Abstract
Although multiple lifestyle exposures simultaneously impact blood pressure (BP) and cardiovascular health, most analysis so far has considered each single lifestyle exposure (e.g., smoking) at a time. Here, we exploit gene-multiple lifestyle exposure interactions to find novel BP loci. For each of 6,254 Framingham Heart Study participants, we computed lifestyle risk score (LRS) value by aggregating the risk of four lifestyle exposures (smoking, alcohol, education, and physical activity) on BP. Using the LRS, we performed genome-wide gene-environment interaction analysis in systolic and diastolic BP using the joint 2 degree of freedom (DF) and 1 DF interaction tests. We identified one genome-wide significant (P-value < 5 × 10−8) and 11 suggestive (P-value < 1 × 10−6) loci. Gene-environment analysis using single lifestyle exposures identified only one of the 12 loci. Nine of the 12 BP loci detected were novel. Loci detected by the LRS were located within or nearby genes with biologically plausible roles in the pathophysiology of hypertension, including KALRN, VIPR2, SNX1, and DAPK2. Our results suggest that simultaneous consideration of multiple lifestyle exposures in gene-environment interaction analysis can identify additional loci missed by single lifestyle approaches.
Keywords: Blood pressure, gene-environment interaction, lifestyle risk score, loci discovery, multiple lifestyle exposures
Introduction
Blood pressure (BP) is a heritable (Ehret, 2010) but modifiable risk factor for cardiovascular diseases (CVD) with major global health and economic burden (Forouzanfar et al., 2017). Characterizing the genetic architecture of BP is critical to advancing our understanding of underlying biological mechanisms, enhancing risk prediction, and developing targeted lifestyle interventions and drugs (Timpson, Greenwood, Soranzo, Lawson, & Richards, 2018). Genome-wide association studies (GWAS) have identified at least 901 loci associated with BP, which are estimated to account for 11.2% of the genetic variance in BP (Evangelou et al., 2017); a large proportion of heritability in BP remains unexplained. This leaves room for enhanced locus discovery by exploring alternative approaches.
One such approach is to consider gene-lifestyle interactions in BP, given that several lifestyle exposures (e.g., smoking status) have been shown to modulate genetic effects on complex traits such as BP (Parnell et al., 2014; Rao et al., 2017). By accounting for gene-lifestyle interactions in BP, one can find genetic loci whose effects vary by lifestyle exposure. Such genetic loci may otherwise be undetected in traditional genetic main effect GWAS (Rao et al., 2017). For example, recent genome-wide interaction analyses performed by the CHARGE Gene-Lifestyle Interactions Working Group identified several novel loci with biologic plausibility for their involvement in BP and lipid homeostasis (Bentley et al., 2019; de Vries et al., 2019; Feitosa et al., 2018; Kilpeläinen et al., 2019; Sung et al., 2019, 2018). Finding such loci is of value for gaining insight to the complex biological underpinnings of BP (Laville et al., 2019).
To date, most genome-wide gene-lifestyle interaction analyses have primarily considered gene interactions with a single lifestyle exposure, such as alcohol (de Vries et al., 2019; Feitosa et al., 2018; Simino, Sung, Kume, Schwander, & Rao, 2013), smoking (Bentley et al., 2019; Sung, de las Fuentes, Schwander, Simino, & Rao, 2015; Sung et al., 2018), and education (Basson et al., 2014). Yet, the genetic effects of some loci may be simultaneously influenced by multiple lifestyle exposures given that cardiovascular health (including BP) is concurrently impacted by multiple lifestyle exposures (Folsom et al., 2011; van Dam, Li, Spiegelman, Franco, & Hu, 2008). In fact, the current strategy for improving cardiovascular health in the general population promotes multiple healthy lifestyle behaviors simultaneously (Lloyd-Jones et al., 2010). We may be able to further enhance discovery of novel BP loci whose effects are modulated by multiple lifestyle exposures by using genome-wide approaches that account for interactions with more than one lifestyle exposure.
To identify loci that interact with multiple environmental factors, a Bayesian approach has been developed, but its implementation is burdened by high computational requirements and an inability to account for family relatedness (Moore et al., 2018). An alternative approach would be to aggregate multiple lifestyle exposures into a composite score that captures overall risk for unfavorable cardiovascular outcomes due to unhealthy lifestyles. This approach has been applied in epidemiological studies (Nayor, Enserro, Vasan, & Xanthakis, 2016; Ogunmoroti et al., 2017), used to examine how genetic risk for CVD and BP is modified by a composite score derived from multiple lifestyle exposures (Pazoki et al., 2018), and recently compared to other approaches for jointly testing multiple interactions involving several candidate genetic variants and exposures (Kim et al., 2019). However, it is yet to be used in genome-wide gene-lifestyle interaction analysis for finding BP loci.
In this study, we report on our findings using a risk score aggregating multiple lifestyle exposures, hereafter ‘Lifestyle risk score’ (LRS), when identifying BP loci. We selected four lifestyle exposures with well-documented effects on BP and cardiovascular health (Chobanian et al., 2003): smoking status, alcohol intake, level of educational attainment (a proxy for socio-economic status), and physical activity. We performed genome-wide interaction studies on BP using both quantitative and discretized LRS. Additionally, we examined whether our analysis using the LRS can identify additional loci not detected through analysis using each of single lifestyle exposures separately.
Methods
Study Population
In this study, we used the Framingham Heart Study (FHS) data, obtained from the database of genotypes and phenotypes (dbGaP). FHS study participants are made up of three cohorts; the Original Cohort recruited in 1948, the Offspring Cohort recruited in 1971, and the Third Generation Cohort recruited in 2002. Across these three cohorts, we selected contemporaneous visits, namely, the 26th visit for the Original Cohort, the 7th visit for the Offspring Cohort, and the 1st visit for the Third Generation Cohort.
In this study, we considered FHS participants of European ancestry that were between 18 and 80 years (to avoid growth or age related changes to BP) (Shankar, Eckert, Saha, Tu, & Pratt, 2005; Wei, 1992), and had non-missing genotype data and phenotype data, including age, sex, SBP and/or DBP, anti-hypertensive medication status, and the four component lifestyle exposures (smoking status, alcohol intake, education, and physical activity). After applying these study inclusion criteria, there were 6,254 and 6,253 participants for SBP and DBP, respectively, and participants belonged to either the Offspring or Third Generation Cohort.
Genotype data
We removed genotyped single nucleotide polymorphisms (SNPs) from FHS data with Hardy-Weinberg equilibrium P-values < 10−6 or call rates < 90%, and imputed SNPs with imputation quality measures < 0.3. We further removed SNPs with minor allele frequency ≤ 1% or, if the product of minor allele count and imputation quality was ≤ 20. Appendix Table S1 shows a summary of the resulting analysis SNP sets, composed of ∼2.4 million SNPs.
Phenotype data
SBP and DBP were measured with a consistent protocol. For each individual, BP values represent an average of three measurements, one taken by a nurse/technician and two taken by a physician. For individuals on anti-hypertensive medications, BP values were adjusted by adding 15 mmHg to SBP and 10 mmHg to DBP (Tobin, Sheehan, Scurrah, & Burton, 2005).
Lifestyle exposures and LRS
We selected the following four lifestyle exposures: smoking status, alcohol intake, level of educational attainment (a proxy for socio-economic status), and physical activity.
We considered two dichotomous variables of each of the first three lifestyle exposures and one variable for physical activity. Table 1 shows the derivation and coding for each of the dichotomous variables representing the four lifestyle exposures. For example, for smoking status, the variable “Smoke now” (Smk_now), reflects whether the individual was a smoker at the time of the clinic visit, while “Smoke ever” (Smk_ever), reflects whether an individual has ever smoked (in the past or at the time of the clinic visit).
Table 1.
Lifestyle exposure variables used in this study
| Lifestyle exposure | Variable | Original variable(s) in FHS | Modification/Remark | Values* | |
|---|---|---|---|---|---|
| Smoking | 1. | Smk_now | Smoke regularly in the past year? | 0: former and never smokers 1: current smokers only |
Smk_now (0) Smk_now (1) |
| 2. | Smk_ever | Ever smoke cigarettes regularly? | 0: never smokers only 1: former and current smokers |
Smk_ever (0) Smk_ever (1) |
|
| Alcohol | 3. | Alc_heavy | Total number of alcoholic drinks/week | 0: consumed 0 or 1 – 7 drinks/week | Alc_heavy (0) |
| 1: consumed > 7 drinks/week | Alc_heavy (1) | ||||
| 4. | Alc_moderate | Total number of alcoholic drinks/week | 0: consumed 0 or > 7 drinks/week | Alc_moderate (0) | |
| 1: consumed 1 – 7 drinks/week | Alc_moderate (1) | ||||
| Education | 5. | Edu_somecol | Post-secondary education? (Education level in categories/years) | 0: if no post-secondary education (i.e., no schooling, grades 1 – 11, high school grad or GED or years of education ≤ 12). | Edu_somecol (0) |
| 1: if there is some form of post-secondary education (i.e., some college, technical school, associate degree, bachelors degree, or graduate/professional degree or years of education ≥ 13) | Edu_somecol (1) | ||||
| 6. | Edu_gradcol | Graduate college? (Education level in categories/years) | 0: if no graduate college degree (i.e., all the levels below bachelors degree defined above). | Edu_gradcol (0) | |
| 1: if bachelors/graduate/professional degree or ≥ 16 years of education. | Edu_gradcol (1) | ||||
| Physical activity | 7. | PA_active | Hours spent in moderate activity, and hours spent in heavy activity | Sum of time spent in moderate activity and twice the time spent in heavy activity, then split by lower quartile (Q1=3); 0: ≥ 3 hours of time equivalent in moderate and heavy physical activity 1: < 3 hours of time equivalent in moderate and heavy physical activity. |
PA_active (0) PA_active (1) |
Value assignment across variables was done in one of two ways; for Smk_now, Smk_ever, Alc_heavy and PA_active, 0 was assigned to the unexposed group and 1 to the exposed group, while for Alc_moderate, Edu_somecol and Edu_gradcol, the unexposed group was assigned a value of 1 and the exposed group a value of 0.
For analyses involving a composite of multiple lifestyle exposures, the LRS was computed in two main steps:
First, risk was assigned to each value of a lifestyle exposure based on its well-established effect on BP or cardiovascular health (Chobanian et al., 2003; Loucks, Abrahamowicz, Xiao, & Lynch, 2011; WHO, 2013). As presented in Table 2, three risk scores were used, no risk (0), low risk (1), and high risk (2). High risk scores are associated with unfavorable cardiovascular health outcomes. For alcohol intake, risk assignment reflects reported J-shaped associations indicating the cardio-protective effect of moderate alcohol consumption compared to abstinence, based on extensive data including healthy individuals and CVD patients (Costanzo et al., 2019; O’Keefe, Bybee, & Lavie, 2007). Unlike the other lifestyle exposures, physical activity has only two risk levels (no risk and low risk) because the effect of any level of activity on BP (as compared to sedentariness) is much more pronounced than amount of activity among active individuals (Diaz & Shimbo, 2013). Moreover, there is no consensus on a cutoff to distinguish between active and very active individuals (with discernible differences in the effects on BP); this problem is compounded by the known overestimation (or underestimation) of self-reported physical activity values (Prince et al., 2008).
Table 2.
Component lifestyle exposures and risk assignment for computing Lifestyle risk score (LRS)
| Component variable | No risk (0) | Low risk (1) | High risk (2) |
|---|---|---|---|
| Smoking | Never | Former | Current |
| Alcohol intake (drinks/week) | Modest (1 – 7) | Abstinence (0) | Heavy (> 7) |
| Education | College degree | Some college | None |
| Physical activity | Active | Inactive |
Subsequently, the quantitative LRS (QLRS) was computed by summing across risk scores of all four lifestyle exposures. We also dichotomized the LRS, as is common practice to harmonize heterogeneous quantitative exposure variables in multi-cohort collaborations (Palla, Higgins, Wareham, & Sharp, 2010). The dichotomous LRS (DLRS), was derived by splitting QLRS by the median threshold of 2; individuals with QLRS values < 2 were assigned to the unexposed (favorable lifestyle) group, while individuals with QLRS values ≥ 2 were assigned to the exposed (unfavorable lifestyle) group. The favorable lifestyle group includes individuals who were at no risk for any of the four exposures or at low risk for only one of the exposures. Individuals with all other combinations of risk levels for the four exposures would fall into the unfavorable lifestyle group.
Statistical analysis
Genome-wide gene-LRS interaction scan: we fit a linear mixed effect model that jointly models the genetic, lifestyle, and interaction effects (Kraft, Yen, Stram, Morrison, & Gauderman, 2007), while adjusting for covariates and potential confounders (e.g., lifestyle-covariate interactions) (Keller, 2014). The model is
| (1) |
where y is the BP trait (SBP or DBP), E is the lifestyle exposure (QLRS or DLRS), G is the dosage of the genetic variant, and Xcov is the vector of covariates including age, sex, lifestyle exposure interactions with age (age*E) and sex (sex*E). is the intercept, is the environmental main effect, is the genetic main effect, is the gene-environment interaction effect, are the covariate effects, and is the error term. From the model output, we calculated two test statistics which follow a χ2 distribution under the null; a 2 degree of freedom (DF) joint test of and (H0: ) and a 1 DF test of (H0: ).
Genome-wide genetic effect scans: to further assess the extent to which signals detected using Model (1) above would be missed in the absence of the interaction term, we performed two additional genome-wide scans using main effects models that adjust for the effect of exposures but do not include a gene-environment interaction term, namely, Models (2) and (3) as defined below:
| (2) |
| (3) |
where model terms are as defined for Model (1), with the exception that there is no gene-environment interaction term in Models (2) and (3). Moreover, in Model (3), in lieu of the aggregate LRS, each exposure variable (i.e., smoking status, alcohol intake, education, and physical activity) is separately adjusted (i.e., ). Also, in Model (3), covariates adjusted for include age, sex, as well as age interactions with each lifestyle exposure (i.e., age*smoking status, age*alcohol intake, age*education, and age*physical activity) and sex interactions with each lifestyle exposure (i.e., sex*smoking status, sex*alcohol intake, sex*education, and sex*physical activity). Here, based on the output from the models, we calculated a 1 DF test of the genetic main effect (H0: ).
Genome-wide gene-single lifestyle exposures scan: we performed genome-wide scans fitting Model (1) as defined above, using each of seven single lifestyle exposure variables (defined in Table 1) as E in Model (1). This genome-wide scan was performed to determine whether the LRS identifies additional loci not detected through analysis using each of single lifestyle exposures separately. As described above in the genome-wide gene-LRS scan, we calculated a 2 DF joint test and 1 DF interaction test.
We accounted for family relatedness in the FHS dataset by applying the genome-wide rapid association using mixed model and regression (GRAMMAR) approach (Aulchenko, de Koning, & Haley, 2007). This approach involves obtaining pedigree-adjusted residuals by fitting a polygenic model on the phenotype (BP traits) using the kinship matrix as a random component. The GRAMMAR approach was applied using the GenABEL package in R (Aulchenko et al., 2007). The resulting pedigree-adjusted residuals were used as the phenotype in the models described above. We obtained effect estimates using the ProbABEL package from the GenABEL suite of programs (Aulchenko, Struchalin, & van Duijn, 2010).
In all, we performed 18 genome-wide analyses using Model (1) to determine whether using an aggregate of multiple lifestyle exposures can identify additional BP loci relative to using individual lifestyle exposures: 2 phenotypes (SBP and DBP) x 9 lifestyle exposure variables (7 individual components of the LRS + QLRS + DLRS). Further, we performed six more genome-wide scans, using Models (2) and (3) to assess the extent to which the interaction term contributed to discovering signals.
We identified SNPs as significant using the threshold of P-value < 5 × 10−8, and as suggestive using the threshold of P-value < 1 × 10-6. For each suggestive/significant association, a locus was defined as a cluster of SNPs within 500 kb of the index SNP (i.e., the SNP with the lowest P-value in the region). We considered loci as novel if component SNPs were not within 500 kb of previously known BP loci in published literature between 2011 and 2019 (reference list in Supporting information S1), and were not detected in the CHARGE Gene-Lifestyle Interactions Working Group’s large-scale studies on gene-smoking (Sung et al., 2018) and gene-alcohol (Feitosa et al., 2018) interactions for BP. Further, we annotated suggestive or significant SNPs using FUMA GWAS (Functional Mapping and Annotation of Genome-Wide Association Studies) (Watanabe, Taskesen, Bochoven, & Posthuma, 2017). Annotation was focused on RegulomeDB categorical scores (Boyle et al., 2012), which indicate evidence for the presence of regulatory elements (e.g., enhancers, promoters, insulators or transcription binding factors), and CADD (Combined Annotation Dependent Depletion) scores (Kircher et al., 2014), which predict whether the functional consequence of a variant is likely to be deleterious. Lastly, using the GWAS atlas tool (https://atlas.ctglab.nl/; Watanebe et al. 2019), we explored whether suggestive or significant SNPs detected by the gene-LRS genome-wide scan have been previously reported in the literature as associated with a BP or other cardiovascular trait at nominal significance (i.e., P-value < 0.05 for the marginal genetic effect).
Results
Descriptive summary statistics for the study sample are presented in Table 3. For QLRS that ranged from 0 – 7, most individuals had intermediate values of the score and with fewer individuals in the tails (Figure 1). Less than a third of the 6,254 eligible participants in the study sample fell into the favorable lifestyle (unexposed) group of the DLRS and were on average younger, more likely female, and had lower BP than those in the exposed group.
Table 3.
Descriptive statistics by lifestyle risk score in FHS study sample
| QLRS | DLRS | ||
|---|---|---|---|
| Unexposed | Exposed | ||
| Sample size | 6254 | 1924 | 4330 |
| Age (years) | 48.4±13.6 | 44.1±12.5 | 50.3 ±13.6 |
| % Male | 46.9 | 43.0 | 48.6 |
| % Taking anti-hypertensive medications | 18.1 | 10.9 | 21.3 |
| SBP (mmHg) | 122.8±18.9 | 117.7±16.7 | 125.0±19.4 |
| DBP (mmHg) | 76.7±10.4 | 75.3±10.0 | 77.3±10.5 |
Sample size shown is for individuals with SBP phenotype data and is the basis for other descriptive statistics shown. Descriptive statistics are very similar for DBP phenotype data with sample size differing by a single individual (N=6,253).
Figure 1.
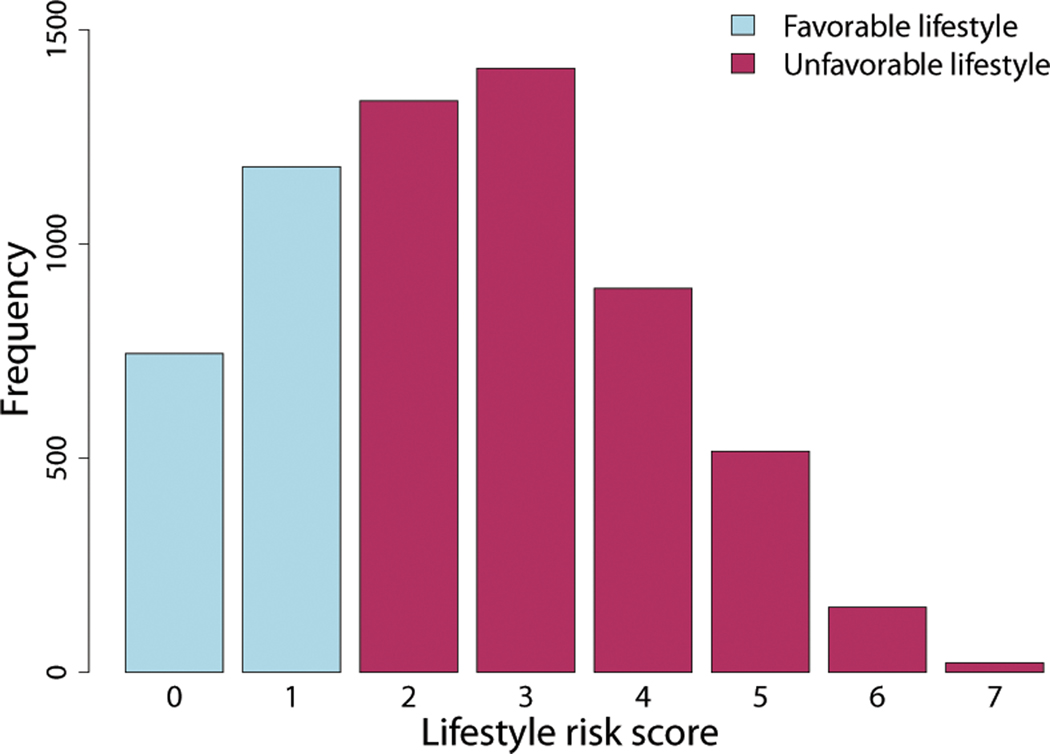
Distribution of lifestyle risk score (LRS) in the FHS data. The quantitative LRS (QLRS), created by summing risks across the four lifestyle exposures, ranged between 0 and 7. The dichotomous LRS (DLRS), created by splitting QLRS by the median of its distribution, was composed of two groups: favorable lifestyle group (QLRS < 2) and unfavorable lifestyle group (QLRS ≥ 2).
QQ plots are presented in appendix Figures S1 and S2. For SBP, genomic control values (λ) ranged from 0.962 – 1.027 for the joint 2 DF test and from 0.952 – 1.036 for the interaction 1 DF test. For DBP, λ ranged from 0.882 – 0.923 for the joint 2 DF test and from 0.966 – 1.022 for the interaction 1 DF test. There was no indication of genomic inflation in any of the 18 analysis sets.
Figure 2 shows a Venn diagram summarizing the numbers of detected loci that were unique to the QLRS, the DLRS, and the single lifestyle exposures. While single lifestyle exposures detected substantially more loci (26 loci in total compared to seven by the QLRS and five by the DLRS), the QLRS and DLRS detected non-overlapping loci that would have been missed in the absence of an aggregate LRS analysis. Association results from the LRS analysis are summarized in Table 4 (Manhattan plots in appendix Figure S3). Also, for associations detected using the LRS analysis, Table 5 presents P-values for the genetic effect when the marginal effect of exposures are adjusted for but interaction terms are absent (i.e., based on Models (2) and (3)). The LRS analysis identified three genome-wide significant and 18 suggestive SNPs, representing 12 loci in all. Eleven of these 12 loci detected by the QLRS and DLRS analyses were not captured by analyses using single lifestyle exposures. With the exception of the one locus that was found in common by QLRS and Smk_ever analysis for SBP, there was generally no overlap in loci found by analysis using each LRS and the seven single lifestyle exposures.
Figure 2.
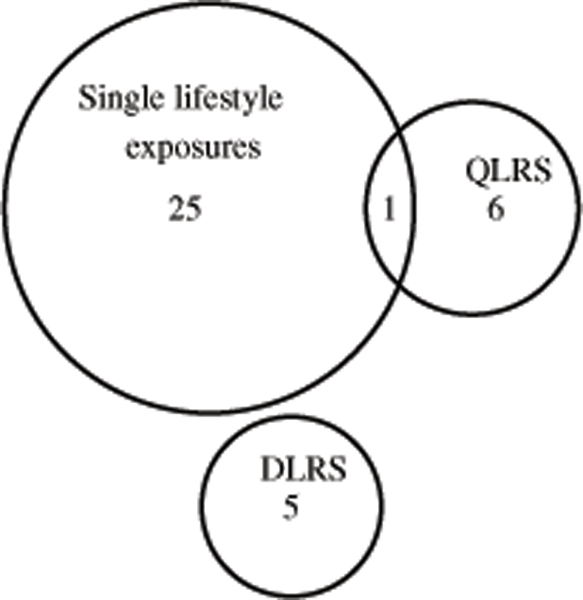
Venn diagram showing the number of loci detected, either uniquely or in common, by analysis using the QLRS, DLRS, and seven single lifestyle exposures.
Table 4.
Twenty-one SNPS that showed suggestive/significant associations with BP in gene-LRS analysis in FHS
| Locus | Chr:position† | SNP | Genomic location¶ | Effect Allele | EAF | BP trait | Exposure | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2:134312960 | rs1368091 | Intronic NCKAP5 | G | 0.281 | DBP | DLRS | −1.15 | 0.28 | 2.85E-05 | 1.73 | 0.35 | 7.23E-07 | 4.16E-06 |
| 2 | 3:123923233 | rs7652065 | Intronic KALRN | A | 0.376 | SBP | DLRS | −1.67 | 0.38 | 1.48E-05 | 2.43 | 0.50 | 8.82E-07 | 4.03E-06 |
| 3:123923922 | rs1444768 | Intronic KALRN | G | 0.376 | SBP | DLRS | −1.66 | 0.38 | 1.48E-05 | 2.43 | 0.49 | 8.74E-07 | 4.01E-06 | |
| 3:123927014 | rs880000 | Intronic KALRN | G | 0.377 | SBP | DLRS | −1.62 | 0.38 | 2.00E-05 | 2.40 | 0.49 | 8.63E-07 | 4.32E-06 | |
| 3:123930542 | rs1444757 | Intronic KALRN | C | 0.377 | SBP | DLRS | −1.62 | 0.38 | 1.99E-05 | 2.40 | 0.49 | 9.13E-07 | 4.54E-06 | |
| 3 | 3:143028187 | rs17636599 | Intronic SLC9A9 | T | 0.011 | SBP | QLRS | 3.39 | 1.61 | 3.59E-02 | −2.41 | 0.51 | 2.28E-06 | 1.80E-07 |
| 4 | 4:180139179 | rs10520420 | Intergenic | T | 0.988 | DBP | DLRS | 3.98 | 0.74 | 9.23E-08 | −4.06 | 1.03 | 7.71E-05 | 6.34E-07 |
| 4:180151019 | rs4146838 | Intergenic | G | 0.988 | DBP | DLRS | 4.07 | 0.73 | 3.01E-08 | −4.16 | 1.02 | 4.63E-05 | 2.13E-07 | |
| 4:180164081 | rs969219 | Intergenic | A | 0.988 | DBP | DLRS | 4.19 | 0.72 | 6.32E-09 | −4.29 | 1.01 | 2.31E-05 | 4.69E-08 | |
| 4:180178378 | rs11131920 | Intergenic | G | 0.010 | DBP | DLRS | −5.10 | 0.67 | 4.01E-14 | 5.29 | 0.99 | 8.95E-08 | 3.71E-13 | |
| 4:180179576 | rs11131921 | Intergenic | T | 0.010 | DBP | DLRS | −5.09 | 0.67 | 4.19E-14 | 5.29 | 0.99 | 9.14E-08 | 3.90E-13 | |
| 5 | 7:158848821 | rs3793217 | Intronic VIPR2 | A | 0.865 | DBP | QLRS | −1.14 | 0.33 | 6.55E-04 | 0.62 | 0.12 | 1.44E-07 | 3.34E-07 |
| 6 | 8:25374910‡ | rs2012485 | Intergenic CDCA2 | C | 0.961 | SBP | QLRS | −4.31 | 1.02 | 2.62E-05 | 1.80 | 0.37 | 8.23E-07 | 4.51E-06 |
| 7 | 8:54274100 | rs7001769 | Intergenic OPRK1 | T | 0.828 | SBP | QLRS | 2.23 | 0.51 | 1.18E-05 | −0.35 | 0.19 | 6.20E-02 | 7.63E-07 |
| 8:54277247 | rs7465458 | Intergenic OPRK1 | A | 0.828 | SBP | QLRS | 2.24 | 0.51 | 1.03E-05 | −0.35 | 0.19 | 5.81E-02 | 6.94E-07 | |
| 8 | 11:11395489§ | rs10047474 | Intronic GALNT18 | T | 0.457 | DBP | QLRS | 1.81 | 0.34 | 0.00E+00 | −0.48 | 0.12 | 5.80E-05 | 4.47E-07 |
| 9 | 11:49202187‡ | rs7124497 | Intronic FOLH1 | G | 0.950 | SBP | DLRS | −1.06 | 0.97 | 2.73E-01 | 4.41 | 1.16 | 1.47E-04 | 7.40E-07 |
| 10 | 12:133441378 | rs4303268 | Intronic CHFR | G | 0.892 | SBP | QLRS | −3.68 | 0.83 | 9.67E-06 | 1.43 | 0.29 | 7.73E-07 | 4.09E-06 |
| 11 | 15:64315336 | rs12442060 | Intronic DAPK2 | C | 0.635 | SBP | DLRS | 1.68 | 0.40 | 2.53E-05 | −2.55 | 0.51 | 5.39E-07 | 3.15E-06 |
| 15:64316458 | rs172995 | Intronic DAPK2 | A | 0.715 | SBP | DLRS | 1.80 | 0.44 | 3.35E-05 | −2.76 | 0.55 | 6.52E-07 | 3.93E-06 | |
| 12 | 18:30053150 | rs11081767 | Intergenic GAREM | A | 0.466 | DBP | QLRS | 1.11 | 0.24 | 2.00E-06 | −0.43 | 0.09 | 7.14E-07 | 2.24E-06 |
Chr:position: based on human genome build 37.
Within boundaries of known loci previously reported to be associated with BP (in published literature between 2011 and 2019; see S?).
Within boundaries of locus detected in CHARGE Gene-Lifestyle Interactions Working Group’s large-scale study on gene-alcohol interactions on BP (Feitosa et al., 2018).
Genomic location: sourced from FUMA GWAS (https://fuma.ctglab.nl). Nearest mapped gene is indicated for intergenic SNPs, while gene located within is indicated for intronic SNPs. No gene is specified for 5 SNPs because only unmapped/uncharacterized genes are located within 500 kb of these SNPs.
Genome-wide significant SNPs at P-value < 5 × 10−8 are in bold font. All other SNPs are suggestive at P-value < 1 × 10−6.
Abbreviations: Chr, chromosome; QLRS, quantitative lifestyle risk score; DLRS, dichotomous lifestyle risk score; EAF, effect allele frequency; , genetic main effect; , standard error for genetic main effect; , P-value for genetic main effect; , gene-environment interaction effect; , standard error for gene-environment interaction effect; , P-value of 1 degree of freedom (DF) interaction test; , P-value of 2 DF Joint test.
Table 5.
P-values for the genetic effect using refined main effects models, which omits the interaction term but adjusts for the marginal effect of the LRS (Model (2)) or the marginal effects of the four component lifestyle variables (Model (3)).
| Chr:position† | SNP | BP trait | Exposure | Model 2 | Model 3 |
|---|---|---|---|---|---|
| 2:134312960 | rs1368091 | DBP | DLRS | 7.59E-01 | 8.37E-01 |
| 3:123923233 | rs7652065 | SBP | DLRS | 9.56E-01 | 9.13E-01 |
| 3:123923922 | rs1444768 | SBP | DLRS | 9.54E-01 | 9.11E-01 |
| 3:123927014 | rs880000 | SBP | DLRS | 8.75E-01 | 8.34E-01 |
| 3:123930542 | rs1444757 | SBP | DLRS | 8.87E-01 | 8.49E-01 |
| 3:143028187 | rs17636599 | SBP | QLRS | 1.20E-02 | 1.42E-02 |
| 4:180139179 | rs10520420 | DBP | DLRS | 9.21E-02 | 8.20E-02 |
| 4:180151019 | rs4146838 | DBP | DLRS | 8.77E-02 | 7.75E-02 |
| 4:180164081 | rs969219 | DBP | DLRS | 8.20E-02 | 7.21E-02 |
| 4:180178378 | rs11131920 | DBP | DLRS | 5.01E-02 | 4.25E-02 |
| 4:180179576 | rs11131921 | DBP | DLRS | 5.03E-02 | 4.27E-02 |
| 7:158848821 | rs3793217 | DBP | QLRS | 5.71E-02 | 7.41E-02 |
| 8:25374910 | rs2012485 | SBP | QLRS | 8.19E-01 | 7.91E-01 |
| 8:54274100 | rs7001769 | SBP | QLRS | 3.36E-06 | 5.78E-06 |
| 8:54277247 | rs7465458 | SBP | QLRS | 3.36E-06 | 5.61E-06 |
| 11:11395489 | rs10047474 | DBP | QLRS | 1.06E-03 | 1.64E-03 |
| 11:49202187 | rs7124497 | SBP | DLRS | 1.92E-04 | 1.22E-04 |
| 12:133441378 | rs4303268 | SBP | QLRS | 7.20E-01 | 6.04E-01 |
| 15:64315336 | rs12442060 | SBP | DLRS | 6.93E-01 | 8.14E-01 |
| 15:64316458 | rs172995 | SBP | DLRS | 6.30E-01 | 7.83E-01 |
| 18:30053150 | rs11081767 | DBP | QLRS | 6.21E-01 | 6.10E-01 |
Chr:position: based on human genome build 37.
Regional association plots for loci detected and their corresponding significant/suggestive SNPs are shown in appendix Figures S4 – S15. Specifically, QLRS found four loci associated with SBP and three loci associated with DBP. Similarly, DLRS found three loci associated with SBP and two loci associated with DBP.
The genome-wide significant locus, intergenic on chromosome 4, was represented by five low-frequency variants, three of which reached genome-wide significance based on the Joint 2 DF test. Three other loci with suggestive associations were also represented by low-frequency variants (rs17636599, rs2012485, and rs7124497). While rs17636599 showed no supporting evidence from surrounding SNPs, rs2012485 and rs7124497 are located within 500 kb of two known BP loci identified through a main effect GWAS (Hoffmann et al., 2017): EBF2 and OR4A47-TRIM51GP, respectively. Similarly, a common variant on chromosome 11, rs10047474, is located within the boundaries of locus GALNT18, which was detected in the CHARGE Gene-Lifestyle Interactions Working Group’s study on gene-alcohol interactions as being associated with SBP in African ancestry (Feitosa et al., 2018).The remaining suggestive associations are novel and were observed for common variants located on chromosomes 2 (NCKAP5 locus), chromosome 3 (SLC9A9 and KALRN loci), chromosome 7 (VIPR2 locus), chromosome 8 (near the OPRK1 locus), chromosome 11 (near the GAREM loci), chromosome 12 (CHFR locus), and chromosome 15 (DAPK2 locus).
All of the suggestive/significant associations identified using LRS (Table 4) were strongly driven by qualitative interactions (opposite signs on and ) between the SNP and the LRS with the exception of SNPs rs7001769 and rs7465458, which have non-significant > 0.05. Consistent with this result, the addition of the interaction term made significant contributions to the signal for 19 out of 21 suggestive/significant SNPs; only associations for SNPs rs7001769 and rs7465458 attained nearly suggestive P-values for the genetic effect in the absence of the interaction term (Table 5). For the QLRS, five SNPs showed a BP decrease while three SNPs showed a BP increase for each unit increase in the QLRS. For the DLRS, eight SNPs showed a BP decrease in individuals with a favorable lifestyle and a BP increase in those with an unfavorable lifestyle, while five SNPs showed the reverse pattern.
Six of the detected variants (rs880000, rs12442060, rs7465458, rs2012485, rs7124497, rs11081767) showed regulatory functional evidence. SNP rs880000 in the KALRN locus on chromosome 3 is likely to affect binding (RegulomeDB categorical score of 2b), with the histone modification evidence suggesting that it is located in a region of enhancers in diverse tissues including heart and brain. Intergenic SNP rs7465458, mapped near OPRK1 on chromosome 8, had a CADD score of 22.1, greater than the recommended threshold for deleteriousness (>12.37). All six SNPs were in strong LD with at least one other SNP that had a CADD score greater than the threshold for deleteriousness and/or a RegulomeDB categorical score of 2b or 2c (Table S2). Moreover, our interrogation of the GWAS atlas showed that 16 of 21 variants detected by the gene-LRS analysis had nominal associations with BP traits (including SBP, DBP, and pulse pressure) and/or cardiovascular traits (e.g., coronary artery disease, heart rate recovery, angina pectoris, etc.), while four genetic variants had nominal associations with other cardiovascular traits (e.g., non-ischemic cardiomyopathy). Only one genetic variant had no record of an at least nominal association with a BP or cardiovascular trait (Table S3).
Discussion
In this study, we performed gene-lifestyle interactions in BP traits using a lifestyle risk score (LRS) aggregating four lifestyle exposures (smoking status, alcohol intake, education, and physical activity) in 6,254 FHS participants. Through our gene-LRS interaction approach, we identified one genome-wide significant locus and 11 suggestive loci in this moderate sample size. Among these 12 loci, only one was detected by analyses that used single lifestyle exposures. Considering the LRS as both quantitative and dichotomous variables was complementary, identifying distinct panels of loci. Strikingly, association between BP traits and all but one of the detected loci was driven by qualitative interactions (showing genetic and interaction effects in opposite signs). Also, nine of the 12 loci detected by the LRS are novel, not previously known to be associated with BP. These results point to a valuable, and complementary, role for the multiple lifestyle exposure approach for the discovery of BP loci.
Our results suggest that the nature of some genetic effects on BP, whether harmful or protective, depends on an individual’s overall lifestyle exposure profile as represented by the LRS. Despite a modest sample size, we found evidence of cumulative interaction effects on BP, at suggestive or significant levels, for 21 SNPs in 12 loci. The fact that 19 of the 21 SNPs (in 11 loci) were not detected in the gene-single lifestyle exposure interaction analysis highlights the potential for increased yield of association when cumulative interaction effects are modeled. This potential for finding additional BP loci is noteworthy because, despite its much smaller sample size and statistical power, the LRS analysis found loci that was distinct from loci detected by well powered gene-single lifestyle exposure interaction studies with sample sizes > 500,000. A classic illustration of this potential is for the locus in the KALRN genomic region that we found to be associated with SBP when accounting for interactions with the DLRS. The effect of the KALRN locus on SBP is typified by the index SNP rs880000 for which one copy of the G allele in individuals with a favorable lifestyle decreased SBP by 1.6 mmHg, yet increased SBP by 0.8 mmHg in those with unfavorable lifestyles. In previous GWAS, this region was reported to be associated with cardiovascular diseases such as coronary artery disease and ischemic stroke (Boroumand et al., 2014; Li et al., 2017), but not with BP even when gene-single lifestyle interactions are accounted for. It is known that combined effects of multiple healthy lifestyle exposures on cardiovascular health yield additional benefits not obtained from individual lifestyle exposures, lending credence to the idea that ‘the whole is greater than the sum of the parts’ (Egan, 2018; Lloyd-Jones, 2014). Moreover, a high genetic risk for BP and cardiovascular diseases can be offset by a favorable lifestyle defined by multiple lifestyle exposures (Pazoki et al., 2018). Along with loci identified through the current practice of using the single lifestyle exposure in genome-wide gene-environment interaction scans, the use of the LRS approach may identify additional loci not detected by single lifestyle exposures. Therefore, an effective strategy for maximizing BP loci discovery would be to apply the two approaches in concert, using the LRS as a complement to the single lifestyle exposure approach.
Whether qualitative interactions, characterized by opposite signs for the genetic main and interaction effects, are prevalent but not detected due to inadequate power of existing approaches remains a relevant unanswered question (Winkler et al., 2017). Remarkably, the gene-LRS interaction analysis consistently detected only qualitative interactions, whereas gene-single lifestyle exposure interaction analysis detected both qualitative and quantitative interactions. Given the relevance of finding qualitative interactions to inform personalized medicine and identify high-risk groups, it may be useful to further investigate whether using a measure of overall lifestyle exposure such as the LRS in gene-environment interaction GWAS provides an advantage for detecting opposite-effect loci.
All of the suggestive/significant SNPs that gene-LRS interaction analysis detected are intronic or intergenic, with some showing evidence of regulatory function. This suggests that the resulting influence on BP may be through regulatory activities on nearby protein coding genes, many with effects on neurohormonal regulation, which may play important roles in the pathophysiology of hypertension (Beevers, Lip, & O’Brien, 2001). For example, there were four SNPs that were intronic to the KALRN gene on chromosome 3. KALRN encodes a guanine nucleotide exchange factor that inhibits vascular inducible nitric oxide, an important regulator of vascular hypertrophy and tone (Zhang et al., 2003), and pituitary secretion of adrenocorticotropic hormone (ACTH) (Ratovitski et al., 1999). Associations between variants in the KALRN genomic region and cardiovascular diseases, including early-onset coronary artery disease and ischemic stroke, have been reported (Boroumand et al., 2014; Li et al., 2017). KALRN protein has also been identified as a biomarker of endothelial dysfunction in hypertensive patients with albuminaria (de la Cuesta et al., 2017).
Similarly, there were two SNPs intronic within DAPK2, a gene that plays a primary role in apoptosis. DAPK2 has been shown to reduce blood pressure in rat models by mediating the mTOR signaling pathway, which prevents oxidative stress and apoptosis in hypertensive disorder complicated pregnancy (Wang et al., 2019). Approximately 100 kb upstream of this region is the sorting nexin 1 gene (SNX1). Sorting nexins play a major role in the regulation of trafficking and signal transduction of G-protein coupled receptors (including the renal dopamine receptor, D5R), a key determinant of water, electrolyte, and BP homeostasis (Yang et al., 2014). An intronic SNP, rs3793217 identifies the gene VIPR2, which encodes a receptor for vasoactive intestinal peptide (VIP), a neurotransmitter and neuromodulator expressed in nearly all tissues (Asnicar et al., 2002), and with known cardiovascular effects including coronary vasodilation, lowering of arterial BP, and regulation of circadian rhythmicity in the heart (Henning & Sawmiller, 2001). Based on evidence from studies of VIPR2 knockout mice, VIPR2 increases insulin sensitivity and regulates circadian rhythm and immune functions (Harmar et al., 2002).
The only locus that had a genome-wide significant association with BP in this study, represented by the index SNP rs11131920, appears to be located in a genomic wilderness, being ∼2 MB away from the nearest mapped gene AGA. Interestingly, this locus has been reported to be associated with the use of several treatment medications (appendix Figures S16), particularly, for hypertension (eprosartan, an angiotensin II receptor blocker; P-value = 4.7 × 10−8) and for pulmonary arterial hypertension and erectile dysfunction (adalafil, a vasodilator; P-value = 1.5 × 10−4) (Churchhouse & Neale, 2017). Nevertheless, this genome-wide significant association with BP requires further investigation.
Despite our innovative application of the LRS, there are some limitations. First, our approach to computing the LRS, while simple, is difficult to interpret. This is because it ignores features like correlations among the individual lifestyle exposures (e.g., heavy drinking and current smoking tend to co-occur) and different magnitude of effects on BP traits. Availability of a richer data resource with larger sample size would allow differences in magnitude of effects of component variables to be addressed by deriving the LRS analogous to polygenic risk scores, i.e., the LRS would be computed as a weighted sum of lifestyle exposures, using beta coefficients as weights for each exposure from BP regression on multiple lifestyle exposures (Kim et al., 2019; Park, Tao, Meeker, Harlow, & Mukherjee, 2014). Second, the LRS identified different set of loci depending on whether it was quantitative or dichotomous, which may be due to statistical power. In general, our approach with LRS seems promising and worth pursuing with larger sample sizes and in large consortium settings.
In summary, by considering lifestyle risk score aggregating multiple lifestyle exposures in gene-lifestyle interactions, our limited proof-of-concept investigation appears to have identified several novel and biologically plausible loci. The genetic effects of these loci on BP may have been modulated by the combined effect of multiple lifestyle exposures. Our findings suggest that the LRS approach can contribute to novel discoveries over and above those from single lifestyle exposure analysis, and can complement traditional approaches that use single lifestyle exposures.
Supplementary Material
Acknowledgements
We thank all participants of the Framingham Heart Study for their dedication to cardiovascular health research. The Framingham Heart Study is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with Boston University (Contract No. N01-HC-25195and HHSN268201500001I). This manuscript was not prepared in collaboration with investigators of the Framingham Heart Study and does not necessarily reflect the opinions or views of the Framingham Heart Study, Boston University, or NHLBI.
This research work was supported by grants R01HL118305 and T32HL091823 from the National Heart, Lung, and Blood Institute. Funding for SHARe Affymetrix genotyping was provided by NHLBI Contract N02-HL-64278. SHARe Illumina genotyping was provided under an agreement between Illumina and Boston University. P.S.D. was additionally supported by American Heart Association grant number 18CDA34110116.
Grant numbers:
Grants R01HL118305 and T32HL091823 from the National Heart, Lung, and Blood Institute, and NIH grants R21 AA024888–01 (SMH), and UL1 TR002345 (LJB & SMH). Funding for SHARe Affymetrix genotyping was provided by NHLBI Contract N02-HL-64278. SHARe Illumina genotyping was provided under an agreement between Illumina and Boston University. PSD was additionally supported by American Heart Association grant number 18CDA34110116.
Data availability
The data that support the findings of this study are available in dbGaP at https://www.ncbi.nlm.nih.gov/gap/, reference number phs000007.v3.p2.
Footnotes
Conflict of Interest
None of the authors have any conflicts of interest to declare.
References
- Asnicar MA, Köster A, Heiman ML, Tinsley F, Smith DP, Galbreath E, … Hsiung HM (2002). Vasoactive intestinal polypeptide/pituitary adenylate cyclase-activating peptide receptor 2 deficiency in mice results in growth retardation and increased basal metabolic rate. Endocrinology, 143(10), 3994–4006. 10.1210/en.2002-220354 [DOI] [PubMed] [Google Scholar]
- Aulchenko YS, de Koning D-J, & Haley C (2007). Genomewide rapid association using mixed model and regression: A fast and simple method for genomewide pedigree-based quantitative trait loci association analysis. Genetics, 177(1), 577–585. 10.1534/genetics.107.075614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulchenko YS, Struchalin MV, & van Duijn CM (2010). ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics, 11, 134 10.1186/1471-2105-11-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson J, Sung YJ, Schwander K, Kume R, Simino J, de las Fuentes L, & Rao D (2014). Gene-education interactions identify novel blood pressure loci in the Framingham Heart Study. American Journal of Hypertension, 27(3), 431–444. 10.1093/ajh/hpt283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beevers G, Lip GYH, & O’Brien E (2001). The pathophysiology of hypertension. BMJ : British Medical Journal, 322(7291), 912–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley AR, Sung YJ, Brown MR, Winkler TW, Kraja AT, Ntalla I, … Cupples LA (2019). Multi-ancestry genome-wide gene-smoking interaction study of 387,272 individuals identifies new loci associated with serum lipids. Nature Genetics, 51(4), 636–648. 10.1038/s41588-019-0378-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroumand M, Ziaee S, Zarghami N, Anvari MS, Cheraghi S, Abbasi SH, … Pourgholi L (2014). The Kalirin Gene rs9289231 Polymorphism as a Novel Predisposing Marker for Coronary Artery Disease. Laboratory Medicine, 45(4), 302–308. 10.1309/LMLS813ZDPHRFLUU [DOI] [PubMed] [Google Scholar]
- Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, … Snyder M (2012). Annotation of functional variation in personal genomes using RegulomeDB. Genome Research, 22(9), 1790–1797. 10.1101/gr.137323.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, Izzo JL, … National High Blood Pressure Education Program Coordinating Committee. (2003). The Seventh Report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure: The JNC 7 report. JAMA, 289(19), 2560–2572. 10.1001/jama.289.19.2560 [DOI] [PubMed] [Google Scholar]
- Churchhouse C, & Neale. (2017, September 20). Rapid GWAS of thousands of phenotypes for 337,000 samples in the UK Biobank. Retrieved from http://www.nealelab.is/blog/2017/7/19/rapid-gwas-of-thousands-of-phenotypes-for-337000-samples-in-the-uk-biobank
- Costanzo S, de Gaetano G, Di Castelnuovo A, Djoussé L, Poli A, & van Velden DP (2019). Moderate alcohol consumption and lower total mortality risk: Justified doubts or established facts? - ScienceDirect. Nutrition, Metabolism & Cardiovascular Diseases. 10.1016/j.numecd.2019.05.062 [DOI] [PubMed] [Google Scholar]
- de la Cuesta F, Baldan-Martin M, Moreno-Luna R, Alvarez-Llamas G, Gonzalez-Calero L, Mourino-Alvarez L, … Barderas MG (2017). Kalirin and CHD7: Novel endothelial dysfunction indicators in circulating extracellular vesicles from hypertensive patients with albuminuria. Oncotarget, 8(9), 15553–15562. 10.18632/oncotarget.14948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries PS, Brown MR, Bentley AR, Sung YJ, Winkler TW, Ntalla I, … Morrison AC (2019). Multiancestry Genome-Wide Association Study of Lipid Levels Incorporating Gene-Alcohol Interactions. American Journal of Epidemiology, 188(6), 1033–1054. 10.1093/aje/kwz005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz KM, & Shimbo D (2013). Physical Activity and the Prevention of Hypertension. Current Hypertension Reports, 15(6), 659–668. 10.1007/s11906-013-0386-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan BM (2018). Is life’s simple 7 a practical paradigm for promoting healthy blood pressure, preventing cardiovascular disease and improving total health? Journal of the American Society of Hypertension, 12(5), 324–326. 10.1016/j.jash.2018.04.002 [DOI] [Google Scholar]
- Ehret GB (2010). Genome-wide association studies: Contribution of genomics to understanding blood pressure and essential hypertension. Current Hypertension Reports, 12(1), 17–25. 10.1007/s11906-009-0086-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelou E, Warren H, Mosen-Ansorena D, Mifsud B, Pazoki R, Gao H, … Witkowska K (2017). Genetic analysis of over one million people identifies 535 novel loci for blood pressure. BioRxiv. Retrieved from https://www.biorxiv.org/content/early/2017/10/11/198234 [Google Scholar]
- Feitosa MF, Kraja AT, Chasman DI, Sung YJ, Winkler TW, Ntalla I, … Levy D (2018). Novel genetic associations for blood pressure identified via gene-alcohol interaction in up to 570K individuals across multiple ancestries. PLOS ONE, 13(6), e0198166 10.1371/journal.pone.0198166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folsom AR, Yatsuya H, Nettleton JA, Lutsey PL, Cushman M, Rosamond WD, & ARIC Study Investigators. (2011). Community prevalence of ideal cardiovascular health, by the American Heart Association definition, and relationship with cardiovascular disease incidence. Journal of the American College of Cardiology, 57(16), 1690–1696. 10.1016/j.jacc.2010.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forouzanfar MH, Liu P, Roth GA, Ng M, Biryukov S, Marczak L, … Murray CJL (2017). Global Burden of Hypertension and Systolic Blood Pressure of at Least 110 to 115 mm Hg, 1990–2015. JAMA-Journal of the American Medical Association, 317(2), 165–182. [DOI] [PubMed] [Google Scholar]
- Harmar AJ, Marston HM, Shen S, Spratt C, West KM, Sheward WJ, … Hastings MH (2002). The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell, 109(4), 497–508. [DOI] [PubMed] [Google Scholar]
- Henning RJ, & Sawmiller DR (2001). Vasoactive intestinal peptide: Cardiovascular effects. Cardiovascular Research, 49(1), 27–37. 10.1016/S0008-6363(00)00229-7 [DOI] [PubMed] [Google Scholar]
- Hoffmann TJ, Ehret GB, Nandakumar P, Ranatunga D, Schaefer C, Kwok P-Y, … Risch N (2017). Genome-wide association analyses using electronic health records identify new loci influencing blood pressure variation. Nature Genetics, 49(1), 54–64. 10.1038/ng.3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MC (2014). Gene × environment interaction studies have not properly controlled for potential confounders: The problem and the (simple) solution. Biological Psychiatry, 75(1), 18–24. 10.1016/j.biopsych.2013.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpeläinen TO, Bentley AR, Noordam R, Sung YJ, Schwander K, Winkler TW, … Loos RJF (2019). Multi-ancestry study of blood lipid levels identifies four loci interacting with physical activity. Nature Communications, 10(1), 1–11. 10.1038/s41467-018-08008-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Ziyatdinov A, Laville V, Hu FB, Rimm E, Kraft P, & Aschard H (2019). Joint Analysis of Multiple Interaction Parameters in Genetic Association Studies. Genetics, 211(2), 483–494. 10.1534/genetics.118.301394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, & Shendure J (2014). A general framework for estimating the relative pathogenicity of human genetic variants. Nature Genetics, 46(3), 310–315. 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft P, Yen Y-C, Stram DO, Morrison J, & Gauderman WJ (2007). Exploiting Gene-Environment Interaction to Detect Genetic Associations. Human Heredity, 63(2), 111–119. 10.1159/000099183 [DOI] [PubMed] [Google Scholar]
- Laville V, Majarian T, Sung YJ, Feitosa MF, Chasman D, Bentley AR, … Aschard H (2019). Genome-wide Interaction Studies by the CHARGE Gene-Lifestyle Interactions Working Group: What we have learned and what is coming next. BioRxiv. 10.1101/562157 [DOI] [Google Scholar]
- Li H, Yu S, Wang R, Sun Z, Zhou X, Zheng L, … Sun Y (2017). Genetic Variant of Kalirin Gene Is Associated with Ischemic Stroke in a Chinese Han Population. BioMed Research International, 2017, 6594271 10.1155/2017/6594271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Jones DM (2014). Cardiovascular Health and Protection Against CVD: More Than the Sum of the Parts? Circulation, 130(19), 1671–1673. 10.1161/CIRCULATIONAHA.114.012869 [DOI] [PubMed] [Google Scholar]
- Lloyd-Jones DM, Hong Y, Labarthe D, Mozaffarian D, Appel LJ, Horn LV, … Rosamond WD (2010). Defining and Setting National Goals for Cardiovascular Health Promotion and Disease Reduction: The American Heart Association’s Strategic Impact Goal Through 2020 and Beyond. Circulation, 121(4), 586–613. 10.1161/CIRCULATIONAHA.109.192703 [DOI] [PubMed] [Google Scholar]
- Loucks EB, Abrahamowicz M, Xiao Y, & Lynch JW (2011). Associations of education with 30 year life course blood pressure trajectories: Framingham Offspring Study. BMC Public Health, 11, 139 10.1186/1471-2458-11-139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R, Casale FP, Bonder MJ, Horta D, Franke L, Barroso I, & Stegle O (2018). A linear mixed-model approach to study multivariate gene–environment interactions. Nature Genetics, 1 10.1038/s41588-018-0271-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayor M, Enserro DM, Vasan RS, & Xanthakis V (2016). Cardiovascular Health Status and Incidence of Heart Failure in the Framingham Offspring Study. Circulation. Heart Failure, 9(1), e002416 10.1161/CIRCHEARTFAILURE.115.002416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogunmoroti O, Oni E, Michos ED, Spatz ES, Allen NB, Rana JS, … Nasir K (2017). Life’s Simple 7 and Incident Heart Failure: The Multi-Ethnic Study of Atherosclerosis. Journal of the American Heart Association, 6(6). 10.1161/JAHA.116.005180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe JH, Bybee KA, & Lavie CJ (2007). Alcohol and Cardiovascular Health: The Razor-Sharp Double-Edged Sword. Journal of the American College of Cardiology, 50(11), 1009–1014. 10.1016/j.jacc.2007.04.089 [DOI] [PubMed] [Google Scholar]
- Palla L, Higgins JPT, Wareham NJ, & Sharp SJ (2010). Challenges in the Use of Literature-based Meta-Analysis to Examine Gene-Environment Interactions. American Journal of Epidemiology, 171(11), 1225–1232. 10.1093/aje/kwq051 [DOI] [PubMed] [Google Scholar]
- Park SK, Tao Y, Meeker JD, Harlow SD, & Mukherjee B (2014). Environmental Risk Score as a New Tool to Examine Multi-Pollutants in Epidemiologic Research: An Example from the NHANES Study Using Serum Lipid Levels. PLOS ONE, 9(6), e98632 10.1371/journal.pone.0098632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnell LD, Blokker BA, Dashti HS, Nesbeth P-D, Cooper BE, Ma Y, … Ordovás JM (2014). CardioGxE, a catalog of gene-environment interactions for cardiometabolic traits. BioData Mining, 7, 21 10.1186/1756-0381-7-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazoki R, Dehghan A, Evangelou E, Warren H, Gao H, Caulfield M, … Tzoulaki I (2018). Genetic Predisposition to High Blood Pressure and Lifestyle Factors: Associations With Midlife Blood Pressure Levels and Cardiovascular Events. Circulation, 137(7), 653–661. 10.1161/CIRCULATIONAHA.117.030898 [DOI] [PubMed] [Google Scholar]
- Prince SA, Adamo KB, Hamel ME, Hardt J, Gorber SC, & Tremblay M (2008). A comparison of direct versus self-report measures for assessing physical activity in adults: A systematic review. International Journal of Behavioral Nutrition and Physical Activity, 5, 56 10.1186/1479-5868-5-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao DC, Sung YJ, Winkler TW, Schwander K, Borecki I, Cupples LA, … CHARGE Gene-Lifestyle Interactions Working Group*. (2017). Multiancestry Study of Gene-Lifestyle Interactions for Cardiovascular Traits in 610 475 Individuals From 124 Cohorts: Design and Rationale. Circulation. Cardiovascular Genetics, 10(3). 10.1161/CIRCGENETICS.116.001649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratovitski EA, Alam MR, Quick RA, McMillan A, Bao C, Kozlovsky C, … Lowenstein CJ (1999). Kalirin inhibition of inducible nitric-oxide synthase. The Journal of Biological Chemistry, 274(2), 993–999. 10.1074/jbc.274.2.993 [DOI] [PubMed] [Google Scholar]
- Shankar RR, Eckert GJ, Saha C, Tu W, & Pratt JH (2005). The Change in Blood Pressure during Pubertal Growth. The Journal of Clinical Endocrinology & Metabolism, 90(1), 163–167. 10.1210/jc.2004-0926 [DOI] [PubMed] [Google Scholar]
- Simino J, Sung YJ, Kume R, Schwander K, & Rao DC (2013). Gene-alcohol interactions identify several novel blood pressure loci including a promising locus near SLC16A9. Frontiers in Genetics, 4, 277 10.3389/fgene.2013.00277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YJ, de las Fuentes L, Schwander KL, Simino J, & Rao DC (2015). Gene–Smoking Interactions Identify Several Novel Blood Pressure Loci in the Framingham Heart Study. American Journal of Hypertension, 28(3), 343–354. 10.1093/ajh/hpu149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YJ, de las Fuentes L, Winkler TW, Chasman DI, Bentley AR, Kraja AT, … Morrison AC (2019). A multi-ancestry genome-wide study incorporating gene–smoking interactions identifies multiple new loci for pulse pressure and mean arterial pressure. Human Molecular Genetics, 28(15), 2615–2633. 10.1093/hmg/ddz070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YJ, Winkler TW, de Las Fuentes L, Bentley AR, Brown MR, Kraja AT, … Chasman DI (2018). A Large-Scale Multi-ancestry Genome-wide Study Accounting for Smoking Behavior Identifies Multiple Significant Loci for Blood Pressure. American Journal of Human Genetics, 102(3), 375–400. 10.1016/j.ajhg.2018.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpson NJ, Greenwood CMT, Soranzo N, Lawson DJ, & Richards JB (2018). Genetic architecture: The shape of the genetic contribution to human traits and disease. Nature Reviews Genetics, 19(2), 110–124. 10.1038/nrg.2017.101 [DOI] [PubMed] [Google Scholar]
- Tobin MD, Sheehan NA, Scurrah KJ, & Burton PR (2005). Adjusting for treatment effects in studies of quantitative traits: Antihypertensive therapy and systolic blood pressure. Statistics in Medicine, 24(19), 2911–2935. 10.1002/sim.2165 [DOI] [PubMed] [Google Scholar]
- van Dam RM, Li T, Spiegelman D, Franco OH, & Hu FB (2008). Combined impact of lifestyle factors on mortality: Prospective cohort study in US women. BMJ (Clinical Research Ed.), 337, a1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liu L-L, Tian Y, Chen Y, Zha W-H, Li Y, & Wu F-J (2019). Upregulation of DAPK2 ameliorates oxidative damage and apoptosis of placental cells in hypertensive disorder complicating pregnancy by suppressing human placental microvascular endothelial cell autophagy through the mTOR signaling pathway. International Journal of Biological Macromolecules, 121, 488–497. 10.1016/j.ijbiomac.2018.09.111 [DOI] [PubMed] [Google Scholar]
- Watanabe K, Taskesen E, Bochoven A. van, & Posthuma D (2017). Functional mapping and annotation of genetic associations with FUMA. Nature Communications, 8(1), 1826 10.1038/s41467-017-01261-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Stringer S, Frei O, Mirkov MU, de Leeuw C, Polderman TJC, … Posthuma D (2019). A global overview of pleiotropy and genetic architecture in complex traits. Nature Genetics, 51, 1339–1348. 10.1038/s41588-019-0481-0 [DOI] [PubMed] [Google Scholar]
- Wei JY (1992). Age and the Cardiovascular System. New England Journal of Medicine, 327(24), 1735–1739. 10.1056/NEJM199212103272408 [DOI] [PubMed] [Google Scholar]
- WHO. (2013). A global brief on hypertension: Silent killer, global public health crisis: World Health Day 2013; Retrieved from http://apps.who.int/iris/handle/10665/79059 [Google Scholar]
- Winkler TW, Justice AE, Cupples LA, Kronenberg F, Kutalik Z, Heid IM, & Consortium, the G. (2017). Approaches to detect genetic effects that differ between two strata in genome-wide meta-analyses: Recommendations based on a systematic evaluation. PLOS ONE, 12(7), e0181038 10.1371/journal.pone.0181038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Armando I, Jones JE, Zeng C, Jose PA, & Villar VAM (2014). Sorting Nexins: New Determinants for the Development of Hypertension. Annals of Clinical and Experimental Hypertension. Retrieved from https://www.jscimedcentral.com/ExperimentalHypertension/experimentalhypertension-2-1008.php [Google Scholar]
- Zhang W, Kuncewicz T, Yu Z-Y, Zou L, Xu X, & Kone BC (2003). Protein-protein interactions involving inducible nitric oxide synthase. Acta Physiologica Scandinavica, 179(2), 137–142. 10.1046/j.1365-201X.2003.01119.x [DOI] [PubMed] [Google Scholar]
- [dataset] DbGaP | phs000007.v3.p2 | Framingham SNP Health Association Resource (SHARe). (2008). Retrieved from https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000007.v3.p2
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


