00005558). The inclusion criterion was diagnosis of sporotrichosis by culture of Sporothrix spp. in the Mycology Laboratory of the University Hospital of Brasília. Diagnoses were performed by growing the fungus from biological samples (skin lesions, sputum, broncho-alveolar lavage, and cerebrospinal fluid) on solid culture media (i.e. Sabouraud and Mycosel) followed by microscopic characterization of colonies. The following data were anonymously retrieved from patient medical records: clinical form of sporotrichosis (cutaneous, mucosal and extracutaneous forms), location, year of diagnosis, age, sex, gender, race, occupation, treatment, and disease outcome.
Animal cases
Three cats and one dog with suspected sporotrichosis from the Veterinary Hospital of the University of Brasília, the Directorate for Environmental Surveillance (zoonoses division) of the Federal District Health Department and a private veterinary clinic were referred to the Mycology Laboratory of the University Hospital of Brasília for diagnosis between 2015 and 2018. Direct microscopic observation of patient samples was completed, and skin lesion specimens were cultured as above, followed by microscopic identification of the fungus. Relevant clinical and epidemiological data were collected. Animal observations and tests were previously approved by the University of Brasília Ethics Committee on the Use of Animals (CEUA-UnB), protocol: 66716/2016.
Morphological studies
Seven Sporothrix spp. isolates from humans (recovered from prolonged storage), and four isolates from animal cases were studied for morphological and molecular characteristics. For the identification of the mycelial phase, monosporic cultures were grown on Sabouraud dextrose agar at room temperature. We used the slide culture method to characterize microscopic filamentous features of the Sporothrix spp. isolates. We inoculated mycelia fragments into 1 × 1 cm Sabouraud dextrose agar and malt extract agar blocks, which were then mounted on a slide, covered with a coverslip and incubated for 14–21 days at room temperature. To induce the yeast phase, we inoculated the mycelial fragments into Brain-Heart Infusion broth and incubated for 7 days at 37°C and 150 rpm agitation. Both mycelial and yeast preparations were stained with lactophenol blue and visualized with a Zeiss AxioObserver Z1 microscope equipped with a 40X objective. Images were collected using the Zeiss ZEN software.
Molecular typing and identification
Genomic DNA was extracted using the PureLink Genomic DNA Mini Kit (Thermo Fisher Scientific, Waltham, MA, USA) from 500 mg of mycelial tissue as input material using a glass bead protocol with a Precellys 24 instrument (Bertin Instruments, Montigny-le-Bretonneux, France). DNA integrity was assessed by running 5 μL aliquot of the extraction on a 0.8% agar agarose-gel electrophoresis stained with 0.5 µg/ml ethidium bromide. To estimate the DNA yield we used a NanoDrop 1000c Instrument (Thermo Fisher Scientific, Waltham, MA, USA). We then amplified a segment of the calmodulin gene by PCR and Sanger sequenced it in an ABI 3130xl Instrument (Applied Biosystems). PCR setting and cycling conditions were done as previously described [6]. We performed initial nucleotide BLAST analysis (BLASTn) for each amplicon to retrieve the best hit results for each strain on the NCBI database. All extractions (N=11) showed Sporothrix spp. as the closest BLAST match.
Whole genome sequencing
We selected two S. brasiliensis (A001 and A005) and one S. schenckii (A003) animal isolates from the Mycology Laboratory of the University Hospital of Brasília for Illumina short read whole-genome sequencing. For comparison, we have also sequenced four S. brasiliensis isolates obtained from humans in Rio de Janeiro, s15677, s28606, s34180 and s48605 (see supplementary Table 1). A001 and A005, were labelled as SbFD (S. brasiliensis, Federal District), whereas isolates s15677, s28606, s34180 and s48605, from the Collection of Pathogenic Fungi at the Oswaldo Cruz Foundation, were designated as SbRJ. From each isolate, we extracted 1 µg of DNA, as described above. Next, we prepared paired-end libraries using the NEBNext® Ultra™ DNA Library Prep Kit for Illumina. The libraries were quantified using the NEBNext® Library Quant Kit for Illumina® (New England Biolabs). We pooled equimolar amounts of each library together and sequenced them in an Illumina NovaSeq 6000 Instrument using the NovaSeq 6000 S4 kit v1.5 (300 cycles). The run was performed in a high output mode. We used Trimmommatic v. 0.36 [27] to remove adapters and filter low-quality bases.
Public data
We used three additional sets of Illumina paired-end reads from S. schenckii isolates from Colombia (EM7 and MS1 - [28]) and Puerto Rico (ATCC 58251 - [29]). To root phylogenetic trees (see below), we retrieved additional genomic assemblies from other Sporothrix species from the NCBI genome browser (Supplementary Table 1).
Read mapping and variant calling
We mapped the newly-sequenced strains and the publicly available data to the S. brasiliensis 5110 reference genome using bwa-mem v 0.7.7 [30]. To identify mismatch intervals and indels, we used GATK v3.3-0RealignerTargetCreator and IndelRealigner [31,32]. To identify polymorphic sites (single nucleotide polymorphisms, SNP), we used the GATK UnifiedGenotyper module using the parameter het = 0.01 to account for a haploid organism. We used the following filters to obtain SNPs calls: QD = 2.0 || FS_filter = 60.0 || MQ_filter = 30.0 || MQ_Rank_Sum_filter = −12.5 || Read_Pos_Rank_Sum_filter = −8 [33]. We excluded polymorphic sites with less than 10X coverage, with less than 90% variant allele calls, or that were identified by Nucmer [34] as located in duplicated regions in the S. brasiliensis reference genome. We identified 115,488 SNPs across five Sporothrix species (16 isolates).
Phylogenetic tree and whole-genome alignments
We used the WGS datasets we generated plus publicly available sequences described above to build a phylogenetic tree of the 16 Sporothrix spp. isolates. We used IQTREE v 1.6.1 [35] to build a Maximum Likelihood (ML) tree with the –m TEST function to determine the best nucleotide substitution model under the ascertainment bias correction (+ASC) model. To assess the strength of the support for each clade present in the tree, we found the consensus phylogenomic tree of 1,000 ultrafast bootstraps coupled with 1,000 Shimodaira–Hasegawa-like approximate likelihood ratio tests (SH-aLRT) [36,37]. We visualized the consensus topology and branch support with FigTree v1.4.2 – http://tree.bio.ed.ac.uk/software/figtree/. We used the Automatic Assembly For The Fungi (AAFTF) pipeline to assemble the newly-sequenced S. brasiliensis genomes, with default parameters [38]. The S. brasiliensis assemblies with the highest N50 (Supplementary Table 1) were aligned to the strain 5110 reference genome using the DGENIES online tool [39] via minimap2 algorithm [40] for comparative genomic purposes.
Timing analysis
Next, we estimated the approximate divergence time of the splits observed in the phylogenomic analysis using Bayesian inference in BEAST v1.10.4 [41]. Evolutionary rates become artificially larger when only SNP sites are included in the alignment, which in turn could underestimate dating estimates (as time and rate are inversely correlated). In order to minimize such bias, we implemented a correction that takes into account invariable sites as well. This was done by inputting in the.xml files the total counts of A, C, G and T across the whole genomes sequenced, minus their counts in the SNP alignment alone (therefore avoiding counting the latter twice). We used a GTR model with gamma variation among sites (using four discrete classes). Priors for the different dating parameters included: a Normal distribution in which a conservative 90% confidence interval was within 3.8 - 4.9 MYA (following [26]) marking the separation between S. brasiliensis and S. schecnkii; a rate interval of [0.9E-3, 16.7E-3] substitutions/site/branch/MY for the parameter ucld.mean (mean of the uncorrelated lognormal distribution of rates across branches, instead of assuming a more simplistic global clock prior), based on evolutionary rate estimation based on different groups of fungi [42]; and a birth-death tree prior with incomplete sampling [43]. We used two independent Markov Chain Monte Carlo (MCMC) chains each run until convergence; the marginal distributions and traces from the two replicate chains were inspected in Tracer v1.7 [44]. We summarized the two MCMC runs using LogCombiner (BEAST package, [45]) after discarding the burnin region. Finally, we used Treeannotator (BEAST package, [45]) to annotate clade posteriors, divergence times, and rate variation across branches.
Population genetic analysis
Finally, we studied the amount of genetic diversity in each Sporothrix group. We estimated the average nucleotide diversity (π) within species or populations and absolute differentiation (Dxy) between S. brasiliensis and S. schenckii, and between the S. brasiliensis SbFD and SbRJ using a series of Python scripts available at https://github.com/simonhmartin/genomics_general.
Results
Both sapronotic and zoonotic transmissions of sporotrichosis occur in Brasília and surrounding areas
The study population involved 91 human cases ranging from 1 to 82 years of age with sporotrichosis confirmed by isolation of Sporothrix spp. in culture at the Mycology Laboratory of the University Hospital of Brasília. All patients were diagnosed either by direct exam or by the microbiological isolation of the pathogen from 1993 to 2018. We reported 64 cases (70%) in males and 27 (30%) females. The years with most cases reported were 1999, 2000, and 2009 with 12 (13%), 7 (8%), and 8 (9%) cases, respectively. Regarding age groups, the 21-30- and 51-60-year ranges have more cases in this retrospective study, corresponding to 17 (18.7%) and 16 (17.6%) cases, respectively (Table 1). Most patients were treated with itraconazole alone or in association with saturated potassium iodide. In severe cases, patients were treated with fluconazole and/or amphotericin B.
Table 1.
Epidemiological characteristics of sporotrichosis in Brasília, Brazil.
| Gender (n = 91) | |
| Male | 64 (70.32%) |
| Female | 27 (29.68%) |
| Profession (n = 91) | |
| Students | 26 (28.57%) |
| Rural Worker | 14 (15.38%) |
| Household/retired | 13 (14.28%) |
| Veterinary | 2 (2.2%) |
| Other | 4 (4.39%) |
| Not reported | 32 (35.18%) |
| HIV condition (n = 91) | |
| positive | 3 (3.29%) |
| negative | 88 (96.71%) |
| Age Group (n = 91) | |
| 0–10 | 12 (13.19%) |
| 11–20 | 10 (10.99%) |
| 21–30 | 17 (18.68%) |
| 31–40 | 11 (12.09%) |
| 41–50 | 11 (12.09%) |
| 51–60 | 16 (17.58%) |
| >61 | 14 (15.38%) |
| Sporotrichosis forms (n = 48) | |
| Cutaneous fixed | 6 (12.5%) |
| Lymphocutaneous | 34 (70.83%) |
| Disseminated | 5 (10.42%) |
| Unknown | 3 (6.25%) |
| Source of infection (n = 48) | |
| Living with a cat or dog | 6 (12.25%) |
| Cat/dog bite or scratch | 5 (10.42%) |
| Trauma with plant/wood material | 5 (10.42%) |
| Unknown | 32 (66.91%) |
We were able to analyze 48 full medical records of the included patients. Among them, 34 were presumably infected in Brasília (70.9%) and six in the neighboring state of Goiás (12.5%). Most patients had the lymphocutaneous form (n = 34; 70.8%, 95% confidence interval [CI] = 55.8%–81.6%), followed by fixed cutaneous form (n = 6; 12.6%, 95% CI=5.1%–23.4%). Five extra-cutaneous cases were detected in the central nervous system (n = 1; 2%, 95% CI = 0.2%–9.6%) and or the upper airway tract (n = 4; 8.3%, 95% CI = 2.7–18.21%). The clinical presentation of three patients (6.3%) was inconclusive or not reported. The most frequent profession/occupation was student (n = 26; 29%, 95% CI = 39.2%–67.0%), followed by farmers (n = 14; 15%, 95% CI = 17.2%–42.3%) and retirees (n = 13, 14%, 95% CI = 15.5%–40.0%), (Table 1). Some comorbidities we found were prostate and breast cancer, high blood pressure, diabetes, cirrhosis, tuberculosis, leishmaniosis, hepatitis and other chronic diseases. Chagas disease and paracoccidioidomycosis co-infections were found in two sporotrichosis patients. Three cases of HIV coinfection were also diagnosed. Three cases were fatal, two of which in patients with cancer and one with chronic cirrhosis.
Additionally, a thorough investigation of the clinical records suggested two main forms of Sporothrix infections in Brasília: (1) the classical sapronotic form acquired from a direct inoculation of plant-derived material harbouring the fungus, which was more observed in rural and construction workers, and (2) the zoonotic transmission triggered by skin injuries provoked by infected cat or dog bite/scratches. Sapronotic sporotrichosis was reported in 5 patients (10%, 95% CI = 3.8%–20.1%) after injuries with plant-derived material. Five patients (10%, 95% CI = 10.8%–33.2%), including two veterinarians, reported cat/dog bites or scratches. Six other patients (12.6%, 95% CI = 5.1%–23.4%) reported living and having frequent contact with dogs or cats with skin lesions. Other patients either did not report any previous injuries or reported a different exposure source (i.e. sporotrichosis acquired after injuries with barbed wire and metal cans).
In parallel, we diagnosed, from 2015 to 2018, 4 animals (3 cats and 1 dog) with sporotrichosis in Brasília. These animals had classical clinical presentations such as multiple suppurative granulomatous cutaneous lesions with a high load of yeast cells, generally with involvement of mucous membranes. Lesions were commonly found at parts such as the head, especially on the nose, as well as on the chest and legs. Two felines were euthanized and the other cat and the dog were treated with itraconazole and potassium iodide. The pet owner of the cat from which the A001 strain was recovered, was also diagnosed with cutaneous sporotrichosis. The diagnosis and treatment were made in another hospital in Brasília, so we do not have medical records or a clinical isolate obtained from the owner.
Sporothrix brasiliensis isolates from Brasília and the epicentre of the zoonotic sporotrichosis epidemics, Rio de Janeiro, are genetically distinct
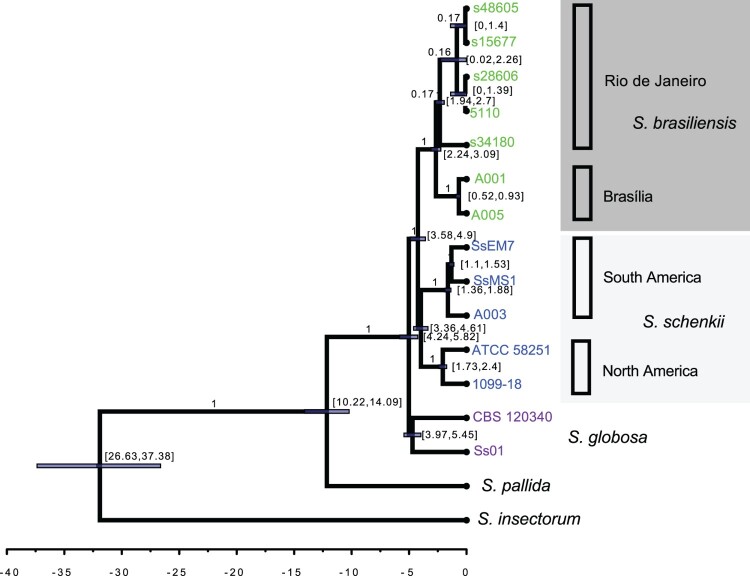
In order to distinguish between two different hypotheses for S. brasiliensis presence in Brasília, that it was recently introduced here from Southeast Brazil or that the Brasília isolates are actually endemic to that region, we performed a number of genetics and genomics analyses. First, we inferred the genealogical relationships between isolates of different species of Sporothrix using whole genome sequencing. By rooting our tree with S. insectorum, we observed that S. schenckii and S. brasiliensis are sister species and form a triad along with S. globosa (Figure 1). The Most Recent Common Ancestor (tMRCA) of Sporothrix emerged between 26.63 and 37.38 Million Years Ago (MYA) (Figure 1). Unlike S. insectorum and S. pallida, this triad of species cause sporotrichosis in humans and animals and form the “Sporothrix pathogenic clade”. The crown age of this pathogenic group (TMRCA) is between 4.24 - 5.28 MYA. The divergence between the pathogenic clade and S. pallida (a non-pathogenic species [46], but see [47]) occurred 10.22 - 14.09 MYA, suggesting that the pathogenesis syndrome must have arisen in the last 14.1 million years.
Figure 1.
Whole-genome Sporothrix genealogy using BEAST analyses. The phylogenomic tree was inferred using Bayesian inference under the GTR model with gamma variation among sites and S. insectorum was set as the outgroup. The 90% confidence interval marking the separation between S. brasiliensis and S. schenckii was set to 3.8 - 4.9 MYA, a rate interval of [0.9E-3, 16.7E-3] based on evolutionary rate estimation based on different groups of fungi and a birth-death tree prior with incomplete sampling were set as priors. The clade distribution, posterior probabilities and divergence times are shown.
Our WGS phylogenetic tree also allowed us to assess the partition of the genetic variation within S. schenckii. We found support for the existence of two clades, one harbouring two Colombian isolates (SsEM7 and SsMS1) and a cat-derived isolate from Brasília (A003) and a clade composed by one strain from the USA (1099-18) and an additional strain from Puerto Rico (ATCC 58251). We estimate that divergence of these two groups occurred between 3.36 and 4.61 MYA. These results suggest that South American and North American populations of S. schenckii are genetically differentiated and call for the question of whether there has been speciation associated with geographical distribution within S. schenckii.
Next, we studied the relationships between the S. brasiliensis isolates from Brasília and Rio de Janeiro. The five Rio de Janeiro isolates (5110, s15677, s28606, s34180 and s48605) form a monophyletic group that does not include the two Brasília isolates, A001 and A005, which in turn are clustered in their own group. We also compared genomic similarities between the two S. brasiliensis cluster variants by aligning one individual from each cluster to the 5110 reference strains using the minimap2 tool. By aligning the S. brasiliensis strain s15677 to the S. brasiliensis 5110 reference strain (both belonging to the Rio de Janeiro population), we observed that the genomes are 99.81% similar. By aligning the S. brasiliensis A001 strain to the same reference, the genomic similarity dropped to 96.31% suggesting that those two groups are genetically unrelated (Supplementary Figure 1).
We calculated the approximate divergence time between S. brasiliensis isolates from Brasília and Southeastern Brazil. The TMRCA of the S. brasiliensis species was estimated to be 2.24 - 3.09 MYA, the TMRCA for the strains from Rio de Janeiro is 1.94 - 2.7 MYA, and the TMRCA for the strains from Brasília was 0.52-0.93 MYA. The S. brasiliensis isolates A001 and A005 are clustered in a single branch and are phylogenetically different from those isolated in Rio de Janeiro (5110, s15677, s28606, s34180 and s48605 – Figure 1). This result suggests that those infections were likely to be locally acquired in Brasília, not in Rio de Janeiro. These results suggest that the two populations are genetically distinct and that the infections observed in Brasília are not the result of a small group of Rio de Janeiro isolates that migrated. Since the causal agent of these infections is genetically differentiated from the causal agent of the Rio de Janeiro infections, it seems more likely that infections caused by S. brasiliensis in Brasília happened autochthonously.
We evaluate a second prediction of the hypothesis that Rio de Janeiro isolates caused the Brasília outbreak. If this hypothesis is correct, then the source population of the outbreak, RJ, should have a larger genetic diversity than the outbreak population, Brasília, because the former has not undergone a bottleneck. We measured the magnitude of genetic variation in each Sporothrix group using pairwise calculations of heterozygosity. Sporothrix schenckii has the highest nucleotide diversity (π = 1.67%), which is in line with the measurements of other fungal species with complex population composition and worldwide distribution [48,49]. Sporothrix brasiliensis has a much lower nucleotide diversity (π = 0.52%) than that of its sister species, probably due to its endemicity to South America. We next calculated the nucleotide diversity within the two Brazilian S. brasiliensis groups. The Brasília group has a nucleotide diversity on the same order of magnitude as S. brasiliensis as a whole (π = 0.20%). On the other hand, the Rio de Janeiro population has an extremely low genetic diversity (π = 9.78 × 10−4%), suggesting an extreme bottleneck. In agreement with our inference from the phylogenomic tree, these results indicate that the Brasília population is extremely unlikely to be derived from the Rio de Janeiro one.
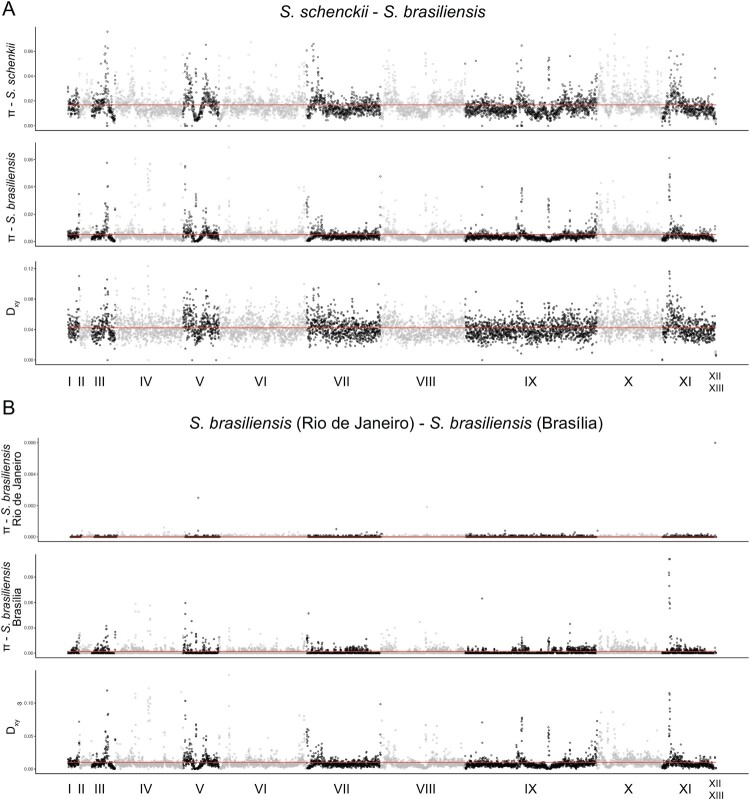
Our phylogenetic tree suggested genetic differentiation between S. brasiliensis isolates from Rio de Janeiro and Brasília. We calculated the pairwise absolute genetic distance (Dxy) between S. schenckii and S. brasiliensis, and within S. brasiliensis. The Dxy between those two species was 0.042 (95% CI=0.0419–0.0425) while between the Brasília and Rio de Janeiro populations it was 0.010 (95% CI = 0.01–0.01). Figure 2 shows the mean pairwise differences per site within populations (nucleotide diversity or π) and mean pairwise differences per site between populations (Dxy) along the 13 scaffolds of the Sporothrix genome. Dxy, a proxy of genome differentiation, between S. schenckii and S. brasiliensis shows high mean values along the whole genome. We observed a strikingly different pattern when we compare the two populations of S. brasiliensis. As expected, Rio de Janeiro S. brasiliensis has little genetic variation along the whole genome (Figure 2A). Dxy along most of the genome is low suggesting low differentiation between Rio de Janeiro and Brasília despite almost 1 MY of divergence (See above, Figure 1). Nonetheless, scaffolds I, III, V, IX and XI, show peaks of high differentiation. Notably, the Dxy peaks in scaffolds I, III, and XI show fixed variants between Rio de Janeiro and Brasília S. brasiliensis and between S. brasiliensis (as a whole) and S. schenckii. These results suggest that the divergence between Brazilian populations of S. brasiliensis is localized to a few genome regions and not as pronounced as between S. schenckii and S. brasiliensis. This pattern might simply be explained by the difference in divergence time between the Sporothrix species and the S. brasiliensis populations.
Figure 2.
Population genetics plots along the S. brasiliensis 5110 reference genome. We have calculated the mean nucleotide diversity (π) within species or populations and absolute differentiation (Dxy) between S. brasiliensis and S. schenckii, and between the S. brasiliensis SbFD and SbRJ (red line) and plotted along each scaffold of the S. brasiliensis 5110 genome using a 5 kb window.
Discussion
Sporotrichosis due to S. brasiliensis has become one of the most important fungal diseases in Brazil [18]. The disease has gone from a localized outbreak in Rio de Janeiro state to a country-wide epidemic during the past 20 years and has affected thousands of humans and animals. In Brasília, located in Midwestern Brazil, and roughly 900 km from Rio de Janeiro, we observed indications that suggest cat/dog-transmitted sporotrichosis might occur. Moreover, both cats and dogs are affected by pathogenic Sporothrix spp. In Rio de Janeiro, both fungal species have been isolated from dogs [19], but S. brasiliensis has been identified in almost all cats tested so far [17]. The etiologic agent of this disease was recently found in clinical and environmental samples in Argentina as well in animal samples in Paraguay [24,25].
In this study, two different hypotheses for the expansion of this disease throughout South America were evaluated, namely that S. brasiliensis recently (< 20 years) spread from Rio de Janeiro to other areas in Brazil and other countries via human, animal hosts or fomites; or that different groups of S. brasiliensis occur in different locations of the South American continent. We focused on a recent sporotrichosis case series in Brasília. If the former hypothesis is correct, one would expect that the Rio de Janeiro population would have a larger genetic variation than the Brasília population. Additionally, one would expect the Rio de Janeiro population to encompass the genetic variants observed in Brasília. Neither of these were observed in our data. The nucleotide diversity (π) in Rio de Janeiro is 200 times lower than in Brasília. Additionally, the Rio de Janeiro and Brasília populations are reciprocally monophyletic, which contradicts the idea that Brasília strains are a subset of the Rio de Janeiro group. Finally, the split time between Brasília and Rio de Janeiro occurred between 0.52-0.93 MYA, predating the human colonization of the Americas and thus precluding the possibility that S. brasiliensis migrated associated with humans.
The observation that the surge of sporotrichosis in Brasília is not caused by recent migration from Rio de Janeiro poses additional questions. First, the reasons behind the surge remain unclear. Other reasons, besides human migration must be responsible for the recent uptick of cases. One possibility is that deforestation and human expansion into previously uninhabited areas has put humans (and their pets) in greater contact with fungi. A second possibility is that in the last 20 years, S. brasiliensis became more virulent. Both novel mutations and introgression from a virulent lineage could be responsible. Finally, it is possible that the improvement in diagnostics has led to the identification and reporting of sporotrichosis due to S. brasiliensis.
Second, it remains unclear why the number of cases in Brasília is lower than that in Rio de Janeiro, one of the most hyperendemic areas in the world. One potential explanation for the higher incidence of sporotrichosis in Rio de Janeiro as compared to Brasília is higher population density and poor sanitation in the former area [18]. Additionally, the two lineages of S. brasiliensis might also differ in their virulence. Finally, our study shows that cats can acquire sporotrichosis caused by both S. schenckii and S. brasiliensis, which indicates that feral and stray cats can be reservoirs of the disease. Brasília and Rio de Janeiro may differ in their number of outdoor cats. Studies have shown a high prevalence of parasitosis in cats from both locations (Brasília: [50], RJ:[51]) but the number of felines and their relative health conditions remain unknown. Currently, we have no support for these three non-mutually exclusive hypotheses.
Our work suggests the existence of at least two groups of S. brasiliensis in South America. The increase in sporotrichosis incidence, along with the increments on the disease burden of other mycoses across the world, and the outbreak of Cryptococcus gattii – originated in the Amazon region – in the Pacific Northwest [52] are sobering indications of the importance of efforts to detect and mitigate or prevent the spread of fungal emerging diseases, both in South America and elsewhere.
Supplementary Material
Acknowledgments
The authors would like to thank to Jéssica S. Boechat and Manoel M. E. Oliveira (Fiocruz) for the technical support. A.M.N was funded by FAP-DF awards 0193.001048/2015-0193.001561/2017 and the CNPq grant 437484/2018-1. B.M.B. was supported by National Institutes of Health/National Institute of Allergy and Infectious Diseases award R21AI28536. D.R.M. was supported by National Institutes of Health/National Institute of General Medical Sciences award R01GM121750. J.E.S. was supported by National Institutes of Health/National Institute of Allergy and Infectious Diseases awards R01AI130128 and R01AI127548 and is a CIFAR Fellow in the program Fungal Kingdom: Threats and Opportunities. M.M.T was supported by CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) award 43460/2018-2. M.S.S.F was supported by FAP-DF (Fundação de Apoio a Pesquisa do Distrito Federal) award 193.001.533/2016. S.A.P. was supported by FAPERJ, grant number E-26/202.737/2019.
Funding Statement
The authors would like to thank to Jéssica S. Boechat and Manoel M. E. Oliveira (Fiocruz) for the technical support. A.M.N was funded by FAP-DF awards 0193.001048/2015-0193.001561/2017 and the CNPq grant 437484/2018-1. B.M.B. was supported by National Institutes of Health/National Institute of Allergy and Infectious Diseases award R21AI28536. D.R.M. was supported by National Institutes of Health/National Institute of General Medical Sciences award R01GM121750. J.E.S. was supported by National Institutes of Health/National Institute of Allergy and Infectious awards R01AI130128 and R01AI127548 and is a CIFAR Fellow in the program Fungal Kingdom: Threats and Opportunities. M.M.T was supported by CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) award 43460/2018-2. M.S.S.F was supported by FAP-DF (Fundação de Apoio a Pesquisa do Distrito Federal) award 193.001.533/2016. S.A.P. was supported by FAPERJ, grant number E-26/202.737/2019. Conselho Nacional de Desenvolvimento Científico e Tecnológico. Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro. Fundação de Apoio à Pesquisa do Distrito Federal. National Institute of Allergy and Infectious Diseases. National Institute of General Medical Sciences.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Fisher MC, Gurr SJ, Cuomo CA, et al. Threats posed by the fungal Kingdom to humans, wildlife, and agriculture. mBio. 2020 May 5;11(3). doi: 10.1128/mBio.00449-20. PubMed PMID: 32371596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao L, Ma Y, Zhao W, et al. Three new species of cyphellophora (Chaetothyriales) associated with sooty blotch and flyspeck. PLoS One. 2015;10(9):e0136857. doi: 10.1371/journal.pone.0136857. PubMed PMID: 26398347; PubMed Central PMCID: PMCPMC4580582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakrabarti A, Bonifaz A, Gutierrez-Galhardo MC, et al. Global epidemiology of sporotrichosis. Med Mycol. 2015 Jan;53(1):3–14. doi: 10.1093/mmy/myu062. PubMed PMID: 25526781. [DOI] [PubMed] [Google Scholar]
- 4.Marimon R, Cano J, Gene J, et al. Sporothrix brasiliensis, S. globosa, and S. mexicana, three new Sporothrix species of clinical interest. J Clin Microbiol. 2007 Oct;45(10):3198–3206. doi: 10.1128/JCM.00808-07. PubMed PMID: 17687013; PubMed Central PMCID: PMCPMC2045377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodrigues AM, de Hoog GS, de Camargo ZP.. Sporothrix species causing outbreaks in animals and humans driven by animal-animal transmission. PLoS Pathog. 2016 Jul;12(7):e1005638. doi: 10.1371/journal.ppat.1005638. PubMed PMID: 27415796; PubMed Central PMCID: PMCPMC4945023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teixeira Mde M, Rodrigues AM, Tsui CK, et al. Asexual propagation of a virulent clone complex in a human and feline outbreak of sporotrichosis. Eukaryot Cell. 2015 Feb;14(2):158–169. doi: 10.1128/EC.00153-14. PubMed PMID: 25480940; PubMed Central PMCID: PMCPMC4311920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orofino-Costa R, Macedo PM, Rodrigues AM, et al. Sporotrichosis: an update on epidemiology, etiopathogenesis, laboratory and clinical therapeutics. An Bras Dermatol. 2017 Sep-Oct;92(5):606–620. doi: 10.1590/abd1806-4841.2017279. PubMed PMID: 29166494; PubMed Central PMCID: PMCPMC5674690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Della Terra PP, Rodrigues AM, Fernandes GF, et al. Exploring virulence and immunogenicity in the emerging pathogen Sporothrix brasiliensis. PLoS Negl Trop Dis. 2017 Aug;11(8):e0005903. doi: 10.1371/journal.pntd.0005903. PubMed PMID: 28854184; PubMed Central PMCID: PMCPMC5595342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Almeida-Paes R, Oliveira MME, Freitas DFS, et al. Refractory sporotrichosis due to Sporothrix brasiliensis in humans appears to be unrelated to in vivo resistance. Med Mycol. 2017 Jul 1;55(5):507–517. doi: 10.1093/mmy/myw103. PubMed PMID: 27771622. [DOI] [PubMed] [Google Scholar]
- 10.Galhardo MC, De Oliveira RM, Valle AC, et al. Molecular epidemiology and antifungal susceptibility patterns of Sporothrix schenckii isolates from a cat-transmitted epidemic of sporotrichosis in Rio de Janeiro, Brazil. Med Mycol. 2008 Mar;46(2):141–151. doi: 10.1080/13693780701742399. PubMed PMID: 18324493. [DOI] [PubMed] [Google Scholar]
- 11.Makri N, Paterson GK, Gregge F, et al. First case report of cutaneous sporotrichosis (Sporothrix species) in a cat in the UK. JFMS Open Rep. 2020 Jan-Jun;6(1):2055116920906001. doi: 10.1177/2055116920906001. PubMed PMID: 32110427; PubMed Central PMCID: PMCPMC7025424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Govender NP, Maphanga TG, Zulu TG, et al. An outbreak of lymphocutaneous sporotrichosis among mine-workers in South Africa. PLoS Negl Trop Dis. 2015 Sep;9(9):e0004096. doi: 10.1371/journal.pntd.0004096. PubMed PMID: 26407300; PubMed Central PMCID: PMCPMC4583532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coles FB, Schuchat A, Hibbs JR, et al. A multistate outbreak of sporotrichosis associated with sphagnum moss. Am J Epidemiol. 1992 Aug 15;136(4):475–487. doi: 10.1093/oxfordjournals.aje.a116521. PubMed PMID: 1415167. [DOI] [PubMed] [Google Scholar]
- 14.Queiroz-Telles F, Fahal AH, Falci DR, et al. Neglected endemic mycoses. Lancet Infect Dis. 2017 Nov;17(11):e367–e377. doi: 10.1016/S1473-3099(17)30306-7. PubMed PMID: 28774696. [DOI] [PubMed] [Google Scholar]
- 15.de Lima Barros MB, Schubach TM, Galhardo MC, et al. Sporotrichosis: an emergent zoonosis in Rio de Janeiro. Mem Inst Oswaldo Cruz. 2001 Aug;96(6):777–779. doi: 10.1590/s0074-02762001000600006. PubMed PMID: 11562701. [DOI] [PubMed] [Google Scholar]
- 16.Gremiao IDF, Oliveira MME, de Miranda LH Monteiro, et al. Geographic expansion of sporotrichosis, Brazil. Emerg Infect Dis. 2020 Mar;26(3):621–624. doi: 10.3201/eid2603.190803. PubMed PMID: 32091376; PubMed Central PMCID: PMCPMC7045854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boechat JS, Oliveira MME, Almeida-Paes R, et al. Feline sporotrichosis: associations between clinical-epidemiological profiles and phenotypic-genotypic characteristics of the etiological agents in the Rio de Janeiro epizootic area. Mem Inst Oswaldo Cruz. 2018 Mar;113(3):185–196. doi: 10.1590/0074-02760170407. PubMed PMID: 29412358; PubMed Central PMCID: PMCPMC5804311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gremiao ID, Miranda LH, Reis EG, et al. Zoonotic epidemic of sporotrichosis: cat to human transmission. PLoS Pathog. 2017 Jan;13(1):e1006077. doi: 10.1371/journal.ppat.1006077. PubMed PMID: 28103311; PubMed Central PMCID: PMCPMC5245785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodrigues AM, de Melo Teixeira M, de Hoog GS, et al. Phylogenetic analysis reveals a high prevalence of Sporothrix brasiliensis in feline sporotrichosis outbreaks. PLoS Negl Trop Dis. 2013;7(6):e2281. doi: 10.1371/journal.pntd.0002281. PubMed PMID: 23818999; PubMed Central PMCID: PMCPMC3688539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silva GM, Howes JCF, Leal CAS, et al. Surto de esporotricose felina na região metropolitana do Recife. Pesquisa Veterinária Brasileira. 2018;38:1767–1771. [Google Scholar]
- 21.Nunes GDL, Carneiro RS, Filgueira KD, et al. ESPOROTRICOSE FELINA NO MUNICÍPIO DE ITAPORANGA, ESTADO DA PARAÍBA, BRASIL: RELATO DE UM CASO. Arquivos de Ciências Veterinárias e Zoologia da UNIPAR. 2011;14(2):5. [Google Scholar]
- 22.Marques-Melo EH, Lessa DFdS, Nunes ACBT, et al., editors. Felino doméstic como agente transmissor de esprotricose para humano: relato ddo primeiro caso no estado de alagoas; 2014.
- 23.Filgueira KD. Esporotricose na espécie canina: relato de um caso na cidade de Mossoró, RN. Ciência Animal Brasileira. 2009;10(2):5. [Google Scholar]
- 24.Cordoba S, Isla G, Szusz W, et al. Molecular identification and susceptibility profile of Sporothrix schenckii sensu lato isolated in Argentina. Mycoses. 2018 Jul;61(7):441–448. doi: 10.1111/myc.12760. PubMed PMID: 29500853. [DOI] [PubMed] [Google Scholar]
- 25.García Duarte JM, Wattiez Acosta VR, Fornerón Viera PML, et al. Esporotricosis trasmitida por gato doméstico. Reporte de un Caso Familiar. Revista del Nacional (Itauguá). 2017;9:67–76. [Google Scholar]
- 26.Teixeira MM, de Almeida LG, Kubitschek-Barreira P, et al. Comparative genomics of the major fungal agents of human and animal sporotrichosis: Sporothrix schenckii and Sporothrix brasiliensis. BMC Genomics. 2014;15:943. doi: 10.1186/1471-2164-15-943. PubMed PMID: 25351875; PubMed Central PMCID: PMC4226871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bolger AM, Lohse M, Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014 Aug 1;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. PubMed PMID: 24695404; PubMed Central PMCID: PMCPMC4103590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomez OM, Alvarez LC, Munoz JF, et al. Draft genome sequences of two sporothrix schenckii clinical isolates associated with human sporotrichosis in Colombia. Genome Announc. 2018 Jun 14;6(24). doi: 10.1128/genomeA.00495-18. PubMed PMID: 29903814; PubMed Central PMCID: PMCPMC6003735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuomo CA, Rodriguez-Del Valle N, Perez-Sanchez L, et al. Genome sequence of the pathogenic fungus sporothrix schenckii (ATCC 58251). Genome Announc. 2014 May 22;2(3). doi: 10.1128/genomeA.00446-14. PubMed PMID: 24855299; PubMed Central PMCID: PMCPMC4031338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009 Jul 15;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. PubMed PMID: 19451168; PubMed Central PMCID: PMCPMC2705234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011 May;43(5):491–498. doi: 10.1038/ng.806. PubMed PMID: 21478889; PubMed Central PMCID: PMCPMC3083463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKenna A, Hanna M, Banks E, et al. The genome analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010 Sep;20(9):1297–1303. doi: 10.1101/gr.107524.110. PubMed PMID: 20644199; PubMed Central PMCID: PMCPMC2928508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teixeira MM, Alvarado P, Roe CC, et al. Population structure and genetic diversity among isolates of Coccidioides posadasii in Venezuela and Surrounding regions. mBio. 2019 Nov 26;10(6). doi: 10.1128/mBio.01976-19. PubMed PMID: 31772050; PubMed Central PMCID: PMCPMC6879716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurtz S, Phillippy A, Delcher AL, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5(2):R12. doi: 10.1186/gb-2004-5-2-r12. PubMed PMID: 14759262; PubMed Central PMCID: PMCPMC395750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen LT, Schmidt HA, von Haeseler A, et al. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015 Jan;32(1):268–274. doi: 10.1093/molbev/msu300. PubMed PMID: 25371430; PubMed Central PMCID: PMCPMC4271533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minh BQ, Nguyen MA, von Haeseler A.. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 2013 May;30(5):1188–1195. doi: 10.1093/molbev/mst024. PubMed PMID: 23418397; PubMed Central PMCID: PMCPMC3670741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guindon S, Dufayard JF, Lefort V, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010 May;59(3):307–321. doi: 10.1093/sysbio/syq010. PubMed PMID: 20525638. [DOI] [PubMed] [Google Scholar]
- 38.Stajich JE, Palmer J. Automatic Assembly For The Fungi (AAFTF). Zenodo; 2019.
- 39.Cabanettes F, Klopp C.. D-GENIES: dot plot large genomes in an interactive, efficient and simple way. PeerJ. 2018;6:e4958. doi: 10.7717/peerj.4958. PubMed PMID: 29888139; PubMed Central PMCID: PMCPMC5991294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018 Sep 15;34(18):3094–3100. doi: 10.1093/bioinformatics/bty191. PubMed PMID: 29750242; PubMed Central PMCID: PMCPMC6137996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suchard MA, Lemey P, Baele G, et al. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018 Jan;4(1):vey016. doi: 10.1093/ve/vey016. PubMed PMID: 29942656; PubMed Central PMCID: PMCPMC6007674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kasuga T, White TJ, Taylor JW.. Estimation of nucleotide substitution rates in Eurotiomycete fungi. Mol Biol Evol. 2002 Dec;19(12):2318–2324. doi: 10.1093/oxfordjournals.molbev.a004056. PubMed PMID: 12446823. [DOI] [PubMed] [Google Scholar]
- 43.Stadler T. On incomplete sampling under birth-death models and connections to the sampling-based coalescent. J Theor Biol. 2009 Nov 7;261(1):58–66. doi: 10.1016/j.jtbi.2009.07.018. PubMed PMID: 19631666. [DOI] [PubMed] [Google Scholar]
- 44.Rambaut A, Drummond AJ, Xie D, et al. Tracer v1.7. 2018.
- 45.Bouckaert R, Heled J, Kuhnert D, et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2014 Apr;10(4):e1003537. doi: 10.1371/journal.pcbi.1003537. PubMed PMID: 24722319; PubMed Central PMCID: PMCPMC3985171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choappa RM Cruz, Oyarzo PI Vieille, Silva LC Carvajal. [Isolation of Sporothrix pallida complex in clinical and environmental samples from Chile]. Rev Argent Microbiol. 2014 Oct-Dec;46(4):311–314. doi: 10.1016/S0325-7541(14)70088-4. PubMed PMID: 25576414. [DOI] [PubMed] [Google Scholar]
- 47.Morrison AS, Lockhart SR, Bromley JG, et al. An environmental Sporothrix as a cause of corneal ulcer. Med Mycol Case Rep. 2013 Apr 10;2:88–90. doi: 10.1016/j.mmcr.2013.03.002. PubMed PMID: 24432225; PubMed Central PMCID: PMCPMC3885947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matute DR, Sepulveda VE.. Fungal species boundaries in the genomics era. Fungal Genet Biol. 2019 Jul 4;131:103249. doi: 10.1016/j.fgb.2019.103249. PubMed PMID: 31279976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leffler EM, Bullaughey K, Matute DR, et al. Revisiting an old riddle: what determines genetic diversity levels within species? PLoS Biol. 2012;10(9):e1001388. doi: 10.1371/journal.pbio.1001388. PubMed PMID: 22984349; PubMed Central PMCID: PMCPMC3439417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aquino LC, Hicks CA, Scalon MC, et al. Prevalence and phylogenetic analysis of haemoplasmas from cats infected with multiple species. J Microbiol Methods. 2014 Dec;107:189–196. doi: 10.1016/j.mimet.2014.10.013. PubMed PMID: 25447887; PubMed Central PMCID: PMCPMC4263531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Macieira DB, de Menezes Rde C, Damico CB, et al. Prevalence and risk factors for hemoplasmas in domestic cats naturally infected with feline immunodeficiency virus and/or feline leukemia virus in Rio de Janeiro–Brazil. J Feline Med Surg. 2008 Apr;10(2):120–129. doi: 10.1016/j.jfms.2007.08.002. PubMed PMID: 17905624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hagen F, Ceresini PC, Polacheck I, et al. Ancient dispersal of the human fungal pathogen Cryptococcus gattii from the Amazon rainforest. PLoS One. 2013;8(8):e71148. doi: 10.1371/journal.pone.0071148. PubMed PMID: 23940707; PubMed Central PMCID: PMC3737135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.